Abstract
In eukaryotes, RNA-binding proteins that contain multiple K homology (KH) domains play a key role in coordinating the different steps of RNA synthesis, metabolism and localization. Understanding how the different KH modules participate in the recognition of the RNA targets is necessary to dissect the way these proteins operate. We have designed a KH mutant with impaired RNA-binding capability for general use in exploring the role of individual KH domains in the combinatorial functional recognition of RNA targets. A double mutation in the hallmark GxxG loop (GxxG-to-GDDG) impairs nucleic acid binding without compromising the stability of the domain. We analysed the impact of the GDDG mutations in individual KH domains on the functional properties of KSRP as a prototype of multiple KH domain-containing proteins. We show how the GDDG mutant can be used to directly link biophysical information on the sequence specificity of the different KH domains of KSRP and their role in mRNA recognition and decay. This work defines a general molecular biology tool for the investigation of the function of individual KH domains in nucleic acid binding proteins.
INTRODUCTION
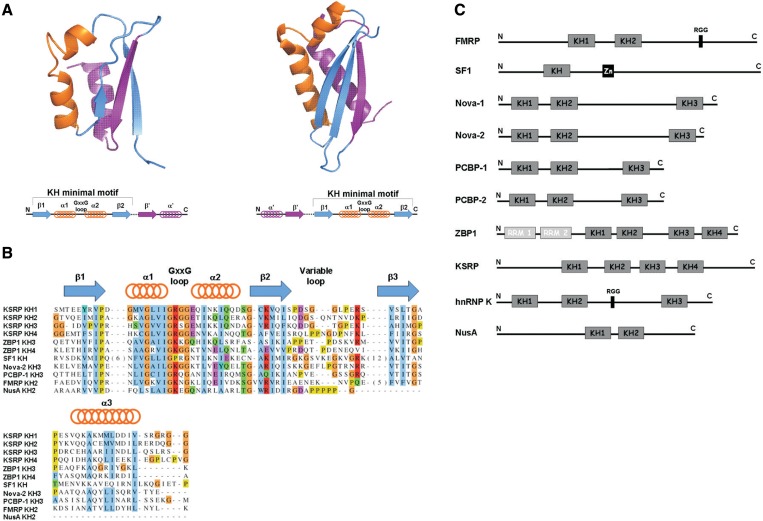
hnRNP K homology (KH) domains are small (∼70 amino acids) αβ nucleic acid (NA) recognition domains (1,2). They are found both in eukaryotes and prokaryotes, albeit with a different topology (Figure 1A and B) (3). KH domains interact with their single stranded NA (ssNA) targets with different affinity and specificity and they have been shown to recognize up to four nucleotides, although non-specific contacts can be made with additional flanking nucleotides (4,5). KH domain-containing proteins perform a wide range of cellular functions and several diseases, including paraneoplastic syndromes and some cancers, are associated with the loss of function of specific KH domains (5). Here we focus on eukaryotic type I KH domains, which are normally found in multiple copies within the same protein (Figure 1C) with RNA recognition normally being achieved by the synergistic contribution of the different domains to RNA binding. KH domains fold as a three stranded anti-parallel β-sheet on the surface of which pack three α-helices (1) (Figures 1A and 2A). ssNA binding is mediated by a hydrophobic groove or cleft formed on one side by two short consecutive α-helices (α1 and α2) and the intervening GxxG loop and, on the other side, by the inner surface of the domain’s β-sheet and the attached variable loop (Figure 2). In the complex, the NA molecule is in an extended conformation and the bases are distributed along the hydrophobic groove, with the Watson–Crick edge pointing towards the β-sheet. Nucleobase recognition is mediated by base-pair-like H-bonding between the moieties on the Watson–Crick edges of the RNA bases and the backbone and side chain of the protein (4,6). The phosphates of the first two RNA nucleotides are instead docked against the GxxG loop by means of electrostatic interactions, H-bonding and inter-molecular Van der Waals interactions, depending on the specific complex (4,6–13). The conserved GxxG loop (Figure 1B) is indeed a hallmark of the KH domain. Although the structures of a few KH domains without a canonical GxxG loop have been solved, the absence of the loop is linked to a loss of NA-binding capability and is accompanied by extensive intra-molecular protein–protein contacts (14,15).
Figure 1.
(A) KH fold. (Top) Cartoon representation of the structure of a type I KH domain (Vigilin KH6, PDBID: 1VIH) and a type II KH domain (ERA C-terminal domain PDBID: 1EGA). (Bottom) Topology of type I KH domains (eukaryotic) and type II KH domains (prokaryotic). (B) Sequence alignment of the KH domains discussed in this manuscript. (C) Domain organization of the KH domain-containing proteins discussed in this manuscript.
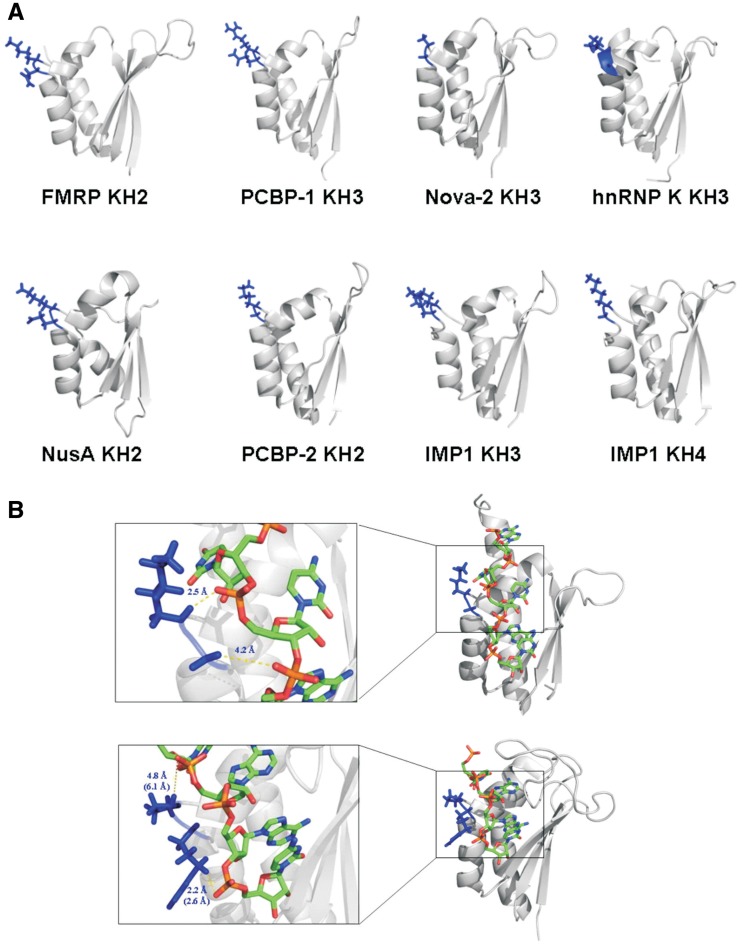
Figure 2.
The GxxG loop is solvent exposed in the free protein and contacts the backbone of the RNA ligands. (A) Cartoon representation of the structure of eight KH domains. The side chains of amino acids of the GxxG loop are displayed in blue. (B) The Nova-2 KH3–RNA (top) and SF1 KH–RNA (bottom) complexes. The protein structure is displayed as a cartoon while the RNA is represented by a stick model. For SF1 the lowest energy NMR structure of the bundle is displayed. On the left are blow ups of the structures that detail the GxxG–RNA interaction. For the X-ray structure of Nova-2 we report the shortest distances between the RNA phosphates and the first and second variable amino acid of the GxxG loop. For the NMR structure of SF1 we report the equivalent distances measured over the bundle of ten structures and in the lowest energy structure (brackets).
