Abstract
Vaccinia virus transcription is regulated in three stages. An intermediate transcription factor, comprised of virus-encoded polypeptides A8 and A23, was previously identified by in vitro analyses. To investigate its role, we engineered cells that stably expressed both subunits and complemented the replication of A8 and A23 deletion mutant viruses. Without A8 or A23, viral early gene expression and DNA replication occurred but intermediate and late gene expression and resolution of genome concatemers were not detected.
TEXT
Poxviruses are large double-stranded DNA viruses that reproduce within the cytoplasm of eukaryotic cells (14). Their ability to replicate outside the nucleus is dependent on the expression of viral proteins that enable DNA synthesis and transcription. The transcriptional program is divided into early, intermediate, and late stages regulated by stage-specific promoters and transcription factors in concert with a multisubunit RNA polymerase. Recent studies suggest that there are 118 early, 53 intermediate, and 38 late genes, although some have multistage promoters (22–24). The enzymes and factors required for early transcription are synthesized late in infection and packaged within the infectious virus particle (9, 16, 21), allowing early gene expression to occur soon after the core enters the cytoplasm (2). Early genes encode the multisubunit RNA polymerase and proteins required for viral DNA replication and intermediate transcription; intermediate genes encode late transcription factors. Intermediate and late gene transcription require viral DNA replication and take place within cytoplasmic factories (11).
Most of the DNA replication proteins of vaccinia virus (VACV), the prototype poxvirus, were first recognized by isolation of conditional lethal, temperature-sensitive mutants and subsequently further characterized (10, 15). In contrast, VACV transcription factors were identified by in vitro complementation assays (1, 5, 12, 17). Therefore, it is important to establish the roles of VACV transcription factors in vivo by genetic methods. In general, genes regulated by intermediate and late promoters can be conditionally repressed (25), a procedure that was effectively used to demonstrate the role of the G8 late transcription factor (26). However, the intermediate transcription factor subunits A8 and A23 are regulated by early promoters (17) and are not amenable to the technology that has been used for stringent repression, and no temperature-sensitive mutants have been mapped to these genes. An alternative approach involving the construction of a complementing cell line to isolate a deletion mutant was successfully used for VACV in only one instance, to demonstrate the role of the D4 protein in VACV DNA replication (8). The goal of the present study was to construct a cell line that expresses both the A8R and A23R genes, which encode the A8 and A23 proteins, and that would be capable of complementing VACV A8R or A23R deletion mutants for further analysis.
Construction of a cell line that expresses the VACV A8 and A23 proteins.
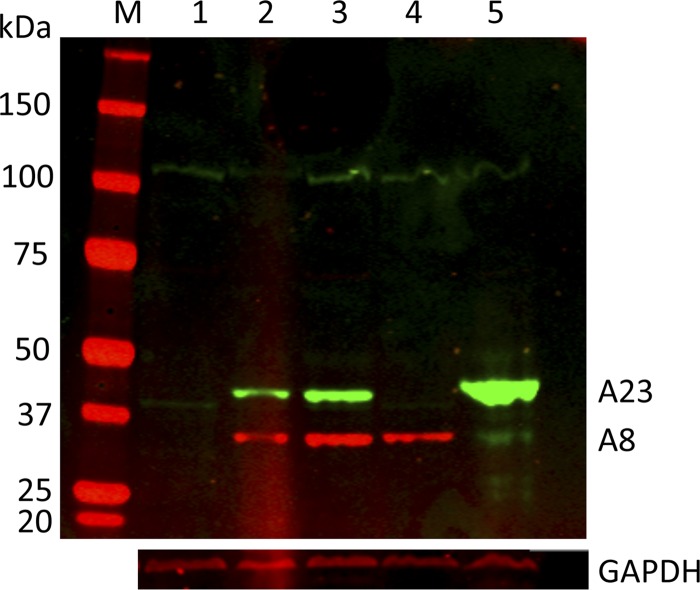
Since it is convenient to have an epitope tag on a protein, we investigated whether A8 and A23 function would be impaired by such an addition. In initial experiments, homologous recombination was used to add V5 and hemagglutinin (HA) epitope tags following the initiation codons of the VACV A8R and A23R genes, respectively, using methods described for other constructs (19). The recombinant viruses vV5-A8 and vHA-A23 replicated normally (data not shown) and expressed the tagged proteins, as determined by Western blotting with specific antibodies (Fig. 1). The expected masses of the A8 and A23 proteins are 33.5 kDa (plus 1.4 kDa for the V5 tag) and 44.6 kDa (plus 1.1 kDa for the HA tag), respectively. Assuming that A8 and A23 are required for VACV replication, the above-mentioned result indicated that the tags did not interfere with protein function.
