Abstract
Streptococcus pneumoniae is a facultative pathogen inhabiting the nasopharynx of humans where it is exposed to a range of antimicrobial peptides (AMPs) of the innate immune response. It is possible therefore that the susceptibility of strains to AMPs plays a role in determining their ability to colonize, and furthermore, that AMPs could mediate competitive interactions between co-colonizing genotypes. However, little is known about patterns of natural variation in AMP susceptibility of S. pneumoniae, and it is unclear whether the susceptibilities of an isolate to multiple human AMPs are correlated. We tested this by characterizing the susceptibility of 31 S. pneumoniae natural isolates to human neutrophil peptide (HNP-1) (α-defensin) and LL-37 (cathelicidin). We observed significant variation in susceptibility between isolates to both AMPs, and in the majority of isolates, susceptibilities to HNP-1 and LL-37 were uncorrelated. Clinical isolates were more susceptible to AMPs than were carriage isolates. The polysaccharide capsule of S. pneumoniae is thought to protect cells against AMPs. However, serotype alone could not explain the observed variation in susceptibility suggesting that genetic background plays an equally important role. We tested directly whether AMPs could mediate competition between isolates using competition experiments in the presence and absence of AMPs. These experiments demonstrated that AMPs could indeed reverse the outcome of competition between selected isolates. AMP-mediated competition could therefore contribute to the maintenance of intraspecific genetic diversity in S. pneumoniae.
Keywords: antimicrobial peptides, diversity, Streptococcus pneumoniae
1. Introduction
Streptococcus pneumoniae is a facultative pathogen inhabiting the nasopharynx of humans; it causes a wide range of infections, from otitis media and acute sinusitis to life-threatening pneumonia, septicaemia and meningitis. Despite significant advances in our understanding of the mechanisms of pneumococcal pathogenesis and epidemiology, together with widely successful antibiotic treatment and vaccination programmes, this species remains responsible for high rates of morbidity and mortality worldwide [1,2]. A crucial first step in invasive disease for S. pneumoniae is host colonization. Colonization rates of S. pneumoniae are high, especially in children where up to 55 per cent of children age six months to 3 years are colonized [3,4]. Carriage duration of isolates ranges from days to months [5,6]. Successful colonization requires competing for resources and space with resident micro-organisms [7,8] as well as evading the host immune response [3]. Whether carriage can be established and for how long thus depends on the immunological composition of the host, the genetic makeup of the invading bacteria and the interactions between them and the resident micro-flora [9].
Pneumococcal populations are both serotypically and genetically diverse [5,10–12]. Several lines of evidence suggest that S. pneumoniae strains may compete to establish carriage within hosts. Multiple serotypes co-circulate within host populations [1]; epidemiological modelling suggests that serotypes vary in their ability to displace, and to prevent displacement by, competing serotypes [3,7,10,13,14]. Instances of co-colonization by multiple S. pneumoniae strains have been observed during carriage in humans [15]. In addition, serotype replacement has been observed in experiments in a mouse model [14], and in humans following vaccination programmes [16]. Such findings suggest that resident strains can prevent colonization of the nasopharynx by invading strains, and that vaccination may open an otherwise occupied ecological niche for non-vaccine serotypes [14].
Intraspecific competition to establish carriage may occur by several mechanisms. These include interactions between strains, such as, exploitative competition for resources, or interference competition via bacteriocins [17]. Alternatively, competition could be mediated by the host's immune response, whereby only those strains better at tolerating the specific environment of the host can persist; so-called immune-mediated competition [18]. Both the innate and adaptive immune systems attempt to control infections in many different ways. Differential responses between serotypes to the selection pressure of the innate immune response can be mediated by mucus clearance [19], neutrophils extracellular traps [20] and non-opsonic phagocytosis [12]. The capsule seems to have a prominent role in evading the host immune system, and it is thus probable that success of colonization is partly dependent on serotype [1,3,5,6,10,11]. The capsule prevents clearance from the adaptive immune system by inhibiting complement-mediated opsonophagocytosis [21]. In addition, epidemiological data suggest that different pneumococcal serotypes are cleared at different rates [5,6] although the mechanisms underlying these differences are poorly understood.
In this study, we examine whether antimicrobial peptides (AMPs) of the innate immune response can mediate competitive interactions between strains of S. pneumoniae. AMPs are short cationic peptides that bind to negatively charged teichoic acids of Gram-positive bacterial membranes through electrostatic bonding [22,23] disrupting the cell membrane after attachment and forming pores. They therefore form a first line of defence against a wide range of micro-organisms and are produced by phagocytes and in tissues that first come into contact with microbes. The composition and expression-level of AMPs produced varies spatially, between sites and tissues in the body, and temporally, with the deployment of immune responses [24]. Moreover, there is evidence that overall AMP expression levels vary between individuals in humans [25].
In order for AMPs to play a role mediating competition between pneumococcal strains requires variation in susceptibility to AMPs between S. pneumoniae strains. Moreover, pneumococcal strains should show an uncorrelated response to multiple AMPs, that is, isolates more susceptible to one AMP are less susceptible to another. Although studies have shown increased resistance to AMPs by several mechanisms, surprisingly little is known about patterns of natural variation and co-variation in AMP susceptibilities of S. pneumoniae carriage and clinical isolates.
To examine natural variation in AMP susceptibility, we measured growth of 15 carriage and 16 clinical S. pneumoniae isolates, in the presence and absence of two AMPs; α-defensin human neutrophil peptide (HNP-1) is exclusively produced in neutrophils and the cathelicidin LL-37 is synthesized as a preproprotein, hCAP-18, which is constitutively expressed in inflammatory and epithelial cells [23]. hCAP-18 is also expressed in neutrophils. We observed significant variation in susceptibility of isolates to these AMPs that could not be explained by serotype alone suggesting an equally important role for genetic background. This was confirmed by comparison with a set of isogenic strains that only varied in capsular type. Furthermore, we show that, for most isolates, susceptibility to HNP-1 and LL-37 were uncorrelated such that, in direct competition assays, the presence of HNP-1 or LL-37 reversed the outcome of competition between selected strains. Our results therefore suggest that immune-mediated competition could play a role in the maintenance of genetic diversity in S. pneumoniae.
