Abstract
The human T – cell leukemia virus type 1 (HTLV-1) was the first retrovirus discovered to be causative of a human cancer, Adult T-cell leukemia (ATL). The transforming entity of HTLV-1 has been attributed to the virally-encoded oncoprotein, Tax. Unlike the v-onc proteins encoded by other oncogenic animal retroviruses that transform cells, Tax does not originate from a c-onc counterpart. In this article, we review progress in our understanding of HTLV-1 infectivity, cellular transformation, anti-sense transcription, and therapy, thirty years after the original discovery of this virus.
Keywords: human T-cell leukemia virus type 1 (HTLV-1), adult T cell leukemia (ATL), Tax, HTLV-1 bZIP factor (HBZ), aneuploidy
Introduction
HTLV-1 was the first human retrovirus identified 30 years ago as a causative agent of adult T-cell leukemia (ATL, see review (Takatsuki 2005)). It was isolated in 1980 in the United Sates (Poiesz et al. 1980) and then in Japan (Yoshida et al. 1982). The events of virus discovery in the United States and Japan (Gallo 2005; Yoshida 2005) have been reviewed well elsewhere. Currently, HTLV-1 infects approximately 20 million individuals world-wide (Proietti et al. 2005). Besides ATL, HTLV-1 infection can also cause a chronic inflammatory disease termed HTLV-1 associated myelopathy (HAM)/tropical spastic paraparesis (TSP) (Gessain et al. 1985) (Osame and Igata 1989). The role of HTLV-1 in HAM/TSP will not be discussed here. Below we summarize and update insights relevant to human leukemogenesis learned over the past 30 years from HTLV-1.
HTLV-1 infectivity and spread in vivo
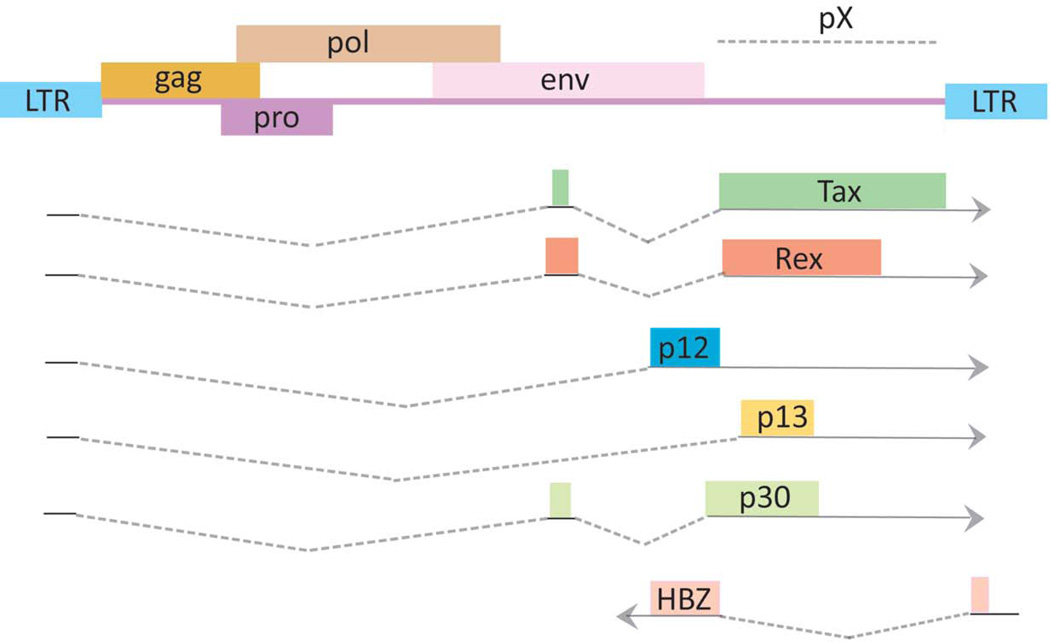
Like other animal retroviruses, the HTLV-1 proviral genome has structural genes, gag, pol and env, bracketed by long terminal repeat (LTR) sequences (Figure 1). The 5’ LTR serves as the viral promoter for transcription. The Pol open reading frame encodes reverse transcription, protease, and integrase functions. Gag provides the virion core proteins, and Env is used for viral infectivity. The HTLV-1 genome has a pX region located between env and the 3’-LTR. pX contains sequences for regulatory viral factors, Tax, Rex, p12, p13, p30 and p21. More recently, the minus strand of pX has been found to encode an antisense transcript, Hbz (Gaudray et al. 2002; Satou et al. 2006; Cavanagh et al. 2006; Matsuoka and Green 2009) discussed in further detail below).
Figure 1. Genome structure of the HTLV-1 provirus.
The gag, pol, and env structural genes are flanked by 5′ and 3′ long terminal repeats (LTRs). The pX region, Tax, Rex, and the antisense HTLV-1 basic leucine zipper factor (HBZ) open reading frame are shown. Drawing is intended to be illustrative and not to exact scale. This drawing is modified after (Matsuoka and Jeang 2007).
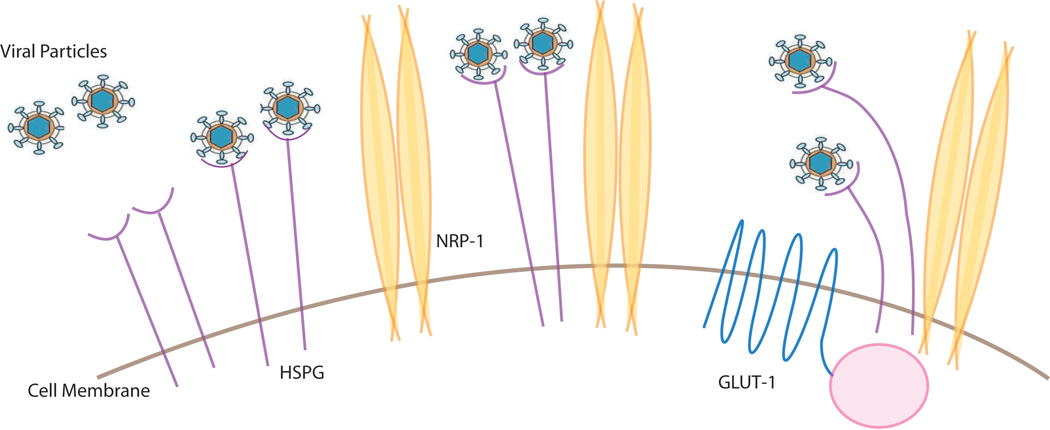
HTLV-1 can infect a variety of cell types, including T-cells, B-cells, fibroblasts and macrophages (Jones et al. 2008; Koyanagi et al. 1993) indicating that the receptor is commonly expressed on these cells. Recent studies show that glucose transporter 1, heparan sulfate proteoglycans (HSPGs), and neuropilin-1 (NRP-1) are three proteins important for the entry of HTLV-1 (Jones et al. 2005; Lambert et al. 2009) (Manel et al. 2003). One way to view the entry events is to consider that the virus could first contact HSPG and then form complexes with NRP-1 followed by Glut1 association on the cell surface before final membrane fusion and entry into the cell (Figure 2). Currently, how all these factors cooperate with each other and with a newly described extracellular matrix mediated transmission (Pais-Correia et al. 2010) or via dendritic cell assisted infection (Jones et al. 2008) requires further delineation. Moreover it should be noted that despite the ubiquitous distribution of these factors, the HTLV-1 provirus is detected mainly in CD4+ T-cells and to a lesser extent in CD8+ T-cells (Yasunaga et al. 2001). This asymmetry in detection may be because HTLV-1 infection recruits CD4+ T-cells into proliferative cell cycling while it seems to simply delay cell death in CD8+ T-cells (Sibon et al. 2006). Thus, the differential outcome of the virus on CD4+ and CD8+ cell proliferation may be more important than receptor-binding and cellular entry differences in dictating the apparent specificity for CD4+ cells (Zane et al. 2010). However, there is also evidence that cellular receptors play an important role in determining the cellular tropism of HTLV-1 (Jones et al. 2006).
Figure 2. Cell surface proteins that participate in HTLV-1 virion infection.
Interaction of HTLV-1 particles with HSPG, NRP-1 and Glut-1 proteins is schematized. Please see text for more detail.
