Abstract
Acyl coenzyme A (acyl-CoA) thioesterases hydrolyze thioester bonds in acyl-CoA metabolites. The majority of mammalian thioesterases are α/β-hydrolases and have been studied extensively. A second class of Hotdog-fold enzymes has been less well described. Here, we present a structural and functional analysis of a new mammalian mitochondrial thioesterase, Them5. Them5 and its paralog, Them4, adopt the classical Hotdog-fold structure and form homodimers in crystals. In vitro, Them5 shows strong thioesterase activity with long-chain acyl-CoAs. Loss of Them5 specifically alters the remodeling process of the mitochondrial phospholipid cardiolipin. Them5−/− mice show deregulation of lipid metabolism and the development of fatty liver, exacerbated by a high-fat diet. Consequently, mitochondrial morphology is affected, and functions such as respiration and β-oxidation are impaired. The novel mitochondrial acyl-CoA thioesterase Them5 has a critical and specific role in the cardiolipin remodeling process, connecting it to the development of fatty liver and related conditions.
INTRODUCTION
Thioesterases participate in lipid metabolism by hydrolyzing thioester bonds in a variety of substrates, including fatty acid-coenzyme A (CoA) esters and palmitoylated proteins (17, 19). Based on structural folds and the catalytic reaction mechanism, thioesterases are subdivided into α/β-hydrolases and Hotdog-fold thioesterases. The majority of mammalian thioesterases are the well-studied α/β-hydrolases; the second class of Hotdog-fold enzymes has been described to a lesser extent (19). Of the eight Hotdog-fold proteins identified thus far in mammals, only a few participate in biological processes involving hydrolysis of acyl-CoAs (5). These are cytoplasmic Acot7 involved in eicosanoid metabolism, Acot11 and Acot12, which have a START domain, and cytoplasmic/mitochondrial Them2, which is induced by peroxisome proliferator-activated receptor α (PPARα) and has been recently shown to regulate hepatic metabolism (5, 8, 16, 18). The mammalian Hotdog thioesterases identified thus far have tetrameric or hexameric arrangements (26).
Them5 is part of the mitochondrial proteome and is predicted to be a member of the Hotdog-fold family (5, 24). We show here that Them5, a novel thioesterase with strong specificity for long and unsaturated fatty acid-CoA esters, has the classical Hotdog-fold with six β-sheets wrapped around a central α-helix. Them4 and Them5 form independent homodimers in crystals, suggesting that they represent a new group of mammalian hotdog thioesterases.
Them5−/− mice exhibit an increase in monolysocardiolipin, one of the specific metabolites of cardiolipin (CL). CL is a mitochondrion-specific phospholipid characterized by three to four linoleic acid chains (C18:2) in its mature forms, particularly in mammals. Proper synthesis and remodeling (i.e., maintenance of the right acyl composition) of these highly unsaturated CL molecules is crucial for mitochondrial physiology, since CL interacts with many mitochondrial proteins. In the Barth syndrome, a CL-associated disorder in humans, the levels of CL are dramatically decreased (31). Moreover, changes in CL remodeling that are associated with fat-rich diets result in mitochondrial dysfunction, thereby implicating CL in metabolic disorders (7, 20). We show that Them5 has a strong substrate preference for C18 polyunsaturated fatty acids. This specificity correlates with the in vivo profile of fatty acids in Them5-deficient mitochondria and strongly suggests a role for this novel thioesterase in CL remodeling and lipid metabolism. Indeed, Them5 knockout mice develop fatty liver, as well as altered mitochondrial morphology and function.
Given the importance of cardiolipin synthesis and remodeling pathways in mitochondrial function, our work has not only uncovered a new biochemical pathway, but the results also suggest that Them5 is involved in human diseases such as metabolic syndrome, diet-induced obesity, and diabetes.
MATERIALS AND METHODS
Crystallization, X-ray data collection, and structure determination.
