Abstract
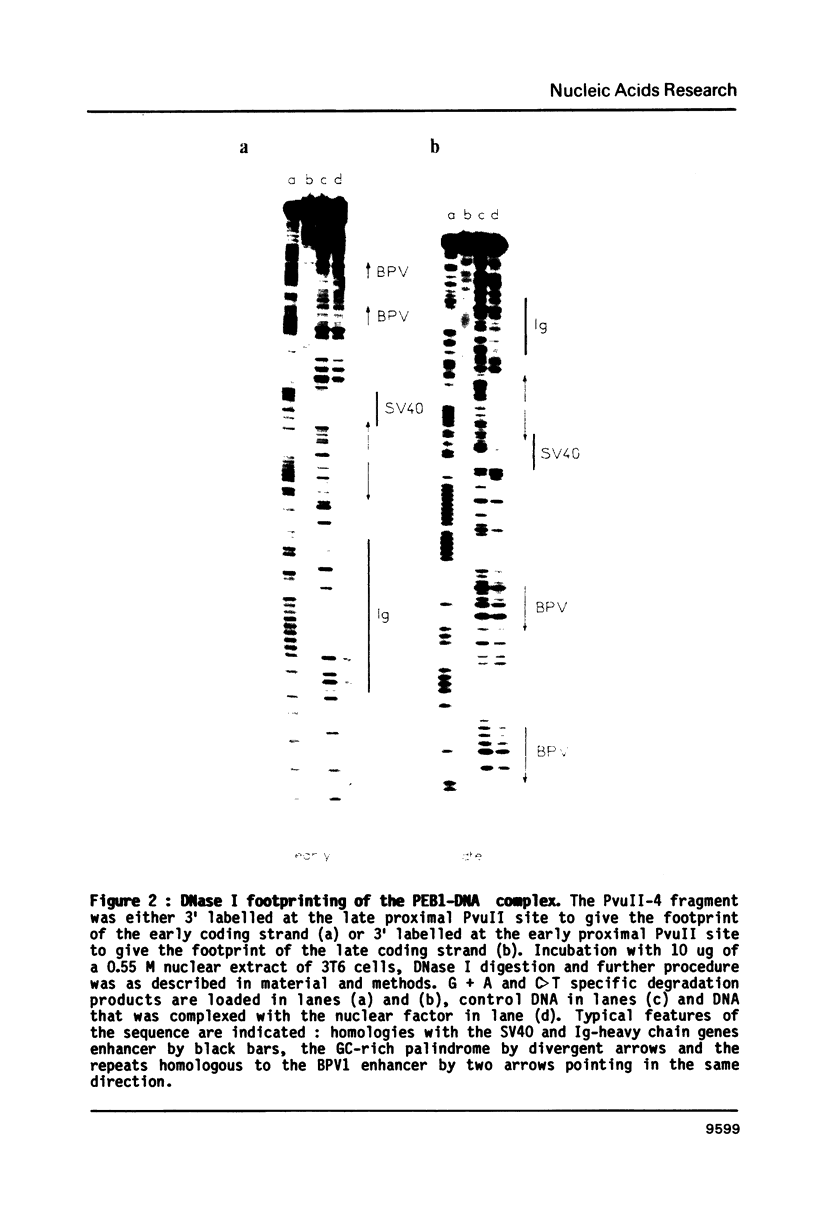
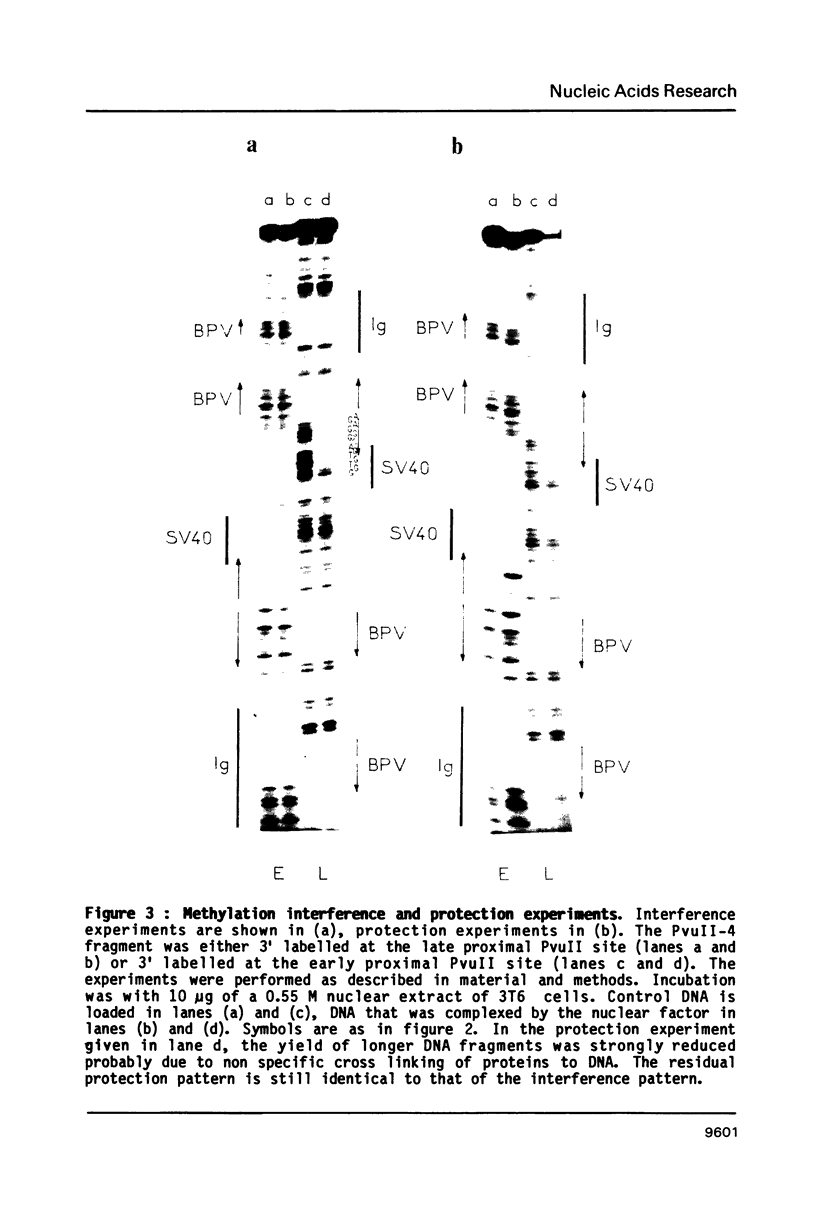
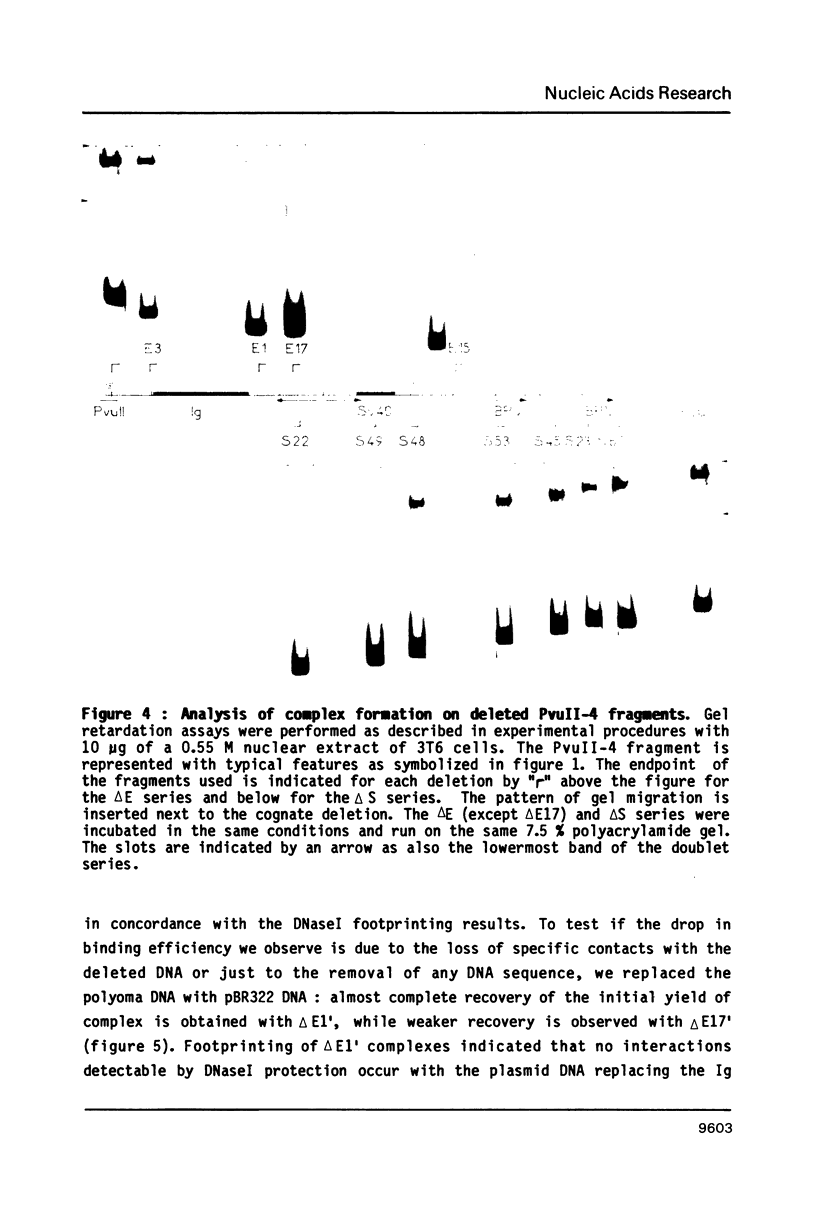
The fine contacts of a mouse nuclear factor, called PEB1, with the B enhancer of polyoma virus were analyzed. It protects against DNaseI attack a region of about 50 base pairs that can be divided in two domains. The first contains a GC-rich palindrome and the homology to the SV40 enhancer. The second is homologous to a sequence in the immunoglobulin (Ig) heavy chain gene enhancer. Methylation interference and protection experiments reveal strong specific contacts only with a purine rich track on the late coding strand of the early proximal part of the palindrome. Deletion analysis show that the minimal sequences necessary for binding include only the first domain. The Ig homology contributes only weakly to the binding. The minimal core is similar to the core of the B enhancer defined in vivo. The interactions we observe here are reminiscent of those of TFIIIA positive transcription factor and the 5SRNA gene of Xenopus.
Full text
PDF





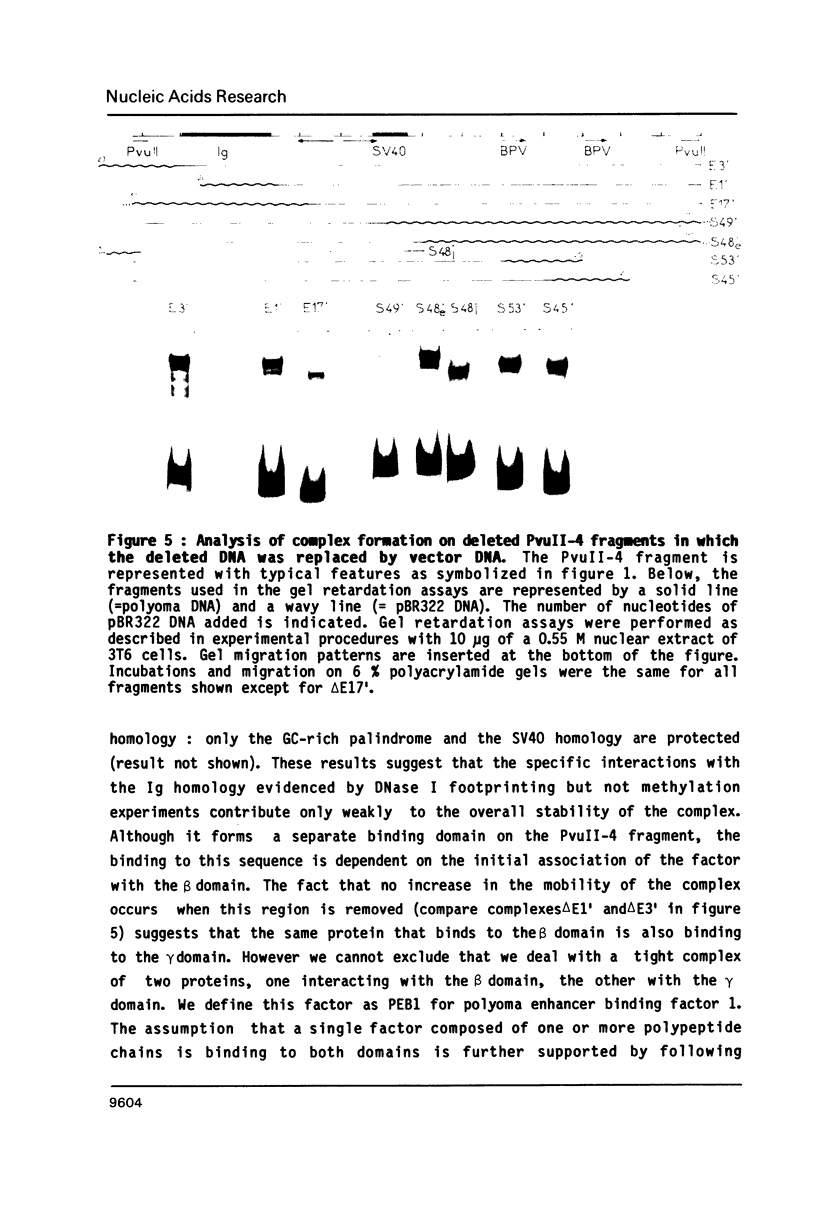







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Hall I. H., Puigjaner L. C., Walker J. K., Wang M. DNA secondary structures: helices, wrinkles, and junctions. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):53–65. doi: 10.1101/sqb.1983.047.01.008. [DOI] [PubMed] [Google Scholar]
- Borgmeyer U., Nowock J., Sippel A. E. The TGGCA-binding protein: a eukaryotic nuclear protein recognizing a symmetrical sequence on double-stranded linear DNA. Nucleic Acids Res. 1984 May 25;12(10):4295–4311. doi: 10.1093/nar/12.10.