Abstract
MK5, a member of the MAPK-activated protein kinase family, is highly expressed in heart. Whereas MK2 and MK3 are activated by p38 MAPK, MK5 has also been shown to be activated by ERK3 and ERK4. We studied the regulation of MK5 in mouse heart. mRNA for 5 splice variants (MK5.1-5.5), including the original form (MK5.1), was detected. MK5 comprises 14 exons: exon 12 splicing was modified in MK5.2, MK5.3, and MK5.5. MK5.2 and MK5.5 lacked 6 bases at the 3′-end of exon 12, whereas MK5.3 lacked exon 12, resulting in a frame-shift and premature termination of translation at codon 3 of exon 13. MK5.4 and MK5.5 lacked exons 2-6, encoding kinase subdomains IVI, and were kinase-dead. All 5 MK5 variants were detected at the mRNA level in all mouse tissues examined; however, their relative abundance was tissue-specific. Furthermore, the relative abundance of variant mRNA was altered both during hypertrophy and postnatal cardiac development, suggesting the generation or the stability of MK5 variant mRNAs is subject to regulation. When expressed in HEK293 cells, MK5.1, MK5.2 and MK5.3 were nuclear whereas MK5.4 and MK5.5 were cytoplasmic. A p38 MAPK activator, anisomycin, induced the redistribution of each variant. In contrast, MK5 co-immunoprecipitated ERK3, but not ERK4 or p38α, in control and hypertrophying hearts. GST pull-down assays revealed unbound ERK4 and p38α but no free MK5 or ERK3 in heart lysates. Hence, 1) in heart MK5 complexes with ERK3 and 2) MK5 splice variants may mediate distinct effects thus increasing the functional diversity of ERK3-MK5 signaling.
Keywords: MK5/PRAK, p38 MAP kinase, ERK3, ERK4, alternative splicing, heart, hypertrophy
1. Introduction
Mitogen-activated protein kinases (MAPKs)1 are protein serine/threonine kinases that respond to extracellular stimuli and regulate a broad range of essential cellular activities, including mitosis, metabolism, motility, survival, apoptosis, and differentiation. Five distinct MAPK pathways have been identified in mammals: extracellular signal-regulated kinases (ERKs) 1/2, ERK3/4, ERK5, c-jun amino-terminal kinases (JNKs) (1–3), and p38 MAPKs (α, β, γ, and δ) [1]. Each pathway displays a characteristic hierarchical organization wherein MAPK kinase kinases (MAPKKK) phosphorylate and activate MAPK kinases (MAPKK), which in turn activate their respective MAPKs. Downstream of certain MAPKs lies an additional tier of protein kinases, the MAP kinase-activated protein kinases (MAPKAPKs): these include MK2, MK3 and MK5 (also known as PRAK) [2].
MK2 is a major target for stress-activated p38α and p38β MAPKs. MK2-deficient mice show resistance to endotoxic shock due to impaired production of pro-inflammatory cytokines such as interleukin-6 and tumor necrosis factor-alpha (TNFα) [3]. MK2 and MK3 show 75% identity at the amino acid level [4]. However, although MK2 and MK3 show similarity in their consensus phosphorylation sites [5], not all MK2 substrates have been identified as substrates for MK3. Recent studies in MK2/MK3 double knockout mice show that MK3 further reduces TNF production, expression of p38α, and tristetraprolin (TTP), compared to MK2−/− mice [6], suggesting MK3 and MK2 have overlapping function. MK5, originally identified as a p38 regulated and activated protein kinase (PRAK) [7, 8], was shown to mediate senescence in response to induction of oncogenic ras and upon activation by p38 [9]. Unlike MK2−/− and MK3−/− mice, MK5-deficient mice show no destabilization of p38α [6]. Furthermore, atypical MAPKs ERK3 and ERK4 are binding partners of MK5 [10–12]. ERK3 and ERK4 show 50% homology with ERK1/2 within the kinase domain but have an S-E-G rather than a T-E-Y motif within the activation loop and contain a unique C-terminal extension [13]. Increased expression of ERK3 retains MK5 in cytoplasm and facilitates the activation of MK5 by cis-autophosphorylation [11]. In contrast, ERK4 directly phosphorylates MK5 [14]. Hence, p38, ERK3 and ERK4 may all be involved in regulating MK5 activity in vivo. Little is known about the downstream effectors or physiological function of MK5.
Our present study demonstrates 4 novel splice variants of MK5, referred to herein as MK5.2-MK5.5, in murine cardiac ventricular myocardium. These variants result from both exon skipping and alternative splice site activation during pre-mRNA processing. MK5.4 and MK5.5 lacked a significant part of the kinase catalytic domain whereas MK5.3 lacked the putative ERK3/ERK4 binding domain. When expressed in HEK293 cells, MK5.1, MK5.2, and MK5.3 localized to the nucleus. In contrast, despite retaining NLS and NES sequences, MK5.4 and MK5.5 localized to the cytoplasm in resting cells. Furthermore, the relative abundance of MK5.3 mRNA decreased whereas MK5.4+MK5.5 mRNA increased during cardiac hypertrophy. Finally, we demonstrate that MK5 co-immunoprecipitated ERK3, but not ERK4 or p38α, in both control hearts and those exposed to pressure overload resulting from 3 d of transverse aortic constriction (TAC). Hence, in heart, MK5 may lie downstream of ERK3, rather than p38 or ERK4, and the novel MK5 splice variants may mediate distinct effects downstream of ERK3, thus increasing the functional diversity of the ERK3-MK5 signaling pathway.
2. Materials and methods
2.1. Materials
Membrane grade (reduced) Triton X-100 (TX-100), leupeptin, and PMSF were from Roche Molecular Biochemicals. SDS-Polyacrylamide gel electrophoresis reagents, nitrocellulose, and Bradford protein assay reagents were from Bio-Rad Laboratories. The cAMP-dependent protein kinase inhibitor peptide (PKI) was from the University of Calgary Peptide Synthesis Core Facility. Canine hsp27, cloned into the pET24a expression vector [15], was a gift from Dr. William Gerthoffer (Reno, NV). His6-ERK3-GST, cloned into the pHGST.1 expression vector was a gift from Dr. Sylvain Meloche [16]. Anti-EGFP antibodies were from BD Biosciences. Anti-MK5 antibodies (PRAK A-7) were from Santa Cruz Biotechnology and anti-V5 antibodies were from Invitrogen. HRP-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories. All other reagents were of analytical grade or best grade available. Plasmids were transformed to E. coli competent strain BL21 (DE3) and expression induced by the addition of 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG). GST fusion proteins were purified by affinity chromatography on glutathione Sepharose.
2.2. Cloning and purification of fusion proteins
Total cellular RNA was isolated using RNeasy® Mini kits (Qiagen Inc.) with minor modifications. Briefly, tissue was homogenized in 2 ml of TRIzol reagent (Sigma) using a Polytron at 10,000 RPM (2 × 15 s), 0.2 ml chloroform added and phase separation induced by centrifugation for 15 min at 18,300 g and 4°C. The aqueous phase was reserved and RNA was precipitated by addition of an equal volume of 70% ethanol, and then purified on Qiagen columns according to the manufacturer’s instructions. RNA was quantified by determining absorbance (A) at 260 nm and only samples having an A260/A280 ratio greater than 1.8 were used. First strand cDNA synthesis was performed in a 20 μl reaction volume containing 1 μg of total RNA, 100 ng of random primers, 1X First Strand buffer (50 mM Tris-HCl pH 8.3, 75 mM KCl, 3 mM MgCl2), 0.5 mM dNTP, 10 mM DTT, 40 U RNaseOUT recombinant ribonuclease inhibitor, and 200 U of M-MLV reverse transcriptase (Invitrogen), according to the manufacturer’s protocol. For PCR amplification of MK5, 2 μl aliquots of cDNA were amplified in 50 μl reactions comprising 60 mM Tris-SO4 (pH 9.1), 18 mM (NH4)2SO4, 1.8 mM MgSO4, 200 μM dNTP, 1 μl Elongase (Invitrogen), and 400 nM primers (FWD: CGT GGA TCC ATG TCG GAG GAC AGC GAC ATG GAG; REV: ACG AAT TCC TAC TGG GGC TCG TGG GGA AGG GTC). PCR reactions were: 1 cycle at 94°C for 30 sec, 25 cycles at 94°C for 30 s, 58°C for 30 s, 68°C for 2 min, and 1 cycle at 72°C for 10 min. The forward and reverse primers used for PCR amplification of MK5 were designed according to the mouse MK5 cDNA sequence in the NCBI database (NM_010765). PCR products appeared as a diffuse band when electrophoresed on 1% agarose gels. The bands were excised, extracted using QIAquick Gel Extraction Kits (Qiagen), ligated into the pCR®2.1-TOPO® (Invitrogen) plasmid vector, and transformed into TOP10 competent cells. Several independent clones were sequenced. RT-PCR and cloning were performed 3 times using RNA prepared independently from 3 different animals. In total, 5 distinct mRNAs were obtained. Full length inserts of MK5.1, MK5.2, MK5.3, MK5.4, and MK5.5, in addition to MK5.1-T182A, were subcloned in phase 3′ to the GST coding sequence in pGEX-2T vector (GE Healthcare). Each pGEX-2T-MK5 variant construct was transformed into E. coli competent strain BL21. Transformed E. coli were then selected on DYT agar media contained 100 μg/ml ampicillin. The transformed bacteria were grown in DYT media contained 100 μg/ml ampicillin to an optical density of 0.6 at 600 nm and then expression of the GST-fusion protein was induced by addition of 1 mM IPTG. Following induction, bacteria were maintained at 37°C with shaking (200 rpm) for 6–7 h. Each GST-MK5 variant was purified by affinity chromatography on GST-Trap columns (GE Healthcare). The integrity and purity of each fusion protein was determined by SDS-PAGE.
