SUMMARY
Release factor 3 (RF3) is a GTPase found in a broad range of bacteria where it is thought to play a critical “recycling” role in translation by facilitating the removal of class 1 release factors (RF1 and RF2) from the ribosome following peptide release. More recently, RF3 was shown in vitro to stimulate a retrospective editing reaction on the bacterial ribosome wherein peptides carrying mistakes are prematurely terminated during protein synthesis. Here we examine the role of RF3 in the bacterial cell and show that the deletion of this gene sensitizes cells to other perturbations that reduce the overall fidelity of protein synthesis. We further document substantial effects on mRNA stability and protein expression using reporter systems, native mRNAs and proteins. We conclude that RF3 plays a primary role in vivo in specifying the fidelity of protein synthesis thus impacting overall protein quantity and quality.
INTRODUCTION
Translation of the genetic code into functional protein sequences is accomplished in all domains of life by the ribosome. Protein synthesis is a complex multistep process that can be divided into four phases: initiation, elongation, termination and recycling. During the elongation cycle, the ribosome selects the cognate aminoacyl-tRNA (aa-tRNA) whose anticodon matches the mRNA codon poised in the aminoacyl (A) site of the small ribosomal subunit from a large competing pool of aa-tRNAs. In bacteria, this decoding process is fast (with an amino acid incorporation rate of ~20 s−1) and accurate (with an in vivo missense error rate in the range of 10−4-10−3) (reviewed in (Zaher and Green, 2009a)), with these two features balanced for evolutionary fitness (Thompson and Karim, 1982). Termination occurs when one of three nearly universal stop codons enters the A site and is specifically recognized by a class 1 release factor (RF1 or RF2) that facilitates a hydrolytic reaction to release the completed peptide. Like elongation, stop-codon recognition is highly accurate such that the RFs rarely act on sense codons (a frequency of ~1 in 105 events) (Jorgensen et al., 1993). Both the tRNA and RF selection processes are driven by conformational rearrangements which ensure that cognate species are preferentially accepted by the ribosome (reviewed in (Zaher and Green, 2009a)).
In addition to this stringent pre-peptidyl-transfer selection process, we have recently identified a post-peptidyl-transfer-quality-control (post PT QC) mechanism by which the ribosome, like DNA and RNA polymerases, retrospectively (after peptide bond formation) evaluates whether mistakes have been made (Zaher and Green, 2009b). We showed that the ribosome monitors the quality of the mRNA:tRNA interaction in the P site, and when the interaction is mismatched, subsequent decoding events in the A site proceed with substantial losses in fidelity (Zaher and Green, 2010). Additional mismatches ultimately lead to premature termination of the miscoded polypeptide by promiscuous release factor activity.
Our initial in vitro characterization of post PT QC revealed an unexpected dependence of premature termination on the class II release factor 3 (RF3) where its presence substantially enhanced (as much as 50 fold) the rate of the reaction (Zaher and Green, 2009b). RF3 is a non-essential GTPase found in a subset of bacteria and is most similar to elongation factor G (EFG) (Margus et al., 2007). The factor was initially seen to stimulate the hydrolytic activities of RF1 and RF2 in vitro (Capecchi and Klein, 1969; Caskey et al., 1969; Milman et al., 1969), a feature later exploited to isolate and identify the protein (Grentzmann et al., 1994). Another study identified RF3 in a nonsense suppressor screen for loss of function mutations (Mikuni et al., 1994). Both studies implicated this factor in termination-related functions.
Early speculation focused on a potential role for RF3 in enhancing fidelity during release, akin to the role of EFTu in tRNA selection (Ito et al., 1996; Nakamura et al., 1996). Biochemical analysis failed to support such models (Freistroffer et al., 2000; Freistroffer et al., 1997). Rather, Ehrenberg and colleagues showed that RF3 had no effect on the kcat for peptide release on authentic stop codons, but stimulated release when RF1/2 concentrations were limiting relative to the ribosome. From these data, the authors proposed that RF3 plays a primary role in the “recycling” of class 1 RFs during translation (Freistroffer et al., 1997). In addition to this well documented role in recycling, RF3 has also been shown to promote the dissociation of short peptidyl-tRNAs from the ribosome during the translation of short “mini-genes” in a reaction that also depends on Ribosome Recycling Factor (RRF) and Elongation Factor G (EFG) (Heurgue-Hamard et al., 1998). This role was uncovered with the observation that inactivation of RF3 restored growth to a thermosensitive peptidyl-hydrolase (PTH) strain (Heurgue-Hamard et al., 1998).
Given the very substantial contribution that RF3 makes to post PT QC in vitro, and the more modest effects documented in recycling, we wondered whether the principal role of RF3 in vivo is in quality control rather than in termination. Such a view might provide a more cogent explanation for the nonsense suppression phenotype observed in the absence of this factor. We note that the actual nonsense suppression event produces a mismatched ribosome complex that should be an ideal target of post PT QC (Figure 1); this target, however, would be overlooked in the context of RF3 deletion. The non-essential nature of RF3 in E. coli (Mikuni et al., 1994) and its somewhat sparse distribution throughout the bacterial kingdom (Margus et al., 2007) are features that seem consistent with this speculation.
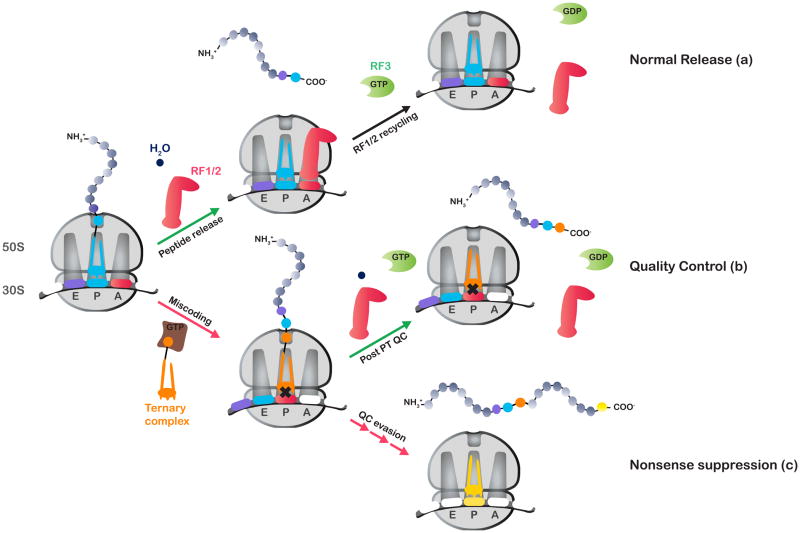
Figure 1.
A model for the action of RF3 in controlling nonsense suppression. During protein synthesis, stop codons are typically recognized by class I release factors, leading to termination (a). Occasionally, stop codons are misread by near-cognate tRNAs, leading to a mismatched pairing between the P-site codon and the anticodon of the peptidyl-tRNA. Under normal conditions, this ribosomal complex would be recognized by the post PT QC machinery (RF1/2 and RF3), leading to premature termination of protein synthesis (b). In the absence of RF3, recognition of the mismatched complex is compromised leading to QC evasion and hence elevated levels of nonsense suppression (c).
Here we explore a potential primary role for RF3 in vivo in maintaining fidelity during translation elongation. To this end, we show that prfC deletion strains are more sensitive to mutations or drugs that increase miscoding. We also find evidence for iterated miscoding and premature termination, and the latter event is significantly reduced in a prfC deletion strain; these data are consistent with our earlier in vitro results. At a more global level, our data hint at a significantly altered transcriptome and proteome in prfC deletion strains. Our findings support a primary role for RF3 in maintaining high-fidelity protein synthesis rather than in termination per se.
