Abstract
RNA interference (RNAi) is an evolutionary conserved mechanism by which small double-stranded RNA (dsRNA) – termed small interfering RNA (siRNA) – inhibits translation or degrades complementary mRNA sequences. Identifying features and enzymatic components of the RNAi pathway have led to the design of highly-effective siRNA molecules for laboratory and therapeutic application. RNA activation (RNAa) is a newly discovered mechanism of gene induction also triggered by dsRNAs termed small activating RNA (saRNA). It offers similar benefits as RNA interference (RNAi), while representing a new method of gene overexpression. In the present study, we identify features of RNAa and explore chemical modifications to saRNAs that improve the applicability of RNAa. We evaluate the rate of RNAa activity in order to define an optimal window of gene induction, while comparing the kinetic differences between RNAa and RNAi. We identify Ago2 as a conserved enzymatic component of both RNAa and RNAi implicating that saRNA may tolerate modification based on Ago2 function. As such, we define chemical modifications to saRNAs that manipulate RNAa activity, as well as exploit their effects to design saRNAs with enhanced medicinal properties. These findings reveal functional features of RNAa that may be utilized to augment saRNA function for mechanistic studies or the development of RNAa-based drugs.
Keywords: Argonaute 2 (Ago2), cancer therapeutics, E-cadherin, gene promoter, p21, RNA activation (RNAa), RNA interference (RNAi), small activating RNA (saRNA), small interfering RNA (siRNA), strand modifications
INTRODUCTION
RNA interference (RNAi) is an evolutionally conserved mechanism of gene regulation by which small double-stranded RNA (dsRNA) molecules inhibit translation or degrade complementary mRNA sequences [1-3]. Synthetic dsRNA duplexes, termed small interfering RNAs (siRNAs), mimic naturally processed dsRNAs [e.g. microRNAs (miRNAs)] to exploit endogenous enzymatic machinery and enter the RNAi pathway.
More recently, short dsRNAs have also been shown to induce gene expression in a phenomenon referred to as RNA activation (RNAa) [4-6]. This class of dsRNA molecule – termed small activating RNAs (saRNAs) to distinguish from siRNAs – triggers an effect opposite to that of RNAi. Several models of RNAa have been described including transcriptional activation by targeting promoter-derived sequences [5-8] and/or gene antisense transcripts [9,10]. Regardless of the target, it is becoming clear that RNAa has potential to induce the expression of a variety of genes. As such, RNAa offers a promising new therapeutic strategy for diseases that can be corrected by stimulating gene expression.
Identifying features of the RNAi pathway have improved its therapeutic application and development as a laboratory tool. Early studies evaluating the rate of RNAi activity defined the optimal window for target knockdown and functional gene analysis [11,12]. Medicinal chemistry also enabled usage and identification of modifications to siRNAs that improved mechanistic analysis and pharmacological properties [13-15]. As such, it is equally important to understand the functional nuances of RNAa. In this report, we evaluate the rate of RNAa activity to define an optimal window of gene induction, as well as identify kinetic differences between RNAa and RNAi. We also identify chemical modifications to saRNAs that manipulate RNAa function and specificity. These findings reveal functional features of RNAa that may be utilized in mechanistic studies, as well as benefit its medicinal development for in vivo application.
RESULTS
The Kinetics of Gene Induction by RNAa
We have previously shown that E-cadherin and p21 are susceptible to RNAa in a variety of cells lines including PC-3 (prostate adenocarcinoma) cells [5,16]. In order to evaluate the rate of gene induction, we transfected PC-3 cells with saRNAs targeting E-cadherin (dsEcad-215) or p21 (dsP21-322) and monitored gene induction throughout a 72-hour time course. As shown in Figs. (1A-B), induction of E-cadherin and p21 expression began to emerge following 48 hours of saRNA transfection with levels continuing to increase by 72 hours. To compare the kinetics of RNAa to RNAi, we also transfected PC-3 cells with siRNA targeting the MOF (siMOF) or E2F1 (siE2F1) gene transcripts and observed the rate of mRNA knockdown. Unlike RNAa, knockdown by RNAi was observed as early as 6 hours with levels almost maximally subsiding by ~24 hours following siRNA transfection (Figs. 1C-D). MOF and E2F1 were selected as suitable targets to monitor RNAi activity based on specific and efficient knockdown by their corresponding siRNAs; MOF and E2F1 were downregulated ≥80% following 72 hours of siRNA transfection (Supplementary Fig. 1).
Fig. (1). The kinetics of RNAa and RNAi activity.