Recognition between KH-containing proteins and their RNA targets is crucial to establish multi-layered post-transcriptional regulatory networks. Modelling these networks requires a molecular understanding of the underlying protein–RNA recognition events, and a way of correlating biophysical data on the domain–RNA interaction with the role of individual domains in a cellular environment (16). The two most common strategies to evaluate the contribution of single domains to RNA recognition and protein function are to either delete the domain in toto or, if a better understanding of the domain structure and RNA-binding properties exists, to mutate single amino acids within the domain in order to eliminate its RNA-binding capability. Both of these methods have potential drawbacks. Deleting a NA-binding domain can eliminate inter-molecular protein–protein interactions and/or destabilize neighbouring domains. Further it can perturb the general structure of the protein. The mutation of an amino acid known to be important for NA binding represents a more conservative strategy that, in principle, can effectively decouple RNA binding from protein structure. However, the mutation of even a single amino acid can severely destabilize a domain and it is often difficult to eliminate the RNA-binding capability of a domain with a single mutation. Because of this, the folding, stability and RNA-binding capability of mutants are normally examined in vitro prior to functional testing. This laborious procedure may involve the screening of several single or multiple mutants before a suitable one is found, and it is not suited to the functional analysis of large numbers of domains. In order to apply mutagenesis efficiently to the hundreds of multi-domain RNA-binding proteins involved in gene regulation, it is important to possess mutants which can reliably impair RNA binding in the domains sharing the same fold without changing their structure or decreasing their stability, i.e. mutants of general use.
Here we report the rational design of a mutant to probe RNA recognition by KH domains. We believe this mutant represents a useful tool in the molecular investigation of KH proteins involved in the regulation of mRNA metabolism. Using our understanding of the structure and RNA-binding properties of KH domains we have designed a GxxG-to-GDDG double mutant (the GDDG mutant) to be used as a general tool to probe the role of RNA binding by individual KH domains in the cellular environment. We have tested the mutation on six different KH domains known to bind to very different RNA sequences and we show both that the mutated proteins have no detectable RNA-binding activity (Kd >> 1 mM) and that the mutation does not cause significant structural changes or domain de-stabilization. We have then used the protein KSRP, a regulator of mRNA metabolism, to exemplify how individual GDDG mutants can provide an accurate insight into domain contribution to functional RNA recognition. We present here an effective tool for the functional analysis of networks of regulatory proteins containing KH domains.
MATERIALS AND METHODS
Constructs
The sequences of all protein constructs used in this study are reported in Supplementary Table S1. Wild-type constructs for the expression of single domain KSRP samples are as described in (17–19). GDDG mutants (GxxG-to-GDDG) were prepared using the Quikchange Site-Directed Mutagenesis Kit (Stratagene) according to manufacturer’s instructions. The Y396F ZBP1 mutant (that is henceforth referred to as Y396F ZBP1 for simplicity), and the two derived constructs with GDDG mutations on KH3 and KH4 (ZBP1 KH3 mutant and ZBP1 KH4 mutant respectively) were also prepared using the Directed Mutagenesis Kit (Stratagene).
For in vitro binding studies, a coding sequence that includes the four-KH domains of human KSRP, (amino acids G68-Q525, NM_003685) was cloned into pETM-30 (EMBL-Heidelberg, Protein Expression Facility). The vector includes a TEV protease cleavable HisTag-GST fusion N-terminal to the insert. GDDG mutant versions of each domain were prepared as described above for the single domain protein.
For cross-linking and functional studies the GDDG mutations were introduced into full-length KSRP cloned into pCMV-Tag2B as described previously (20).
Protein and RNA samples
Unlabelled and 15N-labelled single domain KSRP and double domain ZBP1 proteins were expressed and purified as previously described for KSRP KH4 (17). Briefly, all proteins were expressed as HisTag-GST fusion proteins in Escherichia coli BL21 (DE3) cells (Invitrogen) as reported (17). The fusion proteins were purified from the soluble fraction of the cell extract by nickel-affinity chromatography (Qiagen) according to manufacturer’s instructions, followed by TEV cleavage and dialysis back into loading buffer. Tags were then removed by a second round of nickel affinity chromatography and the domain further purified using a size exclusion column and dialysed in the final buffer.
A different purification strategy was used for the four-KH domain proteins, all of which co-purified with NAs. After TEV cleavage, the protein was further purified using first a MonoQ 5/50GL anion exchange column (GE Healthcare) and then a 1 ml HiTrap Heparin column (GE Healthcare). The KSRP samples were stored in 10 mM Tris–HCl pH 7.4, 50 mM NaCl, 2 mM TCEP, while the ZBP1 samples were stored in 10 mM NaPi pH 6.5, 50 mM NaCl, 0.05% (w/v) NaN3 and protease inhibitors (Roche) at −20°C were added to all of the samples. Protein concentrations were determined using UV absorbances at 280 nm, and their molecular weights and purity were confirmed by Electrospray Mass Spectrometry and SDS–PAGE.
Chemically synthesized RNA oligos were purchased from Dharmacon.
CD spectroscopy
All CD spectra were recorded on Jasco J-715 spectropolarimeter (Jasco) equipped with a PTC-348 Peltier temperature-control system. To assess the thermal stability of the six mutants and compare it with the one of the wild-type proteins, we monitored the changes in the 220 nm CD signal of wild-type and mutant proteins between 20°C and 85°C. The temperature was increased at a rate of 2°C/min and decreased at the same rate, to assess reversibility. The KSRP KH1, KH2, KH3 and KH4 samples were at a concentration of 1–2 µM in 10 mM Tris–HCl pH 7.4, 100 mM NaCl, 0.5 mM TCEP while the ZBP1 samples were at 10–15 µM in 10 mM NaPi pH 6.5, 50 mM NaCl. The data were processed and fitted to a two-state folding-to-unfolding model using in-house software, as described in (17).
NMR
1H–15N correlation spectra were recorded on Bruker Avance spectrometers at 600 and 700 MHz proton frequencies with typical experiment times of 25 min. Protein concentration was 50–70 µM and experiments were recorded in Tris–HCl pH 7.4, 100 mM NaCl, 0.5 mM TCEP buffer (KSRP KH domains) and 10 mM NaPi pH 6.5, 50 mM NaCl (ZBP1 constructs). NMR data were processed using NMRPipe (21), and analyzed using Sparky (22). Assignments of the KSRP and ZBP1 KH domains were obtained using standard methodologies (17–19) and are not reported here.
BioLayer interferometry
BLI experiments were performed in 10 mM Tris–HCl pH 7.4, 150 mM NaCl, 2 mM TCEP, 0.5 mg/ml BSA on an Octet Red Instrument (ForteBio, Inc.) operating at 25°C. Streptavidin coated biosensors with immobilized biotinylated TNF-α 42-mer were exposed to different concentrations of KSRP-WT, KH1, KH2, KH3 and KH4 mutants, as described in (23). Kinetic curves were analyzed using a double exponential fit which allowed us to account for a minor very fast binding event, most likely associated with non-specific binding. The observed rate constant for the major component of the association phase (kOBS) is linearly dependent on protein concentrations and its analysis yielded association rate constants for the five proteins, while we derived the dissociation rate constant(s) by direct analysis of the dissociation of the complex. The length of our RNA construct and the number of KH domains engaged in the interaction (3) indicates that only one protein molecule can bind with high affinity to the RNA and therefore it is reasonable to assume that stoichiometry is 1:1.
Immunoprecipitation of KSRP mutants expressed in HEK-293 cells upon UV-crosslinking to RNA
Total-cell extracts (200 μg protein) and 32P-labelled RNA were incubated at room temperature for 20 min in RNA-binding buffer (200 μl total volume) containing 10 mM HEPES (pH 7.6), 3 mM MgCl2, 100 mM KCl, 2 mM DTT, 5% glycerol, 0.5% NP-40, yeast RNA (1 μg) and heparin (1 μg). Reaction mixtures were transferred to a 24-well plate and irradiated at 4°C for 10 min with a UV crosslinker (254 nm wavelength at a distance of 5 cm). After subsequent digestion with RNase A (200 ng per reaction) for 10 min at 37°C, samples were immunoprecipitated (overnight at 4°C under rotation) using anti-FLAG (M2 Sigma)-bound Protein-G-coated magnetic beads (Dynal). Immunocomplexes were extensively washed with 1× RNA-binding buffer buffer and protein analyzed by SDS–PAGE. Proteins were transferred to nylon membranes (Immobilon, Millipore), and 32P-labelled proteins were visualized by autoradiography. The same membranes were subsequently incubated with anti-Flag antibody in order to normalize the results for the expression of the Flag-tagged proteins.
