Abstract
Bacteria and yeast possess one RecQ helicase homolog whereas humans contain five RecQ helicases, all of which are important in preserving genome stability. Three of these, BLM, WRN and RECQL4, are mutated in human diseases manifesting in premature aging and cancer. We are interested in determining to which extent these RecQ helicases function cooperatively. Here, we report a novel physical and functional interaction between BLM and RECQL4. Both BLM and RECQL4 interact in vivo and in vitro. We have mapped the BLM interacting site to the N-terminus of RECQL4, comprising amino acids 361–478, and the region of BLM encompassing amino acids 1–902 interacts with RECQL4. RECQL4 specifically stimulates BLM helicase activity on DNA fork substrates in vitro. The in vivo interaction between RECQL4 and BLM is enhanced during the S-phase of the cell cycle, and after treatment with ionizing radiation. The retention of RECQL4 at DNA double-strand breaks is shortened in BLM-deficient cells. Further, depletion of RECQL4 in BLM-deficient cells leads to reduced proliferative capacity and an increased frequency of sister chromatid exchanges. Together, our results suggest that BLM and RECQL4 have coordinated activities that promote genome stability.
INTRODUCTION
RecQ helicases are a family of evolutionarily conserved DNA unwinding proteins involved in DNA metabolic pathways to preserve genome stability in the cell (1–3). In bacteria and yeast, there is only one RecQ homolog whereas in humans and mice there are five different RecQ homologs, namely: RECQL1, WRN, BLM, RECQL4 and RECQL5. Among these, WRN, BLM and RECQL4 have been linked to autosomal recessive diseases. Werner syndrome is caused by defects in WRN, Bloom syndrome (BS) is caused by defects in BLM, and three diseases: Rothmund–Thomson syndrome, RAPADILLINO syndrome and Baller Gerold syndrome (BGS) are caused by mutations in RECQL4 [reviewed in (4)]. RecQ helicase members share common biochemical functions including 3′ → 5′ helicase, ATPase, strand annealing and DNA binding activities. However, WRN also has a unique 3′ → 5′ exonuclease activity (5). Among the human RecQ helicases WRN and BLM are well studied whereas much less is known about the cellular functions of RECQL4.
The RecQ helicase members function in different DNA metabolic processes including DNA replication, DNA repair and transcription (6–9). It is likely that they possess both unique and overlapping functions, depending upon specific circumstances. Thus, multiple RecQ helicases may be present in the same DNA repair complexes, and could modulate each others activities. For instance, WRN and BLM are known to work in several overlapping biological processes, and physical and functional interactions between these two RecQ helicases have been reported (10). BLM modulates WRN’s activity by inhibiting the unique exonuclease function of WRN, but the biological significance of their interaction is not well understood. In chicken DT40 cells, a redundant function of BLM and RECQL5 has been proposed to account for the higher frequency of sister chromatid exchanges (SCEs) in BLM−/−/RECQL5−/− cells (11,12). However, contradictory results have been seen in mouse embryonic stem cells, which led to the proposal of a non-redundant function for BLM and RECQL5 in the suppression of crossovers (13). Thus, the RecQ helicase functional interdependency varies and needs to be studied in a system-specific manner.
In this report, we report a novel cooperation between BLM and RECQL4 in human cells. The functions of BLM in the multiple steps of the homologous recombination (HR) repair pathway are well documented (1). Recently, we showed that RECQL4 is also involved in DNA double-strand break (DSB) repair (14). Also, both of the proteins are in complex with Rad51, a key protein in the HR pathway (15,16). Moreover, both BLM and RECQL4 are expressed abundantly during the S-phase of the cell cycle. RECQL4 is involved in initiation and assembly of the DNA replication machinery at the replication fork (6–8,17). In addition, BS cells exhibit replication abnormalities, including retarded replication fork elongation (18) and abnormal replication intermediates (19). Recently, BLM and Rif1 have been shown to work together in the recovery of stalled replication forks and to promote normal replication (20). Therefore, we hypothesized that BLM and RECQL4 might function together in the resolution of various secondary structures during DNA replication, or after stress induced with DNA damaging agents.
We show here that BLM and RECQL4 interact physically and functionally in vivo and in vitro. The interaction domains have been mapped to the N-termini of both BLM and RECQL4. RECQL4 specifically stimulates the unwinding activity of BLM on substrates resembling DNA replication forks with or without internal DNA damage. It is a specific interaction as RECQL4 does not stimulate the unwinding activity of other RECQ helicases. The characterization of RECQL4 and BLM double-deficient cells showed retarded growth and increased frequency of SCEs. Further, the retention of RECQL4 at the site of DNA DSBs is dependent on BLM function. Together, these findings demonstrate that BLM and RECQL4 interact in suppression of genomic instability.
MATERIALS AND METHODS
Cell lines
HeLa, SV40 transformed normal (GM00637) and BLM (GM08505) cell lines were obtained from Coriell Cell Repositories (Camden, NJ, USA). The HeLa cells were cultured in DMEM medium (Life Technologies, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) (Sigma, St Louis, MO, USA) and 1% penicillin–streptomycin at 37°C in a humidified atmosphere with 5% CO2. SV40 transformed normal and BLM fibroblasts were cultured in MEM medium (Life Technologies, Grand Island, NY, USA) supplemented with 10% FBS, 2 mM l-Glutamine, 1X vitamins, 1X non-essential amino acids (Life Technologies, Grand Island, NY, USA), and 1% penicillin–streptomycin at 37°C in a humidified atmosphere with 5% CO2.
Cell synchronization
HeLa cells were grown overnight then arrested in G0-G1 phase by serum starvation for 48 h. The cells were arrested in S-phase by treatment with 2 mM thymidine for 18 h followed by release in normal media containing 10% FBS for 6 h, and again treating the cells with thymidine for another 18 h before final release in normal media containing 10% FBS for 0, 2, 4, 6 and 8 h. The G2/M block was achieved by treating the cells with 75 mM nocodazole for 18 h. Synchronization was checked by fluorescence activated cell sorting (FACS) analysis on an Accuri C6 (BD Accuri Cytometers, Ann Arbor, MI, USA) with propidium iodide staining and analyzed by FlowJo software.
short hairpin RNA and siRNA-mediated RECQL4 knockdown
Stable RECQL4 knockdown (RECQL4 KD) and scrambled control were generated in HeLa cells according to standard protocols using Mission short hairpin RNA (shRNA) lentiviral construct TRCN0000051169 from Sigma-Aldrich (St. Louis, MO, USA). Briefly, the shRNA vector was co-transfected with packing plasmid pCMV-dr8.2 DVPR (Addgene, Cambridge, MA, USA) and envelope vector pCMV-VSV-G (Addgene, Cambridge, MA, USA) into human embryonic kidney 293T cells in DMEM medium with Hyclone FBS (Thermo Fisher Scientific Inc., Rockford, IL, USA) using FuGENE®6 transfection reagent (Roche Applied Sciences, IN, USA) and OptiMEM (Life Technologies, Grand Island, NY, USA). Medium was collected 48 h after transfection and filtered using 0.45-micron PVDF membrane filter (Millipore, Billerica, MA, USA). Virus titer was estimated using Lenti-X™ Go stix (Clontech, Mountain View, CA, USA). Lentivirus was added to the HeLa cells and infections were allowed to proceed for 48 h after which, puromycin was applied to select for lentiviral transduction. Q-PCR was performed showing that the RECQL4 knockdown was ∼90%. Knockdown was also confirmed by western blotting prior to use of the cells in experiments.
SiRNA-mediated RECQL4 KD was achieved by transfecting GM00637 and GM08505 cells (∼40–50% confluent) with 100 pmol of either control (ON-TARGET plus non-targetting siRNA #1, Dharmacon, Chicago, IL, USA) or RECQL4-targeted siRNA (target sequence CAAUACAGCUUACCGUACA, Dharmacon, IL, USA) and Lipofectamine 2000 transfection reagent (Life Technologies, Grand Island, NY, USA) for 6 h. Cells were then grown overnight and again treated with same siRNAs for 6 h. Cells were then grown in antibiotic-containing medium and harvested after 48 h. The RECQL4 level was checked by western blotting.
Growth assay
Four days post-infection of the fibroblasts with either scrambled or RECQL4 lentiviral shRNA, 5000 cells were seeded in 60 mm petri dish in triplicate. The cells were grown at 37°C in a humidified atmosphere with 5% CO2 for the indicated number of days. At the indicated times cells were trypsinized, and counted by a Z1 coulter particle counter (Beckman, Brea, CA, USA). The data were analyzed and the graph was plotted with the number of cells against time after normalization to controls. Experiments were performed in triplicate and the standard deviations were calculated.
Proteins
Wild-type (Wt) human RECQL4 and helicase-dead RECQL4 with a C-terminal 9-histidine tag in the pGEX6p1 vector (GE Healthcare, Piscataway, NJ, USA) were expressed and purified from Escherichia coli as described previously (21). Recombinant histidine-tagged BLM was overexpressed in Saccharomyces cerevisiae and purified as described previously (22). Recombinant histidine-tagged Wt WRN protein was purified using baculovirus expression system, as previously described (23). Recombinant histidine tagged RECQL5 was purified from E. coli (24). RECQL1 was a kind gift of Dr Robert Brosh (National Institute on Aging, Baltimore, USA). Protein concentrations were determined by the Bradford assay (Bio-Rad, Hercules, CA, USA), and purity was determined by SDS–PAGE and Coomassie staining.
The mutant RECQL4 (1–240) and (1–492) proteins were purified as follows. Bacterial pellets were sonicated in lysis buffer (50 mM Tris–HCl, pH 8.0, 300 mM NaCl, 10% glycerol, 25 mM imidazole, 1 mM PMSF, 1 tablet/50ml Complete EDTA-free Protease Inhibitor Cocktail, Roche, IN, USA), spun 30 min at 37 000 g, and filtered through 45 µm membrane. AKTA FPLC (GE Healthcare, Piscataway, NJ, USA) was employed for all chromatographic steps. Clarified lysate was loaded onto HisTrap FF 1 ml column equilibrated in Buffer A (50 mM Tris–HCl, pH 8.0, 300 mM NaCl, 10% glycerol) supplemented with 25 mM imidazole. The column was washed with 30 column volumes (CV) of equilibration buffer followed by 10 CV of Buffer A + 50 mM imidazole, and the protein eluted with Buffer A + 250 mM imidazole. Fractions with protein of interest were pooled, diluted 6-fold with Buffer B (50 mM Tris–HCl, pH 8.0, 10% glycerol) and loaded onto HiTrap Q HP 1 ml column equilibrated with Buffer B + 50 mM NaCl. The column was washed with 5 CV of equilibration buffer and the protein was eluted with 20 CV linear gradient from 50 mM to 1 M NaCl in Buffer B. Protein fractions were pooled, concentrated in Buffer C (50 mM Tris–HCl, pH 8.0, 200 mM NaCl) using Amicon ultra concentrators (Millipore, Billerica, MA, USA). Concentrated protein was supplemented with glycerol to 50% final concentration and stored at −80°C. All purification steps were carried out at 4°C.