Fig 1.
Expression of epitope-tagged A8 and A23 in recombinant viruses and stably transfected cells. Proteins from uninfected, stably transfected, and infected cell extracts were resolved by electrophoresis in an SDS 4 to 12% polyacrylamide gel, transferred to a nitrocellulose membrane, probed with antibody to the HA and V5 epitope tags, and detected by fluorescence. Lanes: M, marker proteins; 1, uninfected RK13 parent cells; 2, RK13-A8/A23 stable cell line 11 expressing V5-A8 and HA-A23 proteins; 3, RK13-A8/A23 stable cell line 22 expressing V5-A8 and HA-A23 proteins; 4, RK13 cells infected with vV5-A8; 5, RK13 cells infected with vHA-A23. The masses in kDa and electrophoresis positions of marker proteins are indicated on the left. The positions of the tagged A23 and A8 proteins are indicated on the right. The blot was reprobed with antibody to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a loading control.
For the next step, the V5-A8 and HA-A23 genes were chemically synthesized in order to optimize expression of mammalian codons and remove any sequences that might potentially interfere with mRNA processing in the nucleus and transport to the cytoplasm (see Fig. S1 in the supplemental material). The synthetic genes were then inserted into the pBudCE4.1 dual expression vector (Invitrogen Life Technologies). The V5-A8 and HA-A23 open reading frames (ORFs) were regulated by the EF-1-alpha and cytomegalovirus (CMV) promoters, respectively, in the same plasmid to ensure both would be expressed in transfected cells. To generate stable transfectants, RK13 cells (ATCC CCL-37) in 6-well plates were transfected with 1 μg of PvuI-linearized plasmid in Lipofectamine. The medium was changed after an overnight incubation, and the cells were allowed to recover in drug-free medium for 48 h. Cells from each well were seeded into 75-cm2 flasks to adhere overnight. The next morning, medium containing 100 μg of zeocin (shown to kill RK13 cells in preliminary experiments) was added, and the medium was changed every 2 days for a total of 10 days of drug selection. The surviving cells were then diluted and plated in 6-well dishes so that individual wells produced single colonies. The cells were maintained in 100 μg of zeocin (changed every 3 days) until confluence and then expanded in 75-cm2 flasks and further expanded in 150-cm2 flasks in drug-containing medium. The clones were tested for expression of V5-A8 and HA-23 by Western blotting, and two positive clones (numbers 11 and 22) that grew well and could be recovered after freezing were used for further studies. Frozen aliquots of these cells were stored in liquid nitrogen, and after thawing, the cells were expanded in 50 μg/ml of zeocin. Western blots of extracts of clone numbers 11 and 22 are shown in Fig. 1. The proteins comigrated with authentic V5-A8 and HA-A23 synthesized by recombinant VACV, although the HA-A23 band was weaker in intensity, suggesting a lower level of expression or stability.
Confocal microscopy was carried out to evaluate the uniformity of V5-A8 and HA-A23 expression in the stably transfected cells. Of 208 cells counted, 95% had detectable V5-A8 expression above background, though the intensity varied greatly between cells and the fluorescence was distributed throughout the cytoplasm and nucleus. In contrast, HA-A23 was difficult to detect above background by confocal microscopy, precluding accurate quantitation.
Construction of VACV A8R and A23R deletion mutants.
The first step in engineering deletion mutants was to construct knockout transfer plasmids. In one plasmid, the gene encoding enhanced green fluorescent protein that was regulated by the VACV P11 late promoter (4) was flanked by sequences of the A7L and A9L genes that retained 50 bp at the N terminus and 54 bp at the C terminus of the A8R open reading frame so as not to interfere with regulatory sequences of A7L or A9L. In the other plasmid, the gene encoding DsRed that was regulated by the P11 promoter was flanked by sequences from the A22R and A24R genes that retained 50 and 75 bp at the N and C termini, respectively, of the A23 open reading frame. The next step was to infect the RK13-A8/A23 cells with VACV and separately transfect the above-described plasmids. In each case, the infected and transfected cells were harvested after 3 days and then the lysates were diluted and inoculated into fresh RK13-A8/A23 monolayers. After 3 days, the monolayers were examined with an inverted fluorescence microscope, and plaques formed by the deletion mutants were recognized by green or red fluorescence. The plaques were picked, and the procedure was repeated until all plaques exhibited green or red fluorescence. Deletion of the A8R and A23R genes in the vA8Δ and vA23Δ viruses was confirmed by PCR using internal and external primers.