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan P. N., Folk W. R. Enhancer sequences responsible for DNase I hypersensitivity in polyomavirus chromatin. Mol Cell Biol. 1986 Jun;6(6):2249–2252. doi: 10.1128/mcb.6.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhnlein E., Gruss P. Interaction of distinct nuclear proteins with sequences controlling the expression of polyomavirus early genes. Mol Cell Biol. 1986 May;6(5):1401–1411. doi: 10.1128/mcb.6.5.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowie A., Kamen R. Guanine nucleotide contacts within viral DNA sequences bound by polyomavirus large T antigen. J Virol. 1986 Feb;57(2):505–514. doi: 10.1128/jvi.57.2.505-514.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. 1985 Aug 29-Sep 4Nature. 316(6031):774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Fujimura F. K. Nuclear activity from F9 embryonal carcinoma cells binding specifically to the enhancers of wild-type polyoma virus and PyEC mutant DNAs. Nucleic Acids Res. 1986 Apr 11;14(7):2845–2861. doi: 10.1093/nar/14.7.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidoni D., Dynan W. S., Tjian R. Multiple specific contacts between a mammalian transcription factor and its cognate promoters. 1984 Nov 29-Dec 5Nature. 312(5993):409–413. doi: 10.1038/312409a0. [DOI] [PubMed] [Google Scholar]
- Gidoni D., Kadonaga J. T., Barrera-Saldaña H., Takahashi K., Chambon P., Tjian R. Bidirectional SV40 transcription mediated by tandem Sp1 binding interactions. Science. 1985 Nov 1;230(4725):511–517. doi: 10.1126/science.2996137. [DOI] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Gronostajski R. M., Adhya S., Nagata K., Guggenheimer R. A., Hurwitz J. Site-specific DNA binding of nuclear factor I: analyses of cellular binding sites. Mol Cell Biol. 1985 May;5(5):964–971. doi: 10.1128/mcb.5.5.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss P. Magic enhancers? DNA. 1984;3(1):1–5. doi: 10.1089/dna.1.1984.3.1. [DOI] [PubMed] [Google Scholar]