2. Material and methods
(a). Strains and media
Carriage isolates used in this study were isolated from the nasopharynx of healthy children attending several day-care centres in the Netherlands and have been described previously [26]. Clinical isolates were provided by the Centers for Disease Control (CDC) and the University of Liverpool. Capsule switch variants were constructed on the TIGR4 genetic background, and have been described previously [12,27]. Of the set of carriage isolates, 15 random clones were picked, and these were examined together with 16 clinical isolates for susceptibility to two AMPs; Defensin HNP-1 (Sigma) and the synthetic human cathelicidin LL-37 (Peptanova). The strains used in this study and their associated multi-locus sequence type(ing) (MLST) sequence type (ST) types, determined using standard methods [28], are given in table 1. Both peptides were dissolved in 0.01 per cent acetic acid—0.1 per cent bovine serine albumin (BSA) [29].
Table 1.
Serotype and sequence type of strains used in this study.
| serotype | strain | ST | source |
|---|---|---|---|
| 1 | 605 | 5316 | carriage isolate [4] |
| 1 | B69 | 228 | bacteraemia study adults Atlanta |
| 1 | D1 | 306 | Biomedical research centre, Liverpool |
| 3 | 1535 | 298 | carriage isolate [4] |
| 4 | 5996-5 | 695 | CDC ABC surveillance |
| 4 | 2341 | CDC ABC surveillance | |
| 4 | D4 | 6103 | Biomedical research centre, Liverpool |
| 5 | Columbia5-19 | 289 | PMEN collection |
| 6B | 68 | 6221 | carriage isolate [4] |
| 6B | 162 | 176 | carriage isolate [4] |
| 6B | 1343 | 5317 | carriage isolate [4] |
| 7F | 9741-04 | 191 | CDC ABC surveillance |
| 8 | 2826-94 | 53 | CDC ABC surveillance |
| 8 | P8-10 | 1110 | Biomedical research centre, Liverpool |
| 9V | Spain 9V-3 | 156 | PMEN collection |
| 9V | D9V | 162 | Biomedical research centre, Liverpool |
| 10 | 1379 | 3258 | carriage isolate [4] |
| 10 | 1277 | 6225 | carriage isolate [4] |
| 10 | 1012 | 6224 | carriage isolate [4] |
| 11 | 472 | 6223 | carriage isolate [4] |
| 14 | Spain14-5 | 18 | PMEN collection |
| 14 | England14-9 | 9 | PMEN collection |
| 14 | Tennessee14-18 | 67 | PMEN collection |
| 15 | 1455 | 5328 | carriage isolate [4] |
| 18C | 7205-04 | 3066 | CDC ABC surveillance |
| 19F | 275 | 5297 | carriage isolate [4] |
| 19F | 210 | 6222 | carriage isolate [4] |
| 19F | 458 | 644 | carriage isolate [4] |
| 23F | 7 | 439 | carriage isolate [4] |
| 23F | 471 | 439 | carriage isolate [4] |
| 23F | Tennessee23F-4 | 37 | PMEN collection |
| no capsule | Rx1 | 128 | laboratory strain |
| 4 | TIGR4 | 205 | laboratory strain |
| no capsule | KT 3 | 205 | unencapsulated TIGR4 [27] |
| 1 | KT 4 | 205 | capsule variant of TIGR4 [12] |
| 3 | KT 5 | 205 | capsule variant of TIGR4 [12] |
| 4 | KT 6 | 205 | capsule revertant of TIGR4 [12] |
| 5 | KT 7 | 205 | capsule variant of TIGR4 [12] |
| 6B | KT 8 | 205 | capsule variant of TIGR4 [27] |
| 7F | KT 9 | 205 | capsule variant of TIGR4 [27] |
| 14 | KT 11 | 205 | capsule variant of TIGR4 [27] |
| 19F | KT 12 | 205 | capsule variant of TIGR4 [27] |
| 23F | KT 13 | 205 | capsule variant of TIGR4 [12] |
All strains were grown prior to the experiment in Todd-Hewitt broth with 0.5 per cent yeast extract (pH 7.4). Susceptibility experiments were conducted following established protocols using cation-adjusted Mueller-Hinton broth [30–32], and colony counts were obtained from tryptic soy agar plates with 3 per cent blood, at 37°C in a 5 per cent CO2 incubator. Cationic adjustment was used because this is believed to bring media ionic composition closer to the physiological levels found in human tissues, whereas AMP activity in cation-free medium can lead to an overestimation of AMP activity [30–32]. Because the cation concentration of the test medium (with or without adjustment) may influence the absolute response of cells to each AMP, we focus on the relative susceptibility of strains to each peptide and on the correlation in susceptibility of each strain to different AMPs. Each strain was grown in broth until an optical density of approximately 0.3 (600 nm) whereupon they were frozen in 15 per cent glycerol in 200 µl aliquots at −80°C.
