Abstract
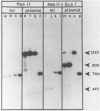
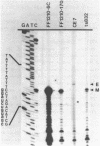
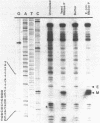
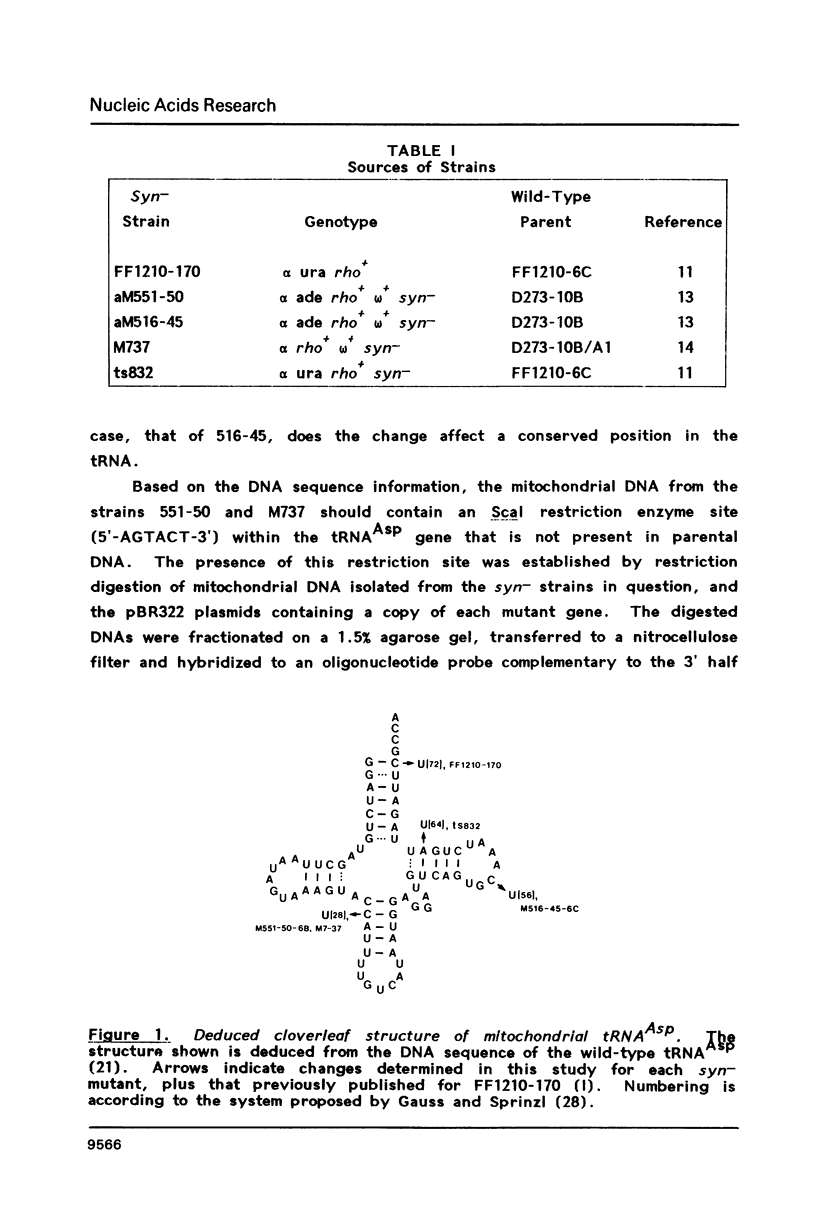
Expression of the mitochondrial tRNAAsp gene of Saccharomyces cerevisiae has been examined in five syn- mutants known to affect tRNAAsp function, and in a rho- mutant which accumulates precursor tRNAs. By comparison of wild-type versus mutant DNA sequence, the lesion in each syn- mutant has been identified as a single base change within the mitochondrial tRNAAsp structural gene. The mutant tRNAAsp genes are transcribed, and the transcripts can be processed to mature 4S-size tRNAAsp. The steady-state level of each mutant tRNAAsp is lower than that of wild-type tRNAAsp. The RNA from two of the syn- mutants contained a second, slow-migrating form of mitochondrial tRNAAsp which is correctly processed at the 5' end. We conclude that the lesions in the syn- mitochondrial tRNAAsp genes block neither transcription of these genes, nor 5'-end processing of the transcripts. The effect of each point mutation must be manifested at the level of 3'-end processing, or at a functional level.