- Hartshorne T. A., Blumberg H., Young E. T. Sequence homology of the yeast regulatory protein ADR1 with Xenopus transcription factor TFIIIA. Nature. 1986 Mar 20;320(6059):283–287. doi: 10.1038/320283a0. [DOI] [PubMed] [Google Scholar]
- Herbomel P., Saragosti S., Blangy D., Yaniv M. Fine structure of the origin-proximal DNAase I-hypersensitive region in wild-type and EC mutant polyoma. Cell. 1981 Sep;25(3):651–658. doi: 10.1016/0092-8674(81)90172-0. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K., Buc H., Spassky A., Wang J. C. Mapping of single-stranded regions in duplex DNA at the sequence level: single-strand-specific cytosine methylation in RNA polymerase-promoter complexes. Proc Natl Acad Sci U S A. 1983 May;80(9):2544–2548. doi: 10.1073/pnas.80.9.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughon A., Scott M. P. Sequence of a Drosophila segmentation gene: protein structure homology with DNA-binding proteins. Nature. 1984 Jul 5;310(5972):25–31. doi: 10.1038/310025a0. [DOI] [PubMed] [Google Scholar]
- Malissen B., Price M. P., Goverman J. M., McMillan M., White J., Kappler J., Marrack P., Pierres A., Pierres M., Hood L. Gene transfer of H-2 class II genes: antigen presentation by mouse fibroblast and hamster B-cell lines. Cell. 1984 Feb;36(2):319–327. doi: 10.1016/0092-8674(84)90225-3. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthew J. B., Ohlendorf D. H. Electrostatic deformation of DNA by a DNA-binding protein. J Biol Chem. 1985 May 25;260(10):5860–5862. