Abstract
Transcription factor IIS (TFIIS) stimulates RNA cleavage by RNA polymerase II by allowing backtracked enzymes to resume transcription elongation. Yeast cells do not require TFIIS for viability, unless they suffer severe transcriptional stress due to NTP-depleting drugs like 6-azauracil or mycophenolic acid. In order to broaden our knowledge on the role of TFIIS under transcriptional stress, we carried out a genetic screening for suppressors of TFIIS-lacking cells’ sensitivity to 6-azauracil and mycophenolic acid. Five suppressors were identified, four of which were related to the transcriptional regulation of those genes encoding ribosomal components [rRNAs and ribosomal proteins (RP)], including global regulator SFP1. This led us to discover that RNA polymerase II is hypersensitive to the absence of TFIIS under NTP scarcity conditions when transcribing RP genes. The absence of Sfp1 led to a profound alteration of the transcriptional response to NTP-depletion, thus allowing the expression of RP genes to resist these stressful conditions in the absence of TFIIS. We discuss the effect of transcriptional stress on ribosome biogenesis and propose that TFIIS contributes to prevent a transcriptional imbalance between rDNA and RP genes.
INTRODUCTION
Gene transcription is played by RNA polymerases, which require a large set of auxiliary factors ranging from pre-initiation complex formation to transcription termination to complete the synthesis of their final RNA transcripts. A relevant category of auxiliary elements is formed by RNA cleavage factors, which stimulate the cleavage of the 3′-end of nascent RNA by RNA polymerases when they are arrested after backtracking (1). Transcription factor IIS (TFIIS) is the eukaryal and archaeal type of the RNA cleavage factor, although other proteins ubiquitously perform similar functions in bacteria (1). Transcript cleavage by RNA polymerase II is catalysed in vitro and in vivo by its intrinsic cleavage activity, which is strongly stimulated by TFIIS (2,3).
In agreement with its RNA-cleavage stimulation activity, TFIIS plays an important role during transcription elongation (4). In addition, TFIIS has also been found to be involved in both transcription initiation regulation (5,6) and the transition from transcription initiation to elongation (7).
Genetic approaches have shown that TFIIS factors are essential for cell viability, as with trypanosome (8), and are required for either correct embryonic development, as in mouse and Xenopus (9,10), or plant seeds dormancy (11). In contrast, TFIIS is not essential through the Saccharomyces cerevisiae life cycle (12). This is particularly striking because RNA polymerase II arrest and backtracking are very frequent phenomena (13,14), suggesting that spontaneous non-stimulated RNA cleavage is sufficient for sustaining gene transcription under standard yeast growing conditions (3).
However, the yeast dst1Δ mutants lacking TFIIS are highly sensitive to drugs that impair the de novo synthesis of NTP, such as 6-azauracil (6AU) and mycophenolic acid (MPA) (15). These drugs inhibit IMP dehydrogenase, the key enzyme in the GMP biosynthetic pathway (15). In response to 6AU or MPA, yeast cells up-regulate the expression of IMD2, a gene encoding an IMP dehydrogenase isoenzyme that is resistant to such drugs (16). dst1Δ mutants are unable to up-regulate IMD2 (16), which exhibits a sophisticated transcriptional attenuation mechanism in response to GTP levels (17). The over-expression of IMD2 suppresses the MPA sensitivity of yeast MPA-sensitive mutants (18). The transcriptional stress caused by NTP-depleting drugs is partially transient in wild-type yeast cells due to the up-regulation of IMD2, whereas it is more intense and permanent in dst1Δ (16).
Most of the studies conducted on the TFIIS function have focused on its role during RNA pol II–dependent transcription. However, it has been shown that TFIIS and TFIIS-like cleavage factors are also important for RNA polymerase I- and RNA polymerase III-dependent transcription (19–23). All this information suggests that TFIIS generally contributes to the biogenesis of ribosomes, whose structural elements are concertedly transcribed by the three nuclear RNA polymerases (24).
This work addresses the role of TFIIS during transcriptional stress by isolating mutations that are capable of suppressing yeast dst1Δ sensitivity to 6AU and MPA. Most suppressor genes functionally relate to the transcriptional regulation of ribosome synthesis. This fact led us to discover that TFIIS is required to maintain the transcriptional activity of RNA polymerase II when transcribing ribosomal protein (RP) genes under transcriptional stress conditions, and that the absence of TFIIS impairs the balanced transcription of rDNA and RP genes. These results are consistent with previous findings from our lab indicating a regulation of RNA polymerase II activity during the transcription elongation of RP genes (25).
MATERIALS AND METHODS
Strains, plasmids and media
The table listing the yeast strains used in this study is included in the Supplementary Data (Supplementary Table S1). Suppression screening was performed with the SChY66.9D strain. Further analyses of simple and double mutants were performed using the complete deletion mutants obtained from EUROSCARF, which were isogenic to BY4741. Tagged strains were also constructed in the BY4741 background. HA-TFIIS strains’ functionality was assayed by checking resistance to 6AU. When RAP1 partial deletions were utilized, they were studied in the corresponding original background, as described elsewhere (26).
For the 6AU treatments, all the strains were transformed with a centromeric plasmid that expresses URA3 and were grown in a complete minimal medium lacking uracil (SC-URA). MPA and 6-AU (Sigma) were dissolved directly in growth media up to the desired concentrations.
Growth assays
For the plate assays, yeast cultures were diluted to the same OD600, and serial dilutions (1:10) were spotted onto plates. At least three independent experiments were carried out in all cases.
For calculating the duplication times in the liquid culture, 250 ml flasks, containing 50 ml SC-Ura cultures, were inoculated in the presence or the absence of 6AU (25 µg/ml) and incubated at 30°C under vigorous agitation (200 rpm).
For the microcolony assays, cells were microencapsulated in calcium alginate by means of a Cellena microencapsulator system (Ingeniatrics) following the manufacturer’s instructions. A microcapsule population, mainly containing single cells, was incubated for 20 h in SC-Ura in the presence or absence of 6AU. After confirming microcolony development by optical microscopy, the population microcolonies was analysed in a COPAS SELECT flow cytometer (Union Biometrica). Relative microcolony size was monitored by measuring self-fluorescence under the following photomultiplier tube settings: Green (1100), Yellow (600) and Red (1100). The time of flight minimum was fixed at 10 and the extinction signal was 1. Sheath fluid pressure was 4.40–5.19, while sample fluid pressure was set to maintain a frequency of 15–25 events per second (2.13–3.63). A detailed protocol of microbial analysis combining microencapsulation and flow cytometry will be published elsewhere.
Genetic screening
Suppressor screening was performed by transforming the SChY66.9D strain with a yeast genomic library which had been mutagenized by the random insertion of an mTn3-lacZ/LEU2 transposon (27). Three independent transformations with 0.25 µg of a Not1-digested library were plated onto 10, 20 and 25 µg/ml 6AU plates to allow the isolation of 14 suppressors of the almost 20 000 transformants. Only those suppressors that were able to grow on both the 6AU and MPA plates, and which presented a single insertion ligated to their suppression phenotype, were selected for further analyses (Supplementary Table S2). Transposon insertion sites (Supplementary Figure S1) were identified by inverse PCR.
Transcriptional run-on and chromatin immunoprecipitation
Run-on assays were performed as formerly described (28). Double-strand immobilized probes were obtained by PCR using the primers listed in Supplementary Table S3. Membranes were exposed on Fuji BAS screens for 5–7 days and were developed with a FUJIX FLA3000 device. ChIP experiments were performed as previously described (28). The primers utilized are listed in Supplementary Table S3.
RESULTS
Mutations suppressing dst1Δ sensitivity to NTP-depleting drugs
Fourteen 6-AU-resistant derivatives from a dst1Δ strain of the 19 500 cells mutagenized with an insertion library were selected as described in the ‘Materials and Methods’ section. Eight of the 6AU-resistant clones were found to also grow in the presence of MPA. Double-resistant mutants were crossed to check monogenicity and were PCR-analysed to map the insertion locus. Five DAS (after dst1Δ 6AU sensitivity) genes were finally identified (Supplementary Figure S1). DAS1 (YJL149W) encodes a putative SCF ubiquitin ligase F-box protein of unknown function (29). The other DAS genes (YLR403W, YHR205W, YIL148W and YDR020C) encode four proteins related to ribosome biogenesis and its control: ribosomal regulators Sfp1 and Sch9 (30), structural RP Rpl40A and an uncharacterized protein, which we called Das2, which has been previously related to rDNA transcription (31).