Crystals of Δ36Them4 were obtained via the vapor diffusion method by mixing 100 nl of protein solution [Δ36Them4 (4.3 mg/ml), 50 mM CHES (pH 9.5), 200 mM NaCl, 5 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP)] with 100 nl of reservoir solution containing 100 mM Tris (pH 7.0), 200 mM NaCl, and 30% polyethylene glycol (PEG) 3000. Crystals were cryoprotected with 20% ethylene glycol and frozen in liquid nitrogen before data collection at the Swiss Light Source (SLS) synchrotron (Villigen, Switzerland). The structures of Δ36Them4 and Δ34Them5 were solved by the molecular replacement method using PHASER (21). A homology model of the conserved thioesterase Hotdog-fold was constructed with the MODELLER software and used as a search model (29). Clear solutions for four molecules per asymmetric unit forming together two homodimers of the thioesterase fold were obtained. Phases calculated from this initial model were used for automated model building of the full molecule in PHENIX (2).
Crystals of Δ34Them5 were obtained by mixing 100 nl of protein solution (Δ34Them5 [4.0 mg/ml], 20 mM Tris [pH 7.5], 200 mM NaCl, 5 mM TCEP) with 100 nl of reservoir solution (100 mM phosphate-citrate [pH 4.2], 10% PEG 3000, 200 mM NaCl). Crystals were cryoprotected with 30% ethylene glycol and frozen in liquid nitrogen before data collection at the SLS. The structure of Δ34Them5 was solved by the molecular replacement method using PHASER with the structure of Δ36Them4 as a search model (21). One Them5 chain per asymmetric unit was found representing half of the homodimeric thioesterase molecule. Phases from this solution were calculated and used for automatic model building with PHENIX (2). Them4 and Them5 models were further improved by the crystallographic simulated annealing routine followed by individual B-factor refinement in PHENIX (2). All structures were refined by several rounds of manual rebuilding in COOT (14), followed by refinement in PHENIX (2). Final structures were validated using the molprobity server (http://molprobity.biochem.duke.edu/) and COOT (14). The structural images for the figures were prepared with PyMOL (http://pymol.sourceforge.net/).
Analytical gel filtration studies.
Human THEM4 (amino acids 37 to 240) and THEM5 (amino acids 35 to 247) proteins were expressed with C-terminal His6 tags using a pOPINE vector and Escherichia coli Rosetta2 DE3 cells (4). The target proteins were purified on nickel-nitrilotriacetic acid beads (Qiagen), followed by gel filtration on a HiLoad 16/60 Superdex 200 column (GE Healthcare) and concentrated to 6 mg/ml in gel filtration buffer (50 mM Tris [pH 8.5], 200 mM NaCl, 10 mM TCEP, 0.02% NaN3). Analytical gel filtration runs were performed in the same buffer on a Tricorn Superdex 200 10/300 GL (GE Healthcare) column, previously calibrated with molecular mass standards (ferritin, 440 kDa; catalase, 232 kDa; aldolase, 158 kDa; albumin, 67 kDa; ovalbumin, 43 kDa; chymotrypsinogen A, 25 kDa; RNase A, 13.7 kDa). A total volume of 100 μl containing THEM proteins (at a final concentration of 0.9 mg/ml) was injected onto the Superdex 200 10/300 column and separated at a flow rate of 0.75 ml/min.
Sequences.
The sequences were as follows: human THEM4 37-240 (24,252.2 Da), MSSEEVILKDCSVPNPSWNKDLRLLFDQFMKKCEDGSWKRLPSYKRTPTEWIQDFKTHFLDPKLMKEEQMSQAQLFTRSFDDGLGFEYVMFYNDIEKRMVCLFQGGPYLEGPPGFIHGGAIATMIDATVGMCAMMAGGIVMTANLNINYKRPIPLCSVVMINSQLDKVEGRKFFVSCNVQSVDEKTLYSEATSLFIKLNPAKSLTKHHHHHH; and human THEM5 35-247 (24,894.5 Da), MGSSTDSMFSRFLPEKTDLKDYALPNASWCSDMLSLYQEFLEKTKSSGWIKLPSFKSNRDHIRGLKLPSGLAVSSDKGDCRIFTRCIQVEGQGFEYVIFFQPTQKKSVCLFQPGSYLEGPPGFAHGGSLAAMMDETFSKTAFLAGEGLFTLSLNIRFKNLPVDSLVVMDVEVDKIEDQKLYMSCIAHSRDQQTVYAKSSGVFLQLQLEEESPQKHHHHHH.
