Abstract
The vast majority of mutations are deleterious and are eliminated by purifying selection. Yet in finite asexual populations, purifying selection cannot completely prevent the accumulation of deleterious mutations due to Muller’s ratchet: once lost by stochastic drift, the most-fit class of genotypes is lost forever. If deleterious mutations are weakly selected, Muller’s ratchet can lead to a rapid degradation of population fitness. Evidently, the long-term stability of an asexual population requires an influx of beneficial mutations that continuously compensate for the accumulation of the weakly deleterious ones. Hence any stable evolutionary state of a population in a static environment must involve a dynamic mutation–selection balance, where accumulation of deleterious mutations is on average offset by the influx of beneficial mutations. We argue that such a state can exist for any population size N and mutation rate U and calculate the fraction of beneficial mutations, ε, that maintains the balanced state. We find that a surprisingly low ε suffices to achieve stability, even in small populations in the face of high mutation rates and weak selection, maintaining a well-adapted population in spite of Muller’s ratchet. This may explain the maintenance of mitochondria and other asexual genomes.
Keywords: Muller’s ratchet, asexual evolution, dynamic equilibrium
PURIFYING selection maintains well-adapted genotypes in the face of deleterious mutations (Haigh 1978). Yet in asexual populations, random genetic drift in the most-fit class of individuals will occasionally lead to its irreversible extinction, a process known as Muller’s ratchet (Muller 1964; Felsenstein 1974). The repetitive action of the ratchet leads to the accumulation of deleterious mutations, despite the action of purifying selection. This ratchet effect has been extensively analyzed (Gessler 1995; Charlesworth and Charlesworth 1997; Gordo and Charlesworth 2000a,b; Stephan and Kim 2002; Jain 2008) and has been observed in experiments (Chao 1990; Duarte et al. 1992; Andersson and Hughes 1996; Zeyl et al. 2001) and in nature (Rice 1994; Lynch 1996; Howe and Denver 2008). In small populations when deleterious mutation rates are high or selection pressures are weak, the ratchet can proceed quickly, causing rapid degradation of asexual genomes (Gabriel and Burger 1993; Lynch et al. 1993, 1995). Hence Muller’s ratchet has been described as a central problem for the maintenance of asexual populations such as mitochondria (Loewe 2006). Avoiding this mutational catastrophe is thought to be a major benefit of sex and recombination (see Barton and Charlesworth 1998 and De Visser and Elena 2007 for reviews).
However, new studies indicate that natural and laboratory asexual populations do not always melt down as predicted. For example, Silander et al. (2007) recently showed that even very small laboratory populations of phage with high mutation rates tend toward fitness plateaus. In addition, a recent comparison of human, chimpanzee, and rhesus Y chromosomes demonstrated that after an initial period of degradation following the halt of recombination, gene loss in the human Y chromosome effectively stopped (Hughes et al. 2012). Finally, theoretical work by Loewe (2006) has revealed several “genomic decay paradoxes” (i.e., species that persist despite predicted unsustainable genomic decay), including human mitochondria.
These results argue for a reexamination of the assumptions behind the classic ratchet model. In the absence of recombination (Bell 1998) or epistasis (Kondrashov 1994), the only forces that can check the deterioration of fitness due to the accumulation of deleterious mutations are back and compensatory mutations (Schultz and Lynch 1997). These mutations are typically assumed to be rare and are either neglected in models of the ratchet or assumed to only slightly slow its rate (Haigh 1978; Kondrashov 1995; Lande 1998; Bachtrog and Gordo 2004). However, as most recently discussed by Charlesworth (2012), it is natural to expect that these back and compensatory mutations become more common as a population declines in fitness. At a minimum, back mutations that revert deleterious point mutations will cause the proportion of available beneficial mutations to increase linearly with the accumulation of these deleterious point mutations. Recent experimental work suggests that the pool of available compensatory mutations in fact increases even faster than this (Silander et al. 2007; Barrick et al. 2010). Hence, based on experimental evidence it is reasonable to assume that the probability that a random mutation is beneficial increases with decreasing absolute fitness (Escarmis et al. 1999; Estes and Lynch 2003; Poon and Chao 2005; Poon et al. 2005; Schoustra et al. 2009). Equivalently, but perhaps more intuitively, one expects the probability that a random mutation is beneficial to decrease with increasing fitness.
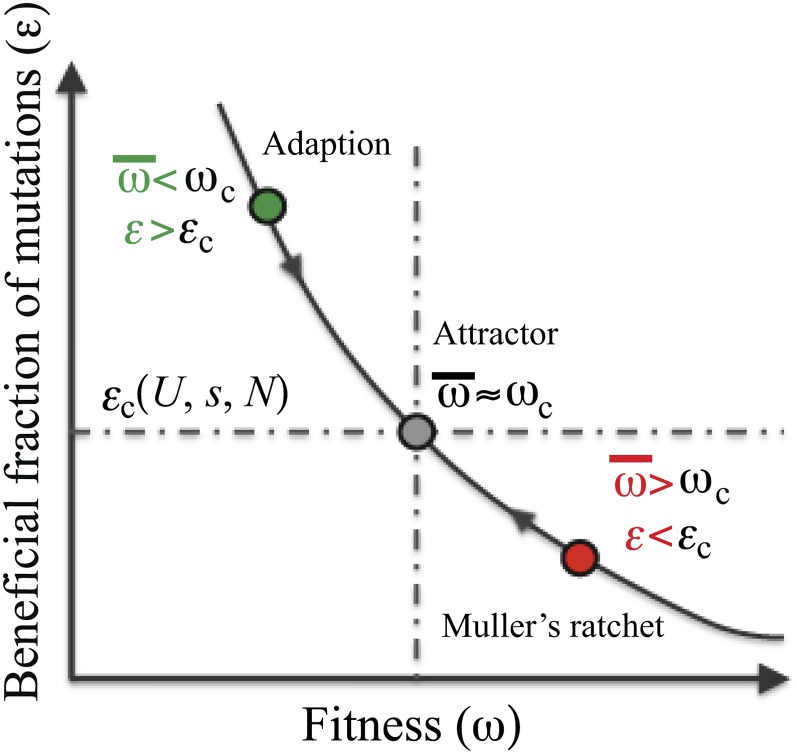
Provided that beneficial (back and compensatory) mutation rates do increase as fitness declines, Muller’s ratchet will eventually come to a halt. Once the fraction of beneficial mutations is high enough to counter the ratchet, the population will remain in a stable dynamic equilibrium state, as illustrated in Figure 1. Qualitatively, we can see that the state is stable from the following argument: let the critical fraction of beneficial mutations needed to precisely counter the ratchet and maintain the equilibrium be εc. In a poorly adapted population, a higher fraction of mutations will be beneficial, ε > εc. This excess of beneficial mutations will push the population toward higher fitness. At the same time, adaptation will deplete the available pool of beneficial mutations, until εc is reached, as shown in Figure 1. Conversely, in an “overadapted” population, we expect ε < εc, so that deleterious mutations dominate, reducing population fitness until εc is recovered. Thus we expect the dynamic mutation–selection balance point to be a stable evolutionary “attractor.” Furthermore, since back and compensatory mutations are selectively favored, they can balance deleterious mutations even while they are relatively rare, thereby maintaining a well-adapted population. This mechanism may be responsible for the fitness plateaus observed in the natural and laboratory populations mentioned above. Moreover, we expect these “treadmill” dynamics, in which continual fixation of deleterious mutations due to the ratchet is exactly offset by fixation of back and compensatory mutations, to be the generic null state of an asexual population under purifying selection. Characterizing this state is an important step toward predicting the patterns of genetic diversity these populations maintain.