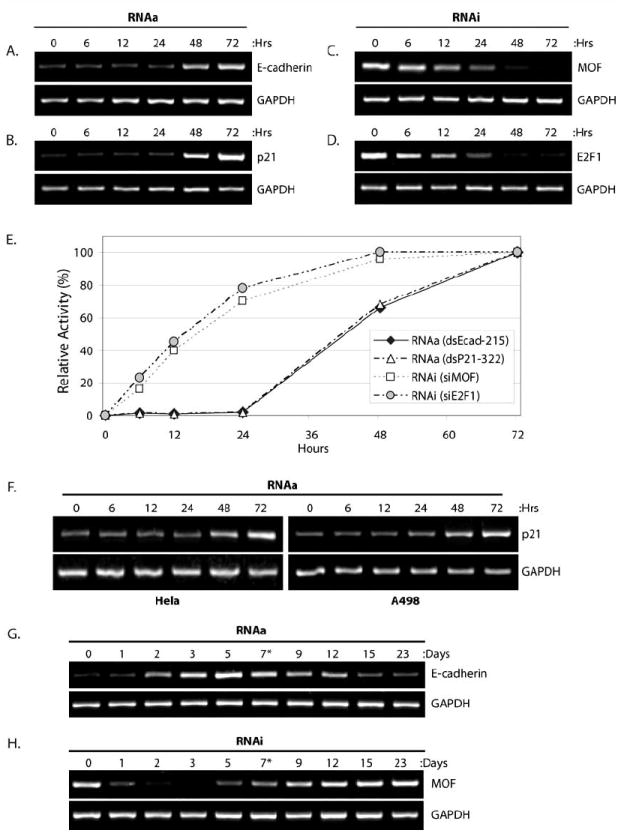
(A-B) PC-3 cells were transfected at 50 nM dsEcad-215 (A) or dsP21-322 (B) for the indicated lengths of time in order to monitor target gene induction via RNAa. Expression levels of E-cadherin (A) and p21 (B) were assessed by standard RT-PCR. GAPDH was also evaluated and served as a loading control. (C-D) PC-3 cells were transfected at 50 nM siMOF (C) or siE2F1 (D) in order to monitor RNAi at the indicated time points. Expression levels of MOF (C) and E2F1 (D) were assessed by standard RT-PCR. (E) Expression levels of E-cadherin, p21, MOF, and E2F1 following saRNA/siRNA treatments were quantified by optical densitometry at each time point. Maximal RNAa activity (100%) correlates to E-cadherin and p21 levels at 72 hours of saRNA transfection, while maximal RNAi activity (100%) correlates to MOF and E2F1 levels at 72 hours following siRNA treatments. Target expression levels at 0 hours designated no activity (0%) for both RNAa and RNAi. Rate of gene induction or target knockdown signify RNAa (dsEcad-215 and dsP21-322) and RNAi (siMOF and siE2F1) kinetics, respectively. (F) HeLa and A498 cells were transfected with dsP21-322 at the indicted time points. Expression of p21 and GAPDH were evaluated by RT-PCR. (G-H) PC-3 cells were transfected at 50 nM dsEcad-215 (G) or siMOF (H) for the indicated lengths of time. Cells were passed following day 7 as denoted by an asterisk (*). Expression levels of E-cadherin (H) or MOF (I) were assessed by standard RT-PCR. GAPDH was also amplified to serve as a loading control.
Quantifying the expression levels of each transcript following saRNA or siRNA transfection can be utilized to calculate the relative activity of RNAa and RNAi, respectively, at each individual time point to allow for the direct comparison of RNAa and RNAi kinetics in PC-3 cells. As shown in (Fig. 1E), the rate at which RNAa activity emerges is delayed by ~24-48 hours in comparison to RNAi.
We also transfected HeLa (cervix adenocarcinoma) and A498 (kidney carcinoma) cell lines with dsP21-322 and monitored p21 induction. In both cell lines, induction of p21 was detectable by ~48 hours (Fig. 1F). These results indicate that the delayed response in RNAa activity in comparison to RNAi is a general feature of RNAa and not specific to any particular cell line. Taken together, these results indicate that the emergence of RNAa activity occurs at a different rate than RNAi emerging ~48 hours after initial treatments.
To compare the duration of RNAa and RNAi activity, we transfected PC-3 cells with dsEcad-215 or siMOF for up to 23 days and monitored E-cadherin and MOF transcript levels, respectively. As shown in (Fig. 1G), induction of E-cadherin was observed at days ~2-12 with optimal levels of induction between days 3-7. Remarkably, E-cadherin induction was detectable through cell passage, which occurred after day 7. Knockdown of MOF was observed at days ~1-7 with optimal activity between days 1-3 (Fig. 1H). MOF levels quickly rebounded following passage of cells at day 7. This data indicates that the optimal window for RNAa activity is between days ~3-7 and generally lasted longer (~10 days) than RNAi activity (~7 days) in PC-3 cells.
Ago2 is Required for RNAa Activity
Argonaute (Ago) family of proteins are key regulators of RNAi that function, in part, by recruiting small dsRNAs [17-20]. Since duplex RNA is the trigger for both RNAa and RNAi, we transfected PC-3 cells with siRNAs targeting Ago1-4 (siAgo1, siAgo2, siAgo3, or siAgo4) in combination with dsEcad-215 or siMOF to compare the functional role of each Ago family member on RNAa and RNAi activity, respectively. As shown in Fig. (2A), the expression of each Ago family member was knocked down by its corresponding siRNA; however, only siAgo2 prevented E-cadherin induction. In support, we have previously reported that selective knockdown of Ago2 also prevented dsP21-322 activity [5]. Since Ago2 is the catalytic core to conventional RNAi [17], depletion of Ago2 also abolished the RNAi-mediated knockdown of MOF transcript (Fig. 2B). Taken together, this data suggests that Ago2 is a conserved factor required by both RNAa and RNAi.