2.3. RNA analysis
Total RNA was isolated from mouse cardiac ventricles and ventricular myocytes using RNeasy® Mini kits (Qiagen Inc.) with minor modifications. Total cellular RNA was extracted by homogenizing tissue in TRIzol reagent and then purified on RNeasy mini kit columns as per the manufacturer’s protocol. Synthesis of cDNA was as described above. Quantitative real-time PCR (qPCR) was performed using a MX3000P QPCR system (Stratagene). Each amplification reaction mixture (25 μl) contained 6.25 ng cDNA equivalent to reverse transcribed RNA, 300 nM forward and reverse primers, 30 nM ROX, 12.5 μl platinum SYBR green mix (2X) (Invitrogen). qPCR reactions were: 1 cycle at 95°C for 10 min, 40 cycles at 95°C for 30 s, 55°C (68°C for MK5.4 + MK5.5) for 30 s, and 1 cycle at 72°C for 1 min. SYBR green fluorescence was measured at the end of the annealing and extension phases of each cycle. The specificity of each primer pair for the amplicon of interest was verified using the dissociation curve. In addition, amplicons were gel purified and sequenced. The amplification efficiency for each primer pair was determined from a standard curve of 50–3.25 ng reverse transcribed RNA isolated from the heart after two-fold serial dilutions. The efficiency for each primer pair was between 90–110%. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was employed as an internal control. Hence, all samples were normalized to GAPDH, which was amplified in parallel in the same run, with MxPro software (Stratagene). Primers specific for MK5 splice variants were designed using Clone Manager 6 (Sci Ed Software, USA) and based upon the sequence of murine MK5 cDNA (NM_010765). The forward and reverse primers used are shown in Table I.
Table I.
Primers used during real-time qPCR
| Target | Primers |
|---|---|
|
| |
| Total MK5 S | 5′-GAC AGA GGC CCT GGA TAA TGT GC-3′ |
| Total MK5 AS | 5′-GAG TGC AGG GGT TTG AGG CTG-3′ |
|
| |
| MK5.2+5.5 S | 5′-TAT CCT CCC CCA GGC TGG AG-3′ |
| MK5.2+5.5 AS | 5′-CTT CGA TCA CCT GCT TTA CCA CC-3′ |
|
| |
| MK5.3 S | 5′-GCG GGG ATC CAG CAG GCG CAC GCC GAG-3′ |
| MK5.3 AS | 5′-CTT CTC ATC TTC ATT CTC TCC CCA-3′ |
|
| |
| MK5.4+5.5 S | 5′-AGA AAG CCA TCA AGG ACG CCC-3′ |
| MK5.5+5.5 AS | 5′-CCA CAA GTC ACA GCT CTT GTT G-3′ |
|
| |
| ANP S | 5′-GTG CGG TGT CCA ACA CAG A-3′ |
| ANP AS | 5′-TTC TAC CGG CAT CTT CTC CTC-3′ |
|
| |
| β-MHC S | 5′-AGG GTG GCA AAG TCA CTG CT-3′ |
| β-MHC AS | 5′-CAT CAC CTG GTC CTC CTT CA-3′ |
|
| |
| GAPDH S | 5′-CTG CAC CAC CAA CTG CTT AGC-3′ |
| GAPDH AS | 5′-ACT GTG GTC ATG AGC CCT TCC A-3′ |
S, sense; AS, anti-sense; GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; MK, MAPK-activated protein kinase; qPCR, quantitative polymer chain reaction; ANP, atrial natriuretic peptide; β-MHC, β-myosin heavy chain.
2.4. Immunocytofluorescence
The subcellular localization of each MK5 variant was determined by confocal fluorescence microscopy. AgeI-BSRgI inserts from pCR2.1-TOPO were ligated to AgeI-BSRgI digested pIRES-V5-EGFP vector. A total of 3 × 105 HEK293 cells were seeded onto laminin-coated glass coverslips in 12-well culture plates with Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal calf serum and allowed to adhere for 24 h. Cells were then rinsed once with DMEM and transfected with 1.6 μg of the indicated pIRES-MK5-V5-EGFP variant construct using OptiMEM II (Gibco) and Lipofectamine 2000 (Invitrogen) as per manufacturer’s protocol. After 5–6 h of transfection, the medium was replaced with DMEM containing 10% fetal bovine serum for 24 h. At the end of this 24 h period, cells were serum starved for 6 h, treated with or without agonist, rinsed with ice-cold PBS, and fixed for 20 min in ice-cold PBS containing 2% paraformaldehyde (pH 7.2). The fixative was removed by washing three times with ice-cold PBS. Cells were blocked and permeabilized by incubating for 30 min in PBS containing 2% donkey serum and 0.1% (w/v) TX-100. Coverslips were rinsed once with PBS and incubated overnight in a humidified chamber with anti-MK5 antibody diluted (1:100) in PBS containing 1% donkey serum and 0.5% (w/v) TX-100. To remove excess primary antibody, cover slips were washed three times with ice-cold PBS. Coverslips were then incubated for 1 h in PBS containing 1% donkey serum, 0.5% (w/v) TX-100, Alexa fluor 555-conjugated donkey anti-mouse IgG antibody (1:400) and 1.5 μM TO-PRO 3. Finally, coverslips were rinsed three times with ice-cold PBS, drained, and mounted onto glass slides using 15 μl of DABCO/glycerol medium. The intracellular localization of MK5, TO-PRO 3, and EGFP were visualized using an LSM 510 confocal fluorescence microscope (Carl Zeiss, Oberkochen, Germany) as described previously [17–19].
2.5. Immunoprecipitation of endogenous MK5
Mice were sedated with pentobarbital and then sacrificed. Hearts were rapidly removed, snap frozen in liquid nitrogen, and pulverized under liquid nitrogen. The powdered tissue was resuspended in 1.2 ml of ice-cold lysis buffer (50 mM Tris-Cl (pH 7.5 at 4°C), 20 mM β-glycerophosphate, 20 mM NaF, 5 mM EDTA, 10 mM EGTA, 1.0% (w/v) TX-100, 1 mM Na3VO4, 1 μM microcystin LR, 5 mM DTT, 10 μg/ml leupeptin, 0.5 mM PMSF, and 10 mM benzamidine) using a 2 ml Potter-Elvehjem tissue grinder (15 passes, on ice). Homogenates were then cleared of cellular debris by centrifugation for 30 min at 100,000 g and 4°C in a Beckman TLA-100.3 rotor. The supernatants were aliquoted, snap-frozen using liquid nitrogen, and stored at −80°C until use. Prior to immunoprecipitation, 2 μg of anti-MK5 antibody was precoupled to 50 μl of a 25% slurry of protein A/G+-Agarose beads (Santa Cruz Biotechnology) by incubating overnight at 4°C with constant mixing. To remove uncoupled primary antibody, antibody-coated beads were washed three times with lysis buffer prior to immunoprecipitation reactions. Supernatants (2 mg) were added to the antibody-coated beads and incubated overnight at 4°C with constant mixing. Immunoprecipitations were washed three times with lysis buffer and twice with 50 mM Tris-Cl (pH 7.5 at 4°C). The bead pellet was suspended in 20 μl of 2X SDS sample buffer and heated at 70°C for 90 s. The beads were pelleted by a brief centrifugation and the supernatants loaded immediately onto 10–20% acrylamide-gradient SDS-PAGE gels.
2.6. GST-pull down assay
HEK293 cells (6 × 105/35 mm dish) were seeded in DMEM containing 10% fetal bovine serum and allowed to adhere for 24 h. Cells were then rinsed once with DMEM and transfected with 3.2 μg of the indicated pIRES-MK5-V5-EGFP construct using OptiMEM II (Gibco) and Lipofectamine 2000 (Invitrogen) as per the manufacturers’ protocol. After 5–6 h of transfection, the OptiMEM II media was replaced with DMEM containing 10% fetal bovine for 24 h. At the end of this 24 h period, cells were rinsed with ice-cold PBS, lysed in 200 μl lysis buffer (see above) on ice, scraped, transferred to 1.5 ml microcentrifuge tubes, and passed 30–40 times through a 21-gauge needle. Cell lysates were then cleared of cellular debris by centrifugation for 30 min at 100,000 g and 4°C in a Beckman TLA-100.3 rotor. The supernatants were aliquoted, snap-frozen using liquid nitrogen, and stored at −80°C until use. His-ERK3-GST or GST alone (1 μg) was mixed with 150 μg cell lysate and incubated for 1 h at 4°C with constant mixing. Next, 20 μl of a 50% slurry of glutathione Sepharose beads was added and samples incubated for 30 min at 4°C with constant mixing. Pull downs were washed three times with lysis buffer and twice with 50 mM Tris-Cl (pH 7.5 at 4°C). The bead pellet was suspended in 20 ml of 2X SDS sample buffer and heated at 70°C for 90 s. The beads were removed by a brief centrifugation and the supernatants loaded immediately onto 10–20% acrylamide-gradient SDS-PAGE gels.
2.7. Immunoblotting
SDS-PAGE and immunoblotting were performed as described previously [17].
2.8. Transverse aortic constriction
Pressure-overload hypertrophy was induced by transverse aortic constriction (TAC) in 10–12 wk-old male C57BL/6 mice (20–30 g body weight; Charles River), as previously described [20]. Sham-operated mice underwent identical interventions except for the constriction of the aorta. All animal experiments were performed according to the guidelines of the Canadian Council on Animal Care.
2.9. Miscellaneous
Protein concentrations were determined by the Bradford method [21] using bovine serum albumin (BSA) as a standard.
The sequences for the new murine MK5 splice variants shown in supplementary Fig. 1 have been deposited in GeneBank under the accession number; MK5.2 (AY533679), MK5.3 (AY533680), MK5.4 (AY533681), and MK5.5 (AY533682).