RESULTS
Loss of RF3 makes cells more sensitive to errors in protein synthesis
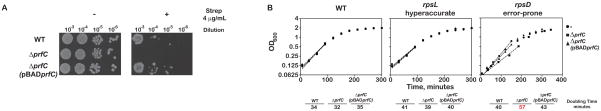
We hypothesized that if RF3 is important in maintaining high fidelity during translation, then deleting the prfC gene should render cells more sensitive to drugs such as streptomycin that increase miscoding. We first compared the growth of the prfC null JW5873 strain (ΔprfC) to its parental wild-type strain (BW25113) in the absence and presence of streptomycin. In the absence of antibiotic, we observe no discernible effect on growth rate (Figure 2A). However, in the presence of antibiotic concentrations tolerated by the wild-type strain, the ΔprfC strain failed to grow at all. The synthetic phenotype was rescued by introduction of plasmid-borne prfC into the deletion strain (Figure 2A).
Figure 2.
Loss of RF3 appears to make cells sensitive to perturbations that reduce the accuracy of protein synthesis. (A) A spotting assay of a dilution series showing that deletion of the prfC gene confers streptomycin-sensitivity. WT refers to the BW25113 strain, whereas ΔprfC refers to the JW5873 strain. (B) Growth curves of the indicated strains showing that the deletion of the prfC gene results in growth defects in an error-prone strain. WT refers to the Xac strain, rpsL refers to the hyperaccurate US157 strain and rpsD refers to the error-prone UD131 strain. See also Figures S1 and S2.
As an alternative approach for demonstrating a genetic interaction between RF3 and fidelity pathways, we deleted the prfC gene in the error-prone (ram) rpsD12 (UD131), the hyperaccurate (restrictive) rpsL141 (US157), and the isogenic wild-type (Xac) strains (Andersson et al., 1982). The deletion of RF3 was detrimental to growth only in the error-prone ram strain (Figure 2B). As above, the slow growth is eliminated when plasmid-borne prfC is introduced into the deletion strain. Together, the effects of streptomycin and the ram mutation on the growth of the prfC null mutants provide evidence for a critical role of RF3 in accurate decoding during protein synthesis.
In addition to their known or potential roles in modulating fidelity, both streptomycin and RF3 have been implicated in increasing the rate of dissociation of peptidyl-tRNA from actively-translating ribosomes (Heurgue-Hamard et al., 1998; Karimi and Ehrenberg, 1996). We performed several additional genetic experiments which indicate that the genetic interaction that we document for RF3 with streptomycin reports on compromised fidelity for the prfC null mutant and is independent of a role for RF3 in peptidyl-tRNA dissociation (Supplemental Results and Figure S1, Supplemental Information online).
In an effort to identify novel properties of the prfC null strain in an unbiased manner, we utilized the phenotypic microarray service provided by Biolog (Hayward, CA) to identify chemical-sensitivities (PM11–20) or differences in metabolite-utilization (PM1–10). These data broadly suggested that RF3 plays a critical role in cells in various stress responses, (presented in detail in Figure S2A, B). We further provide mRNA expression analysis of prfC under various stresses to support these broad conclusions (Figure S2C).
Premature termination following starvation depends on RF2 and RF3
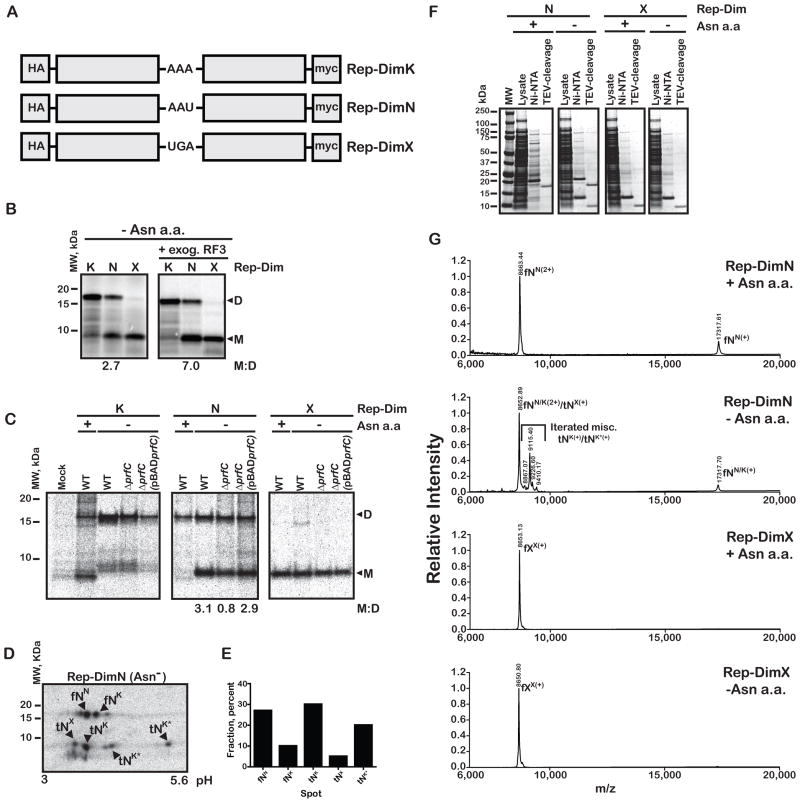
Our initial characterization of the post PT QC process involved ribosome complexes with either a dipeptidyl- or tripeptidyl-tRNA bound in the P site (Zaher and Green, 2009b). Given the limited scope of these in vitro experiments, we were eager to know what the role of post PT QC might be during authentic elongation. We addressed this with a series of synthetic reporter genes (Figure 3A), each encoding two versions of the stable CspE protein where all asparagine residues were replaced by aspartate. Control reporter constructs carry either a lysine (Rep-DimK) or a stop codon (Rep-DimX) in the linker sequence between the two CspE monomers; the experimental construct carries an asparagine codon (Rep-DimN) at this position. For Rep-DimN, it was anticipated that under normal conditions, where asparagine is abundant, full-length dimer protein would be produced. In contrast, under starvation for asparagine, miscoding of the sole AAU codon by Lys-tRNALys would occur (Figure S3A), setting up a situation where premature termination brought about by post PT QC could result in production of stable monomeric protein. For these experiments, the Rep-DimX construct should produce a monomer sized protein, while the Rep-DimK construct should produce a dimer sized protein. In addition, the Rep-DimK reporter should provide some sense of how much natural stalling and drop-off there might be in the linker region.
Figure 3.
Post PT QC takes place in vivo and depends on RF3. (A) Schematic of reporter dimers (Rep-Dim) used for following premature termination as a function of asparagine starvation. (B) An autoradiograph of an SDS-PAGE gel of an S30 in vitro translation assay demonstrating that premature termination takes place during bona fide elongation and is stimulated by the addition of RF3. The ratio of the monomer to the dimer is indicated at the bottom of the gel. (C) An autoradiograph of a Tris-Tricine gel used to follow the status of in vivo pulse-labeled reporters. Constructs were transformed into the asparagine auxotroph JK463 (labeled WT), its ΔprfC derivative HZ001 (labeled ΔprfC), and a derivative of the latter carrying a copy of the prfC gene on a plasmid (labeled pBADprfC). The cells were pulse-labeled with [35S]-methionine in the presence and absence of asparagine, and protein products affinity purified using the N-terminal HA-tag. (D) 2D-PAGE analysis of the prematurely terminated product from Rep-DimN indicates the substitution of a positively-charged amino acid (most likely lysine) in the product. fNN represents a correctly decoded full-length product, fNK represents a full-length product with a lysine substitution, tNX represents a monomer-length product with no additional amino acids, tNK indicates a monomer-length product with one lysine residue added, and tNK* indicates monomer-length products containing additional positively charged residues. (E) Quantitative analysis of the 2D-gel in D. The fractional radioactivity corresponding to the indicated spot is plotted. (F) A coomassie-stained SDS-PAGE gel used to follow the purification of the indicated reporter protein products for mass-spec analysis. Ni-NTA lane denotes proteins that bound to the resin, while TEV-cleavage denotes the flow-through over Ni-NTA resin after TEV-cleavage of the His-tag. (G) MALDI-TOF mass spectra of the indicated sample in the presence or absence of asparagine. In the presence of asparagine, the Rep-DimN sample generated an m/z M+ peak (labeled fNN(+))as well as a corresponding M2+ peak (labeled fNN(2+)). In the absence of asparagine, a collection of peaks corresponding to masses slightly greater than the fNN(2+) and stalled M+ monomer peak (tNX(+)) were additionally observed. In contrast, the Rep-DimX sample generated almost identical spectra in the presence and absence of asparagine, with one prominent m/z peak (fXX(+)). See also Figure S3.