(b). Susceptibility assay
Susceptibilities to AMPs were determined using time-kill assays (note that owing to prohibitive costs it is not feasible to perform minimum inhibitory concentration (MIC) assays for human AMPs). Frozen aliquots of cells were grown to an optical density of OD600 = 0.3. Cells were washed in fresh medium and resuspended in 10 mM phosphate buffer and diluted 100-fold in medium (final cell density approx. 3 × 106 colony forming units (CFU) ml−1). We added 114 μl of this culture to a well of a polypropylene 96-well plate, whereupon 10 μl was taken and used to estimate cell density by plating appropriate dilutions on blood agar plates. Six µl of either a 250 µg ml−1 stock solution of HNP-1 (end concentration 14.4 µg ml−1) or a 229 µg ml−1 stock solution of LL-37 (end concentration 13.2 µg ml−1) or a solution of 0.01 per cent acetic acid–0.1 per cent BSA was added to the well, and cultures were incubated for 4 h at 37°C in a 5 per cent CO2 incubator. After incubation, cell densities were again estimated by plating serial dilutions of the culture on blood agar plates. The chosen concentrations of both HNP-1 and LL-37 lie within the range observed in nasal fluids (HNP-1: approx. 9.2 µg ml−1 [33]; LL-37:approx. 1.2 to more than 80 µg ml−1 [34]).
The relative growth or death of strains was measured by dividing the difference of the log of the total amount of cells between t = 4 and 0 in the presence of the AMP by the difference in log total amount of cells in the absence of AMPs. This was done in order to compare the susceptibility of strains to each other despite not being isogenic. A relative growth of 1 would indicate no inhibition of growth owing to the AMP, whereas growth between 0 and 1 corresponds to net positive growth but at a reduced rate compared to the control assays. Strains that have no net growth, but instead are dying in the presence of AMP have a negative relative growth. All assays were carried out with fourfold replication.
(c). Growth rates
The aliquots that were used for susceptibility assays were also used to obtain growth rate data of the strains. Briefly, aliquots were taken from the freezer and grown until they reached an OD of 0.3. The cells were diluted 100-fold in 200 µl cation-adjusted Mueller-Hinton. The growth rate of each strain is thus measured at the same time and from the same starting culture as its susceptibility measurement. Cells were grown at 37°C with continuous shaking in a 96-well plate reader (BioTec ELx808) and OD 600 was taken every 5 min. The maximum growth rate was estimated during exponential growth from log-transformed values. Several clinical strains were not able to grow under these conditions, perhaps owing to the absence of CO2; we therefore have no data on their growth rate.
(d). Competition experiments
Three sets of strains with differential susceptibility to HNP-1 and LL-37 were chosen and were competed directly in cation-adjusted Mueller-Hinton in the presence of HNP-1, LL-37 or 0.01 per cent acetic acid—0.1 per cent BSA. Conditions in competitions were similar to the conditions of the susceptibility assay. Strain 15353-ST298 can be distinguished from strain 47123F-ST439 by colony morphology, as can strain 23414 from 45819F-ST644. To distinguish Spain9V-3 from England14-9, we isolated Streptomycin resistant clones of both and reciprocally marked strains were competed.
At the beginning and at the end of the competition, the frequency of both competitors was determined by plating onto blood agar plates or blood agar plates supplemented with Streptomycin. From these frequencies, the relative performance of the two competitors was estimated as the selection rate constant [35], which is the difference of their realized Malthusian parameters. The Malthusian parameter is the average rate of increase and is calculated by dividing the natural log of the end density of the competitor by the natural log of the starting density [36]. Competitions were replicated threefold.
3. Results
(a). Patterns of natural variation in susceptibility to AMPs
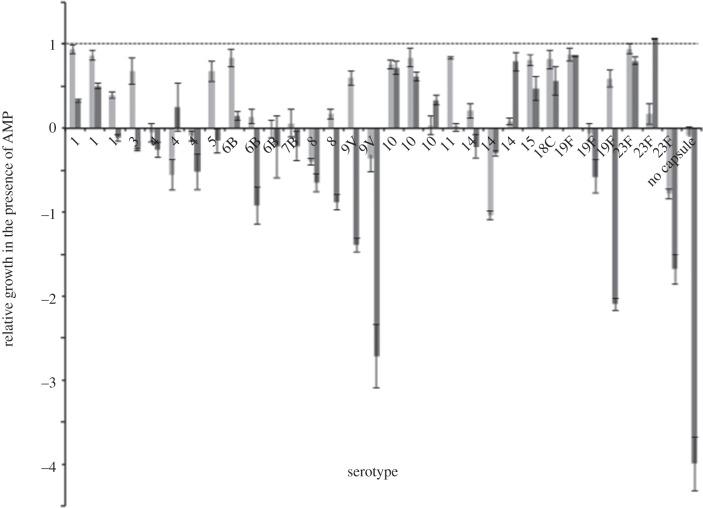
We first examined whether S. pneumoniae isolates varied in susceptibility to HNP-1 and LL-37. Growth of all isolates was significantly inhibited by the AMPs (HNP: t136 = −15.700, p < 0.001; LL-37: t132 = −11.495, p < 0.001), confirming previous reports that S. pneumoniae is susceptible to these AMPs [29,37]. Overall, HNP-1 and LL-37 had dissimilar effects on the growth of the examined isolates (ANOVA: F1,270 = 20.464, p < 0.001) (figure 1), with LL-37 killing isolates more effectively at the concentrations used over the 4-h assay duration. In addition, we found that isolates varied in their AMP susceptibility (ANOVA: F31,270 = 34.162, p < 0.001), and that this difference was not predicted by growth rate in the absence of AMP (analysis of covariance: F1,270 = 0.012, p = 0.911). Importantly, we observed a significant interaction between isolate and AMP (ANOVA: F31,270 = 19.899, p < 0.001). This implies that the susceptibility of a given isolate to either HNP-1 or LL-37 was not only independent of susceptibility to the other AMP, but in some strains was reversed (a possibility we directly test below). This suggests that susceptibility or resistance to a given AMP is somewhat specific, and that generalists with multiple resistances may be relatively rare, perhaps owing to costs associated with resistance or trade-offs between resistance mechanisms.
Figure 1.
