Abstract
Flotillin/reggie proteins are membrane-associated proteins present in all kinds of cells and belong to the family of proteins carrying the SPFH (stomatin, prohibitin, flotillin, and HflK/HflC) domain. In addition to this domain of unknown function, flotillin proteins are characterized by the flotillin domain, which is rich in heptad repeats. Bacterial flotillin orthologs have recently been shown to be part of lipid rafts, like their eukaryotic counterparts, and to be involved in signaling events. Double deletions of floT and the gene encoding the second flotillin-like protein in Bacillus subtilis, floA, show strong synthetic defects in cell morphology, motility, and transformation efficiency. The lack of FloT resulted in a marked defect in motility. Using total internal reflection fluorescence (TIRF) microscopy, we show that both proteins localize in characteristic focal structures within the cell membrane, which move in a highly dynamic and random manner but localize independently of each other. Thus, flotillin paralogs act in a spatially distinct manner. Flotillin domains in both FloA and FloT are essential for focal assemblies and for the proper function of flotillins. Both flotillin genes are situated next to genes encoding NfeD proteins. FloT dramatically affects the localization of NfeD2: FloT apparently recruits NfeD2 into the focal assemblies, documenting a close interaction between flotillins and NfeDs in bacteria. In contrast, the localization of NfeD1b is not affected by FloA, FloT, or NfeD2. FloA does not show a spatial connection with the upstream-encoded NfeD1b (YqeZ). Our work establishes that bacterial flotillin-like proteins have overlapping functions in a variety of membrane-associated processes and that flotillin domain-mediated assembly and NfeD proteins play important roles in setting up the flotillin raft-like structures in vivo.
INTRODUCTION
It has become clear in recent years that membranes of many organisms, from bacteria to humans, do not contain uniformly distributed and randomly positioned proteins but have discontinuities in lipid composition and contain distinct protein assemblies (18, 24). The family of flotillin-like proteins has been implicated in the organization of so-called detergent-resistant microdomains (DRMs) or lipid rafts in eukaryotic cells (1, 3, 4, 16). DRMs represent fractions of the membrane in which cholesterol and sphingolipids are specifically enriched. How these lipids contribute to protein function remains unclear, but differences in fluidity, three-dimensional (3D) structure, and net charge likely play a critical role. Flotillins generally serve as marker proteins for DRMs and have been shown to be involved in signal transduction, endocytosis, and cytoskeletal rearrangements. They are thought to act as scaffolding proteins and to couple the membrane with the actin cytoskeleton (20, 22).
Recent progress in the visualization of specific lipids enabled the discovery of lipid subdomains in bacterial membranes. Cardiolipin was shown to be especially enriched in the region of the cell pole, whereas other negatively charged lipids appear to localize in a helical pattern along the longitudinal axis of Bacillus subtilis (2, 21, 23). Biochemical experiments have shown that, as in eukaryotes, detergent-resistant membrane fractions also exist in bacteria (19). Like DRMs, flotillin-like proteins are not restricted to the eukaryotic kingdom. Homologs have been identified in almost all the chromosomes completely annotated so far (9). They usually share a tripartite domain core structure with an N-terminal transmembrane domain, a central characteristic SPFH (named after the proteins stomatin, prohibitin, flotillin, and HflK/HflC) motif, and variable heptad repeat-rich sequences that are predicted to form inter- and/or intramolecular coiled-coil structures, called the flotillin domain. Interestingly, the analysis of the protein content of the detergent-resistant membrane fraction of B. subtilis revealed the presence of a flotillin-like protein, FloT, suggesting a conserved mechanism of DRM-associated protein localization. Further analysis of these bacterial DRMs revealed the accumulation of proteins involved in signaling during biofilm formation in this structure and implied functions for flotillin paralogs in signaling and raft formation (19).
In a study using Escherichia coli cells, the overproduction of a flotillin ortholog has been reported to compensate for the lack of an important membrane-associated protease (6). For B. subtilis flotillin-like proteins, a mild defect in the initiation of the developmental process of sporulation has been described (7), in addition to the effect on biofilm formation (19). Because sporulation is controlled at multiple levels, it is still unclear how flotillins may act in the bacterial cell.
The Gram-positive model organism B. subtilis encodes several SPFH domain-containing proteins, two of which are clearly related to flotillins (9). FloA (YqfA) resembles a flotillin-like protein containing the three core domains, whereas FloT additionally possesses a conserved C-terminal domain of unknown function. In many bacteria, genes encoding flotillin-like proteins often share an operon structure with an NfeD domain-harboring gene (9). This conserved group of proteins is so far restricted to prokaryotes and is characterized by a hydrophobic membrane-spanning region and the soluble beta-rich NfeD domain of unknown function. Recently, the 3D structures of two NfeD domains have been determined by nuclear magnetic resonance (NMR) spectroscopy (14, 27). According to these high-resolution structures and further evidence from secondary-structure predictions, presumably all SPFH-associated NfeD proteins adopt an oligosaccharide/oligonucleotide-binding (OB) fold consisting of a five-stranded β-barrel but lacking conserved residues that are usually necessary for oligonucleotide/oligosaccharide binding of other OB fold proteins. Therefore, SPFH-associated NfeD proteins are thought to mediate their functions via protein-protein interactions.
Some NfeD proteins also contain a serine protease domain preceding the NfeD motif. In B. subtilis, the NfeD domain-harboring proteins YqeZ and YuaF are encoded directly upstream of yqfA (FloA) and yuaG (FloT), respectively. This grouping of flotillin-like proteins and NfeD domain proteins is not restricted to firmicutes but is observed all over the prokaryotic kingdom (9), so a conserved interaction between these two proteins is possible.
We investigated the localization of FloA and FloT, as well as that of NfeD proteins, using total internal reflection fluorescence (TIRF) microscopy. We demonstrate that YuaF (NfeD2) colocalizes with FloT and that the two proteins influence each other's localization behavior, whereas YqeZ (NfeD1b) and FloA seem to act independently. We also show that double deletions of floA and floT have strong synthetic effects on cell shape and motility. Our data reveal that bacterial flotillin-like proteins have nonredundant and highly important functions in cell shape determination and motility and that the coiled-coil domains are essential for the proper localization and function of both flotillin-like proteins.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains are listed in Table 1. B. subtilis and E. coli strains were routinely cultivated at 37°C in Luria-Bertani (LB) medium and were transferred to S750 minimal medium (containing 0.1% glucose, 0.1% glutamate, 0.01% Casamino Acids) (12) prior to microscopy. The growth rate was determined by the optical density at 600 nm (OD600). Antibiotics were added to the media, and transcription of downstream genes was ensured by the addition of 0.5% (wt/vol) xylose when appropriate.
Table 1.