Unlike the human immunodeficiency virus (HIV-1), HTLV-1 is primarily transmitted through cell-to-cell contact, not by cell-free virions (Igakura et al. 2003). HTLV-1 infected cells form virological synapses with uninfected cells. Tax and the intercellular adhesion molecule-1 (ICAM-1) play important roles in the formation of the virological synapse (Nejmeddine et al. 2009). Enveloped viral particles can transfer through this synapse, thus propagating infection (Majorovits et al. 2008). Recently, it has been reported that HTLV-1 cell-to-cell transmission is ten thousand times more efficient than cell-free infection, while for HIV-1 cell-cell co-culturing enhances infection only two fold (Mazurov et al. 2010). The importance of in vivo cell-to-cell spread is tempered by findings that the administration of reverse transcriptase inhibitors (RTI) to HTLV-1 infected patients with HAM/TSP does not markedly influence the provirus load (Taylor et al. 2006), and that RTI treatment immediately after infection by HTLV-1 in vivo does not change subsequent proviral load. Thus, viral replication itself appears not to be critical for the maintenance of persistent infection; rather, the proliferation of HTLV-1 infected cells seems to determine the viral burden during the carrier state. In this regard, the virus’ strategy to increase the number of infected cells by promoting cellular proliferation is purposeful. Indeed, a long standing observation is that HTLV-1 induces clonal proliferation of infected cells in vivo (Cavrois et al. 1998) (Etoh et al. 1997) (Zane et al. 2010).
Host immune and inflammatory responses to HTLV-1
A persistent virus infection establishes an equilibrium between viral virulence and the host immunity (Virgin et al. 2009). Viruses that maintain chronic infection should evade the host’s attempt at sterilizing immunity. A balance is reached when the extent of inflammation and viral replication avoid the creation of excessive tissue damage. Accordingly, viruses that cause chronic infection have evolved strategies to moderate the host immune system and temper viral replication.
For HTLV-1, it has been reported that an accessory viral protein, P12, physically interacts with the human major histocompatibility complex class I heavy chains, leading to the latter’s degradation (Johnson et al. 2001); this process then facilitates viral escape from the host immune system. In the same vein, moderate HTLV-1 replication appears to arise from in vivo suppression of tax expression (Hanon et al. 2000). CD8+ cell mediated CTLs are in part responsible for this phenomenon because their depletion enhances Tax expression in vivo (Hanon et al. 2000). These CD8-cell dependent CTLs appear to target directly the Tax protein because when histone deacetylase inhibitor, valproate, is used to activate tax transcription, the HTLV-1 proviral load in HAM/TSP individuals becomes reduced (Lezin et al. 2007). Thus the host’s CTL-response targets Tax-expressing cells thereby reducing the number of infected cells in vivo. On the other hand, the role played by antibodies to HTLV-1 remains largely unknown.
During ATL progression, opportunistic infections can be frequent complications (Matsuoka and Jeang 2007). Cell-mediated immunity becomes progressively impaired in ATL patients as indicated by lowered reactivity to the purified protein derivative (PPD) of Mycobacterium tuberculosis recall antigen (Welles et al. 1994). In this setting, Strongyloides stercoralis infection has been reported to become widely disseminated (Gotuzzo et al. 1999). Alternatively, Strongyloides infection in HTLV-1 carriers (Nakada et al. 1987) who have yet to develop frank immunodeficiency has been correlated with increased propensity for ATL. This finding may relate to observations that co-infection of HCV in HTLV-1 carriers increases the risk of liver cancers (Boschi-Pinto et al. 2000) suggesting that chronic inflammation (Rauch et al. 2009) elicited by multiple agents and T cell activation (Swaims et al. 2010) as well as Tax-induced T cell proliferation (Maruyama et al. 1987) contribute to the in vivo promotion of ATL.
It has been reported that ATL cells express FoxP3 in approximately two of thirds of ATL cases (Karube et al. 2004). Because foxp3 is a master gene that controls regulatory functions of regulatory T cells, this finding indicates that ATL could be a neoplastic disease of regulatory T cells. Such immunosuppressive phenotypes of ATL cells may account for the immunodeficiency observed in ATL patients. Regulatory T cells express chemokine receptor, CCR4, on the surface, and ligands for CCD4 and CCL22; these cells are increased in HTLV-1 infected individuals (Toulza et al. 2010). Enhanced production of CCL22 might contribute mechanistically to increased regulatory T cells in HTLV-1 infected individuals (Toulza et al. 2008).
Multifaceted processes in the transformation of virus infected cells
A subset (6.6% for males and 2.1% for females in Japan) of HTLV-1 infected individuals will develop adult T cell leukemia (ATL) after an extended period of time (Matsuoka and Jeang 2007). HTLV-1 encodes a Tax oncoprotein (Figure 1) (Grassmann et al. 2005) (Higuchi and Fujii 2009) which confers survival and proliferative properties to infected cells. Tax is post-translationally modified by phosphorylation, ubiquitination, and acetylation (Durkin et al. 2006; Peloponese, Jr. et al. 2004; Peloponese, Jr. et al. 2009; Chiari et al. 2004; Lodewick et al. 2009; Jeong et al. 2009; Marriott and Semmes 2005). These post-translational modifications have been experimentally shown to be important for Tax function. Expression of Tax-alone has been postulated to be sufficient for the immortalization, but not the transformation, of human T cells (Rosin et al. 1998; Robek and Ratner 1999). Tax’s in vivo transforming capacity has been investigated extensively using transgenic mouse models; all results suggest that the sole expression of Tax can capably drive in vivo tumor formation (Grossman et al. 1995; Hasegawa et al. 2006; Ohsugi et al. 2007). However, because mice have a more relaxed transformation threshold than humans (Haller et al. 2006), whether Tax expression alone is sufficient for human ATL leukemogenesis remains to be clarified. The finding that mouse primary cells are substantially more easily transformed than human primary cells has been well characterized by others (Hahn et al. 1999). Nevertheless, several biological properties of Tax as outlined below are compatible with its transforming potential in human cells.
a) Activation of cellular survival and proliferative pathways
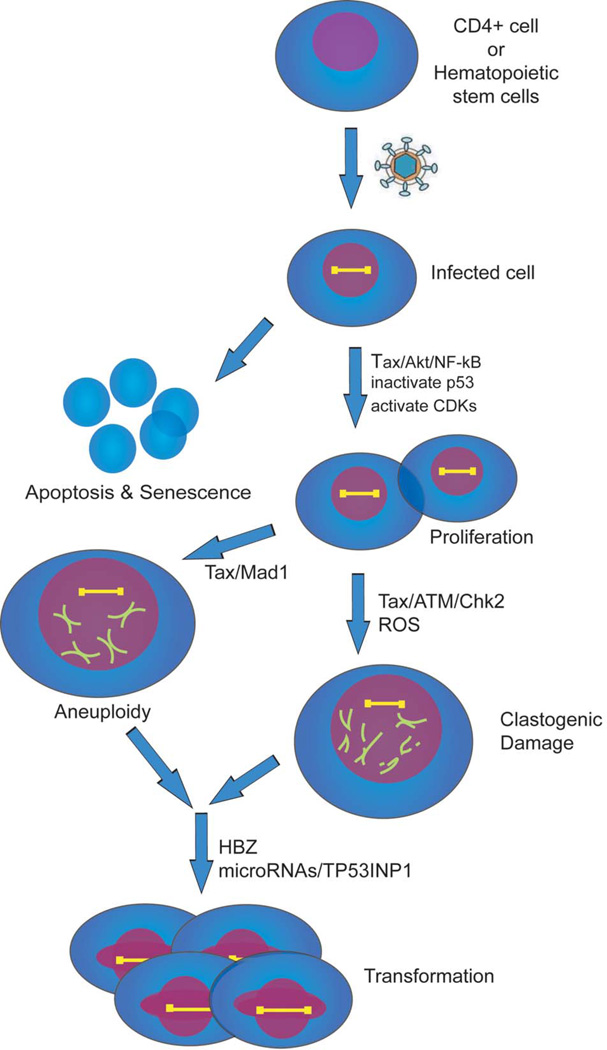
Virus infected cells need to evade apoptosis and commence proliferation as a prelude to immortalization and transformation (Figure 3). In the setting of HTLV-1 infection, the NF-κB and Akt pathways are two major cellular prosurvival routes activated by Tax. NF-κB is constitutively active in HTLV-1-infected cells (Peloponese et al. 2006). There are at least three ways that Tax can modulate cellular NF-κB activity. First, Tax can bind the IKKγ protein and activate the IKKαIKKβ/IKKγ complex (Jin et al. 1999; Chu et al. 1999; Harhaj and Sun 1999) leading to NF-κB (p50/p65) migration into the nucleus, where it activates the transcription of NF-κB-responsive genes (Iha et al. 2003). Second, Tax can stimulate a non-canonical NF-κB pathway through the IKKα-dependent processing of the NF-κB p100 precursor protein to its active p52 form (Xiao et al. 2001) (Shoji et al. 2009). Third, Tax can inactivate the cellular Tax1BP1 adaptor protein which promotes deubiquitination and represses TRAF6-NF-κB signaling. By attenuating Tax1BP1 function, Tax enhances signaling through the TRAF6-NF-κB pathway (Iha et al. 2008; Shembade et al. 2007).
Figure 3. Putative cellular events that are important for ATL development.