DNA substrates
All the oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA, USA). A list of oligonucleotides used in this study is shown in Supplementary Table S1. For each substrate, a single oligonucleotide was 5′-end-labeled with [γ-32P]ATP (PerkinElmer Life Sciences, Waltham, MA, USA) using T4 polynucleotide kinase (New England Biolabs, Ipswich, MA, USA). The kinase reaction was performed in PNK buffer [70 mM Tris–HCl (pH 7.6), 10 mM MgCl2, 5 mM DTT] at 37°C for 1 h. The labeled substrate was purified using Micro Spin columns G-25 (GE healthcare, Piscataway, NJ, USA). The [γ32P]ATP-labeled oligonucleotides were then annealed to a 2-fold excess of the unlabeled complementary strands in annealing buffer [40 mM Tris–HCl (pH 8.0), 50 mM NaCl] by heating at 95°C for 5 min and then cooling slowly to room temperature. The G4 DNA substrates (25,26), D-loop (27), and HJ substrates (28) were prepared as described previously.
Helicase assays
Helicase reactions were performed in 20 µl reaction volume in helicase buffer containing 20 mM Tris–HCl (pH 7.4), 20 mM NaCl, 25 mM KCl, 1 mM DTT, 5% glycerol, 2.5 mM MgCl2, 2.5 mM ATP and 100 µg/ml BSA. Indicated amounts of protein were incubated with 0.5 nM of substrate, and the reactions were carried out for 30 min at 37°C. Proteins were diluted in buffer containing 20 mM Tris–HCl (pH 7.4), 50 mM NaCl, 1 mM DTT and 25% glycerol. Helicase reactions were stopped by adding 10 μl of 3× native stop dye (50 mM EDTA, 40% glycerol, 0.9% SDS, 0.05% bromophenol blue and 0.05% xylene cyanol). Products were separated by electrophoresis on 8% native polyacrylamide gels and exposed to a PhosphorImager plate (GE Healthcare, Piscataway, NJ, USA). Results were analyzed and quantified using ImageQuant version 5.2 (GE Healthcare, NJ, USA). The error bars represent the standard deviation of the three independent experiments.
In vivo co-immunoprecipitation
For co-immunoprecipitation experiments, HeLa cells were treated with either 10 Gy of ionizing radiations (IR), 5 mM hydroxyurea (HU) for 16 h, 50 nM camptothecin (CPT) for 48 h or 20 J/m2 of UV irradiation. HeLa cells were lysed in buffer containing 50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 2 mM EDTA, 1% TritonX-100 and 1× protease inhibitor cocktail (Roche Biosciences, IN, USA) for 30 min at 4°C. The lysate was spun down at 14 000 g for 20 min at 4°C and supernatant was collected. The lysate was then pre-cleared with 20 µl protein G agarose beads for 3 h in presence of ethidium bromide (50 µl/ml). The supernatant was mixed with 4 µg of either rabbit anti-RECQL4 (21) or normal rabbit IgG (sc 2027; Santacruz Biotechnology Inc., Santacruz, CA, USA) and incubated overnight at 4°C with rotation. The lysate was then mixed with 20 µl of protein G agarose beads for additional 2 h before washing the beads 4× with buffer containing 50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 2 mM EDTA, 0.1% Triton X-100. The washed beads were resuspended in 30 µl of 1× SDS sample loading buffer (Life Technologies, Grand Island, NY, USA) and resolved by 6% SDS–PAGE followed by western analysis as described below. The BLM, RECQL5 and RECQL1 were detected by rabbit anti-BLM (ab2179, Abcam, Cambridge, MA, USA), rabbit anti-RECQL5 (29), rabbit anti-RECQL1 (sc25547, Santa Cruz Biotechnology Inc., Santacruz, CA, USA) using the rabbit Trueblot system (eBiosciences, San Diego, CA, USA) and ECL plus detection kit.
In vitro co-immunoprecipitation
For the in vitro protein interactions, purified RECQL4 (10 pmol) and purified BLM (20 pmol) were mixed in the helicase buffer (described in helicase assay section) and incubated for 90 min at 4°C with rotation. Meanwhile, antibody coated protein G agarose beads were prepared. For this, 2 µg each of polyclonal anti-RECQL4 and normal rabbit IgG as control (sc-2027; Santa Cruz Biotech, CA, USA) were mixed with agarose beads and incubated with rotation at 4°C. Then pre-coated antibody agarose beads were added to the purified protein mix, and incubated again for 2 h at 4°C with rotation. After incubation the beads were washed 4× with wash buffer (same as for in vivo pull down). The washed beads were resuspended in 30 µl of 1× SDS sample loading buffer and resolved by 4–15% gradient SDS–PAGE followed by western analysis.
Yeast two-hybrid assay
Bait proteins were expressed as fusions with LexA in the yeast two-hybrid vector pLexA-Km (Dualsystems, Zurich, Switzerland). Preys were expressed as fusion to the activation domain of GAL4 in pACT2 (Clontech, Palo Alto, CA, USA). All plasmids were generated by using internal restriction sites of BLM or RECQL4, or by PCR-based method. Plasmids are available upon request. The S. cerevisiae strain NMY-70 (Dualsystems, Zurich, Switzerland) was co-transformed with bait and prey plasmids using lithium acetate method and selected on minimal medium SD-LW (2% glucose, 6.7% yeast nitrogen base, complete amino acid mix lacking leucine and tryptophan). For β-gal plate test, several colonies were picked from the selection plate and streaked onto SD-LW X-gal plate (containing additionally 75 mM KH2PO4, 0.05% (v/v) β-mercaptoethanol and 40 µg/ml X-Gal) and SD-LW plate as a control.
SCE analysis
Cells were incubated with 100 µM BrdU in MEM medium (described above) for 72 h. Cells were then treated with 0.1 µg/ml Colcemid for 5 h, harvested and immediately incubated in 75 mM KCl for 20 min at 37°C, followed by fixation in ice-cold (3:1) methanol and glacial acetic acid (30). Cells were dropped on slides and dried. Slides were then stained with 0.5 µg/ml Hoechst 33 258 in phosphate buffered saline for 10 min. Slides were exposed to 352 nm black light for 20 min followed by incubation in 2× SSC for 20 min. They were then stained using 3% Giemsa solution for 10 min. The slides were imaged with microscope under bright field and analyzed for SCEs (31).
Laser irradiation and confocal microscopy
The laser irradiation was done as described previously (14). In brief, the confocal microscope system integrated a Stanford Research Systems (SRS) NL100 nitrogen laser by Micropoint ablation system (Photonics Instruments, St. Charles, IL, USA). Site-specific DNA damage was induced using the SRS NL100 nitrogen laser. The power of the laser as a percent intensity was controlled through Improvison’s Volocity software 4.3.1 (Improvision/PerkinElmer, Coventry, UK). Positions internal to the nuclei of transfected cells with GFP-tagged plasmids were targeted with 21% laser intensity via a 40× oil objective lens. Images were captured at various time points and analyzed using Volocity version 4.3.1 build 6 (Improvision). Experiments were performed in an environmental chamber attached to the microscope to maintain the normal atmosphere for the cells (i.e. 37°C and 5% CO2).
Fluorescence recovery after photobleaching
Fluorescence recovery after photobleaching (FRAP) analysis was carried out with live cells transfected with GFP-tagged RECQL4 in normal fibroblasts and BLM-deficient fibroblasts. The same set up as for the microirradiation was used to perform the photobleaching. Fluorescence recovery was monitored as described in the figure legends, and the data for recovery were corrected for the background intensity and the loss of total fluorescence. Each experiment was performed at least 3×, and the data presented are mean intensity values obtained in a given experiment.
RESULTS
BLM and RECQL4 interact in vivo and in vitro
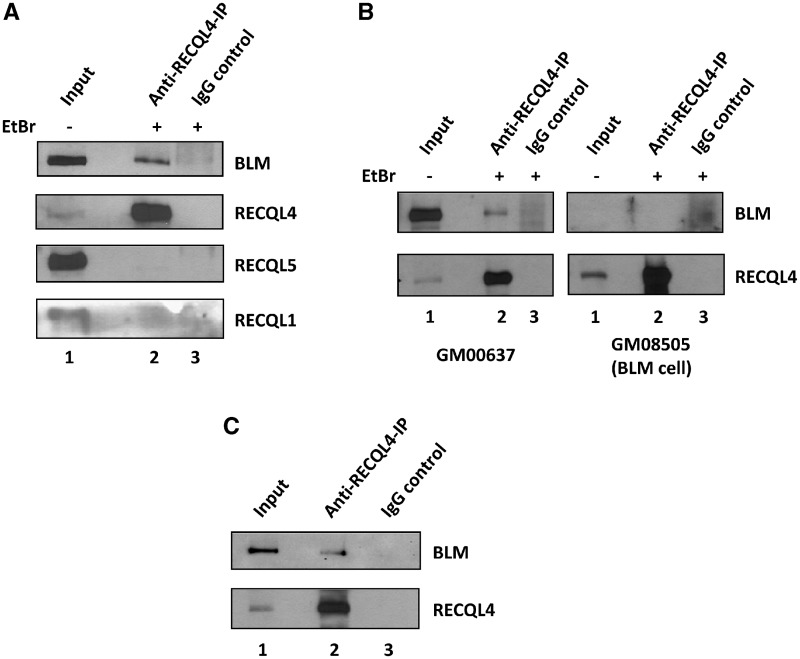
There is emerging, but very limited information about cooperativity among the human RECQ helicases. Using a yeast two-hybrid screen for proteins interacting with the N-terminal region of human RECQL4 (amino acids 1–471 fused to the LexA DNA-binding domain), we isolated a clone from a human peripheral blood cell cDNA library that encoded the full-length BLM, suggesting that BLM might cooperate with RECQL4. To investigate whether RECQL4 and BLM form a complex in human cells, anti-RECQL4 antibody was used for immunoprecipitation from HeLa cell extract and the blot was probed for BLM, RECQL4, RECQL1 and RECQL5 proteins, using specific antibodies against each protein. The results showed that endogenous RECQL4 specifically co-immunoprecipitated endogenous BLM, but not RECQL1 or RECQL5 (Figure 1A, lane 2). BLM was not detected in the IgG control (Figure 1A, lane 3). To further examine the specificity of this interaction, co-immunoprecipitation was carried out under similar conditions using cell extracts from normal (GM00637) and BLM-deficient fibroblasts (GM08505) (Figure 1B). A band corresponding to BLM was not apparent on the blot of RECQL4 immunoprecipitate from BLM-deficient cells, confirming that RECQL4 associates with BLM in vivo.
