Abstract
Human peripheral blood contains antigen-presenting cells (APC), including dendritic cells (DC) and monocytes, that may encounter microbes that have translocated from the intestine to the periphery in disease states like HIV-1 infection and inflammatory bowel disease. We investigated the response of DC and monocytes in peripheral blood mononuclear cells (PBMC) to a panel of representative commensal enteric bacteria, including Escherichia coli, Enterococcus sp., and Bacteroides fragilis. All three bacteria induced significant upregulation of the maturation and activation markers CD40 and CD83 on myeloid dendritic cells (mDC) and plasmacytoid dendritic cells (pDC). However, only mDC produced cytokines, including interleukin-10 (IL-10), IL-12p40/70, and tumor necrosis factor alpha (TNF-α), in response to bacterial stimulation. Cytokine profiles in whole PBMC differed depending on the stimulating bacterial species: B. fragilis induced production of IL-23, IL-12p70, and IL-10, whereas E. coli and Enterococcus induced an IL-10-predominant response. mDC and monocyte depletion experiments indicated that these cell types differentially produced IL-10 and IL-23 in response to E. coli and B. fragilis. Bacteroides thetaiotaomicron did not induce levels of IL-23 similar to those of B. fragilis, suggesting that B. fragilis may have unique proinflammatory properties among Bacteroides species. The addition of recombinant human IL-10 to PBMC cultures stimulated with commensal bacteria abrogated the IL-23 response, whereas blocking IL-10 significantly enhanced IL-23 production, suggesting that IL-10 controls the levels of IL-23 produced. These results indicate that blood mDC and monocytes respond differentially to innate stimulation with whole commensal bacteria and that IL-10 may play a role in controlling the proinflammatory response to translocated microbes.
INTRODUCTION
Commensal bacteria contribute to the normal microflora that reside on epithelial surfaces such as the gastrointestinal tract, which is a major area of bacterial colonization (20). Enteric bacteria are sequestered within the lumen of the intestine by the intestinal epithelial barrier, as well as by factors such as secretory IgA (50), mucus (32), and bacterium-reactive CD4+ T cells in the lamina propria (LP) (25). Enteric bacteria within the intestinal lumen are well tolerated. However, if bacteria escape from the lumen into the systemic circulation, they may proliferate and cause disease, such as sepsis in immunocompromised hosts (35). Therefore, the host immune system must maintain a delicate balance in which commensal bacteria are tolerated in certain locations, like the gut lumen, yet are quickly contained if found beyond these environments.
The process of bacterial escape from the lumen of the intestine into the underlying mucosal tissue and ultimately into the periphery is known as microbial translocation. This process occurs in diseases characterized by intestinal inflammation and epithelial barrier breakdown, such as inflammatory bowel disease (IBD) and human immunodeficiency virus type 1 (HIV-1) infection, allowing microbial products to enter the LP, lymph nodes, and peripheral blood (6, 8). Indeed, bacterial products, such as bacterial lipopolysaccharide (LPS) and bacterial 16S ribosomal DNA, are increased in the blood of patients with HIV-1 infection and are associated with T cell activation (7, 29, 39). Translocation of whole enteric bacteria and bacterial products from the gut into the LP, peripheral lymph nodes, and liver has been demonstrated in simian immunodeficiency virus (SIV)-infected macaques, an animal model of HIV infection (16). Thus, immune cells in the periphery may encounter whole, translocated bacteria as well as bacterial products, and the subsequent response could be a factor contributing to the systemic immune activation and inflammation characteristic of HIV-1 infection. However, the manner in which circulating microbes and microbial products induce systemic inflammation and, in particular, activation of the innate immune cells in peripheral blood remains poorly characterized.
Dendritic cells (DC) are the most potent antigen-presenting cells (APC) in the immune system. They are unique in their ability to bridge innate and adaptive immunity by sensing antigen through innate receptors and presenting this antigen to adaptive immune cells (5, 27). Innate recognition of microbes by DC occurs when conserved viral or bacterial motifs, known as microbe-associated molecular patterns (MAMP), bind to specific pattern recognition receptors (PRR), such as Toll-like receptors (TLR) expressed by the DC. The two major subsets of human blood DC are plasmacytoid DC (pDC) and myeloid DC (mDC). pDC express TLR7 and -9 and produce mainly alpha interferon (IFN-α) in response to stimulation (26, 27). mDC express TLR1 to TLR8 (except TLR7) and produce multiple cytokines, including those of the interleukin-12 (IL-12) family, upon stimulation (27, 28, 30). Human blood monocytes, another class of APC, also express TLR1 to TLR8 (except TLRs 3 and 7) (27, 30) and are able to produce cytokines in response to stimulation, including tumor necrosis factor alpha (TNF-α), IL-6, IL-10, and IL-12 (2, 27, 30).
Previous studies utilizing monocytes, monocyte-derived DC, purified DC, total peripheral blood mononuclear cells (PBMC), or whole blood have demonstrated that pathogenic (15, 17, 40, 48, 53) and commensal bacteria, both probiotic (18, 21, 33, 41, 49) and nonprobiotic (3, 4, 22–24, 31, 54), can induce cytokines that include IL-10 and IL-12p70. However, few studies have examined and compared the responses of human blood DC and monocytes within total PBMC to whole, commensal enteric bacteria, which may be present during microbial translocation. Understanding the capacity of blood APC subsets to respond to bacterial stimulation under normal conditions could help guide future studies investigating the response of these cells in different disease states.
In this report, we measured the cytokine response of total PBMC, as well as specific DC subsets and monocytes within PBMC, from healthy human subjects to stimulation with a panel of whole, enteric commensal bacteria (14). The panel of bacteria used in these studies constitute phyla found to be overrepresented in human mucosal biopsy specimens and therefore might be most likely to translocate into the periphery in the setting of intestinal epithelial barrier breakdown (19). We observed that each of the enteric bacteria tested induced distinct cytokine profiles from blood mDC and monocytes, as well as in total PBMC. Finally, we observed that the ability of PBMC to respond in a proinflammatory manner to bacteria was controlled by the production of the anti-inflammatory cytokine IL-10, produced largely by monocytes.
MATERIALS AND METHODS
Study participants.
Peripheral blood samples were obtained from 25 healthy adults (16 females and 9 males) with a median age of 31 years (range, 23 to 53 years) who voluntarily gave written informed consent. Collection of peripheral blood samples was approved by the Colorado Multiple Institutional Review Board (COMIRB) at the University of Colorado, Anschutz Medical Campus.
Collection and preparation of human PBMC.
After collection of peripheral blood into heparinized Vacutainers, whole blood was spun down at 1,700 rpm for 10 min in order to remove plasma. PBMC were isolated after the removal of plasma by standard Ficoll-Hypaque (Amersham Biosciences, Piscataway, NJ) density gradient centrifugation as described previously (11, 12, 25). PBMC were used in assays immediately following isolation.
Whole-bacterium preparations.
Intact Escherichia coli (Proteobacteria phylum), Enterococcus species (sp.) (Firmicutes phylum), and Bacteroides fragilis and Bacteroides thetaiotaomicron (Bacteroidetes phylum) were used to stimulate PBMC. E. coli stocks were obtained from the American Type Culture Collection (ATCC 25922; Manassas, VA). Enterococcus sp. was isolated from human mucosal tissue and typed by the Clinical Microbiology Laboratory at the University of Colorado Hospital as previously described (25). Enterococcus sp. was expanded by incubation in RPMI medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO) at 37°C with a 5% CO2 atmosphere. E. coli was either expanded as with Enterococcus sp. or by plating on brain heart infusion (BHI) agar (BD Diagnostics, Sparks, MD) followed by incubation at 37°C for 1 day. B. fragilis and B. thetaiotaomicron stocks (ATCC strains 25285 and 29741, respectively) were obtained from the Clinical Microbiology Laboratory or directly from the ATCC and were expanded by culturing on brucella plates (BD Diagnostics) and incubated at 35° to 37°C for 2 days under anaerobic conditions, achieved by using a BD GasPak EZ Anaerobe Pouch System (BD Diagnostics). In addition, a mutant B. fragilis strain defective in polysaccharide A (PSA) expression (B. fragilis ΔPSA) and the corresponding wild-type (WT) strain (both generous gifts from S. Mazmanian, California Institute of Technology, Pasadena, CA) were expanded at 35°C under anaerobic conditions on brucella plates. All bacteria were heat treated for 2 h at 56°C and then washed and resuspended in Dulbecco's phosphate-buffered saline (DPBS) in order to inhibit their growth prior to incubation with cells. The total number of bacterial cells was assessed by diluting samples in trypan blue (Sigma-Aldrich) and determining counts on a hemocytometer at least twice. The bacteria were then frozen at stock concentrations of 3 × 109/ml in DPBS.