Using these reporters, we asked whether any significant premature termination products were observed in an E. coli translation extract (S30), and whether their abundance was increased when asparagine levels were diminished. In the presence of all amino acids, the three constructs generated mostly protein of anticipated length, monomeric product for Rep-DimX and dimeric product for Rep-DimK and Rep-DimN (data not shown), though we note that Rep-DimN (with an AAU codon that is inefficiently read in vitro) already generates some truncated product. In the absence of asparagine, the control reporters (Rep-DimX and Rep-DimK) again produced protein products of the expected lengths, while the test reporter (Rep-DimN) now produced an increased amount of monomer relative to dimer (Figure 3B). The fraction of monomer produced was reduced when RF2 was immuno-depleted from the extract or in the presence of the release inhibitor paramomycin (Youngman et al., 2007) (Figure S3B) and increased by the addition of exogenous RF3 (Figure 3). These data are consistent with the idea that the monomeric truncated protein results from a premature termination event that depends on both class 1 (RF2) and class II (RF3) release factors.
We next asked whether the same features of premature termination were recapitulated in an in vivo setting. For this, we generated a prfC deletion strain in the background of the E. coli asparagine auxotroph JK643 (Precup and Parker, 1987). The ΔprfC strain (HZ001), with or without pBADprfC, and the parental JK643 strain were transformed with the various reporter constructs (described above) expressed at low levels from the constitutive CAT promoter. Total cellular proteins were labeled by pulsing the growing cells with [35S]-methionine, in the presence or absence of the amino acid asparagine in the media. The products from the reporter constructs were then affinity-purified (using the HA epitope at the N-terminus) and resolved on Tris-Tricine SDS-PAGE.
As expected, the Rep-DimX reporter yielded only monomer product while the Rep-DimK reporter yielded primarily dimer product, both in the presence and absence of free asparagine in all strains tested. We note the presence of several bands for Rep-DimK that copurified in the HA-pulldown that we establish to be non-specific using 2D-PAGE (Figure S3C). In the presence of asparagine, the test construct, Rep-DimN, yielded the expected full-length dimeric product, while in its absence, the Rep-DimN construct also yielded a truncated product of a size similar to that of the monomer. We evaluated the ratio of monomer to full-length product for the various strains (assuming monomer contains half the methionine residues found in dimer product). Consistent with a role for RF3 in the QC mechanism, the fraction of monomeric product was substantially decreased (3-fold) in the prfC null strain (Figure 3C); moreover, the fraction of monomeric product was restored on re-introduction of plasmid-borne prfC.
While these data were consistent with post PT QC being responsible for production of the truncated protein product, there are other mechanisms that might lead to premature termination. As noted previously, RF3, in cooperation with RRF and EFG, has been implicated in promoting peptidyl-tRNA drop-off (Heurgue-Hamard et al., 1998). We note that stimulation of peptidyl-tRNA dissociation by RF3/RRF/EFG is limited to relatively short (<7 amino acids) ribosomal nascent chains (Heurgue-Hamard et al., 2000), much shorter than the 90 amino acid long products characterized here. Moreover, in other studies, RF3 did not contribute to peptidyl-tRNA dissociation from stalled ribosome complexes (Janssen and Hayes, 2009). Despite these already compelling arguments, we further ruled out a contribution of peptidyl-tRNA drop-off in a series of biochemical experiments using S30 extracts and our in vitro reconstituted system (Supplemental Results and Figure S3).
Prematurely truncated proteins exhibit heterogeneity consistent with iterated miscoding
The prematurely-terminated products were further analyzed to ask what was substituted at the sensitive asparagine codon under conditions of starvation, and to evaluate any C-terminal heterogeneity that might recapitulate the iterated miscoding documented in vitro (Zaher and Green, 2009b). We hoped to distinguish between those peptides that were released at the stall site prior to the incorporation of asparagine or lysine (with Rep-DimX product for size comparison), and those that had incorporated asparagine or lysine, or potentially additional miscoded amino acids. For this, we used 2D-gel electrophoresis to further resolve the apparent single species isolated from the 1D gels (Figure 3C).
When looking at full-length products, because starvation is not complete, a fraction of the full-length (f) product results from correct decoding of the asparagine codon (N) and the remainder likely corresponds to miscoding by Lys-tRNALys given the basic shift (fNN and fNK in Figure 3D, respectively, where the superscript refers to the likely amino acid incorporated at the sensitive site). As anticipated, the majority of the truncated (t) products in the Rep-DimN sample in the absence of asparagine were distinct from the full-length Rep-DimX product (fXX) (see Figure S3C panel V, where the two critical samples were resolved on the same gel), and migrate in a manner that suggests that at least one positively charged amino acid (likely lysine) has been added to the prematurely terminated product (e.g. tNK). Most interestingly, consistent with our previous in vitro analysis of post PT QC (Zaher and Green, 2009b), the 2D-PAGE analysis hinted at iterated events of miscoding before termination, as seen in the heterogeneous population of prematurely terminated products (multiple species labeled as tNK* in Figure 3D).
After identification of the various distinct products found in the monomeric and dimeric bands isolated from the 1D gel (by analysis of panels I-V in Figure S3G), we performed a quantitative analysis by determining the ratio of prematurely terminated products tNK and tNK* (i.e. those representing truncated products where miscoding has occurred, and not the stalled tNx product) to mistranslated full-length product fNK (i.e. representing full-length miscoded product that evaded post PT QC) (Figure 3E). After taking account the expected number of methionine residues in the various products (2 and 4 for the short and long products, respectively), the contribution of post PT QC to overall fidelity is estimated to be ~10 under the chosen cellular conditions. These findings are broadly consistent with our previously reported value for the contribution of post PT to QC to fidelity in a purified in vitro system (Zaher and Green, 2009b).
Post PT QC associated iterated miscoding documented by mass spectrometry
Next we set out to explore potential iterated miscoding using mass-spectrometry. To increase expression levels of the proteins of interest for MALDI-TOF, we moved the Rep-DimX and Rep-DimN encoding sequences into an IPTG-inducible over-expression vector pPROEX-HTb. As before, in the presence and absence of asparagine, monomeric product of ~9 kDa was produced with the Rep-DimX reporter (Figure 3F); when these products were examined by MALD-TOF, the molecular weight of the products from the two conditions was essentially the same (8653.13 and 8650.80 Da, respectively) (Figure 3G), agreeing well with the predicted value of 8653.75 Da. For the Rep-DimN reporter, in the presence of asparagine only full-length dimer was seen, while in the absence of asparagine two products were seen, one monomeric and the other dimeric (Figure 3F). For full length Rep-DimN product generated in the presence of asparagine, we observed two m/z peaks, a peak at 17317.61, close to the predicted molecular weight of the dimer of 17321.47, the M1+ peak, while the second peak at 8663.45 corresponds to the M2+ peak. In the absence of asparagine, in addition to these same two peaks that derive from full-length dimer, we observe a heterogeneous collection of peaks in the m/z spectra that is overall larger than both the monomer M+1 and the dimer M2+ peaks of 8653 and 8663 (these peaks were not resolved on the spectra), respectively (Figure 3G). These peaks correspond to additional mass of 150–750 Da (~1–7 amino acid residues in mass) relative to the monomer, with the most prominent peak at 9114 Da adding ~450 Da (~ 4 amino acid residues) to the monomer (Figure 3G). These findings are in striking agreement with our previous in vitro predictions (Zaher and Green, 2009b) and provide our first insights into the likely number of miscoding events that might occur in vivo.