3. Results
3.1 Molecular cloning and sequencing of murine MK5 splice variants
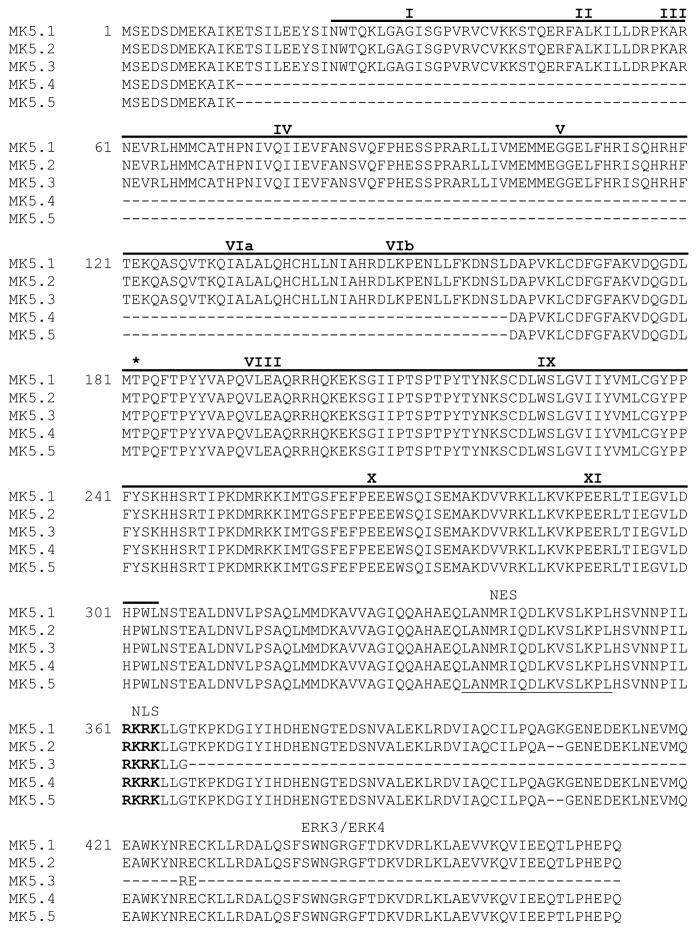
MK5 was cloned from cDNA prepared from murine heart total RNA. Forward and reverse primers were based upon the sequence of murine MK5 cDNA in the NCBI database (NM_010765). PCR products were cloned and sequenced, revealing 5 different products: the form of MK5 originally found in the database and 4 others. An mRNA-to-genomic sequence (MGI:1333110) alignment (http://www.ncbi.nlm.nih.gov/IEB/Research/Ostell/Spidey/) suggested the four products resulted from alternative splicing during RNA maturation. On the basis of their relative size, we refer to the form encoding full-length MK5 as MK5.1 and the novel shorter forms as MK5.2 (AY533679), MK5.3 (AY533680), MK5.4 (AY533681), and MK5.5 (AY533682). Figure 1A shows alignments of the putative splice sites in MK5.1-MK5.5 with the mouse genomic sequence. MK5.1 comprises 14 exons. MK5.4 and MK5.5 lack exons 2, 3, 4, 5, and 6 (supplementary Fig. 1a,b). As shown in supplementary Fig. 1c, MK5.2 and 5.5 lack 6 bases at the 3′ end of exon 12, presumably due to the use of an alternative 3′ splice site within this exon. MK5.3 lacks exon 12 (supplementary Fig. 1c), resulting in a frame shift in translating exon 13 and an in-frame stop codon in the third position. MK5.1 has a 1422 base open reading frame (ORF) and is predicted to encode a 473 amino acid protein (Fig. 1) whereas MK5.2 has a 1416 base ORF and encodes a 471 amino acid protein. MK5.3 has an ORF of 1300 bases, and encodes a 369 amino acid protein. MK5.4 has a 975 base ORF, encoding a 324 amino acid protein. MK5.5 has an ORF of 969 bases and encodes a 322 amino acid protein. Aligning the predicted primary amino acid sequences for all 5 variants revealed that MK5.3 lacks the binding domain for ERK3 [11], and ERK4 [10] whereas MK5.4 and MK5.5 lack conserved protein kinase domains I, II, III, IV, V, VIa, and VIb (Fig. 1, supplementary Fig. 2). In each case, both the nuclear localization sequence (NLS) and nuclear export sequence (NES) remain intact. Furthermore, each variant retained a threonine residue corresponding to Thr-182, the p38/ERK3/ERK4 MAPK phosphorylation site in MK5.1.
Figure 1. Sequence comparison of murine MK5 splice variants.
Figure showns the predicted amino acid sequences. Roman numerals above sequence refer to subdomains (I–IX) conserved in all protein kinases. The catalytic domain (residues 22-304) is indicated by a solid line above the sequence. The consensus nuclear export sequence (NES) is underlined with a solid line and the nuclear localization sequence (NES) is in bold letters. Thr-182, the site of phosphorylation by p38 MAPK is indicated by an asterisk. The ERK3/4 binding site is indicated. Dashes represent gaps.
3.2. In vivo expression of MK5 splice variants in different tissue and MK2 deficient mice
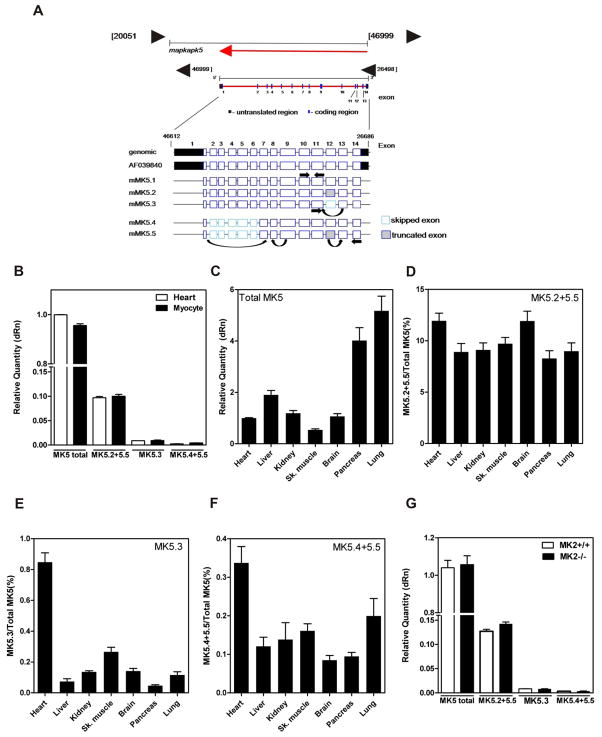
We next wanted to determine the relative quantity of each MK5 variant in heart. Expression of MK5 variants in vivo at the mRNA level was quantified by real time quantitative PCR (qPCR). Primers (Fig. 2A, Table I) were designed to specifically amplify amplicons of equal size from total MK5 (e.g., all 5 variants), MK5.2+MK5.5, MK5.3, and MK5.4+MK5.5. MK5.2+MK5.5 and MK5.4+MK5.5 were quantified together as they shared common deletions, thus precluding selective quantification. A forward primer complementary to sequence in exon 10 and reverse primer in exon 11, which are present in all variants, were employed to quantify total MK5 mRNA. Both MK5.2 and MK5.5 have a 6 base deletion at the exon 12/13 junction. Hence, to quantify mRNA for MK5.2+MK5.5, the forward primer was designed to be selective for the exon 12/13 junction but with a 6-base deletion, whereas the reverse primer was complementary to a region in exon 14. For MK5.3, which lacked exon 12, a forward primer in exon 11 and reverse primer spanning the exon 11/13 junction were employed. MK5.4 and MK5.5 both lack exons 2-6, inclusively, and hence were quantified using a forward primer to the exon 1/7 junction and reverse primer to the exon 8/9 junction. Amplicons for each variant were confirmed by sequencing. In addition, the specificity of each primer was validated by assessing their ability to amplify a product from MK5.1 cDNA in a pGEX-2T plasmid. With the exception of the ‘total’ MK5 primers, none of the primer pairs could amplify products from MK5.1 (results not shown). Using these variant-specific primers, the relative abundance of splice variant mRNAs was measured in mouse heart and isolated ventricular myocytes and expressed relative to the total abundance of MK5 transcripts (Fig. 2B). The variants MK5.2+MK5.5 represented about 10% of total MK5 mRNA, MK5.3 was 1%, and MK5.4+MK5.5 was 0.4%. Hence, in both mouse heart and isolated ventricular myocytes, the most abundant MK5 variant, following MK5.1, was MK5.2.
Figure 2. Detection of MK5 variants in murine tissues.
(A) Schematic representation of the intron-exon structure of MK5 showing the location of the primers used for quantification of MK5 variant mRNAs. (B) The relative abundance of spliced MK5 mRNAs was measured in total RNA isolated from murine heart and cardiac ventricular myocytes by qPCR. (C) The relative abundance of total MK5 in various murine tissues. Abundance of (D) MK5.2+MK5.5, (E) MK5.3, and (F) MK5.4+MK5.5 mRNA expressed as percentage of total MK5. (G) Detection of MK5 variants in hearts from 12-wk old MK2−/− versus MK2+/+ littermate mice. Shown are the mean ± S.E. (n=3).
Liver, kidney, skeletal muscle, brain, pancreas, and lung express high levels of MK5 [22]. Hence, we sought to determine if MK5 splicing was tissue-specific. Alternatively spliced variants of MK5 were observed in all tissue examined. However, the abundance of total MK5 mRNA (Fig. 2C), as well as the relative abundance of each splice variant, was tissue-specific (Fig. 2D–F). In all tissues studied, the most abundant variant, following MK5.1, was MK5.2 (Fig. 2D). Interestingly, in murine heart, the relative abundance of MK5.3 and MK5.4+MK5.5 mRNA was greater in comparison to other tissues examined (Fig. 2E,F), although the expression of total MK5 was lower (Fig. 2C).
MK5 shows a 42% amino acid sequence similarity with MK2 and both are substrates for p38 MAPK. Furthermore, in certain tissues, including the heart, an MK2-deficiency results in reduction in p38α protein levels [4]. Hence we examined the possibility that there may be compensatory changes in expression MK5 in MK2-deficient mice. However, no changes were detected in either MK5 splicing or total MK5 mRNA in hearts from MK2−/− mice (Fig. 2G). This is consistent with previous results showing no change in MK5 expression in MEFs from MK2-deficient mice [23].