Full text
PDF





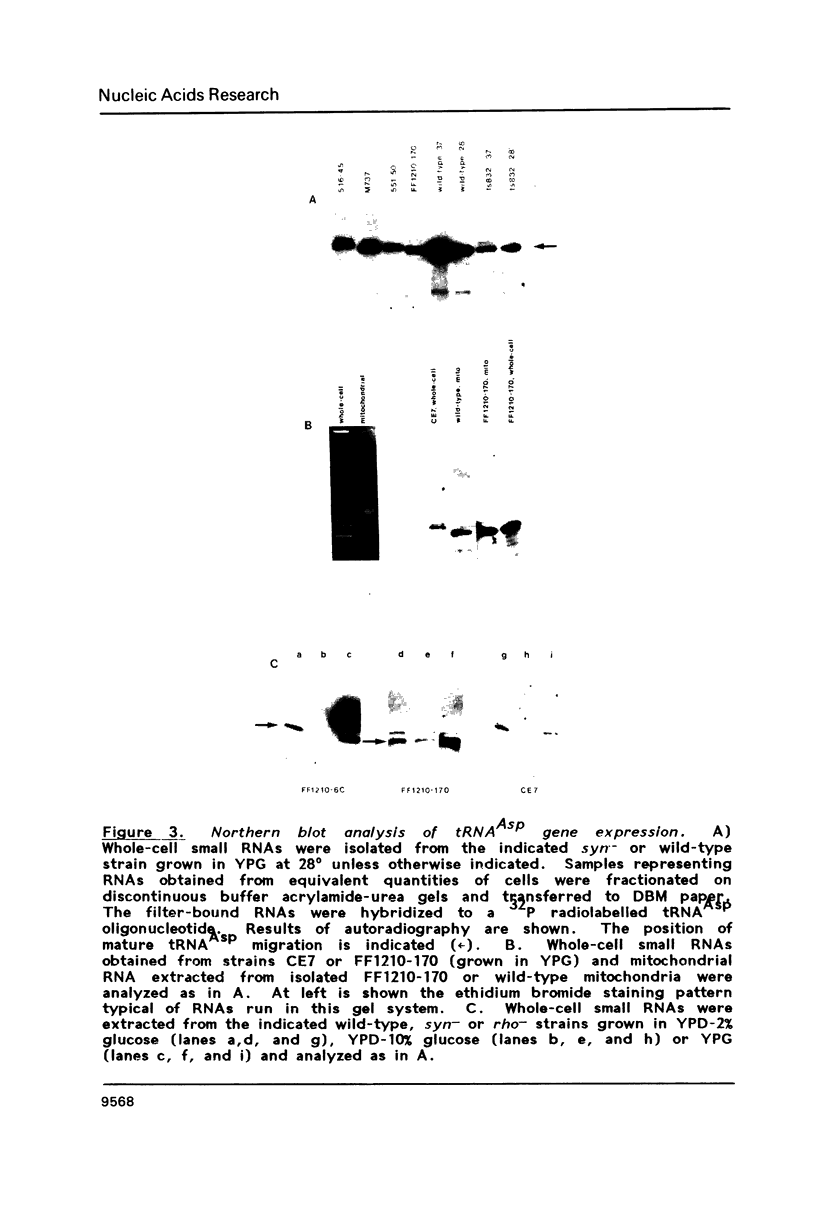


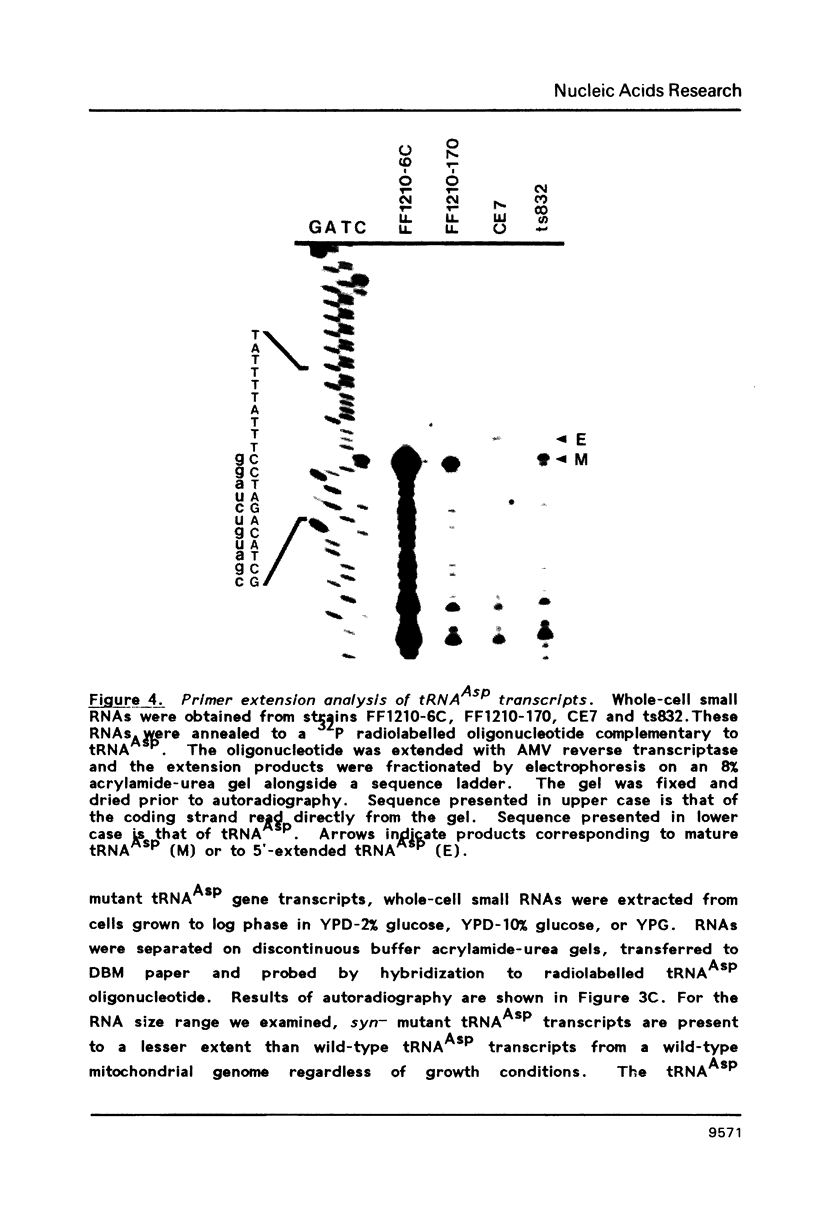

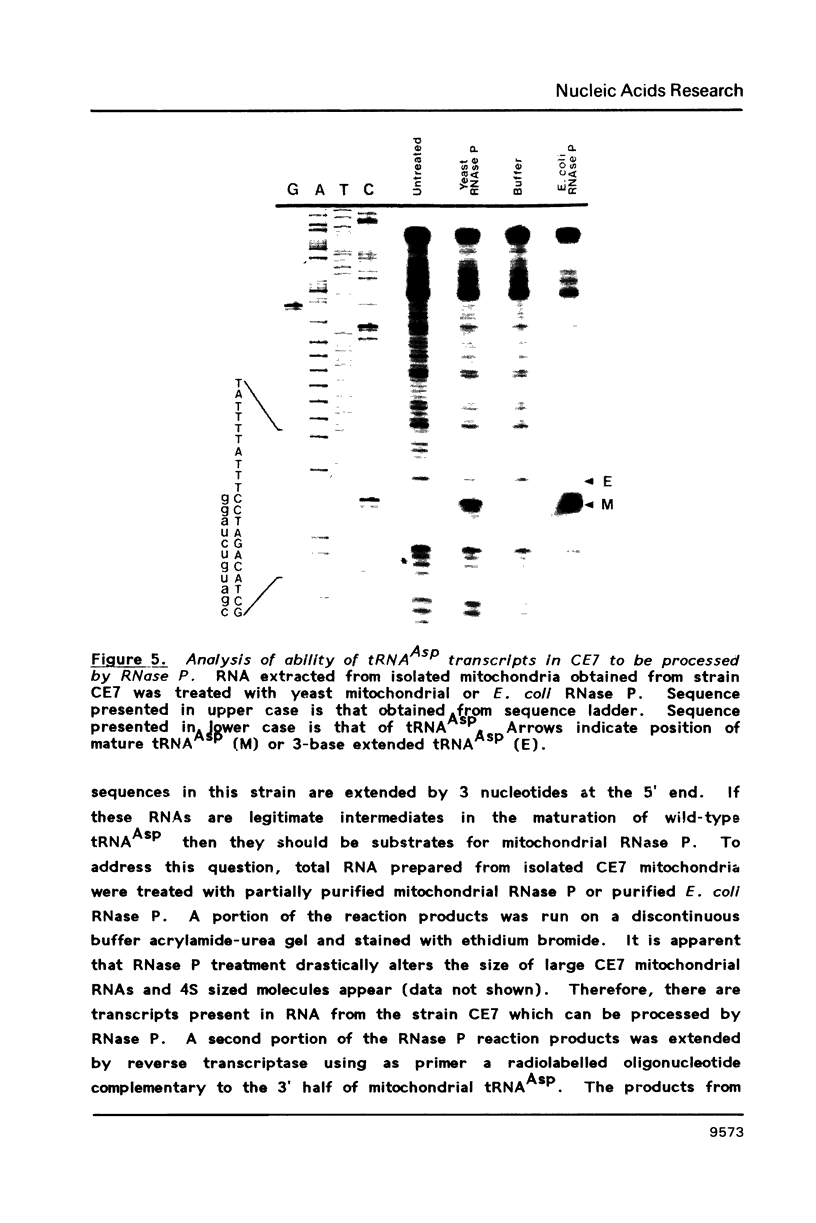





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berlani R. E., Pentella C., Macino G., Tzagoloff A. Assembly of the mitochondrial membrane system: isolation of mitochondrial transfer ribonucleic acid mutants and characterization of transfer ribonucleic acid genes of Saccharomyces cerevisiae. J Bacteriol. 1980 Mar;141(3):1086–1097. doi: 10.1128/jb.141.3.1086-1097.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonitz S. G., Tzagoloff A. Assembly of the mitochondrial membrane system. Sequences of yeast mitochondrial tRNA genes. J Biol Chem. 1980 Oct 10;255(19):9075–9081. [PubMed] [Google Scholar]
- Celis J. E. Collection of mutant tRNA sequences. Nucleic Acids Res. 1979 Jan;6(1):r21–r-27. doi: 10.1093/nar/6.1.419-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T., Edwards J. C., Mueller D. M., Rabinowitz M. Identification of a single transcriptional initiation site for the glutamic tRNA and COB genes in yeast mitochondria. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5564–5568. doi: 10.1073/pnas.80.18.5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T., Rabinowitz M. Identification of multiple transcriptional initiation sites on the yeast mitochondrial genome by in vitro capping with guanylyltransferase. J Biol Chem. 1983 Nov 25;258(22):14025–14033. [PubMed] [Google Scholar]
- Conde J., Fink G. R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna K. J. Determination of fragment order through partial digests and multiple enzyme digests. Methods Enzymol. 1980;65(1):449–467. doi: 10.1016/s0076-6879(80)65055-1. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Gauss D. H., Sprinzl M. Compilation of tRNA sequences. Nucleic Acids Res. 1981 Jan 10;9(1):r1–23. [PMC free article] [PubMed] [Google Scholar]