Strains used in this study
| Strain | Relevant genotype | Reference |
|---|---|---|
| PY79 | Wild type | |
| DML1541 | Wild type | |
| Derivatives of DML1541 | ||
| HiHO114 | In-frame deletion of floT | |
| HiHO115 | In-frame deletion of yuaI | |
| HIHO111 | In-frame deletion of yuaF | |
| FD192 | yqfA-yfp (Cmr) ΔfloT in-frame deletion | This study |
| FD174 | floT-yfp (Cmr) ΔyuaI in-frame deletion | This study |
| FD359 | yqfA-yfp (Cmr) ΔyuaF in-frame deletion | This study |
| FD331 | ΔyqfA::Tet ΔfloT in-frame deletion | This study |
| Derivatives of PY79 | ||
| AKR08 | ΔfloT::Spc ΔyqfA::Tet comFA-mCherry (Cmr) | This study |
| FD313 | ΔfloT::Spc | This study |
| FD256 | ΔyqfA::Tet | This study |
| FD296 | ΔyqfA::Tet ΔfloT::Spc | This study |
| FD295 | floT-yfp (Cmr) | This study |
| FD133 | floT-cerulean cfp (Cmr) | This study |
| FD274 | yqeZ-yfp (Cmr) | This study |
| FD350 | floT-cfp yuaF-yfp (Cmr Tetr) | This study |
| FD124 | yuaF-yfp (Cmr) | This study |
| FD127 | yuaF-yfp Cm::Tet | This study |
| FD158 | floT ΔC-yfp (Cmr) | This study |
| FD148 | floT ΔfloT ΔC-yfp (Cmr) | This study |
| FD328 | floT ΔfloT ΔC-yfp (Cmr) yqfA::Tet | This study |
| FD346 | floT-yfp (Cmr) ΔyqfA::Tet | This study |
| FD339 | yqeZ::psG1164 (Cmr) | This study |
| FD357 | yqeZ::psG1164 Cm::Tet | This study |
| FD341 | yqeZ::psG1164 Cm::Tet floT-cfp (Cmr) | This study |
| FD312 | ΔyuaF::Spc | This study |
| FD175 | ΔyuaF::Spc floT-yfp (Cmr) | This study |
| FD329 | floT ΔC-yfp (Cmr) yqfA::Tet | This study |
| FD191 | yqfA-yfp (Cmr) | This study |
| FD343 | yqfA-yfp (Cmr) yqeZ::psG1164 Cm::Tet | This study |
| FD195 | yqfA ΔfloT-yfp (Cmr) | This study |
| FD302 | yqfA ΔfloT-yfp (Cmr) ΔfloT::Spc | This study |
Motility assays.
To monitor differences in spreading on semisolid surfaces, 30 μl of cells of an exponentially growing culture (OD600 = 0.5) were spotted on a solid LB plate without antibiotics containing 0.3% or 0.5% (wt/vol) agar. The plates were incubated at 37°C for 18 h, and colony size was analyzed.
Construction of strains.
Table 1 lists strains. A deletion mutant of yqfA was constructed by applying long flanking sequence homology PCR (LFH-PCR), replacing the gene with a tetracycline (Tet) resistance cassette, thereby giving rise to strain FD256. In-frame deletions of yuaG or yuaF or replacements of yuaG or yuaF with a spectinomycin (Spc) resistance cassette were generated in the B. subtilis DML1541 background (wild type [wt]) and were kindly provided by M. Hinderhofer (University of Konstanz). To transfer the deletions, chromosomal DNA of the Spc-marked strains was purified and B. subtilis PY79 (wt) was transformed with the corresponding DNA selecting for spectinomycin resistance, generating strains FD313 (ΔyuaG::Spc) and FD312 (ΔyuaF::Spc). The ΔyuaG ΔyqfA double-deletion mutant (FD296) was generated by transformation of strain FD313 with chromosomal DNA of strain FD256 (ΔyqfA::Tet), selecting for Spc and Tet resistance. Strain FD331 (ΔyuaG ΔyqfA::Tet) was generated by transformation of B. subtilis DML 1541 harboring the in-frame deletion of yuaG floT (HIHO114) with chromosomal DNA of FD256. Strains FD256 and FD296 behaved identically in all assays, ruling out downstream effects; results are shown for FD296 (PY79 background).
To obtain C-terminal yellow fluorescent protein (YFP) or cyan fluorescent protein (CFP) fusions, the last 500 bp of the coding sequence of the gene of interest were amplified using the oligonucleotides listed in Table S1 in the supplemental material and chromosomal DNA of B. subtilis PY79 as the template. The resulting fragments were cloned into plasmid pSG1164y or pSG1164c, respectively, using the restriction endonucleases shown in Table S1 in the supplemental material. Competent B. subtilis cells were transformed with the plasmids generating the strains listed in Table 1. For dual labeling of YuaG (FloT) and YuaF (NfeD2) (resulting in strain FD350), strain FD133 (FloT-CFP) was transformed with chromosomal DNA of strain FD127 (NfeD2-YFP), selecting for chloramphenicol (Cm) and Tet resistance. To change the resistance from Cm to Tet in vivo, the strains of interest were transformed with plasmid pcm::tet (http://www.BGSC.org) (this applies to strains FD127, FD357, FD118, and FD343), and the resulting colonies were checked for Tet resistance and simultaneous Cm sensitivity.
YqeZ was truncated by insertion of the plasmid pSG1164, which carried 500 bp internal to yqeZ between ApaI and EcoRI sites, into the coding sequence of yqeZ so that the yfp gene in 1164 is not translated. Transcription of downstream genes of the operon was accomplished by addition of 0.5% (wt/vol) xylose to the medium. We generated several truncated versions of FloT and FloA. Five hundred base pairs upstream of the designated junction between the coding sequence of the truncated protein and yfp was amplified and cloned into the plasmid psG1164. PY79 was transformed with the resulting vectors, and clones were selected for Cm resistance. Using this method, strain FD158 (yuaG ΔC-yfp), expressing a fusion protein consisting of the N-terminal 484 amino acids (aa) of FloT and YFP; strain FD148 (yuaG Δflo ΔC-yfp), expressing the first 202 aa of FloT and YFP; and strain FD195 (yqfA ΔfloT-yfp), expressing the N-terminal 235 aa of FloA fused to YFP, were constructed. Strain FD328 (floTΔ floT ΔC-yfp Cmr yqfA-floA::Tet) was generated by transformation of strain FD148 with chromosomal DNA of strain FD256. Strain FD329 (yuaG ΔC-yfp Cmr yqfA::Tet) was constructed by transformation of strain FD158 (yuaG ΔC1-yfp Cmr) with chromosomal DNA of strain FD256 (ΔyqfA::Tet).
For the localization of YuaG-YFP in the absence of FloA (strain FD346), strain FD295 was transformed with chromosomal DNA of strain FD256. To visualize the effect of the absence of YqeZ on FloT, strain FD124 was transformed with chromosomal DNA of strain FD357, giving rise to strain FD341. To observe the effect of the absence of potential interactor proteins of FloT, strain FD295 (FloT-YFP) was transformed with chromosomal DNA of deletion mutant strain FD312 (ΔyuaF-nfeD2), or of HiHo115 (ΔyuaI), giving rise to strains FD175 and FD174, respectively.
The effects of deletions in the yuaFGI operon on the localization of FloA/YqfA-YFP were investigated by transformation of strain HiHo114 (ΔyuaG) or HiHo111 (ΔyuaF) with chromosomal DNA from strain FD191 (FloA/YqfA-YFP), giving rise to strain FD192 (yqfA-yfp Cmr ΔyuaG) and FD359 (yqfA-yfp Cmr ΔyuaF), respectively. Strain FD302 (yqfA ΔfloT-yfp Cmr ΔyuaG::Spc) was generated by transformation of strain FD195 (yqfA ΔfloT-yfp Cmr) with chromosomal DNA of strain FD313 (ΔyuaG::Spc). To elucidate the effects of the absence of flotillin-like proteins on the localization of proteins involved in competence, strain FD313 (ΔyuaG::Spc) was first transformed with chromosomal DNA of strain MiK4 (13) (comFA-mCherry Cmr), and positive clones were subsequently transformed with chromosomal DNA of FD256 (ΔyqfA-floA::Tet), giving rise to strain AKR08.
Immunoblotting.