Suppressors were confirmed by crossing the corresponding deleted mutants with dst1Δ in the BY4741 background. In most cases, double mutants resisted the 6AU concentrations that were inhibitory for the single dst1Δ mutant (Supplementary Figure S2A). The only exception was rpl40AΔ, which was unable to suppress dst1Δ in the BY4741 background, although a plasmid expressing RPL40A was able to revert the suppression by the original rpl40A insertional mutation (not shown). In the presence of the drugs, double mutants were never able to grow as fast as the wild-type, indicating that the suppression degree was always partial (Supplementary Figure S2A).
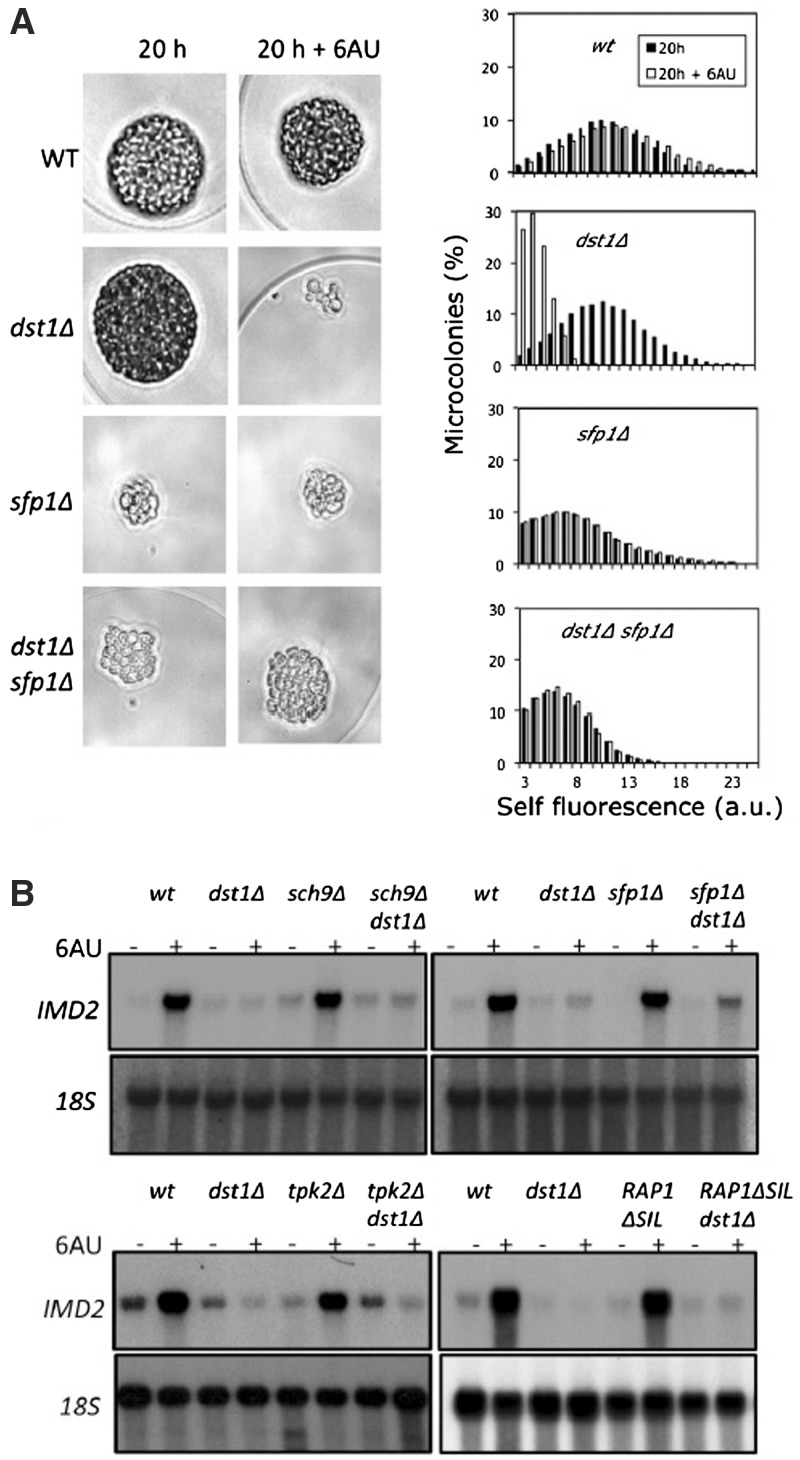
We focused in dst1Δsfp1Δ for characterizing the genetic interaction between DST1 and SFP1. The difference in growth between dst1Δsfp1Δ and the wild-type seemed to be shorter in either 6AU or MPA than in the absence of these drugs (Supplementary Figure S2A), indicating a functional connection between these two genes. The doubling times calculated from liquid cultures further support this indication (Supplementary Figure S2B). Nevertheless, the long doubling time of sfp1Δ under any condition complicated the interpretation of the results and implied the requirement of a large number of generations. In order to overcome this inconvenience, we developed a growth assay based on microencapsulation and flow cytometry, which cuts the observation time from 4 days (visible sfp1Δ colonies on plates) to 20 h (detectable sfp1Δ microcolonies in microcapsules) (Figure 1A and Supplementary Figure S3A). As shown in Figure 1A, 6AU had no significant effect on the wild-type proliferation capacity and the effect was also minimal for the sfp1Δ and dst1Δsfp1Δ cells, whereas the dst1Δ cells underwent a dramatic growth arrest by the drug, resulting in aberrant microcolonies. Despite the intrinsic defect in cell growth that the SFP1 deletion generated, half the cells from the double mutant incubated in 6AU were able to develop bigger microcolonies than any of those produced by the dst1Δ cell population under the same conditions (Supplementary Figure S3B). We conclude that a functional interaction takes place between DST1 and SFP1, which allows a partial suppression of the 6AU-sensitivity phenotype of dst1Δ by sfp1Δ.
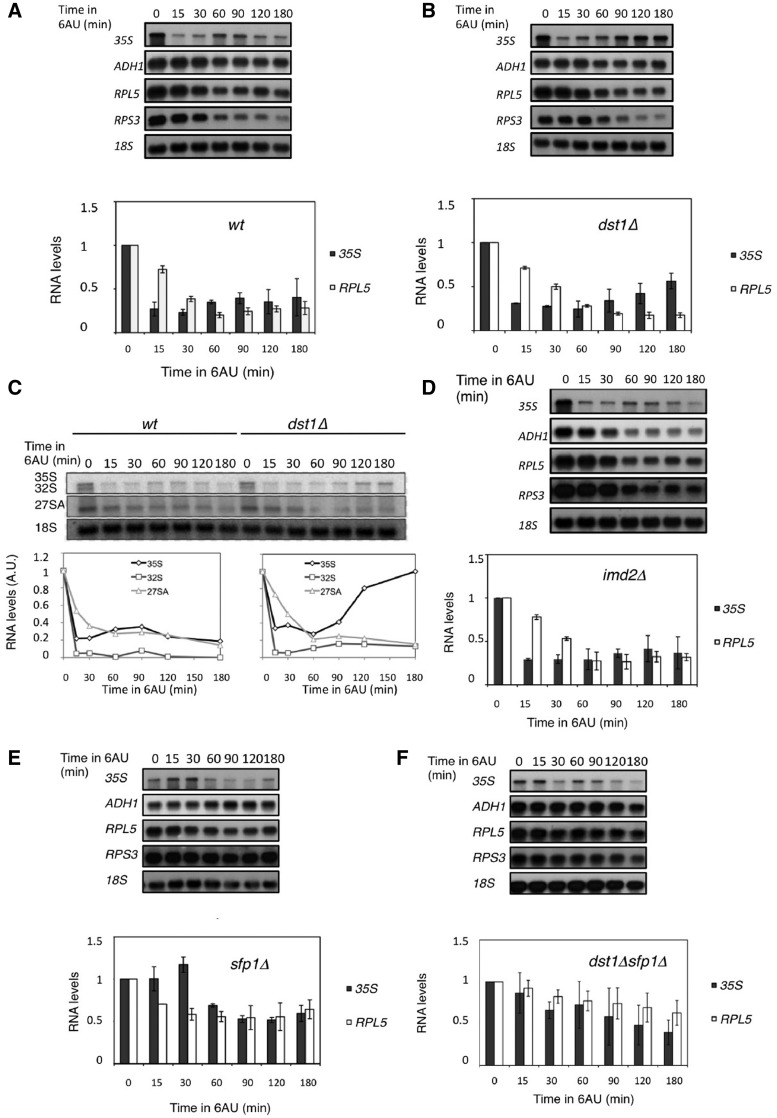
Figure 1.