Fig. (2). Knockdown of Ago2 inhibits RNAa and RNAi function.
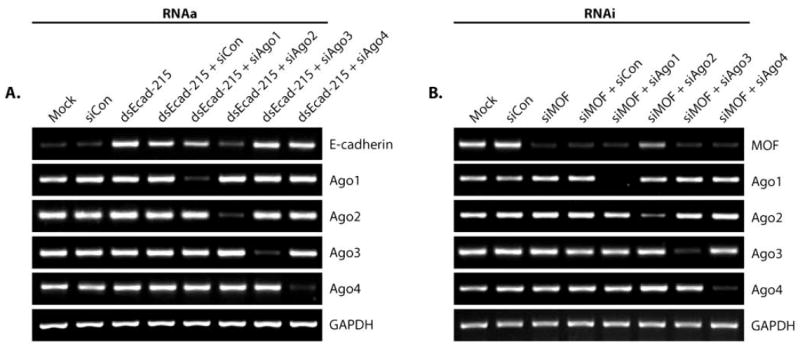
(A) PC-3 cells were transfected at 50 nM siCon or dsEcad-215 for 72 hours. Combination treatments of dsEcad-215 and Ago-specific siRNAs (siAgo1, 2, 3, or 4) were performed using 25 nM concentrations of each RNA duplex. E-cadherin, Ago1-4, and GAPDH expression levels were assessed by standard RT-PCR. GAPDH served as a loading control. (B) PC-3 cells were transfected at 50 nM siCon or siMOF for 72 hours. Co-treatments of siMOF with siAgo1-4 were performed at 25 nM each siRNA. mRNA expression levels were assessed by standard RT-PCR.
Strand Modifications to Manipulate RNAa Activity
Modification to the 5’-terminus of the guide strand in siRNA duplexes is known to interfere with Ago2 function and abolish RNAi activity [21,22]. Since Ago2 is also required for RNAa, we decided to test if blocking the 5’-termini in saRNA modulates RNAa activity. We synthesized modified saRNA molecules derived from dsEcad-215 and dsP21-322 that were covalently linked to biotin at either the 5’-end of the antisense (dsRNA-AS-5’Bio) or sense (dsRNA-S-5’Bio) strand (Fig. 3A). As shown in Fig. (3B), transfection of dsEcad-215 modified at its 5’-end of the antisense strand (dsEcad-215-AS-5’Bio) completely blocked induction of E-cadherin, while 5’-modification to the sense strand (dsEcad-215-S-5’Bio) retained RNAa activity. Likewise, dsP21-322-AS-5’Bio abolished activation of p21, while dsP21-322-S-5’Bio induced p21 levels equivalent to unmodified dsP21-322 (Fig. 3C).
Fig. (3). Strand modifications that manipulate RNAa activity.
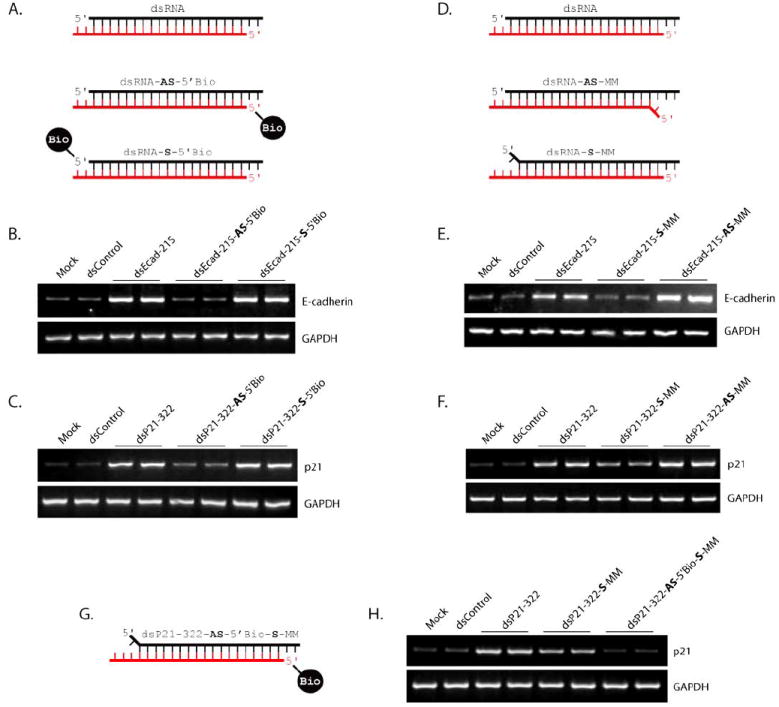
(A) Schematic representation of modified saRNA covalently linked to biotin at the 5’-end of the antisense (dsRNA-AS-5’Bio) or sense (dsRNA-S-5’Bio) strand. Unmodified saRNA (dsRNA) is also depicted. The antisense strand in each duplex is red, while the sense strand is in black. (B-C) PC-3 cells were transfected at 50 nM concentrations of the indicated saRNAs for 72 hours. Mock samples were transfected in the absence of saRNA. Expression levels of E-cadherin (B) or p21 (C) were assessed by standard RT-PCR. GAPDH levels were also evaluated to serve as loading controls. (D) Schematic representation of modified saRNA possessing a mismatched base opposite the 5’ most nucleotide of either the antisense (dsRNA-AS-MM) or sense (dsRNA-S-MM) strand. (E-F) PC-3 cells were transfected with the indicated saRNAs for 72 hours. Expression levels of GAPDH and E-cadherin (E) or p21 (F) were assessed by standard RT-PCR. (G) Schematic depiction of dsP21-322-AS-5’Bio-S-MM. (H) PC-3 cells were transfected with the indicated saRNAs for 72 hours. Expression of p21 and GAPDH were evaluated by standard RT-PCR.