To compare protein expression of FloT-YFP and truncation variants, cells were grown in LB medium at 37°C until stationary phase. Equal amounts of cells were resuspended in lysis buffer (50 mM EDTA, 100 mM NaCl, 2.5 mg/ml lysozyme, pH 7.5) and incubated for 20 min at 37°C. SDS sample buffer (final concentration, 1×) was added to the lysate and boiled at 95°C for 10 min. Proteins were separated via SDS-PAGE on a 10% phosphonoacetic acid (PAA) gel. The proteins were transferred to a nitrocellulose membrane, applying the semidry Western blotting method. YFP-fused proteins were visualized by a primary polyclonal anti-green fluorescent protein (α-GFP) antiserum (dilution, 1:500) and a secondary goat α-rabbit antiserum (dilution, 1:3,000) that was fused to horseradish peroxidase.
Fluorescence microscopy.
Fluorescence microscopy was performed using a Zeiss Axio Observer Z1 microscope and a 100× objective with a numerical aperture (NA) of 1.45, using a TIRF setup from Visitron (Munich, Germany). Cells were mounted on 1% (wt/vol) agarose pads containing S750 minimal medium on object slides. Images were acquired with an Evolve EM-CCD camera (Photometrix) and were processed with Metamorph 6.3 software (Universal Imaging Corp.). Membranes were stained with FM4-64 (final concentration, 1 nM; Molecular Probes), and DNA was visualized with DAPI (4′,6-diamidino-2-phenylindole) (final concentration, 0.2 ng/ml). YFP fluorophores were excited by exposure to a 488-nm laser beam. Other fluorophores were excited using an appropriate filter set.
RESULTS
Synthetic defects of floT and floA double-mutant cells.
B. subtilis contains at least three genes whose products have high similarity to flotillin/reggie-like proteins: yuaG (FloT), yqfA (FloA), and yqfB. All three proteins contain a predicted N-terminal membrane-spanning alpha helix and the flotillin domain, comprising several heptad repeat-rich regions (see Fig. S1 in the supplemental material). FloT possesses an additional distinct heptad repeat region at the very C terminus. FloA and FloT, but not YqfB, also contain the characteristic SPFH domain between the membrane span and the flotillin domain. However, FloT and FloA are grouped into different SPFH domain subclasses: FloT is related to the SPFH2 subfamily and FloA to the class of stomatin-like SPFH5 proteins (9). The floT gene lies downstream of yuaF, and yqfA (encoding the protein we propose be named FloA) downstream of yqeZ. YuaF and YqeZ are NfeD-like proteins anchored within the cell membrane by several membrane-spanning helices, according to structure prediction. YqeZ also contains a ClpP-like protease domain. The yqfB gene is located downstream of yqfA (floA) and encodes a protein with a single predicted membrane span and a flotillin domain (but no SPFH domain). We generated yuaF (we proposed to name the gene product NfeD2, because it belongs to subfamily 2 of the NfeD proteins) (9) and floT in-frame deletions and a truncation of yqeZ (whose product is named NfeD1b) via the integration of a plasmid containing an internal fragment of yqeZ; the construct includes a xylose-inducible promoter that drives the expression of downstream yqfA-floA and yqfB genes. yqfA-floA and yqfB were deleted by the replacement of the genes with a resistance cassette via double crossover (Table 1).
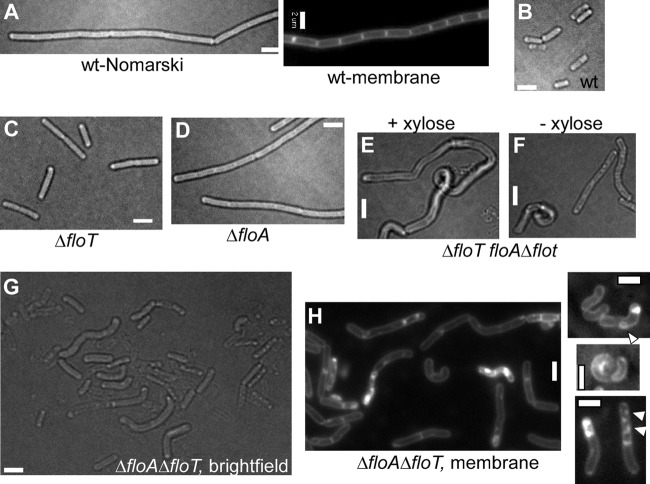
B. subtilis PY79 cells grow as chains of cells during exponential phase (Fig. 1A) or split into single or double cells during stationary phase (Fig. 1B), which was also true for all single-deletion strains. A deletion of floT or floA did not affect culture doubling times, as was reported before (7, 19), or cell size or shape (Fig. 1C and D). Deletions of yuaF, yqeZ, or yqfB also did not show any detectable phenotype (data not shown).
Fig 1.
Effects of deletions of flotillin-encoding genes on cell shape and cell division. (A and B) Chain of growing B. subtilis cells during vegetative growth (membrane stain FM4-64) (A) or in stationary phase (B). (C) ΔfloT cells during exponential growth. (D) ΔfloA cells during exponential growth. (E and F) Cells carrying a floT deletion and a floA truncation, in which the transcription of the yqfB gene is under the control of the xylose promoter, growing in the presence (E) or absence (F) of xylose. Note that there is no visual difference between the shape defects under the two conditions. (G) ΔfloT ΔfloA double-mutant cells during exponential growth. Note that lysed cells appear more transparent than living cells. (H) Membrane stain of exponentially growing ΔfloT ΔfloA double-mutant cells. The arrowheads on the right indicate membrane abnormalities. Bars, 2 μm.
In contrast to the single-mutant cells, floT floA double-mutant cells showed a variety of striking phenotypes. First, double-mutant cells grew more slowly (at least 2-fold) than wild-type cells, and instead of straight chains, the cells grew mostly (>90%) as single, twisted, and irregularly shaped cells during exponential phase (Fig. 1G and H), as well as in stationary phase (data not shown). This striking effect on cell morphology has not been reported before and reveals that flotillin-like proteins are implicated in cell shape maintenance in bacteria. Membrane staining of double-mutant cells also revealed visible irregularities in membrane structure, from vesicle-like structures to extensive spots (Fig. 1H), showing that flotillins affect membrane integrity. To rule out the possibility that the effect of the yqfA floA deletion is caused by a possible downstream effect on yqfB, we combined a yqfB deletion with a floT deletion. Such double-mutant cells did not show any growth or shape defects (data not shown); the cells grew indistinguishably from wild-type cells or from floT mutant cells. We further generated a strain in which floA is truncated and the yqfB gene is driven by the inducible xylose promoter (see below). Cells carrying the floT deletion and the floA truncation grew like and looked similar to cells carrying the double deletion (Fig. 1E and F) irrespective of the absence or presence of xylose, showing that the absence or presence of YqfB does not affect the phenotypes of the floA floT deletion/truncation (this is true for all other phenotypes described below).
Second, flotillin double-mutant cells were strongly impaired in motility, as seen by soft-agar assays (Fig. 2). In a time frame in which PY79 was able to spread over the complete surface of low-percentage (0.3%) agar plates (corresponding to swimming motility), ΔfloT cells showed markedly reduced motility, whereas the floA floT double-deletion strain displayed almost no propagation over the plate (Fig. 2, top). On 0.5% agar plates (corresponding to swarming motility), floT mutant cells showed strongly reduced motility compared to wild-type cells, while double-mutant cells did not show any spreading. Cells lacking FloA showed only a marginal effect on motility (data not shown). Thus, even the single floT deletion leads to reduced motility, and this effect is exacerbated by a double flotillin deletion.