Sensitivity of dst1Δ to 6AU and MPA can be partially suppressed by mutations that alter the transcriptional regulation of RP genes. (A) Size distribution of the microcolonies developed by isogenic yeast cells with the indicated genotypes (BY4741 genetic background), in the absence or presence of 25 µg/ml 6AU. Individual cells were microencapsulated in alginate, incubated for 20 h and analysed in a flow cytometer utilizing self-fluorescence as an indicator of cell size, as described in the ‘Materials and Methods’ section. Images of representative microcolonies are shown. Additional microcolonies are shown in Supplementary Figure S3A. (B) Northern blots showing the effect of 6AU (100 µg/ml, 90 min) on the mRNA levels of IMD2 in the indicated isogenic mutants [strains as in (A)]. See quantification in Supplementary Figure S4. Note that a mutant’s ability to suppress the growth defect of dst1Δ in the presence of 6AU does not involve the suppression of the IMD2 up-regulation defect.
In our growth assays, 6AU and MPA generally produced similar results, although minor differences were detected between the two drugs (Supplementary Figure S2A). These differences might be related to the secondary targets of these drugs, like OMP decarboxylase, which is specifically inhibited by 6AU (32). In addition to these secondary targets, the common metabolic effect of 6AU and MPA is the inhibition of IMP dehydrogenases. IMD2 encodes an IMP dehydrogenease isoenzyme which is resistant to MPA (33). Sensitivity of dst1Δ to NTP-depleting drugs has been connected to the incapacity of dst1Δ to induce IMD2 in response to NTP depletion (16). We measured the mRNA levels of IMD2 in the single and double mutants after a 90-min incubation with 6AU. Similarly to the single dst1Δ mutant, the sfp1Δ dst1Δ and sch9Δ dst1Δ double mutants were never able to reach a 2-fold induction of IMD2, which clearly contrasts with the 17-fold induction of the wild-type (Figure 1B and Supplementary Figure S4A). Similar results were obtained with the other das mutations (not shown). sch9Δ was even able to alleviate the marked sensitivity of imd2Δ to MPA (Supplementary Figure S5). Taken together, these results suggest that the ability of the das mutations to suppress dst1Δ does not correlate with the induction of IMD2, and that das cells can survive and proliferate in the absence of TFIIS under very stringent NTP stress.
Since most DAS genes were functionally related to the structural components of ribosomes, we checked whether other regulators of the genes encoding RPs were able to suppress dst1Δ sensitivity to NTP-depleting drugs. We found that an RAP1 mutant, which lacks the silencing domain of this transcription factor involved in RP transcription (25), was able to partially suppress dst1Δ (Supplementary Figure S6A) under conditions that do not induce IMD2 (Figure 1B and Supplementary Figure S4B). Other RAP1 mutants (26) did not show a suppressed phenotype of dst1Δ, except RAP1ΔC, which also lacked the silencing domain of RAP1 (Supplementary Figure S6A). Likewise, deletion of the PKA-encoding gene, TPK2, which controls RP transcription in response to nutritional signals (30), partially suppressed dst1Δ sensitivity to NTP-depleting drugs without inducing IMD2 (Figure 1B, Supplementary Figures S4C and S6B). We conclude that the alteration of the transcriptional control of RP genes produced by das mutations allows those cells lacking TFIIS to resist NTP stress.
Effect of 6AU on TFIIS binding and RNA polymerase II activity on RP genes
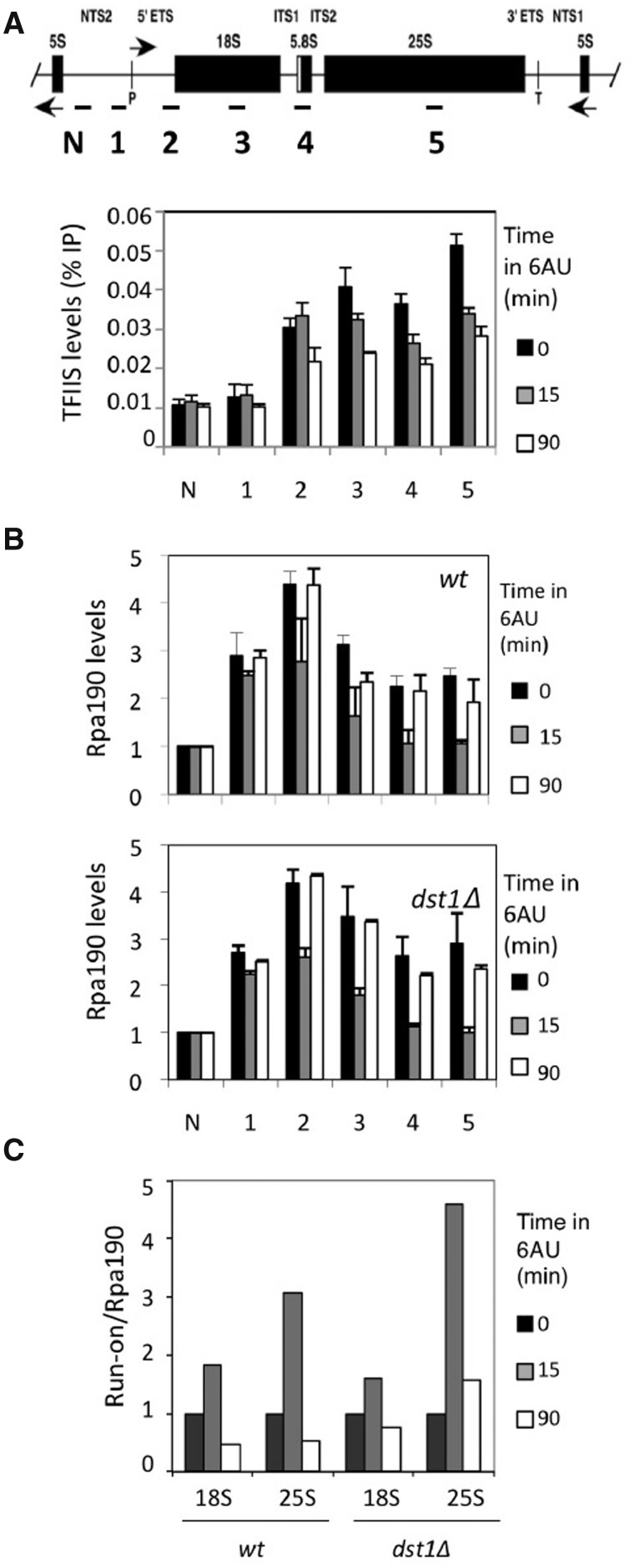
In order to evaluate the contribution of TFIIS to transcriptional activity under transcriptional stress, we monitored its occupancy by performing ChIP experiments. TFIIS was originally described as an RNA pol II-specific factor, although TFIIS and TFIIS-like functions have been reported to also impact transcription by RNA polymerases I and III (19–22). Moreover, a recent work has demonstrated that TFIIS is bound to any transcribed locus of the nuclear genome, including the rDNA regions transcribed by RNA pol I (23). We confirmed the reported binding of TFIIS to rDNA (Supplementary Figure S7A) and noted a significant decrease in this binding upon 6AU treatment (Figure 2A). rDNA occupancy by RNA pol I was transiently influenced by 6AU, as expected for a stressful situation. However, this response was almost identical in a dst1Δ mutant (Figure 2B), suggesting that TFIIS does not play a major role in rDNA transcription during NTP depletion.
Figure 2.