Figure 1.
BLM and RECQL4 interact in vivo and in vitro. (A) The RECQL4 co-immunoprecipitates endogenous BLM (top panel) and not RECQL1 or RECQL5 from HeLa total cell extract. RECQL4 antibody was used for co-immunoprecipitation (lane 2) and the blot was probed with antibodies against indicated proteins. The input and the IgG controls are shown in lanes 1 and 3, respectively. (B) The co-immunoprecipitation was performed with RECQL4 antibody from total cell lysate of normal control fibroblasts (GM00637) and BLM-deficient fibroblasts (GM08505), the blot was probed with BLM antibody. (C) The purified BLM and RECQL4 proteins interact in vitro. The in vitro pull down was performed with anti RECQL4 antibody (lane 2) and normal IgG control (lane 3), as described in ‘Materials and Methods’ section. The BLM was detected by western blotting with anti-BLM antibody (top) and RECQL4 was detected by anti-RECQL4 antibody (bottom).
The immunoprecipitation was also carried out in vitro using purified BLM and RECQL4 proteins. The two recombinant proteins were mixed, then pulled down with the antibody specific for RECQL4 and subsequently blots were probed for BLM. The presence of BLM in the RECQL4 pull down showed that the two proteins are in a complex (Figure 1C, lane 2). A similar band was not detected in the IgG control (Figure 1C, lane 3). The purified BLM and RECQL4 proteins used in this study are shown in Supplementary Figure S1.
RECQL4 stimulates the intrinsic helicase activity of BLM on a DNA fork substrate
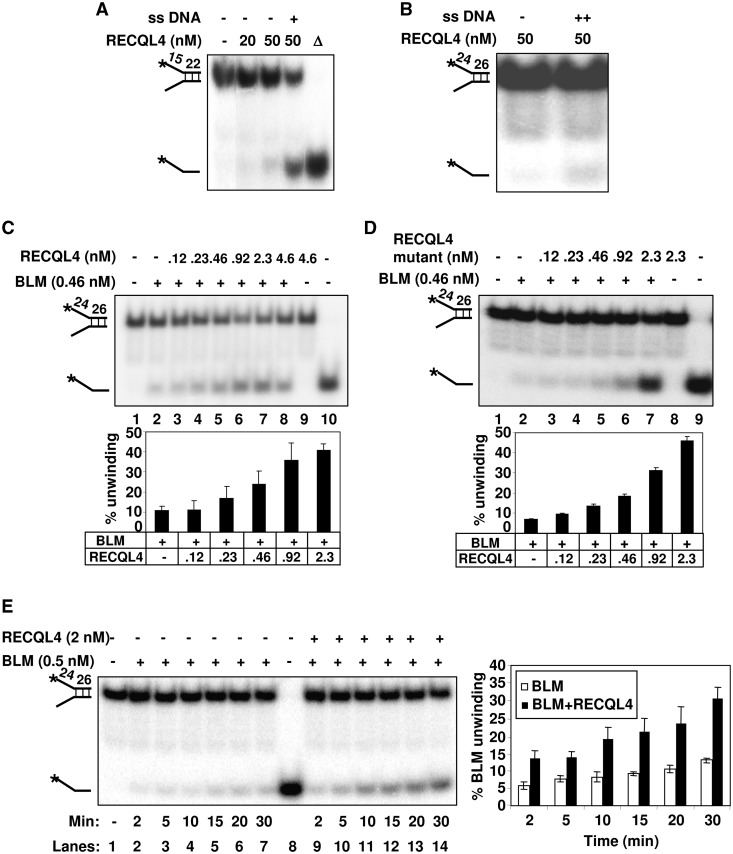
Both BLM and RECQL4 possess intrinsic helicase activity; however, RECQL4 has limited substrate specificity (21). Recently, we have shown that RECQL4 alone can only unwind very short DNA duplexes [Figure 2A; (21)]. On long duplex fork substrates, the strand annealing activity of RECQL4 is more prominent than the helicase activity, and thus it re-anneals the substrate rapidly after it is unwound. Revealing the intrinsic helicase activity on a longer DNA duplex requires the addition of excess complementary single-stranded DNA (ssDNA) to trap the newly separated strands (21,32). When we performed the helicase assays in the presence of excess ssDNA, RECQL4 alone was able to unwind the long duplex fork substrate (Figure 2B).
Figure 2.
The helicase activity of BLM is stimulated in the presence of RECQL4. (A) and (B) show the helicase activity of the wild type RECQL4 on DNA fork substrate with 22-mer and 26-mer duplexes in the absence or the presence of 50-fold excess of single-strand complimentary oligonucleotide as indicated. (C) The stimulation of BLM helicase activity in the presence of increasing concentrations of Wt RECQL4 (lanes 3–8). Helicase activities of BLM or RECQL4 alone are shown in lane 2 and 9, respectively. Substrate alone and heat denatured substrate are shown in lanes 1 and 10, respectively. (D) The BLM helicase activity is stimulated in the presence of increasing concentrations of RECQL4 helicase-dead mutant protein (lanes 3–7). Helicase activities of BLM or RECQL4 helicase-dead mutant protein alone are shown in lanes 2 and 8, respectively. The bar graph representing the percent substrate unwinding activity of BLM in the presence of increasing concentrations of RECQL4 Wt or RECQL4 helicase-dead mutant proteins is shown below panels A and B, respectively. (E) The RECQL4 increases the kinetics of BLM unwinding on DNA fork substrate. The helicase reactions were terminated at 2, 5, 10, 15, 20 and 30 min, and analyzed on 8% polyacrylamide gel. The unwinding activity of the BLM protein alone at different time points (as indicated) is shown in lanes 2–7. The unwinding activity of BLM in the presence of Wt RECQL4 protein (1:4 molar ratio) at different time points is shown in lanes 9–14. The bar graph compares the kinetics of substrate unwinding by BLM in the absence or the presence of RECQL4 (1:4 molar ratio) at different time point as indicated.
To strengthen the evidence for physical cooperativity between BLM and RECQL4, we investigated whether these two proteins also modulate each other functions. We tested BLM’s helicase activity in the presence of increasing concentrations of RECQL4 on a fork DNA duplex substrate upon which RECQL4 alone do not show any activity. Under the reaction conditions mentioned in ‘Materials and Methods’ section, 0.46 nM BLM was able to partially unwind (10%) the helicase substrate (0.5 nM) (Figure 2C, lane 2). Interestingly, this unwinding activity was enhanced up to ∼2-fold in the presence of 0.46 nM RECQL4 and increased to ∼4-fold at a 1:5 molar ratio of BLM to RECQL4 (Figure 2C, lanes 3–7). Even at the highest concentration used in this assay, RECQL4 helicase was not able to unwind the substrate by itself (Figure 2C, lane 9). Under these experimental conditions, the heat denatured RECQL4 was unable to stimulate BLM unwinding activity (Supplementary Figure S2A). RECQL4 was able to stimulate BLM unwinding activity on a DNA duplex substrate longer than 26 bp (Supplementary Figure S2B). These results support the notion that RECQL4 is indeed able to stimulate BLM’s helicase activity on substrates where it has no detectable helicase activity by itself.
Additionally, to rule out the possibility that BLM enhances RECQL4 helicase activity, we next performed the BLM helicase assay in the presence of a helicase-dead mutant of RECQL4. This RECQL4 mutant contains a site-specific K508M substitution in the Walker A motif of RECQL4 rendering it unable to hydrolyze ATP or to function as a helicase (21). The results showed that BLM helicase activity is still enhanced in the presence of the catalytically inactive RECQL4 (∼6–7-fold) (Figure 2D, lanes 3–7).
Further, we also tested RECQL4 unwinding activity in the presence of a helicase-dead mutant of BLM on short DNA duplex. The results showed that helicase-dead BLM was unable to stimulate the unwinding activity of RECQL4 (Supplementary Figure S2C).
Taken together, these results indicate that RECQL4 is able to enhance BLM’s intrinsic helicase activity (∼2-fold) at a biologically relevant molar concentration ratio (1:1) of BLM and RECQL4. This stimulation does not require the ATPase/helicase activity of RECQL4, and the catalytically inactive BLM is not able to stimulate RECQL4 unwinding activity.
We also examined the kinetics of BLM-mediated unwinding in the absence or presence of Wt as well as helicase-dead RECQL4 protein. BLM protein was incubated with or without RECQL4 at a 1:4 molar ratio. The helicase reactions were terminated at 2, 5, 10, 15, 20 and 30 min, and the products were analyzed on a polyacrylamide gel. The results showed that RECQL4 enhances the kinetics of BLM-mediated unwinding of a DNA fork substrate by about 2.5-fold in a time-dependent manner (Figure 2E). Under these experimental conditions, RECQL4 does not show any detectable unwinding activity by itself (Supplementary Figure S2D). Additionally, RECQL4 helicase-dead mutant protein was also able to stimulate BLM unwinding of a DNA fork substrate in a time-dependent manner (Supplementary Figure S2E).