3.3 Expression and characterization of recombinant MK5 variants
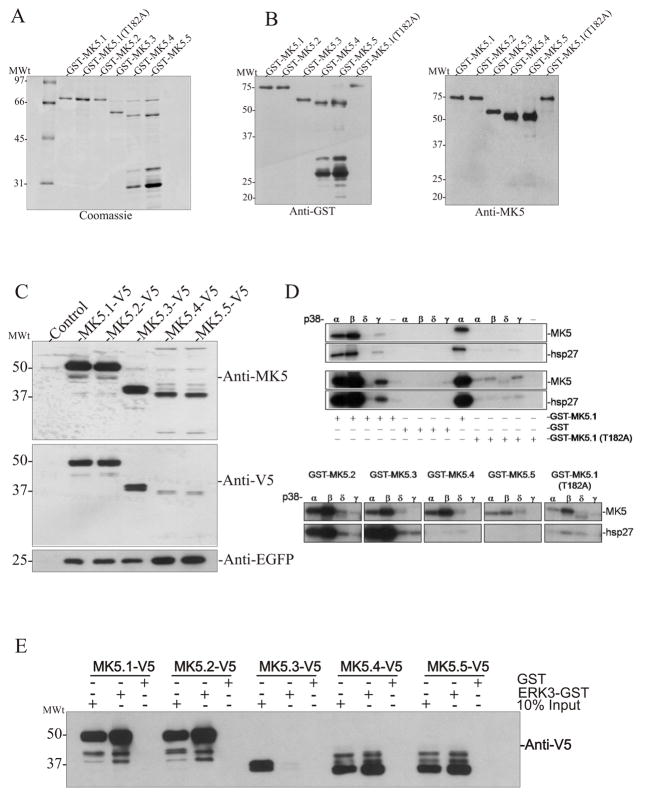
We then sought to characterize the individual MK5 variants at the protein level. cDNAs for all MK5 variants were cloned into the bacterial expression vector pGEX-2T to generate fusion proteins with GST at the N-terminus, expressed in E. coli, and purified by affinity chromatography on glutathione Sepharose. Following purification, SDS-PAGE revealed GST-MK5.1, GST-MK5.2, GST-MK5.3, GST-MK5.4, GST-MK5.5, and GST-MK5.1-T182A fusion proteins of approximately 80-kDa, 80-kDa, 68-kDa, 63-kDa, 63-kDa, and 80-kDa, respectively (Fig. 3A). The identity of each band was confirmed by immunoblotting with both anti-GST and anti-MK5 antibodies (Fig. 3B). Purified preparations of GST-MK5.4 and GST-MK5.5 contained several proteins of lower molecular weight (e.g., 30–35-kDa; Fig. 3A) that were recognized by anti-GST but not by a C-terminal directed anti-MK5 antibody (Fig. 3B). Since the molecular weight of the GST moiety itself is 28-kDa, this suggests that GST-MK5.4 and GST-MK5.5 were expressed as full-length protein in E coli, but were subsequently partially degraded. Similarly, when pIRES-MK5.4-V5-EGFP and pIRES-MK5.5-V5-EGFP were expressed in HEK293 cells, immunoblotting for the V5 epitope tag or MK5 in the lysates of HEK293 cells revealed decreases in band intensity for MK5.4-V5 and MK5.5-V5 (Fig. 3C), whereas EGFP expression was not decreased. Taken together, these results suggest that, although all 5 variants were translated, MK5.4 and MK5.5 may be less stable proteins than the other variants. A similar reduced stability was observed in MK5 exon 6-deficient macrophages [23].
Figure 3. Expression of recombinant MK5 splice variants.
GST-MK5 fusion proteins were expressed in E. coli and purified by chromatography on glutathione Sepharose. Aliquots of purified protein separated on 10–20% acrylamide-gradient SDS-PAGE and either (A) visualized using Coomassie Brilliant blue R250 stain or (B) transferred to nitrocellulose and probed with anti-GST (left) or anti-MK5 (right) antibodies. (C) HEK293 cells were transfected with the indicated pIRES-MK5-V5-EGFP construct. After 24 h, lysates were prepared, separated on 10–20% acrylamide-gradient SDS-PAGE, transferred to nitrocellulose, and probed with an anti-MK5 (top), anti-V5 (middle), or anti-EGFP (bottom) antibodies. (D) Activated p38 was incubated along with purified GST-MK5, hsp27, and radiolabeled [γ32P]ATP. Proteins were separated on 10–20% SDS-PAGE and 32P incorporation was detected by autoradiography. (E) GST pull-down assays were performed on lysates from HEK293 cells transfected with the indicated expression vectors using ether recombinant ERK3-GST or GST alone. Bound MK5 was detected by immunoblotting using an anti-V5 antibody. Equal expression of the MK5 variants was verified by loading 10% total cell lysate input. Numbers at the left indicate the positions of the molecular mass marker proteins (in kDa).
To determine the ability of each MK5 variant to support catalytic activity, we incubated each purified recombinant MK5 with phosphorylated, active p38α, p38β, p38δ, or p38γ plus purified recombinant hsp27 (Fig. 3D). Whereas p38α and p38β phosphorylated each MK5 variant, p38γ and p38δ were much less able to do so. p38-mediated phosphorylation of MK5.1 was virtually absent when Thr-182, the site phosphorylated by p38, was replaced with Alanine. As MK5.4 and MK5.5 lack protein kinase domains I–VI, they were predicted to be catalytically inactive. This was confirmed by in vitro phosphorylation assay where it was observed that, whereas MK5.1, MK5.2, and MK5.3, catalyzed the incorporation of 32PO4 into hsp27, MK5.4 and MK5.5 showed no kinase activity (Figure 3D).
Returning to Figure 1C, an alignment of the primary amino acid sequences of all 5 variants of MK5 reveals that MK5.3 lacks the binding domain for ERK3 [11] and ERK4 [10]. GST-ERK3 pull down assays confirmed that all MK5 variants except MK5.3 bound to GST-ERK3 (Fig. 3E). These results also suggested that the apparent lability of MK5.4 and MK5.5 was likely not a result of these variants’ inability to adopt a native conformation.
3.4 Subcellular localization of exogenously expressed MK5 variants
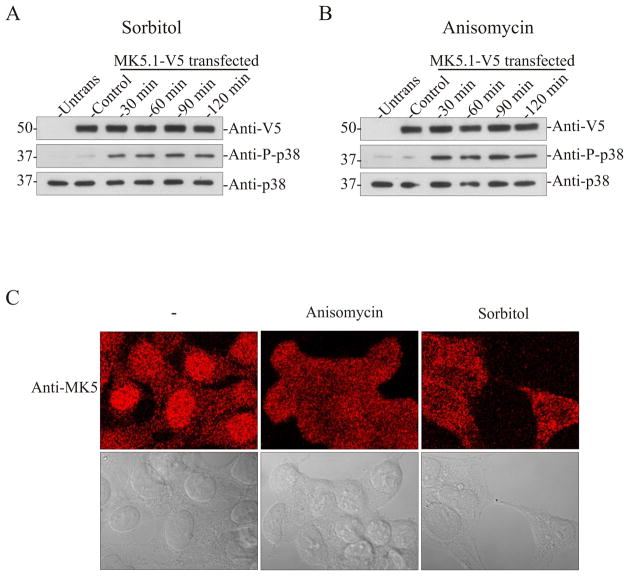
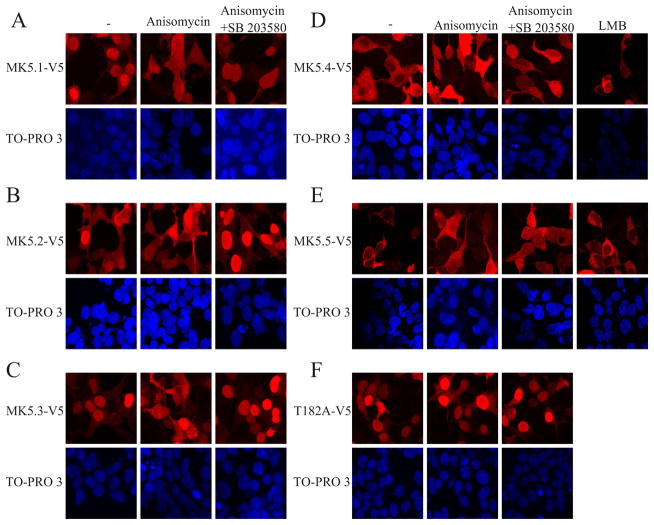
A crucial determinant of the relevance of the MK5 variants in vivo is that they be translated, whereas subcellular localization is a determinant of function. Hence, each MK5 variant was transiently expressed in HEK293 cells. Previous studies have shown that MK5 localizes to the nucleus and is exported to the cytoplasm upon activation of p38 MAPK [12, 22, 24]. First we studied the time-dependence of p38 activation by sorbitol and anisomycin and p38-dependent nuclear export of MK5 by confocal immunofluorescence microscopy. As shown by the increased levels of phospho-p38, the p38 pathway was activated by sorbitol (Fig. 4A) and anisomycin (Fig. 4B) within 30 min and phospho-p38 levels remained elevated for at least 2 h. Hence, HEK293 cells were used to study the p38-dependent changes in the subcellular localization of endogenous MK5. In unstimulated HEK293 cells, endogenous MK5 localized to the nucleus. Following a 2 h stimulation with either sorbitol or anisomycin, MK5 immunoreactivity had translocated into the cytosol (Fig. 4C). As this prolonged stimulation was not associated with breakdown of heterologously expressed MK5.1-V5 (Fig. 4A,B) and induced translocation of endogenous MK5 (Fig. 4C), 2 h stimulations were employed in our subsequent studies of the localization and translocation of each MK5 variant. As with endogenous MK5, heterologously expressed MK5.1-V5 localized to the nucleus in unstimulated cells and was translocated to the cytoplasm upon activation of p38 MAPK with anisomycin (Fig. 5A). Thus, endogenous MK5 and exogenously expressed MK5-V5 behaved in the same manner in HEK293 cells. MK5.1-T182A localized to nucleus and did not translocate to the cytoplasm upon activation of p38 (Fig. 5F), which is consistent with previous results [22] showing that phosphorylation at Thr-182 by p38 is required for export of nuclear MK5. Furthermore, nuclear export of MK5.1-V5 was prevented by preincubation with a p38α/β inhibitor, SB203580 (Fig. 5A). Similar cellular localization and p38α/β-dependent nuclear export were observed for MK5.2-V5 and MK5.3-V5 (Fig. 5B,C). Interestingly, although each of the MK5 variants possessed both nuclear localization sequence (NLS) and nuclear export sequence (NES; Fig. 1, supplementary Fig. 2), MK5.4-V5 and MK5.5-V5 were retained in the cytoplasm in unstimulated cells whereas small amounts of MK5.4-V5 and MK5.5-V5 immunoreactivity was detected within the nucleus following p38 activation (Fig. 5D,E). Leptomycin B, which blocks CRM1 (chromosome region maintenance 1)-dependent nuclear export, and prevents the activation-dependent translocation of both MK2 [25] and MK5.1 [24], did not alter the localization of either MK5.4-V5 or MK5.5-V5, suggesting that their cytoplasmic localization in unstimulated cells was not due to enhanced nuclear export (Fig. 5D,E).