Growth of carriage and clinical isolates of Streptococcus pneumoniae in the presence of α-defensin HNP-1 or LL-37, shown in light and dark grey bars, respectively, relative to their growth without any antimicrobial peptide. Strains are organized by serotype and in order corresponding to table 1. A value of more than or equal to 1 would indicate no reduction in growth owing to the antimicrobial peptide while a value less than 1 indicates an AMP-associated killing. Negative values indicate a decrease in cell numbers during incubation. Values represent the mean ± s.e.
We observed a significant difference in AMP susceptibility between isolates from carriage and those derived from a clinical setting (mixed design ANOVA: F1,28.868 = 6.909, p = 0.014). Surprisingly, clinical isolates were more susceptible to AMPs. This difference could be attributable to the difference in serotypes between groups; the carriage strains examined in this study have serotypes prevalent in carriage, and are so-called ‘colonizers’ and the clinical isolates have serotypes associated with infections, or ‘invasive’ serotypes [38,39].
We next evaluated whether variation in susceptibility is dependent on capsular serotype. Serotypes 1, 4, 6B, 10, 14, 19F and 23F were represented by three strains and serotype 8 and 9V by two strains in our screen for susceptibility (figure 1). We again found a significant interaction between AMP and serotype (mixed design ANOVA: F8,163.872 = 16. 316, p < 0.001), suggesting that serotypes susceptible to HNP-1 are less susceptible to LL-37 and vice versa. However, for each AMP, there was only a marginally significant effect of serotype on susceptibility (mixed design ANOVA: F8,24.742 = 2.305, p = 0.053). Therefore, protection of S. pneumoniae against AMPs could not be explained solely by capsular serotype, indicating that the genotype of the isolates was also important in determining susceptibility. The genotype of our strains, as defined by MLST, was different in all but two strains (table 1). Therefore, any similarity found between isolates within a serotype was independent of genotype, at least with the resolution of these markers. Remarkably, the two strains that shared serotype and sequence type, 723F-ST439 and 27523F-ST439, behaved differently in the presence of the two peptides. 723F-ST439 showed high resistance to HNP-1, but was more susceptible to LL-37. In contrast, 27523F-ST439 was the only strain that was completely resistant to LL-37 but was highly susceptible to HNP-1. As S. pneumoniae is naturally transformable, genetic diversity could still be high despite strains having similar MLST genotypes.
(b). Patterns of susceptibility to AMPs in capsule switch variants
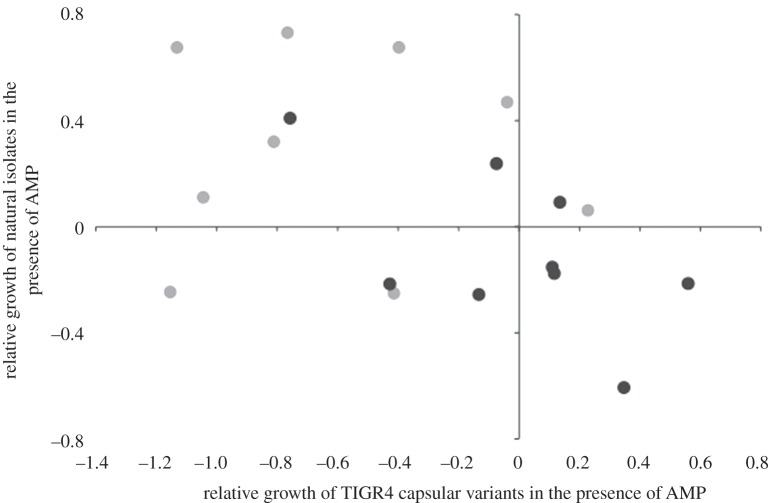
To further explore our findings that both serotype and genetic background contribute to AMP susceptibility, we examined a panel of TIGR4 capsule switch variants that are isogenic except for the capsule locus [27]. Consistent with the idea that susceptibility is a function of capsular type, we found that the susceptibility of these isogenic TIGR4 strains varied depending on the capsule serotype expressed (ANOVA: F9,40 = 59.055, p < 0.001). Equally, consistent with our findings that genetic background also plays a critical role we found no correlation between AMP susceptibilities of natural isolates and TIGR4 capsule switch variants expressing the same capsular serotype (HNP-1: ρ = −0.13, p = 0.973; LL-37: ρ = −0.596, p = 0.090) (figure 2). This is further supported by the observation that the TIGR4 capsule switch variants were more susceptible overall to HNP-1 than were our natural variants, regardless of serotype. The original TIGR4 clone that gave rise to this isogenic set is highly susceptible (data not shown), and accordingly, the genotype of the TIGR4 clone remains relatively susceptible, regardless of the capsule type it expresses.
Figure 2.
Relative growth in the presence of AMP of serotypes of the wild-type strains as a function of the relative growth of serotypes of the isogenic capsule-switch variants. Light grey circles represent susceptibility of serotypes to HNP-1 and black circles represent LL-37 data.