In vitro stimulation of PBMC.
Immediately after Ficoll separation, PBMC were resuspended at 2 × 106 cells/ml in complete medium (CM) consisting of RPMI medium (Invitrogen) supplemented with 1% penicillin-streptomycin-l-glutamine (Sigma-Aldrich) and 10% human AB serum (HS, Gemini Bioproducts, West Sacramento, CA). Cells were incubated at 37°C in a humidified 5% CO2 atmosphere for 18 to 24 h with either heat-treated E. coli, Enterococcus sp., B. fragilis, B. fragilis ΔPSA, or B. thetaiotaomicron at a ratio of 1 PBMC to 5 bacterial cells or with 5 μg/ml CL097, a derivative of the imidazoquinoline compound R848 (TLR7/TLR8 ligands; InvivoGen, San Diego, CA), as a positive control or with medium alone. For assessment of intracellular cytokines, 1 μg/ml Golgi plug (brefeldin A; BD Biosciences, San Jose, CA) was added 1 h after PBMC were placed into culture. For assessment of secreted cytokines, culture supernatants were collected after 20 to 24 h of cell culture in the absence of Golgi plug and frozen at −20°C. Prior to cell collection, 10 μg/ml DNase I (Sigma-Aldrich) was added for 5 min at 37°C to dissociate cell clumps.
In vitro stimulation of mDC-depleted and monocyte-depleted cultures.
The majority of blood mDC have been shown to be BDCA-1+ (CD1c+), and a small population was shown to be BDCA-3+ (CD141+) (13). Therefore, to deplete PBMC of mDC, PBMC were labeled with a cocktail of biotin–BDCA-1 and biotin–BDCA-3, followed by antibiotin microbeads (all from Miltenyi Biotec, Auburn, CA). FcR blocking reagent (Miltenyi Biotec) was used to limit nonspecific binding through Fc receptors. The cells were run over LS Columns (Miltenyi Biotec) twice to increase the purity of the mDC-depleted fraction. In order to control for the effect of the magnetic column, unlabeled PBMC were also run through an LS Column twice and used as control total PBMC. The median percentage of mDC (negative for a lineage cocktail consisting of CD3, CD14, CD16, CD19, CD20, and CD56 [lineage cocktail−] and expressing HLA-DR+, CD11c+, and low levels of CD123 [CD123low]) in the total PBMC control was 0.37% (range, 0.24 to 0.56%). The median percentage of mDC within the mDC-depleted fraction was 0.01% (range, 0.01 to 0.03%). Total PBMC and the mDC-depleted fraction were cultured at 2 × 106 cells/ml in CM and stimulated with E. coli and B. fragilis as previously described. Culture supernatants were collected after 18 to 24 h in culture and stored at −20°C.
PBMC were depleted of monocytes by positive selection using CD14+ microbeads (Miltenyi Biotec), as per the manufacturer's recommended protocol. As with the mDC depletion, the monocyte-depleted and unlabeled total PBMC control fractions were run over LS Columns twice. The median percentage of CD14+ monocytes within the total PBMC control was 13.04% (range, 6.82 to 14.57%). The median percentage of CD14+ monocytes within the monocyte-depleted fraction was 0.1% (range, 0.03 to 0.18%). Total PBMC and the monocyte-depleted fraction were cultured at 2 × 106 cells/ml in CM and stimulated with E. coli and B. fragilis as previously described. Culture supernatants were collected after 18 to 24 h in culture and stored at −20°C.
In vitro stimulation of monocyte-enriched cultures.
PBMC were enriched for monocytes by negative selection using a magnetic bead kit (Monocyte Isolation Kit II; Miltenyi Biotec), as per the manufacturer's recommended protocol with the addition of a biotin–BDCA-1 antibody (Miltenyi Biotec) to the provided antibody-biotin cocktail in order to remove BDCA-1+ mDC. In order to increase the purity of the enriched monocytes, labeled cells were run over the LS Column twice. The median percentage of CD14+ monocytes within total PBMC prior to enrichment was 13.4% (range, 8.29 to 21.74%). The monocyte-enriched fraction contained a median of 64.39% CD14+ monocytes (range, 52.47 to 80.18%). Enriched monocyte cultures were plated at 2 × 106 cells/ml in CM and stimulated with E. coli and B. fragilis as previously described. Culture supernatants were collected after 18 to 24 h of culture and stored at −20°C.
Addition of recombinant human proteins and anti-human IL-10 antibody.
In some experiments, 20 ng/ml of recombinant human IL-10 ([rhIL-10] R&D Systems, Minneapolis, MN) was added to PBMC cultures for 2 h. E. coli or B. fragilis was then added at a ratio of 1 PBMC to 5 bacterial cells for an additional 1 h. Finally, 1 μg/ml Golgi plug was added, and cultures were incubated for a further 16 to 19 h before collection and used in flow cytometry assays. For supernatant collection, 20 ng/ml of rhIL-10, rhIL-12, rhIL-23, or 10 μg/ml anti-human IL-10 antibody or matched isotype (all from R&D systems) was added to PBMC cultures concurrently with E. coli or B. fragilis. In dose titration experiments, a range of 0.1 ng/ml to 20 ng/ml of rhIL-10 was added to the cultures. Supernatants were collected after 22 to 24 h and stored at −20°C.
Antibodies for flow cytometry.
In order to identify peripheral blood DC in PBMC, the following antibodies were used: lineage cocktail (fluorescein isothiocyanate [FITC]-labeled anti-CD3/CD14/CD16/CD19/CD20/CD56), HLA-DR (allophycocyanin [APC]-Cy7 label), CD11c (phycoerythrin [PE]-Cy5 label) (all from BD Biosciences), and CD123 (peridinin chlorophyll protein [PerCP]-Cy5.5 [BD Biosciences] or APC [Miltenyi Biotec] label). mDC were identified as lineage cocktail− HLA-DR+ CD123low CD11c+, whereas pDC were identified as lineage cocktail− HLA-DR+ CD123high CD11c− (where CD123high indicates high levels of CD123). Representative flow plots of the DC gating strategy are shown in Fig. S1 in the supplemental material. This gating strategy was also used to assess the purity of mDC-depleted fractions. To assess the purity of monocyte-enriched fractions and monocyte-depleted fractions, cells were stained with an antibody against CD14 (V450; BD Biosciences). To identify maturation markers expressed on peripheral blood DC, the following antibodies were used: biotinylated CD40 (Ancell, Bayport, MN), streptavidin-PE-Texas Red (energy-coupled dye [ECD]; Beckman Coulter, Fullerton, CA), and CD83 (PE label; BD Biosciences). To identify intracellular cytokines, the following antibodies were used: IL-10 (PE label) and TNF-α (Alexa-Fluor 700) (both from BD Biosciences), IL-12p40/70 (Pacific Blue or eFluor 450) and IL-23p19 (PE) (both from eBiosciences, San Diego, CA), and IFN-α (APC; Miltenyi Biotec). The recommended, appropriate isotype control antibody for each cytokine antibody was used in each assay.
Flow cytometry protocol.