The presence of TFIIS in rDNA did not influence the distribution of RNA polymerase I along rDNA. (A) The ChIP experiments reveal a constant binding of TFIIS along rDNA, which partially decreased upon 100 µg/ml 6AU addition. Location of the amplicons utilized for quantitative PCR are shown in Graph. (B) Variation in the distribution of RNA polymerase I along rDNA upon 6AU addition, as measured by the ChIP experiments, using antibodies against a HA-tagged version of the biggest subunit of the enzyme. Rpa190 ChIP data are represented as being normalized to a non-transcribed amplicon within NTS2 (N). Note that addition of 6AU (100 µg/ml) led to a transient decrease in the amount of RNA polymerase I bound to the transcribed region, and that this decrease was equally transient in the absence of TFIIS. All the values represent the average of three independent experiments. Error bars indicate standard deviation. (C) Variation in the specific activity of the RNA polymerases sitting on rDNA caused by 6AU addition (100 µg/ml) to the wild-type and to an isogenic dst1Δ strain. RNA polymerase I specific activity was expressed as the ratio between variation in the transcriptional run-on signal (shown in Supplementary Figure S10A) and variation in the Rpb3 ChIP signal [shown in (B)]. In the latter, amplicons 3 and 5 were used to achieve the ratios in combination with the 18S and 25S run-on signals, respectively.
We also monitored the amount of transcriptionally active polymerases in dst1Δ sitting on rDNA by following transcriptional run-on techniques. The run-on results did not show lower levels of active RNA pol I molecules in dst1Δ (Supplementary Figure S10A). Moreover, the run-on/Rpa190 ratios, calculated by combining the data from Figure 2B and Supplementary Figure S10A, confirm that TFIIS does not play a relevant role in preserving the activity of elongating RNA pol I under NTP depletion since similar profiles were observed in dst1Δ and in the wild-type (Figure 2C).
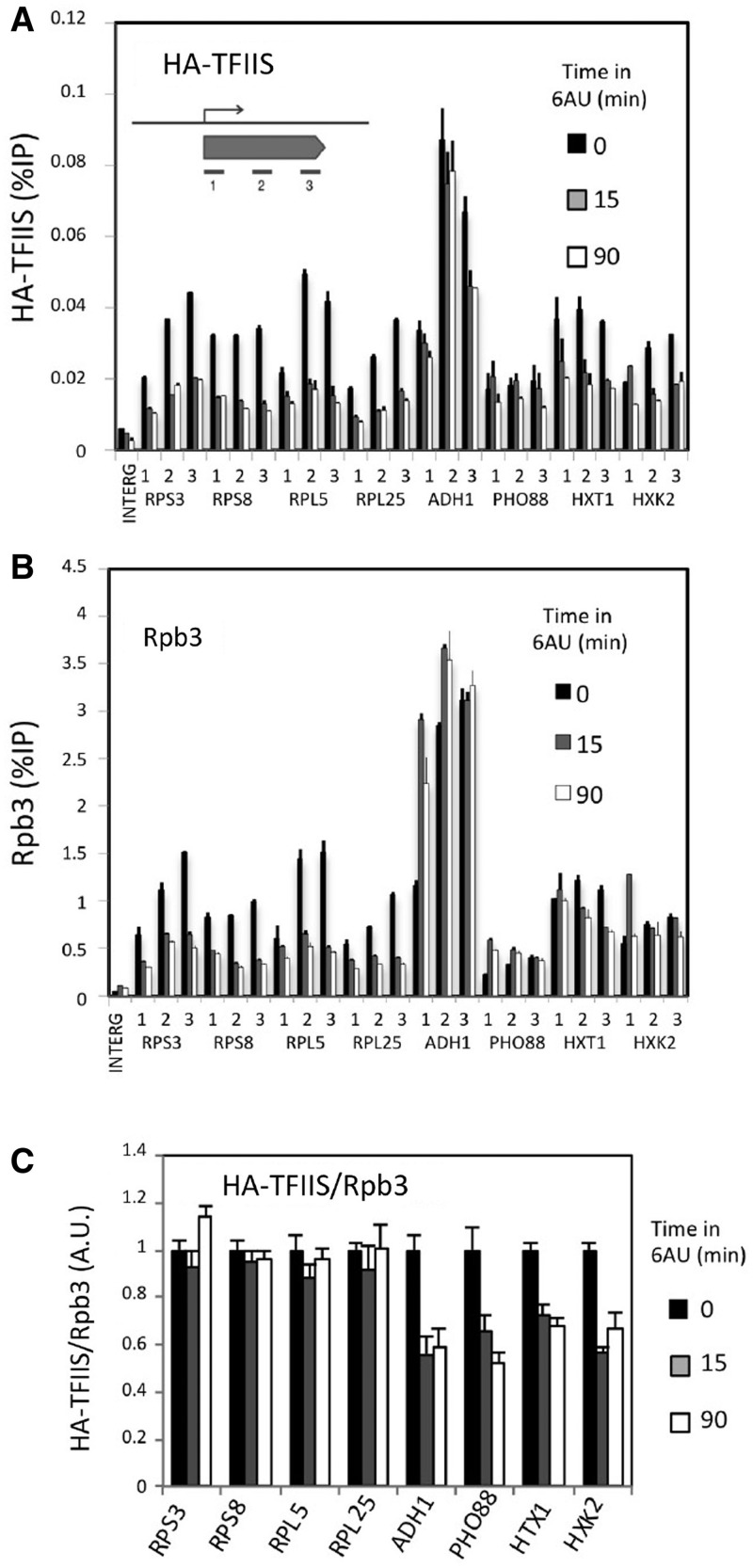
We also detected the presence of TFIIS in the genes encoding RPs and other highly expressed RNA pol II-dependent genes (Supplementary Figure S7B). We measured TFIIS binding at three different positions within the transcribed region of 10 different genes: four RP genes (RPS3, RPS8, RPL5 and RPL25), two RiBi genes functionally related to ribosomes, but which do not encode RPs (RPA43 and RRP12), and four genes with no direct functional link to ribosomes (ADH1, PHO88, HXT1 and HXK2) (Figure 3A and Supplementary Figure S8A). TFIIS ChIP signals were consistent and showed intensities that were highly proportional to the amount of RNA polymerase II present in the genes, as measured by Rpb3 ChIP (Figure 3B and Supplementary Figure S8B). Addition of 6AU to cultures led to a decrease of TFIIS through the four RP genes, HXT1 and HXK2, and to minor effects in the other four genes (Figure 3A and Supplementary Figure S8A). 6AU also brought about a rapid decrease of the RNA polymerase II occupancy of the RP genes and RRP12 across their entire length (Figure 3B and Supplementary Figure S8B). This was not the case for the other tested genes, which showed an accumulation of RNA polymerase II at the 5′-end, as previously described (4), with no significant decrease noted along the transcribed region (Figure 3B and Supplementary Figure S8B). The comparison between the TFIIS and the Rpb3 ChIP results uncovers a striking difference between ribosome-related genes and the rest. Whereas RP genes, and to a lesser extent the two RiBi genes, showed parallel changes of TFIIS and Rpb3 signals in response to 6AU, the other four genes exhibited an imbalance between Rpb3 and TFIIS upon 6AU addition (Figure 3A–B and Supplementary Figure S8A–S8B).
Figure 3.
TFIIS and RNA polymerase II occupancy in response to 6AU. Changes in HA-TFIIS (A) and Rpb3 (B) binding to RNA polymerase II-dependent genes in response to 6AU (100 µg/ml). All the values represent the average of three independent experiments at three different amplicons distributed along the genes. Samples were taken from the same extracts to analyse HA-TFIIS and Rpb3 in parallel. ChIP signals were quantified in relation to the input material. The results of an untranscribed intergenic region (Chromosome V, co-ordinates 9716–9863) are also shown. Error bars indicate standard deviation. (C) Variation of TFIIS/RNA polymerase II ratios upon 6AU (100 µg/ml) addition, as measured by ChIP experiments utilizing antibodies against HA-TFIIS and Rpb3. All the values represent the average of three independent experiments and three different amplicons [shown in (A) and (B)].