In vitro RNA degradation assays
β-catenin 3′-UTR fragment subcloned into pCY2 to produce 32P-labelled RNA substrate was previously described (20). S100 extracts were prepared from αT3-1 cells in which KSRP was knocked-down as previously described (20). Total extracts (200 μg protein) from HEK-293 cells either mock-transfected or transfected with Flag-tagged WT KSRP or individual GDDG KH mutants were immunoprecipitated using anti-FLAG-bound Protein-G-coated magnetic beads. Immunocomplexes were incubated with S100 extracts and in vitro degradation experiments were performed as previously described (20).
RESULTS
Design of a general use KH mutant with impaired RNA-binding capability
An important general feature we observe in KH–NA interactions is the key role played by the GxxG loop in the positioning of the NA molecule. The details of the interaction between the GxxG loop and the NA backbone vary in different KH–RNA complexes (Figure 2). However, in most KH domains at least one of the variable amino acids in the loop has a positively charged side chain. This and the proximity of the RNA phosphate groups indicate that addition of one or more negative charges is likely to have a general effect on RNA binding. Indeed, it has been reported that mutation of an R to an E in the GxxG loop of the KH domain of SF1 protein leads to substantial impairment of RNA binding (7).
The distance between the two variable amino acids in the loop and the RNA phosphate groups varies in different structures. In some cases (e.g. in Nova-1) the first variable amino acid is closer than the second to the RNA phosphate group, while in others (e.g. in SF1) the opposite is observed (Figure 2B). We reasoned that two side-by-side negatively charged amino acids in the central part of the loop would create a wide negative surface and hinder the contacts with the RNA backbone that are necessary for the positioning of the ssNA in the KH domain groove. In order to obtain a mutant of general use for KH domains, we mutated both variable amino acids in the GxxG loop to Aspartate.
An important consideration when designing a lack-of-function mutant is whether the mutation is likely to destabilize the protein. The analysis of the structures of KH domains reveals that the side chains of the two central amino acids of the GxxG loop are not packed against other protein side chains (Figure 2A). Furthermore, the NMR relaxation data published for a number of KH domains show that the backbone amide groups of the two central amino acids of the loop experience significant motions (9,17,18,24–27). In crystal structures of KH domains, high B factors are observed for the GxxG loop (Supplementary Figure S1), contributing to an overall picture of the GxxG loop as a flexible element that locks into position upon RNA binding. It is therefore unlikely that the GxxG mutation will significantly destabilize the protein fold. Finally, as our purpose is to design a mutant that can decouple RNA binding from other functional interactions, it is important to point out that the GxxG loop is not involved in protein–protein interaction.
The GDDG mutation does not destabilize the KH domain
We tested the GDDG mutation on six different KH domains whose structure, stability and RNA-binding properties have been extensively characterized by our group (the four KH domains of the KSRP protein and the third and fourth KH domain of the ZBP1 protein, Figure 1B and C) and are here used to represent the ensemble of KH domains. The affinities of the KH domains used in this study span the micromolar range of Kds (28). It is worth mentioning that, as far as we know, all single KH–ssRNA interactions fall in this range. The domains also have different sequence specificities, as described in detail in the following paragraph. Importantly they have different thermal stabilities (the Tms for the folded-to-unfolded transition of the isolated domains of KSRP vary between ∼50°C and ∼70°C, Figure 3). The six domains have a canonical type I KH fold that in KSRP KH1 and KH4 are flanked by additional structural elements and in KH3 and KH4 of ZBP1 are engaged in intra-molecular inter-domain interactions. We have independently expressed and purified the wild-type and mutant versions of the four KH domains of KSRP. The third and fourth domains of ZBP1 protein interact, burying a large surface and creating a continuous six-strand β-sheet (29). These two domains are unstable in isolation even at room temperature but we could purify a stable version of the two domain construct (Figure 3 and Supplementary Table S1) that is known to bind RNA in vitro and in the cell (30). To dissect the role of ZBP1 KH3 and KH4 domains, we compared RNA binding and thermal stability of the recombinant protein (which we will henceforth refer to as ZBP1 KH34, see ‘Materials and Methods’ section) with the ones of the two distinct KH3-KH4 proteins bearing the GDDG mutations in respectively KH3 and KH4 (indicated as ZBP1 KH3 mutant and ZBP1 KH4 mutant, respectively).
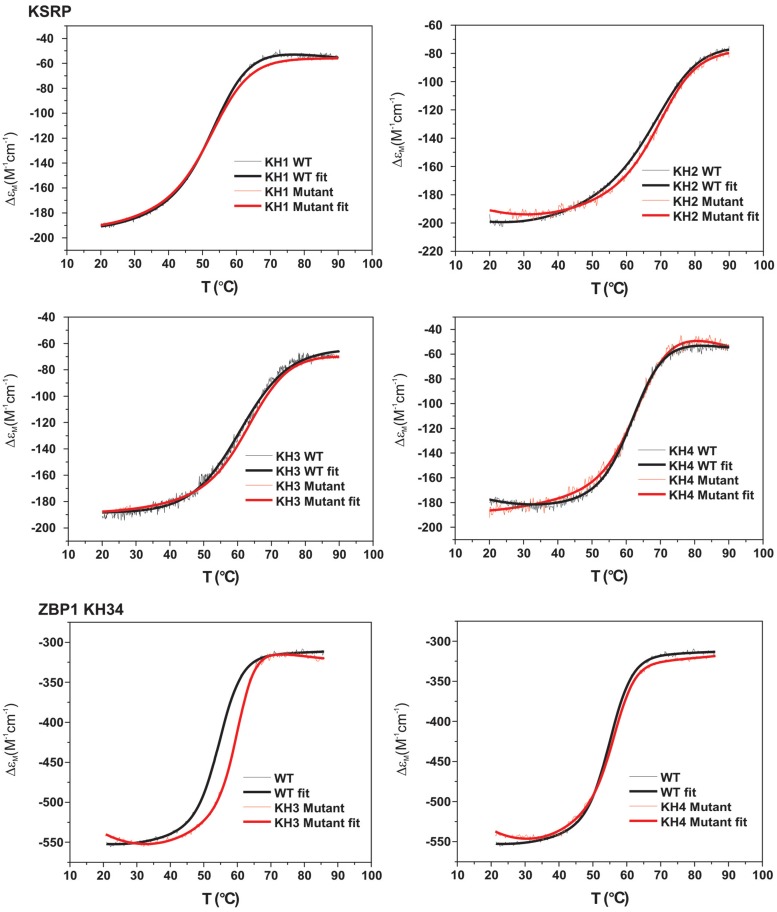
Figure 3.
The GDDG mutation does not destabilize the KH domain. Thermal stability of the GDDG mutants. 220-nm CD signal of wild-type (black) and mutant(s) (red) domains as a function of temperature (thin lines). The data are fitted using a two-state folded-to-unfolded model (thick line). The midpoint of the transition between the folded and unfolded state is indicated as a dashed line and reported in Supplementary Table S2. Five of the six mutants show only negligible changes in stability. The sixth mutant (KH3 of ZBP1) shows a modest stabilization, with a ∼5°C shift in the measured Tm.