(c). AMPs can reverse the outcome of competition between isolates
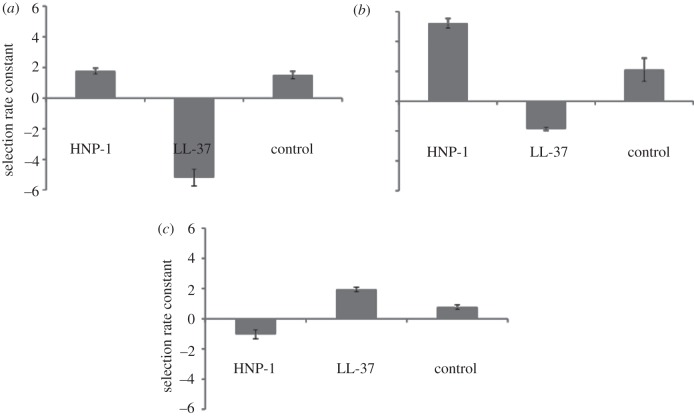
Having identified that susceptibility to HNP-1 and LL-37 were uncorrelated for pneumococcal isolates, we next tested whether AMPs could mediate competition between isolates. Strain 15353-ST298 outcompetes strain 47123F-ST439 both in the control as well as in the presence of HNP-1 (figure 3a). However, competitiveness is reversed when these strains are competed in the presence of LL-37 (ANOVA: F2,9 = 116.279, p < 0.001). The competitive outcome for Spain9V-3 was also dependent upon the AMP present (ANOVA: F2,15 = 126.726, p < 0.001) (figure 3b). For this set of clones, a marker was necessary for the competition, in order to distinguish the strains on plates for counting CFUs. Streptomycin resistance was used as a marker, which had an effect on outcome of competition (ANOVA: F1,15 = 92.041, p < 0.001), however, no interaction between AMP and the marker was found (ANOVA: F2,15 = 0.312, p = 0.739), and thus the reciprocal competition is not shown. In the third competition, strain 23414 is the superior competitor in the presence of LL-37, but is outcompeted by strain 45819F-ST644 in the presence of HNP-1 (ANOVA: F2,9 = 50.073, p < 0.001) (figure 3c). Overall, these assays reveal that AMPs can be important drivers of competitive differences between pneumococcal strains.
Figure 3.
Selection rate constant in the presence of HNP-1 and LL-37 and in the absence of AMP of (a) strain 15353-ST298 when in competition with strain 47123F-ST439 and (b) Spain9V-3 when in competition with strain England14 -9 and (c) strain 23414 when in competition with strain 45819F-ST644. Selection rate constant is the difference of the realized Malthusian parameters.
4. Discussion
The experiments reported here suggest that AMPs of the innate immune system may play a previously unrecognized role in mediating intraspecific competitive interactions of S. pneumoniae in the nasopharynx. We found that different pneumococcal genotypes vary in their susceptibility to 14.4 ug ml−1 of HNP-1 and 13.2 ug ml−1 LL-37 following 4 h of drug challenge. Additionally, the lower susceptibility of a strain to one particular AMP is sometimes opposed by higher susceptibility to a second AMP and vice versa. We next examined whether AMPs could mediate competition in vitro by directly competing selected isolates in the presence or absence of either peptide. The peptides were indeed able to reverse the outcome of competition between isolates, confirming a possible role of AMPs in immune-mediated competition. A necessary requirement for this hypothesis is that composition and levels of AMPs differ between human hosts and/or between different sites within human hosts. There is evidence of natural variation in the levels of AMPs in humans from studies linking low levels of AMPs to increased risk of infectious disease [23,25]. Interestingly, nasal fluid from carriers of Staphylococcus aureus is impaired in antimicrobial activity [40] when compared with non-carriers.
The interaction between susceptibility of the colonizer and immunological composition of AMPs in the individual host potentially plays a role in the maintenance of variation of S. pneumoniae. Explaining the maintenance of phenotypic and genetic variation in populations of bacteria is a major challenge in microbial ecology. Both host and bacterial factors can be responsible for this maintenance [7,41]. For S. pneumoniae, studying maintenance of serotypes is of importance, as the pneumococcal conjugate vaccine targets only up to 13 of the 91 known serotypes. Even though the vaccine is effective and is seen to reduce the carriage rates and disease of the serotypes it targets, serotype replacement has been recorded in a number of studies [1]. Obtaining information about maintenance of variation in serotypes could help make predictions about the increase of non-vaccine type carriage. We speculate that because of host variation in composition and concentration of AMPs, selection pressure on colonizers differs between hosts. If trade-offs in susceptibilities to AMPs exist between serotypes, diversity can be maintained.
A remarkable finding was the overall higher resistance to both AMPs of serotypes more prevalent in carriage. This was unexpected as it is generally accepted that resistance to AMPs is a virulence factor [42]. We had therefore anticipated that clinical isolates would be more resistant to AMPs, preventing them from being killed by the innate immune system. However, from an evolutionary point of view, this result is perhaps not surprising, as the carriage isolates tend to colonize hosts for longer periods of time and are thus exposed to AMPs for a longer duration and at a higher population density, during which time selection for resistance can take place [43]. Alternatively, it is possible that strains resistant to AMPs have an ecological advantage over susceptible strains and are thus more prevalent owing to their higher resistance. Our competition assays demonstrate that clonal take-over by this mechanism is possible in vitro. Differences in serotype prevalence among carriage isolates may therefore be partly explained by AMP susceptibility.
(a). Genotype and serotype contribute to differences in susceptibility
Possible explanations for the observed variation in susceptibility of serotypes are differences in the charge, size and structure of the polysaccharide capsule. Using information on capsule charge compiled from the literature [19,37,42], we detected that charge plays a role in AMP susceptibility. Strains that are isogenic, except for the capsular loci, expressing neutral and positively charged capsules are more resistant than cells expressing highly negatively charged capsules (F1,28 = 39.063, p < 0.001). As negatively charged capsules are more likely to attract the positively charged AMPs, this was anticipated. However, we do not find this relationship for our natural isolates, suggesting that genetic differences between strains override the effect of capsule charge. Since capsule size and structure are not only determined by serotype [12], but also by genetic background as well as by intra strain phase variation [44], further studies, that are beyond the scope of this work, will be required to explore whether these factors affect AMP susceptibility.
While the capsule hides the negative charge of the membrane, AMP susceptibility can also be altered by changes to the membrane [45,46] that modify its charge. For example, increasing the positive charge of the membrane by D-alanylation of lipoteichoic acids, catalysed by the dlt operon, [20,45] decreases AMP susceptibility. Similarly, deletion of choline-binding proteins, such as LytA, increases pneumococcal susceptibility to AMPs [47]. In addition, pneumococcus can alter its gene-expression as a result of exposure to AMPs and can physiologically increase resistance [48]. All but two strains used in this study have different genetic backgrounds as defined by MLST (table 1), and those strains that do share serotype and ST type vary markedly in their response to AMPs. It is thus clear that both serotype and genetic background jointly play important roles in determining susceptibility to AMPs.