Complementation of vA8Δ and vA23Δ virus infectivity in RK13-A8/23 cells.
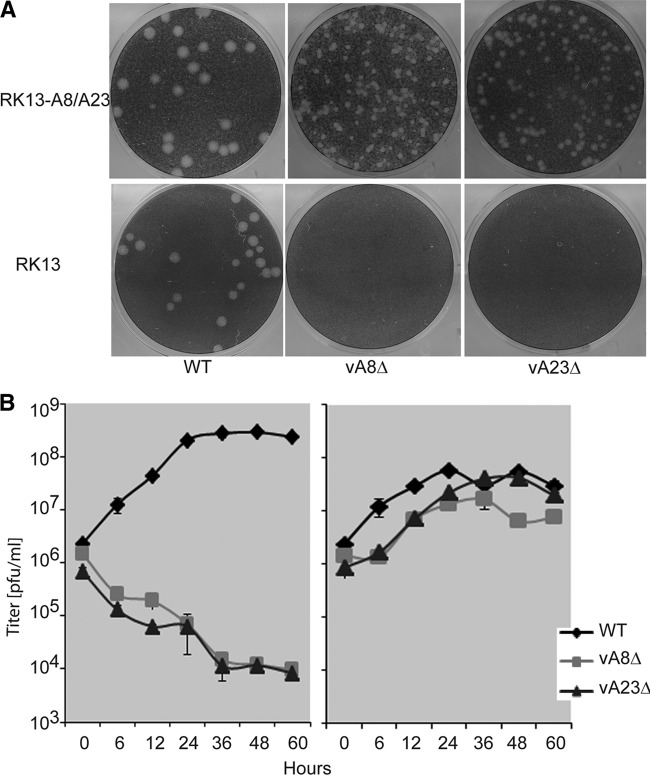
Further experiments demonstrated that the vA8Δ and vA23Δ viruses were unable to form plaques on the parental RK13 cells (Fig. 2A). However, both mutant viruses formed plaques on RK13-A8/A23 cells, although the plaques were smaller at 48 h than those of the wild-type (WT) VACV. The specific ability of the vA8Δ and vA23Δ viruses to replicate in RK13-A8/A23 cells was also demonstrated by one-step growth curves (Fig. 2B). The yields and titers of the virus stocks were 168 PFU/cell and 3.4 × 109/ml for the vA8Δ mutant and 223 PFU/cell and 4.5 × 109/ml for the vA23Δ mutant, values which are comparable to those of WT VACV.
Fig 2.
Replication of A8R and A23R gene deletion mutant viruses in the complementing RK13-A8/A23 cell line. (A) Plaque formation. Suitable dilutions of WT VACV and mutant vA8Δ and vA23Δ VACV were added to RK13 and RK13-A8/A23 cell (clone number 11) monolayers and covered with a methylcellulose overlay. After 48 h at 37°C, the cells were stained with crystal violet. Plaques appear as round white areas against a dark background. (B) One-step growth curve. RK13 cells (left panel) or RK13-A8/A23 cells (right panel) were infected with 10 PFU/cell WT VACV or mutant vA8Δ or vA23Δ virus. The cells were harvested at the indicated times, and the virus titers were determined by plaque assay.
A8 and A23 are required for intermediate and late gene expression.
We carried out a variety of experiments to define the stage at which replication of the vA8Δ and vA23Δ viruses was arrested in RK13 cells. Abundant viral proteins can be readily detected by pulse-labeling infected cells with radioactive amino acids at successive hours postinfection (hpi) and analyzing the lysates by SDS-polyacrylamide gel electrophoresis followed by autoradiography. RK13 and RK13-A8/23 cells were infected with 10 PFU/cell of WT, vA8Δ, or vA23Δ VACV. At the indicated hpi, the cells were washed and incubated with methionine- and cysteine-free medium for 30 min at 37°C and then for another 30 min in the same medium with 100 μCi of a mixture of [35S]methionine and [35S]cysteine at 37°C. The cells were then washed and lysed, and the proteins were resolved by SDS-polyacrylamide gel electrophoresis and detected by autoradiography. Prominent viral proteins were readily discerned in autoradiographs of RK13 cells at 6, 12, and 24 hpi with WT virus but not with either vA8Δ or vA23Δ virus (Fig. 3A). With the mutant viruses, host protein synthesis continued in RK13 cells. In contrast, the labeling patterns of WT VACV and the deletion mutants were similar in RK13-A8/A23 cells except for a lower intensity of bands in the case of the ΔA8R virus (Fig. 3B).