The comparison of the 15N–1H HSQC spectra of the six GDDG mutants with the corresponding spectra of the wild-type proteins (Figure 4) shows that all of the mutants are stable at room temperature and that no major structural change takes place upon mutation. To better evaluate the overall stability of the domain, we investigated the temperature-mediated unfolding of the six mutant domains using CD and again compared it with the one of the wild-type domains. For KSRP KH domains and the ZBP1 KH4 mutant, the differences between the folded-to-unfolded transition midpoints of wild-type and mutant proteins are within the experimental error (Figure 3 and Supplementary Table S2). In the ZBP1 KH3 the mutation actually increases the stability of the domain, with a transition midpoint 5°C higher than the wild-type. It is not immediately obvious why the mutation in the GxxG loop of KH3 increases stability as the loop is exposed to the solvent. The thermal denaturation studies reported here only show the net effect of the mutation on the stability; they do not provide information on whether the native state, the ensemble of denatured states, or both are affected. The analysis of the structure(s) indicates that the stabilizing effect of the mutation in KH3 is unlikely to result from a decrease in the free energy of the native state, and therefore is likely to stem from an increase in the free energy of the denatured state. Given that the mutation involves changing two residues to aspartic acid the most likely interpretation of the increased stability reported is that electrostatic interactions in the mutant are energetically less favourable in the denatured state. CD and NMR data indicate that the GDDG mutation does not change the fold of the domain and nor does it compromise its stability.
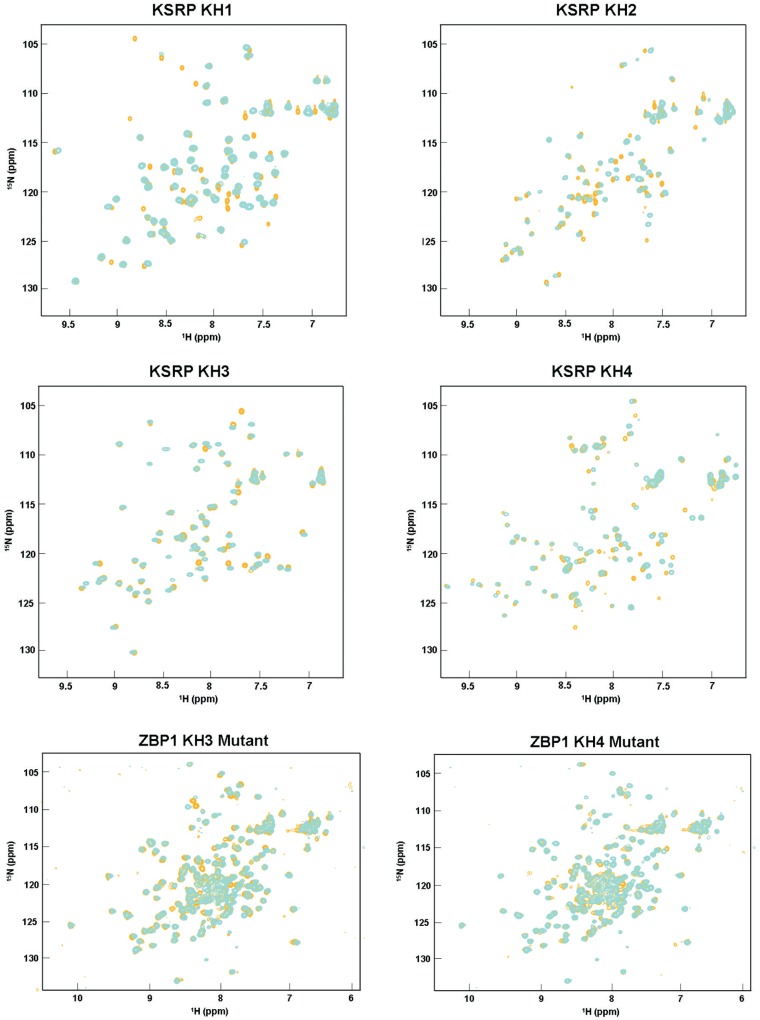
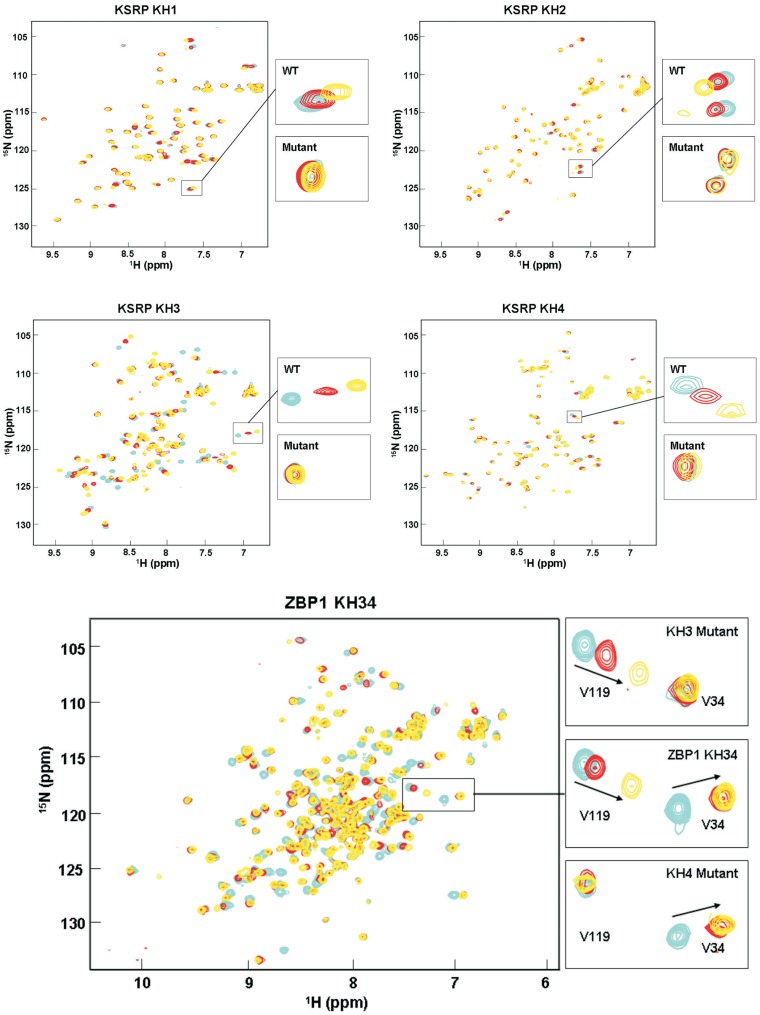
Figure 4.
The GDDG mutation does not change the fold of the KH domain. Superimposition of 1H–15N correlation spectra of wild-type (cyan) and mutant (orange) for the KH1, KH2, KH3, KH4 domains of KSRP and KH34 di-domain of ZBP1 proteins. The limited changes we observe confirm that the domains maintain their folded state.
The GDDG mutation impairs RNA binding by KH domains
Next we wanted to establish whether the GDDG mutation impairs RNA binding in all six KH domains described above. The specificity of the six domains is very different and the sequences tested include canonical C/A rich RNAs (ZBP1) as well as U/G, U/A and G-rich ones (KSRP). The binding affinities of the six domain for the RNA targets are also very different, spanning nearly three orders of magnitude (28).
The RNA-binding specificity of the KH domains of KSRP has been established by Scaffold Independent Analysis (SIA) and further tested in follow-up studies (28). Using NMR, we monitored a titration of each KH domain, wild-type and GDDG mutants, with the preferred RNA sequence and show that the RNA-binding capability of the domains is abolished by the mutation (Figure 5). In the case of ZBP1 we use a known CACACCC target (29,31), which we show here is bound by both domains, albeith with different affinity (Figure 5) and therefore this RNA can be used to test the RNA-binding capability of the two mutants. Our assays show that mutation of either KH3 or KH4 resulted in the loss of any detectable RNA-binding activity. Further tests with several other RNA sequences confirmed the generality of this result (data not shown). The six GDDG mutated domains shows no detectable RNA binding at 50 µM concentration and 1:10 protein:RNA ratio, indicating that the Kd is >>1 mM (Figure 5), and that binding is too weak to be relevant in functional protein–RNA interaction assays.
Figure 5.
