Abstract
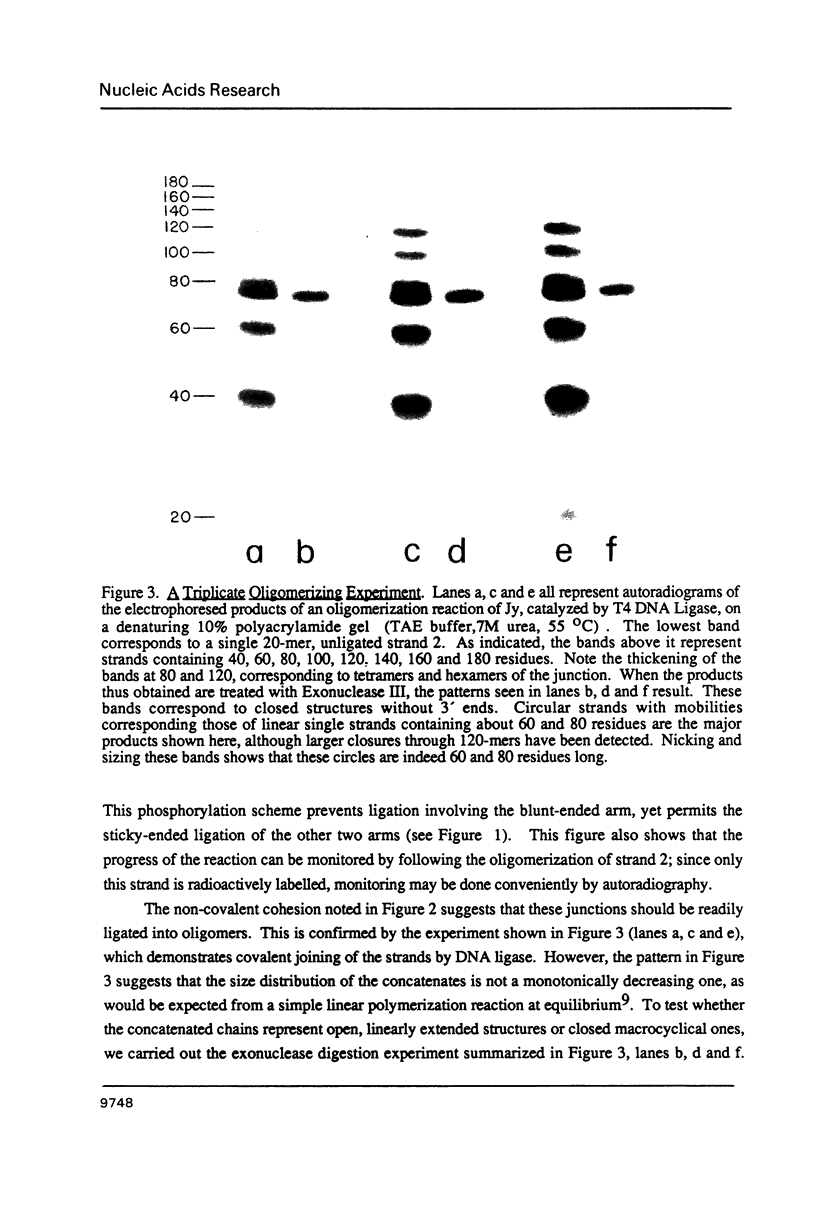
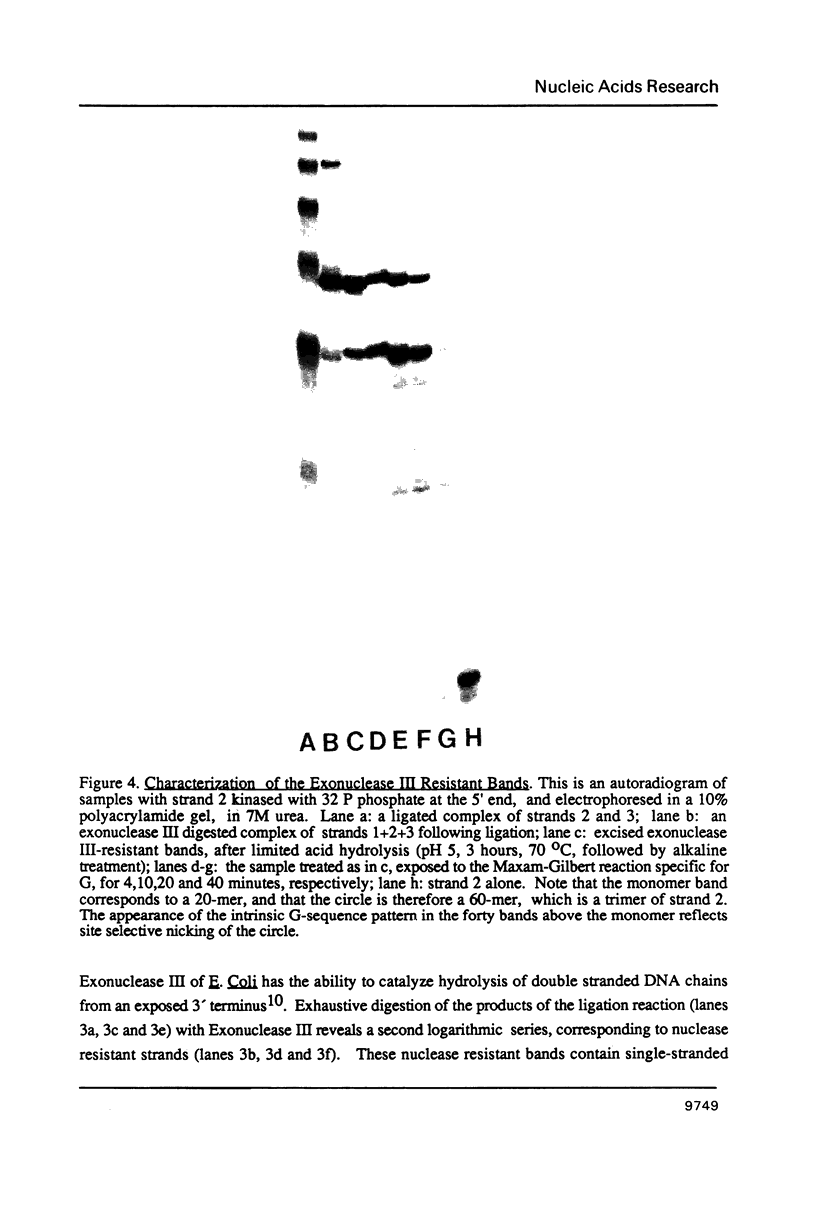
Nucleic acid junctions are stable analogs of branched DNA structures which occur transiently in living systems. We show here that junctions which contain three double helical arms can be enzymatically oligomerized, using conventional sticky-ended ligation procedures, to create larger complexes. The products consist of a series of linked junctions separated by 20 base pairs. Junction dimers are formed that have free termini only, whereas trimers and larger species are found to be both unclosed and cyclized. The formation of a series of macrocyclic products which, surprisingly, begins with trimers and tetramers indicates that this junction is flexible about a bending axis, and perhaps twist-wise as well. We have obtained the same results from three different 3-arm junctions, two in which the junction is flanked by a 3 Watson-Crick base pairs, and one in which a G-G base pair flanks the junction.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Kallenbach N. R., Ma R. I., Wand A. J., Veeneman G. H., van Boom J. H., Seeman N. C. Fourth rank immobile nucleic acid junctions. J Biomol Struct Dyn. 1983 Oct;1(1):159–168. doi: 10.1080/07391102.1983.10507432. [DOI] [PubMed] [Google Scholar]
- Shore D., Baldwin R. L. Energetics of DNA twisting. II. Topoisomer analysis. J Mol Biol. 1983 Nov 15;170(4):983–1007. doi: 10.1016/s0022-2836(83)80199-5. [DOI] [PubMed] [Google Scholar]
- Shore D., Langowski J., Baldwin R. L. DNA flexibility studied by covalent closure of short fragments into circles. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4833–4837. doi: 10.1073/pnas.78.8.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]