Fig 2.
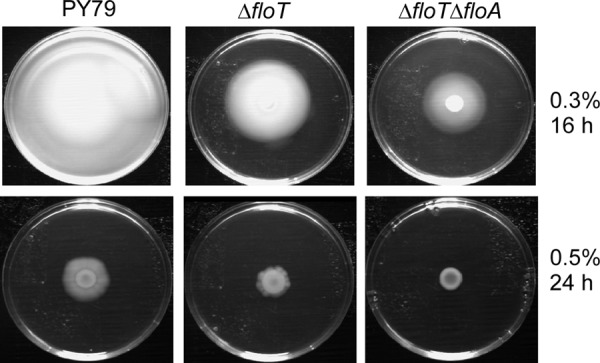
Motility of flotillin mutant cells. Shown are sizes of colonies on solid plates containing 0.3% or 0.5% agar that were grown at 37°C for 16 h or 24 h, respectively.
Third, floT floA double-mutant cells showed a strong defect in gene transfer via competence. Transformation efficiency in cells grown to competence was more than 100-fold reduced compared to wild-type cells or compared to floA or floT single-mutant cells. During the state of competence, 10 to 20% of wild-type cells express the DNA uptake and recombination machineries, which assemble at one or both cell poles (8, 13). We analyzed if the defect in transformation is caused by a general mislocalization of the competence machinery, or by a failure to assemble the complex, by visualizing ComFA-mCherry as a marker for the DNA uptake machinery. ComFA-mCherry localized to the cell poles in 5% of the double-mutant cells grown to competence (see Fig. S2 in the supplemental material) and to 10 to 15% in floA or floT single-mutant cells (data not shown). This finding shows that the assembly of the DNA uptake machinery is 3- to 4-fold reduced in the absence of the two flotillins. This defect cannot explain the marked drop (>100-fold) in transformation efficiency. However, we cannot rule out the possibility that other components of the uptake or recombination machineries fail to assemble within the complexes. In any event, our findings reveal that flotillin double-mutant cells show several severe and pleiotropic phenotypes.
FloT and FloA localize differentially and independently of each other.
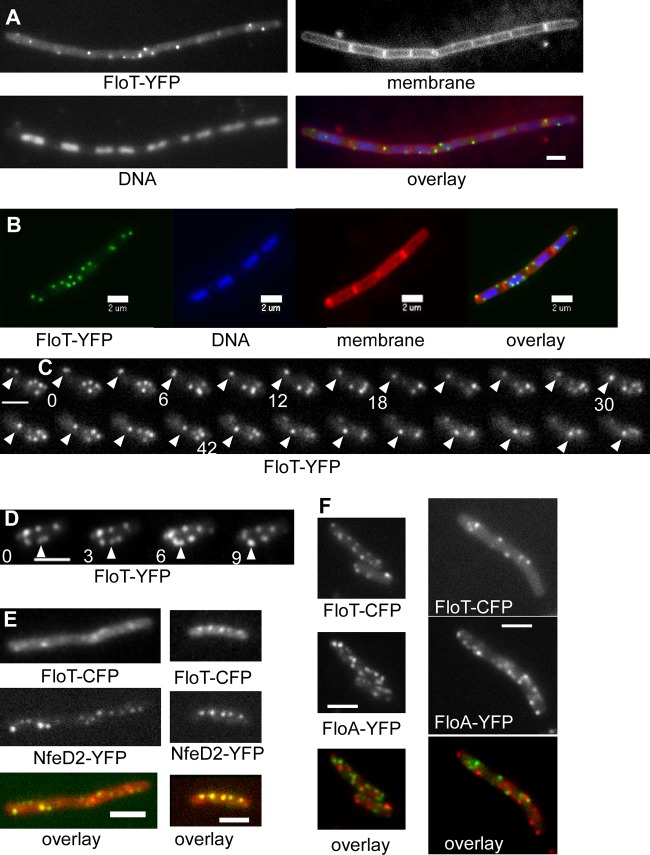
FloT and FloA have been reported to be localized in clustered structures within the cell membrane (19). We generated C-terminal fusions for FloT and FloA that also localize as foci within the cell membrane (Fig. 3A and F). FloT-YFP ΔfloA cells (Fig. 4B) or FloA-YFP ΔfloT cells (Fig. 4M) did not show any of the phenotypes described above and were as motile as cells lacking the fluorescent tags, demonstrating that both fusions are fully functional. In one report, FloT was characterized as forming helical band-like structures (7); in a second report, a random localization pattern could be observed (19). We employed time lapse TIRF microscopy to gain a more detailed picture of the true localization of FloT. We did not find any evidence for helical structures but could observe highly dynamic and random movement of FloT-YFP (Fig. 3C; see Movies S1 and S2 in the supplemental material) and of FloA-YFP (see Movies S3 and S4 in the supplemental material; note that Movie S4 is in epifluorescence mode because the many foci in minimal medium are difficult to follow in TIRF mode). Movies S1 to S4 in the supplemental material show that for both proteins, static as well as highly dynamic foci are present in the same cell, and that movement in any direction is apparent. In Fig. 3C, A FloT-YFP focus can be seen to traverse almost the entire cell length along a lateral path, a pattern frequently observed. In Fig. 3D, two foci can be seen to fuse, as the fluorescence intensity of the resulting focus is higher than that of the two individual foci before fusion. Splitting of foci was also observed (see the movies in the supplemental material), suggesting that flotillin foci consist of interchangeable fractions of FloT-YFP (or FloA-YFP for its structures) that move within the membrane at very different speeds. To obtain more information on the mode of movement of flotillin assemblies, we treated cells with carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) to dissipate the membrane potential and lower intracellular ATP levels. One hour after addition of the uncoupler, foci still moved with dynamics indistinguishable from those of untreated cells (see Movie S5 in the supplemental material), suggesting that flotillin movement is based on Brownian motion rather than on active movement.
Fig 3.
Localization of FloT by TIRF microscopy. (A and B) Localization of FloT-YFP (fully functional fusion) during exponential growth (A) and in stationary phase (B). In the overlays, FloT-YFP is green, the membrane is red, and DNA is blue. (C) Time lapse images of a cell expressing FloT-YFP. A single accumulation (indicated by the arrowheads) moves laterally within the membrane, while polar accumulations show random dynamics. (D) FloT-YFP foci represent structures that are able to fuse; note that the fused focus indicated by the arrowheads is much brighter than the two foci before fusion. The images were taken every 3 s; selected time intervals are labeled in panel C. (E) FloT-CFP and NfeD2-YFP frequently colocalize. For most FloT-CFP foci (red in overlay), there is a corresponding NfeD2-YFP focus (green in overlay; colocalizing proteins appear yellow in the overlay). (F) FloT-CFP and FloA-YFP both localize as dynamic foci, but colocalization (FloT-CFP, red, and FloA-YFP, green in overlay) is rarely observed. Bars, 2 μm.
Fig 4.