RECQL4 specifically stimulates the unwinding activity of BLM on DNA fork substrate
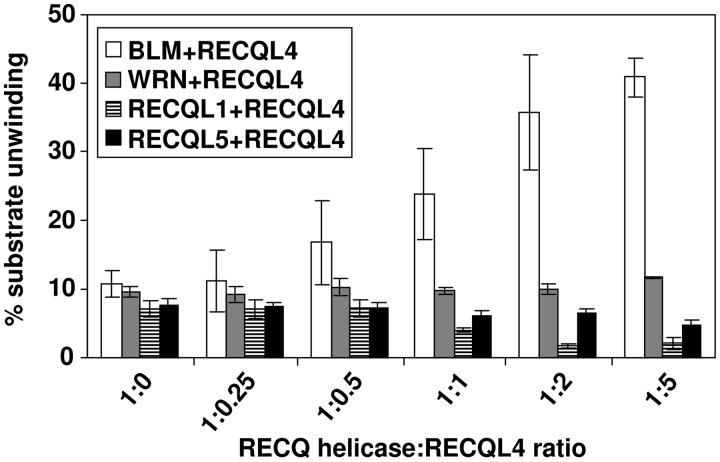
We further investigated the functional specificity of the RECQL4 and BLM interaction by testing whether RECQL4 could stimulate the helicase activity of other human RecQ helicase family members. Using similar experimental conditions, our results showed that RECQL4 was unable to stimulate unwinding activity of WRN, RECQL1 or RECQL5 (Figure 3 and Supplementary Figure S3), suggesting that RECQL4 specifically stimulates BLM helicase activity.
Figure 3.
The RECQL4 specifically stimulates BLM unwinding activity. Bar graph represents the unwinding activities of different RecQ helicases (BLM, WRN, RECQL1 and RECQL5) in the presence of increasing concentrations of RECQL4 as indicated. The molar ratios of RECQL4 with respect to different RecQ helicases are indicated on x-axis. The experiments were performed in triplicate and the error bars represent the standard deviation (±).
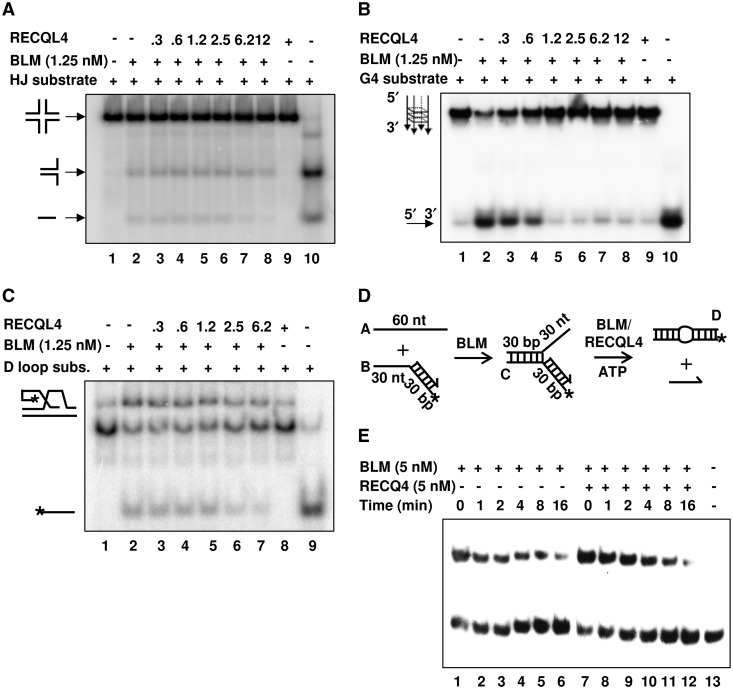
The substrate specificity of RECQL4-mediated stimulation of BLM’s helicase activity was then tested by assaying if RECQL4 could stimulate BLM helicase activity on other biologically relevant and preferred BLM substrates, such as: Holliday junctions (HJ), telomeric D-loops, G-quadruplexes (G4) or stalled replication forks. It was reported previously that BLM efficiently unwinds these substrates (25,33–35). However, our results showed that RECQL4 was not able to stimulate BLM’s helicase activity on any of these substrates (Figure 4). We observed inhibition of BLM unwinding activity when higher concentrations of RECQL4 were used in the reaction. This could be mainly due to competition between BLM and RECQL4 for susbstrate binding. Since, both BLM and RECQL4 possess strong DNA binding ability, the use of higher concentrations of RECQL4 might compete with BLM binding to the DNA substrate.
Figure 4.
BLM’s helicase activity is not stimulated by RECQL4 on HJ, D-loop, G4 quadruplex or stalled replication fork substrates. (A) and (B) show BLM helicase activity in the presence of increasing concentrations of the RECQL4 on HJ and G4 substrates in lanes 3–8 of each panel, respectively. Helicase activities of the BLM and the RECQL4 proteins alone on the HJ and G4 substrates, respectively, are shown in lanes 2 and 9 of each panel, respectively. The heat denatured substrates are shown in lane 10 of each panel. (C) shows BLM helicase activity in the presence of increasing concentrations of RECQL4 on a D-loop substrate (lanes 3–7). Helicase activities of the BLM and the RECQL4 proteins alone on D-loop substrate are shown in lanes 2 and 8. The heat denatured substrate is shown in lane 9. (D) shows the scheme for the BLM-mediated strand exchange activity on a synthetic stalled replication fork in presence of RECQL4. The lengths of the individual arms are represented in nucleotides (nt) or base pairs (bp). The 3′-end of the lagging oligonucleotide is indicated by an arrow and the position of the 5′-32P label is marked by an asterisk. The substrate was prepared as described in Supplementary Materials and Methods 1 section. In (E), a 10 μl aliquot of the reaction was taken out and mixed with 10 μl stop buffer at the start of the reaction (lane 1). Then 2 mM ATP and buffer (lanes 2–6) or RECQL4 (lanes 7–12) were added. Then 10 μl aliquots were taken out at different time points (1, 2, 4, 8 and 16 min) from each reaction vial and mixed with stop buffer. The product was analyzed by non-denaturing PAGE followed by phosphorimaging and analysis by ImageQuant software.
Taken together, these results suggest that RECQL4 specifically stimulates the helicase activity of BLM, but not of any other human RecQ helicase, and only on a forked DNA substrate, indicating a possible interaction during replication-associated DNA repair.
Mapping the interaction domains of RECQL4 and BLM
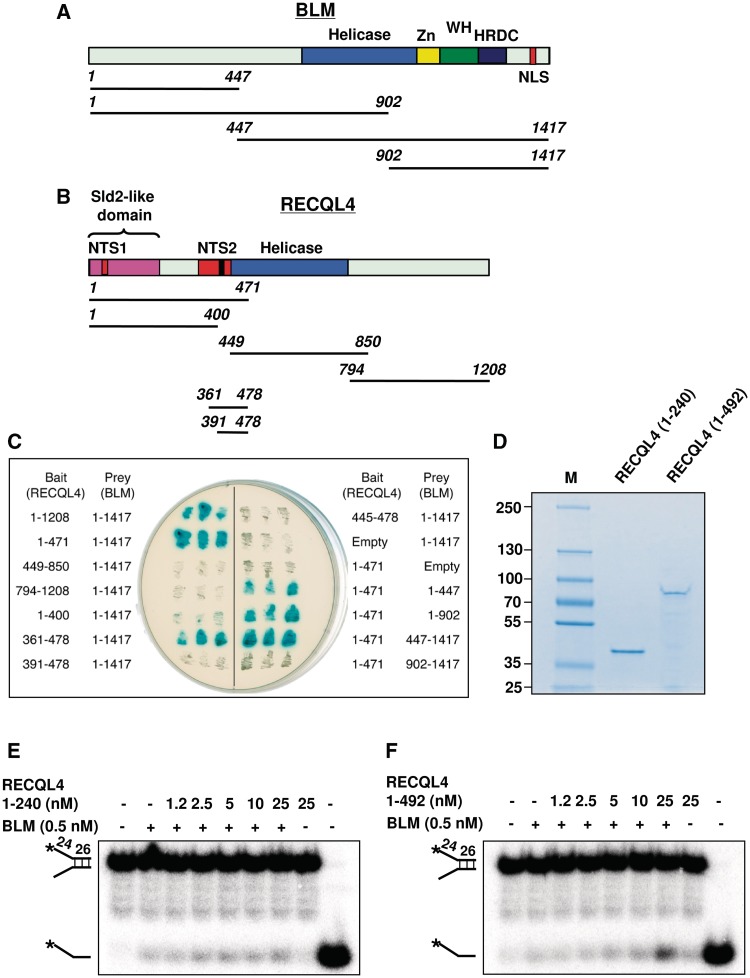
To precisely map the region of RECQL4 mediating its interaction with BLM, we employed a yeast two-hybrid β-galactosidase assay. Both BLM and RECQL4 contain a conserved helicase domain in the center of the protein (Figure 5A and B). BLM possesses a zinc binding domain (Zn), a winged-helix (WH) domain, a helicase and RNaseD C-terminal domain (HRDC) and a nuclear localization domain (NLS) located at the C-terminus. The HRDC domain has been implicated in the localization of BLM to the site of the DNA DSB and is required for the dissolution of double HJs (36,37) (Figure 5A). In contrast, RECQL4 lacks the WH and HRDC domains. RECQL4 possesses two nuclear-targeting sequences (NTS1 and NTS2) in the N-terminus. Interestingly, amino acids 1–200 of RECQL4 show similarity with Sld2, a yeast replication factor, implicated in initiation of DNA replication (Figure 5B) (7). It has been shown that the N-terminus of RECQL4 is essential and sufficient for the viability of vertebrate cells (38).
Figure 5.
Mapping of interaction domains of RECQL4 and BLM. (A) Schematic representation of BLM and its deletion variants. Yellow box, zinc-binding domain (ZN); green box, WH domain; blue box, helicase domain; purple box, HRDC domain; and red box, nuclear localization domain (B) Schematic representation of RECQL4 and its deletion variants. Blue box, helicase domain; black box, 376-KQAWKQKWRKK-386 motif; red box, nuclear-targeting sequences 1 and 2; pink box, sld2-like domain. Numbers in italics in both panels (A) and (B) indicate terminal amino acid positions. (C) β-gal plate test. RECQL4 and its deletion variants were expressed as fusions with a LexA DNA binding domain (bait). BLM and its deletion variants were expressed as fusions with an activation domain (prey). Indicated bait and prey constructs were co-transformed into the S. cerevisiae strain NMY-70 and the obtained clones were subjected to the β-galactosidase assay as described in ‘Materials and Methods’ section. Blue coloration of the yeast colonies is an indicative of interaction. (D) Coomassie stained gel of purified RECQL4 truncated proteins (1–240) and (1–492). (E) and (F) represents the unwinding activity of BLM in the absence and the presence of RECQL4 (1–240) and (1–492) proteins, respectively.