In order to quantify the impact of 6AU on the transcriptional availability of TFIIS, we calculated the average TFIIS/Rpb3 ratio for each gene, normalized to the initial value (before 6AU addition). We omitted the two RiBi genes because of their very low Rpb3 ChIP values, close to those of a non-transcribed control (Supplementary Figure S8B). We found that the TFIIS/Rpb3 ratios remained unchanged in the four RP genes upon the addition of 6AU, whereas the RNA polymerases transcribing the four non-ribosomal genes became TFIIS-impoverished (Figure 3C). This difference detected between RP and the non-ribosomal genes was observed along the entire length of the genes (Supplementary Figure S8C).
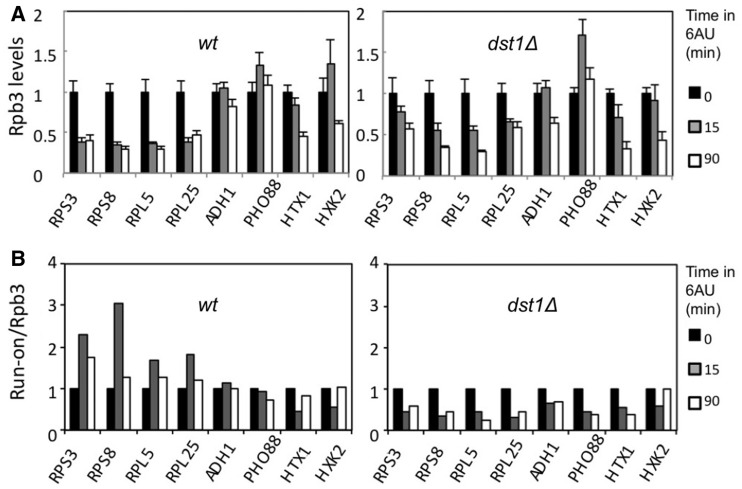
These results suggest a preferential binding of TFIIS to those RNA polymerases transcribing RP genes upon 6AU addition. In order to investigate whether this phenomenon had any functional influence on RP transcription, we monitored the variation of RNA polymerase II occupancy in response to 6AU in the dst1Δ mutant. We found that the RP genes underwent a slower RNA polymerase II decrease in the mutant than in the wild-type, whereas the non-RP genes behaved more similarly than the RP genes in the two strains (Figure 4A). The difference for RP genes between the dst1Δ and the wild-type became particularly clear for the 5′-end; the differential effect of 6AU on the Rpb3 profiles (from 5′ to 3′) that we detected between RP and non-RP genes was milder in dst1Δ than in the wild-type (Supplementary Figure S9A and S9B).
Figure 4.
TFIIS sustains RNA polymerase II activity in RP genes under transcriptional stress. (A) Variation in the levels of the RNA polymerase II bound to the indicated genes, caused by the addition of 6AU (100 µg/ml) to both the wild-type and an isogenic dst1Δ strain. All the values represent the average of three independent experiments and three different amplicons distributed along the indicated genes (Supplementary Figure S9A and S9B) normalized to time 0. (B) Variation in the specific activity of RNA polymerases sitting on the indicated genes caused by 6AU addition (100 µg/ml) to the wild-type and to an isogenic dst1Δ strain. RNA polymerase II specific activity was expressed as the ratio between the variation in transcriptional run-on signal (Supplementary Figure S10A) and the variation in the Rpb3 ChIP signal [shown in (A)].
We measured the amount of the transcriptionally active polymerases on the different genes tested by transcriptional run-on. Unlike ChIP, which can detect any RNA pol II molecule bound to DNA independently of its transcriptional status, transcriptional run-on allows the specific detection of those RNA polymerases actively engaged in transcription, which do not take a backtracked configuration. We assumed that each active polymerase produces a similar signal in the run-on assay irrespectively of its position within the genome (34); accordingly, any variation in the run-on signal of a gene would involve a change in the number of active RNA polymerases actually transcribing such a gene. In the wild-type, most of the genes analysed were able to maintain their run-on signals unchanged or close to initial values after 15 min in 6AU (Supplementary Figure S10A). In dst1Δ, all the genes showed decreased run-on signals upon 6AU addition, but this decrease was especially intense in the four RP genes, whose profiles showed the largest difference when compared to the wild-type (Supplementary Figure S10A).
The comparison made between Rpb3 ChIP and the run-on results offer additional clues to interpret these experiments. The specific reduction in RNA pol II occupancy exhibited in the wild-type by RP genes after 15 min in the presence of 6AU (Figure 4A) was not reflected in the density of the active polymerases measured by transcriptional run-on (Supplementary Figure S10A). The same comparison made in dst1Δ offers the opposite outcome for RP genes: a slower decrease of Rpb3 than the run-on (Figure 4A and Supplementary Figure S10A). These results suggest that TFIIS plays an important role in RP genes during NTP depletion by maintaining their RNA polymerase II population fully active. The run-on/Rpb3 ratios calculated for each gene confirm a marked increase of RNA polymerase II specific-activity (active transcription/total RNA polymerase II) in the four RP genes upon 6AU addition, which was absent in non-RP genes (Figure 4B). After 90 min in 6AU, a condition under which the up-regulation of IMD2 must diminish NTP stress, RP genes still exhibited higher run-on/Rpb3 ratios than the four control genes, although the difference had diminished considerably (Figure 4B). In contrast, RNA polymerase II specific-activity sharply dropped in the dst1Δ cells in all the genes tested upon 6AU addition (Figure 4B), thus confirming that the higher run-on/Rpb3 ratios exhibited by the four RP genes in the wild-type depend on TFIIS. We conclude that the sustained TFIIS/RNA polymerase II ratio exhibited by RP genes after NTP depletion is responsible for the high RNA polymerase II-specific activity detected in these genes.
Consequences of the absence of TFIIS in the expression of rDNA and RP genes
The differential effect of the absence of TFIIS in rDNA and RP genes under NTP stress predicts an imbalance between newly synthesized ribosome components (rRNAs and RPs) and a general disarrangement in ribosome biogenesis. We followed the changes in the levels of the rRNA precursor 35S and two RP mRNAs in response to 6AU addition. We noted a transient decrease of 35S at 10–30 min after 6AU addition, which is consistent with the detected down-regulation of rDNA transcription (Figure 2B); a subsequent transient decrease of RPS3 and RPL5 mRNAs (60 min) and a final equilibrium between 35S and RP mRNAs after 3 h in 6AU (Figure 5A). The mRNA levels of a non-RP gene like ADH1 scarcely varied (Figure 5A).
Figure 5.
TFIIS is required for the balance between rDNA transcripts and RPs mRNAs. Northern blots showing the variation of the 35S primary rRNA precursor and the mRNAs of the indicated genes upon 6AU addition (100 µg/ml) in the wild-type (A) and in the dst1Δ (B), imd2Δ (D), sfp1Δ (E) and dst1Δsfp1Δ (F) isogenic strains. The quantifications for 35S and RPL5 mRNA are shown in order to visualize the 35S/RP mRNA balance. (C) Levels of the 35S, 32S and 27SA rRNA precursors measured by Northern blot upon 6AU addition (100 µg/ml) in the wild-type and in an isogenic dst1Δ strain. Note that the accumulation of 35S detected in dst1Δ is not coupled with a similar accumulation of either 32S or 27SA, as would be expected for a general increase in rDNA transcription. 18S rRNA is shown as the loading control in all the panels.
In contrast to the wild-type, 35S rRNA accumulated and produced a significantly altered 35S/RP mRNAs ratio in the absence of TFIIS (Figure 5B). This imbalance could be the result of either the over-activation of RNAP I transcription or the consequence of rRNA processing impairment. In order to distinguish between these two possibilities, we measured the levels of two different intermediates downstream of 35S, 32S and 27SA. Under standard conditions, the 32S and 27SA rRNAs levels mirror those of 35S. Any accumulation of 35S over 32S or 27SA indicates a defect in current ribosome biogenesis (35). We found that the dst1Δ cells did not accumulate significant levels of 32S or 27SA rRNAs in the presence of 6AU (Figure 5C), indicating that 35S accumulation was not due to a late up-regulation of the rDNA transcription, but was directly owing to a failure in ribosome biogenesis.