Figure 1 .
The relationship between the fraction of mutations that are beneficial, ε, and the absolute fitness of the population, ω. A poorly adapted population (green) with and ε < εc will adapt toward the dynamic equilibrium “attractor” state (gray) with higher fitness and lower ε. Conversely, an “overadapted” population (red) with and ε < εc will decline in fitness toward the dynamic equilibrium state due to Muller’s ratchet.
Several earlier studies have considered aspects of these treadmill dynamics in which both beneficial and deleterious mutations accumulate, including simulations by Wagner and Gabriel (1990), Antezana and Hudson (1997), and Schultz and Lynch (1997); experimental work by Silander et al. (2007); and a combination of simulations and analytical work by Rouzine et al. (2003, 2008) and Manrubia et al. (2003). Theoretical work by Poon and Otto (2000) also considered compensatory mutations using Fisher’s geometrical model, and analytical work by Lande (1998) described alternate fixation of deleterious and beneficial mutations in small sexual populations. These studies have all shown that finite populations settle to fitness plateaus in the presence of purifying selection. However, the nature of this dynamic equilibrium state remains poorly understood: the critical fraction of beneficial mutations εc, the mean fitness of the population at equilibrium, and the distribution about that mean all have yet to be characterized.
In this article, we present a detailed analytical description of the nature of the dynamic equilibrium mutation–selection balance. Crucially, we decouple the problem of the global stability of the dynamic balance state (which depends on how the rate of compensatory mutations changes with absolute fitness) from the properties of the dynamic balance state itself. We accomplish this by treating the probability that a mutation is beneficial as an independent parameter ε. The dependence of ε on absolute fitness becomes important for the evolutionary dynamics of the population away from the equilibrium point, but not at the fixed point itself. This allows us to identify the dynamic balance condition ε = εc and determine how it depends on population size, mutation rate, and strength of selection, independent of any assumptions regarding the dependence of ε on absolute fitness. We show that even a very modest rate of compensatory mutations is sufficient to forestall Muller’s ratchet. Even for remarkably high mutation rates and small population sizes, dynamic balance can be maintained with most mutations being deleterious. Muller’s ratchet notwithstanding, selection enables rare beneficial mutations to compensate for more frequent deleterious mutations and to maintain a well-adapted population.
Materials and Methods
Simulation methods
Our simulations are done using a custom-written Python code available on request. We implement a discrete-time Wright–Fisher model where the population is represented by a vector nk with elements corresponding to the number of individuals in fitness class k. Each generation consists of separate selection and mutation steps. To implement selection, the vector nk is multiplied by to obtain a vector containing the expected number of offspring in class k. Here is the fitness of class k relative to the population mean, and maintains an approximately constant population size around N.
Our model is simplified by using beneficial and deleterious mutation with effect ±s and mutation rate Uε and U(1 − ε), respectively. To implement mutation, we calculate the probability P(i, j) of a genome being hit by i beneficial and j deleterious mutations, which are Poisson distributed. This mutation matrix is then applied to ; i.e., the parts of are moved up or down according to the net number of mutations they accrued in this time step. Having constructed the expected number of individuals in fitness class k after selection and mutation, we draw a population sample from each class from a Poisson distribution. If necessary, the population is recentered in the discrete vector nk. This prevents occupied classes from running off the grid due to accumulated increase or decrease in their absolute fitness.
The above process is repeated for a specified number of generations. The speed of adaptation, v (i.e., the rate of change of population-averaged fitness), and other features of the dynamics are measured after an equilibration time to remove transient effects from the initial conditions. In the parameter regimes studied, we found that 104 generations were generally sufficient for establishing a steady “traveling wave” with velocity v dependent on ε and other parameters.
To solve for εc(U, N, s), we rerun the simulation while iteratively adjusting ε to get v as close to zero as possible. Since v is a fluctuating quantity, εc can be determined only with limited accuracy, but this can be improved by increasing the number of generations for each run.
We have also run simulations in which ε increases with decreasing fitness (i.e., assuming εk = ε0 − ck, where c is a small constant) to demonstrate that the population indeed evolves toward the values of k such that the nose of the distribution is at the appropriate εc for local dynamical balance.
Model
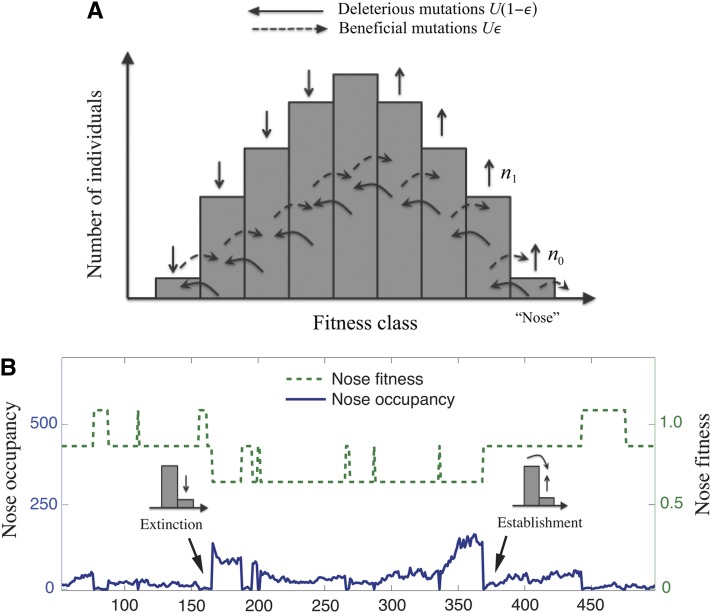
Our analysis is based on the standard discrete-generation Wright–Fisher model with population size fixed at N. For the bulk of this article, we assume that beneficial and deleterious mutations increase or decrease fitness by the same constant amount s (where s > 0 by convention). In a section below, we use simulations and bounding analysis to show that similar behavior occurs when beneficial and deleterious mutations have different fitness effects. In our simple model with a single s, each individual can be described by its number, k, of deleterious mutations relative to the perfectly adapted state. We define to be the mean number of deleterious mutations per individual, so that an individual with k deleterious mutations has fitness relative to the mean, as shown in Figure 2. The population can be characterized by the number of individuals nk in each of these fitness classes. We define U to be the total mutation rate and ε to be the fraction of mutations that are beneficial, so that the beneficial mutation rate is Ub = Uε and the deleterious mutation rate is Ud = U(1 − ε).