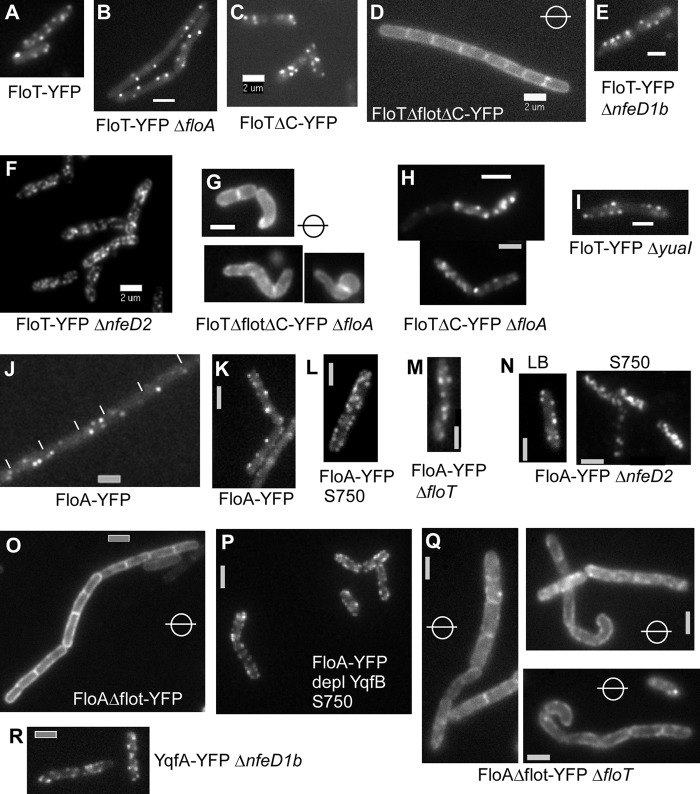
Localization (YFP fluorescence) of FloA and full-length and truncated versions of FloT in mutant backgrounds. Note that the circles with bars in the central plane indicate images acquired in epifluorescence rather than in TIRF mode. (A) FloT-YFP. (B) FloT-YFP in the absence of FloA. (C and D) The localization of FloT-YFP is not altered by the absence of the C-terminal heptad repeat-rich domain of unknown function (C), but FloT-YFP is uniformly distributed upon loss of the flotillin domain (D). (E) FloT-YFP in the absence of NfeD1b. (F) FloT-YFP in the absence of NfeD2 (note the higher number of foci). (G and H) Combination of the FloT-flotillin domain truncation with the floA deletion results a severe cell shape maintenance defect (G), which is not observed in the floA deletion background combined with FloT ΔC-terminal domain construct (H). (I) FloT-YFP in the absence of the downstream yuaI gene. (J and K) FloA-YFP localizes in cells that are incubated in rich medium as distinct accumulations at the membrane during exponential growth (septa between cells are indicated by white lines) (J) and in stationary phase (K). (L) The number of FloA-YFP accumulations is significantly higher in cells growing in minimal medium. (M and N) FloA-YFP localization is not dependent on FloT (M) or on NfeD2 (N). (O) The depletion of the downstream-encoded yqfB gene does not affect the localization of FloA-YFP. (P) The deletion of the flotillin domain of FloA leads to uniform distribution of the truncated YFP-fused protein. (Q) Combination of the FloA-YFP truncation with a deletion of floT alters cell shape. (R) FloA-YFP in cells with nfeD1b deleted. If nothing else is indicated, cells were grown in LB medium. Bars, 2 μm.
The expression of FloT is upregulated during stationary phase in response to the induction of alternative sigma factor W, which regulates many genes, most of which are involved in cell wall-related stress response (5, 10). Indeed, during stationary phase, the cells contained many FloT-YFP foci (Fig. 3B), about 3 to 5 times more than in exponentially growing cells (Fig. 3A). However, the protein was also detectable in exponentially growing cells, albeit only one to a few foci per cell (Fig. 3A), showing that the protein is also expressed in nonstressed cells. Our data thus show that the number of FloT assemblies increases during stationary phase and that the structures are highly dynamic, even during exponential growth.
For both FloT and FloA, we found that the patterns of localization are different in rich and poor media. While in LB rich medium, few (2 to 5) FloA-YFP foci per cell (i.e., on the top surface, because TIRF microscopy only detects signals at the top membrane) were detectable (Fig. 4J), cells growing in minimal medium contained many more foci (from 10 to innumerable) (Fig. 4L), showing that the medium composition affects the quantity of FloA-YFP assemblies. For FloT, this effect was also seen, but to a much lesser extent than for FloA. The number of FloT-YFP foci per cell in exponentially growing cells in rich medium (0.35 foci/cell; 184 cells analyzed) was lower than that for FloA-YFP, and most cells showed only a single FloT-YFP assembly (Fig. 3A); the number increased when the cells entered stationary phase (Fig. 3B). The number of FloT-YFP foci per cell in minimal medium was highly heterogeneous (3.35 on average; 160 cells analyzed) (data not shown).
Next, we generated a strain in which both FloT and FloA could be simultaneously visualized. We used TIRF microscopy to gain fast dual-labeling acquisitions. We found that while 5 to 10% of the foci colocalized, 90% of the signals were clearly separate (Fig. 3F). We analyzed a total of 200 cells to find that, generally, FloT and FloA do not colocalize. This finding shows that the functions of FloA and FloT are spatially separated, although the deletions are synthetic with respect to cell shape, growth, and motility. This agrees with the finding that the pattern of FloT-YFP was not visibly altered in floA mutant cells (Fig. 4B), and vice versa (Fig. 4M). Thus, the two flotillin/reggie proteins do not influence each other's patterns of localization.
The function and specific localization of FloA and FloT depend on their flotillin domains.
Heptad repeat-rich sequences are a hallmark of flotillin-like proteins in prokaryotic and eukaryotic cells (4, 16, 25). We investigated whether these domains play a role in the localization and function of bacterial flotillin-like proteins. FloT contains two discernible regions of heptad repeats, the flotillin domain and the C-terminal domain (see Fig. S1 in the supplemental material). We generated a YFP fusion that lacks the C-terminal domain (ΔC) and a strain in which both the flotillin and C-terminal domains are deleted (ΔfloT ΔC) (see Fig. S1 in the supplemental material). Both truncations were expressed as full-length proteins, as seen by Western blotting (Fig. 5). The ΔC protein was expressed to a lesser extent than the full-length protein (note that equal loading can be seen from the nonspecific bands). The pattern of localization of the single-domain mutant was not visibly different from that of the wild-type protein (Fig. 4C); however, the mutant version lacking both domains was completely uniformly distributed throughout the membrane (Fig. 4D). FloA does not possess a C-terminal domain similar to that of FloT. The lack of the flotillin domain in FloA also resulted in uniform membrane localization (Fig. 4O). The fact that all fusions are produced as full-length proteins and are still targeted to the membrane indicates that there is no problem with protein degradation or folding. These experiments suggest that the assemblies formed by bacterial flotillin paralogs depend on flotillin domain-mediated (most likely coiled-coil) interactions, rather than on lipid structures. It will be interesting to investigate if the lipid composition of the membrane may affect the localization of flotillins.
Fig 5.