We first tested a series of RECQL4 deletion mutants for their ability to interact with the full-length BLM protein in the yeast two-hybrid assay. Our results indicated that the BLM interaction domain of RECQL4 is located within the region spanning amino acids 361–478 (Figure 5C). Interestingly, this region contains a basic motif, 376-KQAWKQKWRKK-386, which is acetylated at lysine residues by p300, leading to accumulation of RECQL4 in the cytoplasm (39). Using the same approach, we also mapped the RECQL4 interaction site on BLM. Various fragments of BLM expressed as prey were tested for their interaction with the N-terminal domain of RECQL4 spanning amino acids 1–471. The data suggested that BLM interacts with RECQL4 via at least two domains (Figure 5C). One domain is located between amino acids 1–447, and the second domain is located in the central part of the protein.
Additionally, we purified two recombinant fragments of RECQL4: (i) RECQL4 (1–240) containing the Sld2 domain and (ii) RECQL4 (1–492) lacking the C-terminal and helicase domains but including the putative BLM interaction domain (Figure 5D). We tested their ability to stimulate BLM unwinding activity. Our results showed that RECQL4 (1–492), but not RECQL4 (1–240), was able to stimulate BLM helicase activity (Figure 5E and F). These results further support the findings from the yeast two-hybrid screen.
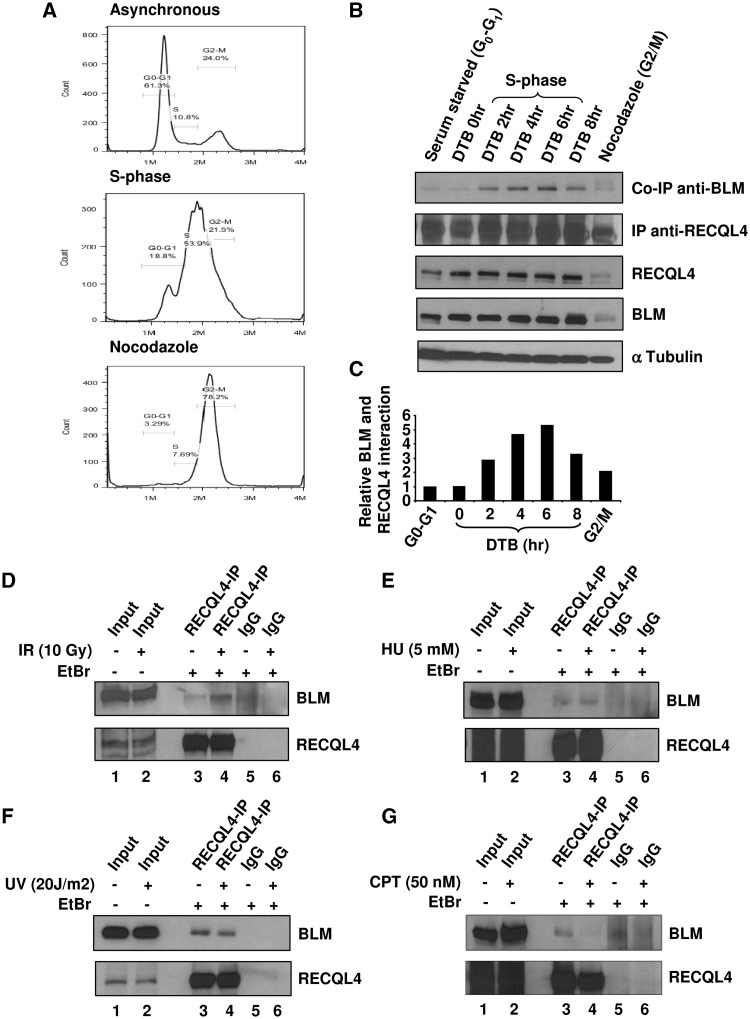
Interaction of BLM and RECQL4 is stimulated during S-phase
We next investigated whether the BLM and RECQL4 interaction was cell cycle regulated. HeLa cells were synchronized in different phases of the cell cycle by serum starvation, double thymidine block, or nocodazole (see ‘Materials and Methods’ section for details). The synchronized HeLa cells were analyzed by FACS, and the results are shown in Figure 6A. The FACS data indicated that the majority of serum starved or the double thymidine blocked HeLa cells were synchronized in G0–G1 phase of the cell cycle. After 4 h of release from the double thymidine block into the normal media, the majority of the HeLa cells were in S-phase of the cell cycle. The extracts from these cells were used for co-immunoprecipitation with anti-RECQL4 antibody, and the blot was probed for BLM. Results showed that the interaction between BLM and RECQL4 was cell cycle regulated, and was maximal during the S-phase (Figure 6B, top panel). The quantification of the signal intensity of the western blots showed that the interaction is enhanced ∼5-fold during S-phase compared with cells synchronized in G0–G1 phase of the cell cycle (Figure 6C). The expression patterns of BLM and RECQL4 were also checked by analyzing 10 µg of cell extract by western blotting with BLM- and RECQL4-specific antibodies (Figure 6B). Similar to previous reports, the expression of both BLM and RECQL4 was abundant during the S-phase of the cell cycle.
Figure 6.
Interaction between BLM and RECQL4 is stimulated during S phase of the cell cycle. (A) FACS analysis of the representative cell populations of the HeLa cells synchronized in different phases of cell cycle as described in ‘Materials and Methods’ section. (B) The RECQL4 antibody was used for co-immunoprecipitation from cell cycle synchronized HeLa cell lysates (panel 2) and the blot was probed with antibody against BLM protein (panel 1). The expression levels of RECQL4 (panel 3) and BLM (panel 4) in different phases of cell cycle is detected by blotting membrane with anti RECQL4 and anti-BLM antibody. α-Tubulin was probed as a loading control (bottom panel). (C) The histogram represents the relative interaction between BLM and RECQL4 in different phases of the cell cycle. The values are normalized after background subtractions. (D–G) Interaction of BLM and RECQL4 after various genotoxic stresses. Co- immunoprecipitation was performed from HeLa cell extracts prepared from cells treated with 10 Gy IR (D), 5 mM HU (E), 20 J/m2 UV radiation (F) and 50 nM CPT (G), using RECQL4 antibody and the blot was probed for BLM protein using anti BLM antibody.
To further determine if the interaction between BLM and RECQL4 was affected after DNA damage, we treated the HeLa cells with variety of DNA damaging agents: 10 Gy IR that predominantly causes DNA DSBs; 5 mM HU that induces replication stress; 50 nM CPT that creates replication blockage; and 20 J/m2 UV light introducing pyrimidine dimers that are repaired by nucleotide excision repair. Our results showed that there was an increase in BLM–RECQL4 interaction after IR, suggesting that the interaction could be a response to DNA DSBs. The level of interaction remained unchanged in the presence of other stresses (Figure 6D–G) and it did decrease to some extent after CPT treatment (Figure 6G).
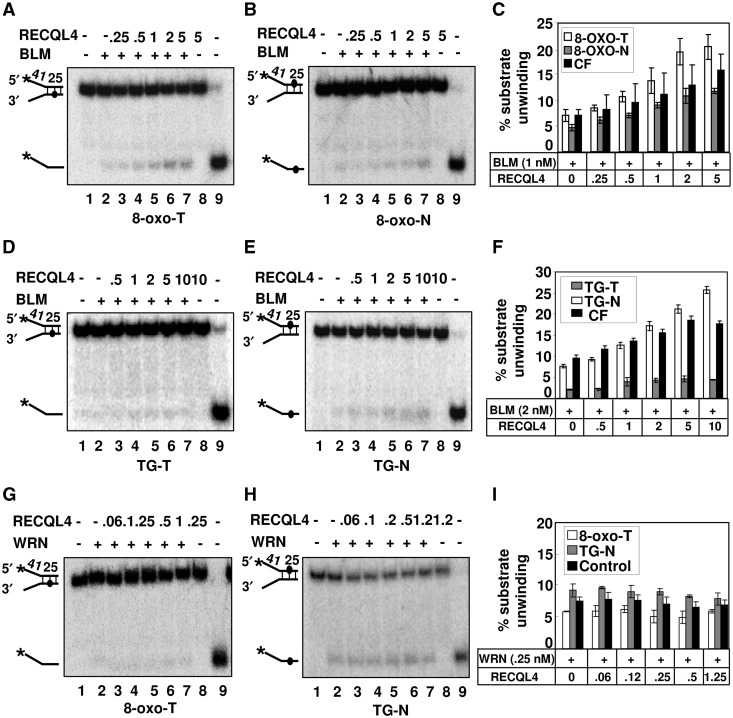
RECQL4 stimulates BLM’s helicase activity on DNA fork substrates containing 8-oxoguanine and thymine glycol
Our co-immunoprecipitation results showed that BLM and RECQ4 association is slightly enhanced after IR. The IR exposure to the cells, produces a variety of DNA damages including single-strand breaks (SSBs), DNA DSBs and base damages (40). Hence, we investigated the effect of RECQL4 on BLM-mediated unwinding on DNA fork substrate containing 8-oxoguanine (8-oxoG) and thymine glycol (TG) DNA lesions. These base modifications are produced either endogenously during normal DNA metabolic processes, or exogenously after exposure to IRs. If unrepaired, these lesions can be mutagenic or lethal to the cells. Interestingly, RECQL4 showed specificity in stimulating BLM’s helicase activity depending on whether the lesion was positioned on the translocating (T) or on the non-translocating (N) strand of the DNA fork (Figure 7). When a single 8-oxoG lesion was positioned on the translocating strand (8-oxo-T), RECQL4 stimulation of BLM’s helicase activity was ∼2-fold higher than when the 8-oxoG damage was present on the non-translocating strand (8-oxo-N) at a 1:2 molar ratio of BLM to RECQL4 (Figure 7A–C). We also tested the ability of RECQL4 helicase-dead mutant to stimulate BLM’s helicase activity on 8-oxoG-containing DNA substrate, and observed that the RECQL4 mutant protein was also able to stimulate BLM unwinding activity on 8-oxoG lesion containing DNA substrate (Supplementary Figure S4).
Figure 7.