Fig 3.
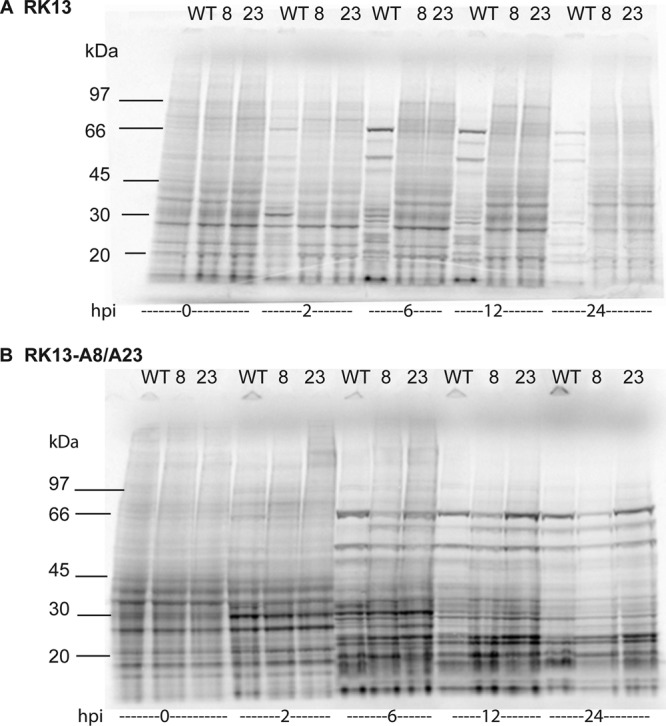
Analysis of polypeptide synthesis following infection with WT and deletion mutant viruses. RK13 (A) and RK13-A8/A23 (B) cells were infected with WT, vA8Δ (8), or vA23Δ (23) VACV and pulse-labeled for 30 min at 0, 2, 6, 12, or 24 hpi with a mixture of [35S]methionine and [35S]cysteine. Proteins were resolved by SDS-polyacrylamide gel electrophoresis and detected by autoradiography. The positions and masses of marker proteins are indicated on the left.
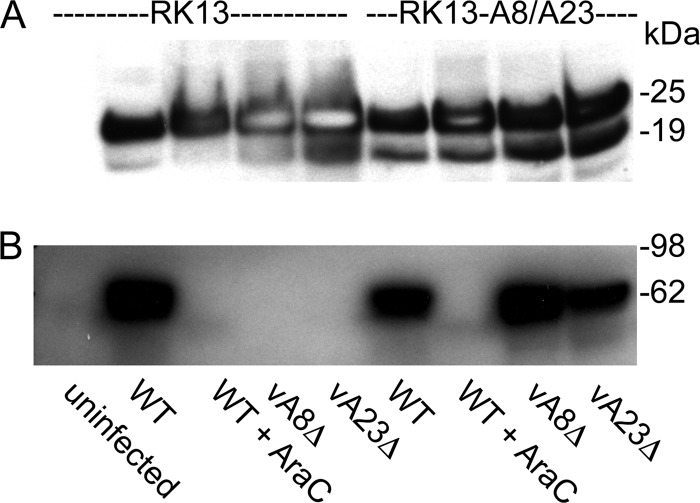
It is difficult to discern early proteins by metabolic labeling because of their low abundance and synthesis of host proteins. However, Western blot experiments demonstrated that the early E3 protein was synthesized by the vA8Δ and vA23Δ viruses in both RK13 and RK13-A8/A23 cells (Fig. 4A), whereas the late A3 (P4B) protein was made only in RK13-A8/A23 cells (Fig. 4B). The overloaded gel emphasizes the total absence of the viral late protein in RK13 cells infected with the deletion mutants despite the robust expression of the viral early protein.
Fig 4.
Synthesis of representative early and late viral proteins. RK13 and RK13-A8/A23 cells were uninfected or infected with WT, vA8Δ, or vA23Δ VACV at a multiplicity of 10 PFU/cell. After approximately 24 h, the cells were lysed and the proteins were analyzed by Western blotting with mouse monoclonal antibody to the early E3 protein (20) (A) and rabbit polyclonal antibody to the late A3 (R. Doms and B. Moss, unpublished) (P4B) protein (B) and visualized by chemiluminescence. Where indicated, cells were infected with WT virus in the presence of AraC. The positions and masses of marker proteins are indicated on the right. Note that there are two translation start sites for E3, accounting for the double band.