Surface marker and intracellular cytokine expression levels by DC subsets within total PBMC were assessed using standard flow cytometric protocols as previously described (11, 12) on an LSR-II flow cytometer (BD Immunocytometry Systems, San Jose, CA). Briefly, to assess surface expression of activation and maturation markers, the cells were stained with biotin anti-CD40 and FcR blocking reagent. Next, the cells were stained with streptavidin-ECD, PE-labeled anti-CD83, and the DC-identifying antibodies described above. The cells were then stained with a Live/Dead Fixable Dead Cell Stain (aqua-fluorescent reactive dye; Invitrogen), fixed in 4% paraformaldehyde (PFA; Sigma-Aldrich), and resuspended in DPBS containing 1% bovine serum albumin (BSA) supplemented with 2 mM EDTA (fluorescence-activated cell sorting [FACS] buffer) for acquisition.
To identify intracellular cytokines, PBMC were surface stained with the DC-identifying antibodies and then the Live/Dead Fixable Dead Cell Stain. Next, the cells were fixed and permeabilized in Cytofix/Cytoperm buffer (BD Biosciences) and incubated with antibodies for IL-10, IL-12p40/70, IL-23p19, TNF-α, and IFN-α and resuspended in 0.5% PFA for acquisition.
To assess the purity of the monocyte-enriched or -depleted fractions, total PBMC, enriched monocytes, and monocyte-depleted cells were stained with anti-CD14 and FcR block, stained with Live/Dead Fixable Cell Stain, fixed in 4% PFA, and resuspended in FACS buffer for acquisition. To assess the purity of the mDC-deleted fractions, total PBMC and mDC-depleted fractions were stained with the DC-identifying antibodies, stained with Live/Dead Fixable Cell Stain, fixed in 4% PFA, and resuspended in FACS buffer for acquisition.
Quality control on the LSR-II was performed daily through the Cytometer Setup and Tracking component of the BD FACSDiva software (BD Biosciences), as previously described (10). A median of 401987.5 total events (range, 67,589 to 896308 total events) was collected from FACS experiments.
Detection of IL-23, IL-12p70, and IL-10 within culture supernatants.
IL-12p70, IL-23, and IL-10 enzyme-linked immunosorbent assays ([ELISAs] all from eBioscience) were completed using thawed culture supernatants following the manufacturer's recommended protocols. Lower detection limits were 4 pg/ml for IL-12p70, 15 pg/ml for IL-23, and 2.3 pg/ml for IL-10.
Data and statistical analysis.
All flow cytometry data analysis was performed using FACSDiva software, version 6.1.2 (BD Biosciences). Isotype control values were subtracted from the antibody values as appropriate. Net values of the percentage of cytokine-positive cells and amount of soluble cytokine production were calculated by subtracting the background (medium-only values) from the stimulation condition values. A Wilcoxon matched-pairs signed rank test was used to assess induction of cytokine production or expression of surface markers under the stimulated condition relative to values with medium only. A Friedman test was used for matched paired comparisons across multiple groups with Dunn's multiple comparison test performed for P values of <0.05. Statistical analysis was performed using GraphPad Prism statistical software (version 5.0; GraphPad Software, San Diego, CA).
RESULTS
Surface activation markers are upregulated on blood pDC and mDC in response to stimulation with bacteria.
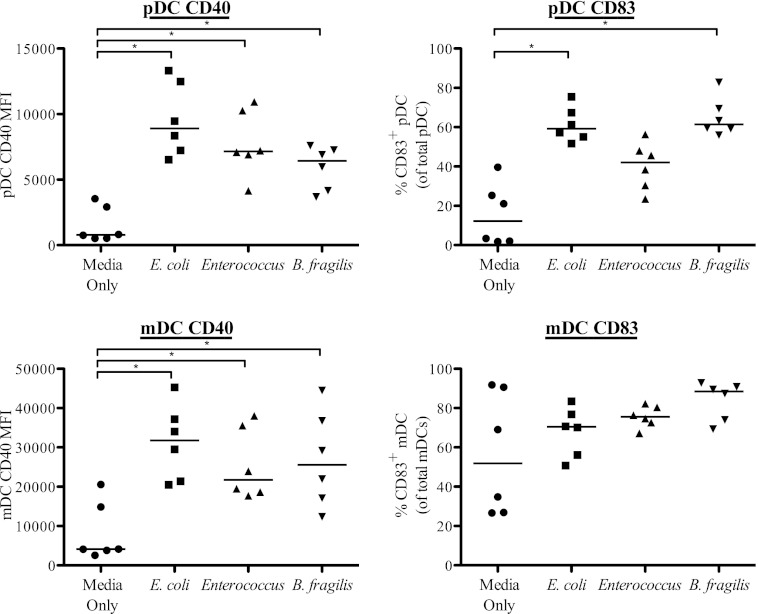
To study the effect of enteric commensal bacteria on the maturation profile of the two blood DC subsets, PBMC were cultured with heat-treated E. coli, Enterococcus, or B. fragilis or with medium alone, and surface expression levels of the maturation markers CD40 and CD83 were analyzed on pDC and mDC within total PBMC by multiparameter flow cytometry. An example of the gating strategy used to identify the DC subsets in viable PBMC and representative flow plots showing CD40 and CD83 expression on both mDC and pDC after B. fragilis stimulation are shown in Fig. S1 in the supplemental material. All bacterial stimuli induced statistically significant upregulation of CD40 expression on both pDC and mDC relative to that observed in unstimulated cultures (Fig. 1). The extent of the increase of CD40 varied with the species of bacteria, with E. coli generally inducing the highest CD40 expression on both DC subsets. All three bacteria induced increased frequencies of CD83+ pDC relative to unstimulated pDC although this did not reach statistical significance for Enterococcus stimulation (P = 0.06). Conversely, although the median percentage of mDC expressing CD83 was increased in response to bacteria compared to the level in the medium-only control, the increase did not reach statistical significance for any of the bacteria (E. coli, P = 0.44; Enterococcus, P = 0.31; B. fragilis, P = 0.09). It was noted that the frequency of CD83+ mDC markedly increased in cultures without exogenous stimulation (medium-only control) relative to that measured directly ex vivo, resulting in high background expression, as reported by others (36).
Fig 1.
Blood pDC and mDC upregulate activation and maturation markers in response to stimulation with commensal bacteria. Maturation marker expression on pDC and mDC in response to stimulation with E. coli, Enterococcus, or B. fragilis (n = 6) was determined. Results are shown as net CD40 mean fluorescence intensity (MFI) (the net value was calculated by subtracting out isotype staining) or as the percentage of cells expressing CD83 within the total pDC or mDC population (percent positive) minus the isotype control. Horizontal bars indicate median values. Statistical significance for each stimulation condition compared to the medium-only condition was evaluated using a Wilcoxon matched-pairs signed rank test. *, P < 0.05.
Bacterial stimulation of PBMC induces differential mDC cytokine production.
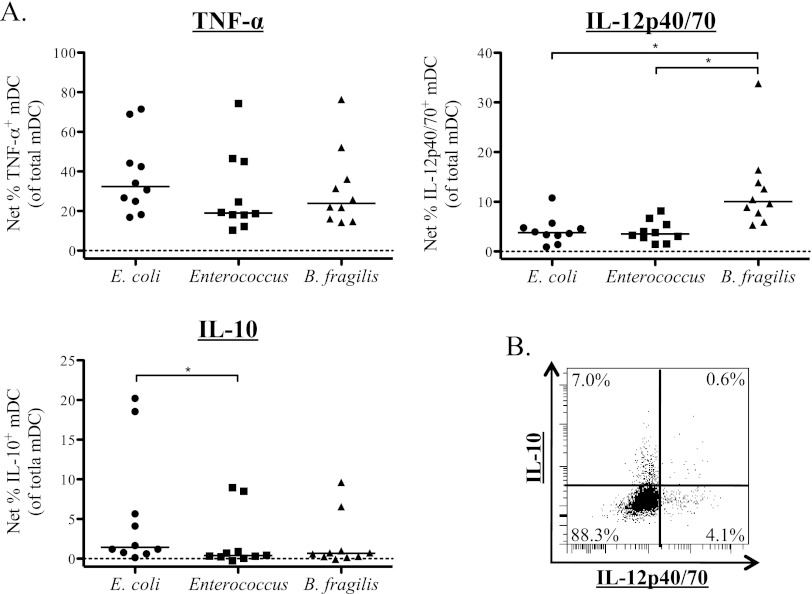
Bacterial stimulation failed to induce significant frequencies of pDC to produce cytokine (see Fig. S2 in the supplemental material). Thus, all further experiments in this report focused on cell types that were found to produce cytokines in response to bacterial stimulation. Indeed, stimulation with each of the three bacterial species induced significant frequencies of mDC to produce TNF-α, IL-12p40/70, and IL-10 (see Table S1 in the supplemental material) as measured by intracellular cytokine staining (ICS). Representative flow plots of the mDC ICS assay are shown in Fig. S3. Comparison of net cytokine frequencies, calculated by subtracting the medium-only value from the stimulation value showed that there were no significant differences between the net percentages of TNF-α+ mDC induced by the three bacteria (overall P value, = 0.60) (Fig. 2A). However, IL-12p40/70+ mDC frequencies in response to B. fragilis were significantly greater than those induced by E. coli (P < 0.01) or Enterococcus (P < 0.01) (Fig. 2A). E. coli induced greater frequencies of IL-10+ mDC than the other bacteria tested although this only reached statistical significance between E. coli and Enterococcus (Fig. 2A). When the coproduction of cytokines by bacteria-stimulated mDC was analyzed, it was noted that mDC production of IL-10 and IL-12p40/70 occurred in distinct mDC populations, regardless of the bacteria used to stimulate the cells (Fig. 2B).