A broad impact of RF3 on protein expression
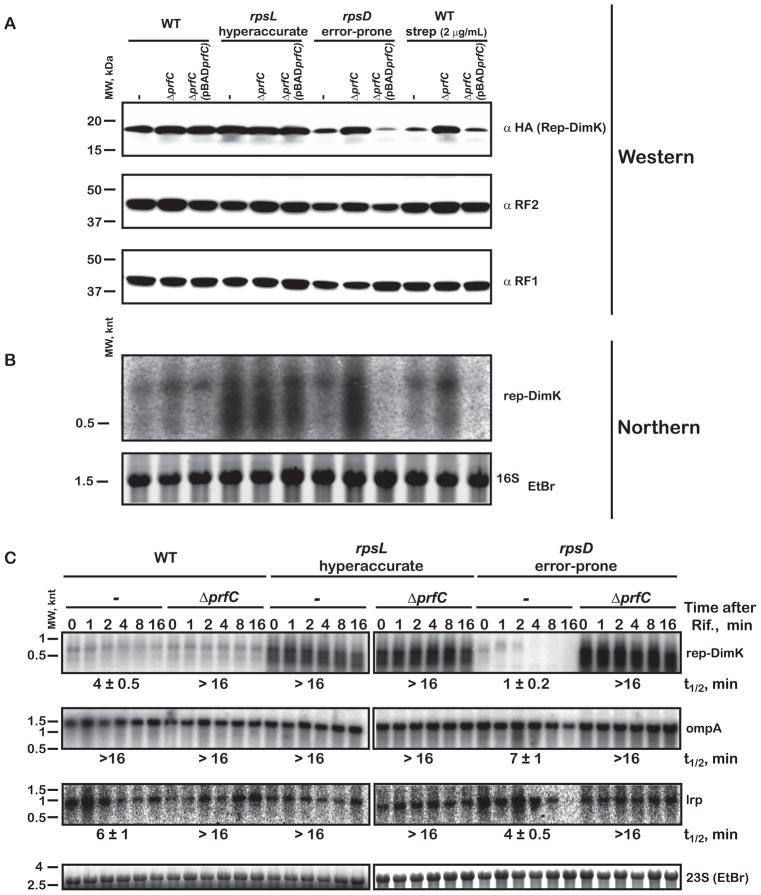
In the previous sections, we used a reporter protein and a programmed miscoding event to specifically visualize and study RF3-mediated premature termination. We were next interested in looking more globally at the effects of RF3 function on protein output in a strain with general and unpredictable miscoding events. We first evaluated by western analysis the overall yield of Rep-DimK reporter in the wild-type Xac strain, the hyperaccurate rpsL141 strain (US157) and the error-prone rpsD12 strain (UD131). We chose this construct because we knew that the protein was generally poorly expressed and anticipated that it might be generally subject to post PT QC. In the wild-type and hyperaccurate background, where relatively little miscoding takes place, the prfC null mutation had no discernible effect on the yield of full-length protein (Figure 4A). In contrast, in the error-prone background (rpsD12) where significantly more miscoding takes place, the prfC null strains produced considerably more full-length protein. These observations are consistent with the proposed role of RF3 in post PT QC; increased miscoding results in decreased product since RF3 prematurely terminates proteins where errors occur, and conversely, yield increases when post PT QC is inactivated (in ΔprfC strain). As anticipated, the increased yield of full-length protein seen in the ΔprfC strain was lost on re-introduction of plasmid-borne prfC (Figure 4A). Unlike our previous analysis of the Rep-DimN construct under conditions of asparagine starvation where a stable prematurely terminated product was produced (Figure 3C), our analysis here relied on unpredictable miscoding and hence unpredictable premature termination events that are expected to produce heterogeneous and labile products that are not easily detected. We also note that although more full-length protein is produced in the absence of RF3, the protein is expected to be mistake-ridden.
Figure 4.
Protein and mRNA levels are impacted by RF3 in a fidelity-dependent manner. (A) Western blot used to assess the expression of Rep-DimK in the indicated strains (WT, rpsL and rpsD here refer to the Xac, hyperaccurate US157 and error-prone UD131 strains, respectively). (B) Northern blot of the same samples as in A used to assess the level of the reporter transcripts in the different strains. (C) Northern blot used to determine the stability of mRNAs by measuring the levels of transcripts following the inhibition of transcription initiation using rifampicin (Rif). The half-lives, indicated at the bottom of the blots, were determined by plotting the relative amount of the transcript to the 23S rRNA and were fit to a single-exponential decay function. See also Figures S4 and S5.
As an alternative approach, we evaluated the expression of the Rep-DimK reporter in the wild-type Xac strain, but now in the presence of the miscoding aminoglycoside streptomycin. Consistent with the rpsD12 data, streptomycin had substantial effects on protein yield which were alleviated by the prfC deletion (Figure 4A).
A potential general role for RF3 in protein output was further evaluated using 2D-PAGE analysis of newly synthesized proteins as detected by pulse-labeling with [35S]-methionine. Direct comparisons of protein output from wild-type and the prfC deletion strain reveal widespread differences in the protein output of the cell (Figure S4A); the visual differences were quantitatively evaluated using unbiased line analysis (Figure S4B). Again, consistent with the connection that we have established between RF3 and fidelity, differences in the protein expression pattern were most pronounced in the rpsD12 strain (with and without RF3).
A broad impact of RF3 on mRNA stability
Because we had documented substantial effects on protein expression in the ΔprfC strain, we next asked whether these observed effects correlated with RNA levels. Steady-state levels of the Rep-DimK transcripts were largely independent of the presence of prfC both in the wild-type and hyperaccurate strains (rpsL141), while the RNA levels varied widely as a function of prfC in the error-prone strain (rpsD12) and in the wild-type strain incubated with streptomycin (Figure 4B). In these error-prone situations (rpsD12 and streptomycin), the mRNA levels appear to broadly correlate with the amount of protein produced in the corresponding strain.
As these results could reflect transcriptional or post-transcriptional effects, we asked whether the presence of RF3 has any impact on the rate of transcription in these strains. For this, we developed a modified “run-on” assay for E. coli, and observed no differences in the amount of nascent Rep-DimK and rpoB transcripts in the ΔprfC strains relative to their parental strains (Figure S5) indicating that RF3 plays no direct role in transcription.
The increased steady-state mRNA levels observed for the reporter in the absence of RF3 likely reflects a decreased rate of mRNA decay. This idea was tested by comparing the rate of decay for the Rep-DimK transcripts together with a subset of randomly-chosen endogenous transcripts (ompA, lrp and msrA) in the ΔprfC and parental strains. Rifampicin was added to the cultures to inhibit transcription initiation, aliquots were taken at different time points and the level of various transcripts analyzed by northern blot. In the wild-type ΔprfC strain, the half-life of the Rep-DimK transcript was at least fourfold higher than in the parental strain (Figure 4C); we observed equivalent effects on the stability of other mRNAs (Figure 4C and data not shown). Interestingly, in the hyperaccurate strain, all tested mRNAs appeared to be substantially stabilized relative to the wild-type Xac strain; indeed, the transcripts were so stable that we were unable to determine their actual half-lives (Figure 4C). The effects of RF3 on mRNA stability were most profound in the error-prone rpsD12 strain where, for example, the half-life of the Rep-DimK transcript increased from 1 minute in the parental strain to >16 minutes in the equivalent ΔprfC strain (Figure 4C).