Figure 4. Activation of p38 MAPK induces translocation of endogenous MK5 in HEK293 cells.
HEK293 cells were transfected with pIRES-MK5.1-V5-EGFP. After 24 h cells were serum starved for 6 h and then treated with or without sorbitol (0.3 M) (A) or anisomycin (15 μg/ml) (B) for the indicated times and then lysates were prepared. Activation of p38 was detected by immunoblotting using an anti-phospho p38 antibody. The stability of exogenous MK5 and endogenous p38α during prolonged stress was verified by immunoblotting using anti-V5 and anti-p38α antibodies. (C) Untransfected HEK293 cells were grown for 48 hours, serum starved for 6 h, and then treated with or without sorbitol (0.3 M) or anisomycin (15 μg/ml) for 2 h. Cells were then fixed and MK5 was visualized by staining with an anti-MK5 antibody and an Alexa 555-coupled secondary (anti-mouse) antibody. The upper panels are the fluorescence images whereas the lower panels contain differential interference contrast images of the same cells. Images were acquired using a 63x/1.4 plan-apochromat oil DIC objective lens.
Figure 5. p38 MAPK activation alters the subcellular localization of MK5-V5 variants.
HEK293 cells were transfected with (A) pIRES-MK5.1-V5-EGFP, (B) pIRES-MK5.2-V5-EGFP, (C) pIRES-MK5.3-V5-EGFP, (D) pIRES-MK5.4-V5-EGFP, (E) pIRES-MK5.5-V5-EGFP, or (F) pIRES-MK5.1-T182A-V5-EGFP. After 24 h of transfection cells were serum starved for 6 h and then treated with or without anisomycin (15 μg/ml, 2 h), SB203580 (20 μM), or LMB (3 ng/ml, 10 min). Where indicated, cells were preincubated with SB203580 for 10 min prior to the addition of anisomycin. Following treatment, cells fixed and MK5 was visualized by staining with an anti-V5 antibody and Alexa 555-coupled secondary (anti-mouse) antibody (upper panels). Nuclei were visualized by TO-PRO 3 iodide (1.5 μM) staining (lower panels). Several fields of cells were examined and representative images are shown.
3.5. Effect of cardiac hypertrophy and post-natal development on the expression and splicing of MK5
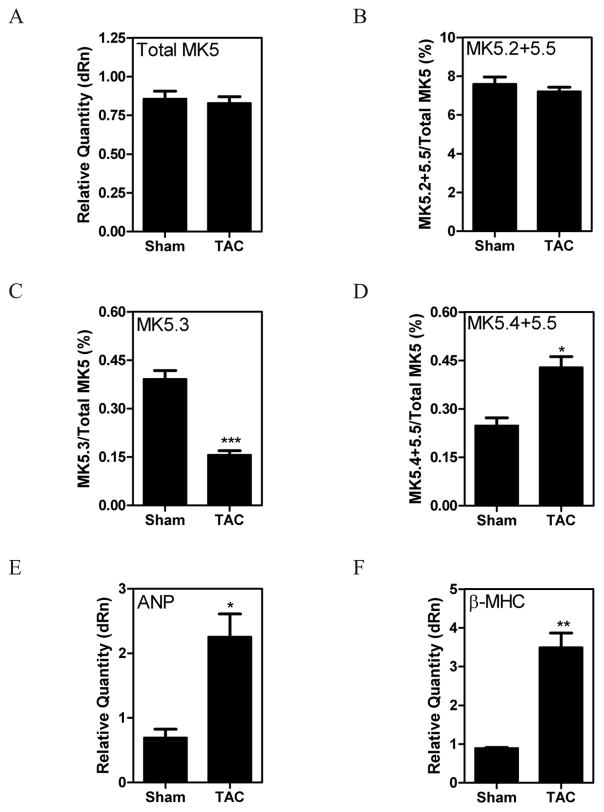
The relative abundance of the MK5 variants differed in a tissue-specific manner (Fig. 2). This suggested that the splicing of MK5 may be subject to regulation. Hence, to determine if cardiomyopathy altered MK5 expression and/or splicing, we examined the effect of cardiac hypertrophy induced by 1 wk of TAC. Hypertrophy is associated with a reexpression of fetal genes, including atrial and B-type natriuretic peptides (ANP, BNP), β-myosin heavy chain (β-MHC) and a-skeletal actin (α-SKA) [26]. The establishment of a hypertrophic response induced by 1-wk TAC was confirmed by the increased expression of atrial natriuretic peptide and β-myosin heavy chain (Fig. 6E,F). After 1 wk of TAC, there was no significant change in total MK5 mRNA (Fig. 6A); however, the abundance of MK5.3 mRNA had decreased whereas that of MK5.4 and MK5.5 had increased (Fig. 6C,D) relative to sham-operated mice. These results demonstrate that the pattern of MK5 splicing was altered during left ventricular hypertrophy, however this change was limited to the lowest abundance variants.
Figure 6. Pressure overload-induced hypertrophy alters MK5 splicing.
Pressure overload hypertrophy was induced in mice by transverse aortic constriction (TAC). Sham animals underwent the identical surgical procedure, however the aortae were not constricted. After 1 wk of TAC, mice were sacrificed, the hearts isolated, and total RNA isolated. MK5 variants were quantified at the mRNA level by qPCR. GAPDH mRNA was employed as an internal control and the abundance of total MK5 mRNA (A) is expressed relative to GAPDH. The abundance of MK5.2+MK5.5 (B), MK5.3 (C), and MK5.4+MK5.5 (D) mRNAs are expressed as a percentage of total MK5 mRNA. Expression of cardiac fetal genes atrial natriuretic peptide (ANP) (E) and β-myosin heavy chain (β-MHC) (F) are expressed relative to GAPDH. Shown are the mean ± S.E. (n=6). ***, p < 0.001: **, p < 0.01: *, p < 0.05: one-way ANOVA with Newman-Keuls post-hoc analysis.
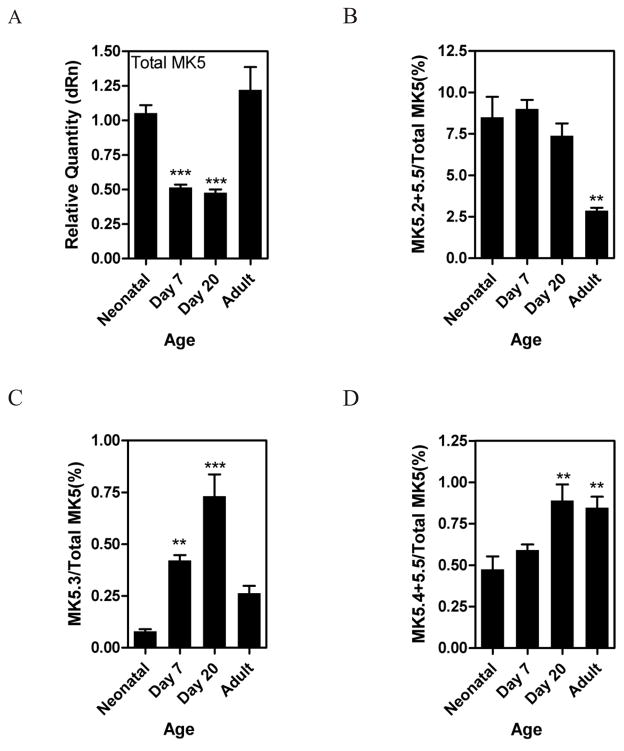
We next investigated whether the expression and/or splicing of MK5 was altered in murine heart during post-natal development. Total MK5 mRNA decreased within the first 7 d, but returned to a level similar to that of the neonatal heart by 8 wk (Fig. 7A). The relative abundance of MK5.2+MK5.5 did not change in young mice, but was significantly lower in the adult, indicating that when total MK5 mRNA increased, the generation of MK5.2+MK5.5 did not rise proportionately (Fig. 7B). A comparison of the data shown in Figures 7B and 7D indicates that the variant changing in Figure 7B, and accounting for up to 8–9% of the total MK5 mRNA, was MK5.2. Interestingly, while the abundance of total MK5 mRNA decreased during the first weeks of life, that of MK5.3 actually increased several-fold during this period (Fig. 7C). However, even at its peak of abundance, the actual amounts of MK5.3 mRNA were low in comparison to the total quantity of MK5 mRNA. A significant 2-fold increase in the abundance of MK5.4+MK5.5 mRNA was also noted during postnatal development, but, as with MK5.3, the actual amount of MK5.4+MK5.5 mRNA remained comparatively low (Fig. 7D).
Figure 7. Regulation of MK5 splicing in heart during postnatal maturation.