Selection of the guide strand in RNAi is determined by the terminal thermodynamic characteristics within siRNA molecules. The strand with lower thermodynamic stability at its 5’-end is preferentially loaded into Ago2 to become the guide strand [23]. Given that RNAa is also dependent on Ago2, we synthesized modified saRNA molecules that possessed mismatched bases opposite the 5’ most nucleotide of either the antisense (dsRNA-AS-MM) or sense (dsRNA-S-MM) strand (Fig. 3D). Presumably, the intentional mismatch would lower the thermodynamic stability of the 5’-terminus at either strand and forcibly select it as the guide strand. As shown in Fig. (3E), transfection of dsEcad-215 with a mismatch at the 5’-end of the antisense strand (dsEcad-215-AS-MM) resulted in robust activity, while the 5’ mismatch of the sense strand (dsEcad-215-S-MM) inhibited E-cadherin induction. Similarly, dsP21-322-AS-MM enhanced p21 expression, while dsP21-322-S-MM sequestered RNAa activity (Fig. 3F). It is also important to note that mismatches at the 5’-end of the antisense strand of both dsEcad-215 (dsEcad-215-AS-MM) and dsP21-322 (dsP21-322-AS-MM) further enhanced RNA activity (Fig. 3E-F).
Interestingly, dsP21-322-S-MM possessed some residual RNAa function (Fig. 3F). To determine if the remaining RNAa activity may have resulted from the sense strand, we synthesized a modified dsP21-322 molecule that possessed both a mismatch at the 5’-end of the sense strand and a biotin modification at the 5’-terminus of the antisense strand to forcibly load the sense strand and block any residual activity of the antisense strand, respectively (Fig. 3G). As shown in Fig. (3H), transfection of the modified dsP21-322 molecule (dsP21-322-AS-5’Bio-S-MM) completely suppressed p21 gene activation. Utilizing both modifications in combination clearly defined strand function in dsP21-322; the antisense strand of dsP21-322 is responsible for RNAa activity. As such, the residual activity of dsP21-322-S-MM most likely resulted from the occasional selection of the antisense strand to guide p21 activation.
Modifying the 2’-backbone in siRNA molecules is known to increase endonuclease resistance and abrogate immune stimulation [15,24,25]. In order to determine the tolerance of saRNA molecules to 2’-backbone modification, we designed two dsP21-322 variants that contained 2’-O-methyl (2’-OMe) modifications in either the sense (dsP21-322-S-2’OMe) or antisense (dsP21-322-AS-2’OMe) strand. As shown in Fig. (4A-B), 2’-OMe modification within the sense strand suppressed RNAa activity of dsP21-322; p21 induction by dsP21-322-S-2’OMe was ~50% less than dsP21-322 or dsP21-322-AS-2’OMe in both PC-3 and HeLa cells. This data suggests that excessive modification to the 2’-backbone of the passenger strand in saRNA molecules may interfere with RNAa activity.
Fig. (4). Modification to the 2’-backbone and 3’-termini in saRNA.
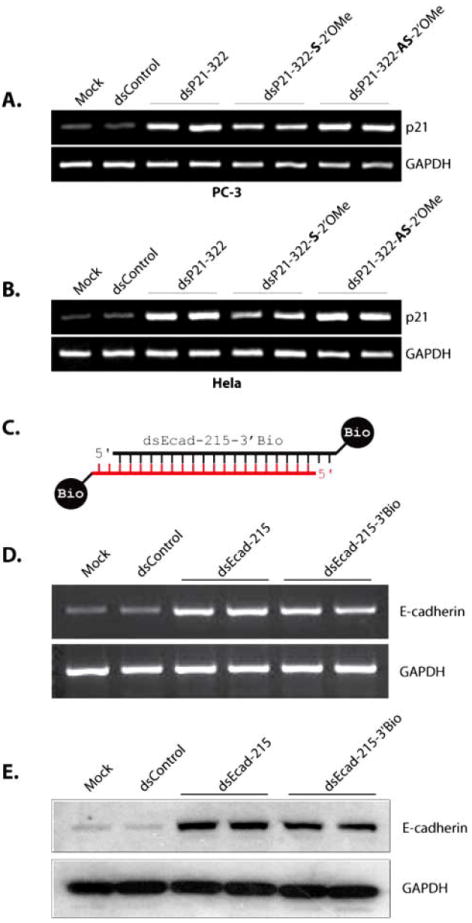
(A-B) PC-3 (A) and HeLa (B) cells were transfected with 50 nM concentrations of dsControl, dsP21-322, dsP21-322-S-2’OMe, or dsP21-322-AS-2’OMe for 72 hours. Mock samples were transfected in the absence of saRNA. Expression of p21 and GAPDH were evaluated by standard RT-PCR. (C) Schematic representation of dsEcad-215-3’Bio possessing biotin covalently linked to both 3’-termini. The antisense strand is in red, while the sense strand is black. (D) PC-3 cells were transfected at 50 nM dsControl, dsEcad-215, or dsEcad-215-3’Bio for 72 hours. Expression of E-cadherin and GAPDH mRNA levels were evaluated by standard RT-PCR. (E) Induction of E-cadherin protein was confirmed by immunoblot analysis. GAPDH was also detected and served as a loading control.