These findings provide evidence for a relationship between translational fidelity and mRNA stability modulated by RF3. While we observe significant differences in mRNA stability among the wild-type, hyperaccurate and error-prone strains (rpsL > WT > rpsD), mRNAs in all strains were stabilized in the absence of RF3, especially notable given the normal range of mRNA half-lives in E. coli ranging from 2 to 8 minutes.
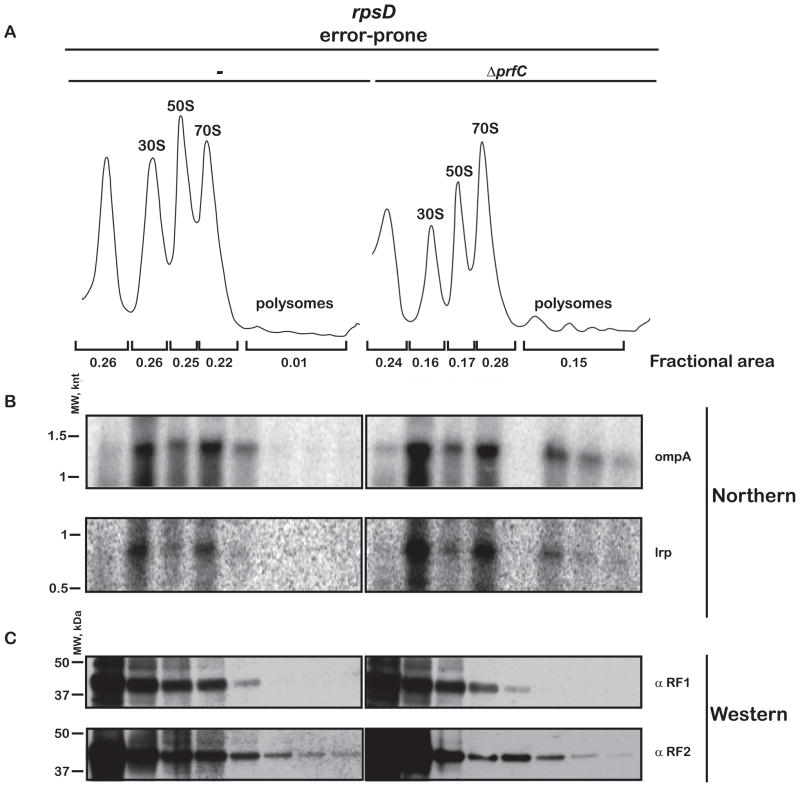
The changes that we observe in protein and mRNA levels in the absence of RF3 suggest that we might see differences in the polysomal profiles of the various strains that reflect changes in ribosomal occupancy. Such a correlation between translational inefficiency and compromised mRNA stability has been well-documented (for example, (Nogueira et al., 2001)), and likely depends on direct protection of transcripts by the ribosome. If this were the case, we would anticipate that mRNAs would be enriched in the heavy polysomal fractions in the ΔprfC strain, where mismatched ribosome complexes are no longer effectively cleared. Indeed, analysis of the wild-type strain and hyperaccurate strains revealed modest effects of the prfC deletion on the total amount of polysomes (Figure S6A, B), while more substantial effects were observed in the error-prone strain (Figure 5A). In the error-prone strain, while only a small fraction of ribosomes was found in the heavy polysome region, in the ΔprfC error-prone strain, the fraction of ribosomes in polysomes increased by as much as a factor of 15 (Figure 5A). As before, we expect that these differences are easier to observe in an error-prone strain where RF3 plays a more significant role (Figure 5 and Figure S6A, B). We note that the distribution of two randomly chosen endogenous transcripts (ompA and lrpA) is enriched in the polysomal fractions in the ΔprfC strain (Figure 5B). While the increased polysome density in the ΔprfC strain might be explained by defects in recycling as a result of slower dissociation of RF1/2 from the ribosomes (Freistroffer et al., 1997), it is not clear why increased polysome density would correlate with compromised fidelity. Moreover, the distributions of both RF1 and RF2 across the gradient appear to be largely unaffected by the prfC deletion (Figure 5C).
Figure 5.
Deletion of RF3 increases ribosome occupancy of the mRNAs in an error-prone background. (A) Polysome profile of the error-prone rpsD strain (UD131) and its prfC deletion derivative. (B) Northern blot of the same fractions follows the distribution of two endogenous transcripts, ompA and lrpA, across the gradient. (C) Western blot of fractions from the sucrose-density gradient in A used to follow the distribution of RF1 and RF2 across the gradient. See also Figure S6.
To extend these results, we compared the polysome profile of a WT and a ΔprfC strain under conditions of general starvation (Figure S6C). Here, we chose the asparagine auxotrophic JK463 strain and its ΔprfC derivative. Consistent with the results from the error-prone strain, under amino acid starvation (and hence elevated error levels), the heavy polysome density was substantially higher in the ΔprfC strain (Figure S6C).
RF3 minimizes frameshifting
While post PT QC appears to reduce missense errors during translation, these errors are typically less deleterious than frameshifting errors which inevitably eliminate protein function and often produce toxic proteins. At a molecular level, the likely outcome of any frameshifting event will be mismatched pairings between the codon and anticodon in both the P and E sites of the ribosome. These complexes are expected to be prime targets for recognition by RF3-dependent post PT QC (Zaher and Green, 2009b).
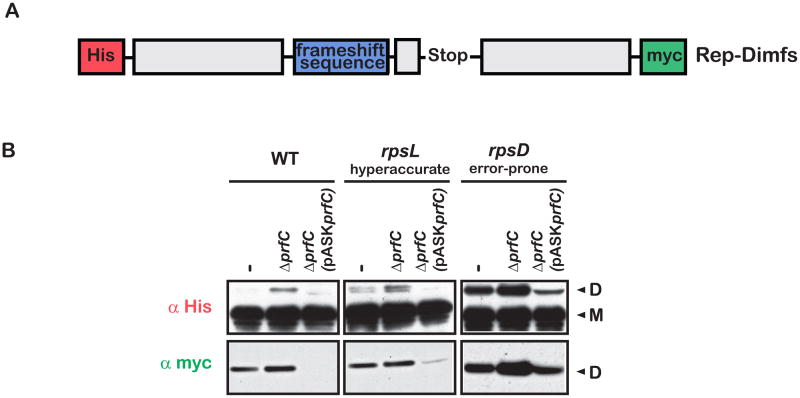
We set out to explore the role of RF3 in frameshifting quality control on the ribosome. We first constructed a reporter (rep-Dimfs) that contains a modified version of the efficient and well-studied programmed prfB frameshifting sequence (Craigen and Caskey, 1986). In particular, the stop codon that naturally occupies the A site just before frameshifting occurs was changed to the rare arginine codon AGG to disentangle effects that RF3 might have on canonical release (Figure 6A). A mutant version of this construct (rep-Dimfsm) that disrupts the prfB Shine-Dalgarno frameshifting trigger was used to confirm the mechanism of frameshifting in these experiments (Figure S7). In the absence of frameshifting, a smaller protein (monomer, M, in Figure 6B) will be produced; when frameshifting occurs, a larger protein (dimer, D, in Figure 6B) will be produced. The reporter was designed to have a His tag at the N-terminus and an out of frame C-terminal myc tag (which is only expressed if +1 frameshifting takes place). The reporter was expressed in the same strains used throughout this study (WT, rpsL and rpsD with and without prfC) and western analysis was performed, probing with anti-His and anti-myc antibodies. In all strains, deletion of prfC resulted in a marked increase in dimer production (frameshifting) when the samples were probed for the N-terminal His tag, with the highest level of frameshifting seen in the error-prone strain (Figure 6B); similar effects were observed when the same samples were probed for the C-terminal myc-tag (especially in the rpsD12 strain). In all strains, over-expression of RF3 from a plasmid-borne copy greatly reduces the amount of frameshifting (Figure 6B). These data argue that RF3 plays a crucial role in controlling the frequency of frameshifting in E. coli.