Mice were sacrificed at birth, 7 d, 20 d, or 8 wk (adult) of age and mRNA levels of MK5 variants quantified by qPCR. The abundance of total MK5 mRNA (A) is expressed relative to GAPDH. The abundance of MK5.2+MK5.5 (B), MK5.3 (C), and MK5.4+MK5.5 (D) mRNAs are expressed as a percentage of total MK5 mRNA. Shown are the mean ± S.E. (n=3). ***, p < 0.001: **, p < 0.01: one-way ANOVA with Newman-Keuls post-hoc analysis.
3.6 MK5 signalling heart
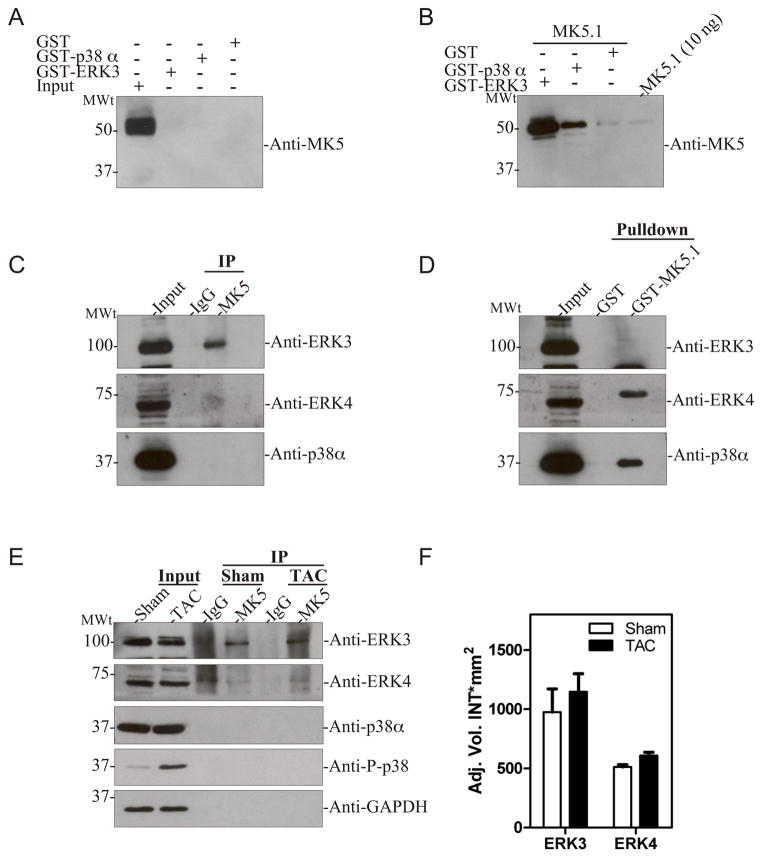
In further experiments, we sought to determine the expression of MK5 splice variants in mouse heart and mouse cardiac ventricular myocytes at the protein level. However, none of the currently available commercial antibodies were able to detect MK5 immunoreactivity in heart lysates or cytosolic extracts (data not shown). Furthermore, attempts to raise a rabbit polyclonal antibody against intact MK5.1 did not result in antisera that was useful in heart. Given that all MK5 variants retained the p38 and, with the exception of MK5.3, ERK3/ERK4 binding sites (Fig. 3E), we decided to investigate endogenous expression of MK5 variants in heart after pull-down with either GST-ERK3 or GST-p38α. Surprisingly, none of endogenous MK5 variants were pulled down with either exogenously added GST-ERK3 or GST-p38α (Fig. 8A). The band detected at 50-kDa in the input lane was also present in controls where the primary antibody was omitted (data not shown) and hence represents a non-specific interaction with the anti-mouse secondary antibody. One plausible explanation for the lack of MK5 binding to exogenous GST-ERK3 or GST-p38α is that endogenous MK5 is bound in a manner such that the docking motifs are occluded and unable to interact with exogenous ERK3 or p38α. This was further studied by adding 1 μg of exogenous MK5.1 (treated with thrombin to remove the GST moiety) to heart lysates (2 mg) prior to the pull-down assay. In this case, MK5 immunoreactivity was detected in the both GST-ERK3 and GST-p38α pull-down assays, with GST-ERK3 being more efficient, suggesting higher affinity binding (Fig. 8B). These data suggest that MK5 is expressed at the protein level in mouse heart, but that it is present in complexes and, hence, unavailable to interact with exogenously added ERK3 or p38α. To determine if endogenous MK5 was complexed with ERK3, ERK4, or p38α, MK5 was immunoprecipitated and the immune complexes probed with anti-ERK3, anti-ERK4, or anti-p38α antisera. ERK3 immunoreactivity was present in MK5 immunoprecipitates, but not the pre-immune serum control (Fig. 8C). In contrast, ERK4 and p38α immunoreactivity were not detected in MK5 immunoprecipitates. We then sought to determine if endogenous ERK3, ERK4, or p38α were free to interact with exogenous GST-MK5. Exogenously added GST-MK5.1 was able to pull down ERK4 and p38α but not ERK3 (Fig. 8D). The slower migrating band observed in the MK5 pull-down is consistent with the hyperphosphorylated form of ERK4 showing higher affinity for MK5 [27, 28]. Based upon our results showing a stable interaction between ERK3 and MK5 in heart, we attempted to demonstrate the expression of MK5 splice variants at the protein level by immunoprecipitating ERK3 and then doing an MK5 immunoblot on the immune complexes; however, the ERK3 antibody was unsuitable for immunoprecipitating endogenous MK5 from heart lysates (data not shown).
Figure 8. MK5 interacts with ERK3 in heart.
(A) GST pull-down assays were performed on murine ventricular myocardium lysates (2 mg) using either GST-p38α, ERK3-GST, or GST alone. Bound MK5 was detected by immunoblotting using an anti-MK5 monoclonal antibody. As a control, 50 μg lysate (input) was loaded in lane 1. (B) To validate the GST-pull-down assay, lysates (2 mg) were ‘spiked’ with 1 μg of thrombin-cleaved MK5.1 prior to performing the pull-down assay using either GST-p38α, ERK3-GST, or GST alone. Bound MK5 was detected by immunoblotting using an anti-MK5 monoclonal antibody. As a positive control, 10 ng of MK5.1 was loaded onto the gel. (C) Murine ventricular myocardium lysates were prepared and MK5 immunoprecipitated. Purified rabbit IgG was employed in control immunoprecipitations. Co-immunoprecipitated ERK3, ERK4, or p38α was detected by immunoblotting. (D) GST pull-down assays were performed on murine ventricular myocardium lysates (2 mg) using either GST-MK5.1 or GST alone. Bound ERK3, ERK4, or p38α was detected by immunoblotting. As a control, 50 μg of lysate (input) was loaded in lane 1. (E) Pressure-overload hypertrophy was induced in mice by transverse aortic constriction (TAC). Sham animals underwent the identical surgical procedure, however the aortae were not constricted. After 3-d of TAC, mice were sacrificed, the ventricular myocardium isolated, lysates prepared, and MK5 immunoprecipitated. Purified rabbit IgG was employed in control immunoprecipitations. Co-immunoprecipitated ERK3, ERK4, p38α, phospho p38, and GAPDH were detected by immunoblotting. Aliquots (50 μg) of lysate from TAC and sham hearts were included as controls (Input). Results shown are representative of 3 animals in each group. (F) Lysates (50 μg) from 3-d TAC and sham hearts were analyzed by SDS-PAGE and immunoblotting using an anti-ERK3 or anti-ERK4 antibodies. Shown are the mean ± S.E. (n=3).
The results shown in Figure 8A-D were obtained in lysates prepared from unstressed hearts. TAC-induced pressure overload results in activation of p38 MAPK that peaks after 3-d of TAC and cardiac hypertrophy within 2-wk (Dingar et al., In Preparation). Hence, to determine if MK5 dissociates from ERK3 and associates with p38 upon activation of the p38 MAPK pathway, we assessed if ERK3, ERK4 or p38α co-immunoprecipitated with MK5 in lysates from 3-d TAC hearts. In both TAC- and sham-operated hearts, ERK3 co-immunoprecipitated with MK5 whereas p38α immunoreactivity was not detected (Figure 8E). Similarly, no association of MK5 with ERK4 was observed in stressed hearts. Immunoblotting using an anti-phospho-p38 antibody confirmed activation of the p38 pathway following 3-d of TAC (Fig. 8E).
ERK3 is an unstable protein and down-regulation of MK5 expression results in reduction of ERK3 protein levels [11, 12]. Hence, if ERK3 were to be displaced from MK5 upon activation of the p38 pathway, one may expect to detect a decrease in ERK3 immunoreactivity. However, following 3-d TAC, no change in ERK3 levels was detected by immunoblotting (Fig. 8F). ERK4 expression was also unaffected (Fig. 8F). These data suggest that in heart, most or all endogenous ERK3 and MK5 were in complexes, whereas at least some of the endogenous pool of ERK4 and p38α was available to interact with the exogenous GST-MK5.1. Furthermore, MK5 complexes included ERK3 but not ERK4 or p38α, and this was independent of the activation status of the p38 MAPK pathway, indicating that, in heart, MK5 is regulated by ERK3 and not ERK4 or p38.