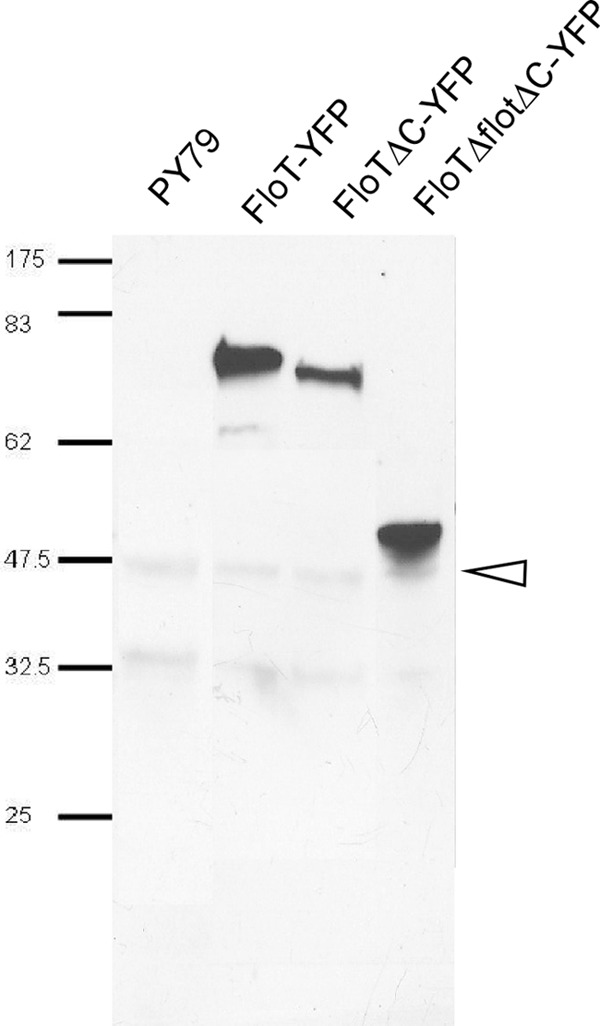
Immunodetection of FloT-YFP and truncation mutants using α-GFP antiserum. The signals present in all lanes corresponding to sizes of 48 (indicated by the arrowhead) and 33 kDa are cross-reactions. The calculated sizes of the three fusion proteins were as follows: FloT-YFP, 82 kDa; FloT ΔC-YFP (with deletion of the C-terminal domain), 79 kDa; FloT ΔfloT ΔC-YFP (with deletion of flotillin and the C-terminal domain), 50 kDa.
We also investigated the important question of whether flotillin assemblies are important for the physiology of the cells. We therefore combined a FloT-flotillin domain truncation with a floA deletion and a FloA-FloT domain truncation with a floT deletion. In both cases, the resulting cells showed slow growth and aberrant cell shape (Fig. 4G and Q, respectively), similar to the double-deletion strain (Fig. 1G and H). These results show that the loss of assembly into the dynamic foci renders FloT and FloA nonfunctional and that the assemblies are important for growth and the physiology of the cells.
FloT and its corresponding NfeD protein, NfeD2, affect each other's patterns of localization.
We investigated the connection between FloT and NfeD2, whose genes are adjacent within the same operon. Like FloT, NfeD2-YFP expressed from the original gene locus formed dynamic membrane-associated foci (Fig. 6A; see Movie S6 in the supplemental material). The number of foci was low during exponential phase, increased in stationary phase, and was lower in rich medium than in minimal medium, similar to FloT (data not shown). We generated a strain that expresses NfeD2-YFP and FloT-CFP from their original gene loci (the FloT-CFP fusion is expressed through a xylose-inducible promoter). Figure 3E shows that the two fusions colocalized at more than 90% frequency. Bearing in mind that many FloT-CFP or NfeD2-YFP proteins are highly dynamic, a change in the filters or between excitation lasers results in a shift of foci that originally colocalized, causing underestimation of the proportion of colocalized proteins. Our results are thus consistent with NfeD2 being present in large amounts within FloT assemblies.
Fig 6.
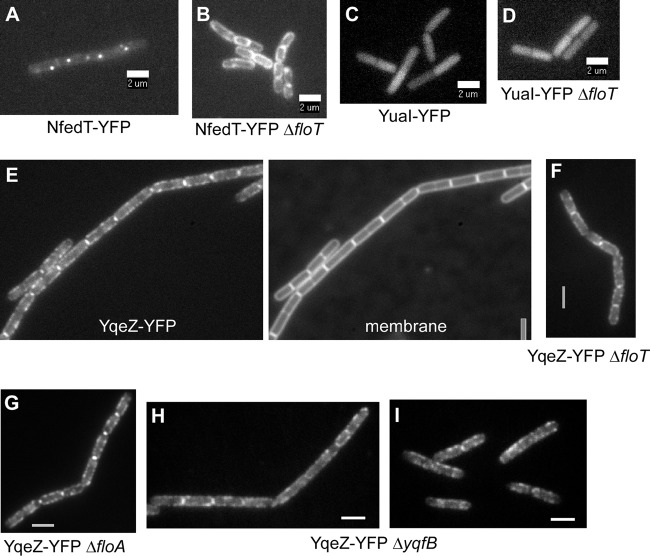
Localization of NfeD2 and NfeD1b. (A) NfeD2-YFP. (B) NfeD2-YFP in cells with floT deleted. (C) NfeD1b-YFP. (D to G) NfeD1b-YFP localization is not affected by the absence of FloT (D), FloA (E), or YqfB (F and G). (F) Exponential growth. (G) Stationary phase. Bars, 2 μm.
To test if FloT and NfeD2 affect each other's patterns of localization, we monitored the localization of FloT-YFP in a yuaF (nfeD2) in-frame deletion strain, and vice versa. Interestingly, in the absence of NfeD2, FloT-YFP formed visibly more foci (Fig. 4F), which were difficult to number and which never reached the fluorescence of FloT-YFP foci in wild-type cells (Fig. 4A) but were also dynamic (data not shown). Therefore, the lack of NfeD2 has an effect on the proper formation of FloT assemblies. However, an nfeD2 floA double-deletion strain did not show a considerable effect on growth or on cell morphology (data not shown), showing that the altered FloT assemblies are still largely functional. Conversely, NfeD2-YFP changed its pattern of localization dramatically in the absence of FloT, because the fusion was completely delocalized throughout the membrane in floT mutant cells (Fig. 6B). Thus, flotillin T and its upstream NfeD protein affect each other's patterns of localization, and FloT recruits—directly or indirectly—NfeD2 into the FloT assemblies.
NfeD1b has a different pattern of localization than FloA and FloT and does not affect their localization.
Interestingly, a spatial interaction was not seen between FloA and YqeZ/NfeD1b, whose genes are also present next to each other in an operon-like structure. The patterns of localization of FloA-YFP were indistinguishable between wild-type (Fig. 4J) and yqeZ nfeD1b mutant cells (Fig. 4N). Strikingly, NfeD1b-YFP showed a different distribution within the cell membrane than FloT and FloA. NfeD1b-YFP expressed from the original gene locus formed many patch-like structures all around the membrane, many of which were not as distinct from each other as the assemblies formed by FloT and FloA (Fig. 6C). A further distinction was the fact that NfeD1b-YFP fluorescence was associated with all completed septa (Fig. 6C), while FloT and FloA were occasionally present at the septum, but not as regularly as NfeD1b. We combined FloT, FloA, or NfeD1b fluorescent protein fusions with gene deletions in all combinations. None of the respective patterns of localization changed in a visually detectable way. NfeD1b localized normally in the absence of FloT (Fig. 6D), FloA (Fig. 6E), or YqfB (Fig. 6F and G), and FloT and FloA did not localize differently in the absence of NfeD1b (Fig. 4E and N), showing that FloT is spatially linked to NfeD2, but not to NfedD1b, while FloA and NfeD1b do not affect each other's localization patterns. Lastly, we visualized FloA in yuaF (nfeD2) mutant cells. FloA formed dynamic foci in the absence of NfeD2 in a manner indistinguishable from that of wild-type cells (Fig. 4R), showing that the interaction between FloT and NfeD2 is not related to FloA, in agreement with a differential localization of FloA and FloT.