The results described so far support a direct and specific role of TFIIS in the transcription of RP genes. According to this view, the malfunction of RP transcription under NTP stress may explain the impairment of ribosome biogenesis. However, there is an alternative interpretation of these data; we may argue that the role of TFIIS might be indirect as it contributes to the recovery of NTP pools via IMD2 induction. To test this hypothesis, we followed the effect of 6AU on the RP mRNAs and 35S levels in an imd2Δ mutant. We found that the RNA kinetics were surprisingly similar to the wild-type, with a decay of all the RNAs and a final 35S/RP mRNAs equilibrium after 2 h (Figure 5D). In this mutant, ADH1 mRNA underwent a severe, sustained decrease (Figure 5D). These results support the direct role of TFIIS, not mediated by the expression of IMD2, in balancing the transcription of those genes encoding ribosomal components.
sfp1Δ overcomes the requirement of TFIIS in RP genes’ response to NTP depletion
We also followed the levels of RP mRNAs and 35S upon 6AU addition in sfp1Δ and in the double dst1Δ sfp1Δ mutant. We observed that the 35S and the RP mRNA levels in sfp1Δ lowered less intensely than in the wild-type (Figure 5E). Accordingly, an imbalanced 35S/RP mRNAs ratio upon 6AU addition was never observed in the double mutant (Figure 5F). These results suggest that sfp1Δ is able to prevent the deleterious effects produced by 6AU on RP genes transcription in the absence of TFIIS.
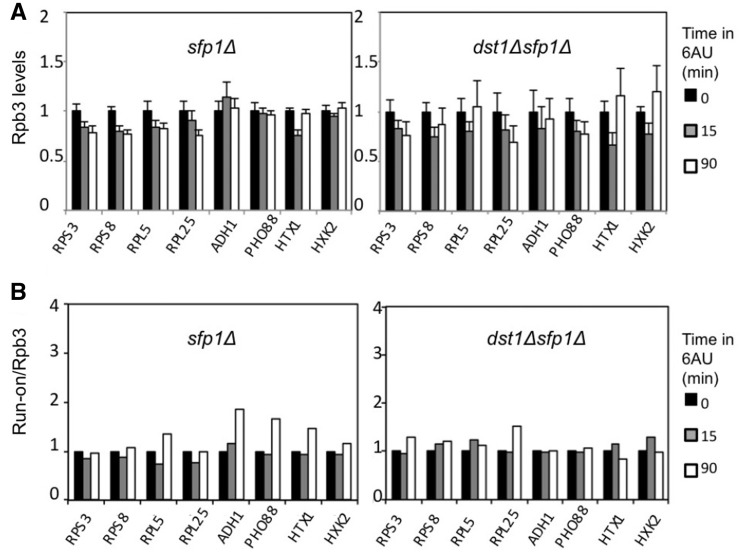
In order to confirm this hypothesis, we analysed the transcriptional response of both RP and non-RP genes to 6AU in both sfp1Δ and dst1Δ sfp1Δ. Unlike the marked decrease detected in the RNA pol II occupancy of RP genes upon 6AU addition in the wild-type (Figure 4A), a reduction of only 20% was observed in sfp1Δ (Figure 6A). Likewise, the RNA pol II decrease of the four RP genes in dst1Δ (Figure 4A) was absolutely abolished in dst1Δ sfp1Δ (Figure 6A). Similar results were obtained with the run-on assay (Supplementary Figure S10B). Consequently upon 6AU addition, the run-on/Rpb3 ratios of the RP genes in sfp1Δ and in dst1Δ sfp1Δ did not undergo any significant variation that was comparable to those underwent by the same genes in the wild-type or in dst1Δ (compare Figures 4B and 6B). We conclude that RP genes’ transcriptional behaviour changes in the absence of Sfp1, to such an extent that it enables them to remain active in the presence of 6AU, independently of TFIIS.
Figure 6.
Deletion of SFP1 almost eliminates the response of RNA polymerase II-dependent genes to 6AU, even in the absence of TFIIS. (A) Variation in the levels of the RNA polymerase II bound to the indicated genes, caused by the addition of 6AU (100 µg/ml) to sfp1Δ and dst1Δsfp1Δ cells. All the values represent the average of three independent experiments and three different amplicons distributed along the indicated genes (Supplementary Figure S9C and S9D). (B) Variation in the specific activity of the RNA polymerases sitting on the indicated genes caused by the addition of 6AU (100 µg/ml) to the sfp1Δ and dst1Δsfp1Δ cells. RNA polymerase II-specific activity was expressed as the ratio between variation in the transcriptional run-on signal (Supplementary Figure S10B) and variation in the Rpb3 ChIP signal [shown in (A)]. Scales were set to facilitate a comparison to Figure 4A and B, respectively.
This change might merely consist in a general down-regulation of the number of initiating RNA pol II molecules, as deduced from the low Rpb3 ChIP signals of the RP genes in sfp1Δ and dst1Δsfp1Δ (Supplementary Figure S9C and S9D). However, our results provide some clues to suggest that Sfp1’s transcriptional role could go beyond merely regulating RP genes’ RNA pol II initiation. One intriguing finding is that the absence of Sfp1 not only abolishes RP and non-RP genes’ differential response to 6AU, but also seems to modify non-RP genes’ transcriptional response to 6AU. Several lots of data support this view. Firstly, non-RP genes also showed lower levels of RNApol II in sfp1Δ and dst1Δsfp1Δ (Supplementary Figure S9C and S9D). No accumulation of RNA polymerase II at the 5′ of non-RP genes was observed in either sfp1Δ or dst1Δ sfp1Δ (Supplementary Figure S9C–D). Moreover, no decrease in either RNA pol II occupancy or the run-on signal was observed for non-RP genes in dst1Δsfp1Δ, even after 90 min in the presence of 6AU (Figure 6A and Supplementary Figure S10B). Accordingly, the run-on/Rpb3 ratios of the non-RP genes in dst1Δ sfp1Δ did not change upon 6AU addition (Figure 6B), which contrasts to their significant variation in dst1Δ (Figure 4D). Finally, the run-on signals and run-on/Rpb3 ratios of the non-RP genes in sfp1Δ increased after 90 min in the presence of 6AU (Figure 6B and Supplementary Figure S10B). All these data suggest a general effect of Sfp1 on elongation by RNA polymerase II, which is especially relevant for the transcription of RP genes.
DISCUSSION
This work shows that sensitivity to NTP-depleting drugs caused by the absence of TFIIS can be suppressed by the mutation of some of the genes affecting the regulation of ribosome biogenesis. This suppression even takes place under conditions in which IMD2 is not significantly up-regulated, indicating that a change in RP genes’ expression allows RNA pol II-dependent transcription to support cell growth in the absence of TFIIS and under severe NTP stress. Accordingly, we propose that TFIIS is required particularly for the transcription of RP genes under NTP-depletion conditions.
The detailed transcriptional analysis of some RP genes supports this view. We found that, upon 6AU treatment, the TFIIS/RNA pol II ratios of RP genes are substantially higher than those exhibited by non-ribosomal genes, including strongly transcribed ADH1 (Figure 3C). As a recent structural work reveals, the TFIIS–RNA pol II complex is incompatible with the RNA polymerization reaction (36). Accordingly TFIIS, recruitment likely responds to a previous backtracked configuration, and should involve the reactivation of arrested RNA polymerases. In fact, the preferential occupancy of RP genes by TFIIS in response to 6AU (Figure 3C) agrees with the strongly increased RNA pol II specific-activity found in RP genes, as reflected by the run-on/Rpb3 ratios (Figure 4B). The simplest explanation for this correlation is that TFIIS is preferentially recruited to those elongating RNA pol II molecules which become backtracked in RP genes when NTPs are scarce. This hypothesis involves a greater tendency of RNA pol II to become backtracked in RP genes than in non-RP genes. This is, in fact, one of the conclusions drawn by a previous study from our lab, which revealed that RP genes exhibit the highest RNA pol II ChIP/run-on ratios throughout the genome, interpreted as the highest proportion of arrested RNA pol II (25). Similarly, a nascent elongating transcript sequencing (NET-seq) database has revealed a high frequency of pausing in RP genes (14). Alternatively, the relatively high TFIIS/Rpb3 ratios exhibited by RP genes upon 6AU treatment could well be the result of a longer resident time of the TFIIS–RNApol II complexes under down-regulation conditions. Reactivated RNApol II (after RNA cleavage) has been described to stay in place for a time before resuming transcription (14). One possible cause for this delay may be the slow kinetics of TFIIS dissociation. In a down-regulation situation of RP genes, provoked by NTP stress, this slow TFIIS off-rate could involve a transient enrichment in the TFIIS–RNA pol II complexes, these being competent complexes for run-on. In either of the alternative explanations we offer herein, the higher TFIIS/Rpb3 ratios exhibited by RP genes and the differential effect of dst1Δ indicate a crucial role for TFIIS in the transcription of RP genes.