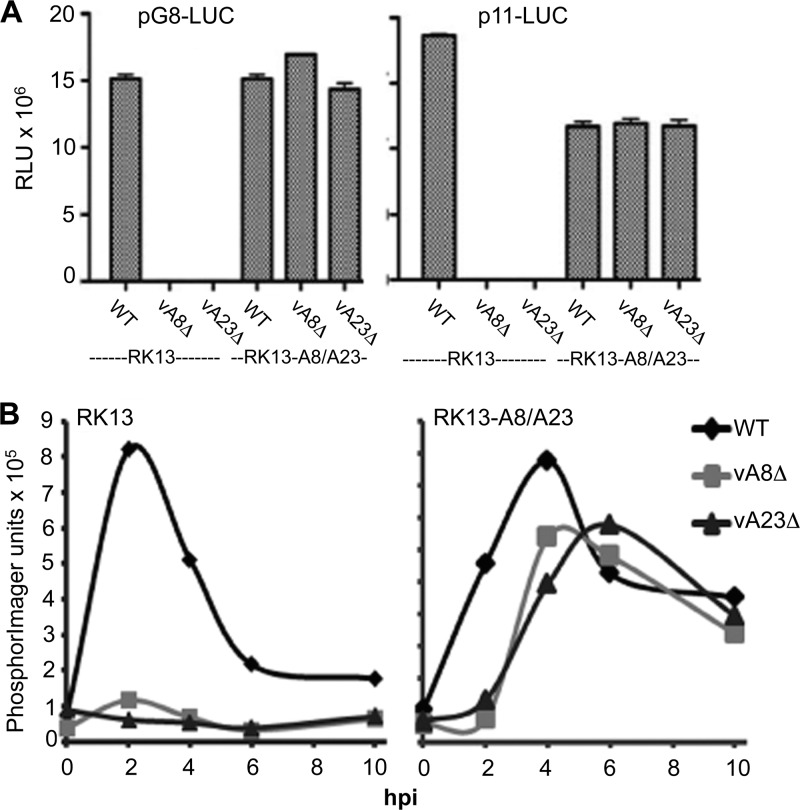
A quantitative assessment of intermediate- and late-stage expression was made by transfecting plasmids that express firefly luciferase regulated by the intermediate G8R promoter (pG8-LUC) and the late F17R promoter (p11-LUC) (3) into RK13 and RK13-A8/A23 cells infected with 1 PFU/cell of WT or mutant virus. At 10 hpi, luciferase activity was measured with a luminometer. As expected, WT VACV enabled the expression of pG8-LUC and p11-LUC in either cell type (Fig. 5A). In contrast, the vA8Δ and vA23Δ viruses enabled expression of pG8-LUC and p11-LUC only in complementing RK13-8/23 cells (Fig. 5A).
Fig 5.
Reporter gene assays and Northern blotting. (A) Firefly luciferase reporter gene expression. RK13 cells were infected with WT, vA8Δ, or vA23Δ VACV and then transfected with plasmids containing the firefly luciferase gene under the control of the intermediate G8R (pG8-LUC) or the late p11 (p11-LUC) promoter. After 10 hpi, lysates were prepared and luciferase activity was measured in relative light units (RLU). (B) Northern blot analysis of G8R intermediate-stage transcription. RK13 and RK13-A8/A23 cells were infected with 10 PFU/cell of WT, vA8Δ, or vA23Δ VACV and harvested at 0, 2, 4, 6, and 10 hpi. Following electrophoresis and transfer to a membrane, RNA was detected with a 32P-labeled G8R riboprobe and quantified in PhosphorImager units.
A further experiment was carried out to determine whether the block in intermediate gene expression occurred at the transcriptional level in RK13 cells infected with vA8Δ and vA23Δ VACV. RK13 and RK13-A8/A23 cells were infected with 10 PFU/cell of WT, vA8Δ, or vA23Δ virus, incubated at 37°C, and harvested at 0, 2, 4, 6, and 10 hpi. Following electrophoresis and transfer to a membrane, RNA was detected with a 32P-labeled, 200-nucleotide riboprobe complementary to the G8R mRNA sequence and quantified with a Typhoon PhosphorImager. In RK13 and RK13-A8/A23 cells infected with WT VACV, the G8 transcript peaked at 2 h and decreased thereafter (Fig. 5B), in agreement with previous studies of WT VACV infection of HeLa cells (2). In contrast, vA8Δ or vA23Δ VACV transcribed G8R only in RK13-A8/23 cells, although the peak was slightly lower and delayed compared to that of WT VACV (Fig. 5B).