We also analyzed whether the YqfB protein, whose deletion also does not show any detectable phenotype, is related to FloA or FloT. First, FloA-YFP did not change its pattern of localization in rich or minimal medium in yqfB-depleted cells (Fig. 4P), and second, the deletion of yqfB in floT mutant cells or the depletion of YqfB in floA mutant cells did not result in any synthetic phenotype (data not shown). A YqfB-YFP fusion showed extremely weak fluorescence within the membrane, so the exact pattern of localization could not be determined (data not shown). However, based on the absence of a detectable phenotype or of a mislocalization of FloA or FloT, our findings suggest that YqfB does not confer a function related to FloA or FloT, in spite of its possession of a flotillin-like domain.
FloT forms membrane patches in S2 cells.
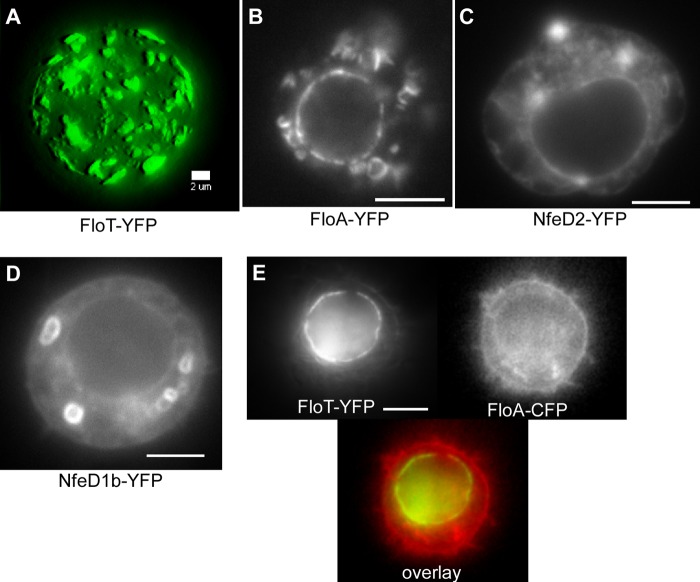
We wondered if FloT may also form membrane patches in a heterologous cell system lacking bacterial protein interaction partners. To use a system that is far diverged from B. subtilis, we expressed the functional FloT-YFP fusion in S2 cells from Drosophila flies (6a). Although FloT-YFP did not contain an added membrane-sorting sequence, it accumulated in the cell membrane (Fig. 7A; see Movie S7 in the supplemental material), but also at internal membrane structures, such as the nucleus (Fig. 7E) (note that this is a rather rare case in which nuclear staining is stronger than that of the cell membrane). Within all membranes, FloT-YFP formed striking patch structures of various sizes. These experiments show that flotillin T does not require protein cofactors from Bacillus to form raft-like structures. For FloA-YFP, we also found membrane decoration that was nonhomogeneous (Fig. 7B), while NfeD2 and NfeD1b showed rather homogeneous membrane localization (Fig. 7C and D). Coexpression of FloT-YFP and FloA-CFP did not show any striking colocalization; often, the proteins showed accumulation in different parts of the S2 cells (Fig. 7E). These experiments reinforce the idea that FloA and FloT form independent assemblies within the cell membrane. Coexpression of FloT-CFP and NfeD2-YFP yielded ambiguous results, as NfeD2 localized throughout the membranes, so that clear colocalization could not be deduced from the images (data not shown).
Fig 7.
Localization of flotillin proteins in S2 cells. (A) FloT-YFP forms extensive patches on the cell membrane upon expression in Schneider S2 cells. (B) FloA-YFP also localizes to all membranes, where it shows a nonuniform distribution. (C and D) NfeD2-YFP (C) and NfeD1b-YFP (D) show homogeneous localization to vesicles and membranes. (E) FloT-YFP and FloA-CFP do not colocalize when coexpressed. Bars, 5 μm (2 μm in panel A).
DISCUSSION
One major contribution of our work is the finding that bacterial flotillin/reggie-like proteins play important roles in a wide range of cellular functions. Previous studies have shown that FloT affects the timing of initiation of sporulation (7) and that two flotillin-like proteins (FloT and FloA) from B. subtilis colocalize with signaling kinase KinC into distinct patches within the cell membrane. In the absence of both flotillin paralogs, KinC mislocalizes and signaling (biofilm formation) is impaired (19). We provide evidence that the function of flotillin-like proteins is not restricted to signaling/differentiation systems but affects cell morphology and cell growth, as well as motility during exponential growth and DNA transformation. Cell shape and membrane structure are highly perturbed after a loss of FloT and FloA, and growth is severely compromised. Moreover, even cells carrying single deletions have strongly reduced motility, which also affects the ability to form biofilms, because motility is an important factor for biofilm formation and maintenance (26). Further, the state of competence, one of the differentiation processes allowing B. subtilis cells to take up external DNA, is affected in that cells are no longer able to become transformed by extracellular DNA. The effect on competence is not caused by an apparent mislocalization of the DNA uptake machinery, which localizes to one or both cell poles (8, 13). Therefore, flotillins do not appear to be general localization factors, such as TipN in Caulobacter crescentus (11, 15). This is in agreement with their dynamic localization: through the use of TIRF microscopy, we demonstrate that FloT and FloA structures are highly dynamic and move at random speeds and in random directions throughout the cell membrane. Based on the rapid and random movement, flotillins may be present at any position within the membrane at any time, in accord with their implication in raft-like structures.
Interestingly, we found that the number of FloA and FloT assemblies is higher in minimal medium than in rich medium. The transcription of floT yuaG is at least partially under the control of a sigma factor W-responsive promoter, and sigma W plays a key role in the response to cell envelope stress (10). The activity of sigma W increases during the transition to stationary phase (5), and indeed, stationary-phase cells contain more FloT and FloA assemblies than exponentially growing cells. Therefore, the number of flotillin assemblies is responsive to the growth phase, as well as to the medium composition.
A second major finding of the work is that FloT and FloA do not colocalize to the same focal assemblies but to distinct dynamic membrane assemblies. Previous studies had hinted that both proteins may be present in the same raft-like structures that contain KinC, and probably other proteins (19), but TIRF microscopy unequivocally shows that FloT and FloA generally localize to different focal assemblies. This finding is important in order to understand that FloT and FloA present distinct structures that may contain only partially overlapping additional proteins and that FloA and FloT do not affect each other in terms of localization and function. The fact that floA and floT deletions are synthetic in all aspects we have investigated suggests that they indeed confer partially redundant functions and thus organize functionally overlapping protein species into differential raft-like structures.
A third important contribution of our work is the demonstration that the loss of flotillin assemblies results in a loss of function and has severe consequences for the physiology of the cells. In the absence of flotillin domains, FloT and FloA localized uniformly throughout the membranes, suggesting that flotillin domains are essential for the formation of focal assemblies and that the formation of focal assemblies depends on protein-protein interactions, possibly mediated through coiled-coil interactions. In the absence of one flotillin, the lack of the flotillin domain in the other paralog resulted in a flotillin “null” phenotype, revealing the importance of flotillin domain-mediated assemblies for several fundamental cellular functions.