HTLV-1 infects either CD4+ cells or a hematopoietic progenitor cell. Infected cells activate survival pathways NF-κB and Akt to evade apoptosis and senescence. Inactivation of p53 and activation of CDKs accelerate cellular proliferation. Tax attenuation of the spindle assembly checkpoint protein, Mad1, Tax reduction of DNA-damage repair, and Tax activation of reactive oxygen species (ROS) lead to aneuploidy and clastogenic damage. Late in ATL development when Tax expression is extinguished HBZ, microRNA changes, and TP53INP1 inactivation may be important to achieve a fully transformed phenotype.
Akt is a serine/threonine kinase regulated by phosphatidylinositol 3-kinase (PI3K) through site-specific phosphorylation on Ser473 (Song et al. 2005). Tax binds PI3K and promotes Akt phosphorylation and its activation (Peloponese, Jr. and Jeang 2006). Activated Akt can then enhance cellular survival in part through the induction of activator protein 1, AP1, responsive genes (Jeong et al. 2008). This route is an additionally significant cellular prosurvival/proproliferative path stimulated by Tax.
In addition to Akt and NF-κB, Tax has been shown to increase cyclin D2 expression through direct activation of its promoter (Santiago et al. 1999), to activate CDKs, such as CDK4, through direct protein binding (Fraedrich et al. 2005; Haller et al. 2002), and to repress CKIs such as INK4A, INK4B (Low et al. 1997; Suzuki et al. 1996), INK4C (Akagi et al. 1996), INK4D (Riou et al. 2000; Suzuki et al. 1999)and KIP1 (Iwanaga et al. 2001). These interactions result in accelerated cell cycling of HTLV-1 infected cells (Neuveut et al. 1998) (Boxus et al. 2008) promoting in vivo clonal expansion (Zane et al. 2010).
b) Neutralization of cellular checkpoints
Aberrant cellular proliferation is physiologically reined in by a multitude of cellular checkpoint functions. A key cellular checkpoint is enforced by the p53 protein. Accordingly, in order to achieve successful transformation, ~ 50% of human cancers have function-disabling mutations in p53. Many cancer causing viruses have evolved multifaceted mechanisms to inactivate p53 (Soria et al. 2010). Curiously, mutations which inactivate p53 are infrequent in ATL cells (Yasunaga and Matsuoka 2007). Nevertheless, in the absence of genetic mutation of p53, the p53 checkpoint is functionally inactivated in HTLV-1 infected cells by the Tax protein (Reid et al. 1993) (Tabakin-Fix et al. 2006). How p53 is mechanistically inactivated in ATL remains incompletely clarified. Several redundant mechanisms (Ariumi et al. 2000) (Pise-Masison et al. 2000; Miyazato et al. 2005) appear to be employed by Tax. It is also currently unanswered whether cellular phosphatases such as Wip1 (Lindqvist et al. 2009) which counteract p53 activity are co-opted by Tax.
ATL cells are highly aneuploid (Yasunaga and Jeang 2009). A spindle assembly checkpoint (SAC) normally operates in mitosis to preserve euploidy by monitoring the fidelity of mitotic chromosomal segregation (Chi and Jeang 2007). SAC proteins such as MAD1, MAD2, MAD3/BUBR1, BUB1, BUB3, and MSP1 function at kinetochores (Chi and Jeang 2007). It has been noted that SV40 (Hein et al. 2009), EBV (Pan et al. 2009) and HPV (Patel and McCance 2010) infected cells have attenuated SAC function. Similarly, the loss of SAC function has also been verified in several ATL cells (Kasai et al. 2002). Mechanistically, the inactivation of SAC in ATL occurs through Tax binding to the checkpoint protein MAD1 (Jin et al. 1998) whose loss of function leads to tumorigenesis (Iwanaga et al. 2007) (Figure 3). Tax binding to and inactivation of the anaphase-promoting complex/cyclosome (APC/C) which functions downstream of the SAC (Liu et al. 2005), and Tax promotion of multipolar mitosis (Peloponese, Jr. et al. 2005) have also been postulated to contribute to the prevalent aneuploidy observed in ATLs.
c) DNA-damage and attenuation of repair
DNA-damage induced by Tax (Majone and Jeang 2000; Majone et al. 1993) (Majone et al. 2005) and other oncoproteins (Felsher and Bishop 1999) (Ramadan et al. 2008) (Vafa et al. 2002) is well-documented. In principle, damaged DNA can be created in two ways. First, the oncoprotein can attenuate damage sensing checkpoint and repair proteins. Second, the oncoprotein can directly elicit DNA lesions. It has been shown that Tax represses the cell’s DNA polymerase β enzyme which is employed for base excision repair (BER) (Jeang et al. 1990; Philpott and Buehring 1999). Similarly, Tax also suppresses nucleotide excision repair (NER) (Kao and Marriott 1999; Lemoine et al. 2000) and the expression of human DNA mismatch repair (MMR) genes (Morimoto et al. 2005). Furthermore, while it was suggested originally that Tax weakens the action of ATR/CHK1 DNA-damage signaling (Park et al. 2004), many subsequent studies have found that Tax more potently inactivates the ATM/CHK2 axis (Figure 3) and downstream repair factors (Park et al. 2006; Durkin et al. 2008) (Ramadan et al. 2008) (Chandhasin et al. 2008; Gupta et al. 2007) (Belgnaoui et al. 2010). Collectively then, through Tax’s inactivation of DNA-damage sensing and repair proteins, ambient DNA lesions that might have been repaired in cells are permitted to accumulate.
In addition to allowing the persistence of genetic lesions, Tax was recently found to also induce reactive oxygen species (ROS) which can directly create damaged DNA (Kinjo et al. 2010). Tax induction of ROS is consistent with similar ROS-induction by other viral transforming proteins such as Ras (Lee et al. 1999), c-Myc (Vafa et al. 2002), and Epstein-Barr virus (EBV) EBNA-1 protein (Gruhne et al. 2009). Collectively, the creation of new DNA-damage and the prevention of ambient repair lead to a picture of significant clastogenic damage as reported for Tax expressing cells (Majone and Jeang 2000).
d) Changes in microRNAs (miRNAs) with oncogenic potentials
Various cancers have different microRNA signatures (Croce 2009) (Lu et al. 2005) (Bouzar and Willems 2008). Three recent reports have described changes in miRNA expression in ATL cells (Yeung et al. 2008; Pichler et al. 2008; Bellon et al. 2009). Because the experimental settings were different, there was considerable discordance in the individual miRNA changes that were reported (Ruggero et al. 2010). However, a salient consensus (i.e. TP53INP1) did emerge from the studies. Thus, Yeung et al. reported that the tumor suppressor protein TP53INP1 in HTLV-1-infected/transformed cells was repressed by miR-93 and miR-130b (Yeung et al. 2008) while a second report found that TP53INP1 was targeted in HTLV-1 infected/transformed cells by miR-21, -24, -146a, and -155 (Pichler et al. 2008). A third report described ATL cells with increased expression of miR-155 (Bellon et al. 2009). Interestingly, miR-155 has been reported to be a silencer of TP53INP1 (Gironella et al. 2007), reinforcing the notion that TP53INP1 may be a critical factor in ATL transformation. That oncogenic miRNAs contribute to ATL is further strengthened by a recent finding that small molecular inhibitors of miRNA functions can selectively reverse the dysregulated proliferation of several HTLV-1 transformed cells (Watashi et al. 2010), and by the finding that Tax can induce the expression of some cellular miRNAs (Yeung et al. 2008). Taken together, the perturbation of oncogenic miRNAs in ATL may represent an additional multifaceted factor that needs to be considered for in vivo leukemogenesis.
HTLV-1 antisense transcript HBZ and viral pathogenesis
The 3’ portion of the HTLV-1 genome has been shown to direct sense (plus) and antisense (minus) transcripts that encode different proteins (Figure 1) (Cavanagh et al. 2006; Satou et al. 2006). The plus strand of this region contains transcripts directed from the 5’ LTR, while the 3’ LTR directs a novel antisense transcript termed the HTLV-1 bZIP factor, Hbz (Figure 1). The promoter for the Hbz gene derives from U5 sequence of the 3’LTR. Mutational analyses of this promoter region show that three Sp1 sites are critical for HBZ transcription (Yoshida et al. 2008). Since the expression of Sp1 is relatively constant in most cells, Hbz gene expression is well correlated with the amount of integrated provirus in HTLV-1 infected individuals (Saito et al. 2009).
While tax transcripts are detected in only ~40% of transformed ATL cells, it was recently demonstrated that Hbz RNA is ubiquitously expressed in all ATL cells, and possesses cell proliferative function (Satou et al. 2006). The HBZ protein has been ascribed to interact with CREB, CREB-2, CREM-Ia, ATF-1 (Lemasson et al. 2007), c-Jun (Basbous et al. 2003; Matsumoto et al. 2005), JunB (Hivin et al. 2007), and JunD (Thebault et al. 2004) through its bZIP domain, and was originally reported to suppress Tax-mediated viral transcription (Gaudray et al. 2002). Furthermore, HBZ selectively inhibits the classical NF-κB pathway by inhibiting DNA binding of p65 and promotes the degradation of p65 (Zhao et al. 2009). Tax can activate both classical and alternate NF-κB pathways. The two pathways differentially control genes with anti-apoptotic functions in lymphoma cell lines (Bernal-Mizrachi et al. 2006). Predominant activation of the alternate pathway by Tax and HBZ might be implicated in the proliferation of ATL cells.