- Geck P., Nász I. Concentrated, digestible DNA after hydroxylapatite chromatography with cetylpyridinium bromide precipitation. Anal Biochem. 1983 Dec;135(2):264–268. doi: 10.1016/0003-2697(83)90681-4. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLLEY R. W., APGAR J., DOCTOR B. P., FARROW J., MARINI M. A., MERRILL S. H. A simplified procedure for the preparation of tyrosine and valine-acceptor fractions of yeast "soluble ribonucleic acid". J Biol Chem. 1961 Jan;236:200–202. [PubMed] [Google Scholar]
- Hollingsworth M. J., Martin N. C. RNase P activity in the mitochondria of Saccharomyces cerevisiae depends on both mitochondrion and nucleus-encoded components. Mol Cell Biol. 1986 Apr;6(4):1058–1064. doi: 10.1128/mcb.6.4.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudspeth M. E., Shumard D. S., Tatti K. M., Grossman L. I. Rapid purification of yeast mitochondrial DNA in high yield. Biochim Biophys Acta. 1980 Dec 11;610(2):221–228. doi: 10.1016/0005-2787(80)90003-9. [DOI] [PubMed] [Google Scholar]
- Koski R. A., Clarkson S. G., Kurjan J., Hall B. D., Smith M. Mutations of the yeast SUP4 tRNATyr locus: transcription of the mutant genes in vitro. Cell. 1980 Nov;22(2 Pt 2):415–425. doi: 10.1016/0092-8674(80)90352-9. [DOI] [PubMed] [Google Scholar]
- Kurjan J., Hall B. D., Gillam S., Smith M. Mutations at the yeast SUP4 tRNATyr locus: DNA sequence changes in mutants lacking suppressor activity. Cell. 1980 Jul;20(3):701–709. doi: 10.1016/0092-8674(80)90316-5. [DOI] [PubMed] [Google Scholar]
- Locker J. Analytical and preparative electrophoresis of RNA in agarose-urea. Anal Biochem. 1979 Oct 1;98(2):358–367. doi: 10.1016/0003-2697(79)90154-4. [DOI] [PubMed] [Google Scholar]
- Locker J., Rabinowitz M. Transcription in yeast mitochondria: analysis of the 21 S rRNA region and its transcripts. Plasmid. 1981 Nov;6(3):302–314. doi: 10.1016/0147-619x(81)90038-x. [DOI] [PubMed] [Google Scholar]