The GDDG mutation abolishes RNA binding. Superimposition of 1H–15N correlation spectra recorded during titrations of (top) the KSRP KH1, KH2, KH3 and KH4 with increasing amounts of respectively of UUGGG, UUUAG, UGGGU and UAGGG oligos, the highest affinity SIA-selected RNA sequences known to bind to these domains (28) and (bottom) the ZBP1 KH3/KH4 construct with increasing amounts of the GCACACCC target sequence (32,33). A subset of resonances of the free protein (cyan) shift when 1 (red) and 4 (yellow) equivalents of RNA are added, reporting on the binding. The main panels display the backbone amide region of the spectra, while the insets provide a comparison between the behaviour of representative resonances in wild-type and mutant. In (wild-type) ZBP1 KH34 spectrum resonances of both KH3 and KH4 are shifting upon RNA addition, while in the KH3 and KH4 mutants spectra only resonances of the non-mutated domain (KH4 and KH3 respectively) are shifting.
The effect of the GDDG mutations within single KH domains on KSRP–RNA binding
KSRP is a multifunctional protein that has been linked to several steps of RNA metabolism. Probably the most extensively studied function of KSRP is to promote the rapid degradation of short lived mRNAs through the recruitment of the exosome degradation machinery (34,35). Selectivity in this degradation mechanism is provided by KSRP recognition of AU-rich elements (ARE) in the 3′-UTR of the mRNA, triggering the so-called ARE-mediated mRNA decay (AMD). Using SIA and NMR we have previously explored the different specificities of the domains of KSRP and their affinity for the AU-rich sequences typically found in AREs (UAUUUA and UAUUAU). None of the domains shows strong sequence specificity for AU-rich sequences. The known affinities of the four domains for the short AU-rich RNA oligos (28) follow the trend: KH3 > KH4 ≥ KH2 >> KH1, while KH2, KH3 and KH4 bind the two short RNA oligos with Kds in the 100–400 µM range, KH1 binds with Kd > 1 mM (Table 1). This relatively weak binding of the isolated domains is consistent with the multi-domain mechanism of KSRP–RNA recognition inferred from both cellular and molecular data (28,36). Here we have used the GDDG mutants to directly link RNA binding by the isolated domains to their respective contribution to the binding of the four-domain protein to the physiological RNA targets.
Table 1.
Affinities of the individual KH domains of KSRP for the UAUUUA RNA oligo
*Previously published in (28).
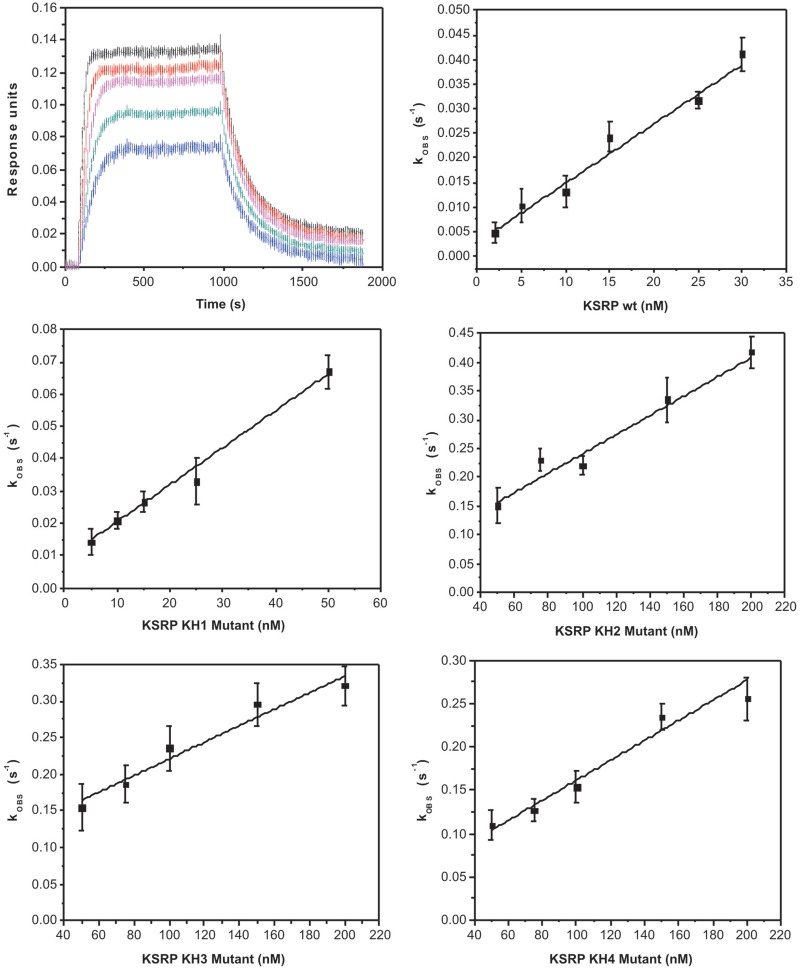
We cloned and purified four KSRP constructs comprising the NA-binding region of the protein (i.e. the four KH domains) each with a GDDG mutation in one of the KH domains. We tested the binding of the wild-type KSRP and the four GDDG mutants to a short 3′-UTR sequence containing the TNFα ARE (TNFα BLI, Supplementary Table S3). The moderate length and low complexity of the sequence allowed us to assay protein–RNA interactions using BLI, a technique that we have successfully used to measure the DNA-binding affinity of FBP, a KSRP paralog (23). BLI experiments show that the Kd of the wild-type KSRP–RNA complex is 3 nM, that mutating one of the four domains decreases the affinity between 2 and 30 times and that the contribution of the domains to the binding affinity can be ranked as KH3 > KH4 ≥ KH2 >> KH1 (Table 2 and Figure 6). Furthermore, we notice that the ranking of the contributions of the domains to RNA-binding recapitulates accurately the domain’s affinity for the short AU-rich target sequences.
Table 2.
Affinity of the four-domain KSRP constructs (wild-type and GDDG mutants) for the 42-mer TNFα ARE RNA
| Kd (nM) | |
|---|---|
| KSRP WT | 3.6 ± 1.4 |
| KSRP KH1 Mutant | 7.3 ± 1.7 |
| KSRP KH2 Mutant | 43 ± 14 |
| KSRP KH3 Mutant | 95 ± 19 |
| KSRP KH4 Mutant | 50 ± 16 |
Figure 6.
RNA binding by the wild-type and mutant four-domain KSRP constructs. Top left—Association and dissociation of the wild-type KSRP protein to a 42-mer TNFα ARE immobilized on a BLI sensor. Protein concentrations are 5, 10, 15, 20 and 30 nM. Other panels—The kOBS measured in five experiments (wild-type and the four GDDG mutants) are plotted against protein concentrations.