A further study suggested that the Hbz RNA, besides its protein form, is also important for the proliferation of HTLV-1-infected cells. Moreover, it has been shown that an HTLV-1 molecular clone with a mutation in the leucine zipper domain of HBZ exhibited reduced proviral load compared to wild type virus when inoculated into rabbits (Arnold et al. 2008) and that HBZ can increase the activity of the human telomerase reverse transcriptase (hTERT) gene (Kuhlmann et al. 2007). Collectively, the extant data support that HBZ protein and Hbz RNA have roles in promoting viral replication and cellular proliferation. One potential way to interpret the interplay between Tax and HBZ is that the former is needed to initiate transformation while the latter is required to maintain the transformed phenotype late in ATL when Tax expression is extinguished. Whether HBZ also plays roles in HTLV-1 associated HAM/TSP disease remains to be clarified (Saito et al. 2009).
Therapeutic approaches for ATL
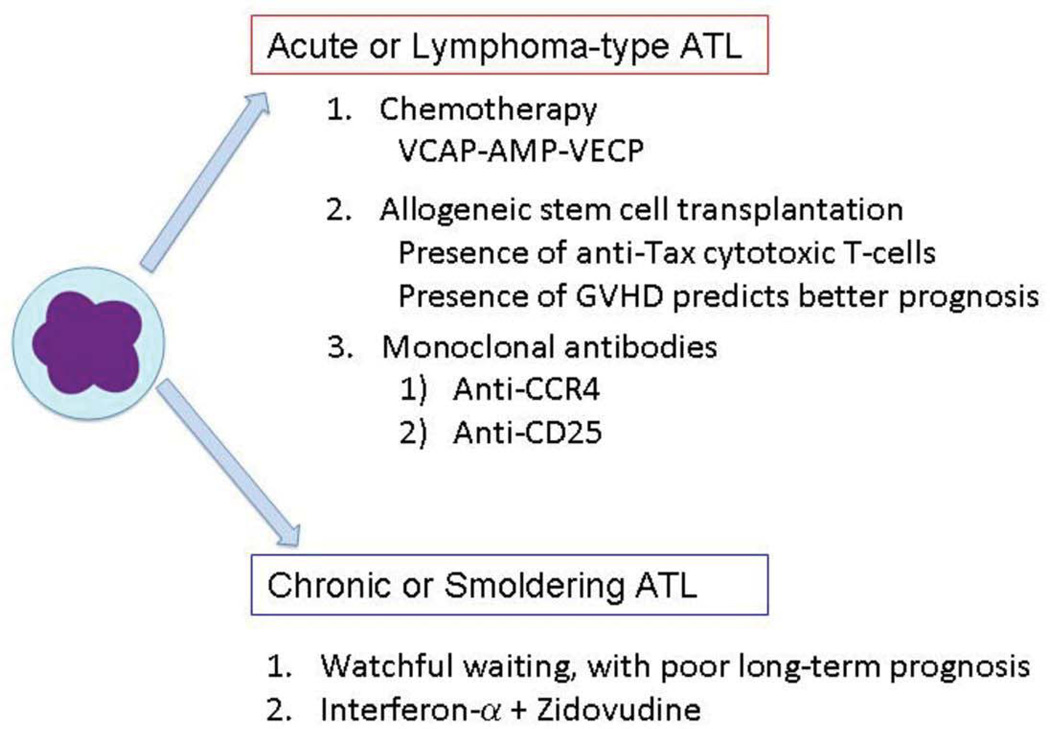
In many ways, ATL is a poorly treatable disease. Patients with acute or lymphoma-type ATL are usually addressed with combination chemotherapy. The representative protocol for ATL patients in Japan is vincristine, cyclophosphamide, doxorubicin, and prednisone (VCAP), doxorubicin, ranimustine, and prednisone (AMP), and vindesine, etoposide, carboplatin, and prednisone (VECP) (VCAP-AMP-VECP). This protocol was found to be superior to biweekly CHOP therapy (Tsukasaki et al. 2007). It has been found that the complete response rate was higher with VCAP-AMP-VECP than biweekly CHOP (40% v 25%). Indeed, the overall survival at 3 years was 24% in the VCAP-AMP-VECP arm and 13% in the CHOP arm. The major obstacles in therapy are the drug resistance of ATL cells to chemotherapeutic agents and the profoundly weakened and immunodeficient state of ATL patients. The cause of ATL immunodeficiency may be from the immunosuppressive function of ATL cells since they appear to arise from regulatory T cells (Karube et al. 2004) and can produce immunosuppressive cytokines (Mori et al. 1996). As noted above, ATL immunodeficiency leads to complications from various opportunistic fungal, viral, protozoal and bacterial infections which can worsen the prognosis.
Allogeneic stem cell transplantation (AlloSCT) has been shown to be effective in ATL patients (Utsunomiya et al. 2001). Patients treated with alloSCT with reduced-intensity conditioning (RIST) had overall survival at 3 years of 36%. (Tanosaki et al. 2008). These studies showed that 30% to 40% of ATL patients, who achieve remission and have suitable donors, became long-term survivors with either conventional alloSCT or RIST (Okamura et al. 2007). It should be noted that provirus load remarkably decreased in many patients who received SCT. The findings suggest that cell mediated immunity to HTLV-1 was augmented in these patients, which might account for the efficacy of this therapy.
The presence of graft-versus-host disease (GVHD) is a good prognostic factor for ATL patients (Okamura et al. 2007), indicating that immune attack by donor lymphocytes is critical for the efficacy of treatment. ATL cells express Fas antigen highly and are susceptible to Fas mediated signaling (Tamiya et al. 1998; Yasunaga et al. 2004). Cytotoxic T-lymphocytes (CTLs) can attack ATL cells in the host, resulting in good therapeutic responses. In allogeneic stem cell transplantation treated individuals, CTLs to Tax peptides were activated in the recipients; and provirus load became profoundly suppressed, indicating the role of anti-HTLV-1 immune responses in the efficacious outcome (Harashima et al. 2004). Nevertheless, it remains unknown whether CTLs to Tax are required for the efficacy of stem cell transplantation therapy.
Of therapeutic implication, it has been noted that most ATL leukemic cells express CC chemokine receptor 4 (CCR4) molecules on their surfaces (Yoshie et al. 2002). Since Treg cells express CCR4, this finding is consistent with one of the origins of ATL cells as Treg cells. KW-0761, a defucosylated humanized anti- CCR4 antibody, has been shown to be effective in ATL patients (Yamamoto et al. 2010). Among fifteen patients treated with KW-0761, five patients achieved positive objective responses: two complete and three partial responses. Potentially, the KW-0761 antibody could be beneficial when combined with rituximab treatment.
It has been reported that the mean survival time of patients with indolent subtypes, chronic and smoldering ATL, was 4.1 years. This finding shows that the prognosis of indolent ATL is poorer than previously thought (Takasaki et al. 2010). Therapy using interferon α combined with zidovudine has been reported to be highly effective to these indolent ATL (Bazarbachi et al. 2010) suggesting that such therapy could be beneficial for this category of patients.
Experiments using animal models have provided important information on therapeutic strategies for ATL patients. For example, monoclonal antibodies to CD25 (Phillips et al. 2000), CD2 (Zhang et al. 2003b), CD52 (Zhang et al. 2003a), and CCR4(Ishii et al. 2010) have been shown to be effective to ATL cells within in vivo model. In addition, it has been reported that a proteasome inhibitor, bortezomib, suppresses tumor formation of ATL cells in vivo (Mitra-Kaushik et al. 2004) (Satou et al. 2004) (Tan and Waldmann 2002). NF-κB inhibitors also effectively induce apoptosis of ATL cells since NF-κB is highly activated in ATL cells (Dewan et al. 2003; Watanabe et al. 2005). These findings suggest the potential efficacy of these compounds and antibodies although they remain to be verified in clinical studies.