4. Discussion
Mapkapk5, the gene encoding MK5, lies on the negative strand of murine chromosome 5 and comprises 14 exons and 13 introns (Fig. 2A). We have demonstrated 4 novel variants of MK5, which we named MK5.2, MK5.3, MK5.5, and MK5.5. These variants appear to have resulted from a combination of exon skipping and activation of alternative splice sites during RNA maturation. MK5.2 had a 6 base deletion at the 3′-end of exon 12, perhaps arising from the selection of an alternative splice site. In MK5.3, exon 12 was totally absent. The deletion of exon 12 in MK5.3 produced a shift in reading frame in exon 13 resulting in an in-frame stop codon rather than an amino acid in position 3 of exon 13. Thus, the predicted amino acid sequence indicates that MK5.3 lacks C-terminal amino acids 370 through 473. MK5.4 resulted from skipping of exons 2 through 6, inclusively. Finally, MK5.5 lacked both exons 2-6 as well as the 6 bases at the 3′-end of exon 12 also absent from MK5.2. As a result of the deletion of exons 2-6, MK5.4 and MK5.5 lack kinase subdomains I-VIb. All MK5 variants retained both a nuclear export sequence (NES) and a nuclear localization sequence (NLS) within the C-terminus. The NLS overlaps with the p38 docking site (D-domain) [29] and p38-mediated nuclear export requires both phosphorylation of Thr-182 and masking of the NLS [22]. The predicted amino acid sequence suggests that all MK5 variants possess the D-domain that interacts with the CD domain in p38 (Fig. 1). MK5 also interacts with atypical MAPKs ERK3 and ERK4. However, while the D domain in MK5 does not interact with the CD domains in ERK3 and ERK4, it does interact with last 50 amino acids of their C-termini [12, 14]. Hence, with the exception of MK5.3, which lacks 106 amino acids in C-terminus, all MK5 variants retain the binding site for ERK3 and ERK4 (Fig. 2E).
Although, as discussed above, one possible explanation for the 6 base deletion found in MK5.2 and MK5.5 is the activation of an alternative splice site at the 3′-end of exon 12, a search of the NCBI murine database revealed a predicted ORF that appears to encode MK5.2. This predicted ORF has been named “similar to MK5 type 2” (XM_001479242, gene ID 100047833) and lies 96.5-Mbp 3′ to Mapkapk5 on the negative strand of chromosome 5. Hence, an alternative explanation would be that there are only 2 splice variants, which are derived from Mapkapk5 and similar to MK5 type 2 primary RNA transcripts. MK5.3, which lacks exon 12, may arise from both gene transcripts, as the 6 bases missing in the similar to MK5 type 2 ORF are found at the 3′-end of exon 12. MK5.4, which lacks exons 2-6, might result from alternative splicing of the Mapkapk5 transcript whereas MK5.5, which lacks exons 2-6 along with the 3′-end 6 bases in exon 12, would be derived from the similar to MK5 type 2 transcript. As both Mapkapk5 and similar to MK5 type 2 have the same intron/exon structure, with the exception of a 6 base deletion at the 3′-end of exon 12 in similar to MK5 type 2, and both are located on chromosome 5, this would be difficult to prove experimentally.
MK5 is expressed in heart, brain, placenta, lung, liver, skeletal muscle, kidney and pancreas [7]. Analysis by qPCR revealed the presence of mRNA for five MK5 variants in each of the above tissues; however, the abundance of each variant relative to the level of total MK5 was tissue-specific. In each of the tissues examined, the most abundant variant after MK5.1 was MK5.2. The relative expression of MK5.3, which lacks the ERK3 and ERK4 binding domain, was highest in the heart compared to the other tissues examined (Fig. 2C–F). MK2 and MK3 show 42% amino acid sequence homology with MK5 and all are substrates for p38 MAPK. Furthermore, in certain tissues, including the heart, an MK2-deficiency results in reduction in p38α protein levels [4]. Therefore, we examined the possibility that a deficiency in MK2 may induce compensatory changes in the expression and/or splicing of MK5. However, no changes in the abundance of MK5 splice variants were detected (Fig. 2G). These results are consistent with previous results suggesting that there is no functional overlap between MK5 and MK2 [23], despite the structural similarity [8] and evolutionary relatedness [30]. In the heart, the p38 cascade is activated by pro-hypertrophic stimuli including endothelin-1, phenylephrine, angiotensin II, mechanical stretch and pressure overload [31, 32]. The role of MK5 in heart, as well as its substrates, remains to be elucidated. Interestingly, the relative abundance of MK5.3 mRNA was reduced after 1 wk of TAC. Further work is needed to understand the mechanism(s) regulating MK5 splicing during hypertrophic cardiomyopathy and the functional role of MK5 in the myocardium.
MK5 has been shown to localize to the nucleus when inactive and to translocate to the cytosol upon overexpression or activation of p38 [7, 22, 24]. All MK5 variants carry both NLS and NES. When exogenously expressed in HEK293 cells, MK5.1-MK5.3 localized to the nucleus and translocated to the cytoplasm upon p38 activation. Translocation was blocked by SB203580, an inhibitor of p38α/β activity. Hence, the deletion of 2 amino acids in MK5.2 and 106 amino acids in the C-terminus of MK5.3 had no effect on their ability to form a stable interaction with p38 and undergo phosphorylation at Thr-182. Unexpectedly, MK5.4 and MK5.5 remained in the cytoplasm in unstimulated cells and a small amount of immunoreactivity was observed within the nucleus upon p38 activation. The translocation of MK5.4 and MK5.5 into the nucleus was not blocked by SB203580. Furthermore, their cytosolic localization was not altered by leptomycin B, suggesting that this altered localization was due to an inability to enter the nucleus rather than enhanced nuclear export. One possible explanation for the appearance of immunoreactivity within the nucleus following activation of p38 is that cytoplasmic MK5.4 and MK5.5 interact with p38 and enter the nucleus as heterodimers with activated p38.
The mechanisms regulating the activation and subcellular localization of MK5 are currently controversial. Both p38 and atypical MAPKs ERK3 and ERK4 have been implicated. MK5 was originally identified in HeLa cells where it was shown to be activated by p38α and p38β in response to stress, including anisomycin, or overexpression of a constitutively activated form of MKK6 [7]. As with MK2 and MK3, MK5 is primarily in the nucleus in non-stressed cells and relocalizes to the cytosol upon activation of p38. However, nuclear export of MK2 in response to arsenite or sorbitol treatment is more rapid than that of MK5, suggesting differences in regulation [24]. In addition, although stress-induced translocation of MK5 involves activation of p38, expression of kinase-dead p38α or p38β [24] as well as non-activatable p38α [22] also cause nuclear export of MK5, suggesting that the binding of p38 to MK5 may be sufficient to induce translocation. However, whereas p38 stability and stress-induced hsp27-kinase activity are reduced in MK2-deficient mice, no such effects are observed in an MK5 knock-out model [3, 23]. Furthermore, no change in the phosphorylation of endogenous hsp25 (the mouse homolog of hsp27) following treatment with arsenite was observed in MK5−/− MEFs [23]. Finally, immune complex assays fail to show activation of MK5 following treatment of MK5+/+ MEFs with either arsenite or sorbitol [23]. These results suggest MK5 activation may not be regulated by p38 in vivo.
Further understanding of the regulation of MK5 activity came with the studies of atypical MAPKs ERK3 and ERK4, which bind MK5 [10–12]. Increased expression of ERK3 retains MK5 in cytoplasm and facilitates the activation of MK5 by cis-autophosphorylation [11]. In contrast, ERK4 directly phosphorylates MK5 [14]. As ERK3 expression increases in terminally differentiated cells and growth-arrested cells [33], perhaps both ERK3/4 and p38 regulate MK5 activity depending upon the differentiation status and nature of the cell system being studied. As adult cardiac ventricular myocytes are terminally differentiated cells, one would predict that ERK3 regulation of MK5 would predominate in these cells whereas p38 may play a significant role in regulating MK5 activation in other cellular systems. In the present study, in HEK cells, translocation of both endogenous and heterologously expressed MK5 was induced by anisomycin and prevented by the p38α/β inhibitor SB203580. In contrast, in heart, under conditions where p38 activity was minimal, no association of MK5 with p38α was observed. Activation of p38 by TAC-induced pressure overload did not alter this association. Furthermore, no free MK5 was detected and at least some of the endogenous MK5 was associated with ERK3. In both ERK3−/− fibroblasts and HeLa cells following an siRNA-mediated knock-down of ERK3, 40–60% of the endogenous MK5 activity remained [12] and this was shown to be due to the interaction of MK5 with ERK4 [14]. Similarly, in HeLa cells, a knock-down of ERK4 reduces endogenous MK5 activity by 50% whereas simultaneous targeting of both ERK3 and ERK4 reduces MK5 activity by 80% [14]. In contrast, no interaction of MK5 with ERK4 was detected in heart. The presence of MK5-ERK3 complexes in heart raises questions concerning the regulation of MK5 activity. Current literature indicates that ERK3 must be phosphorylated within its activation loop in order to bind MK5 and this binding results in activation and cytosolic relocalization of MK5 [27]. In the present study we were unable to determine the subcellular localization of MK5 in heart due to a lack of suitable antibodies.
The regulation, substrate specificity, and physiological functions of ERK3 and ERK4 are currently poorly understood. Neither is regulated by extracellular stimuli that activate the classical MAPKs ERK1/2, p38s, and JNKs but are constitutively phosphorylated within the S-E-G motif in their activation loop [27, 28]. ERK3 activity appears to be regulated both at the level of expression and protein stability: it is an unstable protein that is constitutively degraded by the ubiquitin-proteosome pathway [33–35]. The ERK3 that is expressed in excess of the abundance of certain ERK3 binding partners appears to be degraded. To date, the only protein known to stabilize ERK3 in vivo is MK5 and deletion of MK5 results in a reduction in ERK3 protein levels [11]. In contrast to ERK3, ERK4 is a more stable protein [10, 14]. Furthermore, whereas ERK3 is detected in both the nucleus and the cytosol, ERK4 is strictly detected in the cytosol [14]. The differences in protein stability and subcellular localization suggest different mechanisms regulate the activity of ERK3-MK5 and ERK4-MK5 complexes. Whereas activation of MK5 by ERK4 is absolutely dependent upon ERK4 being catalytically active, catalytically inactive ERK3 is able to phosphorylate and activate MK5, albeit to a lesser extent that wild-type ERK3 [10, 14]. The implication based upon studies in cell expression systems is that ERK3-MK5 signaling would be constitutively active. It is possible that following overexpression, conditions are created where the abundance of the ERK3-MK5 complex exceeds the cellular content of other proteins that would normally bind to, and regulate the activity of, the ERK3-MK5 complex, resulting in apparent constitutive activation. Along these lines, it was recently shown that the phosphoprotein phosphatase Cdc14A both interacts with ERK3 and MK5 and dephosphorylates a C-terminal fragment of ERK3 following phosphorylation by Cdk1 kinase [36]. It remains to be determined if Cdc14A dephosphorylates ERK3 in vivo or at sites other than those phosphorylated by Cdk1 kinase; however, association with a phosphatase could conceivably provide a means whereby the activation of ERK3-MK5 complexes could be regulated in response to physiological stimuli.