Fig 2.
Blood mDC produce IL-12p40/70, IL-10, and TNF-α in response to stimulation with commensal bacteria. Total PBMC were stimulated with bacteria, and multicolor flow cytometry was used to assess expression of cytokines by mDC. (A) Percentage of mDC expressing intracellular TNF-α, IL-12p40/70, and IL-10 in response to stimulation with E. coli, Enterococcus, or B. fragilis (n = 10). Results are shown as the percentage of total mDC positive for expression of the indicated cytokine with background (isotype staining) subtracted from the antibody value. Horizontal bars indicate median values. Statistical significance for each stimulation condition compared to the medium-only condition was evaluated using a Wilcoxon matched-pairs signed rank test. **, P < 0.01. (B) Example of a flow plot demonstrating production of IL-10 and IL-12p40/70 by different mDC populations in response to E. coli stimulation.
Production of IL-12p70, IL-23, and IL-10 by total PBMC in response to bacterial stimulation.
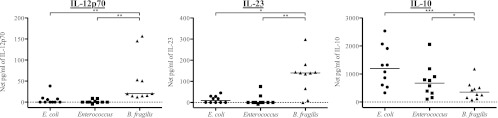
IL-12 and IL-23 share the same p40 subunit (42). Therefore, to determine whether the intracellular IL-12p40/70 staining of mDC was associated with IL-12p70 or IL-23 production, levels of IL-12p70 and IL-23 production within the culture supernatant collected after overnight culture of PBMC in medium only or stimulated with E. coli, Enterococcus, or B. fragilis were assessed (see Table S2 in the supplemental material]). In addition, to confirm IL-10 cytokine production as detected by the ICS assays, IL-10 ELISAs were performed on culture supernatant. Each bacterial species induced statistically significant levels of IL-10 relative to the medium-only condition (see Table S2). Only stimulation with B. fragilis induced significant levels of IL-12p70, whereas both B. fragilis and E. coli induced significant levels of IL-23 compared to the medium-only condition (see Table S2). B. fragilis induced significantly more IL-12p70 and IL-23 than E. coli or Enterococcus (Fig. 3). Conversely, E. coli and Enterococcus induced levels of IL-10 that were significantly higher than the level induced by B. fragilis (Fig. 3). Stimulation of PBMC with E. coli and B. fragilis simultaneously led to production of similar levels of IL-10 compared to the level with E. coli alone. However, there was no statistical difference in IL-23 production in response to stimulation with E. coli and B. fragilis simultaneously compared to stimulation with E. coli or B. fragilis alone (see Fig. S4 in the supplemental material).
Fig 3.
Stimulation of total PBMC with different commensal bacteria induces distinct cytokine profiles. Total PBMC were stimulated with E. coli, Enterococcus, and B. fragilis, and levels of IL-12p70, IL-23, and IL-10 were assessed within the culture supernatant by ELISA (n = 10). Results are shown as the net amount of the indicated cytokine (stimulation condition value minus the medium-only value). Horizontal bars indicate median values. Statistical significance between each stimulation condition was evaluated using a Freidman test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
mDC and monocytes differentially contribute to IL-10 and IL-23 production in response to stimulation with E. coli and B. fragilis.
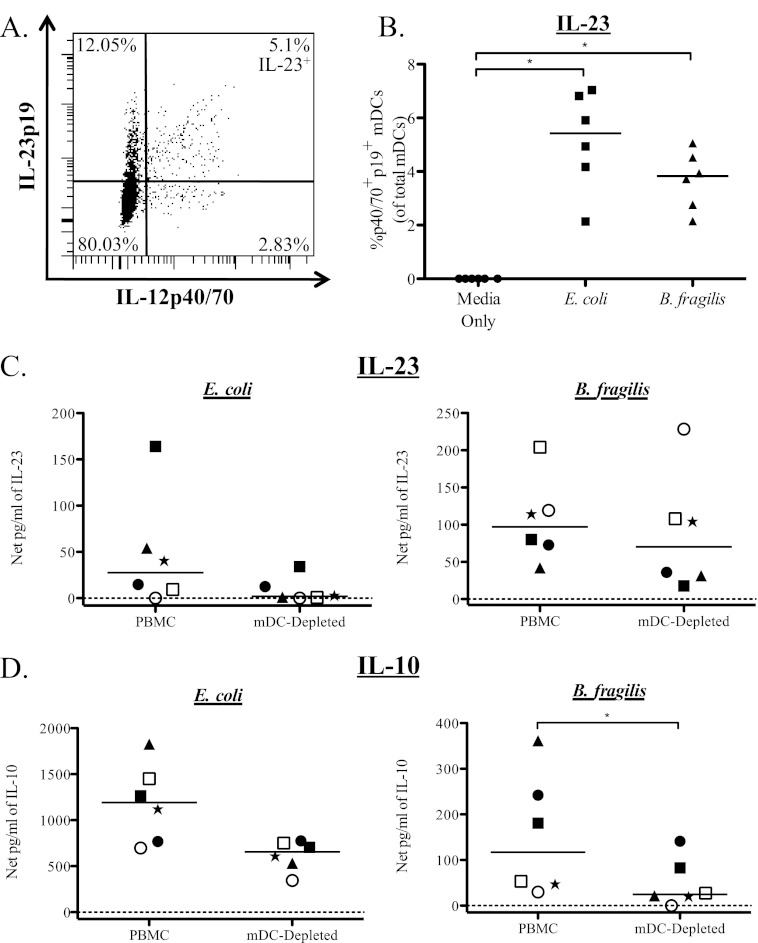
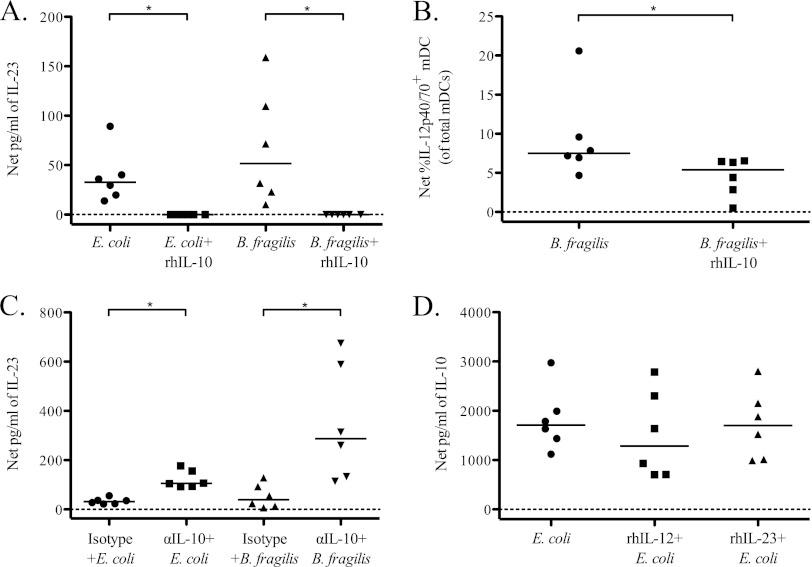
To assess mDC-specific IL-23 production, we used a recently commercially available antibody specific for the human IL-23p19 subunit appropriate for use in an ICS assay in conjunction with the IL-12p40/70 antibody (a representative flow plot shown in Fig. 4A). IL-12p40/70+ IL-23p19+ (IL-23) mDC were significantly induced in response to stimulation with both E. coli and B. fragilis, suggesting that mDC do produce IL-23 in response to bacterial stimulation (Fig. 4B).