Concluding remarks
Recent developments suggest that cancers, whether solid tumors (e.g., breast and lung) or hematological malignancies (e.g., leukemia), are comprised of two categories of cells: those with high- and those with limited- proliferative potential. Cells in the former category are termed cancer stem cells (CSCs). CSCs share several properties with adult stem cells, particularly the abilities to self-renew and differentiate into multiple cell types. CSCs are found in a given cancer as a small subpopulation; and CSCs are reasoned to be the moieties which cause disease relapse and metastasis. They, unlike the bulk of tumor cells, are postulated to be the only cells capable of giving rise to new tumors. Currently, an outstanding question in HTLV research is whether ATL originates from the transformation of a differentiated CD4+ T cells or from virus infection and transformation of a hematopoietic precursor cell (Banerjee et al. 2010) (Banerjee et al. 2008). Many attempts to transform human CD4+ T cells with Tax have failed; and it remains to be seen whether hematopoietic precursor cells or human stem cells can be transformed by Tax. Success in the latter attempts would point to factors that are expressed in undifferentiated, rather than differentiated, cells that may be critical to ATL leukemogenesis. Certainly, we expect these answers to be forthcoming in the next 30 years of HTLV-1 research.
Figure 4. Therapeutic approaches for ATL.
Current therapeutic strategies for Acute or Lymphoma-type ATL and Chronic or Smoldering ATL are listed.
Acknowledgements
Work in KTJ’s laboratory is supported in part by NIAID intramural funds, and work in MM’s laboratory is supported by Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan. We thank Lauren Lee for preparation of manuscript and figures, and Junichiro Yasunaga and Linda Zane for discussions.
Reference List
- Akagi T, Ono H, Shimotohno K. Oncogene. 1996;12:1645–1652. [PubMed] [Google Scholar]
- Ariumi Y, Kaida A, Lin JY, Hirota M, Masui O, Yamaoka S, Taya Y, Shimotohno K. Oncogene. 2000;19:1491–1499. doi: 10.1038/sj.onc.1203450. [DOI] [PubMed] [Google Scholar]
- Arnold J, Zimmerman B, Li M, Lairmore MD, Green PL. Blood. 2008;112:3788–3797. doi: 10.1182/blood-2008-04-154286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee P, Crawford L, Samuelson E, Feuer G. Retrovirology. 2010;7:8. doi: 10.1186/1742-4690-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee P, Sieburg M, Samuelson E, Feuer G. Stem Cells. 2008;26:3047–3058. doi: 10.1634/stemcells.2008-0353. [DOI] [PubMed] [Google Scholar]
- Basbous J, Arpin C, Gaudray G, Piechaczyk M, Devaux C, Mesnard JM. J Biol Chem. 2003;278:43620–43627. doi: 10.1074/jbc.M307275200. [DOI] [PubMed] [Google Scholar]
- Bazarbachi A, Plumelle Y, Ramos JC, Tortevoye P, Otrock Z, Taylor G, Gessain A, Harrington W, Panelatti G, Hermine O. J Clin Oncol. 2010 doi: 10.1200/JCO.2010.28.0669. [DOI] [PubMed] [Google Scholar]
- Belgnaoui SM, Fryrear KA, Nyalwidhe JO, Guo X, Semmes OJ. J Biol Chem. 2010 doi: 10.1074/jbc.M110.146373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellon M, Lepelletier Y, Hermine O, Nicot C. Blood. 2009;113:4914–4917. doi: 10.1182/blood-2008-11-189845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal-Mizrachi L, Lovly CM, Ratner L. Proc Natl Acad Sci U S A. 2006;103:9220–9225. doi: 10.1073/pnas.0507809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschi-Pinto C, Stuver S, Okayama A, Trichopoulos D, Orav EJ, Tsubouchi H, Mueller N. J Infect Dis. 2000;181:35–41. doi: 10.1086/315177. [DOI] [PubMed] [Google Scholar]
- Bouzar AB, Willems L. Retrovirology. 2008;5:101. doi: 10.1186/1742-4690-5-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxus M, Twizere JC, Legros S, Dewulf JF, Kettmann R, Willems L. Retrovirology. 2008;5:76. doi: 10.1186/1742-4690-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh MH, Landry S, Audet B, rpin-Andre C, Hivin P, Pare ME, Thete J, Wattel E, Marriott SJ, Mesnard JM, Barbeau B. Retrovirology. 2006;3:15. doi: 10.1186/1742-4690-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavrois M, Leclercq I, Gout O, Gessain A, Wain-Hobson S, Wattel E. Oncogene. 1998;17:77–82. doi: 10.1038/sj.onc.1201906. [DOI] [PubMed] [Google Scholar]
- Chandhasin C, Ducu RI, Berkovich E, Kastan MB, Marriott SJ. J Virol. 2008;82:6952–6961. doi: 10.1128/JVI.02331-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi YH, Jeang KT. J Cell Biochem. 2007;102:531–538. doi: 10.1002/jcb.21484. [DOI] [PubMed] [Google Scholar]
- Chiari E, Lamsoul I, Lodewick J, Chopin C, Bex F, Pique C. J Virol. 2004;78:11823–11832. doi: 10.1128/JVI.78.21.11823-11832.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu ZL, Shin YA, Yang JM, DiDonato JA, Ballard DW. J Biol Chem. 1999;274:15297–15300. doi: 10.1074/jbc.274.22.15297. [DOI] [PubMed] [Google Scholar]
- Croce CM. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewan MZ, Terashima K, Taruishi M, Hasegawa H, Ito M, Tanaka Y, Mori N, Sata T, Koyanagi Y, Maeda M, Kubuki Y, Okayama A, Fujii M, Yamamoto N. J Virol. 2003;77:5286–5294. doi: 10.1128/JVI.77.9.5286-5294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin SS, Guo X, Fryrear KA, Mihaylova VT, Gupta SK, Belgnaoui SM, Haoudi A, Kupfer GM, Semmes OJ. J Biol Chem. 2008;283:36311–36320. doi: 10.1074/jbc.M804931200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin SS, Ward MD, Fryrear KA, Semmes OJ. J Biol Chem. 2006;281:31705–31712. doi: 10.1074/jbc.M607011200. [DOI] [PubMed] [Google Scholar]
- Etoh K, Tamiya S, Yamaguchi K, Okayama A, Tsubouchi H, Ideta T, Mueller N, Takatsuki K, Matsuoka M. Cancer Res. 1997;57:4862–4867. [PubMed] [Google Scholar]
- Felsher DW, Bishop JM. Proc Natl Acad Sci U S A. 1999;96:3940–3944. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraedrich K, Muller B, Grassmann R. Retrovirology. 2005;2:54. doi: 10.1186/1742-4690-2-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo RC. Retrovirology. 2005;2:17. doi: 10.1186/1742-4690-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudray G, Gachon F, Basbous J, Biard-Piechaczyk M, Devaux C, Mesnard JM. J Virol. 2002;76:12813–12822. doi: 10.1128/JVI.76.24.12813-12822.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessain A, Barin F, Vernant JC, Gout O, Maurs L, Calender A, de TG. Lancet. 1985;2:407–410. doi: 10.1016/s0140-6736(85)92734-5. [DOI] [PubMed] [Google Scholar]
- Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, Garcia S, Nowak J, Yeung ML, Jeang KT, Chaix A, Fazli L, Motoo Y, Wang Q, Rocchi P, Russo A, Gleave M, Dagorn JC, Iovanna JL, Carrier A, Pebusque MJ, Dusetti NJ. Proc Natl Acad Sci U S A. 2007;104:16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotuzzo E, Terashima A, Alvarez H, Tello R, Infante R, Watts DM, Freedman DO. Am J Trop Med Hyg. 1999;60:146–149. doi: 10.4269/ajtmh.1999.60.146. [DOI] [PubMed] [Google Scholar]
- Grassmann R, Aboud M, Jeang KT. Oncogene. 2005;24:5976–5985. doi: 10.1038/sj.onc.1208978. [DOI] [PubMed] [Google Scholar]