(b). Limitations
Our observations need to be complemented by in vivo studies as the bactericidal effect of AMPs may be influenced by physiological factors in the host and by synergisms and antagonisms with other AMPs, like β-defensins, enzymes such as lysozymes, and proteins like lactoferrin [49,50]. Moreover, the AMPs and the bacteria will both modulate the adaptive immune system in numerous ways [51]. Despite these limitations, our data provide support for the hypothesis that AMPs of the innate immune system in vitro influence the competitiveness of S. pneumoniae strains.
Acknowledgements
We are grateful to Mal J. Horsburgh for his contribution in discussions, as well as Tim Cooper and three anonymous reviewers for their helpful comments on a previous version of this manuscript. We would like to thank Daniel Weinberger for sending us his epidemiological dataset. Peter W. M. Hermans provided us with carriage isolates, Lesley McGee, Daniela Ferreira and Paul Roberts with the clinical isolates, and Krzysztof Trzcinski and Marc Lipsitch with the capsular variants. We would like to thank Benjamin Evans for his help in collecting MLST data and acknowledge the use of the pneumococcal MLST database which is located at Imperial College London and is funded by the Wellcome Trust. This work is funded by grants from the Leverhulme Trust (M.A.B.) and the BBSRC (D.E.R.).
References
- 1.Klugman K. P. 2011. Contribution of vaccines to our understanding of pneumococcal disease. Phil. Trans. R. Soc. B 366, 2790–2798 10.1098/rstb.2011.0032 (doi:10.1098/rstb.2011.0032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Brien K. L., et al. 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374, 893–902 10.1016/S0140-6736(09)61204-6 (doi:10.1016/S0140-6736(09)61204-6) [DOI] [PubMed] [Google Scholar]
- 3.Bogaert D., de Groot R., Hermans P. W. M. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4, 144–154 10.1016/S1473-3099(04)00938-7 (doi:10.1016/S1473-3099(04)00938-7) [DOI] [PubMed] [Google Scholar]
- 4.Bogaert D., Sluijter M., Lemmens-den Toom N., Mitchell T. J., Goessens W. H. F., Clarke S. C., de Groot R., Hermans P. W. M. 2006. Dynamics of pneumococcal colonization in healthy Dutch children. Microbiology—SGM 152, 377–385 10.1099/mic.0.28394-0 (doi:10.1099/mic.0.28394-0) [DOI] [PubMed] [Google Scholar]
- 5.Brueggemann A. B., Peto T. E. A., Crook D. W., Butler J. C., Kristinsson K. G., Spratt B. G. 2004. Temporal and geographic stability of the serogroup-specific invasive disease potential of Streptococcus pneumoniae in children. J. Infect. Dis. 190, 1203–1211 10.1086/423820 (doi:10.1086/423820) [DOI] [PubMed] [Google Scholar]
- 6.Sleeman K. L., Griffiths D., Shackley F., Diggle L., Gupta S., Maiden M. C., Richard Moxon E., Crook D. W., Peto T. E. A. 2006. Capsular serotype-specific attack rates and duration of carriage of Streptococcus pneumoniae in a population of children. J. Infect. Dis. 194, 682–688 10.1086/505710 (doi:10.1086/505710) [DOI] [PubMed] [Google Scholar]
- 7.Lysenko E. S., Lijek R. S., Brown S. P., Weiser J. N. 2010. Within-host competition drives selection for the capsule virulence determinant of Streptococcus pneumoniae. Curr. Biol. 20, 1222–1226 10.1016/j.cub.2010.05.051 (doi:10.1016/j.cub.2010.05.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiser J. N. 2010. The pneumococcus: why a commensal misbehaves. J. Mol. Med. 88, 97–102 10.1007/s00109-009-0557-x (doi:10.1007/s00109-009-0557-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Rossum A. M. C., Lysenko E. S., Weiser J. N. 2005. Host and bacterial factors contributing to the clearance of colonization by Streptococcus pneumoniae in a murine model. Infect. Immun. 73, 7718–7726 10.1128/IAI.73.11.7718-7726.2005 (doi:10.1128/IAI.73.11.7718-7726.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sjostrom K., Spindler C., Ortqvist A., Kalin M., Sandgren A., Kuhlmann-Berenzon S., Normark B. H. 2006. Clonal and capsular types decide whether pneumococci will act as a primary or opportunistic pathogen. Clin. Infect. Dis. 42, 451–459 10.1086/499242 (doi:10.1086/499242) [DOI] [PubMed] [Google Scholar]
- 11.Weinberger D. M., et al. 2010. Association of serotype with risk of death due to pneumococcal pneumonia: a meta-analysis. Clin. Infect. Dis. 51, 692–699 10.1086/655828 (doi:10.1086/655828) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinberger D. M., et al. 2009. Pneumococcal capsular polysaccharide structure predicts serotype prevalence. PLoS Pathog. 5, e1000476. 10.1371/journal.ppat.1000476 (doi:10.1371/journal.ppat.1000476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brueggemann A. B., Griffiths D. T., Meats E., Peto T., Crook D. W., Spratt B. G. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 187, 1424–1432 10.1086/374624 (doi:10.1086/374624) [DOI] [PubMed] [Google Scholar]
- 14.Lipsitch M., Dykes J. K., Johnson S. E., Ades E. W., King J., Briles D. E., Carlone G. M. 2000. Competition among Streptococcus pneumoniae for intranasal colonization in a mouse model. Vaccine 19, 598. 10.1016/S0264-410X(00)00236-X (doi:10.1016/S0264-410X(00)00236-X) [DOI] [PubMed] [Google Scholar]