Figure 2 .
(A) Schematic illustration of the fitness distribution within a population. We refer to the most-fit class as the “nose” of the distribution. (B) A typical realization of the stochastic dynamics at the nose. In the event of extinction of the nose, n1 becomes the new nose and relative nose fitness decreases by s. Conversely, when a more-fit nose is established, the relative nose fitness increases by s.
Although mutations and genetic drift are stochastic processes, in the bulk of the distribution—where fitness classes contain many individuals—the population dynamics are well captured by a deterministic approximation. On average, the number of individuals in fitness class k evolves as
| (1) |
The terms on the right-hand side of this equation describe the different processes acting on fitness class k: (I) the effect of selection relative to the mean fitness, (II) mutational load, (III) deleterious mutations from more-fit individuals, and (IV) beneficial mutations from less-fit individuals. We assume that the selective effect of a single mutation is small, s ≪ 1.
Although we expect that the probability a mutation is beneficial will depend on the absolute fitness and hence on k, as discussed in the Introduction, we assume ε to be an independent constant parameter. The neglect of the k dependence of ε is a reasonable approximation because at any given time there will be only a relatively narrow range of k present in the population. The k dependence of ε becomes important away from the dynamic-balance point corresponding to the stationary solution of Equation 1, since on longer timescales and hence the population mean fitness begin to increase or decrease significantly. This dependence—assuming that ε increases with decreasing absolute fitness—ensures the global stability of the dynamic balance point, making it an evolutionary attractor. Yet our approximation that ε is locally constant allows us to accurately compute the εc, and corresponding distribution nk, at which the population stays on average at a constant fitness. We have tested this approximation against simulations that introduce k dependence of ε and verified that our characterization of the dynamic equilibrium remains accurate; these simulations are described in supporting information, File S1.
We also note that by defining fitness relative to the population mean in Equation 1, as is standard in Wright–Fisher dynamics, we neglect the possible dependence of population size on absolute fitness, an effect that could potentially lead to mutational meltdown. We return to this possibility in the Discussion.
Analysis
In steady state, the fitness distribution nk stays constant on average: for all k. Solving Equation 1 for the steady state in the case of ε = 0 leads to the familiar mutation–selection balance (Haigh 1978) with a Poisson distribution of fitness,
| (2) |
where k = 0 corresponds to the most-fit class. Here we have defined
| (3) |
This dimensionless ratio is a key parameter that appears often in the analysis below.
The deterministic approximation (Equation 1) corresponds to an infinite population where Muller’s ratchet (i.e., the stochastic extinction of the fittest genotype) does not operate. In the absence of such stochastic extinction, any beneficial mutation from the fittest class would move the distribution toward higher fitness, in conflict with the steady-state assumption. Hence, in infinite populations, steady state can be achieved only with εc = 0. Conversely, in a finite population where the fittest class can be lost due to genetic drift, Muller’s ratchet will eventually lead to a decrease in fitness if no beneficial mutations are available. Thus we must have εc > 0 to be in the dynamic steady state.
A correct description of the dynamic equilibrium state therefore requires a suitable treatment of genetic drift. These stochastic effects are particularly important in the most-fit edge of the distribution, which we call the “nose,” where the number of individuals with a particular fitness is small. Our analysis is based on matching a stochastic treatment of the nose with a deterministic description of the bulk of the fitness distribution (as governed by Equation 1 with ε > 0). The stochastic dynamics of the nose are determined by two competing processes: the random extinction of the most-fit class vs. the establishment of a new more-fit class due to a beneficial mutation. At stationarity, the rates of these two processes have to be equal; this condition determines the number of individuals in the nose class. We match this stochastic condition with the number of individuals in the nose class calculated based on the deterministic distribution. This determines the critical fraction of beneficial mutations εc as a function of the population parameters.
The Shape of the Fitness Distribution
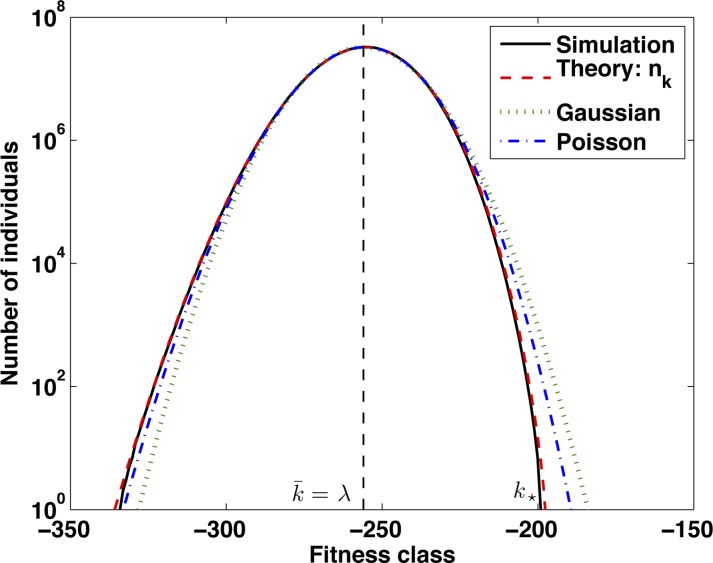
We begin with a deterministic analysis of the shape of the fitness distribution at steady state. Since only relative fitnesses matter, it is convenient to set the origin of k such that the mean of the fitness distribution is . In File S1, we solve for the steady state of Equation 1, using Fourier analysis. We find
| (4) |
where Jk denotes the Bessel function of order k and we have defined . A comparison of this solution to simulation results is shown in Figure 3. For k in the vicinity of , this distribution is approximately Gaussian with variance . While the low-fitness tail of the distribution decays less rapidly than a Gaussian, the high-fitness side decays more rapidly than a Gaussian.
Figure 3 .
Comparison of the fitness distribution obtained in simulations with the analytic result Equation 4 and a Gaussian with equal mean and variance. The simulation result is the median fitness distribution observed over 106 generations with parameters λ = 256, Ns = 1.25 × 106, and N = 109.
It is important to note that in the deterministic limit there is no true stationary state for arbitrary ε > 0; this manifests itself in the loss of positivity of the solution given by Equation 4 for high-fitness classes above the position of the nose class. While the position of the nose, which we call k⋆, requires a more careful treatment, Equation 4 gives a very accurate description of the bulk of the fitness distribution (i.e., nk for k > k⋆).
To determine the position of the nose, k⋆, we observe that if classes with k < k⋆ carry no individuals, fitness class k⋆ does not receive an influx of mutations from the more-fit class k⋆ − 1. That is, term III in Equation 1 is absent, yielding . This, along with Equation 4, gives the following equation for k⋆:
| (5) |
This must in general be solved numerically. However, in File S1 we show that
| (6) |
To gain some intuition into the significance of k⋆, note that k⋆ = 0 in an infinite population with εc = 0. In this case, the fitness distribution is the familiar Poisson distribution and the fittest class contains a fraction e−λ of all individuals. In a finite population, Ne−λ < 1 for sufficiently large λ = U/s, which implies that at steady state the mutation-free genotype is typically absent and therefore k⋆ > 0. This adjustment of k⋆ for finite population size is reflected in Equation 6, which states that the most-fit class gets closer to the population mean as ε increases.