- Grossman WJ, Kimata JT, Wong FH, Zutter M, Ley TJ, Ratner L. Proc Natl Acad Sci U S A. 1995;92:1057–1061. doi: 10.1073/pnas.92.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhne B, Sompallae R, Marescotti D, Kamranvar SA, Gastaldello S, Masucci MG. Proc Natl Acad Sci U S A. 2009;106:2313–2318. doi: 10.1073/pnas.0810619106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SK, Guo X, Durkin SS, Fryrear KF, Ward MD, Semmes OJ. J Biol Chem. 2007;282:29431–29440. doi: 10.1074/jbc.M704110200. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- Haller K, Kibler KV, Kasai T, Chi YH, Peloponese JM, Yedavalli VS, Jeang KT. Oncogene. 2006;25:2137–2147. doi: 10.1038/sj.onc.1209259. [DOI] [PubMed] [Google Scholar]
- Haller K, Wu Y, Derow E, Schmitt I, Jeang KT, Grassmann R. Mol Cell Biol. 2002;22:3327–3338. doi: 10.1128/MCB.22.10.3327-3338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanon E, Hall S, Taylor GP, Saito M, Davis R, Tanaka Y, Usuku K, Osame M, Weber JN, Bangham CR. Blood. 2000;95:1386–1392. [PubMed] [Google Scholar]
- Harashima N, Kurihara K, Utsunomiya A, Tanosaki R, Hanabuchi S, Masuda M, Ohashi T, Fukui F, Hasegawa A, Masuda T, Takaue Y, Okamura J, Kannagi M. Cancer Res. 2004;64:391–399. doi: 10.1158/0008-5472.can-03-1452. [DOI] [PubMed] [Google Scholar]
- Harhaj EW, Sun SC. J Biol Chem. 1999;274:22911–22914. doi: 10.1074/jbc.274.33.22911. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Sawa H, Lewis MJ, Orba Y, Sheehy N, Yamamoto Y, Ichinohe T, Tsunetsugu-Yokota Y, Katano H, Takahashi H, Matsuda J, Sata T, Kurata T, Nagashima K, Hall WW. Nat Med. 2006;12:466–472. doi: 10.1038/nm1389. [DOI] [PubMed] [Google Scholar]
- Hein J, Boichuk S, Wu J, Cheng Y, Freire R, Jat PS, Roberts TM, Gjoerup OV. J Virol. 2009;83:117–127. doi: 10.1128/JVI.01515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Fujii M. Retrovirology. 2009;6:117. doi: 10.1186/1742-4690-6-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hivin P, Basbous J, Raymond F, Henaff D, rpin-Andre C, Robert-Hebmann V, Barbeau B, Mesnard JM. Retrovirology. 2007;4:14. doi: 10.1186/1742-4690-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igakura T, Stinchcombe JC, Goon PK, Taylor GP, Weber JN, Griffiths GM, Tanaka Y, Osame M, Bangham CR. Science. 2003;299:1713–1716. doi: 10.1126/science.1080115. [DOI] [PubMed] [Google Scholar]
- Iha H, Kibler KV, Yedavalli VR, Peloponese JM, Haller K, Miyazato A, Kasai T, Jeang KT. Oncogene. 2003;22:8912–8923. doi: 10.1038/sj.onc.1207058. [DOI] [PubMed] [Google Scholar]
- Iha H, Peloponese JM, Verstrepen L, Zapart G, Ikeda F, Smith CD, Starost MF, Yedavalli V, Heyninck K, Dikic I, Beyaert R, Jeang KT. EMBO J. 2008;27:629–641. doi: 10.1038/emboj.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Ishida T, Utsunomiya A, Inagaki A, Yano H, Komatsu H, Iida S, Imada K, Uchiyama T, Akinaga S, Shitara K, Ueda R. Clin Cancer Res. 2010;16:1520–1531. doi: 10.1158/1078-0432.CCR-09-2697. [DOI] [PubMed] [Google Scholar]
- Iwanaga R, Ohtani K, Hayashi T, Nakamura M. Oncogene. 2001;20:2055–2067. doi: 10.1038/sj.onc.1204304. [DOI] [PubMed] [Google Scholar]
- Iwanaga Y, Chi YH, Miyazato A, Sheleg S, Haller K, Peloponese JM, Jr, Li Y, Ward JM, Benezra R, Jeang KT. Cancer Res. 2007;67:160–166. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- Jeang KT, Widen SG, Semmes OJ, Wilson SH. Science. 1990;247:1082–1084. doi: 10.1126/science.2309119. [DOI] [PubMed] [Google Scholar]
- Jeong SJ, Dasgupta A, Jung KJ, Um JH, Burke A, Park HU, Brady JN. Virology. 2008;370:264–272. doi: 10.1016/j.virol.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SJ, Ryo A, Yamamoto N. Biochem Biophys Res Commun. 2009;381:294–299. doi: 10.1016/j.bbrc.2009.02.024. [DOI] [PubMed] [Google Scholar]
- Jin DY, Giordano V, Kibler KV, Nakano H, Jeang KT. J Biol Chem. 1999;274:17402–17405. doi: 10.1074/jbc.274.25.17402. [DOI] [PubMed] [Google Scholar]
- Jin DY, Spencer F, Jeang KT. Cell. 1998;93:81–91. doi: 10.1016/s0092-8674(00)81148-4. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Nicot C, Fullen J, Ciminale V, Casareto L, Mulloy JC, Jacobson S, Franchini G. J Virol. 2001;75:6086–6094. doi: 10.1128/JVI.75.13.6086-6094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KS, Fugo K, Petrow-Sadowski C, Huang Y, Bertolette DC, Lisinski I, Cushman SW, Jacobson S, Ruscetti FW. J Virol. 2006;80:8291–8302. doi: 10.1128/JVI.00389-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KS, Petrow-Sadowski C, Bertolette DC, Huang Y, Ruscetti FW. J Virol. 2005;79:12692–12702. doi: 10.1128/JVI.79.20.12692-12702.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KS, Petrow-Sadowski C, Huang YK, Bertolette DC, Ruscetti FW. Nat Med. 2008;14:429–436. doi: 10.1038/nm1745. [DOI] [PubMed] [Google Scholar]
- Kao SY, Marriott SJ. J Virol. 1999;73:4299–4304. doi: 10.1128/jvi.73.5.4299-4304.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karube K, Ohshima K, Tsuchiya T, Yamaguchi T, Kawano R, Suzumiya J, Utsunomiya A, Harada M, Kikuchi M. Br J Haematol. 2004;126:81–84. doi: 10.1111/j.1365-2141.2004.04999.x. [DOI] [PubMed] [Google Scholar]
- Kasai T, Iwanaga Y, Iha H, Jeang KT. J Biol Chem. 2002;277:5187–5193. doi: 10.1074/jbc.M110295200. [DOI] [PubMed] [Google Scholar]
- Kinjo T, Ham-Terhune J, Peloponese JM, Jr, Jeang KT. J Virol. 2010;84:5431–5437. doi: 10.1128/JVI.02460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi Y, Itoyama Y, Nakamura N, Takamatsu K, Kira J, Iwamasa T, Goto I, Yamamoto N. Virology. 1993;196:25–33. doi: 10.1006/viro.1993.1451. [DOI] [PubMed] [Google Scholar]
- Kuhlmann AS, Villaudy J, Gazzolo L, Castellazzi M, Mesnard JM, Duc DM. Retrovirology. 2007;4:92. doi: 10.1186/1742-4690-4-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S, Bouttier M, Vassy R, Seigneuret M, Petrow-Sadowski C, Janvier S, Heveker N, Ruscetti FW, Perret G, Jones KS, Pique C. Blood. 2009;113:5176–5185. doi: 10.1182/blood-2008-04-150342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AC, Fenster BE, Ito H, Takeda K, Bae NS, Hirai T, Yu ZX, Ferrans VJ, Howard BH, Finkel T. J Biol Chem. 1999;274:7936–7940. doi: 10.1074/jbc.274.12.7936. [DOI] [PubMed] [Google Scholar]
- Lemasson I, Lewis MR, Polakowski N, Hivin P, Cavanagh MH, Thebault S, Barbeau B, Nyborg JK, Mesnard JM. J Virol. 2007;81:1543–1553. doi: 10.1128/JVI.00480-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine FJ, Kao SY, Marriott SJ. AIDS Res Hum Retroviruses. 2000;16:1623–1627. doi: 10.1089/08892220050193056. [DOI] [PubMed] [Google Scholar]
- Lezin A, Gillet N, Olindo S, Signate A, Grandvaux N, Verlaeten O, Belrose G, de Carvalho BM, Hiscott J, Asquith B, Burny A, Smadja D, Cesaire R, Willems L. Blood. 2007;110:3722–3728. doi: 10.1182/blood-2007-04-085076. [DOI] [PubMed] [Google Scholar]
- Lindqvist A, de BM, Macurek L, Bras A, Mensinga A, Bruinsma W, Voets O, Kranenburg O, Medema RH. EMBO J. 2009;28:3196–3206. doi: 10.1038/emboj.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Hong S, Tang Z, Yu H, Giam CZ. Proc Natl Acad Sci U S A. 2005;102:63–68. doi: 10.1073/pnas.0406424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodewick J, Lamsoul I, Polania A, Lebrun S, Burny A, Ratner L, Bex F. Virology. 2009;386:68–78. doi: 10.1016/j.virol.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low KG, Dorner LF, Fernando DB, Grossman J, Jeang KT, Comb MJ. J Virol. 1997;71:1956–1962. doi: 10.1128/jvi.71.3.1956-1962.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, varez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Majone F, Jeang KT. J Biol Chem. 2000;275:32906–32910. doi: 10.1074/jbc.C000538200. [DOI] [PubMed] [Google Scholar]