Fourth, we show that a member of the NfeD proteins, which are associated with flotillins, based on cooccurrence and frequent genetic linkage on bacterial chromosomes, is indeed associated with flotillins in bacteria and plays a role in the assembly of flotillin structures. In the absence of its cognate NfeD, NfeD2, FloT forms an increased number of assemblies that contain fewer molecules, while in the absence of FloT, NfeD2 is homogeneously dispersed throughout the membrane. Because FloT and NfeD2 colocalize, our data show that NfeD2 is recruited into FloT assemblies and apparently modifies these structures, so that more FloT molecules are present. Thus, there is a visual interplay between a flotillin and an NfeD protein in bacteria, and one function of NfeDs could be the mediation of protein-protein contacts within flotillin assemblies. It remains to be investigated if the interaction between FloT and NfeD2 is a direct or indirect protein-protein interaction. We have not yet identified a corresponding NfeD protein for FloA. NfeD1b, whose gene is adjacent to the floA gene, is different from NfeD2 in that it shows a pattern of localization distinct from those of FloA, FloT, and NfeD2 in the membrane, which is not affected by the lack of any flotillin. Conversely, NfeD1b does not affect the localization of FloA and FloT, indicating that genetic linkage between flotillin and NfeD genes does not necessarily show spatial interaction.
In the absence of NfeD2, FloT still assembles into membrane-associated superstructures. To find out if the structures depend on other proteins in the B. subtilis membrane, we expressed FloT-YFP and FloA-YFP in eukaryotic cells. FloT indeed formed patch structures within the eukaryotic cell membrane, and also within internal membrane systems. FloA also localized nonhomogeneously within S2 cell membranes, but less clearly than FloT, while NfeD2 and NfeD1b showed homogeneous localization. These data support the idea that flotillin-like proteins form raft-like structures that depend on the flotillin domain (likely mediating protein-protein interactions), but not on other bacterial cofactors. Remarkably, FloT assemblies were no longer observed in B. subtilis cells with an altered lipid composition within the cell membrane (upon inhibition of squalene synthesis) (19). Furthermore, overexpression of FloT and of NfeD2 results in altered membrane fluidity (17), revealing an intricate interplay between flotillin rafts and membrane lipids. It will be important to shed light on the function of flotillin/reggie proteins at a molecular level and to elucidate the nature of the effects of membrane lipids on protein localization and assemblies in prokaryotes, as well as in eukaryotes.
Thus, our work sheds important light on the interplay of flotillin-like proteins and NfeD proteins and reveals a wide range of cellular functions for flotillin assemblies, suggesting that lipid raft structures are implicated in many membrane-associated processes in bacteria.
Supplementary Material
ACKNOWLEDGMENTS
We thank Markus Hinderhofer for the generation of the yuaG and yuaF in-frame deletion strains, Anna Katharina Herr for technical assistance, and Stephan Altenburger for Figure S2A in the supplemental material.
This work was supported by the Deutsche Forschungsgemeinschaft (IRTG1474, FOR 929, and MO 1075/3-1).
Footnotes
Published ahead of print 29 June 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Babuke T, Tikkanen R. 2007. Dissecting the molecular function of reggie/flotillin proteins. Eur. J. Cell Biol. 86:525–532 [DOI] [PubMed] [Google Scholar]
- 2. Barak I, Muchova K, Wilkinson AJ, O'Toole PJ, Pavlendova N. 2008. Lipid spirals in Bacillus subtilis and their role in cell division. Mol. Microbiol. 68:1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bickel PE, et al. 1997. Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. J. Biol. Chem. 272:13793–13802 [DOI] [PubMed] [Google Scholar]
- 4. Browman DT, Hoegg MB, Robbins SM. 2007. The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol. 17:394–402 [DOI] [PubMed] [Google Scholar]
- 5. Cao M, et al. 2002. Defining the Bacillus subtilis sigma(W) regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316:443–457 [DOI] [PubMed] [Google Scholar]
- 6. Chiba S, Ito K, Akiyama Y. 2006. The Escherichia coli plasma membrane contains two PHB (prohibitin homology) domain protein complexes of opposite orientations. Mol. Microbiol. 60:448–457 [DOI] [PubMed] [Google Scholar]
- 6a. Dempwolff F, Reimold C, Reth M, Graumann PL. 2011. Bacillus subtilis MreB orthologs self-organize into filamentous structures underneath the cell membrane in a heterologous cell system. PLoS One 6:e27035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Donovan C, Bramkamp M. 2009. Characterization and subcellular localization of a bacterial flotillin homologue. Microbiology 155:1786–1799 [DOI] [PubMed] [Google Scholar]
- 8. Hahn J, Maier B, Haijema BJ, Sheetz M, Dubnau D. 2005. Transformation proteins and DNA uptake localize to the cell poles in Bacillus subtilis. Cell 122:59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hinderhofer M, et al. 2009. Evolution of prokaryotic SPFH proteins. BMC Evol. Biol. 9:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang X, Gaballa A, Cao M, Helmann JD. 1999. Identification of target promoters for the Bacillus subtilis extracytoplasmic function sigma factor, sigma W. Mol. Microbiol. 31:361–371 [DOI] [PubMed] [Google Scholar]
- 11. Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. 2006. Bacterial birth scar proteins mark future flagellum assembly site. Cell 124:1025–1037 [DOI] [PubMed] [Google Scholar]
- 12. Jaacks KJ, Healy J, Losick R, Grossman AD. 1989. Identification and characterization of genes controlled by the sporulation regulatory gene spo0H in Bacillus subtilis. J. Bacteriol. 171:4121–4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaufenstein M, van der Laan M, Graumann PL. 2011. The three-layered DNA uptake machinery at the cell pole in competent Bacillus subtilis cells is a stable complex. J. Bacteriol. 193:1633–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuwahara Y, et al. 2008. The solution structure of the C-terminal domain of NfeD reveals a novel membrane-anchored OB-fold. Protein Sci. 17:1915–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lam H, Schofield WB, Jacobs-Wagner C. 2006. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell 124:1011–1023 [DOI] [PubMed] [Google Scholar]
- 16. Langhorst MF, Reuter A, Stuermer CA. 2005. Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cell Mol. Life Sci. 62:2228–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee YH, Kingston AW, Helmann JD. 2012. Glutamate dehydrogenase affects resistance to cell wall antibiotics in Bacillus subtilis. J. Bacteriol. 194:993–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lingwood D, Simons K. 2010. Lipid rafts as a membrane-organizing principle. Science 327:46–50 [DOI] [PubMed] [Google Scholar]
- 19. Lopez D, Kolter R. 2010. Functional microdomains in bacterial membranes. Genes Dev. 24:1893–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ludwig A, et al. 2010. Flotillin microdomains interact with the cortical cytoskeleton to control uropod formation and neutrophil recruitment. J. Cell Biol. 191:771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matsumoto K, Kusaka J, Nishibori A, Hara H. 2006. Lipid domains in bacterial membranes. Mol. Microbiol. 61:1110–1117 [DOI] [PubMed] [Google Scholar]
- 22. Resnik N, et al. 2011. Desmosome assembly and cell-cell adhesion are membrane raft-dependent processes. J. Biol. Chem. 286:1499–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Romantsov T, et al. 2007. Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol. Microbiol. 64:1455–1465 [DOI] [PubMed] [Google Scholar]
- 24. Simons K, Gerl MJ. 2010. Revitalizing membrane rafts: new tools and insights. Nat. Rev. Mol. Cell Biol. 11:688–699 [DOI] [PubMed] [Google Scholar]
- 25. Solis GP, et al. 2007. Reggie/flotillin proteins are organized into stable tetramers in membrane microdomains. Biochem. J. 403:313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Verstraeten N, et al. 2008. Living on a surface: swarming and biofilm formation. Trends Microbiol. 16:496–506 [DOI] [PubMed] [Google Scholar]
- 27. Walker CA, Hinderhofer M, Witte DJ, Boos W, Moller HM. 2008. Solution structure of the soluble domain of the NfeD protein YuaF from Bacillus subtilis. J. Biomol. NMR 42:69–76 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.