The Stochastic Matching Condition
We seek to determine the ε = εc at which the distribution is stationary for a given finite N. Since the most-fit class is populated by a comparatively small number of individuals, fluctuations due to genetic drift may change its occupancy significantly. In particular, the population at the nose can go extinct, in which case the next fitness class becomes the new nose. Alternatively, a lucky beneficial mutation can cause the nose to advance by one class. Stationarity can therefore be achieved only in an average sense: the rate of advancing the nose has to equal the rate of extinction of the nose.
Since these two processes depend sensitively on the number of individuals in the nose, requiring the equality of extinction and establishment rates determines the average size of the population of the nose, , as a function of U, s, and ε. We match this to the deterministic solution determined above to find εc(N, s, U).
The nature of the dynamics at the nose depends qualitatively on If is large enough that its dynamics are dominated by selection, it is rarely lost and Muller’s ratchet is slow. Conversely, if is small, the class turns over neutrally and is easily lost and reseeded. We begin by considering the slow-ratchet regime, where is relatively large, and then turn to the opposite fast-ratchet regime.
Slow-ratchet regime
In the regime where , the fittest class is only rarely lost due to drift. Note that this regime corresponds to ελ2 < 1, which implies and k⋆ ≈ 0. In this regime the mean extinction rate for the nose, r−, is due to rare large fluctuations that overcome the “restoring force” due to selection trying to preserve mutation–selection balance. Estimates of this extinction rate have been obtained via diffusion theory (Haigh 1978; Gordo and Charlesworth 2000a; Stephan and Kim 2002; Jain 2008; Neher and Shraiman 2012) and have the form (see File S1) where γ is a “phenomenological” parameter characterizing the effective strength of selection on the fittest class, introduced by Haigh (1978) and used in Gordo and Charlesworth (2000a), Stephan and Kim (2002), and Jain (2008) (see below).
To impose our stochastic condition, we must also calculate the rate at which a new more-fit class establishes, r+. This is given by the product of the rate at which new beneficial mutants are generated and the probability they establish, . Since k⋆ ≈ 0 in this regime, we have Pest ≈ 2s. Equating the two rates yields the stochastic condition
| (7) |
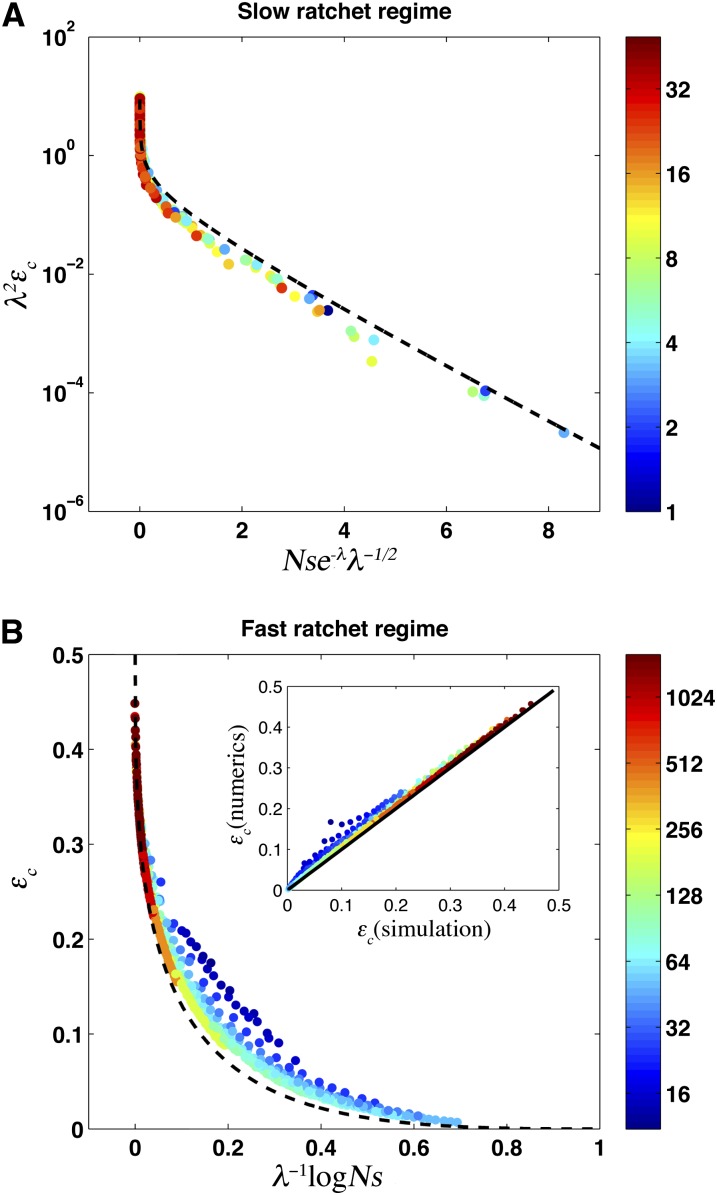
This condition suggests that εc in the slow-ratchet regime is a function of a combination of population parameters: λεc(U, N, s)/γ2 ∼ f (γsNe−λ). We find that contrary to earlier studies (Haigh 1978; Gordo and Charlesworth 2000a), constant γ is not consistent with simulations. Instead, as shown in Figure 4A, the simulation data in the presented range are well explained by , which for values of λ considered, is a good approximation to the more general expression found in Neher and Shraiman 2012. This empirical dependence is, however, consistent with the approximation γ ≈ 0.6 used by Gordo and Charlesworth (2000a), within the range of parameters addressed by their study. Since we consider a much broader range of parameters , accounting for the dependence of γ on U/s becomes important. Our analytical solution Equation 7, shown as a dotted line in Figure 4A, is in good agreement with the simulation data across this wide range of parameters.
Figure 4 .
The critical proportion of beneficial mutations, εc, across both slow- and fast-ratchet regimes for many combinations of U, s, and N for the two regimes. Simulation results are shown as points, with color indicating λ. (A) Slow-ratchet regime. We show εcλ2 as a function of for . The dashed black line is the analytic solution given by Equation 7. (B) Fast-ratchet regime. Note that εc is a function of λ−1log Ns and even in small populations approaches only for very large λ. The dashed black line shows the asymptotic solution εc in the large population size limit , Equation 9. The inset compares the numerical solution from Equation 8 to simulation data.
In this regime, εc depends exponentially on Nse−λ and hence rapidly approaches zero as λ becomes small, approaching the infinite population limit where Muller’s ratchet does not operate.