To determine if modification of the 3’-termini within saRNA interferes with RNAa activity, we synthesized dsE-cad-215 with biotin linked to the 3’-end of both the sense and antisense strands (dsEcad-215-3’Bio; Fig. 4C). As shown in Figs. (4D-E), transfection of dsEcad-215-3’Bio still induced the expression of E-cadherin. This data indicates that saRNA modified at the 3’-termini retain RNAa activity.
Exploiting saRNA Modifications to Optimize RNAa Function
Improper selection of the passenger strand in saRNA duplexes may lead to off-target effects by interacting with non-specific transcripts or gene promoters complementary to the passenger strand. In order to optimize RNAa activity and suppress the off-target effects of the passenger strand, we designed and synthesized dsEcad-215 possessing a blocked 5’-terminus on the sense strand and a mismatched base opposite the 5’ most nucleotide of the antisense strand (dsEcad-215-S-5’Bio-AS-MM) to suppress sense strand activity and enhance selection of the antisense strand, respectively (Fig. 5A). Transfection of dsEcad-215-S-5’Bio-AS-MM readily induced E-cadherin expression with increased activity similar to dsEcad-215-AS-MM, while dsEcad-215-S-5’Bio matched levels achieved by unmodified dsEcad-215 (Figs. 5B-C). In order to validate reduced off-target effects caused by selection of the sense strand, we cloned a target site complementary to the sense strand into a luciferase reporter vector (pOffTar-luc) and quantified luciferase activity in the presence of several modified dsEcad-215 molecules. As shown in Fig. (5D), dsEcad-215 resulted in reduced luciferase activity of pOffTar-luc, while the activity of a non-specific luciferase reporter construct (pNonSpec-luc) was not altered by saRNA treatment. dsEcad-215-S-MM served as a positive control since the mismatched base present in the duplex would forcibly enhance sense strand selection and subsequent off-target activity. The reduction in pOffTar-luc activity by dsEcad-215 and dsEcad-215-S-MM confirms the off-target potential of the sense strand. However, modified saRNAs (dsEcad-S-5’Bio, dsEcad-215-AS-MM, and dsE-cad-215-S-5’Bio-AS-MM) caused luciferase activity of pOffTar-luc to rebound demonstrating inhibition of off-target function (Fig. 5E). Overall, dsEcad-215-S-5’Bio-AS-MM possessed both enhanced RNAa activity toward E-cadherin expression and inhibition of non-specific function of the sense strand. This data indicates that functional modifications to saRNA molecules can be utilized to both enhance RNAa activity and reduce non-specific off-target effects.
Fig. (5). Utilizing strand modifications to improve RNAa function.
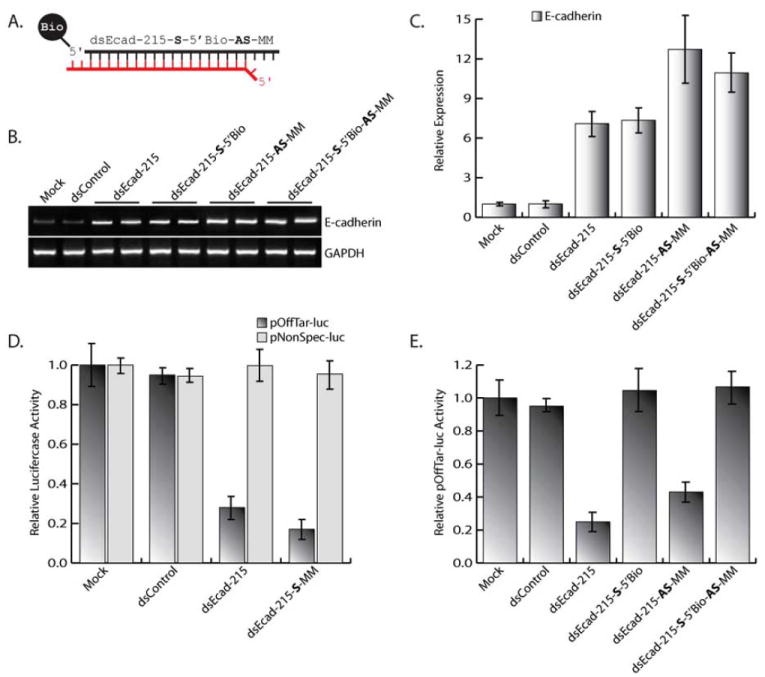
(A) Depiction of dsEcad-215-S-5’Bio-AS-MM possessing biotin covalently linked to the 5’-terminus of the sense strand and a mismatched base opposite the 5’ most nucleotide of the antisense strand. The antisense strand is in red, while the sense strand is black. (B) PC-3 cells were transfected with 50 nM concentrations of the indicated E-cadherin saRNAs for 72 hours. Expression levels of E-cadherin and GAPDH were assessed by standard RT-PCR. GAPDH served as a loading control. (C) Relative expression of E-cadherin was determined by real-time PCR (mean ± standard error from three independent experiments). Values of E-cadherin were normalized to GAPDH. The level of E-cadherin induction signifies the RNAa activity generated by each saRNA molecule. (D) Relative luciferase activity of pOffTar-luc or pNonSpec-luc in PC-3 cells transfected with mock, dsControl, dsEcad-215, or dsEcad-215-S-MM. Treatment with dsEcad-215-S-MM served as a positive control for targeted reduction of pOffTar-luc activity. (E) Luciferase activity of pOffTar-luc in PC-3 cells transfected with the indicated saRNA molecules. Off-target activity was validated by a reduction in pOffTar-luc luciferase activity.