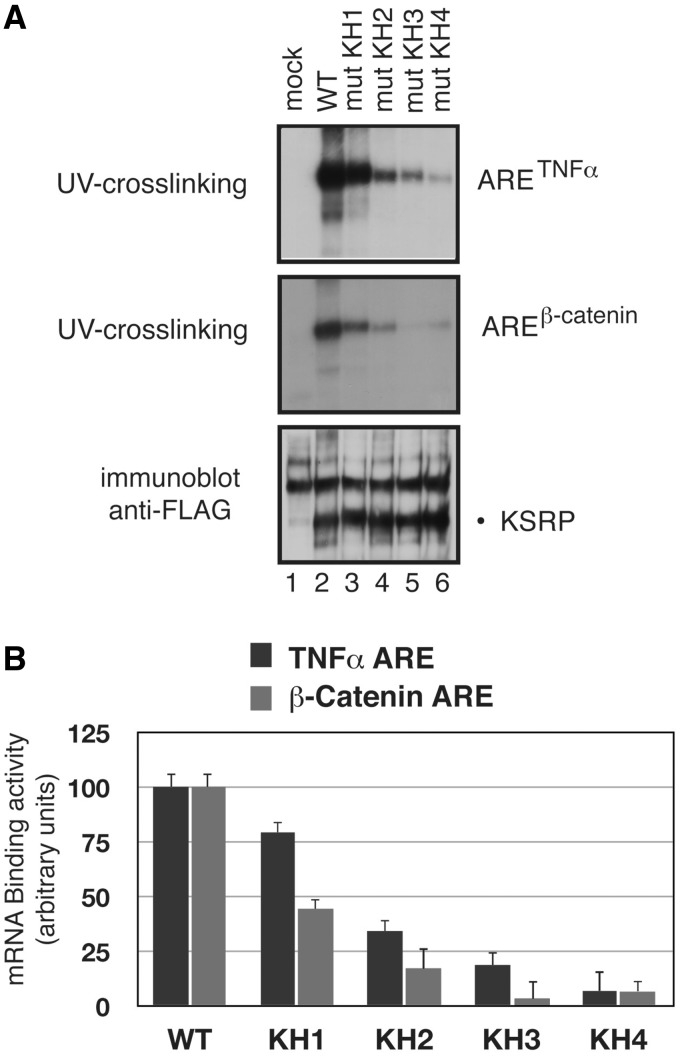
To correlate our in vitro data on RNA binding to the functional effect of the different mutations, we evaluated the capability of KSRP (wild-type and mutants) to cross-link its ARE targets within cell extract. The cross-linking assays were performed on AREs from both the TNFα and β-catenin mRNAs. The TNFα ARE core sequence is a short low complexity sequence (Supplementary Table S3) (34) that lends itself to biophysical studies. In contrast, the β-catenin ARE is ∼200-nt long with six AU-rich motifs interspersed by non U-rich sequences (Supplementary Table S3) and is more unsuitable for our biophysical assays, but is one of the functionally best characterized KSRP targets (20,37). Comparison of cross-linking assays performed on the two AREs allowed us to better link BLI data to the functional data. We expressed wild-type Flag-tagged nearly full-length KSRP (amino acids 47–711) and its GDDG mutant derivatives in HEK-293 cells, prepared total cell lysates and assayed aliquots for binding to in vitro synthesized RNAs. Upon UV-crosslinking, samples were immunoprecipitated by anti-Flag antibody and the relative RNA-binding activity of either wild-type KSRP or its mutants was quantified (Figure 7). Our assays show that eliminating RNA binding by KH1 has only a modest effect on the ability of KSRP to cross-link the two AREs, while eliminating KH2 binding has a substantial effect and eliminating either KH3 or KH4 binding reduces the cross-linking to the threshold of detection (Figure 7). The relative importance of the different KH domains in RNA cross-linking within the cell extract is consistent with their role in securing high-affinity binding by the four domain protein. Interestingly, despite the sequence differences existing between TNFα and β-catenin AREs, the relative ability of each mutant to bind the mRNAs was very similar.
Figure 7.
Interaction of wild-type and KSRP GDDG mutants with TNFα and β-catenin ARE RNA. (A) Aliquots (200 µg) of total extracts from HEK-293 cells transiently transfected with either wild-type Flag-tagged nearly full-length KSRP (amino acids 47–711) or its GDDG mutant derivatives (or with the empty vector, mock) were assayed for binding to in vitro synthesized 32P-labelled TNFα and β-catenin ARE RNAs. After UV-crosslinking, the reactions were immunoprecipitated with anti-Flag antibody, separated by SDS–PAGE, and autoradiographed (top panels). Immunoprecipitates presented in the top panels were analyzed by immunoblot analysis using anti-Flag antibody (the slower-migrating bands in the blot correspond to non-specific immunoreactivity). An experiment representative of the three performed is shown. (B) Bands in the UV-crosslinking experiments displayed in panel a and in the autoradiograms from two other independent experiments were analyzed by densitometric scanning (ImageJ64 software), normalized for the levels of anti-Flag immunoreactivity and presented as average ± SEM.
The effect of the GDDG mutations within single KH domains on KSRP-mediated mRNA degradation
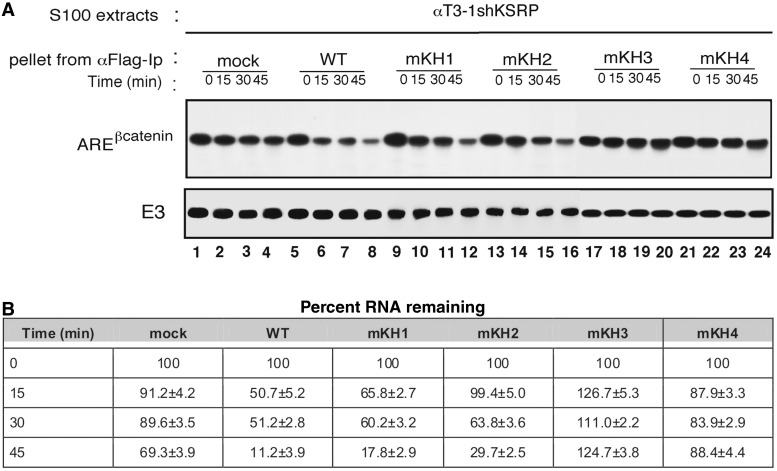
As discussed above, KSRP recruits components of the general enzymatic degradation machinery (exosome, PARN, DCP2) to the target mRNAs promoting rapid mRNA degradation (20,35). RNA in vitro degradation assays have proved to faithfully reproduce the mechanism and the regulation of labile mRNA occurring in intact cells and these assays have been extensively utilized in previous studies (20,34). Here radiolabelled β-catenin ARE RNA was incubated with S100 cytoplasmic extract of αT3-1 cells in which KSRP has been knocked-down (20) in the presence of either wild-type KSRP or its mutant derivatives immunopurified from total extracts of transiently transfected HEK-293 cells. As shown in Figure 8, in the presence of limiting amounts of S100 extracts of KSRP-depleted αT3-1 cells, β-catenin ARE mRNA is stable (lanes 1–4) and the incubation with immunopurified wild-type KSRP promotes its rapid decay (lanes 5–8). While KH1 and KH2 mutants were able to promote mRNA decay, KH3 and KH4 mutants were ineffective (Figure 8). This is in general agreement with the RNA-binding data, except for KH2 that seems to play a less prominent role in mRNA turnover than in protein function, which may relate to the overall structure of the 3′-UTR and will require further investigations.
Figure 8.
β-catenin mRNA degradation assays. (A) In vitro RNA degradation assays performed using 2.5 µg of S100 extracts from αT3-1 shKRP cells (20) incubated with Flag-tagged wild-type KSRP or its GDDG mutant derivatives immunopurified from transiently transfected HEK-293 cells. Internally 32P-labelled, capped RNA substrates were incubated with the extracts for the indicated times, and their decay was analyzed as described in the Experimental Procedures. Please note that for technical reasons samples of KH1 and KH2 and KH3 and KH4 GxxG mutants were analysed on two gels run in parallel and are collated here for clarity. (B) The intensity of the bands corresponding to β-catenin mRNA were quantitated using ImageJ software (http://rsb.info.nih.gov/ij/index.html) and expressed as percentage of the intensity of the mRNA present at 0 time points.
DISCUSSION
Rationally designed mutations that decouple a protein function(s) from its stability and fold are an important tool to probe the molecular basis of physiological processes. Multi-domain RNA-binding proteins coordinate the different steps of mRNA metabolism. Specificity of regulation is provided by the recognition of the mRNA targets by one or more RNA-binding domains [e.g. KH, RRM, ZnF; (16)]. In this article we focus on the KH domain, a common motif mediating RNA binding in hundreds of eukaryotic proteins, and describe the rational design of a GDDG mutant that decouples the RNA-binding activity of the KH domain from its fold and stability. This mutant represents an effective tool to test the role of RNA recognition in KH containing proteins.
We have tested the GDDG mutation on six well-characterized KH domains. Our experiments show that the mutation does not compromise the stability of the domain but does impair its NA-binding capability. It is difficult to quantify precisely the decrease of the RNA-binding affinity, as GDDG mutant proteins have no measureable RNA-binding activity in our NMR assays. However, the affinity of KSRP KH3 for the G-rich RNA target decreases by at least three orders of magnitude, as the Kd of the wild-type protein is ∼1.5 µM, while the Kd of the mutant is >>1 mM. Importantly, a Kd>>1 mM is unlikely to reflect a functionally relevant interaction, i.e. our six mutant domains are, as far the NA-binding activity is concerned, functionally silent.