Fig 4.
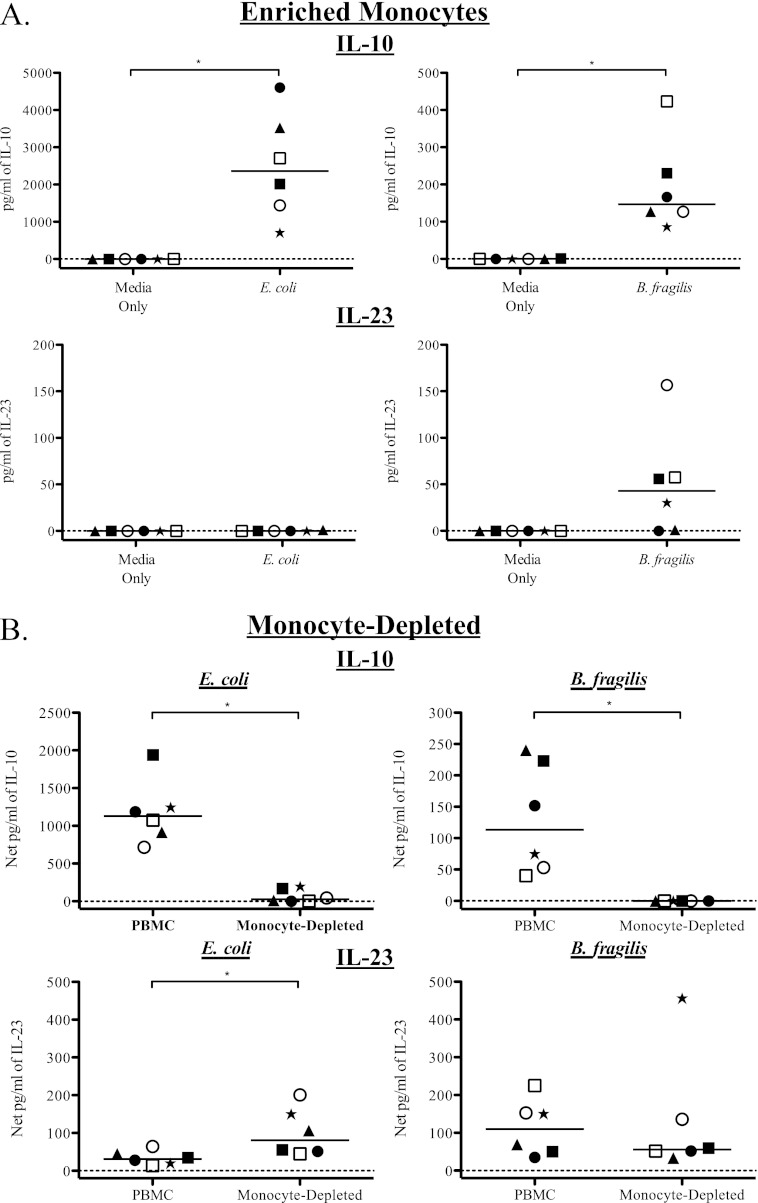
mDC produce IL-10 and IL-23 in response to E. coli and B. fragilis stimulation. Total PBMC were stimulated with bacteria, and mDC were evaluated for expression of IL-12p40/70 and IL-23p19. (A) Example flow plot demonstrating production of IL-12p40/70 and IL-23p19 by mDC in response to B. fragilis stimulation. (B) Percentage of mDC expressing IL-12p40/70 and IL-23p19 (IL-12p40/70+ IL-23p19+) after stimulation with E. coli and B. fragilis (n = 6). Results are shown as the net percentage of IL-12p40/70+ IL-23p19+ mDC, with background (isotype staining) subtracted from the antibody value. To evaluate the contribution of mDC to total cytokine production, mDC were depleted from total PBMC, and levels of IL-23 (C) and IL-10 (D) in response to E. coli and B. fragilis were evaluated in culture supernatants. Results are shown as the net amount of the indicated cytokine (stimulation condition minus the medium-only value). In order to demonstrate the response of individual donors under each condition, different symbols are used to identify each donor. Horizontal bars indicate median values. Statistical significance between each condition was evaluated using a Wilcoxon matched-pairs signed rank test. *, P < 0.05.
To confirm the contribution of mDC to PBMC cytokine production in response to bacterial stimulation, mDC were depleted from total PBMC, and culture supernatants were assessed for cytokine production by ELISA. In five of six donors tested, IL-23 production was reduced in mDC-depleted cultures compared to the level in total PBMC when stimulated with either E. coli or B. fragilis (Fig. 4C). Although reduction of IL-23 did not reach statistical significance for stimulation with either of the bacteria (E. coli, P = 0.06; B. fragilis, P = 0.44), the reduction in IL-23 with mDC depletion in response to E. coli appeared to be more complete than the response to B. fragilis. Depletion of mDC also caused a reduction in the amount of IL-10 produced in response to both bacteria, with the decrease reaching statistical significance only in response to B. fragilis (Fig. 4D).
As mDC did not appear to be responsible for all of the IL-23 and IL-10 produced by PBMC in response to bacteria, we investigated the potential contribution of CD14+ monocytes. PBMC were enriched for monocytes and stimulated with E. coli or B. fragilis. Enriched monocytes produced significant amounts of IL-10 in response to both bacteria compared to the medium-only control (Fig. 5A) although the quantities of IL-10 in response to E. coli were, on average, 15.5-fold greater than in response to B. fragilis. Conversely, the enriched monocytes did not produce IL-23 in response to E. coli but did produce low levels of IL-23 in response to B. fragilis compared to the medium-only control (Fig. 5A) although this only trended toward statistical significance (P = 0.06). Monocyte depletion resulted in a statistically significant decrease in IL-10 production in response to both E. coli and B. fragilis (Fig. 5B), confirming the important role of monocytes in IL-10 production. Notably, the depletion of monocytes from total PBMC resulted in an increase in IL-23 production in response to E. coli stimulation and a trend toward a decrease in IL-23 production in response to B. fragilis (Fig. 6B).
Fig 5.
Monocytes contribute to IL-10 production in response to E. coli and B. fragilis stimulation. To evaluate the contribution of monocytes to cytokine production, total PBMC were enriched or depleted of monocytes and stimulated with bacteria. (A) Monocytes were enriched from total PBMC and stimulated with E. coli and B. fragilis (n = 6). Levels of IL-10 and IL-23 were evaluated within culture supernatants. Results are shown as pg/ml of the indicated cytokine. To evaluate the contribution of monocytes to total cytokine production, monocytes were depleted from total PBMC (B), and levels of IL-10 and IL-23 in response to E. coli and B. fragilis were evaluated in culture supernatants (n = 6). Results are shown as the net amount of the indicated cytokine (stimulation condition minus the medium-only value). In order to demonstrate the response of individual donors under each condition, different symbols are used to identify each donor. Horizontal bars indicate median values. Statistical significance between each condition was evaluated using a Wilcoxon matched-pairs signed rank test. *, P < 0.05.
Fig 6.