Fast-ratchet regime
For larger mutation rate, smaller population size, or weaker selection, the occupancy of the most-fit class decreases, thereby increasing its rate of extinction. Consequently, a higher rate of beneficial mutations (larger ε) is required to match the extinction rate. The resulting rapid turnover of the population at the nose leads to the failure of the quasi-static approximation we used in the slow-ratchet regime.
As the occupancy of the nose decreases, in particular when , the dynamics of the fittest class are governed by drift. The rate of extinction, r−, can therefore be estimated from neutral diffusion: . The rate at which a new more-fit class is established is given by the same formula as before, . However, Pest now refers to the probability that a new mutant lineage reaches individuals. At this point, the lineage crosses over from stochastic to deterministic dynamics, entering the domain described by the deterministic solution Equation 4. Thus Pest = 2(k⋆ + 1)s. Equating r− = r+ yields the stochastic condition
| (8) |
This condition, together with the solution Equation 4 for the distribution, allows us to determine εc(N, s, U). As shown in Figure 4, our solution Equation 8 is in excellent agreement with simulations for most of the data. However, our solution overestimates εc for populations with small population size and small λ that still satisfy . A better description in this regime will require a more careful analysis of fluctuations beyond the nose.
Large population size limit
Large populations can maintain a well-adapted genome with even for moderately large λ. In the limit of large populations and large U/s, with the limit taken so that εcλ2 ≫ 1 while , the matching condition reduces to . This equation has the approximate solution
| (9) |
This result can be made more precise through iteration (see File S1 for derivations and comparisons to simulation data). Note here that εc depends only very weakly on Ns and on λ, in contrast to the slow-ratchet regime where εc declines exponentially with Nse−λ.
High mutation rate or weak purifying selection
In this limit, the dynamic equilibrium tends toward the state where beneficial and deleterious mutations are equally frequent, . This corresponds to a population that can no longer maintain a well-adapted state. In File S1, we derive an approximate expression for εc in the fast-ratchet regime in the limit of large λ:
| (10) |
From this expression we can immediately determine the point at which the population can no longer maintain a well-adapted state, as defined by a maximum fraction of beneficial mutations, εmax. To have εc < εmax, we require
| (11) |
valid for εmax ≳ 0.2. Note that for εmax = 1/4, the right-hand side (RHS) of this expression is 48; for εmax = 1/3, the RHS is 162. Thus even small populations with Ns ≳ 1 can maintain relatively well-adapted genomes in the face of high mutation rates and weak selection (i.e., when U ≫ s so that λ ≫ 1).
Different fitness effects for beneficial and deleterious mutations
Our analysis to this point has assumed that both beneficial and deleterious mutations have the same fitness effect s. However, compensatory mutations often only partially compensate for the cost of a deleterious mutation. Hence beneficial mutations may tend to have a smaller effect on fitness than deleterious mutations. The opposite case, where beneficial mutations have larger effects than deleterious mutations, may also be relevant, as weak-effect deleterious mutations accumulate more rapidly and for some effect distributions may dominate. We now consider the effects of these differences in the fitness effects of beneficial and deleterious mutations. We denote the effect of a deleterious mutation by sd and the effect of a beneficial mutation by sb, where by convention both sd and sb are positive.
When sd ≠ sb, there still exists a critical value of ε = εc corresponding to a dynamic equilibrium state, which (provided as always that ε increases as fitness decreases) is globally stable. However, the shape of the fitness distribution in this equilibrium will be different from that calculated above, and the critical εc will also change. Fortunately, our analysis of the single-s model places bounds on this new critical εc that applies for sd ≠ sb. To see this, we first define the εc that corresponds to a particular single-s model (as calculated above) to be εc(s). Now consider the case sb < sd. The critical εc for this more complex situation must be less than εc(sb), because stronger-effect deleterious mutations can only decrease εc (stronger selection against deleterious mutations cannot speed their accumulation). On the other hand, the critical εc must also be greater than εc(sd), because weaker-effect beneficial mutations can only increase εc. A similar argument leads to analogous bounds for the case sb > sd.
These simple bounding arguments imply that the critical εc is at most εc(sb) or εc(sd), whichever is greater. Since we have seen that εc remains small even for remarkably small s and large U, this can still correspond to a relatively well-adapted state. Provided that deleterious mutations and the corresponding compensatory mutations do not have widely differing fitness effects, these bounding arguments also constrain εc to a narrow range. Thus although the quantitative details of the stable equilibrium state change whenever sb ≠ sd, the main qualitative conclusions of our single-s analysis still apply.
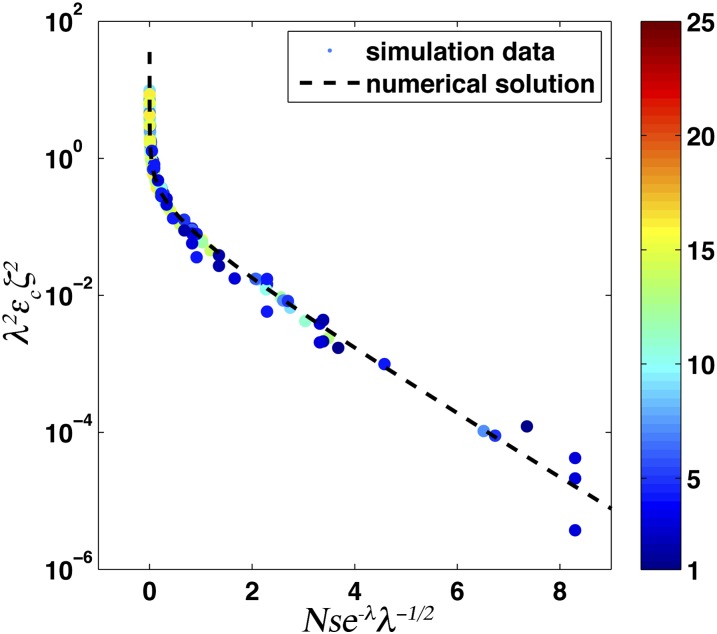
To provide a more precise analysis of how εc depends on both sb and sd, we use simulations to compare εc for sb = sd/ζ with ζ = 0.5, 1, 2 and show that in the slow-ratchet regime εc(ζ) ≈ ζ2εc(1), demonstrated by the collapse of simulation data for different parameter values onto the same curve, as shown in Figure 5. This behavior in the slow-ratchet regime is quite intuitive, as the rate of forward motion of the nose increases as . Here one factor of sb arises from the establishment probability and the other from the size of the forward step due to each established beneficial mutation. Hence if sb → sb/ζ, the rate of beneficial mutations necessary to maintain dynamic balance must increase by ζ2. Remarkably, this simple scaling approximation holds well into the crossover to the rapid-ratchet regime.
Figure 5 .
The critical fraction of beneficial mutations, εc, in the slow-ratchet regime for sb = sd/ζ with ζ = 0.5, 1, 2 and many combinations of U, s, and N. As in Figure 4, simulation results are shown as points, with color indicating λ. The dashed black line is the analytic solution given by Equation 7.