- Majone F, Luisetto R, Zamboni D, Iwanaga Y, Jeang KT. Retrovirology. 2005;2:45. doi: 10.1186/1742-4690-2-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majone F, Semmes OJ, Jeang KT. Virology. 1993;193:456–459. doi: 10.1006/viro.1993.1145. [DOI] [PubMed] [Google Scholar]
- Majorovits E, Nejmeddine M, Tanaka Y, Taylor GP, Fuller SD, Bangham CR. PLoS One. 2008;3:e2251. doi: 10.1371/journal.pone.0002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manel N, Kim FJ, Kinet S, Taylor N, Sitbon M, Battini JL. Cell. 2003;115:449–459. doi: 10.1016/s0092-8674(03)00881-x. [DOI] [PubMed] [Google Scholar]
- Marriott SJ, Semmes OJ. Oncogene. 2005;24:5986–5995. doi: 10.1038/sj.onc.1208976. [DOI] [PubMed] [Google Scholar]
- Maruyama M, Shibuya H, Harada H, Hatakeyama M, Seiki M, Fujita T, Inoue J, Yoshida M, Taniguchi T. Cell. 1987;48:343–350. doi: 10.1016/0092-8674(87)90437-5. [DOI] [PubMed] [Google Scholar]
- Matsumoto J, Ohshima T, Isono O, Shimotohno K. Oncogene. 2005;24:1001–1010. doi: 10.1038/sj.onc.1208297. [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Green PL. Retrovirology. 2009;6:71. doi: 10.1186/1742-4690-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka M, Jeang KT. Nat Rev Cancer. 2007;7:270–280. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- Mazurov D, Ilinskaya A, Heidecker G, Lloyd P, Derse D. PLoS Pathog. 2010;6:e1000788. doi: 10.1371/journal.ppat.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra-Kaushik S, Harding JC, Hess JL, Ratner L. Blood. 2004;104:802–809. doi: 10.1182/blood-2003-11-3967. [DOI] [PubMed] [Google Scholar]
- Miyazato A, Sheleg S, Iha H, Li Y, Jeang KT. J Virol. 2005;79:9346–9350. doi: 10.1128/JVI.79.14.9346-9350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori N, Gill PS, Mougdil T, Murakami S, Eto S, Prager D. Blood. 1996;88:1035–1045. [PubMed] [Google Scholar]
- Morimoto H, Tsukada J, Kominato Y, Tanaka Y. Am J Hematol. 2005;78:100–107. doi: 10.1002/ajh.20259. [DOI] [PubMed] [Google Scholar]
- Nakada K, Yamaguchi K, Furugen S, Nakasone T, Nakasone K, Oshiro Y, Kohakura M, Hinuma Y, Seiki M, Yoshida M. Int J Cancer. 1987;40:145–148. doi: 10.1002/ijc.2910400203. [DOI] [PubMed] [Google Scholar]
- Nejmeddine M, Negi VS, Mukherjee S, Tanaka Y, Orth K, Taylor GP, Bangham CR. Blood. 2009;114:1016–1025. doi: 10.1182/blood-2008-03-136770. [DOI] [PubMed] [Google Scholar]
- Neuveut C, Low KG, Maldarelli F, Schmitt I, Majone F, Grassmann R, Jeang KT. Mol Cell Biol. 1998;18:3620–3632. doi: 10.1128/mcb.18.6.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsugi T, Kumasaka T, Okada S, Urano T. Nat Med. 2007;13:527–528. doi: 10.1038/nm0507-527. [DOI] [PubMed] [Google Scholar]
- Okamura J, Uike N, Utsunomiya A, Tanosaki R. Int J Hematol. 2007;86:118–125. doi: 10.1532/IJH97.07070. [DOI] [PubMed] [Google Scholar]
- Osame M, Igata A. Jpn J Med. 1989;28:412–414. doi: 10.2169/internalmedicine1962.28.412. [DOI] [PubMed] [Google Scholar]
- Pais-Correia AM, Sachse M, Guadagnini S, Robbiati V, Lasserre R, Gessain A, Gout O, Alcover A, Thoulouze MI. Nat Med. 2010;16:83–89. doi: 10.1038/nm.2065. [DOI] [PubMed] [Google Scholar]
- Pan SH, Tai CC, Lin CS, Hsu WB, Chou SF, Lai CC, Chen JY, Tien HF, Lee FY, Wang WB. Carcinogenesis. 2009;30:366–375. doi: 10.1093/carcin/bgn291. [DOI] [PubMed] [Google Scholar]
- Park HU, Jeong JH, Chung JH, Brady JN. Oncogene. 2004;23:4966–4974. doi: 10.1038/sj.onc.1207644. [DOI] [PubMed] [Google Scholar]
- Park HU, Jeong SJ, Jeong JH, Chung JH, Brady JN. Oncogene. 2006;25:438–447. doi: 10.1038/sj.onc.1209059. [DOI] [PubMed] [Google Scholar]
- Patel D, McCance DJ. J Virol. 2010 doi: 10.1128/JVI.00259-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peloponese JM, Jr, Haller K, Miyazato A, Jeang KT. Proc Natl Acad Sci U S A. 2005;102:18974–18979. doi: 10.1073/pnas.0506659103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peloponese JM, Jr, Iha H, Yedavalli VR, Miyazato A, Li Y, Haller K, Benkirane M, Jeang KT. J Virol. 2004;78:11686–11695. doi: 10.1128/JVI.78.21.11686-11695.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peloponese JM, Jr, Jeang KT. J Biol Chem. 2006;281:8927–8938. doi: 10.1074/jbc.M510598200. [DOI] [PubMed] [Google Scholar]
- Peloponese JM, Jr, Yasunaga J, Kinjo T, Watashi K, Jeang KT. J Virol. 2009;83:3238–3248. doi: 10.1128/JVI.01824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peloponese JM, Yeung ML, Jeang KT. Immunol Res. 2006;34:1–12. [PubMed] [Google Scholar]
- Phillips KE, Herring B, Wilson LA, Rickford MS, Zhang M, Goldman CK, Tso JY, Waldmann TA. Cancer Res. 2000;60:6977–6984. [PubMed] [Google Scholar]
- Philpott SM, Buehring GC. J Natl Cancer Inst. 1999;91:933–942. doi: 10.1093/jnci/91.11.933. [DOI] [PubMed] [Google Scholar]
- Pichler K, Schneider G, Grassmann R. Retrovirology. 2008;5:100. doi: 10.1186/1742-4690-5-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pise-Masison CA, Mahieux R, Radonovich M, Jiang H, Duvall J, Guillerm C, Brady JN. AIDS Res Hum Retroviruses. 2000;16:1669–1675. doi: 10.1089/08892220050193128. [DOI] [PubMed] [Google Scholar]
- Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. Proc Natl Acad Sci U S A. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proietti FA, Carneiro-Proietti AB, Catalan-Soares BC, Murphy EL. Oncogene. 2005;24:6058–6068. doi: 10.1038/sj.onc.1208968. [DOI] [PubMed] [Google Scholar]
- Ramadan E, Ward M, Guo X, Durkin SS, Sawyer A, Vilela M, Osgood C, Pothen A, Semmes OJ. Retrovirology. 2008;5:92. doi: 10.1186/1742-4690-5-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch D, Gross S, Harding J, Bokhari S, Niewiesk S, Lairmore M, Piwnica-Worms D, Ratner L. Retrovirology. 2009;6:116. doi: 10.1186/1742-4690-6-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid RL, Lindholm PF, Mireskandari A, Dittmer J, Brady JN. Oncogene. 1993;8:3029–3036. [PubMed] [Google Scholar]
- Riou P, Bex F, Gazzolo L. J Biol Chem. 2000;275:10551–10560. doi: 10.1074/jbc.275.14.10551. [DOI] [PubMed] [Google Scholar]
- Robek MD, Ratner L. J Virol. 1999;73:4856–4865. doi: 10.1128/jvi.73.6.4856-4865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin O, Koch C, Schmitt I, Semmes OJ, Jeang KT, Grassmann R. J Biol Chem. 1998;273:6698–6703. doi: 10.1074/jbc.273.12.6698. [DOI] [PubMed] [Google Scholar]
- Ruggero K, Corradin A, Zanovello P, Amadori A, Bronte V, Ciminale V, D'Agostino DM. Mol Aspects Med. 2010 doi: 10.1016/j.mam.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Saito M, Matsuzaki T, Satou Y, Yasunaga J, Saito K, Arimura K, Matsuoka M, Ohara Y. Retrovirology. 2009;6:19. doi: 10.1186/1742-4690-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago F, Clark E, Chong S, Molina C, Mozafari F, Mahieux R, Fujii M, Azimi N, Kashanchi F. J Virol. 1999;73:9917–9927. doi: 10.1128/jvi.73.12.9917-9927.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou Y, Nosaka K, Koya Y, Yasunaga JI, Toyokuni S, Matsuoka M. Leukemia. 2004;18:1357–1363. doi: 10.1038/sj.leu.2403400. [DOI] [PubMed] [Google Scholar]