IL-10 controls the production of IL-12p70 and IL-23 by total PBMC and IL-12p40/70 by mDC in response to stimulation with commensal bacteria. To evaluate the effect of IL-10 on the total response to bacteria, exogenous recombinant human IL-10 (rhIL-10) was added to total PBMC cultures stimulated with bacteria, and cytokine production was assessed by ELISA and multicolor flow cytometry. (A) Production of IL-23 by total PBMC in response to stimulation with E. coli or B. fragilis in the presence or absence of rhIL-10 within culture supernatants (n = 6). Results are shown as the net amount of IL-23 (stimulation condition value minus the medium-only value). (B) Detection of intracellular IL-12p40/70 within mDC after stimulation with B. fragilis or B. fragilis plus rhIL-10 by flow cytometry (n = 6). Results are shown as the net percentage of total mDC positive for expression of IL-12p40/70. (C) To evaluate IL-23 production in the absence of the suppressive effect of IL-10, total PBMC were stimulated with E. coli or B. fragilis in the presence or absence of anti-IL-10 (α-IL-10) antibody or a matched isotype control, and IL-23 expression was measured within culture supernatants (n = 6). Results are shown as the net amount of IL-23. (D) To evaluate the effect of IL-12p70 and IL-23 on IL-10 production in response to bacterial stimulation, exogenous rhIL-12 and rhIL-23 were added to total PBMC cultures stimulated with E. coli, and production of IL-10 was assessed within culture supernatants (n = 6). Results are shown as the net amount of IL-10. Horizontal bars indicate median values. Statistical significance between different conditions was evaluated using a Wilcoxon matched-pairs signed rank test. *, P < 0.05.
IL-10 regulates the IL-23 response to commensal bacteria stimulation.
Since reciprocal patterns of IL-10 and IL-23 production were observed in response to E. coli versus B. fragilis stimulation, as well as an increase in IL-23 production in response to E. coli when the IL-10-producing monocytes were removed from total PBMC, we evaluated whether either cytokine regulated production of the other in response to bacterial stimulation in vitro. The addition of rhIL-10 at a concentration of 20 ng/ml completely abrogated the IL-23 response to both E. coli and B. fragilis (Fig. 6A). This IL-10 effect was dose dependent, with concentrations of rhIL-10 as low as 0.5 ng/ml causing reduction of IL-23 production in response to B. fragilis (data not shown). The addition of rhIL-10 to PBMC cultures stimulated with B. fragilis led to a significant decrease in the percentage of mDC expressing intracellular IL-12p40/70 (Fig. 6B). Conversely, neutralizing IL-10 resulted in significant increases in the amount of IL-23 produced in response to both E. coli and B. fragilis (Fig. 6C). The hierarchical induction of IL-23, with greater levels observed following B. fragilis stimulation, persisted even in the setting of IL-10 neutralization. Conversely, the addition of rhIL-12 and rhIL-23 to PBMC stimulated with E. coli had no significant impact on the amount of IL-10 produced (rhIL-12, P = 0.22; rhIL-23, P = 0.31) (Fig. 7D).
Fig 7.
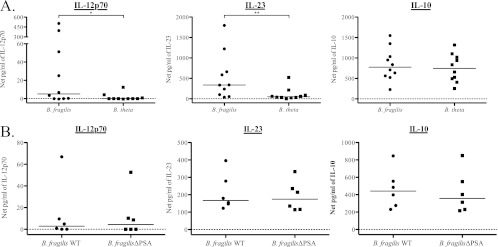
The proinflammatory response by total PBMC to B. fragilis is not genus specific or dependent on expression of PSA. To determine if bacteria within the same genus elicit similar cytokine responses, total PBMC were stimulated with B. fragilis and B. thetaiotaomicron (B. theta). (A) Production of IL-12p70, IL-23, and IL-10 within culture supernatants of total PBMC stimulated with B. fragilis and B. thetaiotaomicron was assessed by ELISA (n = 10). (B) To determine if expression of polysaccharide A (PSA) is responsible for the unique cytokine profile elicited by B. fragilis, total PBMC were stimulated with wild-type B. fragilis (WT) or with a mutant form of B. fragilis (B. fragilis ΔPSA). Production of IL-12p70, IL-23, and IL-10 within culture supernatants was assessed by ELISA (n = 10). Results are shown as the net amount of the indicated cytokine (stimulation condition value minus the medium-only value). Horizontal bars indicate median values. Statistical significance between each bacterial stimulation condition was evaluated using a Wilcoxon matched-pairs signed rank test. *, P < 0.05; **, P < 0.01.
The proinflammatory PBMC profile induced by B. fragilis is not inherent to the Bacteroides genus and is not dependent on expression of PSA.
Although B. fragilis is part of the normal intestinal flora, it is frequently isolated from intra-abdominal abscesses, suggesting that it may have unique pathogenic properties (43). Conversely, a related commensal species, B. thetaiotaomicron, is less frequently identified in clinical isolates (43). In order to determine whether the proinflammatory cytokine profile induced by B. fragilis is unique to this organism or is present in other Bacteroides species, PBMC were stimulated with either B. fragilis or B. thetaiotaomicron, and cytokine production was measured in culture supernatants. B. fragilis induced significantly more IL-12p70 and IL-23 than did B. thetaiotaomicron in total PBMC, whereas the amount of IL-10 induced by each species of bacteria did not differ significantly (P = 0.43) (Fig. 7A).
It has been suggested that B. fragilis may play an important role in the development of the immune system and that its unique properties may be in part due to expression of polysaccharide A (PSA), a zwitterionic molecule present on the surface of the B. fragilis capsule (37, 51). To assess the role of PSA in the proinflammatory PBMC response to B. fragilis, we stimulated PBMC with a mutant B. fragilis defective in PSA expression (B. fragilis ΔPSA) and evaluated IL-12p70, IL-23, and IL-10 cytokine production relative to that induced by wild-type B fragilis. The levels of all three cytokines induced in total PBMC by B. fragilis ΔPSA did not differ significantly from levels induced by wild-type B. fragilis (IL-10, P = 0.31; IL-12p70, P = 0.63; IL-23, P = 0.22) (Fig. 7B). Thus, the unique cytokine profile induced by B. fragilis is not due solely to expression of PSA but may result from B fragilis-specific expression of other MAMP or factors.
DISCUSSION
APC, including DC and monocytes, are crucial in responding to innate microbial stimuli and subsequently influencing adaptive immune responses. Although human DC and monocyte responses to individual TLR ligands and to assorted bacteria have been variably described (27, 30), less is known about the ability of intact, commensal bacteria to induce blood APC cytokine responses. We have previously shown that human intestinal DC, conditioned in the regulatory environment of the gut, have blunted cytokine responses following exposure to individual bacterial products relative to the responses of blood DC (12). In the context of intestinal microbial translocation, it is important to understand how blood APC will respond to commensal bacteria in the absence of gut conditioning factors. We hypothesized that blood DC, in the absence of intestinal conditioning factors, would produce proinflammatory cytokines upon exposure to commensal bacteria. We observed that in vitro stimulation of PBMC with various species of whole commensal enteric bacteria induced distinct pro- and anti-inflammatory cytokine profiles in PBMC, and these distinct profiles originated through differential stimulation of human blood DC and monocytes.
Previous studies showed that Gram-positive bacteria induced higher IL-12 production from monocytes than Gram-negative bacteria, which induced higher levels of IL-10 (22, 31). In contrast to these studies, we observed that Enterococcus, a Gram-positive bacterium, induced significant production of IL-10, but not IL-12p70 or IL-23, from total PBMC. Furthermore, E. coli and B. fragilis, the two Gram-negative bacteria examined, induced differential pro- and anti-inflammatory cytokine responses in PBMC relative to one another. Although our observed cytokine profile in response to E. coli is in agreement with a previous study (23), the differential responses we detected to these two Gram-negative bacteria highlight the fact that defining the peripheral blood cytokine response to different bacterial species may not be as simple as differentiating between Gram-positive and Gram-negative organisms. Thus, our work underscores the importance of identifying translocating bacterial species and better defining the immune response to species that are associated with different diseases.
It is likely that both the method of bacterial recognition by different cell types and the response to the multiple cytokines produced contribute to the unique PBMC responses to different bacteria, which emphasizes the need to evaluate the contributions of the multiple cell types that may encounter these microbes and their products in vivo. For example, in response to E. coli, monocytes contributed the bulk of the IL-10 produced, and only mDC contributed to IL-23 production (with the level of IL-23 produced likely suppressed by large amounts of monocyte-derived IL-10). Conversely, both mDC and monocytes contributed to IL-23 production in response to B. fragilis, and, in the context of the smaller amount of IL-10, a more proinflammatory response to B. fragilis resulted.