A8 and A23 are required for processing of viral DNA concatemers.
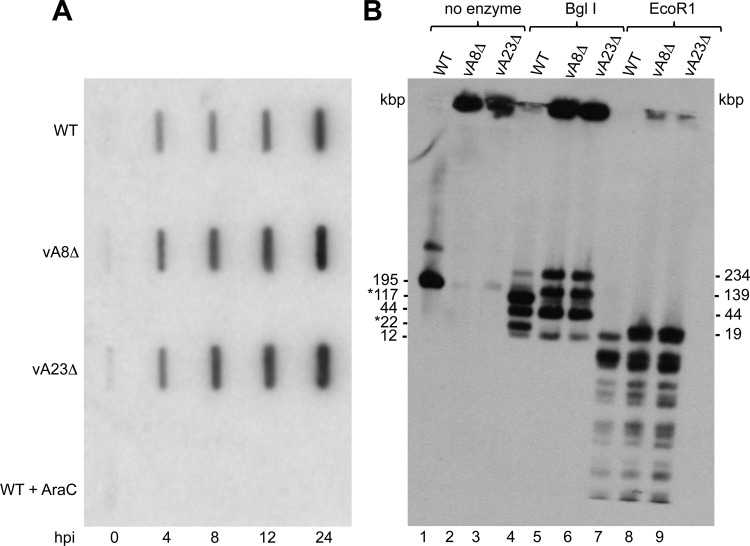
At least seven early genes are directly involved in VACV genome replication (15). Since our data indicated that early gene expression occurred when RK13 cells were infected with vA8Δ or vA23Δ virus, we anticipated that viral DNA would be synthesized. RK13 cells were infected with 10 PFU/cell of WT, vA8Δ, or vA23Δ VACV and harvested at times between 0 and 24 h. Proteinase-treated total DNA was denatured, loaded on a positively charged membrane, cross-linked, and probed with a digoxigenin-dUTP-labeled F17R probe. The results showed that viral DNA was synthesized in RK13 cells infected with WT, vA8Δ, or vA23Δ VACV, indicating that neither A8 nor A23 proteins were required for VACV DNA replication (Fig. 6A).
Fig 6.
Viral DNA synthesis and concatemer resolution. (A) Slot blot analysis. Total DNA samples isolated at 0, 4, 8, 12, and 24 hpi from RK13 and RK13-A8/A23 cells infected with WT, vA8Δ, or vA23Δ VACV or WT VACV in the presence of AraC were subjected to slot blot analysis using a digoxigenin-dUTP-labeled VACV DNA probe. (B) DNA from RK13 cells infected with WT, vA8Δ, or vA23Δ VACV was embedded in molten agarose, deproteinized, and incubated without restriction endonuclease (lanes 1 to 3) or with the restriction endonucleases BglI (lanes 4 to 6) and EcoRI (lanes 7 to 9). Following pulsed-field electrophoresis, the DNA was transferred to a membrane and hybridized to a digoxigenin-labeled VACV DNA probe. The sizes in kbp of full-length VACV genomic DNA and BglI restriction endonuclease fragments are shown on the left. Asterisks indicate end fragments. The sizes of the BglI and EcoRI concatemer junction fragments are shown on the right.
VACV DNA is synthesized as long concatemers that are cleaved into unit-length genomes by the A22 Holliday junction resolvase (6, 7), which has an intermediate promoter (24). Thus, we predicted that resolution of viral DNA concatemers would fail to occur in the absence of intermediate transcription mediated by the A8 and A23 proteins. Pulsed-field gel electrophoresis is a method that has been used to separate ∼200,000-bp VACV genomes from longer, unresolved VACV DNA concatemers that remain at or near the sample well (13). RK13 cells were infected with 10 PFU/cell of WT, vA8Δ, or vA23Δ VACV, the cell pellets were embedded in molten agarose plugs to prevent shearing, and proteins were digested with proteinase K in SDS and EDTA. After washing, the gel plugs were incubated with or without the restriction endonucleases BglI and EcoRI, which have 6- and 4-bp recognition sites, respectively. Electrophoresis was carried out in a 1% agarose gel in a CHEF-DR III apparatus (Bio-Rad) as described previously (18). Following electrophoresis, Southern blotting was performed using digoxigenin-dUTP-labeled DNA from purified virions to specifically hybridize to viral DNA. Undigested viral DNA from RK13 cells infected with wild-type VACV migrated predominantly as a 195-kbp genome-length molecule, and the minor band above it corresponds to an unresolved dimer (18); very little viral DNA remained near the top of the gel (Fig. 6B). In contrast, most of the undigested viral DNA from RK13 cells infected with vA8Δ or vA23Δ virus remained as concatemers near the top of the gel with only a very faint band at the size of unit-length molecules (Fig. 6B).