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson H., Edlund T. Sequence-specific interactions of nuclear factors with the insulin gene enhancer. Cell. 1986 Apr 11;45(1):35–44. doi: 10.1016/0092-8674(86)90535-0. [DOI] [PubMed] [Google Scholar]
- Piette J., Kryszke M. H., Yaniv M. Specific interaction of cellular factors with the B enhancer of polyoma virus. EMBO J. 1985 Oct;4(10):2675–2685. doi: 10.1002/j.1460-2075.1985.tb03987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds W. F., Gottesfeld J. M. 5S rRNA gene transcription factor IIIA alters the helical configuration of DNA. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1862–1866. doi: 10.1073/pnas.80.7.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder K., Silver S., DeLucia A. L., Fanning E., Tegtmeyer P. An altered DNA conformation in origin region I is a determinant for the binding of SV40 large T antigen. Cell. 1986 Mar 14;44(5):719–725. doi: 10.1016/0092-8674(86)90838-x. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D. Contact points between a positive transcription factor and the Xenopus 5S RNA gene. Cell. 1982 Dec;31(2 Pt 1):395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- Sen R., Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986 Aug 29;46(5):705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- Singh H., Sen R., Baltimore D., Sharp P. A. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986 Jan 9;319(6049):154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- Smith D. R., Jackson I. J., Brown D. D. Domains of the positive transcription factor specific for the Xenopus 5S RNA gene. Cell. 1984 Jun;37(2):645–652. doi: 10.1016/0092-8674(84)90396-9. [DOI] [PubMed] [Google Scholar]