- Martin N. C., Miller D. L., Underbrink K., Ming X. Structure of a precursor to the yeast mitochondrial tRNAMetf. Implications for the function of the tRNA synthesis locus. J Biol Chem. 1985 Feb 10;260(3):1479–1483. [PubMed] [Google Scholar]
- Miller D. L., Folse J. R., Benson P. J., Martin N. C. Identification and consequences of a guanosine-15 to adenosine-15 change in the yeast mitochondrial tRNASerUCX gene. Biochemistry. 1983 Mar 29;22(7):1709–1714. doi: 10.1021/bi00276a029. [DOI] [PubMed] [Google Scholar]
- Miller D. L., Martin N. C., Pham H. D., Donelson J. E. Sequence analysis of two yeast mitochondrial DNA fragments containing the genes for tRNA Ser UCR and tRNA Phe UUY. J Biol Chem. 1979 Nov 25;254(22):11735–11740. [PubMed] [Google Scholar]
- Miller D. L., Najarian D. R., Folse J. R., Martin N. C. A mutation in the tRNAAsp gene from yeast mitochondria. Effects on RNA and protein synthesis. J Biol Chem. 1981 Oct 10;256(19):9774–9777. [PubMed] [Google Scholar]
- Myers A. M., Pape L. K., Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985 Aug;4(8):2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K., Kurjan J., Hall B. D., De Robertis E. M. Genetic analysis of the processing of a spliced tRNA. EMBO J. 1982;1(2):263–268. doi: 10.1002/j.1460-2075.1982.tb01157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nóbrega M. P., Nóbrega F. G. Mapping and sequencing of the wild-type and mutant (G116-40) alleles of the tyrosyl-tRNA mitochondrial gene in Saccharomyces cerevisiae. J Biol Chem. 1986 Mar 5;261(7):3054–3059. [PubMed] [Google Scholar]
- Osinga K. A., Tabak H. F. Initiation of transcription of genes for mitochondrial ribosomal RNA in yeast: comparison of the nucleotide sequence around the 5'-ends of both genes reveals a homologous stretch of 17 nucleotides. Nucleic Acids Res. 1982 Jun 25;10(12):3617–3626. doi: 10.1093/nar/10.12.