Data collection and refinement statistics.
Data collection and refinement statistics are given in Table 1.
Table 1.
Data collection and refinement statistics
| Data collection or refinement statistics | Them4 | Them5a |
|---|---|---|
| Data collection | ||
| Space group | P1 | C2221 |
| Cell constants | ||
| a, b, c (Å) | 50.5, 58.2, 69.7 | 45.4, 88.6, 105.3 |
| α, β, γ (°) | 90.0, 71.1, 64.6 | 90.0, 90.0, 90.0 |
| λ (Å) | 0.96 | 1.00 |
| Resolution range (Å) | 30.0–1.6 (1.66–1.60) | 30.0–1.45 (1.49–1.45) |
| No. of unique reflections | 83,501 | 70,847 |
| Completeness (%) | 93.9 (84.0) | 96.8 (96.7) |
| Multiplicity | 1.9 | 2.1 |
| Rsym (%)b | 4.8 (21.6) | 2.8 (37.3) |
| I/σ(I) | 22.7 (3.2) | 17.1 (2.1) |
| Refinement | ||
| Resolution range (Å) | 30.0–1.6 | 30.0–1.45 |
| No. of reflections used | 83,488 | 70,483 |
| R-factor (%) | 18.5 | 14.0 |
| Rfree (%) | 22.3 | 17.1 |
| RMSc bond length (Å) | 0.006 | 0.005 |
| RMS bond angle (°) | 1.02 | 1.02 |
| Wilson B-factor (Å2) | 15.0 | 16.1 |
| Mean B-factor (Å2) | 20.0 | 25.5 |
| Ramachandran plot regions (favored, allowed) (%) | 98.9, 1.1 | 98.4, 1.6 |
Friedel pairs were kept separate for Them5.
Rsym = Σhkl Σj|Ij,hkl − 〈Ihkl〉|/Σhkl ΣjIj,hkl, where 〈Ihkl〉is the average of the intensity Ij,hkl over j = 1, …, N observations of symmetry equivalent reflections hkl. Numbers in parentheses refer to values in thehighest-resolution shell.
RMS, root mean square.
Thioesterase activity assay.
The reaction mixture contained recombinant protein, fatty acyl-CoA substrate (Avanti-Polar Lipids., Inc.) at concentrations of 1 to 200 μM, 50 mM HEPES, 50 mM KCl, and 0.5 mM DTNB [5,5′-dithiobis(2-nitrobenzoic acid)] in 1 ml of total volume. Reactions were observed spectrophotometrically at 412 nm (ε = 13.6 mM−1 cm−1) at 37°C using a UV1800 spectrophotometer controlled by UVPro2.33 software (Shimadzu Schweiz GmbH).
Lipid extraction and MS analysis.
Phospholipids were extracted from purified mitochondria and analyzed by electrospray mass spectrometry (MS) as described previously (15, 22).
RNA extraction, reverse transcription reaction, and qRT-PCR analysis.
Total RNA was extracted from tissues or cells using TRIzol reagent (Invitrogen/Life Technologies) according to the manufacturer's protocol. A 2-μg portion of RNA was used for the reverse transcription reaction. Quantitative reverse transcription-PCR (qRT-PCR) was performed using an Applied Biosystems ABI7000 sequence detection system. Primer sequences are available upon request. Mitochondrial DNA (mtDNA) content is presented as the ratio of qRT-PCR assessed levels of single-copy mitochondrial genes (ND1 and COX1) to nuclear gene GAPDH (glyceraldehyde-3-phosphate dehydrogenase).
Subcellular fractionation.
Subcellular fractionation of cells was performed according to procedures described elsewhere (9, 25).
Immuno-EM.
Immuno-electron microscopy (immuno-EM) was performed according to the protocol described elsewhere, with slight modifications (32). Immunolabeling was carried out with anti-hThem5 at a dilution of 1/50 and antihemagglutinin (anti-HA) at 1/200 (Clontech, catalog no. 631207); 10-nm protein A-gold (UMC-Utrecht University, Utrecht, Netherlands) was used at a 1:50 dilution.