There are three BglI restriction sites in the VACV genome, giving rise to two end fragments of 117 kbp and 22 kbp and two middle fragments of 44 kbp and 12 kbp. Three additional fragments of 234 kbp, 44 kbp, and 139 kbp, representing head-to-head, tail-to-tail, and head-to-tail junction fragments, respectively, can form from BglI digestion of concatemers. BglI digestion of DNA from RK13 cells infected with WT VACV yielded the four fragments expected from a unit genome and only faint 234-kbp and 139-kbp junction bands from concatemeric DNA (Fig. 6B). The 44-kbp junction fragment cannot be resolved from the 44-kbp middle fragment. In contrast, the two end fragments of 117 kbp and 22 kbp were missing from the BglI digests of DNA from RK13 cells infected with vA8Δ and vA23Δ VACV, and in their place were the junction fragments of 234 kbp and 139 kbp that were derived from concatemers, indicating a failure of concatemer resolution.
There are 59 EcoRI restriction sites in the VACV genome, and the two end fragments are 9.4 kbp each. Thus, a single junction fragment of 18.8 kbp, larger than any of the middle fragments, can be derived from a concatemer. A comparison of the EcoRI digests of DNA from RK13 cells infected with the WT and mutant viruses confirmed the presence of the 18.8-kbp junction fragment in only the latter DNAs (Fig. 6B).
Concluding remarks.
The A8 and A23 subunits of the VACV intermediate transcription factor were identified by in vitro transcription complementation studies 12 years ago (17). Until now, however, the role of A8 and A23 in VACV replication had not been reported. In the original study of Sanz and Moss (17), it was shown that A8 and A23 existed as a complex and that coexpression of both proteins in Escherichia coli was required for transcription factor activity in vitro. When the proteins were expressed individually and mixed, no activity was detected. Therefore, we decided to coexpress the two subunits in order to make a complementing cell line. As an additional precaution, the genes were chemically synthesized to optimize translation and to remove potential sequences that might interfere with nuclear mRNA synthesis and transport to the cytoplasm.
We were able to confirm expression of the two subunits by Western blotting. Since the same plasmid contained the genes of both proteins, we assume that all cells expressed both. However, low sensitivity for detection of the A23 protein prevented us from confirming this by either confocal microscopy or flow cytometry. Importantly, however, the strategy was successful, as the stably transfected cells supported the replication of either an A8 or an A23 deletion virus. The isolation of the mutant viruses allowed us to demonstrate that both proteins are essential for expression of intermediate genes and subsequent steps in VACV replication, including late gene expression and resolution of viral DNA concatemers in untransfected cells.
The mutant viruses and complementing cell line may be useful for further studies of structure/function relationships of A8 and A23 and for analyzing viral early gene expression and viral DNA synthesis in the absence of intermediate and late proteins and without a requirement for small-molecule inhibitors. In addition, the mutant viruses may have advantages as gene expression vectors because of the decreased shutdown of host protein synthesis and their inability to replicate, similar to the characteristics of the D4 deletion virus (8). One difference between the D4 deletion mutant and the A8 and A23 deletion mutants is that viral DNA is synthesized with the latter in nonpermissive cells and this DNA may serve as a template for recombinant gene expression. The present study also emphasizes the usefulness of complementing cell lines for the study of essential VACV early genes.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Tatiana Senkevich, P. S. Satheshkumar, and Zain Bengali for providing technical advice and materials. Stuart Isaacs kindly provided a monoclonal antibody to the E3 protein.
The work was performed while R.D.W. was in the graduate program of the Department of Cell Biology and Molecular Genetics at the University of Maryland, College Park.
Support was provided by the Division of Intramural Research, NIAID, NIH. R.D.W. was the recipient of an NIH Intramural Fellowship to Promote Diversity.
