Abstract
Background: Various processes such as annotation and filtering of variants or comparison of variants in different genomes are required in whole-genome or exome analysis pipelines. However, processing different databases and searching among millions of genomic loci is not trivial.
Results: gSearch compares sequence variants in the Genome Variation Format (GVF) or Variant Call Format (VCF) with a pre-compiled annotation or with variants in other genomes. Its search algorithms are subsequently optimized and implemented in a multi-threaded manner. The proposed method is not a stand-alone annotation tool with its own reference databases. Rather, it is a search utility that readily accepts public or user-prepared reference files in various formats including GVF, Generic Feature Format version 3 (GFF3), Gene Transfer Format (GTF), VCF and Browser Extensible Data (BED) format. Compared to existing tools such as ANNOVAR, gSearch runs more than 10 times faster. For example, it is capable of annotating 52.8 million variants with allele frequencies in 6 min.
Availability: gSearch is available at http://ml.ssu.ac.kr/gSearch. It can be used as an independent search tool or can easily be integrated to existing pipelines through various programming environments such as Perl, Ruby and Python.
Contacts:
1 INTRODUCTION
Recent technological advances in next-generation sequencing have made it possible to sequence whole genomes at unprecedented speeds and low costs (Shendure and Ji, 2008). One of the crucial goals of whole-genome sequencing (WGS) is to seek a small number of variants related to a phenotype. To this end, millions of variants are heuristically filtered according to allele frequencies, conservation, gene models and predicted impact on protein function as described in Stitziel et al. (2011). There exist several tools for such analysis (San Lucas et al., 2012; Wang et al., 2010). Additionally, selected variants can be analyzed further or visualized for biological or clinical insights (Teer et al., 2012; Yandell et al., 2011). Even though many annotation and filtering methods for WGS have been proposed, challenges still remain, such as processing sequence variants from many genomes and working with customized reference data. Current methods require a significant amount of time for processing one genome and are hardly scalable to thousands of genomes. Additionally, many tools adopt their own data formats and the use of customized annotation data is supported in a limited way.
We address these issues by providing a search tool that enables flexible annotation and filtering of WGS data from an individual in seconds on normal desktop computers. To this end, we focus on locus comparison, as it is the root functionality of any annotation or filtering tool. Furthermore, locus comparison can be utilized for other tasks, e.g. finding de novo mutation from a family dataset. We developed efficient algorithms for searching variants against a large dataset and implemented multi-threaded versions of them using the C language. It should be noted that our tool is not bound to a specific set of annotation databases. Instead, it facilitates the use of custom and public reference files from UCSC, NCBI and Ensembl by supporting a wide range of formats for genome annotation.
2 METHODS
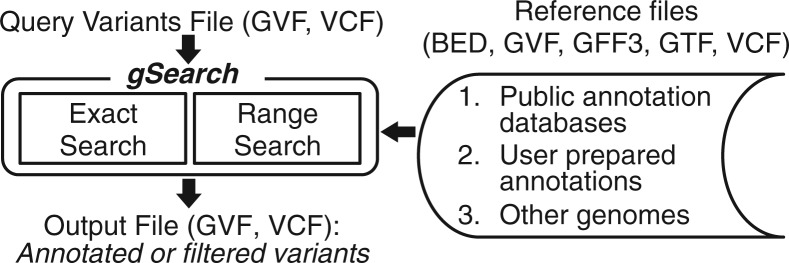
The proposed tool, gSearch, identifies matching or overlapping in two input files based on genomic region. We define a genomic region as a specific DNA segment, represented by its chromosome and start/end positions on that chromosome. A variant is a specific example of a genomic region with additional sequence information, i.e. reference and variant sequences. The basic gSearch workflow is as follows (refer to Fig. 1 for a schematic overview): a query file consisting of variants in a genome is obtained and compared with a reference file drawn from public annotation databases, user-prepared annotations or variants in other genomes. Query files for gSearch should be in Genome Variation Format (GVF) (Reese et al., 2010), which is an extension of the widely used Generic Feature Format version 3 (GFF3) standard for describing genome data or Variant Call Format (VCF), which was developed for the 1000 Genomes Project. For reference files, GVF, GFF3, Gene Transfer Format (GTF), VCF and Browser Extensible Data (BED) format are supported. Users can also specify a tab-delimited reference file having the following seven fields: chromosome, start position, end position, reference sequence (default =.) , variant sequence (default =.) , annotation (default =.) and numerical value (default = 0).
Fig. 1.