We finally note that of course in general not all beneficial mutations will have the same fitness effect sb nor will all deleterious mutations have the same cost sd. Instead, the effect of any individual mutation will be drawn from some distribution of beneficial or deleterious fitness effects. The values of sb and sd then represent some weighted average effect of a beneficial or deleterious mutation. A full analysis of this more complex situation is beyond the scope of this article, but we return to this issue, qualitatively, in the Discussion below.
Discussion
In infinite populations it has long been recognized that the balance between mutational pressure and purifying selection leads to a fitness equilibrium (Eigen 1971; Haigh 1978). Our analysis demonstrates that such an equilibrium also exists in a finite population, despite the action of genetic drift and Muller’s ratchet. Much of the previous work on these finite populations has focused primarily on the rate of Muller’s ratchet in the absence of beneficial mutations and has generally assumed that back and compensatory mutations are rare enough to be neglected (Gordo and Charlesworth 2000a,b; Stephan and Kim 2002; Bachtrog and Gordo 2004; Etheridge et al. 2009). However, both intuition and experimental data (Silander et al., 2007) strongly suggest that beneficial mutations become more common as a population accumulates deleterious mutations. Silander et al. (2007) also provided a simple analysis of the possible dynamic balance, but it did not extend beyond the low mutation rate limit in which mutant alleles fix or go extinct within an otherwise clonal population. The dynamics of diverse populations subject to the combined effect of both deleterious and beneficial mutations have remained understudied (for notable exceptions see Rouzine et al. 2003, 2008 and Pfaffelhuber et al. 2011).
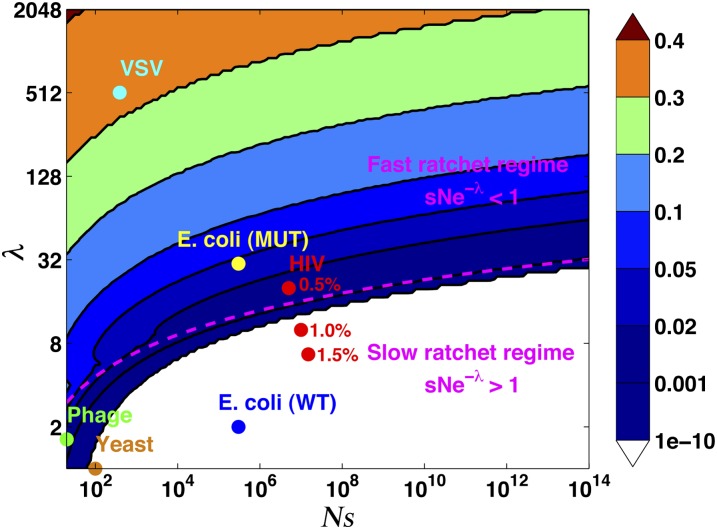
We have shown that for any population size, mutation rate, and selection pressure, there exists a proportion of beneficial mutations, εc, which balances the accumulation of deleterious mutations. Because beneficial mutations are favored by natural selection, a small-fraction beneficial mutation can suffice to maintain stability, εc ≪ 1. We have demonstrated this by explicitly calculating εc as a function of population size, mutation rate, and the strength of selection. There are two qualitatively different regimes for εc, as shown in Figure 6. In the slow-ratchet regime, selection stabilizes the fittest subset of the population at the nose of the distribution, so εc is small and depends exponentially on the population parameters Ns and λ. On the other hand, in the fast-ratchet regime, εc must be larger to balance rapid accumulation of deleterious alleles, and it depends more weakly on the population parameters Ns and λ. The boundary of the two regimes is approximately given by Nse−λ = 1.
Figure 6 .
The fraction of beneficial mutations εc necessary to maintain the dynamic mutation–selection balance in a population with parameters Ns and λ = U/s. The dashed line separates the slow- and fast-ratchet regimes. Experiments with various model organisms with different population parameters are represented as points.
We note that Rouzine et al. (2003, 2008) studied population dynamics under the combined action of purifying and positive selection in a model similar to ours, finding both simulation and analytical evidence for the existence of the dynamic balance state. However, their analysis focuses on the rate of the ratchet (with ε ≪ εc) or the rate of adaptation (with ε ≫ εc) and does not apply as ε → εc. Our work, by contrast, focuses exclusively on the dynamic equilibrium state and the critical εc required to maintain it.
Our analysis treats ε as an independent parameter, even though we expect it to depend on absolute fitness ω. As discussed above, we expect ε(ω) to decrease with increasing ω. In this case the steady state corresponding to dynamic balance at εc(N, λ) is stable and is a global attractor point. For ε > εc mean fitness increases (dω/dt > 0), while for ε < εc mean fitness decreases (dω/dt < 0), which for dε(ω)/dω < 0 guarantees local stability of the ε = εc equilibrium. While our presentation focused on the properties of dynamic equilibrium, our approach also defines the actual rate of change of dω/dt = r+ − r− as a function of instantaneous ε and N (note that, as in Rouzine et al. 2008, in general r± themselves become dependent on this “velocity”). This quantitative understanding of differential dynamics, i.e., the rate of change of mean fitness conditional on a given value of ε, can be integrated for any given ε(ω) defining the corresponding evolutionary trajectory as a function of time. This consideration can be readily generalized (by allowing N to change along ω) to include possible reduction in the size of the population that may follow reduction of absolute fitness. In particular, it is possible that under some conditions the population will collapse—i.e., “mutational meltdown” will occur—before the stable dynamic equilibrium is reached (Lynch et al. 1993) (see File S1 for more extended discussion). The possibility of such a meltdown instability depends critically on precisely how ε depends on ω and hence on details of the ecology of the specific system. However, given any specific set of assumptions about ε(ω), this effect can be analyzed quantitatively in terms of the dynamics formulated above.
Our analytic results for εc in the dynamic equilibrium were made possible by the explicit calculation of the shape of the fitness distribution, given in Equation 4 and illustrated in Figure 3. With increasing λ and decreasing N this distribution deviates from the Poisson distribution for the classic mutation–selection balance (Haigh 1978), most notably reducing the fitness of the top class, k⋆, relative to the mean. Examining the properties of the dynamic mutation–selection equilibrium over the full parameter range, shown in Figure 6, has revealed a strong asymmetry between beneficial and deleterious mutations. For example, for a small population Ns ∼ 103 with high mutation rate λ ∼ 8, just 2% beneficial mutations are enough to counteract the effect of deleterious mutations. This indicates that purifying selection is remarkably effective even for conditions where Muller’s ratchet would proceed extremely quickly in the absence of back and compensatory mutations.
Populations with ε > εc should adapt, while populations with ε < εc should decline in fitness. Experimental evolution of model organisms in controlled laboratory environments appears to be consistent with this expectation. In particular, Silander et al. (2007) showed that bacteriophage ϕX174 converged to a population-size–dependent fitness plateau, as our model would predict. For their population parameters (s ∼ 0.08, U ∼ 0.13) our theory predicts εc ranging from 2% to 0.2% for their experiments (in which N ranged from 100 to 200), consistent with the beneficial mutation rates they infer. Their experiments with lower population sizes (N ≲ 30) are in the fast-ratchet regime, and estimating εc requires further analysis of fluctuations beyond the nose.