Figure 6.
Deletion of RF3 leads to elevated levels of frameshifting. (A) Schematic of the reporter used to assess the levels of frameshifting with relevant elements labeled. The myc-tag is in the −1 frame, and as a result is only expressed when a +1 frameshift occurs. (B) Western blot using antibodies against the N-terminal His-tag and the out of frame C-terminal myc-tag. In all strains, deletion of prfC is accompanied by an increase in expression of the dimer. Conversely, over-expression of RF3 is accompanied by a decrease in expression of the dimer. See also Figure S7.
DISCUSSION
Here we demonstrated that the class II release factor RF3 is principally involved in dictating the overall fidelity of protein synthesis in vivo through its function in post PT QC and not, as previously thought, in recycling class I RFs during normal termination (Freistroffer et al., 1997). We further uncover unanticipated connections between RF3 and global cellular protein and RNA levels.
These insights were initially revealed through the sensitivity of the ΔprfC strain to the induction of miscoding in vivo (Figure 2). Subsequently, by exploring the molecular events of post PT QC in vivo with various reporters, we were able to define the role of RF3 in this process (Zaher and Green, 2009b). We found that: (1) Premature termination following starvation-induced miscoding was dependent on RF3 (Figure 3C). (2) Iterated miscoding does occur following the initial miscoding event (as documented by mass spectrometry, Figure 3G). (3) Post PT QC increases the overall fidelity of protein synthesis by as much as a factor of 10 (Figure 3D,E).
We further document profound connections between RF3 and global protein and mRNA levels. Under conditions that lead to elevated miscoding, the deletion of RF3 leads to general increases in protein yield and decreases in protein quality (Figures 4A and S4). Interestingly, we also observed an intimate relationship between the fidelity of protein synthesis and mRNA stability that appears to be strongly modulated by RF3 (Figure 4C). This relationship between translational efficiency and mRNA stability was accounted for through differential ribosomal occupancy in the various strains (Figure 5, and Figure S6). The overall biochemical and genetic data strongly support the model proposed in Figure 7 where RF3 plays an essential role in maintaining high fidelity protein synthesis in E. coli.
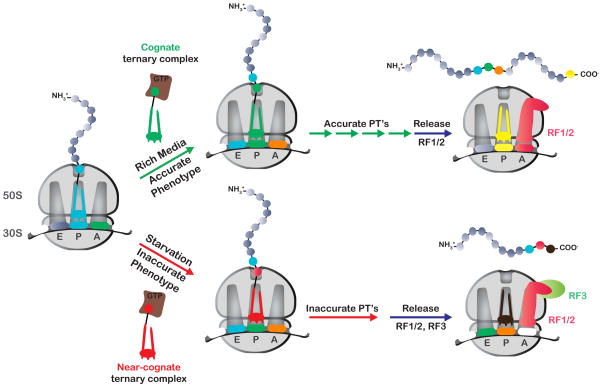
Figure 7.
Schematic of the proposed model for RF3 action in maintaining high-fidelity protein synthesis. Under conditions where the accuracy of protein synthesis is relatively high, few mistakes occur during protein synthesis and translation continues until canonical termination takes place. Under conditions (such as starvation) where the accuracy of protein synthesis is compromised, more mistakes occur during protein synthesis, leading to the generation of mismatched complexes, to iterated miscoding, eventually resulting in recognition of the aberrant complexes by class I RFs and RF3.
In retrospect, early in vivo data provided evidence for a model where RF3 plays a critical role in maintaining high-fidelity protein synthesis (Capecchi, 1967; Caskey et al., 1969; Grentzmann et al., 1994; Mikuni et al., 1994; Milman et al., 1969). In these early studies, RF3 was isolated as a nonsense suppressor (Mikuni et al., 1994), a phenotype classically associated with either a suppressor tRNA (with a compensatory change in the anticodon) or, as was the case with RF3, with changes in the efficiency of the peptide-release reaction. We realized, however, that the efficiency of stop-codon readthrough depends on a miscoding event that results in a mismatched P-site tRNA:mRNA interaction; decreased post PT QC on these complexes will increase readthrough signal. These ideas provide a clear rationale for the isolation of RF3 in the nonsense suppressor screen (Figure 1), despite the rather limited impact of this factor in termination per se (Freistroffer et al., 2000).
Later studies by Buckingham and colleagues similarly documented context-specific effects on the efficiency of stop-codon readthrough on interruption of the prfC gene, again consistent with a role for RF3 in post PT QC (Grentzmann et al., 1995); these differences in efficiency in vivo are consistent with variation that we observed in vitro (Zaher and Green, 2009b). The study by Buckingham and colleagues also showed that interruption of prfC leads to increases in frameshifting at the prfB-programmed frameshift sequence (Grentzmann et al., 1995); these results are consistent with the effects of RF3 on premature termination following frameshifting that we document here (Figure 6). Finally, RF2 synthesis in E. coli depends on an auto-regulatory programmed frameshift where the efficiency of output almost certainly depends on temporarily reduced levels of post PT QC (since [RF2] itself is low) (Craigen and Caskey, 1986).
The role of RF3 in recycling has been well documented and characterized in detail (Mora et al., 2003; Zavialov et al., 2001; Zavialov and Ehrenberg, 2003; Zavialov et al., 2002). The model for its action in recycling (reviewed in (Kisselev et al., 2003)) involves a weak association step between RF3:GDP and the ribosome and exchange of GDP for GTP triggered by this interaction. Strong interactions between the ribosome complex and RF3:GTP trigger conformational changes that lead to dissociation of the class I RF, the hydrolysis of GTP, and ultimately the release of RF3:GDP from the ribosome. While a recycling role for RF3 is not disputed here, we suspect that the relatively modest impact of RF3 on recycling (Freistroffer et al., 1997) is likely to limit its impact on cellular fitness and may not explain the nonsense-suppression phenotype observed in its absence. Consistent with this idea, the total concentration of RF1/2 has been estimated to be in the range of 14–60 μM while that of the ribosome is in the range of 5–55 μM (with 21–33% occupied by an RF) (Adamski et al., 1994); both ranges are orders of magnitude above the in vitro determined Km value for the RF-ribosome interaction. As additional support for this idea, we showed that the deletion of RF3 does not affect the extent of RF1/2 association with the ribosomes in vivo (Figure 5).
Another prediction from a role for RF3 in recycling is that deletion of the factor should lead to stalling, and ultimately in reduced rates of protein synthesis. We might also expect overall reduced growth rates, independent of strain-fidelity background. Contrary to this prediction, the deletion of prfC from the wild-type and hyperaccurate (rpsL141) strains has no apparent effect on growth rate and thus the overall rate of cellular protein synthesis. In the error-prone strain (rpsD12), the deletion of prfC increased protein output for certain proteins, however with significantly reduced growth rate (Figures 2 and 4). These observations are inconsistent with a primary recycling function for RF3 in vivo and are instead consistent with a role in increasing overall fidelity during protein synthesis.