RECQL4 stimulates BLM helicase activity on DNA substrates containing 8-oxoG and thymine glycol lesions. (A) and (B), the stimulation of the BLM helicase activity in the presence of increasing concentrations of RECQL4 (lanes 3–7) on DNA substrate containing 8-oxoG lesions on the translocating (8-oxo-T) and non-translocating (8-oxo-N) strands, respectively. In each panels, the helicase activities of BLM or RECQL4 proteins alone are shown in lane 2 and 8, respectively. Substrate alone and heat denatured substrate are shown in lanes 1 and 9, respectively. Graph representing the percent substrate unwinding on 8-oxo-T (open square), 8-oxo-N (gray square) strand and control fork (CF) (filled square) is shown in (C). Similarly, The BLM helicase activity in the presence of increasing concentrations of RECQL4 (lanes 3–7) on DNA substrate containing thymine glycol (TG) lesions on the translocating (TG-T) and non-translocating (TG-N) strands is shown in (D) and (E), respectively. Graph representing the percent substrate unwinding on TG-T (gray square), TG-N (opened square) strand and control fork (filled square) is shown in (F). The WRN helicase activity in the presence of increasing concentrations of RECQL4 (lanes 3–7) on DNA substrate containing 8-oxo-T and TG-N lesions is shown in panels (G) and (H), respectively. Graph representing the percent substrate unwinding on 8-oxo-T (open square), TG-N (gray square) strand and control fork (filled square) are shown in (I). All the experiments were performed in triplicate and the error bars represent the standard deviation (±).
In contrast, when the thymine glycol was present on the translocating strand, BLM alone failed to unwind the substrate (41), and RECQL4 was unable to stimulate its activity (Figure 7D). However, when the TG was present on the non-translocating strand, RECQL4 was able to stimulate BLM’s helicase activity slightly more than on the control fork substrate (Figure 7E and F).
Interestingly, RECQL4 failed to stimulate WRN’s helicase activity on 8-oxoG-T and TG-N substrates (Figure 7G–I), suggesting that the RECQL4-mediated stimulation of BLM’s helicase activity upon 8-oxoG and TG-containing fork substrates is specific.
Taken together, these results suggest that RECQL4 could stimulate BLM’s helicase activity on damage containing fork DNA substrate and show strand specificity with regard to the position of lesions on the DNA fork substrate.
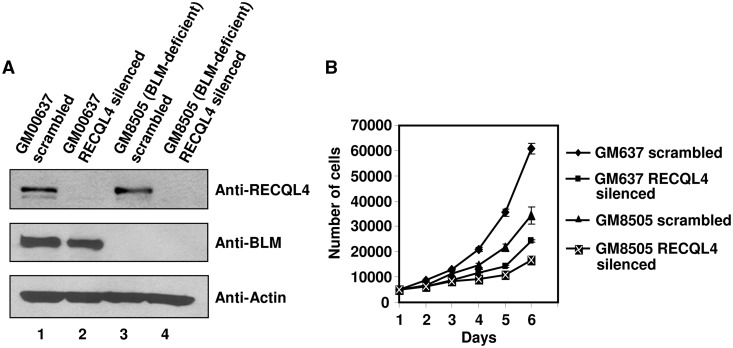
RECQL4-depleted BLM-deficient cells show retarded growth and higher frequency of SCE
Our co-immunoprecipitation and biochemical results were suggestive of a functional cooperation between BLM and RECQL4 during a replication-associated process. Consequently, to characterize the biological significance of the BLM and RECQL4 interaction, we stably depleted RECQL4 from BLM-deficient cells, and tested whether its absence adversely affected cell proliferation and growth properties. For this analysis, RECQL4 was depleted from normal fibroblasts (GM00637) and from BLM-deficient fibroblasts (GM08505) using short hairpin RNA packaged in lentivirus. RECQL4 was effectively depleted from both normal and BLM-deficient fibroblasts 4 days post-infection (Figure 8A). First we analyzed the proliferation rate of RECQL4-depleted normal fibroblasts. Our results showed that the depletion of RECQL4 in normal fibroblasts led to reduced proliferation compared with normal fibroblasts transduced with scrambled shRNA (Figure 8B). This result was in accordance with proposed functions of RECQL4 in DNA replication (6–8). BLM-deficient cells also showed reduced proliferation compared with normal control cells. However, depletion of RECQL4 in BLM-deficient cells drastically reduces the proliferative capacity of BLM and RECQL4 double-deficient cells (∼2 fold) as compared with BLM-deficient cells (Figure 8B). These results suggested that both BLM and RECQL4 have additive effects on cell proliferation and indicated a possible cooperation of the proteins during the replicative phase of the cell cycle. However, FACS analysis of the BLM and RECQL4 double-deficient cells and BLM-deficient cells did not reveal any significant differences in the cell cycle profile between the two cell lines (Supplementary Figure S5). When RECQL4 was depleted with siRNA in BLM cells, the cell proliferation was severely decreased compared with BLM cells transfected with control siRNA (Supplementary Figure S6B).
Figure 8.
The BLM and RECQL4 double-deficient fibroblasts show retarded growth. (A) Western blot showing the lentivirus-mediated scrambled (control) and RECQL4 silencing in normal fibroblasts (GM00637) and BLM-deficient fibroblasts (GM08505) 4 days post-infection. (B) The graph represents the growth assay of various knockdown cells as indicated. The number of cells is plotted with respect to time.
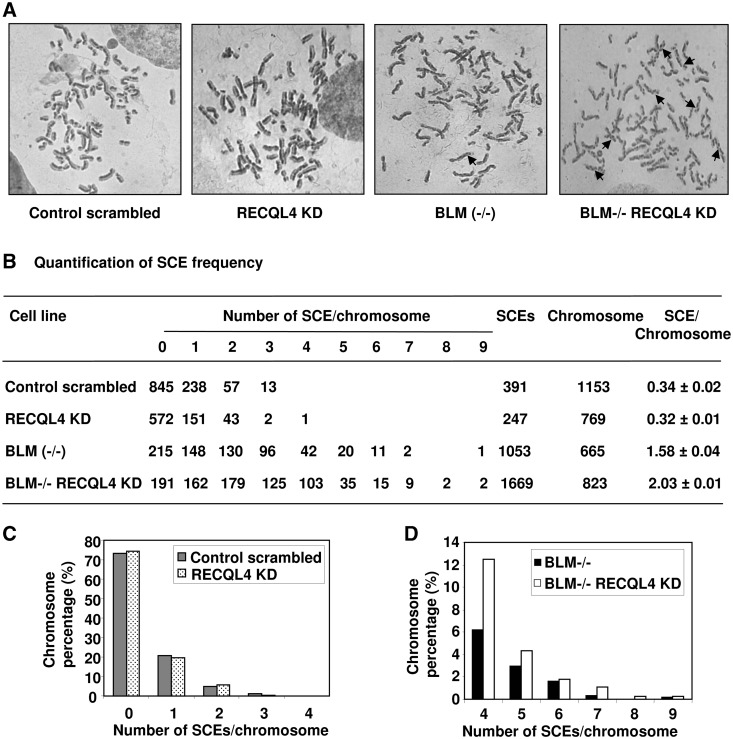
One of the characteristic features of BLM-deficient cells is a high frequency of SCEs (42). SCEs are formed during homologous recombination and require sequence homology. The process occurs during the S-phase of the cell cycle. Since, the BLM and RECQL4 interaction is enhanced during S-phase, we investigated the effect of RECQL4 depletion in BLM-deficient cells on the frequency of SCEs (Figure 9). Our results showed that the lentivirus-mediated knockdown of RECQL4 in normal fibroblasts had a SCE frequency (0.32 SCE/chromosome) comparable with that of normal control fibroblasts transduced with scrambled shRNA (0.34 SCE/chromosome) (Figure 9B). Hence, we observed no significant difference in the frequencies of SCEs in normal (control) and RECQL4-depleted normal fibroblasts, (Figure 9B). This result was consistent with previous studies by Mann et al. (43) who did not find any increase in the frequency of SCE in RECQL4−/− mouse cells. We then depleted RECQL4 in the BLM-deficient fibroblasts and compared the frequencies of SCEs with BLM-deficient cells transduced with scramble shRNA. We found that BLM and RECQL4 double-deficient cells had significantly higher levels of SCEs per chromosome (2.03) than the BLM-deficient cells (1.58) (Figure 9B). Similar trends were also seen with the increase in the frequency of SCEs in BLM and RECQL4 double-deficient cells compared with BLM-deficient cells, when the BLM-deficient cells were transfected with siRNA targeted against RECQL4 (Supplementary Figure S6C). These results suggest that RECQL4 alone does not play a significant role in SCE formation, however, in a SCE-prone background such as BLM−/−, the contribution that RECQL4 makes in SCE formation can be visualized.
Figure 9.
The frequency of SCEs is increased in BLM and RECQL4 double-deficient fibroblasts. (A) Representative images of metaphase spread from different cell lines as indicated. The typical SCEs are marked by arrows. (B) Quantification of frequencies of SCEs/chromosome in different cell lines. (C) and (D), bar graph representation of percentage of chromosomes showing different number of SCEs/chromosome as indicated on x-axis in different cell lines.