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palleschi C., Francisci S., Zennaro E., Frontali L. Expression of the clustered mitochondrial tRNA genes in Saccharomyces cerevisiae: transcription and processing of transcripts. EMBO J. 1984 Jun;3(6):1389–1395. doi: 10.1002/j.1460-2075.1984.tb01982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieczenik G. Multimers of a suppressor transfer RNA: supporting evidence for alternate conformations of the anticodon loop region. J Mol Biol. 1980 Apr 25;138(4):879–884. doi: 10.1016/0022-2836(80)90071-6. [DOI] [PubMed] [Google Scholar]
- Stellwag E. J., Dahlberg A. E. Electrophoretic transfer of DNA, RNA and protein onto diazobenzyloxymethyl (DBM) - paper. Nucleic Acids Res. 1980 Jan 25;8(2):299–317. doi: 10.1093/nar/8.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalenfeld B. E., Bonitz S. G., Nobrega F. G., Macino G., Tzagoloff A. oli1 Transcripts in wild type and in a cytoplasmic "petite" mutant of yeast. J Biol Chem. 1983 Dec 10;258(23):14065–14068. [PubMed] [Google Scholar]
- Thalenfeld B. E., Hill J., Tzagoloff A. Assembly of the mitochondrial membrane system. Characterization of the oxi2 transcript and localization of its promoter in Saccharomyces cerevisiae D273-10B. J Biol Chem. 1983 Jan 10;258(1):610–615. [PubMed] [Google Scholar]
- Tobian J. A., Drinkard L., Zasloff M. tRNA nuclear transport: defining the critical regions of human tRNAimet by point mutagenesis. Cell. 1985 Dec;43(2 Pt 1):415–422. doi: 10.1016/0092-8674(85)90171-0. [DOI] [PubMed] [Google Scholar]
- Trembath M. K., Macino G., Tzagoloff A. The mapping of mutations in tRNA and cytochrome oxidase genes located in the cap-par segment of the mitochondrial genome of S. cerevisiae. Mol Gen Genet. 1977 Dec 14;158(1):35–45. doi: 10.1007/BF00455117. [DOI] [PubMed] [Google Scholar]
- Zassenhaus H. P., Martin N. C., Butow R. A. Origins of transcripts of the yeast mitochondrial var 1 gene. J Biol Chem. 1984 May 10;259(9):6019–6027. [PubMed] [Google Scholar]