RF3 has also been shown to increase the dissociation of short peptidyl-tRNAs from the ribosome in a reaction that depends on RRF and EFG (Heurgue-Hamard et al., 2000; Heurgue-Hamard et al., 1998). This activity for RF3 appears to be distinct from its role in recycling, as recent genetic studies have identified a class of mutations in RF3 that appear to affect the peptidyl-tRNA dissociation activity without affecting recycling (Watanabe et al., 2010). Here we perform several experiments that confirm that the effects of RF3 that we document are the result of post PT QC, rather than of peptidyl-tRNA drop-off. First, the action of RF3 in promoting premature termination was correlated with reduced fidelity conditions (Figures 2, 3 and 4) that appear not to affect the rate of peptidyl-tRNA dissociation (Figures S1 and S3). Second, competition assays show that dissociation of peptidyl-tRNAs from mismatched complexes as catalyzed by RF3/RRF/EFG could at most contribute up to 30% of the observed premature termination (with short dipeptidyl-tRNA ribosomal complexes, Figure S3D), but would minimally contribute to premature termination during more typical translation elongation (with longer ribosomal nascent chains, Figure S3F).
At a molecular level, how might RF3 modulate the activity of the class I RF on mismatched ribosomal complexes? While we have no structural information for ribosomes bound by both a class I RF and RF3, cryoEM reconstructions of post-termination complexes with either RF3:GDP or RF3:GDPNP bound (Gao et al., 2007; Klaholz et al., 2004) offer some clues. These structures suggest that class I and II RFs can be bound simultaneously to the ribosome. The binding of RF3 to a post-termination complex, however, appears to induce conformational changes in the decoding and GTPase activation centers of the ribosome, likely changing the interaction between the class I RF and the ribosome (Gao et al., 2007), and hence could stimulate their catalytic potentials on mismatched complexes. These ideas are supported by earlier studies showing that RF3 stimulates the kcat of RF1 and RF2 on near-stop complexes (Freistroffer et al., 2000).
RF3 is only found in a subset of bacteria, and no ortholog is known to exist in eukaryotes (Margus et al., 2007). Eukaryotic RF3 (eRF3) is a core and essential component of the termination machinery, where it is thought to form a heterodimer with eRF1 in solution before the complex catalyzes peptide release on the ribosome (Pisareva et al., 2006). Consistent with this distinction between RF3 and eRF3, RF3 is more closely related to EFG while eRF3 is more closely related to EFTu/EF1α (Margus et al., 2007). Indeed, a recent in vitro biochemical study from our laboratory found that post PT QC appears not to exist in the eukaryote Saccharomyces cerevisiae, at least with a mechanism similar to that documented in bacteria (Eyler and Green, 2011). Eukaryotes tend to possess multiple ribosome-based quality control processes such as nonsense-mediated decay (NMD), no-go decay (NGD) and no-stop decay (NSD) (Isken and Maquat, 2007), and so it seems possible that a form of post PT QC exists in these organisms, but involves factors yet to be identified. Alternatively, as protein synthesis in eukaryotes proceeds with higher overall accuracy than seen in E. coli (Kramer et al., 2010), and a cost of slower tRNA-selection parameters (Matthews et al., 2007), the additional contribution to fidelity from post PT QC may be unnecessary. Further analysis will better define the similarities and differences in molecular mechanism in the distinct lineages.
EXPERIMENTAL PROCEDURES
Detailed methods are described in the supplemental information.
Bacterial strains and plasmids
E. coli strains used in this study are listed in Table S1. ΔprfC strains were generated through standard P1-phage transduction (Sternberg and Hoess, 1983) using the Keio Collection JW5873 (Baba et al., 2006; Datsenko and Wanner, 2000) as a donor and screening for kanamycin resistance. Deletion of prfC was confirmed using PCR amplification with prfC-specific primers in the presence of prfB-specific primers used as a control. Plasmids used in this study are listed in Table S2.
Cell-free in vitro translation assays (S30 extracts)
S30 extracts were prepared essentially as described earlier (Kigawa et al., 2004). mRNA templates were generated using T7 in vitro transcription from double stranded DNA (Zaher and Unrau, 2004). In a typical 20 μL reaction, translation was initiated by incubating the mRNA (~2 μM) in the presence of 500 μM of each amino acid (minus methionine and where indicated asparagine), 0.5 μCi/μL of [35S]-methionine (specific activity 1175 Ci/mmole), 8 μL S30 premix (Promega) and 6 μL of the S30 extract. The mixture was incubated at 37°C for 30 minutes.
MALDI-TOF analysis
0.5 μL of the matrix (saturated α-cyano-4-hydroxycinnamic acid solution in 50 % acetonitrile containing 0.1 % TFA) was applied to a 100-well stainless steel plate followed by the addition of 0.5 μL of each sample. The mixture was allowed to dry before inserting into a Voyager DE-STR MALDI-TOF mass spectrometer (Applied Biosystems) equipped with a nitrogen laser giving a 337 nm output. The ions were accelerated with a voltage of 25 kV, and measurements made in the delayed extraction mode with a 5000 Da low-mass gate. The spectrometer was used in the positive-ion detection and linear mode. Typically we collected 50–100 shots per spectrum. Prior to data collection, the instrument was externally calibrated with a mixture of insulin, cytochrome c, apomyoglobin and aldolase (all from Sigma).
Supplementary Material
HIGHLIGHTS.