It is likely that several factors contribute to the development of these differential responses. First, as whole bacteria express multiple MAMP, simultaneous engagement of multiple TLR during whole-bacterium stimulation could result in each bacterial species inducing different combinations of signaling cascades, thus initiating distinctive responses. Second, recognition of bacterial motifs is not limited to TLR, and engagement of additional PRR may influence the response to translocated bacteria. Indeed, signaling through NOD2 and Dectin-1 has been shown to induce IL-23 but not IL-12p70 from DC (34, 52). Third, structural differences in the shared MAMP expressed by bacteria may affect the cytokine response induced. Previous work has demonstrated that the shape and charge of lipid A, a portion of the LPS structure, had a direct effect on the induction of IL-6 from human PBMC (46, 47). Lastly, different APC express variable types and levels of PRR, leading to further complexity in the overall response to a given set of MAMP. Further evaluation of the role of specific PRR-MAMP pairs in the overall cytokine response is necessary to determine if one or a combination of these factors is responsible for the observed differences in innate responses to commensal bacteria.
In addition to the initial innate responses of different APC to bacteria, the overall PBMC cytokine profile appears to be further determined by cytokine feedback mechanisms. Our observations (i) that exogenous IL-10 abrogated IL-23 production, (ii) that blocking IL-10 resulted in increased PBMC production of IL-23 in response to B. fragilis and E. coli, and (iii) that removing monocytes from total PBMC led to an increase in IL-23 production in response to E. coli, likely due to a lack of monocyte IL-10 suppression of mDC IL-23 production, suggest that the anti-inflammatory pathway initiated by IL-10 signaling can override the pathways leading to IL-23 production. A previous study investigating the relationship between IL-10 and IL-12 production after LPS stimulation found that autocrine IL-10 dampened IL-12 responses from human monocyte-derived DC (9). In addition, blocking IL-10 led to increased IL-12 cytokine production by human blood mononuclear cells in response to a human rectal commensal bacterium (23). Given that IL-10 signaling has been shown to regulate IL-12 production by inhibiting transcription of the IL-12p40 gene (1) and that IL-12 and IL-23 share the same p40 subunit (42), it is possible that this mechanism of IL-12p70 inhibition by IL-10 also applies to IL-23 inhibition. Thus, IL-23 produced by human mononuclear cells in response to commensal bacteria is controlled by concurrent production of IL-10 in response to the same stimulus. Our findings underscore the fact that the establishment of unique innate immune footprints in response to different bacterial stimulation is the culmination of a set of complex interactions and mechanisms that include not only the initial recognition of the microbes by the immune system and the inherent cytokine response that these organisms elicit but also the effect that these cell types and the various cytokines they produce have on each other.
A noteworthy finding of this work was that B. fragilis induced a more proinflammatory cytokine response in PBMC than the other bacteria tested, including a bacterium within the same genus, B. thetaiotaomicron. Although B. fragilis and B. thetaiotaomicron likely express motifs recognized by the same PRR, our findings suggest that B. fragilis must express other factors that render it uniquely proinflammatory. B. fragilis has previously been reported to have distinct properties related to expression of PSA, a capsular polysaccharide (37, 38, 44, 45). Due to the ability of PSA to induce IL-12 production from bone marrow-derived DC (37), we evaluated whether this molecule was critical to the induction of IL-23 in PBMC by B. fragilis by using a PSA-deleted organism. We observed in our system that the absence of PSA expression did not alter the distinctive proinflammatory response to B. fragilis that we detected in human PBMC. The lack of an effect of PSA on the cytokine response by PBMC could potentially be attributed to differences in cell types, culture conditions, or target species compared to previous studies. Based on these results, it is likely that motifs expressed by B. fragilis other than PSA are responsible for its proinflammatory response in human PBMC. However, more work is necessary to determine the exact mechanism underlying this unique response.
In conclusion, our study demonstrated that unique cytokine responses could be elicited from human blood mononuclear cells after stimulation with whole, commensal enteric bacteria. IL-10 produced by blood monocytes regulated the production of proinflammatory cytokines including IL-23 by mDC and monocytes in response to the same stimulus. Our experimental setup allowed us to examine the effect of bacterial stimulation on monocytes and DC in the context of total PBMC, a more physiologically relevant model than using purified APC alone. Further understanding of the mechanisms by which unique cytokine responses induced by different bacteria are achieved could aid in the designing of better therapies for diseases in which microbial translocation occurs as limiting the innate response to translocated bacteria could reduce systemic inflammation and its clinical sequelae.
Supplementary Material
ACKNOWLEDGMENTS
We thank all of the subjects that participated in this study. We also thank Sarkis Mazmanian for the generous gift of the B. fragilis PSA mutant and corresponding WT strain. We thank Nancy Madinger and Christine McAndrews at the University of Colorado Clinical Microbiology Laboratory for their help in growing B. fragilis and B. thetaiotaomicron stocks. We also thank Toni Schwarz, Mira Estin, and Lisa Rogers for their help during this project. Finally, we thank the Center for AIDS Research Immunology Core for their assistance with flow cytometry.
This work was supported by grants R01 DK088663 (C.W.) and K24 AI074343 (C.W.) from the NIDDK and the NIAID. J.M. was supported by NIH/NCRR Colorado CTSI grant number TL1 RR025778 and by an NIAID training grant to the Integrated Department of Immunology at University of Colorado, Denver (T32-AI07405).
Footnotes
Published ahead of print 13 June 2012
Supplemental material for this article may be found at http://cvi.asm.org/.
REFERENCES
- 1. Aste-Amezaga M, Ma X, Sartori A, Trinchieri G. 1998. Molecular mechanisms of the induction of IL-12 and its inhibition by IL-10. J. Immunol. 160:5936–5944 [PubMed] [Google Scholar]
- 2. Auffray C, Sieweke MH, Geissmann F. 2009. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu. Rev. Immunol. 27:669–692 [DOI] [PubMed] [Google Scholar]
- 3. Baba N, Samson S, Bourdet-Sicard R, Rubio M, Sarfati M. 2008. Commensal bacteria trigger a full dendritic cell maturation program that promotes the expansion of non-Tr1 suppressor T cells. J. Leukoc. Biol. 84:468–476 [DOI] [PubMed] [Google Scholar]
- 4. Baba N, Samson S, Bourdet-Sicard R, Rubio M, Sarfati M. 2009. Selected commensal-related bacteria and Toll-like receptor 3 agonist combinatorial codes synergistically induce interleukin-12 production by dendritic cells to trigger a T helper type 1 polarizing programme. Immunology 128:e523–e531 doi:10.1111/j.1365–2567.2008.03022.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banchereau J, et al. 2000. Immunobiology of dendritic cells. Annu. Rev. Immunol. 18:767–811 [DOI] [PubMed] [Google Scholar]
- 6. Brenchley JM, Douek DC. 2008. HIV infection and the gastrointestinal immune system. Mucosal Immunol. 1:23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brenchley JM, et al. 2006. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 12:1365–1371 [DOI] [PubMed] [Google Scholar]
- 8. Caradonna L, et al. 2000. Enteric bacteria, lipopolysaccharides and related cytokines in inflammatory bowel disease: biological and clinical significance. J. Endotoxin Res. 6:205–214 [PubMed] [Google Scholar]
- 9. Corinti S, Albanesi C, la Sala A, Pastore S, Girolomoni G. 2001. Regulatory activity of autocrine IL-10 on dendritic cell functions. J. Immunol. 166:4312–4318 [DOI] [PubMed] [Google Scholar]