Footnotes
Published ahead of print 27 June 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Ahn B-Y, Gershon PD, Moss B. 1994. RNA polymerase-associated protein RAP94 confers promoter specificity for initiating transcription of vaccinia virus early stage genes. J. Biol. Chem. 269:7552–7557 [PubMed] [Google Scholar]
- 2. Baldick CJ, Jr, Moss B. 1993. Characterization and temporal regulation of mRNAs encoded by vaccinia virus intermediate stage genes. J. Virol. 67:3515–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bengali Z, et al. 2011. Drosophila S2 cells are nonpermissive for vaccinia virus DNA replication following entry via low pH-dependent endocytosis and early transcription. PLoS One 6:e17248 doi:10.1371/journal.pone.0017248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bertholet C, Stocco P, Van Meir E, Wittek R. 1986. Functional analysis of the 5′ flanking sequence of a vaccinia virus late gene. EMBO J. 5:1951–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Broyles SS, Yuen L, Shuman S, Moss B. 1988. Purification of a factor required for transcription of vaccinia virus early genes. J. Biol. Chem. 263:10754–10760 [PubMed] [Google Scholar]
- 6. Garcia AD, Aravind L, Koonin EV, Moss B. 2000. Bacterial-type DNA Holliday junction resolvases in eukaryotic viruses. Proc. Natl. Acad. Sci. U. S. A. 97:8926–8931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garcia AD, Moss B. 2001. Repression of vaccinia virus Holliday junction resolvase inhibits processing of viral DNA into unit-length genomes. J. Virol. 75:6460–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Holzer G, Falkner FG. 1997. Construction of a vaccinia virus deficient in the essential DNA repair enzyme uracil DNA glycosylase by a complementing cell line. J. Virol. 71:4997–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kates JR, McAuslan BR. 1967. Poxvirus DNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 58:134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kato SEM, et al. 2008. Marker rescue mapping of the combined Condit/Dales collection of temperature-sensitive vaccinia virus mutants. Virology 375:213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Katsafanas GC, Moss B. 2007. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2:221–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Keck JG, Baldick CJ, Moss B. 1990. Role of DNA replication in vaccinia virus gene expression: a naked template is required for transcription of three late transactivator genes. Cell 61:801–809 [DOI] [PubMed] [Google Scholar]
- 13. Merchlinsky M, Moss B. 1989. Resolution of vaccinia virus DNA concatemer junctions requires late gene expression. J. Virol. 63:1595–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moss B. 2007. Poxviridae: the viruses and their replication, p 2905–2946 In Knipe DM, et al. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 15. Moss B, De Silva F. 2006. Poxvirus DNA replication and human disease, p 707–727 In DePamphilis ML. (ed), DNA replication and human disease. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 16. Munyon WE, Paoletti E, Grace JT., Jr 1967. RNA polymerase activity in purified infectious vaccinia virus. Proc. Natl. Acad. Sci. U. S. A. 58:2280–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sanz P, Moss B. 1999. Identification of a transcription factor, encoded by two vaccinia virus early genes, that regulates the intermediate stage of viral gene expression. Proc. Natl. Acad. Sci. U. S. A. 96:2692–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Senkevich TG, Koonin EV, Moss B. 2009. Predicted poxvirus FEN1-like nuclease required for homologous recombination, double-strand break repair and full-size genome formation. Proc. Natl. Acad. Sci. U. S. A. 106:17921–17926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Senkevich TG, Weisberg A, Moss B. 2000. Vaccinia virus E10R protein is associated with the membranes of intracellular mature virions and has a role in morphogenesis. Virology 278:244–252 [DOI] [PubMed] [Google Scholar]
- 20. Weaver JR, et al. 2007. The identification and characterization of a monoclonal antibody to the vaccinia virus E3 protein. Virus Res. 130:269–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wei CM, Moss B. 1975. Methylated nucleotides block 5′-terminus of vaccinia virus mRNA. Proc. Natl. Acad. Sci. U. S. A. 72:318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang Z, Bruno DP, Martens CA, Porcella SF, Moss B. 2011. Genome-wide analysis of the 5′ and 3′ ends of vaccinia virus early mRNAs delineates regulatory sequences of annotated and anomalous transcripts. J. Virol. 85:5897–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang Z, Bruno DP, Martens CA, Porcella SF, Moss B. 2010. Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing. Proc. Natl. Acad. Sci. U. S. A. 107:11513–11518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang Z, et al. 2011. Expression profiling of the intermediate and late stages of poxvirus replication. J. Virol. 85:9899–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Y, Moss B. 1991. Inducer-dependent conditional-lethal mutant animal viruses. Proc. Natl. Acad. Sci. U. S. A. 88:1511–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang YF, Keck JG, Moss B. 1992. Transcription of viral late genes is dependent on expression of the viral intermediate gene G8R in cells infected with an inducible conditional-lethal mutant vaccinia virus. J. Virol. 66:6470–6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.