5. Conclusion
We have cloned and expressed 4 novel splice variants of MK5, named MK5.2-MK5.5. These variants were phosphorylated by p38α and p38β in vitro and when exogenously expressed in HEK293 cells each variant was translated and translocated upon activation of endogenous p38 MAPK. The relative abundance of the MK5 variant mRNAs was altered in the ventricular myocardium during pressure-induced hypertrophy and postnatal cardiac development. We demonstrated for the first time that MK5 associates with ERK3, but not ERK4 or p38, in both control hearts and those exposed to pressure-overload resulting from 3-d of TAC. Hence, in heart, MK5 may be downstream of ERK3 signalling, rather than p38 or ERK4. MK5 splice variants may mediate distinct effects downstream of ERK3, thus increasing the functional diversity of the ERK3-MK5 signaling pathway. The physiological significance of ERK3-MK5 signalling in heart remains to be determined.
Supplementary Material
Acknowledgments
We thank Ms. Nadège Moïse for technical assistance and Dr. Maya Khairallah for her thoughtful discussion and critical reading of the manuscript. This work was supported by grants from the Canadian Institutes of Health Research [MOP-77791] and the Fonds de l’Institut de Cardiologie de Montréal (FICM). BGA was a Senior Scholar of the Fondation de la Recherche en Santé du Québec (FRSQ).
Footnotes
Abbreviations: The abbreviations used are: DMSO, dimethylsulfoxide; DTT, dithiothreitol; ERK, extracellular signal-related kinase; FPLC, fast protein liquid chromatography; GST, glutathione S-transferase; MAP kinase, mitogen-activated protein kinase; MK2, MAP kinase-activated protein kinase-2; MK3, MAP kinase-activated protein kinase-3; MK5, MAP kinase-activated protein kinase-5; MBP, myelin basic protein; NES, nuclear export signal; NLS, nuclear localization signal; PKI, cyclic AMP-dependent protein kinase inhibitory peptide; PAGE, polyacrylamide gel electrophoresis; PMSF, phenylmethylsulfonyl fluoride; TAC, transverse arotic constriction; TX-100, Triton X-100.
The authors declare no conflicts of interest.
References
- 1.Chen Z, Gibson TB, Robinson F, Silvestro L, Pearson G, Xu B, Wright A, Vanderbilt C, Cobb MH. Chem Rev. 2001;101(8):2449–2476. doi: 10.1021/cr000241p. [DOI] [PubMed] [Google Scholar]
- 2.Roux PP, Blenis J. Microbiol Mol Biol Rev. 2004;68(2):320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotlyarov A, Neininger A, Schubert C, Eckert R, Birchmeier C, Volk H-D, Gaestel M. Nature Cell Biol. 1999;1:94–97. doi: 10.1038/10061. [DOI] [PubMed] [Google Scholar]
- 4.Gaestel M. Nat Rev Mol Cell Biol. 2006;7(2):120–130. doi: 10.1038/nrm1834. [DOI] [PubMed] [Google Scholar]
- 5.Ludwig S, Engel K, Hoffmeyer A, Sithanandam G, Neufeld B, Palm D, Gaestel M, Rapp UR. Mol Cell Biol. 1996;16(12):6687–6697. doi: 10.1128/mcb.16.12.6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ronkina N, Kotlyarov A, Dittrich-Breiholz O, Kracht M, Hitti E, Milarski K, Askew R, Marusic S, Lin LL, Gaestel M, Telliez JB. Mol Cell Biol. 2007;27(1):170–181. doi: 10.1128/MCB.01456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.New L, Jiang Y, Zhao M, Liu K, Zhu W, Flood LJ, Kato Y, Parry GCN, Han J. EMBO J. 1998;17(12):3372–3384. doi: 10.1093/emboj/17.12.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ni H, Wang XS, Diener K, Yao Z. Biochem Biophys Res Commun. 1998;243:492–496. doi: 10.1006/bbrc.1998.8135. [DOI] [PubMed] [Google Scholar]
- 9.Sun P, Yoshizuka N, New L, Moser BA, Li Y, Liao R, Xie C, Chen J, Deng Q, Yamout M, Dong MQ, Frangou CG, Yates JR, 3rd, Wright PE, Han J. Cell. 2007;128(2):295–308. doi: 10.1016/j.cell.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 10.Kant S, Schumacher S, Singh MK, Kispert A, Kotlyarov A, Gaestel M. J Biol Chem. 2006;281(46):35511–35519. doi: 10.1074/jbc.M606693200. [DOI] [PubMed] [Google Scholar]
- 11.Schumacher S, Laass K, Kant S, Shi Y, Visel A, Gruber AD, Kotlyarov A, Gaestel M. Embo J. 2004;23(24):4770–4779. doi: 10.1038/sj.emboj.7600467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seternes OM, Mikalsen T, Johansen B, Michaelsen E, Armstrong CG, Morrice NA, Turgeon B, Meloche S, Moens U, Keyse SM. Embo J. 2004;23(24):4780–4791. doi: 10.1038/sj.emboj.7600489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coulombe P, Meloche S. Biochim Biophys Acta. 2007;1773(8):1376–1387. doi: 10.1016/j.bbamcr.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Aberg E, Perander M, Johansen B, Julien C, Meloche S, Keyse SM, Seternes OM. J Biol Chem. 2006;281(46):35499–35510. doi: 10.1074/jbc.M606225200. [DOI] [PubMed] [Google Scholar]
- 15.Larsen JK, Gerthoffer WT, Hickey E, Weber LA. Gene. 1995;161(2):305–306. doi: 10.1016/0378-1119(95)00290-m. [DOI] [PubMed] [Google Scholar]
- 16.Coulombe P, Meloche S. Anal Biochem. 2002;310(2):219–222. doi: 10.1016/s0003-2697(02)00319-6. [DOI] [PubMed] [Google Scholar]
- 17.Boivin B, Chevalier D, Villeneuve LR, Rousseau E, Allen BG. J Biol Chem. 2003;278:29153–19163. doi: 10.1074/jbc.M301738200. [DOI] [PubMed] [Google Scholar]
- 18.Boivin B, Lavoie C, Vaniotis G, Baragli A, Villeneuve LR, Ethier N, Trieu P, Allen BG, Hébert TE. Cardiovasc Res. 2006;71(1):69–78. doi: 10.1016/j.cardiores.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 19.Boivin B, Villeneuve LR, Farhat N, Chevalier D, Allen BG. J Mol Cell Cardiol. 2005;38:665–676. doi: 10.1016/j.yjmcc.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 20.Rockman HA, Ross RS, Harris AN, Knowlton KU, Steinhelper ME, Field LJ, Ross J, Jr, Chien KR. Proc Nat Acad Sci USA. 1991;88(18):8277–8281. doi: 10.1073/pnas.88.18.8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradford MM. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 22.New L, Jiang Y, Han J. Mol Biol Cell. 2003;14:2603–2616. doi: 10.1091/mbc.E02-08-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Y, Kotlyarov A, Laaβ K, Gruber AD, Butt E, Marcus K, Meyer HE, Friedrich A, Volk HD, Gaestel M. Mol Cell Biol. 2003;23(21):7732–7741. doi: 10.1128/MCB.23.21.7732-7741.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seternes OM, Johansen B, Hegge B, Johannessen M, Keyse SM, Moens U. Mol Cell Biol. 2002;22(20):6931–6945. doi: 10.1128/MCB.22.20.6931-6945.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engel K, Kotlyarov A, Gaestel M. EMBO J. 1998;17(12):3363–3371. doi: 10.1093/emboj/17.12.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du XJ. Br J Pharmacol. 2007;152(2):169–171. doi: 10.1038/sj.bjp.0707353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deleris P, Rousseau J, Coulombe P, Rodier G, Tanguay PL, Meloche S. J Cell Physiol. 2008;217(3):778–788. doi: 10.1002/jcp.21560. [DOI] [PubMed] [Google Scholar]
- 28.Perander M, Aberg E, Johansen B, Dreyer B, Guldvik IJ, Outzen H, Keyse SM, Seternes OM. Biochem J. 2008;411(3):613–622. doi: 10.1042/BJ20071369. [DOI] [PubMed] [Google Scholar]
- 29.Tanoue T, Maeda R, Adachi M, Nishida E. EMBO J. 2001;20(3):466–479. doi: 10.1093/emboj/20.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 31.Clerk A, Michael A, Sugden PH. J Cell Biol. 1998;142:523–535. doi: 10.1083/jcb.142.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer TA, Ludwig S, Flory E, Gambaryan S, Singh K, Finn P, Pfeffer MA, Kelly RA, Pfeffer JM. Hypertension. 2001;37(5):1222–1228. doi: 10.1161/01.hyp.37.5.1222. [DOI] [PubMed] [Google Scholar]
- 33.Coulombe P, Rodier G, Pelletier S, Pellerin J, Meloche S. Mol Cell Biol. 2003;23(13):4542–4558. doi: 10.1128/MCB.23.13.4542-4558.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coulombe P, Rodier G, Bonneil E, Thibault P, Meloche S. Mol Cell Biol. 2004;24(14):6140–6150. doi: 10.1128/MCB.24.14.6140-6150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmermann J, Lamerant N, Grossenbacher R, Furst P. J Biol Chem. 2001;276(14):10759–10766. doi: 10.1074/jbc.M008567200. [DOI] [PubMed] [Google Scholar]
- 36.Hansen CA, Bartek J, Jensen S. Cell Cycle. 2008;7(3):325–334. doi: 10.4161/cc.7.3.5354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.