- 10. Dillon SM, et al. 2011. Blood myeloid dendritic cells from HIV-1-infected individuals display a proapoptotic profile characterized by decreased Bcl-2 levels and by caspase-3+ frequencies that are associated with levels of plasma viremia and T cell activation in an exploratory study. J. Virol. 85:397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dillon SM, et al. 2008. Plasmacytoid and myeloid dendritic cells with a partial activation phenotype accumulate in lymphoid tissue during asymptomatic chronic HIV-1 infection. J. Acquir. Immune Defic. Syndr. 48:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dillon SM, et al. 2010. Human intestinal lamina propria CD1c+ dendritic cells display an activated phenotype at steady state and produce IL-23 in response to TLR7/8 stimulation. J. Immunol. 184:6612–6621 [DOI] [PubMed] [Google Scholar]
- 13. Dzionek A, et al. 2000. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J. Immunol. 165:6037–6046 [DOI] [PubMed] [Google Scholar]
- 14. Eckburg PB, et al. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elson G, Dunn-Siegrist I, Daubeuf B, Pugin J. 2007. Contribution of Toll-like receptors to the innate immune response to Gram-negative and Gram-positive bacteria. Blood 109:1574–1583 [DOI] [PubMed] [Google Scholar]
- 16. Estes JD, et al. 2010. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog. 6:e1001052 doi:10.1371/journal.ppat.1001052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feezor RJ, et al. 2003. Molecular characterization of the acute inflammatory response to infections with gram-negative versus gram-positive bacteria. Infect. Immun. 71:5803–5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fink LN, Zeuthen LH, Ferlazzo G, Frokiaer H. 2007. Human antigen-presenting cells respond differently to gut-derived probiotic bacteria but mediate similar strain-dependent NK and T cell activation. FEMS Immunol. Med. Microbiol. 51:535–546 [DOI] [PubMed] [Google Scholar]
- 19. Gillevet P, Sikaroodi M, Keshavarzian A, Mutlu EA. 2010. Quantitative assessment of the human gut microbiome using multitag pyrosequencing. Chem. Biodivers. 7:1065–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guarner F, Malagelada JR. 2003. Gut flora in health and disease. Lancet 361:512–519 [DOI] [PubMed] [Google Scholar]
- 21. Hart AL, et al. 2004. Modulation of human dendritic cell phenotype and function by probiotic bacteria. Gut 53:1602–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hessle C, Andersson B, Wold AE. 2000. Gram-positive bacteria are potent inducers of monocytic interleukin-12 (IL-12) while gram-negative bacteria preferentially stimulate IL-10 production. Infect. Immun. 68:3581–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hessle C, Hanson LA, Wold AE. 1999. Lactobacilli from human gastrointestinal mucosa are strong stimulators of IL-12 production. Clin. Exp. Immunol. 116:276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hessle CC, Andersson B, Wold AE. 2005. Gram-positive and Gram-negative bacteria elicit different patterns of pro-inflammatory cytokines in human monocytes. Cytokine 30:311–318 [DOI] [PubMed] [Google Scholar]
- 25. Howe R, et al. 2009. Evidence for dendritic cell-dependent CD4+ T helper-1 type responses to commensal bacteria in normal human intestinal lamina propria. Clin. Immunol. 131:317–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ito T, Wang YH, Liu YJ. 2005. Plasmacytoid dendritic cell precursors/type I interferon-producing cells sense viral infection by Toll-like receptor (TLR) 7 and TLR9. Springer Semin. Immunopathol. 26:221–229 [DOI] [PubMed] [Google Scholar]
- 27. Iwasaki A, Medzhitov R. 2004. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5:987–995 [DOI] [PubMed] [Google Scholar]
- 28. Jarrossay D, Napolitani G, Colonna M, Sallusto F, Lanzavecchia A. 2001. Specialization and complementarity in microbial molecule recognition by human myeloid and plasmacytoid dendritic cells. Eur. J. Immunol. 31:3388–3393 [DOI] [PubMed] [Google Scholar]
- 29. Jiang W, et al. 2009. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. J. Infect. Dis. 199:1177–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kadowaki N, et al. 2001. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. J. Exp. Med. 194:863–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karlsson H, Larsson P, Wold AE, Rudin A. 2004. Pattern of cytokine responses to gram-positive and gram-negative commensal bacteria is profoundly changed when monocytes differentiate into dendritic cells. Infect. Immun. 72:2671–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lamont JT. 1992. Mucus: the front line of intestinal mucosal defense. Ann. N. Y. Acad. Sci. 664:190–201 [DOI] [PubMed] [Google Scholar]
- 33. Latvala S., et al. 2008. Potentially probiotic bacteria induce efficient maturation but differential cytokine production in human monocyte-derived dendritic cells. World J. Gastroenterol. 14:5570–5583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. LeibundGut-Landmann S, et al. 2007. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat. Immunol. 8:630–638 [DOI] [PubMed] [Google Scholar]
- 35. MacFie J, et al. 1999. Gut origin of sepsis: a prospective study investigating associations between bacterial translocation, gastric microflora, and septic morbidity. Gut 45:223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martinson JA, et al. 2007. Dendritic cells from HIV-1 infected individuals are less responsive to toll-like receptor (TLR) ligands. Cell Immunol. 250:75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122:107–118 [DOI] [PubMed] [Google Scholar]
- 38. Mazmanian SK, Round JL, Kasper DL. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453:620–625 [DOI] [PubMed] [Google Scholar]
- 39. Merlini E, et al. 2011. Evidence for polymicrobic flora translocating in peripheral blood of HIV-infected patients with poor immune response to antiretroviral therapy. PLoS One 6:e18580 doi:10.1371/journal.pone.0018580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mogensen TH, Paludan SR, Kilian M, Ostergaard L. 2006. Live Streptococcus pneumoniae, Haemophilus influenzae, and Neisseria meningitidis activate the inflammatory response through Toll-like receptors 2, 4, and 9 in species-specific patterns. J. Leukoc. Biol. 80:267–277 [DOI] [PubMed] [Google Scholar]
- 41. Mohamadzadeh M, et al. 2005. Lactobacilli activate human dendritic cells that skew T cells toward T helper 1 polarization. Proc. Natl. Acad. Sci. U. S. A. 102:2880–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oppmann B, et al. 2000. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity 13:715–725 [DOI] [PubMed] [Google Scholar]
- 43. Polk BF, Kasper DL. 1977. Bacteroides fragilis subspecies in clinical isolates. Ann. Intern. Med. 86:569–571 [DOI] [PubMed] [Google Scholar]
- 44. Round JL, et al. 2011. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332:974–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Round JL, Mazmanian SK. 2010. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. U. S. A. 107:12204–12209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schromm AB, et al. 2000. Biological activities of lipopolysaccharides are determined by the shape of their lipid A portion. Eur. J. Biochem. 267:2008–2013 [DOI] [PubMed] [Google Scholar]
- 47. Schromm AB, et al. 1998. The charge of endotoxin molecules influences their conformation and IL-6-inducing capacity. J. Immunol. 161:5464–5471 [PubMed] [Google Scholar]
- 48. Scott K, et al. 2005. Qualitatively distinct patterns of cytokines are released by human dendritic cells in response to different pathogens. Immunology 116:245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smits HH, et al. 2005. Selective probiotic bacteria induce IL-10-producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin. J. Allergy Clin. Immunol. 115:1260–1267 [DOI] [PubMed] [Google Scholar]
- 50. Suzuki K, et al. 2004. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl. Acad. Sci. U. S. A. 101:1981–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tzianabos AO, et al. 1992. The capsular polysaccharide of Bacteroides fragilis comprises two ionically linked polysaccharides. J. Biol. Chem. 267:18230–18235 [PubMed] [Google Scholar]
- 52. van Beelen AJ, et al. 2007. Stimulation of the intracellular bacterial sensor NOD2 programs dendritic cells to promote interleukin-17 production in human memory T cells. Immunity 27:660–669 [DOI] [PubMed] [Google Scholar]
- 53. Veckman V, Julkunen I. 2008. Streptococcus pyogenes activates human plasmacytoid and myeloid dendritic cells. J. Leukoc. Biol. 83:296–304 [DOI] [PubMed] [Google Scholar]
- 54. Zeuthen LH, Christensen HR, Frokiaer H. 2006. Lactic acid bacteria inducing a weak interleukin-12 and tumor necrosis factor alpha response in human dendritic cells inhibit strongly stimulating lactic acid bacteria but act synergistically with gram-negative bacteria. Clin. Vaccine Immunol. 13:365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.