EM.
Samples were prepared according to standard procedures. Briefly, tissues or cells were fixed in 2.0% paraformaldehyde–2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, washed in 0.1 M cacodylate buffer (pH 7.4), subsequently postfixed in 1% OsO4–1.5% KFeCN and then in 1% OsO4 in cacodylate buffer, rinsed in ddH2O, stained with 1% uranyl acetate in water for 20 min, rinsed in water, gradually dehydrated in ethanol (50 to 100%), and embedded in Durcupan resin. Serial block-face scanning EM was performed using the 3View SBFSEM system (Gatan, Inc.) under low-vacuum conditions. Images were captured at a resolution of 4,096 × 4,096 pixels with 50-nm slices, all controlled by DigitalMicrograph software (Gatan, Inc.), and then processed using Imaris software. For mitochondrial volume quantification, a minimum of 15 mitochondria per cell were reconstructed in a minimum of three cells per mouse per genotype.
Imaging.
For the analysis of mitochondrial morphology, the images were 0.5 μm by 0.5 μm by 0.2 μm and were deconvolved afterward with the Huygens software (Scientific Volume Imaging B.V.) using a Hygens remote manager. Image analysis was performed with the Imaris software (Bitplane, Zurich, Switzerland). For live-cell imaging, Olympus IX81 spinning-disk with Yokogawa CSU-X1 scanhead microscope equipped with piezo stage was used. Fixed cells were analyzed with a Zeiss LSM700 microscope.
Free fatty acid measurement.
The levels of free fatty acids (FFA) in the plasma of Them5−/− and wild-type mice were measured using a ZenBio serum/plasma fatty acid detection kit (ZenBio, Inc.).
Ketone body measurement.
Levels of 3-hydroxybutyrate (3-HB) were determined with the Autokit 3-HB test for quantitative determination of 3-HB in serum or plasma (Wako).
Cellular fatty acid oxidation.
Isolated hepatocytes were incubated for 1 h in scintillation vials in Krebs Ringer buffer with 3% fatty acid-free bovine serum albumin with 1 mM [1-14C]palmitic acid with a specific activity 1 mCi/mmol (Hartmann Analytic GmbH). Radiolabeled CO2 was collected in center wells with Whatman no. 1 filter paper and 200 μl of 1 M methylbenzethonium hydroxide in methanol. At the end of the incubation, 300 μl of 5 M H2SO4 was added to volatilize the remaining CO2, and the solution was incubated for another 30 min. The center wells were then placed in other scintillation vials, 8 ml of scintillation liquid was added, and the vials were counted using a β-counter.
Preparation and culture of primary MEFs.
Mouse embryonic fibroblasts (MEFs) were derived from embryonic day 13.5 embryos. Cells were cultured in Dulbecco modified Eagle medium containing 10% fetal calf serum under low-oxygen conditions (3%) at 37°C (33). All experiments were performed with primary MEFs (passages 3 to 15).
Respirometry analysis.
Respirometry analysis was performed with Seahorse XF24 analyzer (Seahorse Bioscience, Billerica, MA). Mitochondria were isolated from liver of Them5 wild-type (WT) and knockout (KO) mice, protein was quantified using BCA (Pierce), and 5 μg of protein per well was plated on polyethyleneimine-treated plates (1:15,000 dilution from a 50% solution from Sigma-Aldrich). For the coupling assay, an initial mix contained succinate and rotenone (10 mM and 2 μM, respectively). After an oxygen consumption rate (OCR) baseline measurement, ADP, oligomycin, carbonyl cyanide 4-(trifluoromethoxy)-phenylhydrazone (FCCP), and antimycin A were sequentially added to each well to final concentrations of 4 mM, 2.5 μg/ml, 4 μM, and 4 μM, respectively, and changes in the OCR were analyzed. For the electron transfer assay, the initial mix contained malate, pyruvate, and FCCP (2 mM, 10 mM, and 4 μM final concentrations, respectively). After the initial OCR measurement, subsequent injections of rotenone, succinate, antimycin A, and ascorbate–N1,N1,N1,N1-tetramethyl-1,4-phenylene diamine (TMPD) at 2 μM, 10 mM, 4 μM, and 10 mM/100 μM final concentrations, respectively, were performed, and changes in the OCR were analyzed. The measurement was performed accordingly to the protocol provided by manufacturer.