- Takeda Y., Ohlendorf D. H., Anderson W. F., Matthews B. W. DNA-binding proteins. Science. 1983 Sep 9;221(4615):1020–1026. doi: 10.1126/science.6308768. [DOI] [PubMed] [Google Scholar]
- Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986 Aug 15;46(4):567–574. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- Tyndall C., La Mantia G., Thacker C. M., Favaloro J., Kamen R. A region of the polyoma virus genome between the replication origin and late protein coding sequences is required in cis for both early gene expression and viral DNA replication. Nucleic Acids Res. 1981 Dec 11;9(23):6231–6250. doi: 10.1093/nar/9.23.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Lupton S., Kamen R. Polyomavirus enhancer contains multiple redundant sequence elements that activate both DNA replication and gene expression. Mol Cell Biol. 1985 Apr;5(4):649–658. doi: 10.1128/mcb.5.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Wildeman A. G., Zenke M., Schatz C., Wintzerith M., Grundström T., Matthes H., Takahashi K., Chambon P. Specific protein binding to the simian virus 40 enhancer in vitro. Mol Cell Biol. 1986 Jun;6(6):2098–2105. doi: 10.1128/mcb.6.6.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]
- de Villiers J., Schaffner W. A small segment of polyoma virus DNA enhances the expression of a cloned beta-globin gene over a distance of 1400 base pairs. Nucleic Acids Res. 1981 Dec 11;9(23):6251–6264. doi: 10.1093/nar/9.23.6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers J., Schaffner W., Tyndall C., Lupton S., Kamen R. Polyoma virus DNA replication requires an enhancer. Nature. 1984 Nov 15;312(5991):242–246. doi: 10.1038/312242a0. [DOI] [PubMed] [Google Scholar]