The six domains have different stabilities (with temperature unfolding midpoints varying between 50°C and 70°C) and very different RNA sequence specificities. Some of them have a structurally extended KH fold (KSRP KH1 and KH4) while others make stable intra-molecular inter-domain contacts (ZBP1 KH3 and KH4). The differences in structure, stability and RNA target specificity between the domains emphasize that the GDDG mutant is a tool of general applicability for proteins containing the KHs.
Several studies report the mutational analysis of KH domains with the aim to impair RNA-binding activity. The I304N mutation in the second KH domain of FMRP is known to impair RNA binding and protein function (38,39) and mutation of the corresponding hydrophobic amino acid to an asparagine has been shown to significantly decrease NA binding by the KH module of the SF1 protein (7) and by the four KH domains of the PSI protein (40). However, the side chain of this amino acid packs against the hydrophobic core of the protein and its mutation can destabilize the KH domain—as observed in KH domains from the Vigilin, FMRP, PSI and Nova proteins (1,24,40,41). Single mutations within the GxxG loop have been tested in a number of proteins. For example, it has been shown that mutation of R160 and G161 in the GPRG loop of SF1 has a strong effect on RNA binding (7). However, the distance between the side chains of the central amino acids in the GxxG loop and the nearest RNA phosphate group are significantly different in different complexes (Figure 2) indicating that the effect of mutating only one of the two central variable amino acids of the GxxG loop is likely to be different in different proteins. Furthermore, the GxxG backbone is dynamic in the unbound protein (Figure 2A and Supplementary Figure S1) and we would expect that, at least in some cases, the position of the single negatively charged mutant side chain will be able to partially adjust to the NA molecule, allowing some residual binding. Finally, most of the reported mutants in variable amino acids of the GxxG loop have strongly reduced but not completely abolished the RNA-binding ability (10,42). Other mutations across the RNA-binding groove have also been tested, but no consistent effect on RNA binding has been reported. We believe that the GDDG mutation represents a very effective and general tool to impair NA binding, as the two neighbouring negatively charged side chains cover a wide space and prevent the approach of the RNA backbone phosphates to the loop.
Here we dissect the contribution of the four KH domains of the KSRP protein to ARE RNA recognition and protein function providing an example of how this tool can be used to analyse the relation between RNA binding and function. KSRP is a multi-functional protein and its four KH domains show different levels of specificity. KH1 binding to KSRP AU-rich target sequences is significantly weaker than the binding of the other domains (at least 10-fold weaker than KH3), while the differences between KH2, KH3 and KH4 are only 2- to 3-fold (28). One important question is if the relatively small differences we observe between the binding affinities of the isolated domains are maintained in the four domain protein and if they impact on protein function. Our BLI assays show that a significant decrease in binding affinity to the TNFα ARE RNA takes place when a GDDG mutation is inserted in one of the domains. However, the effect is very different for different domains varying from the 2-fold decrease observed for KH1 to the ∼30-fold decrease observed for KH3. Interestingly, the ranking of the four KH domains according to the effect that their mutation has on KSRP RNA-binding affinity recapitulates the affinity of the single domain for AU-rich sequences.
Cross-linking efficiency does not quantitatively relate to RNA-binding ability but can report about binding in the cellular environment. In cross-linking assays a strong effect was observed for the KH3 and KH4 mutations that severely impaired RNA binding. The effect of mutating KH2 was less severe, while only a mild effect was observed upon mutation of KH1. This pattern is maintained for both TNFα and β-catenin AREs. It is worth pointing out that the fact that the cross-linking is nearly completely abolished by mutation of either KH3 or KH4 does not imply that the effect of these two mutations is equivalent, but only that in both cases the effect is strong enough to reach the limit of the binding range explored by the experiment. The modest effect observed upon mutation of KH1 is consistent with the low affinity of the domain for AU-rich sequences. Finally, we used a β-catenin mRNA degradation assay to connect in vitro data on mRNA turnover to binding data. The assays show how mutating KH1 results in a very modest impairment of the RNA degradation capability of KSRP, in agreement with its limited role in RNA binding. This is also consistent with existing data that indicate that the functional unfolding of KH1 upon phosphorylation does not impair the association with the RNA (18,20) and with the existence of a cytoplasmatic KSRP splicing variant that lacks the KH1 domain but maintains its capability to degrade RNA (43). Instead, mutating KH3 or KH4 results in a much more severe impairment i.e. it strongly affects protein function. Mutating KH2 results in a relatively weak impairment of mRNA degradation. In general, our data show a correspondence between RNA binding, cross-linking and functional assays. Interestingly, the effect of a KH2 mutation on cross-linking and particularly mRNA degradation is less robust than expected from the affinity of the domain for RNA in vitro, which could be linked to the structural context provided by the 3′-UTR.
Here we relate biophysical and structural parameters to biological function in order to define the role of specific domains in RNA recognition by the KSRP protein. It is important to point out that the GDDG mutant represents a tool of broad scope that can be used to rationalize the set of RNA targets of the vast ensemble of KH containing proteins where combinatorial binding takes place—and to provide a new angle on the evolution of the function of multi-domain proteins at the single domain level.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figure 1.
FUNDING
Medical Research Council (MRC) [U117574558, U117570592]; European Molecular Biology Organization (EMBO) fellowship number [368-2008 to A.M.C.]; AICR [#10-0527 to R.G.]; AIRC [I.G. #10090 to R.G.]; Regione Liguria [CIPE 2007 to R.G.]. Funding for open access charge: MRC [U117574558].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Drs T. A. Frenkiel, G. Kelly and A. Oreggioni for their help in recording spectra. The plasmid of the original KSRP clone was provided by Dr CY Chen. All NMR spectra were recorded at the Medical Research Council Biomedical NMR centre.