Other experiments with vesicular stomatitis virus found that large populations adapt while small populations melt, also consistent with our analysis (Moya et al. 2000). Similar results have been observed in yeast (Desai et al. 2007; Lang et al. 2011). In the long-term evolution experiments of Lenski and collaborators in Escherichia coli, parameters are such that we expect the critical εc to be very small, εc ≪ 10−10, consistent with their observation of continuous adaptation even after tens of thousands of generations. On the other hand, mutator strains of E. coli (Trindade et al. 2010) and yeast (Desai et al. 2007) sometimes show decrease in fitness, which could be used to further test the model. In Figure 6, we show approximate estimates for the parameter regimes in which each of these experimental systems lie. For example, population parameters for human immunodeficiency virus seem to place it close to the boundary between the slow- and fast-ratchet regimes (Figure 6), so that its evolutionary dynamics depend sensitively on the strength of selection.
While a global “phase diagram” of population dynamics is highly intriguing, our present attempt to construct it highlights the challenge of doing so. First, our analysis has assumed that all mutations have the same selective effect s. In general, however, mutations have a range of selective effects. Mutations with a particular characteristic strength may tend to dominate the dynamics, if more strongly selected mutations are quickly eliminated by selection while more weakly selected mutations are effectively neutral given interference from selection on the other mutations. Thus, our λ parameter must be regarded as an effective parameter corresponding to mutations with effect strength that dominate the balance. The possibility of defining such an effective λ in the model of Muller’s ratchet with deleterious effects of different size has been discussed by Soderberg and Berg (2007) on the basis of numerical simulations. Yet the general description of interference within an arbitrary distribution of (deleterious and beneficial) mutational effects that would provide a precise definition of effective λeff remains an open problem and an important avenue for future work. Second, our analysis has focused on asexual populations and neglected recombination. Since even weak recombination has the potential to significantly slow Muller’s ratchet, it would be interesting to generalize our dynamic balance to include its effects (Bell 1998; Gordo and Campos 2008). Increasing recombination rates would presumably allow the population to maintain better-adapted genotypes in this steady state. Last but not least, better experimental characterization of mutation effect distributions for different organisms will be needed to fully achieve the synthesis attempted in Figure 6.
The dynamic balance state has important implications for patterns of molecular evolution. Recent analysis of the effects of purifying selection on the structure of genealogies has suggested that Muller’s ratchet plays a crucial role in determining the structure of genetic variation in asexual populations or on short distance scales in the genomes of sexual organisms (Gordo et al. 2002; Seger et al. 2010). However, these expectations must be revised if populations exist instead in the dynamic state in which both beneficial and deleterious mutations fix, without any continuous net degradation or growth of fitness. This means that even though no change in fitness occurs, signatures of both positive and negative selection are likely to be found in patterns of molecular evolution, as has been suggested by earlier studies (Hartl and Taubes 1996; Antezana and Hudson 1997). We argue that this state is the natural null expectation for the effects of mutations and purifying selection on patterns of genetic variation; efforts to look for positive selection that represent “true” adaptation should look for deviations from this situation.
Finally, we note that although the dynamic balance we have analyzed is stable, a population will typically fluctuate around this steady state. A few beneficial mutations may become established at the nose by chance, leading to a temporary increase in mean fitness that is later balanced by a reduction in ε at this higher fitness, restoring the population to the equilibrium state. Conversely, a few clicks of Muller’s ratchet will occasionally lead to temporary reductions in population fitness before the corresponding increase in ε restores the steady state. These fluctuations around the dynamic balance may be important to the clonal structure of the population and hence are likely to play a key role in patterns of molecular evolution and in understanding the effects of recombination.
Supplementary Material
Acknowledgments
We thank Pierre Neveu, Adel Dayarian, Aleksandra Walczak, and Paul Sniegowski for many useful discussions. S.G., D.J.B., and B.I.S. were supported by the National Institute of General Medical Sciences GM086793 and the Human Frontier Science Program RFG0045/2010. E.R.J. acknowledges support from a National Science Foundation graduate research fellowship. R.A.N. is supported by the European Research Council through grant StG-2010-260686. M.M.D. acknowledges support from the James S. McDonnell Foundation, the Alfred P. Sloan Foundation, and the Harvard Milton Fund.
Footnotes
Communicating editor: L. M. Wahl
Literature Cited
- Andersson D., Hughes D., 1996. Muller’s ratchet decreases fitness of a dna-based microbe. Proc. Natl. Acad. Sci. USA 93: 906–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antezana M., Hudson R., 1997. Point-mutations, the ratchet, and the initial success of eukaryotic sex: a simulation study. Evol. Theory Rev. 11: 209–235 [Google Scholar]
- Bachtrog D., Gordo I., 2004. Adaptive evolution of asexual populations under Muller’s ratchet. Evol. Int. J. Org. Evol. 58: 1403–1413 [DOI] [PubMed] [Google Scholar]
- Barrick J., Kauth M., Strelioff C., Lenski R., 2010. Escherichia coli rpob mutants have increased evolvability in proportion to their fitness defects. Mol. Biol. Evol. 27: 1338–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N., Charlesworth B., 1998. Why sex and recombination? Science 281: 1986–1990 [PubMed] [Google Scholar]
- Bell G., 1998. Recombination and the immortality of the germ line. J. Evol. Biol. 1: 67–82 [Google Scholar]
- Chao L., 1990. Fitness of RNA virus decreased by Muller’s ratchet. Nature 348: 454–455 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 2012. The effects of deleterious mutations on evolution at linked sites. Genetics 190: 5–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 1997. Rapid fixation of deleterious alleles can be caused by Muller’s ratchet. Genet. Res. 70: 63–73 [DOI] [PubMed] [Google Scholar]
- De Visser J., Elena S., 2007. The evolution of sex: empirical insights into the roles of epistasis and drift. Nat. Rev. Genet. 8: 139–149 [DOI] [PubMed] [Google Scholar]
- Desai M. M., Fisher D. S., Murray A. W., 2007. The speed of evolution and maintenance of variation in asexual populations. Curr. Biol. 17: 385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte E., Clarke D., Moya A., Domingo E., Holland J., 1992. Rapid fitness loss in mammalian RNA virus clones due to Muller’s ratchet. Proc. Natl. Acad. Sci. USA 89: 6015–6019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen M., 1971. Self organization of matter and the evolution of biological macromolecules. Naturwissenschaften 58: 465–523 [DOI] [PubMed] [Google Scholar]