- 15.Brugger S. D., Frey P., Aebi S., Hinds J., Muhlemann K. 2010. Multiple colonization with S. pneumoniae before and after introduction of the seven-valent conjugated pneumococcal polysaccharide vaccine. PLoS ONE 5, e11638. 10.1371/journal.pone.0011638 (doi:10.1371/journal.pone.0011638) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filippo A., Daniela D., Paola C., Paolo D., Matteo B., Giancarlo I. 2010. Serotype replacement in Streptococcus pneumoniae after conjugate vaccine introduction: impact, doubts and perspective for new vaccines. Rev. Med. Microbiol. 21, 56–64 10.1097/MRM.0b013e32833a345f (doi:10.1097/MRM.0b013e32833a345f) [DOI] [Google Scholar]
- 17.Dawid S., Roche A. M., Weiser J. N. 2007. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect. Immun. 75, 443–451 10.1128/IAI.01775-05 (doi:10.1128/IAI.01775-05) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raberg L., de Roode J. C., Bell A. S., Stamou P., Gray D., Read A. F. 2006. The role of immune-mediated apparent competition in genetically diverse malaria infections. Am Nat. 168, 41–53 10.1086/505160 (doi:10.1086/505160) [DOI] [PubMed] [Google Scholar]
- 19.Nelson A. L., Roche A. M., Gould J. M., Chim K., Ratner A. J., Weiser J. N. 2006. Capsule enhances pneumococcal colonization by limiting mucus-mediated clearance. Infect. Immun. 75, 83–90 10.1128/IAI.01475-06 (doi:10.1128/IAI.01475-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wartha F., Beiter K., Albiger B., Fernebro J., Zychlinsky A., Normark S., Henriques-Normark B. 2007. Capsule and D-alanylated lipoteichoic acids protect Streptococcus pneumoniae against neutrophil extracellular traps. Cell. Microbiol. 9, 1162–1171 10.1111/j.1462-5822.2006.00857.x (doi:10.1111/j.1462-5822.2006.00857.x) [DOI] [PubMed] [Google Scholar]
- 21.Magee A. D., Yother J. 2001. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect. Immun. 69, 3755–3761 10.1128/IAI.69.6.3755-3761.2001 (doi:10.1128/IAI.69.6.3755-3761.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brogden K. A. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250 10.1038/nrmicro1098 (doi:10.1038/nrmicro1098) [DOI] [PubMed] [Google Scholar]
- 23.Burton M. F., Steel P. G. 2009. The chemistry and biology of LL-37. Nat. Prod. Rep. 26, 1572–1584 10.1039/b912533g (doi:10.1039/b912533g) [DOI] [PubMed] [Google Scholar]
- 24.Jenssen H., Hamill P., Hancock R. E. W. 2006. Peptide antimicrobial agents. Clin. Microbiol. Rev. 19, 491–511 10.1128/CMR.00056-05 (doi:10.1128/CMR.00056-05) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivas-Santiago B., Serrano C. J., Enciso-Moreno J. A. 2009. Susceptibility to infectious diseases based on antimicrobial peptide production. Infect. Immun. 77, 4690–4695 10.1128/IAI.01515-08 (doi:10.1128/IAI.01515-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bogaert D., Engelen M. N., Timmers-Reker A. J. M., Elzenaar K. P., Peerbooms P. G. H., Coutinho R. A., de Groot R., Hermans P. W. M. 2001. Pneumococcal carriage in children in The Netherlands: a molecular epidemiological study. J. Clin. Microbiol. 39, 3316–3320 10.1128/JCM.39.9.3316-3320.2001 (doi:10.1128/JCM.39.9.3316-3320.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trzcinski K., Thompson C. M., Lipsitch M. 2003. Construction of otherwise isogenic serotype 6B, 7F, 14 and 19F capsular variants of Streptococcus pneumoniae strain TIGR4. Appl. Environ. Microbiol. 69, 7364–7370 10.1128/AEM.69.12.7364-7370.2003 (doi:10.1128/AEM.69.12.7364-7370.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enright M. C., Spratt B. G. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology-UK 144, 3049–3060 10.1099/00221287-144-11-3049 (doi:10.1099/00221287-144-11-3049) [DOI] [PubMed] [Google Scholar]
- 29.Lee H. Y., et al. 2004. Antimicrobial activity of innate immune molecules against Streptococcus pneumoniae, Moraxella catarrhalis and nontypeable Haemophilus influenzae. BMC Infect. Dis. 4, 12. 10.1186/1471-2334-4-12 (doi:10.1186/1471-2334-4-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sánchez-Gómez S., et al. 2008. Comparative analysis of selected methods for the assessment of antimicrobial and membrane-permeabilizing activity: a case study for lactoferricin derived peptides. BMC Microbiol. 8, 196. 10.1186/1471-2180-8-196 (doi:10.1186/1471-2180-8-196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao Y., Cai Y., Bommineni Y. R., Fernando S. C., Prakash O., Gilliland S. E., Zhang G. 2006. Identification and functional characterization of three chicken cathelicidins with potent antimicrobial activity. J. Biol. Chem. 281, 2858–2867 10.1074/jbc.M507180200 (doi:10.1074/jbc.M507180200) [DOI] [PubMed] [Google Scholar]
- 32.Yin L. M., Edwards M. A., Li J., Yip C. M., Deber C. M. 2012. Roles of hydrophobicity and charge distribution of cationic antimicrobial peptides in peptide-membrane interactions. J. Biol. Chem. 287, 7738–7745 10.1074/jbc.M111.303602 (doi:10.1074/jbc.M111.303602) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole A. M., Liao H.-I., Stuchlik O., Tilan J., Pohl J., Ganz T. 2002. Cationic polypeptides are required for antibacterial activity of human airway fluid. J. Immunol. 169, 6985–6991 [DOI] [PubMed] [Google Scholar]