REFERENCES
- 1.Musco G, Stier G, Joseph C, Castiglione Morelli MA, Nilges M, Gibson TJ, Pastore A. Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome. Cell. 1996;85:237–245. doi: 10.1016/s0092-8674(00)81100-9. [DOI] [PubMed] [Google Scholar]
- 2.Siomi H, Matunis MJ, Michael WM, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grishin NV. KH domain: one motif, two folds. Nucleic Acids Res. 2001;29:638–643. doi: 10.1093/nar/29.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backe PH, Messias AC, Ravelli RBG, Sattler M, Cusack S. X-ray crystallographic and NMR studies of the third KH domain of hnRNP K in complex with single-stranded nucleic acids. Structure. 2005;13:1055–1067. doi: 10.1016/j.str.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Valverde R, Edwards L, Regan L. Structure and function of KH domains. FEBS J. 2008;275:2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 6.Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell RB, Burley SK. Sequence-specific RNA binding by a nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Luyten I, Bottomley MJ, Messias AC, Houngninou-Molango S, Sprangers R, Zanier K, Kramer A, Sattler M. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science. 2001;294:1098–1102. doi: 10.1126/science.1064719. [DOI] [PubMed] [Google Scholar]
- 8.Braddock DT, Baber JL, Levens D, Clore GM. Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: solution structure of a complex between hnRNP K KH3 and single-stranded DNA. EMBO J. 2002;21:3476–3485. doi: 10.1093/emboj/cdf352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braddock DT, Louis JM, Baber JL, Levens D, Clore GM. Structure and dynamics of KH domains from FBP bound to single-stranded DNA. Nature. 2002;415:1051–1056. doi: 10.1038/4151051a. [DOI] [PubMed] [Google Scholar]
- 10.Beuth B, Pennell S, Arnvig KB, Martin SR, Taylor IA. Structure of a Mycobacterium tuberculosis NusA-RNA complex. EMBO J. 2005;24:3576–3587. doi: 10.1038/sj.emboj.7600829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du Z, Lee JK, Tjhen R, Li S, Pan H, Stroud RM, James TL. Crystal structure of the first KH domain of human poly(C)-binding protein-2 in complex with a C-rich strand of human telomeric DNA at 1.7 A. J. Biol. Chem. 2005;280:38823–38830. doi: 10.1074/jbc.M508183200. [DOI] [PubMed] [Google Scholar]
- 12.Du Z, Lee JK, Fenn S, Tjhen R, Stroud RM, James TL. X-ray crystallographic and NMR studies of protein-protein and protein-nucleic acid interactions involving the KH domains from human poly(C)-binding protein-2. RNA. 2007;13:1043–1051. doi: 10.1261/rna.410107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenn S, Du Z, Lee JK, Tjhen R, Stroud RM, James TL. Crystal structure of the third KH domain of human poly(C)-binding protein-2 in complex with a C-rich strand of human telomeric DNA at 1.6 A resolution. Nucleic Acids Res. 2007;35:2651–2660. doi: 10.1093/nar/gkm139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakel K, Hartung SA, Bonneau F, Eckmann CR, Conti E. Four KH domains of the C. elegans Bicaudal-C ortholog GLD-3 form a globular structural platform. RNA. 2010;16:2058–2067. doi: 10.1261/rna.2315010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oddone A, Lorentzen E, Basquin J, Gasch A, Rybin V, Conti E, Sattler M. Structural and biochemical characterization of the yeast exosome component Rrp40. EMBO Rep. 2007;8:63–69. doi: 10.1038/sj.embor.7400856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Mayoral MF, Hollingworth D, Masino L, Diaz-Moreno I, Kelly G, Gherzi R, Chou CF, Chen CY, Ramos A. The structure of the C-terminal KH domains of KSRP reveals a noncanonical motif important for mRNA degradation. Structure. 2007;15:485–498. doi: 10.1016/j.str.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Diaz-Moreno I, Hollingworth D, Frenkiel TA, Kelly G, Martin S, Howell S, Garcia-Mayoral M, Gherzi R, Briata P, Ramos A. Phosphorylation-mediated unfolding of a KH domain regulates KSRP localization via 14-3-3 binding. Nat. Struct. Mol. Biol. 2009;16:238–246. doi: 10.1038/nsmb.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diaz-Moreno I, Hollingworth D, Kelly G, Martin S, Garcia-Mayoral M, Briata P, Gherzi R, Ramos A. Orientation of the central domains of KSRP and its implications for the interaction with the RNA targets. Nucleic Acids Res. 2010;38:5193–5205. doi: 10.1093/nar/gkq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gherzi R, Trabucchi M, Ponassi M, Ruggiero T, Corte G, Moroni C, Chen CY, Khabar KS, Andersen JS, Briata P. The RNA-binding protein KSRP promotes decay of beta-catenin mRNA and is inactivated by PI3K-AKT signaling. PLoS Biol. 2006;5:e5. doi: 10.1371/journal.pbio.0050005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 22.Goddard TD, Kneller DG. San Fancisco: Sparky 3. University of California; 2004. [Google Scholar]
- 23.Cukier CD, Hollingworth D, Martin SR, Kelly G, Diaz-Moreno I, Ramos A. Molecular basis of FIR-mediated c-myc transcriptional control. Nat. Struct. Mol. Biol. 2010;17:1058–1064. doi: 10.1038/nsmb.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Musco G, Kharrat A, Stier G, Fraternali F, Gibson TJ, Nilges M, Pastore A. The solution structure of the first KH domain of FMR1, the protein responsible for the fragile X syndrome. Nat. Struct. Biol. 1997;4:712–716. doi: 10.1038/nsb0997-712. [DOI] [PubMed] [Google Scholar]
- 25.Baber JL, Levens D, Libutti D, Tjandra N. Chemical shift mapped DNA-binding sites and 15N relaxation analysis of the C-terminal KH domain of heterogeneous nuclear ribonucleoprotein K. Biochemistry. 2000;39:6022–6032. doi: 10.1021/bi000105e. [DOI] [PubMed] [Google Scholar]
- 26.Ramos A, Hollingworth D, Major SA, Adinolfi S, Kelly G, Muskett FW, Pastore A. Role of dimerization in KH/RNA complexes: the example of Nova KH3. Biochemistry. 2002;41:4193–4201. doi: 10.1021/bi011994o. [DOI] [PubMed] [Google Scholar]
- 27.Du Z, Fenn S, Tjhen R, James TL. Structure of a construct of a human poly(C)-binding protein containing the first and second KH domains reveals insights into its regulatory mechanisms. J. Biol. Chem. 2008;283:28757–28766. doi: 10.1074/jbc.M803046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Mayoral MF, Diaz-Moreno I, Hollingworth D, Ramos A. The sequence selectivity of KSRP explains its flexibility in the recognition of the RNA targets. Nucleic Acids Res. 2008;36:5290–5296. doi: 10.1093/nar/gkn509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 2010;24:148–158. doi: 10.1101/gad.1862910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of [beta]-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 31.Patel VL, Mitra S, Harris R, Buxbaum AR, Lionnet T, Brenowitz M, Girvin M, Levy M, Almo SC, Singer RH, et al. Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev. 2012;26:43–53. doi: 10.1101/gad.177428.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farina KL, Huttelmaier S, Musunuru K, Darnell R, Singer RH. Two ZBP1 KH domains facilitate beta-actin mRNA localization, granule formation, and cytoskeletal attachment. J. Cell Biol. 2003;160:77–87. doi: 10.1083/jcb.200206003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Chou CF, Mulky A, Maitra S, Lin WJ, Gherzi R, Kappes J, Chen CY. Tethering KSRP, a decay-promoting AU-rich element-binding protein, to mRNAs elicits mRNA decay. Mol. Cell Biol. 2006;26:3695–3706. doi: 10.1128/MCB.26.10.3695-3706.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trabucchi M, Briata P, Filipowicz W, Rosenfeld MG, Ramos A, Gherzi R. How to control miRNA maturation? RNA Biol. 2009;6:536–540. doi: 10.4161/rna.6.5.10080. [DOI] [PubMed] [Google Scholar]
- 37.Ruggiero T, Trabucchi M, Ponassi M, Corte G, Chen CY, al-Haj L, Khabar KS, Briata P, Gherzi R. Identification of a set of KSRP target transcripts upregulated by PI3K-AKT signaling. BMC Mol. Biol. 2007;8:28. doi: 10.1186/1471-2199-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibson TJ, Rice PM, Thompson JD, Heringa J. KH domains within the FMR1 sequence suggest that fragile X syndrome stems from a defect in RNA metabolism. Trends Biochem. Sci. 1993;18:331–333. doi: 10.1016/0968-0004(93)90068-x. [DOI] [PubMed] [Google Scholar]
- 39.Siomi H, Choi M, Siomi MC, Nussbaum RL, Dreyfuss G. Essential role for KH domains in RNA binding: Impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 1994;77:33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 40.Chmiel NH, Rio DC, Doudna JA. Distinct contributions of KH domains to substrate binding affinity of Drosophila P-element somatic inhibitor protein. RNA. 2006;12:283–291. doi: 10.1261/rna.2175706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis HA, Chen H, Edo C, Buckanovich RJ, Yang YY, Musunuru K, Zhong R, Darnell RB, Burley SK. Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains. Structure. 1999;7:191–203. doi: 10.1016/S0969-2126(99)80025-2. [DOI] [PubMed] [Google Scholar]
- 42.Nielsen J, Adolph SK, Rajpert-De Meyts E, Lykke-Andersen J, Koch G, Christiansen J, Nielsen FC. Nuclear transit of human zipcode-binding protein IMP1. Biochem. J. 2003;376:383–391. doi: 10.1042/BJ20030943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu M, McCarrey JR, Hecht NB. A cytoplasmic variant of the KH-type splicing regulatory protein serves as a decay-promoting factor for phosphoglycerate kinase 2 mRNA in murine male germ cells. Nucleic Acids Res. 2008;36:7157–7167. doi: 10.1093/nar/gkn800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.