DISCUSSION
The optimal window of RNAa activity was delayed by ~24-48 hours in comparison to RNAi. Perhaps, the delay in RNAa activity reflects a more complicated mechanism with additional rate-limiting steps. In nematode, a special ribonucleoprotein is required to shuttle small siRNAs into nuclei in order to facilitate nuclear RNAi [26]. Although this protein is not conserved in humans, cytoplasmic miRNA has been shown to actively migrate into the nuclear fraction of living human cells [27]. Because RNAa is a nuclear process acting on gene transcription, acquiring access to the nucleus may serve as an additional rate-limiting step for RNAa. Changes in chromatin structure are also associated with RNAa [4,5,28], which may further contribute to the delayed kinetics. Regardless, identifying the delay and defining the optimal window of RNAa activity allows for proper assessment for gene induction and functional analysis of saRNAs. Assessing the rate of RNAa activity in cell culture also gives insight into the anticipated in vivo pharmacological properties of RNAa. For instance, RNAa-based drugs may require several days before target gene induction or beneficial changes in phenotype are evident. Moreover, the longer-lasting effect of RNAa may result in less frequent administration of saRNA; a potential benefit as duplex RNA in excess can have toxic consequences [29].
Identifying features and key factors involved in the RNAa pathway can influence saRNA design. As such, defining Ago2 as an important mechanistic component implicated that chemically-modified saRNAs may function to manipulate RNAa activity in a manner similar to RNAi [21,22]. Utilizing dsP21-322 and dsEcad-215 as functional examples of saRNA molecules revealed that blocking the 5’-end or incorporating intentional mismatches can determine strand function. Studies have revealed an abundance of sense and antisense transcription within the promoters and flanking regions of active genes [30-32]. Furthermore, overlapping noncoding RNAs and upstream cryptic transcripts have been shown to play substantial roles in regulating gene expression [33-37]. As such, models for RNAa have included saRNAs targeting antisense transcripts and/or promoter-derived sequences to facilitate gene activation [5-10]. RNAs transcribed in sense and antisense orientations have already been shown to serve as docking sites for transcriptional gene silencing (TGS) mediated by small duplex RNAs [38-40]. Likewise, nascent sense and antisense transcripts may both serve as the targets for RNAa, as well. Utilizing modified saRNAs can not only improve mechanistic studies by defining strand activity, but also assist in determining orientation of such putative target transcripts.
Identifying functional modifications is also necessary for therapeutic development in order to improve the medicinal properties of saRNAs. In the case dsP21-322 and/or dsEcad-215 (i) blocking the 5’-end of the sense strand completely inhibited its potential off-target effects; (ii) incorporating an intentional mismatch opposite the 5’ most nucleotide in the antisense strand enhanced target gene induction, as well as reduced the off-target activity generated by the sense strand; (iii) 2’Ome modification to the sense strand inhibited RNAa activity, while the same modification to the antisense strand did not interfere with gene induction; (iv) modifying the 3’-end of either the sense or antisense strand had minimal effects on RNAa activity. Although the preferred guide strand may vary between the sense or antisense strand in different saRNAs, each modification may still be applied to manipulate saRNA activity or define strand function. As such, extrapolating these modifications to fit other saRNAs based on strand activity will also improve their medicinal properties.
Development of saRNAs for therapeutic application may also require multiple modifications to optimize medicinal benefits. For instance, we were able to enhance dsEcad-215 activity by blocking both the 5’-end of the sense strand and incorporating a mismatch opposite the 5’ most nucleotide of the antisense strand. The combination of both modifications alleviated any potential off-target effects that would arise from improper use of the sense strand and enhanced gene induction; features needed to manipulate in order to develop RNAa therapeutics. Modification to the sense and antisense backbones (i.e. 2’-OMe, 2’-flouro, etc.) in saRNA duplexes may also improve therapeutic application by increasing endonuclease resistance and serum stability, much as they are utilized to stabilize siRNAs, as long the passenger strand is devoid of inhibitory modifications. Tethering small molecules (i.e. cholesterol) to the 3’-ends of saRNAs could also be used to improve systemic delivery of RNAa-based drugs. Conjugation of other compounds (i.e. flurogenic labels) to the 3’-termini may be effective at providing visual confirmation of saRNA uptake into target cells or tissue, as well.