The dissociation of GFP-tagged RECQL4 from DNA damage is increased in BLM-deficient cells
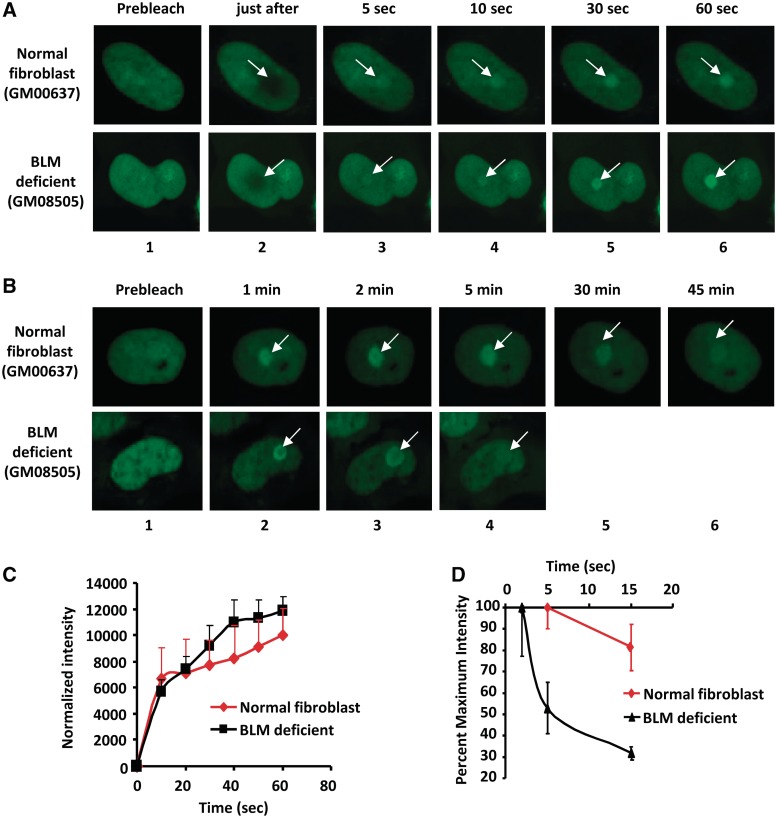
The recruitment and retention kinetics of DNA repair proteins at the site of laser-induced DNA damage are an important component in elucidating the cellular dynamics of DNA repair. Recently, we showed that GFP-RECQL4 is recruited to the site of laser-induced DNA damage (14). Given that BLM and RECQL4 interact physically and functionally in vitro and in vivo, we wanted to determine if the cellular dynamics of GFP-RECQL4 association to the site of laser-induced DNA damage was affected by the absence of BLM protein. Thus, we studied the dynamic association and dissociation kinetics of GFP-RECQL4 at the site of localized laser-induced DNA damage in normal and BLM-deficient cells. Laser microirradiation was carried out using 21% laser intensity, which causes DNA DSBs (14). The GFP-tagged RECQL4 was transiently expressed in SV40 transformed normal fibroblasts (GM00637) and in BLM-deficient fibroblasts (GM08505). The cells were microirradiated and analyzed by live cell imaging (described in ‘Materials and Methods’ section). The results showed that GFP-RECQL4 was recruited efficiently to the microirradiated sites in the absence of functional BLM protein (Figure 10A and C). GFP-RECQL4 started to accumulate at the damaged site within 5–10 s in normal and in BLM-deficient cells (Figure 10A and C). The rate of recruitment, up to 1 min after induction of damage, was slightly higher in BLM-deficient cells than in normal fibroblasts (Figure 10C).
Figure 10.
The dissociation of GFP-RECQL4 at the site of laser-induced DNA damage site is fast in BLM cells. The images of recruitment and dissociation kinetics of transiently transfected GFP-RECQL4 protein at the site of laser-induced DNA damage in SV40 transformed normal fibroblasts (GM00637), and BLM (GM08505) cells are shown in panels (A) and (B), respectively. The site of DNA damage is marked by an arrow. The normalized intensity values of the recruitment kinetics of GFP-RECQL4 in normal fibroblasts and BLM-deficient fibroblasts till 60 s after laser-induced DNA damage have been represented in (C). (D) represents the dissociation kinetics of transiently transfected GFP-RECQL4 protein from the site of laser-induced DNA damage in SV40 transformed normal fibroblasts and BLM-deficient fibroblasts, respectively. The GFP-RECQL4 retention plot showing the dissociation kinetics of GFP-RECQL4 from the maximum intensity values at the site of the DNA damage in normal fibroblasts and BLM-deficient cells with respect to time.
However, the maximal accumulation intensity of GFP-RECQL4 at the site of localized DNA damage occurred at different times in normal fibroblasts and in BLM-deficient cells (Figure 10D). In BLM-deficient cells, GFP-RECQL4 reached its maximum intensity (considered as 100%) within 2 min, compared with at ∼5 min in normal fibroblasts (Figure 10D). Interestingly, the loss of GFP-RECQL4 fluorescence intensity was much more rapid and extensive in cells lacking BLM than in normal cells. Additionally, in BLM-deficient cells, ∼70% of the GFP-RECQL4 dissociated from the sites of DNA damage within 15 min, compared with a 20% decrease in normal cells (Figure 10B and D). These results suggest that BLM is required for optimal retention of GFP-RECQL4 at the site of damage, and that these two proteins may cooperate in the DNA repair process.
DISCUSSION
Higher eukaryotes have multiple RecQ helicases, whereas lower eukaryotes have only one. Mammals have five RecQ helicases, and we are trying to understand whether these proteins act in concert or as soloists. In this study, we sought to investigate the hypothesis that specific RecQ helicases collaborate functionally in DNA metabolic pathways by providing novel cellular and biochemical evidence that BLM and RECQL4 cooperate in replication-associated DNA repair.
The RecQ helicases share common biochemical properties and have common interacting protein partners. For example, all the RecQ helicases interact with Rad51, which plays a key role in the HR pathway of DSB repair, and some have been shown to modulate its function (16,44–47). Therefore, it is likely that they are present in the same DNA repair protein complex, and that they can affect each other’s functions. Consistent with this notion, a physical and functional interaction between BLM and WRN has been shown previously (10), validating the notion that RecQ helicases seem to work cooperatively under certain circumstances.
An association between BLM and RECQL4 was shown in vivo and in vitro (Figure 1). Also, both RECQL4 and BLM are abundant during the S-phase of cell cycle, indicating their likely involvement in DNA replication-associated functions [Figure 6; (7,48)]. Both RECQL4- and BLM-deficient cells show replication abnormalities and defective S-phase progression (19,49). Therefore, we propose that BLM and RECQL4 might cooperate functionally during the replication phase of the cell cycle. To support this hypothesis, we showed that the interaction between BLM and RECQL4 increases during S-phase of the cell cycle (Figure 6). RECQL4 interacts with BLM through its N-terminal region spanning amino acids 361–478, which does not include the helicase domain. The N-terminal region of RECQL4 lacking the helicase domain is both essential and sufficient for the viability of vertebrate cells (38). It has been proposed that the RECQL4 N-terminal region plays a role in recruiting DNA polymerase α to the nascent DNA replication fork in Xenopus egg extract (50). The N-terminus of human RECQL4 interacts with the MCM complex and contains a Sld2-like domain, which may be involved in the initiation of DNA replication, thus RECQL4 is implicated in the initiation phase of DNA replication (7). Therefore, the interaction of BLM with N-terminus of RECQL4 is interesting and might be linked to their involvement during a replication-associated function.
Biochemically, both Wt RECQL4 and the RECQL4 helicase-dead mutant were able to stimulate BLM unwinding activity on a DNA substrate resembling a replication fork. Thus, we propose that BLM and RECQL4 might co-operate to remove various secondary structures or DNA lesions that are encountered during DNA replication. In accordance with this hypothesis, we found increased association between BLM and RECQL4 after IR treatment (Figure 6D). Also, we found that RECQL4 has a shorter retention time at laser-induced DSB sites in BLM-deficient cells than in control cells (Figure 10). These results are suggestive of an interaction between BLM and RECQL4 in response to DNA damage. Biochemically, we tested the stimulatory effect of RECQL4 on BLM unwinding activity on DNA substrates containing 8-oxoG and thymine glycol lesions. These lesions could arise during normal DNA metabolic processes or after exposure to IRs and can potentially block DNA replication. Our results indicated that RECQL4 could stimulate BLM-mediated unwinding on DNA substrates containing these lesions. Interestingly, BLM and RECQL4 also showed specificity in unwinding these substrates, depending on whether the lesions are present on the translocating or the non-translocating strand (Figure 7). Using similar substrates, Suhasini et al. (41) have shown that FANCJ helicase could specifically sense oxidative base modifications and its activity is stimulated by RPA in a strand-specific manner to overcome the inhibitory effect of thymine glycol on non-translocating strand of the DNA substrate.
Interestingly, RECQL4 was not able to stimulate WRN helicase activity on the DNA fork substrate with or without lesions (Figures 3 and 7). This specificity suggests that RECQL4 and BLM work together more closely than RECQL4 and WRN, at least during the replication-associated DNA metabolic steps. One of the ways by which BLM might promote fork restart is by repressing the aberrant formation of recombination intermediates at stalled forks. RECQL4 may then be able to stabilize stalled forks by specific stimulation of BLM helicase activity to efficiently resolve replication intermediates at the stalled fork.
The cell proliferation was dramatically altered when both BLM and RECQL4 were depleted. BLM functions during the S-phase and its expression peaks during the S/G2 phases of the cell cycle (48). Consistent with these observations is the fact that BS cells exhibit replication abnormalities, including retarded replication-fork elongation (18) and abnormal replication intermediates (19). The expression of RECQL4 peaks during transition from G1 to S-phase possibly in accordance with its role in the initiation of DNA replication (7,8). Thus, BLM and RECQL4 may coordinate their activities during resolution of replication intermediates, or in the removal of aberrant DNA structures. The significant elevation in the frequencies of SCEs in BLM and RECQL4 double-deficient cells, as compared with BLM-deficient cells, suggests that RECQL4 functions in suppression of the SCE when BLM function is impaired. A similar observation has been made in chicken DT40 cell where double deletion of BLM and RECQL5 (BLM−/−/RECQL5−/−) resulted in increased frequency of SCE, as compared to BLM−/− cells. The RECQL5−/− cells showed frequencies of SCEs similar to those of Wt cells. Thus, RECQL5 suppresses SCE under conditions of impaired BLM function, which suggests BLM and RECQL5 redundancy (11,12).
We observed that the intrinsic RECQL4 helicase activity did not cooperate with that of BLM on certain recombinogenic structures such as a HJ, a D-loop or a synthetic stalled replication fork. This may be because of the very weak helicase activity of RECQL4. When complimentary ssDNA is added in the in vitro reaction we do observe RECQL4 helicase activity on a D-loop substrate (data not shown). In the cellular context it is likely that other proteins stimulate RECQL4 helicase function that could then be important in the resolution of some of these recombinogenic structures. The stimulation of RECQL4 by RPA in an in vitro assay supports this notion (21). Therefore, in BLM-deficient cells, RECQL4 could help in the prevention of illegitimate recombination events and when both proteins are depleted, it leads to the increase in SCEs as we observe in the double-deficient cells.
In conclusion, our results suggest that two of the RecQ helicases, BLM and RECQL4, coordinate their functions in the maintenance of genome integrity in the cell. Biochemical results indicate that they can function together at a replication fork site and might help in the resolution of certain replication intermediates. Furthermore, the interesting phenotype of the BLM and RECQL4 double-deficient cells is consistent with an important and biologically relevant interaction between the two proteins. Future experiments will be necessary to shed light on the DNA metabolic pathways in which the RecQ helicases participate either individually or collaboratively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–6 and Supplementary Material and Methods 1.