- Satou Y, Yasunaga J, Yoshida M, Matsuoka M. Proc Natl Acad Sci U S A. 2006;103:720–725. doi: 10.1073/pnas.0507631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shembade N, Harhaj NS, Liebl DJ, Harhaj EW. EMBO J. 2007;26:3910–3922. doi: 10.1038/sj.emboj.7601823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Higuchi M, Kondo R, Takahashi M, Oie M, Tanaka Y, Aoyagi Y, Fujii M. Retrovirology. 2009;6:83. doi: 10.1186/1742-4690-6-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibon D, Gabet AS, Zandecki M, Pinatel C, Thete J, fau-Larue MH, Rabaaoui S, Gessain A, Gout O, Jacobson S, Mortreux F, Wattel E. J Clin Invest. 2006;116:974–983. doi: 10.1172/JCI27198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Ouyang G, Bao S. J Cell Mol Med. 2005;9:59–71. doi: 10.1111/j.1582-4934.2005.tb00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria C, Estermann FE, Espantman KC, O'Shea CC. Nature. 2010;466:1076–1081. doi: 10.1038/nature09307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Kitao S, Matsushime H, Yoshida M. EMBO J. 1996;15:1607–1614. [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Narita T, Uchida-Toita M, Yoshida M. Virology. 1999;259:384–391. doi: 10.1006/viro.1999.9760. [DOI] [PubMed] [Google Scholar]
- Swaims AY, Khani F, Zhang Y, Roberts AI, Devadas S, Shi Y, Rabson AB. Blood. 2010 doi: 10.1182/blood-2009-07-231050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakin-Fix Y, Azran I, Schavinky-Khrapunsky Y, Levy O, Aboud M. Carcinogenesis. 2006;27:673–681. doi: 10.1093/carcin/bgi274. [DOI] [PubMed] [Google Scholar]
- Takasaki Y, Iwanaga M, Imaizumi Y, Tawara M, Joh T, Kohno T, Yamada Y, Kamihira S, Ikeda S, Miyazaki Y, Tomonaga M, Tsukasaki K. Blood. 2010;115:4337–4343. doi: 10.1182/blood-2009-09-242347. [DOI] [PubMed] [Google Scholar]
- Takatsuki K. Retrovirology. 2005;2:16. doi: 10.1186/1742-4690-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamiya S, Etoh K, Suzushima H, Takatsuki K, Matsuoka M. Blood. 1998;91:3935–3942. [PubMed] [Google Scholar]
- Tan C, Waldmann TA. Cancer Res. 2002;62:1083–1086. [PubMed] [Google Scholar]
- Tanosaki R, Uike N, Utsunomiya A, Saburi Y, Masuda M, Tomonaga M, Eto T, Hidaka M, Harada M, Choi I, Yamanaka T, Kannagi M, Matsuoka M, Okamura J. Biol Blood Marrow Transplant. 2008;14:702–708. doi: 10.1016/j.bbmt.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Taylor GP, Goon P, Furukawa Y, Green H, Barfield A, Mosley A, Nose H, Babiker A, Rudge P, Usuku K, Osame M, Bangham CR, Weber JN. Retrovirology. 2006;3:63. doi: 10.1186/1742-4690-3-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thebault S, Basbous J, Hivin P, Devaux C, Mesnard JM. FEBS Lett. 2004;562:165–170. doi: 10.1016/S0014-5793(04)00225-X. [DOI] [PubMed] [Google Scholar]
- Toulza F, Heaps A, Tanaka Y, Taylor GP, Bangham CR. Blood. 2008;111:5047–5053. doi: 10.1182/blood-2007-10-118539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulza F, Nosaka K, Tanaka Y, Schioppa T, Balkwill F, Taylor GP, Bangham CR. J Immunol. 2010;185:183–189. doi: 10.4049/jimmunol.0903846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukasaki K, Utsunomiya A, Fukuda H, Shibata T, Fukushima T, Takatsuka Y, Ikeda S, Masuda M, Nagoshi H, Ueda R, Tamura K, Sano M, Momita S, Yamaguchi K, Kawano F, Hanada S, Tobinai K, Shimoyama M, Hotta T, Tomonaga M. J Clin Oncol. 2007;25:5458–5464. doi: 10.1200/JCO.2007.11.9958. [DOI] [PubMed] [Google Scholar]
- Utsunomiya A, Miyazaki Y, Takatsuka Y, Hanada S, Uozumi K, Yashiki S, Tara M, Kawano F, Saburi Y, Kikuchi H, Hara M, Sao H, Morishima Y, Kodera Y, Sonoda S, Tomonaga M. Bone Marrow Transplant. 2001;27:15–20. doi: 10.1038/sj.bmt.1702731. [DOI] [PubMed] [Google Scholar]
- Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM. Mol Cell. 2002;9:1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- Virgin HW, Wherry EJ, Ahmed R. Cell. 2009;138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Ohsugi T, Shoda M, Ishida T, Aizawa S, Maruyama-Nagai M, Utsunomiya A, Koga S, Yamada Y, Kamihira S, Okayama A, Kikuchi H, Uozumi K, Yamaguchi K, Higashihara M, Umezawa K, Watanabe T, Horie R. Blood. 2005;106:2462–2471. doi: 10.1182/blood-2004-09-3646. [DOI] [PubMed] [Google Scholar]
- Watashi K, Yeung ML, Starost MF, Hosmane RS, Jeang KT. J Biol Chem. 2010;285:24707–24716. doi: 10.1074/jbc.M109.062976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welles SL, Tachibana N, Okayama A, Shioiri S, Ishihara S, Murai K, Mueller NE. Int J Cancer. 1994;56:337–340. doi: 10.1002/ijc.2910560307. [DOI] [PubMed] [Google Scholar]
- Xiao G, Cvijic ME, Fong A, Harhaj EW, Uhlik MT, Waterfield M, Sun SC. EMBO J. 2001;20:6805–6815. doi: 10.1093/emboj/20.23.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Utsunomiya A, Tobinai K, Tsukasaki K, Uike N, Uozumi K, Yamaguchi K, Yamada Y, Hanada S, Tamura K, Nakamura S, Inagaki H, Ohshima K, Kiyoi H, Ishida T, Matsushima K, Akinaga S, Ogura M, Tomonaga M, Ueda R. J Clin Oncol. 2010;28:1591–1598. doi: 10.1200/JCO.2009.25.3575. [DOI] [PubMed] [Google Scholar]
- Yasunaga J, Jeang KT. Environ Mol Mutagen. 2009;50:733–740. doi: 10.1002/em.20480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasunaga J, Matsuoka M. Rev Med Virol. 2007;17:301–311. doi: 10.1002/rmv.548. [DOI] [PubMed] [Google Scholar]
- Yasunaga J, Taniguchi Y, Nosaka K, Yoshida M, Satou Y, Sakai T, Mitsuya H, Matsuoka M. Cancer Res. 2004;64:6002–6009. doi: 10.1158/0008-5472.CAN-04-1422. [DOI] [PubMed] [Google Scholar]
- Yasunaga J, Sakai T, Nosaka K, Etoh K, Tamiya S, Koga S, Mita S, Uchino M, Mitsuya H, Matsuoka M. Blood. 2001;97:3177–3183. doi: 10.1182/blood.v97.10.3177. [DOI] [PubMed] [Google Scholar]
- Yeung ML, Yasunaga J, Bennasser Y, Dusetti N, Harris D, Ahmad N, Matsuoka M, Jeang KT. Cancer Res. 2008;68:8976–8985. doi: 10.1158/0008-5472.CAN-08-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M. Oncogene. 2005;24:5931–5937. doi: 10.1038/sj.onc.1208981. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Miyoshi I, Hinuma Y. Proc Natl Acad Sci U S A. 1982;79:2031–2035. doi: 10.1073/pnas.79.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Satou Y, Yasunaga J, Fujisawa J, Matsuoka M. J Virol. 2008;82:9359–9368. doi: 10.1128/JVI.00242-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshie O, Fujisawa R, Nakayama T, Harasawa H, Tago H, Izawa D, Hieshima K, Tatsumi Y, Matsushima K, Hasegawa H, Kanamaru A, Kamihira S, Yamada Y. Blood. 2002;99:1505–1511. doi: 10.1182/blood.v99.5.1505. [DOI] [PubMed] [Google Scholar]
- Zane L, Sibon D, Jeannin L, Zandecki M, fau-Larue MH, Gessain A, Gout O, Pinatel C, Lancon A, Mortreux F, Wattel E. Retrovirology. 2010;7:17. doi: 10.1186/1742-4690-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang M, Goldman CK, Ravetch JV, Waldmann TA. Cancer Res. 2003a;63:6453–6457. [PubMed] [Google Scholar]
- Zhang Z, Zhang M, Ravetch JV, Goldman C, Waldmann TA. Blood. 2003b;102:284–288. doi: 10.1182/blood-2002-11-3601. [DOI] [PubMed] [Google Scholar]
- Zhao T, Yasunaga J, Satou Y, Nakao M, Takahashi M, Fujii M, Matsuoka M. Blood. 2009;113:2755–2764. doi: 10.1182/blood-2008-06-161729. [DOI] [PubMed] [Google Scholar]