RNAi is rapidly developing into a promising new approach for combating disease at the genetic level; however, it can only provide antagonism of specific molecular targets. By utilizing saRNAs as therapeutic compounds, RNAa offers similar benefits as RNAi, while facilitating the exact opposite response – gene activation. This approach addresses a missing void in RNA-based gene therapies and offers a novel solution to provide greater efficacy in disease control. RNAa has already been shown to activate genes capable of suppressing cancer cell growth (e.g. p21, E-cadherin, p53, NKX3.1, ect.), triggering angiogenesis (e.g. VEGF), or influencing stem cell maintenance (e.g. CXCR4) [5,16,28,41]. As such, the ability to selectively up-regulate genes acting against a disease state can have far-reaching impacts in almost every therapeutic realm. However, application of RNAa is not limited to only cancer therapeutics. RNAa also has potential to function as a surrogate tool for vector-based gene overexpression systems. RNAa offers a new approach to enhance endogenous gene expression that may be manipulated to target a variety of genes. As momentum within the biological sciences increases, RNAa may become an important technique to augment gene expression for therapeutics and functional gene studies.
MATERIALS AND METHODS
Cell culture and dsRNA transfection
PC-3, HeLa, and A498 cells were maintained in RPMI 1640 medium supplemented with 10% FBS, L-glutamine (2 mM), penicillin (100 U/ml) and streptomycin (100 μg/ml) in a humidified atmosphere of 5% CO2 at 37°C. The day before transfection, cells were plated in growth medium without antibiotics at a density of ~50-60%. Transfection of saRNA and/or siRNA was carried out using Lipofectamine RNAiMax (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. All siRNAs, unmodified saRNAs, biotin-linked duplexes, and mismatched derivates were synthesized by Invitrogen. The 2’-OMe-modified saRNA molecules were synthesized by Dharmacon (Lafayette, CO). Sequences are listed in Supplementary Table 1.
Analysis and quantification of mRNA expression
Total RNA was extracted using the RNeasy RNA isolation kit (Qiagen, Velencia, CA) according to the manufacturer’s protocol. One microgram of total RNA was reverse transcribed using the Reverse Transcription System (Promega, Madison, WI) with oligo(dT) primers. The resulting cDNA samples were amplified by PCR using primers specific for E-cadherin, p21, MOF, E2F1, Ago1-4, or GAPDH (Supplementary Table 1) and visualized on an agarose gel. Optical densitometry was utilized to quantify relative abundance of each gene transcript in order to evaluate RNAa or RNAi kinetics. GAPDH served as an endogenous control used to normalize data. RNAa activity of the E-cadherin saRNA molecules and efficiency of the MOF and E2F1 siRNAs was quantified by real-time PCR. Gene-specific TaqMan© assay kits (Applied Biosystems, Foster City, CA) for E-cadherin, MOF, E2F1, and GAPDH were used in conjunction with the 7300 Real-Time System (Applied Biosystems) to measure relative transcript levels. Each sample was analyzed in quadruplicate and GAPDH levels were utilized to normalize data. Relative expression and standard error were calculated by the supplied 7300 Real-Time System software.
Immunoblotting
Cultured cells were washed with cold phosphate buffered saline (PBS) buffer and lysed with M-PER protein extraction buffer (Pierce, Rockford, IL) containing protease inhibitors. Cell lysates were centrifuged and supernatants were collected. Equal quantities of protein were resolved by electophoresis on sodium dodecyl sulfate (SDS) polyacrlamide gels and transferred to 0.45 μm nitrocellulose membranes by voltage gradient. The resulting blots were blocked with 5% non-fat dry milk and probed with primary antibodies specific to E-cadherin (Zymed, South San Francisco, CA) or GAPDH (Chemicon, Temecula, CA). Immunodetection occurred by incubating blots with appropriate secondary HRP-linked antibodies and utilizing chemiluminescence to visualize the antigen-antibody complexes. GAPDH served as an internal control.
Analysis of off-target activity
A target site complementary to the sense strand of dsEcad-215 (pOffTar) was cloned into the 3’UTR of the pMIR-Report luciferase reporter vector (Ambion, Foster City, CA) in order quantify the off-target activity of dsEcad-215 and its modified variants (e.g. dsEcad-215-S-5’Bio-AS-MM, etc.). A non-specific site (pNonSpec) was also cloned to serve as a control for specificity. All oligonucleotide sequences used to create the 3’UTR constructs are listed in Supplementary Table 1. PC-3 cells were transfected with 0.6 μg pOffTar or pNonSpec, 0.4 μg pMIR-Report Beta-gal, and 30 nM dsRNA for 24 hours. The pMIR-Report Beta-gal vector served as a control to monitor transfection efficiency. The Dual-Light System® chemiluminescent reporter gene assay (Applied Biosystems) was used to quantify luciferase and β-galactosidase activity. Off-target activity was confirmed by a reduction in luciferase activity.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (1R01GM090293-0109), National Institutes of Health/National Cancer Institute, University of California, San Francisco SPORE Special Program of Research Excellence (P50CA89520), and Department of Defense (W81XWH-08-1-0260).