Our aforementioned work also demonstrated that the RNA pol II ChIP/run-on ratio of RP genes could be regulated. Mutants like tpk2Δ, affecting RP genes’ response to the Ras–PKA pathway, or RAP1DSIL, affecting the main gene-specific transcription factor of RP genes, caused a substantial increase in the proportion of active RNA pol II (run-on signal) without changing the relative amount of bound polymerase (ChIP signal) (25). We thought it most significant that the same mutations could suppress the sensitivity of dst1Δ to 6AU (Supplementary Figure S6). The silencing domain of RAP1 is able to recruit SIR proteins which, in turn, contribute to the formation of a high-order silent chromatin structure (37). Its genetic interaction with TFIIS suggests a role of heterochromatic chromatin in RP genes’ transcriptional behaviour. In a different study, we also described how RP genes display the lowest 3′/5′ ratios of active transcription (measured by run-on) of the yeast genome (28). All these data indicate that the likelihood of RNA pol II molecules pausing and backtracking is greater in RP genes than in most other genes and, therefore, the requirement of TFIIS is maximal in RP genes.
As stated in the ‘Results’ section, we assume that all the RNA polymerase II molecules are able to produce similar run-on signals, at least on average. We base this assumption on the stringent run-on assay conditions, which should provide a homogenous template (nucleosome-free DNA) for each elongating RNA polymerase (34). Alternative interpretations of the run-on signal variation, i.e. a different length of the run-on transcript, would change the molecular meaning of our results, but would not change the main message of this work: the differential transcriptional behaviour of the RNA pol II molecules when transcribing RP genes and their higher dependency on TFIIS in comparison with other highly transcribed genes.
It has been recently demonstrated that backtracking and RNA cleavage are very common phenomena and that they are likely consubstantial to RNA pol II-dependent transcription in vivo (3,14). This is in good agreement with previous in vitro experiments (13). However, life without TFIIS is possible in yeast under standard growing conditions since dst1Δ mutants are viable. This viability is explained by the basal intrinsic cleavage activity of RNA pol II, which takes place even in the absence of the stimulatory influence of TFIIS (2,3). Yet under NTP-depleting conditions, the frequency of backtracking in RP genes in the absence of TFIIS would be so high that cells would undergo a RP shortage and would eventually become unviable. This RP shortage occurring in dst1Δ is reflected by the imbalance between 35S and RP mRNAs. RNA pol I ChIP data do not suggest that this imbalance is caused by the up-regulation of rDNA Transcription (Figure 2B). Other rRNA intermediates, like 32S and 27SA, do not accumulate in parallel to 35S (Figure 5C). All together, these results not only indicate a serious problem in rRNA processing caused by 6AU in dst1Δ, but favour the conclusion that TFIIS contributes to the coordination of the genome fraction which encodes ribosomal elements.
According to the transcriptional analysis that we performed, the role of TFIIS in RP genes’ response to 6AU is sufficient to explain the defect detected in rRNA biogenesis. TFIIS might also be involved in the transcriptional response of ribosome biogenesis factors to NTP depletion. However, the suppression of dst1Δ sensitivity to NTP-depleting drugs by the mutations in RAP1 does not support this possibility since Rap1 is a transcription factor that regulates almost every RP gene, but does not bind most RiBi promoters. We conclude that RP genes are the main targets of TFIIS in the response of ribosome biogenesis to 6AU. Nevertheless, a role of TFIIS in RiBi genes under transcriptional stress cannot be ruled out.
TFIIS seems to be partially dispensable to resist NTP-depleting drugs in the absence of DAS genes, including the RP genes regulators SFP1 and SCH9. Sch9 is the yeast homologue of mammalian Akt/PKB and regulates stress-resistance by the activation of stress-responsive genes (38,39). Sfp1 controls the transcription of ribosomal-related genes in response to nutrients (40). Communication of the growth potential to ribosome synthesis is a common role of Sch9 and Sfp1 (30) since other targets of Sch9 are not influenced by Sfp1 (41), and since sfp1Δ and sch9Δ exhibits opposite phenotypes in response to other environmental signals (42). This fact suggests that das mutations cause major alterations in the regulatory mechanisms of ribosomal genes to the extent that the arrest-prone scenario of RP genes would be prevented, thus alleviating their dependency on TFIIS under NTP stress.
We investigated the case of sfp1Δ and shown that this mutation suppresses the imbalance between 35S and RP mRNAs provoked by 6AU in the absence of TFIIS (Figure 5). This result suggests that sfp1Δ is able to rescue RP transcription from TFIIS-dependency since TFIIS does not play a functional role in the response of rDNA to 6AU (Figure 2B). This is exactly what we found: when Sfp1 is absent, RP transcription is not significantly affected by 6AU in either a dst1Δ or a wild-type background (Figure 6). A simple explanation for these results would involve a transcriptional function of Sfp1 upstream of TFIIS and in an antagonist fashion, which would favour RNA polymerase II backtracking.
One unexpected result we found is the general effect of sfp1Δ. All the tested genes (RP and non-RP) were almost insensitive to 6AU in the absence of Sfp1. So far, all the transcriptional functions of Sfp1 had been restricted to RP genes. The general effects we detected of Sfp1 may prove to be an indirect result of its role in RP regulation. Alternatively, Sfp1 might perform a more general function in RNA pol II-dependent transcription which, in the case of RP genes, would be especially relevant for their expression.
The present work describes how TFIIS plays an important regulatory role for a fraction of the yeast genome (RP genes) whose transcription is modulated by RNA pol II inactivation/reactivation during elongation (25). Gene-expression regulation at the transcription elongation level is, in fact, a very common phenomenon in metazoa. It is tempting to predict a general regulatory role for TFIIS in higher eukaryotes, as already demonstrated for the heat shock genes in Drosophila (43) and for the growth-promoting genes in humans (44).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–10.
FUNDING
Funding for open access charge: Ministry of Economy and Competitiveness [BFU2007-67575-C03-02, BFU-2010-21975-C03-03 to S.Ch. and FPI fellowships to L.deM. and L.D.-R.]; the Andalusian Government [P07-CVI-02623 and P08-CVI-03508]; the European Union (Regional Development European Fund).
Conflict of interest statement. S.Ch. is a shareholder of Ingeniatrics.
Supplementary Material
ACKNOWLEDGEMENTS
The authors dedicate this work to the memory of Pierre Thuriaux. The authors thank Jesús de la Cruz, José E. Pérez-Ortín, Alfonso Rodríguez-Gil, Juan Carlos Rodríguez-Aguilera, Rico Bongaarts and all the members of de la Cruz and Chávez laboratories for their ideas and experimental help, and Michel Werner for his helpful comments. The authors also thank Alistair Chambers and Myke Tyers for their generous gifts of strains and plasmids. The experimental support from the University of Seville research facilities (CITIUS) is also acknowledged.