- Escarmis C., Davila M., Domingo E., 1999. Multiple molecular pathways for fitness recovery of an RNA virus debilitated by operation of Muller’s ratchet. J. Mol. Biol. 285: 495–505 [DOI] [PubMed] [Google Scholar]
- Estes S., Lynch M., 2003. Rapid fitness recovery in mutationally degraded lines of Caenorhabditis elegans. Evolution 57: 1022–1030 [DOI] [PubMed] [Google Scholar]
- Etheridge A., Pfaffelhuber P., Wakolbinger A., 2009. How often does the ratchet click? Facts, heuristics, asymptotics, pp. 365–390 in Trends in Stochastic Analysis, edited by P. M. Jochen Blath and M. Scheutzow. Cambridge University Press, Cambridge, UK
- Felsenstein J., 1974. The evolutionary advantage of recombination. Genetics 78: 157–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel W., Burger R., 1993. Muller’s ratchet and mutational meltdowns. Evolution 47: 1744–1757 [DOI] [PubMed] [Google Scholar]
- Gessler D., 1995. The constraints of finite size in asexual populations and the rate of the ratchet. Genet. Res. 49: 135–146 [DOI] [PubMed] [Google Scholar]
- Gordo I., Campos P., 2008. Sex and deleterious mutations. Genetics 179: 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordo I., Charlesworth B., 2000a The degeneration of asexual haploid populations and the speed of Muller’s ratchet. Genetics 154: 1379–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordo I., Charlesworth B., 2000b On the speed of Muller’s ratchet. Genetics 156: 2137–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordo I., Navarro A., Charlesworth B., 2002. Muller’s ratchet and the pattern of variation at a neutral locus. Genetics 161: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigh J., 1978. The accumulation of deleterious genes in a population. Theor. Popul. Biol. 14: 251–267 [DOI] [PubMed] [Google Scholar]
- Hartl D., Taubes C., 1996. Compensatory nearly neutral mutations: selection without adaptation. J. Theor. Biol. 182: 303–309 [DOI] [PubMed] [Google Scholar]
- Howe D., Denver D., 2008. Muller’s ratchet and compensatory mutation in Caenorhabditis briggsae mitochondrial genome evolution. BMC Evol. Biol. 8: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. F., Skaletsky H., Brown L. G., Pyntikova T., Graves T., et al. , 2012. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature 483: 82–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain K., 2008. Loss of least-loaded class in asexual populations due to drift and epistasis. Genetics 179(4): 2125–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov A., 1994. Muller’s ratchet under epistatic selection. Proc. Natl. Acad. Sci. USA 136: 1469–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov A., 1995. Contamination of the genome by very slightly deleterious mutations: Why have we not died 100 times over? J. Theor. Biol. 175: 583–594 [DOI] [PubMed] [Google Scholar]
- Lande R., 1998. Risk of population extinction from fixation of deleterious and reverse mutations. Genetica 102/103: 21–27 [PubMed] [Google Scholar]
- Lang G. I., Botstein D., Desai M. M., 2011. Genetic variation and the fate of beneficial mutations in asexual populations. Genetics 188: 647–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe L., 2006. Quantifying the genomic decay paradox due to Muller’s ratchet in human mitochondrial DNA. Genet. Res. Camb. 87: 133–159 [DOI] [PubMed] [Google Scholar]
- Lynch M., 1996. Mutation accumulation in transfer RNAs: molecular evidence for Muller’s ratchet in mitochondrial genomes. Mol. Biol. Evol. 13: 209–220 [DOI] [PubMed] [Google Scholar]
- Lynch M., Burger R., Butcher D., Gabriel W., 1993. The mutational meltdown in asexual populations. J. Hered. 84: 339–344 [DOI] [PubMed] [Google Scholar]
- Lynch M., Conery J., Burger R., 1995. Mutation accumulation and the extinction of small populations. Am. Nat. 146: 489–518 [Google Scholar]
- Manrubia S. C., Lazaro E., Perez-Mercader J., 2003. Fitness distributions in exponentially growing asexual populations. Phys. Rev. Lett. 90(18): 188102. [DOI] [PubMed] [Google Scholar]
- Moya A., Elena S. F., Bracho A., Miralles R., Barrio E., 2000. The evolution of RNA viruses: a population genetics view. Proc. Natl. Acad. Sci. USA 97: 6967–6973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H., 1964. The relation of recombination to mutational advance. Mutat. Res. 1: 2–9 [DOI] [PubMed] [Google Scholar]
- Neher R. A., Shraiman B. I., 2012. Fluctuations of fitness distributions and the rate of Muller's ratchet. Genetics 191: 1309–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffelhuber P., Staab P. R., Wakolbinger A., 2011. Muller’s ratchet with compensatory mutations. Ann. Appl. Probab. (in press) [Google Scholar]
- Poon A., Chao L., 2005. The rate of compensatory mutation in the DNA bacteriophage φx174. Genetics 170: 989–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon A., Otto S., 2000. Compensating for our load of mutations: freezing the meltdown of small populations. Evolution 54: 1467–1479 [DOI] [PubMed] [Google Scholar]
- Poon A., Davis B., Chao L., 2005. The coupon collector and the suppressor mutation: estimating the number of compensatory mutations by maximum likelihood. Genetics 170: 1323–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice W., 1994. Degeneration of a nonrecombining chromosome. Science 263: 230–232 [DOI] [PubMed] [Google Scholar]
- Rouzine I., Wakeley J., Coffin J., 2003. The solitary wave of asexual evolution. Proc. Natl. Acad. Sci. USA 100: 587–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzine I., Brunet E., Wilke C., 2008. The traveling-wave approach to asexual evolution: Muller’s ratchet and speed of adaptation. Theor. Popul. Biol. 73: 24–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoustra S., Bataillon T., Gifford D., Kassen R., 2009. The properties of adaptive walks in evolving populations of fungus. PLoS Biol. 7: e1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S., Lynch M., 1997. Deleterious mutation and extinction: the role of variable mutational effects, synergistic epistasis, beneficial mutations, and degree of outcrossing. Evolution 51: 1363–1371 [DOI] [PubMed] [Google Scholar]
- Seger J., Smith W. A., Perry J. J., Hunn J., Kaliszewska Z. A., et al. , 2010. Gene genealogies strongly distorted by weakly interfering mutations in constant environments. Genetics 184: 529–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silander O., Tenaillon O., Chao L., 2007. Understanding the evolutionary fate of finite populations: the dynamics of mutational effects. PLoS Biol. 5: e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg R., Berg O., 2007. Mutational interference and the progression of Muller’s ratchet when mutations have a broad range of deleterious effects. Genetics 177: 971–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W., Kim Y., 2002, Recent Applications of Diffusion Theory to Population Genetics. Oxford University Press, Oxford, pp. 72–93 [Google Scholar]
- Trindade S., Perfeito L., Gordo I., 2010. Rate and effects of spontaneous mutations that affect fitness in mutator Escherichia coli. Philos. Trans. R. Soc. B Biol. Sci. 365: 1177–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G., Gabriel W., 1990. Quantitative variation in finite parthenogenetic populations: What stops Muller’s ratchet in the absence of recombination? Evolution 44: 715–731 [DOI] [PubMed] [Google Scholar]
- Zeyl C., Mizesko M., de Visser J., 2001. Mutational meltdown in laboratory yeast populations. Evolution 55: 909–917. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.