Footnotes
SUPPORTIVE/SUPPLEMENTARY MATERIAL
Supportive/Supplementary material is available on the publishers Web site along with the published article.
References
- 1.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci USA. 2003;100(17):9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3(3):166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 5.Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci USA. 2006;103(46):17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA. 2008;105(5):1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116(6):779–793. doi: 10.1016/s0092-8674(04)00248-x. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454(7200):126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4(11):e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, Janowski BA. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15(8):842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi Y, Yamaoka K, Nishikawa M, Takakura Y. Moment analysis for kinetics of gene silencing by RNA interference. Biotechnol Bioeng. 2006;93(4):816–819. doi: 10.1002/bit.20718. [DOI] [PubMed] [Google Scholar]
- 12.Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci. 2001;114(Pt 24):4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- 13.Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr Opin Chem Biol. 2004;8(6):570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 14.de Fougerolles A, Manoharan M, Meyers R, Vornlocher HP. RNA interference in vivo: toward synthetic small inhibitory RNA-based therapeutics. Methods Enzymol. 2005;392:278–296. doi: 10.1016/S0076-6879(04)92016-2. [DOI] [PubMed] [Google Scholar]
- 15.Layzer JM, McCaffrey AP, Tanner AK, Huang Z, Kay MA, Sullenger BA. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10(5):766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z, Place RF, Jia ZJ, Pookot D, Dahiya R, Li LC. Antitumor effect of dsRNA-induced p21(WAF1/CIP1) gene activation in human bladder cancer cells. Mol Cancer Ther. 2008;7(3):698–703. doi: 10.1158/1535-7163.MCT-07-2312. [DOI] [PubMed] [Google Scholar]
- 17.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15(2):185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131(6):1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 19.Hock J, Meister G. The Argonaute protein family. Genome Biol. 2008;9(2):210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23(3):304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiu YL, Rana TM. RNAi in human cells: basic structural and functional features of small interfering RNA. Mol Cell. 2002;10(3):549–61. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- 22.Chen PY, Weinmann L, Gaidatzis D, Pei Y, Zavolan M, Tuschl T, Meister G. Strand-specific 5’-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA. 2008;14(2):263–74. doi: 10.1261/rna.789808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115(2):209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 24.Czauderna F, Fechtner M, Dames S, Aygun H, Klippel A, Pronk GJ, Giese K, Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31(11):2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sioud M, Furset G, Cekaite L. Suppression of immunostimulatory siRNA-driven innate immune activation by 2’-modified RNAs. Biochem Biophys Res Commun. 2007;361(1):122–126. doi: 10.1016/j.bbrc.2007.06.177. [DOI] [PubMed] [Google Scholar]
- 26.Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An Argonaute transports siR-NAs from the cytoplasm to the nucleus. Science. 2008;321(5888):537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foldes-Papp Z, Konig K, Studier H, Buckle R, Breunig HG, Uchugonova A, Kostner GM. Trafficking of mature miRNA-122 into the nucleus of live liver cells. Curr Pharm Biotechnol. 2009;10(6):569–578. doi: 10.2174/138920109789069332. [DOI] [PubMed] [Google Scholar]
- 28.Turunen MP, Lehtola T, Heinonen SE, Assefa GS, Korpisalo P, Girnary R, Glass CK, Vaisanen S, Yla-Herttuala S. Efficient regulation of VEGF expression by promoter-targeted lentiviral shRNAs based on epigenetic mechanism: a novel example of epigenetherapy. Circ Res. 2009;105(6):604–609. doi: 10.1161/CIRCRESAHA.109.200774. [DOI] [PubMed] [Google Scholar]
- 29.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441(7092):537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 30.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322(5909):1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322(5909):1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322(5909):1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 33.Goodrich JA, Kugel JF. From bacteria to humans, chromatin to elongation, and activation to repression: The expanding roles of noncoding RNAs in regulating transcription. Crit Rev Biochem Mol Biol. 2009;44(1):3–15. doi: 10.1080/10409230802593995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322(5909):1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C. Antisense transcription in the mammalian transcriptome. Science. 2005;309(5740):1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 36.Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127(6):1209–12021. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429(6991):571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 38.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci USA. 2007;104(30):12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahmoudi S, Henriksson S, Corcoran M, Mendez-Vidal C, Wiman G, Farnebo M. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol Cell. 2009;33(4):462–471. doi: 10.1016/j.molcel.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 40.Gonzalez S, Pisano DG, Serrano M. Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs. Cell Cycle. 2008;7(16):2601–2608. doi: 10.4161/cc.7.16.6541. [DOI] [PubMed] [Google Scholar]
- 41.Huang V, Qin Y, Wang J, Wang X, Place RF, Lin G, Lue TF, Li LC. RNAa is conserved in mammalian cells. PLoS One. 5(1):e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