RF3 genetically interacts with components that reduce translational fidelity
Miscoding triggers more miscoding in vivo resulting in RF3-dependent premature termination
Deletion of RF3 increases mRNA stability and protein yield
RF3 plays a primary role in maintaining high-fidelity protein synthesis
Acknowledgments
We thank Beth Rogers for help in preparing reagents, Allen Buskirk, Nick Guydosh and Chris Shoemaker for comments on the manuscript, Tord Hagervall (Umeå University, Sweden) for strain JK463, Phil Farabaugh (University of Maryland) for the hyperaccurate and error-prone strains, and the Keio collection (Japan) for strain JW5873. The work was supported by the NIH and HHMI (R.G.) and an NIH K99/R00 (H.S.Z.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamski FM, McCaughan KK, Jorgensen F, Kurland CG, Tate WP. The concentration of polypeptide chain release factors 1 and 2 at different growth rates of Escherichia coli. J Mol Biol. 1994;238:302–308. doi: 10.1006/jmbi.1994.1293. [DOI] [PubMed] [Google Scholar]
- Andersson DI, Bohman K, Isaksson LA, Kurland CG. Translation rates and misreading characteristics of rpsD mutants in Escherichia coli. Mol Gen Genet. 1982;187:467–472. doi: 10.1007/BF00332630. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi MR. Polypeptide chain termination in vitro: isolation of a release factor. Proc Natl Acad Sci USA. 1967;58:1144–1151. doi: 10.1073/pnas.58.3.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi MR, Klein HA. Characterization of three proteins involved in polypeptide chain termination. Cold Spring Harb Symp Quant Biol. 1969;34:469–477. doi: 10.1101/sqb.1969.034.01.053. [DOI] [PubMed] [Google Scholar]
- Caskey T, Scolnick E, Tompkins R, Goldstein J, Milman G. Peptide chain termination, codon, protein factor, and ribosomal requirements. Cold Spring Harb Symp Quant Biol. 1969;34:479–488. doi: 10.1101/sqb.1969.034.01.054. [DOI] [PubMed] [Google Scholar]
- Craigen WJ, Caskey CT. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322:273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler DE, Green R. Distinct response of yeast ribosomes to a miscoding event during translation. RNA. 2011:17. doi: 10.1261/rna.2623711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistroffer DV, Kwiatkowski M, Buckingham RH, Ehrenberg M. The accuracy of codon recognition by polypeptide release factors. Proc Natl Acad Sci USA. 2000;97:2046–2051. doi: 10.1073/pnas.030541097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistroffer DV, Pavlov MY, MacDougall J, Buckingham RH, Ehrenberg M. Release factor RF3 in E.coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J. 1997;16:4126–4133. doi: 10.1093/emboj/16.13.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Zhou Z, Rawat U, Huang C, Bouakaz L, Wang C, Cheng Z, Liu Y, Zavialov A, Gursky R, et al. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell. 2007;129:929–941. doi: 10.1016/j.cell.2007.03.050. [DOI] [PubMed] [Google Scholar]
- Grentzmann G, Brechemier-Baey D, Heurgue-Hamard V, Buckingham RH. Function of polypeptide chain release factor RF-3 in Escherichia coli. RF-3 action in termination is predominantly at UGA-containing stop signals. J Biol Chem. 1995;270:10595–10600. doi: 10.1074/jbc.270.18.10595. [DOI] [PubMed] [Google Scholar]
- Grentzmann G, Brechemier-Baey D, Heurgue V, Mora L, Buckingham RH. Localization and characterization of the gene encoding release factor RF3 in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:5848–5852. doi: 10.1073/pnas.91.13.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurgue-Hamard V, Dincbas V, Buckingham RH, Ehrenberg M. Origins of minigene-dependent growth inhibition in bacterial cells. EMBO J. 2000;19:2701–2709. doi: 10.1093/emboj/19.11.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurgue-Hamard V, Karimi R, Mora L, MacDougall J, Leboeuf C, Grentzmann G, Ehrenberg M, Buckingham RH. Ribosome release factor RF4 and termination factor RF3 are involved in dissociation of peptidyl-tRNA from the ribosome. EMBO J. 1998;17:808–816. doi: 10.1093/emboj/17.3.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- Ito K, Ebihara K, Uno M, Nakamura Y. Conserved motifs in prokaryotic and eukaryotic polypeptide release factors: tRNA-protein mimicry hypothesis. Proc Natl Acad Sci USA. 1996;93:5443–5448. doi: 10.1073/pnas.93.11.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen BD, Hayes CS. Kinetics of paused ribosome recycling in Escherichia coli. J Mol Biol. 2009;394:251–267. doi: 10.1016/j.jmb.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen F, Adamski FM, Tate WP, Kurland CG. Release factor-dependent false stops are infrequent in Escherichia coli. J Mol Biol. 1993;230:41–50. doi: 10.1006/jmbi.1993.1124. [DOI] [PubMed] [Google Scholar]
- Karimi R, Ehrenberg M. Dissociation rates of peptidyl-tRNA from the P-site of E.coli ribosomes. EMBO J. 1996;15:1149–1154. [PMC free article] [PubMed] [Google Scholar]
- Kigawa T, Yabuki T, Matsuda N, Matsuda T, Nakajima R, Tanaka A, Yokoyama S. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J Struct Funct Genomics. 2004;5:63–68. doi: 10.1023/B:JSFG.0000029204.57846.7d. [DOI] [PubMed] [Google Scholar]
- Kisselev L, Ehrenberg M, Frolova L. Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J. 2003;22:175–182. doi: 10.1093/emboj/cdg017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaholz BP, Myasnikov AG, Van Heel M. Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature. 2004;427:862–865. doi: 10.1038/nature02332. [DOI] [PubMed] [Google Scholar]
- Kramer EB, Vallabhaneni H, Mayer LM, Farabaugh PJ. A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA. 2010;16:1797–1808. doi: 10.1261/rna.2201210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margus T, Remm M, Tenson T. Phylogenetic distribution of translational GTPases in bacteria. BMC Genomics. 2007;8:15. doi: 10.1186/1471-2164-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews MB, Sonenberg N, Hershey JWB. Origins and principles of translational control. In: Matthews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold spring harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 1–40. [Google Scholar]
- Mikuni O, Ito K, Moffat J, Matsumura K, McCaughan K, Nobukuni T, Tate W, Nakamura Y. Identification of the prfC gene, which encodes peptide-chain-release factor 3 of Escherichia coli. Proc Natl Acad Sci USA. 1994;91:5798–5802. doi: 10.1073/pnas.91.13.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman G, Goldstein J, Scolnick E, Caskey T. Peptide chain termination. 3. Stimulation of in vitro termination. Proc Natl Acad Sci USA. 1969;63:183–190. doi: 10.1073/pnas.63.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora L, Zavialov A, Ehrenberg M, Buckingham RH. Stop codon recognition and interactions with peptide release factor RF3 of truncated and chimeric RF1 and RF2 from Escherichia coli. Mol Microbiol. 2003;50:1467–1476. doi: 10.1046/j.1365-2958.2003.03799.x. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Ito K, Isaksson LA. Emerging understanding of translation termination. Cell. 1996;87:147–150. doi: 10.1016/s0092-8674(00)81331-8. [DOI] [PubMed] [Google Scholar]
- Nogueira T, de Smit M, Graffe M, Springer M. The relationship between translational control and mRNA degradation for the Escherichia coli threonyl-tRNA synthetase gene. J Mol Biol. 2001;310:709–722. doi: 10.1006/jmbi.2001.4796. [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Hellen CU, Rodnina MV, Pestova TV. Kinetic analysis of interaction of eukaryotic release factor 3 with guanine nucleotides. J Biol Chem. 2006;281:40224–40235. doi: 10.1074/jbc.M607461200. [DOI] [PubMed] [Google Scholar]
- Precup J, Parker J. Missense misreading of asparagine codons as a function of codon identity and context. J Biol Chem. 1987;262:11351–11355. [PubMed] [Google Scholar]
- Sternberg N, Hoess R. The molecular genetics of bacteriophage P1. Annu Rev Genet. 1983;17:123–154. doi: 10.1146/annurev.ge.17.120183.001011. [DOI] [PubMed] [Google Scholar]
- Thompson RC, Karim AM. The accuracy of protein biosynthesis is limited by its speed: high fidelity selection by ribosomes of aminoacyl-tRNA ternary complexes containing GTP[gamma S] Proc Natl Acad Sci USA. 1982;79:4922–4926. doi: 10.1073/pnas.79.16.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Nakamura Y, Ito K. A novel class of bacterial translation factor RF3 mutations suggests specific structural domains for premature peptidyl-tRNA drop-off. FEBS Lett. 2010;584:790–794. doi: 10.1016/j.febslet.2009.12.048. [DOI] [PubMed] [Google Scholar]
- Youngman EM, He SL, Nikstad LJ, Green R. Stop codon recognition by release factors induces structural rearrangement of the ribosomal decoding center that is productive for peptide release. Mol Cell. 2007;28:533–543. doi: 10.1016/j.molcel.2007.09.015. [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009a;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R. Quality control by the ribosome following peptide bond formation. Nature. 2009b;457:161–166. doi: 10.1038/nature07582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R. Kinetic basis for global loss of fidelity arising from mismatches in the P-site codon:anticodon helix. RNA. 2010;16:1980–1989. doi: 10.1261/rna.2241810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Unrau PJ. T7 RNA polymerase mediates fast promoter-independent extension of unstable nucleic acid complexes. Biochemistry. 2004;43:7873–7880. doi: 10.1021/bi0497300. [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Buckingham RH, Ehrenberg M. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell. 2001;107:115–124. doi: 10.1016/s0092-8674(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Ehrenberg M. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell. 2003;114:113–122. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Mora L, Buckingham RH, Ehrenberg M. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol Cell. 2002;10:789–798. doi: 10.1016/s1097-2765(02)00691-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.