FUNDING
National Institutes of Health Intramural Program of the National Institute on Aging [Z01-AG000726-17]; Czech Science Foundation [GAP305/10/0281]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Drs Monika Aggarwal and Chandrika Chanugovi for critical reading of the manuscript. We also thank Irina Sheveleva for help with yeast two-hybrid experiments. The pET16b-RECQL4 (1-240) and RECQL4 (1-492) plasmids were kind gift from Yilun Liu (City of Hope, CA, USA). The RECQL1 protein, 8-oxoG and TG containing DNA substrates were provided by Dr Robert Brosh, NIA, Baltimore. We would like to thank Robert Wersto and Jade Scheers, NIA Flow Core facility, for assistance with the cell cycle analysis.
REFERENCES
- 1.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 2.Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem. Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh DK, Ghosh AK, Croteau DL, Bohr VA. RecQ helicases in DNA double strand break repair and telomere maintenance. Mutat. Res. 2011 doi: 10.1016/j.mrfmmm.2011.06.002. Jun 13. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh DK, Ahn B, Bohr VA. Roles of RECQ helicases in recombination based DNA repair, genomic stability and aging. Biogerontology. 2009;10:235–252. doi: 10.1007/s10522-008-9205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang S, Beresten S, Li B, Oshima J, Ellis NA, Campisi J. Characterization of the human and mouse WRN 3′→5′ exonuclease. Nucleic Acids Res. 2000;28:2396–2405. doi: 10.1093/nar/28.12.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 7.Xu X, Rochette PJ, Feyissa EA, Su TV, Liu Y. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. EMBO J. 2009;28:3005–3014. doi: 10.1038/emboj.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thangavel S, Mendoza-Maldonado R, Tissino E, Sidorova JM, Yin J, Wang W, Monnat RJ, Jr, Falaschi A, Vindigni A. Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol. Cell Biol. 2010;30:1382–1396. doi: 10.1128/MCB.01290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aygun O, Svejstrup J, Liu Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc. Natl Acad. Sci. USA. 2008;105:8580–8584. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von KC, Karmakar P, Dawut L, Opresko P, Zeng X, Brosh RM, Jr, Hickson ID, Bohr VA. Colocalization, physical, and functional interaction between Werner and Bloom syndrome proteins. J. Biol. Chem. 2002;277:22035–22044. doi: 10.1074/jbc.M200914200. [DOI] [PubMed] [Google Scholar]
- 11.Wang W, Seki M, Narita Y, Nakagawa T, Yoshimura A, Otsuki M, Kawabe Y, Tada S, Yagi H, Ishii Y, et al. Functional relation among RecQ family helicases RecQL1, RecQL5, and BLM in cell growth and sister chromatid exchange formation. Mol. Cell Biol. 2003;23:3527–3535. doi: 10.1128/MCB.23.10.3527-3535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Islam MN, Fox D, III, Guo R, Enomoto T, Wang W. RecQL5 promotes genome stabilization through two parallel mechanisms—interacting with RNA polymerase II and acting as a helicase. Mol. Cell Biol. 2010;30:2460–2472. doi: 10.1128/MCB.01583-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, Lu X, Barnes E, Yan M, Lou H, Luo G. Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol. Cell Biol. 2005;25:3431–3442. doi: 10.1128/MCB.25.9.3431-3442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh DK, Karmakar P, Aamann M, Schurman SH, May A, Croteau DL, Burks L, Plon SE, Bohr VA. The involvement of human RECQL4 in DNA double-strand break repair. Aging Cell. 2010;9:358–371. doi: 10.1111/j.1474-9726.2010.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu L, Davies SL, Levitt NC, Hickson ID. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 2001;276:19375–19381. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- 16.Petkovic M, Dietschy T, Freire R, Jiao R, Stagljar I. The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J. Cell Sci. 2005;118:4261–4269. doi: 10.1242/jcs.02556. [DOI] [PubMed] [Google Scholar]
- 17.Im JS, Ki SH, Farina A, Jung DS, Hurwitz J, Lee JK. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc. Natl Acad. Sci. USA. 2009;106:15628–15632. doi: 10.1073/pnas.0908039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hand R, German J. A retarded rate of DNA chain growth in Bloom's syndrome. Proc. Natl Acad. Sci. USA. 1975;72:758–762. doi: 10.1073/pnas.72.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lonn U, Lonn S, Nylen U, Winblad G, German J. An abnormal profile of DNA replication intermediates in Bloom's syndrome. Cancer Res. 1990;50:3141–3145. [PubMed] [Google Scholar]
- 20.Xu D, Muniandy P, Leo E, Yin J, Thangavel S, Shen X, Ii M, Agama K, Guo R, Fox D, III, et al. Rif1 provides a new DNA-binding interface for the Bloom syndrome complex to maintain normal replication. EMBO J. 2010;29:3140–3155. doi: 10.1038/emboj.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossi ML, Ghosh AK, Kulikowicz T, Croteau DL, Bohr VA. Conserved helicase domain of human RecQ4 is required for strand annealing-independent DNA unwinding. DNA Repair. 2010;9:796–804. doi: 10.1016/j.dnarep.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karow JK, Chakraverty RK, Hickson ID. The Bloom’s syndrome gene product is a 3′-5′ DNA helicase. J. Biol. Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 23.Orren DK, Brosh RM, Jr, Nehlin JO, Machwe A, Gray MD, Bohr VA. Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 1999;27:3557–3566. doi: 10.1093/nar/27.17.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huber MD, Lee DC, Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom’s syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 27.Opresko PL, von KC, Laine JP, Harrigan J, Hickson ID, Bohr VA. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 28.Karow JK, Constantinou A, Li JL, West SC, Hickson ID. The Bloom’s syndrome gene product promotes branch migration of holliday junctions. Proc. Natl Acad. Sci. USA. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramamoorthy M, Tadokoro T, Rybanska I, Ghosh AK, Wersto R, May A, Kulikowicz T, Sykora P, Croteau DL, Bohr VA. RECQL5 cooperates with Topoisomerase II alpha in DNA decatenation and cell cycle progression. Nucleic Acids Res. 2012;40:1621–1635. doi: 10.1093/nar/gkr844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghosh AK, Rossi ML, Singh DK, Dunn C, Ramamoorthy M, Croteau DL, Liu Y, Bohr VA. RECQL4, the protein mutated in Rothmund-Thomson syndrome, functions in telomere maintenance. J. Biol. Chem. 2012;287:196–209. doi: 10.1074/jbc.M111.295063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suhasini AN, Rawtani NA, Wu Y, Sommers JA, Sharma S, Mosedale G, North PS, Cantor SB, Hickson ID, Brosh RM., Jr Interaction between the helicases genetically linked to Fanconi anemia group J and Bloom’s syndrome. EMBO J. 2011;30:692–705. doi: 10.1038/emboj.2010.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu X, Liu Y. Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J. 2009;28:568–577. doi: 10.1038/emboj.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom’s syndrome helicase. Nucleic Acids Res. 2006;34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karmakar P, Seki M, Kanamori M, Hashiguchi K, Ohtsuki M, Murata E, Inoue E, Tada S, Lan L, Yasui A, et al. BLM is an early responder to DNA double-strand breaks. Biochem. Biophys. Res. Commun. 2006;348:62–69. doi: 10.1016/j.bbrc.2006.07.037. [DOI] [PubMed] [Google Scholar]
- 37.Wu L, Chan KL, Ralf C, Bernstein DA, Garcia PL, Bohr VA, Vindigni A, Janscak P, Keck JL, Hickson ID. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 2005;24:2679–2687. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abe T, Yoshimura A, Hosono Y, Tada S, Seki M, Enomoto T. The N-terminal region of RECQL4 lacking the helicase domain is both essential and sufficient for the viability of vertebrate cells. Role of the N-terminal region of RECQL4 in cells. Biochim. Biophys. Acta. 2011;1813:473–479. doi: 10.1016/j.bbamcr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Dietschy T, Shevelev I, Pena-Diaz J, Huhn D, Kuenzle S, Mak R, Miah MF, Hess D, Fey M, Hottiger MO, et al. p300-mediated acetylation of the Rothmund-Thomson-syndrome gene product RECQL4 regulates its subcellular localization. J. Cell Sci. 2009;122:1258–1267. doi: 10.1242/jcs.037747. [DOI] [PubMed] [Google Scholar]
- 40.Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog. Nucleic Acid Res. Mol. Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 41.Suhasini AN, Sommers JA, Mason AC, Voloshin ON, Camerini-Otero RD, Wold MS, Brosh RM., Jr FANCJ helicase uniquely senses oxidative base damage in either strand of duplex DNA and is stimulated by replication protein A to unwind the damaged DNA substrate in a strand-specific manner. J. Biol. Chem. 2009;284:18458–18470. doi: 10.1074/jbc.M109.012229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaganti RS, Schonberg S, German J. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc. Natl Acad. Sci. USA. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mann MB, Hodges CA, Barnes E, Vogel H, Hassold TJ, Luo G. Defective sister-chromatid cohesion, aneuploidy and cancer predisposition in a mouse model of type II Rothmund-Thomson syndrome. Hum. Mol. Genet. 2005;14:813–825. doi: 10.1093/hmg/ddi075. [DOI] [PubMed] [Google Scholar]
- 44.Otterlei M, Bruheim P, Ahn B, Bussen W, Karmakar P, Baynton K, Bohr VA. Werner syndrome protein participates in a complex with RAD51, RAD54, RAD54B and ATR in response to ICL-induced replication arrest. J. Cell Sci. 2006;119:5137–5146. doi: 10.1242/jcs.03291. [DOI] [PubMed] [Google Scholar]
- 45.Sharma S, Brosh RM., Jr Human RECQ1 is a DNA damage responsive protein required for genotoxic stress resistance and suppression of sister chromatid exchanges. PLoS. One. 2007;2:e1297. doi: 10.1371/journal.pone.0001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu L, Davies SL, Levitt NC, Hickson ID. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 2001;276:19375–19381. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- 48.Dutertre S, Ababou M, Onclercq R, Delic J, Chatton B, Jaulin C, Amor-Gueret M. Cell cycle regulation of the endogenous wild type Bloom’s syndrome DNA helicase. Oncogene. 2000;19:2731–2738. doi: 10.1038/sj.onc.1203595. [DOI] [PubMed] [Google Scholar]
- 49.Park SJ, Lee YJ, Beck BD, Lee SH. A positive involvement of RecQL4 in UV-induced S-phase arrest. DNA Cell Biol. 2006;25:696–703. doi: 10.1089/dna.2006.25.696. [DOI] [PubMed] [Google Scholar]
- 50.Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol. Cell Biol. 2006;26:4843–4852. doi: 10.1128/MCB.02267-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.