- 34.Lysenko E. S., Gould J., Bals R., Wilson J. M., Weiser J. N. 2000. Bacterial phospholylcholine decreases susceptibility to the antimicrobial peptide LL-37/hCAP18 expressed in the upper respiratory tract. Infect. Immun. 68, 1664–1671 10.1128/IAI.68.3.1664-1671.2000 (doi:10.1128/IAI.68.3.1664-1671.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Travisano M., Lenski R. E. 1996. Long-term experimental evolution in Escherichia coli. IV. Targets of selection and the specificity of adaptation. Genetics 143, 15–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lenski R. E., Rose M. R., Simpson S. C., Tadler S. C. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138, 1315–1341 10.1086/285289 (doi:10.1086/285289) [DOI] [Google Scholar]
- 37.Beiter K., Wartha F., Hurwitz R., Normark S., Zychlinsky A., Henriques-Normark B. 2008. The capsule sensitizes Streptococcus pneumoniae to alpha-defensins human neutrophil proteins 1 to 3. Infect. Immun. 76, 3710–3716 10.1128/IAI.01748-07 (doi:10.1128/IAI.01748-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandgren A., Sjostrom K., Olsson-Liljequist B. O., Christensson B., Samuelsson A., Kronvall G., Normark B. H. 2004. Effect of clonal and serotype-specific properties on the invasive capacity of Streptococcus pneumoniae. J. Infect. Dis. 189, 785–796 10.1086/381686 (doi:10.1086/381686) [DOI] [PubMed] [Google Scholar]
- 39.Battig P., Hathaway L. J., Hofer S., Muhlemann K. 2006. Serotype-specific invasiveness and colonization prevalence in Streptococcus pneumoniae correlate with the lag phase during in vitro growth. Microbes Infect. 8, 2612–2617 10.1016/j.micinf.2006.07.013 (doi:10.1016/j.micinf.2006.07.013) [DOI] [PubMed] [Google Scholar]
- 40.Cole A. M., Tahk S., Oren A., Yoshioka D., Kim Y. H., Park A., Ganz T. 2001. Determinants of Staphylococcus aureus nasal carriage. Clin. Diagn. Lab. Immunol. 8, 1064–1069 10.1128/CDLI.8.6.1064-1069.2001 (doi:10.1128/CDLI.8.6.1064-1069.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipsitch M., O'Hagan J. J. 2007. Patterns of antigenic diversity and the mechanisms that maintain them. J. R. Soc. Interface 4, 787–802 10.1098/rsif.2007.0229 (doi:10.1098/rsif.2007.0229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Llobet E., Tomas J. M., Bengoechea J. A. 2008. Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiology—SGM. 154, 3877–3886 10.1099/mic.0.2008/022301-0 (doi:10.1099/mic.0.2008/022301-0) [DOI] [PubMed] [Google Scholar]
- 43.de Visser J. A. G. M., Zeyl C. W., Gerrish P. J., Blanchard J. L., Lenski R. E. 1999. Diminishing returns from mutation supply rate in asexual populations. Science 283, 404–406 10.1126/science.283.5400.404 (doi:10.1126/science.283.5400.404) [DOI] [PubMed] [Google Scholar]
- 44.Kim J. O., Weiser J. N. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J. Infect. Dis. 177, 368–377 10.1086/514205 (doi:10.1086/514205) [DOI] [PubMed] [Google Scholar]
- 45.Kovacs M., Halfmann A., Fedtke I., Heintz M., Peschel A., Vollmer W., Hakenbeck R., Bruckner R. 2006. A functional dlt operon, encoding proteins required for incorporation of D-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J. Bacteriol. 188, 5797–5805 10.1128/JB.00336-06 (doi:10.1128/JB.00336-06). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vollmer W., Tomasz A. 2000. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J. Biol. Chem. 275, 20 496–20 501 10.1074/jbc.M910189199 (doi:10.1074/jbc.M910189199) [DOI] [PubMed] [Google Scholar]
- 47.Swiatlo E., Champlin F. R., Holman S. C., Wilson W. W., Watt J. M. 2002. Contribution of choline-binding proteins to cell surface properties of Streptococcus pneumoniae. Infect. Immun. 70, 412–415 10.1128/IAI.70.1.412-415.2002 (doi:10.1128/IAI.70.1.412-415.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Majchrzykiewicz J. A., Kuipers O. P., Bijlsma J. J. E. 2010. Generic and specific adaptive responses of Streptococcus pneumoniae to challenge with three distinct antimicrobial peptides, bacitracin, LL-37, and nisin. Antimicrob. Agents Chemother. 54, 440–451 10.1128/AAC.00769-09 (doi:10.1128/AAC.00769-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan H., Hancock R. E. W. 2001. Synergistic interactions between mammalian antimicrobial defense peptides. Antimicrob. Agents Chemother. 45, 1558–1560 10.1128/AAC.45.5.1558-1560.2001 (doi:10.1128/AAC.45.5.1558-1560.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagaoka I., Hirota S., Yomogida S., Ohwada A., Hirata M. 2000. Synergistic actions of antibacterial neutrophil defensins and cathelicidins. Inflamm. Res. 49, 73–79 10.1007/s000110050561 (doi:10.1007/s000110050561) [DOI] [PubMed] [Google Scholar]
- 51.Moranta D., Regueiro V., March C., Llobet E., Margareto J., Larrate E., Garmendia J., Bengoechea J. A. 2010. Klebsiella pneumoniae capsule polysaccharide impedes the expression of beta-defensins by airway epithelial cells. Infect. Immun. 78, 1135–1146 10.1128/IAI.00940-09 (doi:10.1128/IAI.00940-09) [DOI] [PMC free article] [PubMed] [Google Scholar]