Schematic overview of gSearch
Two search modes are provided in gSearch: exact search and range search. In the exact search mode, gSearch finds reference variants on the same genomic region with the same variant sequence as a query variant. This function is useful when annotating query variants based on a previously reported variant database such as dbSNP and the 1000 Genomes Project. In the range search mode, gSearch finds annotations in a region of a reference file that overlaps a region of a query file. In this mode, sequence information is not considered for search. This function can also be utilized for various purposes such as transcription factor binding site (TFBS) annotation and microRNA host gene annotation.
gSearch provides diverse search options for effective variant annotation and filtering or comparison of variants in different genomes. The usage of gSearch is as follows:
gsearch [mandatory arguments] [optional arguments]
[mandatory arguments]
-i input file
-r reference file
-o output file
-m search mode (exact or range search)
-t annotation type
-a report annotation
-l report overlap
-v report average value
-s separate matched and unmatched variants
[optional arguments]
-if input file type (default = GVF)
-rf reference file type (default = GVF)
-tg annotation tag (default value = tag)
-nt number of threads (default value = 1)
-b big data
-h show help
We implemented the classic binary search algorithm for exact search. The computational complexity of this algorithm is bounded by 0(m · logn), where m is the number of query variants and n is the number of entries in a reference file. For the range search, we developed a novel algorithm. Let us assume that there are m query and n reference regions on the same chromosome. Two genomic regions, [qstart, qend] and [rstart, rend], overlap if and only if qstart ≤ rend and rstart ≤ qend. Because any reference genomic region [rstart, rend] is possible to satisfy the above inequalities for a query region [qstart, qend] regardless of the order based on rstart and rend, all reference regions should be checked against the query region. Thus, the time complexity of this operation for all query regions is bounded by 0(m · n). A general solution for reducing such complexity is to rely on special data structures such as k-dimensional trees. With gSearch, however, this is not necessary as the length of genomic variants has a maximum size for each chromosome. Fast performance with gSearch's range search algorithm is achieved by selecting a small number of reference regions that potentially overlap with a query region based on its length and the length of the longest reference region, as well as by utilizing multi-threading.
3 DISCUSSION
We compared the performance and accuracy of our method with the region-based and filter-based functionalities of ANNOVAR (version March 8, 2012) using the dataset of 52 million variants from the Known VARiants (KAVIAR) (Glusman et al., 2011). The KAVIAR files (hg18 and hg19) were converted to GVF format for gSearch and to the standard input format for ANNOVAR. Each query file (hg18 or hg19) contained ∼ 52 million variants. All analyses were performed on a desktop PC equipped with an Intel Core i5 processor (4 cores at 2.66 GHz) and 4 GB of RAM. To compare exact search, the hg19 Kaviar variant file was annotated with allele frequencies from the 1000 Genomes Project (ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/release/20110521/) that covered over 39 millions of loci. gSearch was run in its exact search mode and ANNOVAR was run in its filter-based mode. gSearch completed the annotation process in ∼ 347 s while ANNOVAR took ∼ 4001 s. The annotation results from both tools were the same. To compare range search, the hg 18 Kaviar variant file was annotated with their overlap with 3.8 million TFBSs prepared with the tfbsConsSites table from the UCSC Table browser. ANNOVAR was run in its region-based mode and gSearch was run in its range search mode. It took 148 s for gSearch to annotate the query file, whereas ANNOVAR took 1276 s. Two methods produced exactly the same result.
Although a majority of previously published WGS analysis tools include novel data formats in their pipelines, our proposed method can be used with diverse annotation resources in standard genome annotation file formats. With notable improvements in search speed and flexibility for multiple annotation file formats, gSearch can be used as a stand-alone genomic search tool or integrated into existing analysis pipelines. Using gSearch, users can expeditiously annotate and filter variants in many genomes with various resources on their desktop computers.
Acknowledgments
Funding: S.W.K. and M.H. are supported, in part, by NIH/NHGRI U01HG006500. S.W.K. and K.L. are supported by NIH/NIMH P50MH094267. S.W.K. is supported, in part, by NIH/NIMH R01MH085143. K.-B.H. is supported by the Soongsil University Research Fund. T.S. and K.-B.H. are supported, in part, by the Proteogenomic Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science, and Technology.
Conflict of Interest: none declared.
REFERENCES
- Glusman G., et al. KAVIAR: an accessible system for testing SNV novelty. Bioinformatics. 2011;27:3216–3217. doi: 10.1093/bioinformatics/btr540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese M.G., et al. A standard variation file format for human genome sequences. Genome Biol. 2010;11:R88. doi: 10.1186/gb-2010-11-8-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Lucas F.A., et al. Integrated annotation and analysis of genetic variants from next-generation sequencing studies with variant tools. Bioinformatics. 2012;28:421–422. doi: 10.1093/bioinformatics/btr667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J., Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Stitziel N.O., et al. Computational and statistical approaches to analyzing variants identified by exome sequencing. Genome Biol. 2011;12:227. doi: 10.1186/gb-2011-12-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teer J.K., et al. VarSifter: visualizing and analyzing exome-scale sequence variation data on a desktop computer. Bioinformatics. 2012;28:599–600. doi: 10.1093/bioinformatics/btr711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., et al. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell M., et al. A probabilistic disease-gene finder for personal genomes. Genome Res. 2011;21:1529–1542. doi: 10.1101/gr.123158.111. [DOI] [PMC free article] [PubMed] [Google Scholar]