REFERENCES
- 1.Fish RN, Kane CM. Promoting elongation with transcript cleavage stimulatory factors. Biochim. Biophys. Acta. 2002;1577:287–307. doi: 10.1016/s0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- 2.Weilbaecher RG, Awrey DE, Edwards AM, Kane CM. Intrinsic transcript cleavage in yeast RNA polymerase II elongation complexes. J. Biol. Chem. 2003;278:24189–24199. doi: 10.1074/jbc.M211197200. [DOI] [PubMed] [Google Scholar]
- 3.Sigurdsson S, Dirac-Svejstrup AB, Svejstrup JQ. Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol. Cell. 2010;38:202–210. doi: 10.1016/j.molcel.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell. 2005;17:83–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 5.Prather DM, Larschan E, Winston F. Evidence that the elongation factor TFIIS plays a role in transcription initiation at GAL1 in Saccharomyces cerevisiae. Mol. Cell Biol. 2005;25:2650–2659. doi: 10.1128/MCB.25.7.2650-2659.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guglielmi B, Soutourina J, Esnault C, Werner M. TFIIS elongation factor and Mediator act in conjunction during transcription initiation in vivo. Proc. Natl Acad. Sci. USA. 2007;104:16062–16067. doi: 10.1073/pnas.0704534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malagon F, Tong AH, Shafer BK, Strathern JN. Genetic interactions of DST1 in Saccharomyces cerevisiae suggest a role of TFIIS in the initiation-elongation transition. Genetics. 2004;166:1215–1227. doi: 10.1534/genetics.166.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uzureau P, Daniels JP, Walgraffe D, Wickstead B, Pays E, Gull K, Vanhamme L. Identification and characterization of two trypanosome TFIIS proteins exhibiting particular domain architectures and differential nuclear localizations. Mol. Microbiol. 2008;69:1121–1136. doi: 10.1111/j.1365-2958.2008.06348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito T, Arimitsu N, Takeuchi M, Kawamura N, Nagata M, Saso K, Akimitsu N, Hamamoto H, Natori S, Miyajima A, et al. Transcription elongation factor S-II is required for definitive hematopoiesis. Mol. Cell Biol. 2006;26:3194–3203. doi: 10.1128/MCB.26.8.3194-3203.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taira Y, Kubo T, Natori S. Participation of transcription elongation factor XSII-K1 in mesoderm-derived tissue development in Xenopus laevis. J. Biol. Chem. 2000;275:32011–32015. doi: 10.1074/jbc.M003920200. [DOI] [PubMed] [Google Scholar]
- 11.Grasser M, Kane CM, Merkle T, Melzer M, Emmersen J, Grasser KD. Transcript elongation factor TFIIS is involved in arabidopsis seed dormancy. J. Mol. Biol. 2009;386:598–611. doi: 10.1016/j.jmb.2008.12.066. [DOI] [PubMed] [Google Scholar]
- 12.Nakanishi T, Nakano A, Nomura K, Sekimizu K, Natori S. Purification, gene cloning, and gene disruption of the transcription elongation factor S-II in Saccharomyces cerevisiae. J. Biol. Chem. 1992;267:13200–13204. [PubMed] [Google Scholar]
- 13.Galburt EA, Grill SW, Wiedmann A, Lubkowska L, Choy J, Nogales E, Kashlev M, Bustamante C. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446:820–823. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- 14.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Exinger F, Lacroute F. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- 16.Shaw RJ, Reines D. Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol. Cell Biol. 2000;20:7427–7437. doi: 10.1128/mcb.20.20.7427-7437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuehner JN, Brow DA. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol. Cell. 2008;31:201–211. doi: 10.1016/j.molcel.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 18.Desmoucelles C, Pinson B, Saint-Marc C, Daignan-Fornier B. Screening the yeast “disruptome” for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. J. Biol. Chem. 2002;277:27036–27044. doi: 10.1074/jbc.M111433200. [DOI] [PubMed] [Google Scholar]
- 19.Schnapp G, Graveley BR, Grummt I. TFIIS binds to mouse RNA polymerase I and stimulates transcript elongation and hydrolytic cleavage of nascent rRNA. Mol. Gen. Genet. 1996;252:412–419. doi: 10.1007/BF02173006. [DOI] [PubMed] [Google Scholar]
- 20.Tschochner H. A novel RNA polymerase I-dependent RNase activity that shortens nascent transcripts from the 3' end. Proc. Natl Acad. Sci. USA. 1996;93:12914–12919. doi: 10.1073/pnas.93.23.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labhart P. Transcript cleavage in an RNA polymerase I elongation complex. Evidence for a dissociable activity similar to but distinct from TFIIS. J. Biol. Chem. 1997;272:9055–9061. doi: 10.1074/jbc.272.14.9055. [DOI] [PubMed] [Google Scholar]
- 22.Chédin S, Riva M, Schultz P, Sentenac A, Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 1998;12:3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghavi-Helm Y, Michaut M, Acker J, Aude JC, Thuriaux P, Werner M, Soutourina J. Genome-wide location analysis reveals a role of TFIIS in RNA polymerase III transcription. Genes Dev. 2008;22:1934–1947. doi: 10.1101/gad.471908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warner JR, Vilardell J, Sohn JH. Economics of ribosome biosynthesis. Cold Spring Harb. Symp. Quant. Biol. 2001;66:567–574. doi: 10.1101/sqb.2001.66.567. [DOI] [PubMed] [Google Scholar]
- 25.Pelechano V, Jimeno-Gonzalez S, Rodriguez-Gil A, Garcia-Martinez J, Perez-Ortin JE, Chavez S. Regulon-specific control of transcription elongation across the yeast genome. PLoS Genet. 2009;5:e1000614. doi: 10.1371/journal.pgen.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graham IR, Haw RA, Spink KG, Halden KA, Chambers A. In vivo analysis of functional regions within yeast Rap1p. Mol. Cell Biol. 1999;19:7481–7490. doi: 10.1128/mcb.19.11.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burns N, Grimwade B, Ross-Macdonald PB, Choi EY, Finberg K, Roeder GS, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez-Gil A, Garcia-Martinez J, Pelechano V, Munoz-Centeno Mde L, Geli V, Perez-Ortin JE, Chavez S. The distribution of active RNA polymerase II along the transcribed region is gene-specific and controlled by elongation factors. Nucleic Acids Res. 2010;38:4651–4664. doi: 10.1093/nar/gkq215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kus BM, Caldon CE, Andorn-Broza R, Edwards AM. Functional interaction of 13 yeast SCF complexes with a set of yeast E2 enzymes in vitro. Proteins. 2004;54:455–467. doi: 10.1002/prot.10620. [DOI] [PubMed] [Google Scholar]
- 30.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hontz RD, Niederer RO, Johnson JM, Smith JS. Genetic identification of factors that modulate ribosomal DNA transcription in Saccharomyces cerevisiae. Genetics. 2009;182:105–119. doi: 10.1534/genetics.108.100313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bell JB, Jones ME. Purification and characterization of yeast orotidine 5'-monophosphate decarboxylase overexpressed from plasmid PGU2. J. Biol. Chem. 1991;266:12662–12667. [PubMed] [Google Scholar]
- 33.Jenks MH, Reines D. Dissection of the molecular basis of mycophenolate resistance in Saccharomyces cerevisiae. Yeast. 2005;22:1181–1190. doi: 10.1002/yea.1300. [DOI] [PubMed] [Google Scholar]
- 34.Hirayoshi K, Lis JT. Nuclear run-on assays: assessing transcription by measuring density of engaged RNA polymerases. Methods Enzymol. 1999;304:351–362. doi: 10.1016/s0076-6879(99)04021-5. [DOI] [PubMed] [Google Scholar]
- 35.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 36.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 37.Sperling AS, Grunstein M. Histone H3 N-terminus regulates higher order structure of yeast heterochromatin. Proc. Natl Acad. Sci. USA. 2009;106:13153–13159. doi: 10.1073/pnas.0906866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- 39.Pascual-Ahuir A, Proft M. The Sch9 kinase is a chromatin-associated transcriptional activator of osmostress-responsive genes. EMBO J. 2007;26:3098–3108. doi: 10.1038/sj.emboj.7601756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- 41.Prusty R, Keil RL. SCH9, a putative protein kinase from Saccharomyces cerevisiae, affects HOT1-stimulated recombination. Mol. Genet. Genomics. 2004;272:264–274. doi: 10.1007/s00438-004-1049-x. [DOI] [PubMed] [Google Scholar]
- 42.Hosiner D, Lempiäinen H, Reiter W, Urban J, Loewith R, Ammerer G, Schweyen R, Shore D, Schüller C. Arsenic toxicity to Saccharomyces cerevisiae is a consequence of inhibition of the TORC1 kinase combined with a chronic stress response. Mol. Biol. Cell. 2009;20:1048–1057. doi: 10.1091/mbc.E08-04-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell. 2005;17:103–112. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 44.Shema E, Kim J, Roeder RG, Oren M. RNF20 inhibits TFIIS-facilitated transcriptional elongation to suppress pro-oncogenic gene expression. Mol. Cell. 2011;42:477–488. doi: 10.1016/j.molcel.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.