Mice.
Mice were housed in groups of 3 with 12-h dark/light cycles and free access to food and water in accordance with the Swiss Animal Protection Ordinance. All procedures were performed with the approval of the appropriate authorities. All experiments were performed with males of mixed backgrounds (C57BL/6 and 129/Ola). For all fasting experiments, mice were fasted overnight.
Statistical analysis.
Data are provided as means ± the standard errors of the mean, where n is the number of independent experiments. All data were subjected to one-way analysis of variance, Student t test, or Mann-Whitney test, where applicable. Significance is indicated by asterisks as follows: ***, P < 0.001; **, P < 0.01; and *, P < 0.05.
Protein structure accession numbers.
The coordinates and structure factors have been deposited in the Protein Data Bank (PDB) with entry codes 4AE7 (THEM5) and 4AE8 (THEM4).
RESULTS
Them5 is a mitochondrial protein.
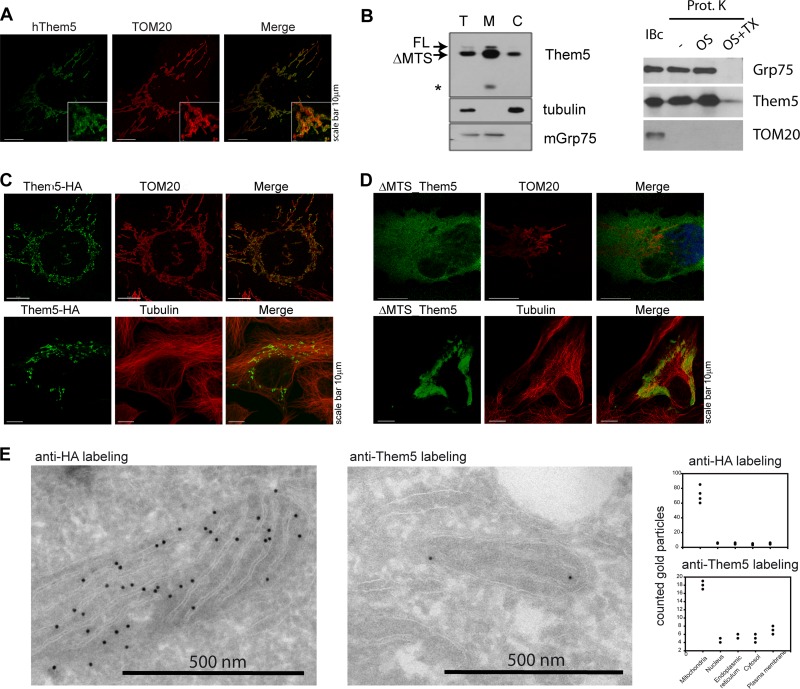
Bioinformatic analysis of Them5 revealed a mitochondrial targeting sequence in the N-terminal part of the human Them5 protein and in its orthologs in other species (see Fig. S1 in the supplemental material). Immunofluorescence analysis showed an overlap of protein staining with the mitochondrial marker TOM20 (Fig. 1A). Like the majority of nucleus-encoded mitochondrial proteins, the mitochondrial targeting signal of Them5 is cleaved by mitochondrial proteases upon protein import into the mitochondria. However, the conserved consensus motif for mitochondrial processing peptidase cleavage [RXX(X)SX] is missing in Them5 (data not shown). Immunoblotting with anti-Them5 antibodies detected the processed (faster-migrating band) and nonprocessed forms of the Them5 protein (Fig. 1B, left). Overexpressed full-length Them5 resides specifically in the mitochondria (Fig. 1C), whereas a mutant lacking the first 37 N-terminal amino acids (ΔMTS) was excluded from mitochondria (Fig. 1D). In contrast to Them4, two-dimensional EM (2D-EM) analysis and mitochondrial fractionation (Fig. 1E, and B, right) showed Them5 to be localized in the matrix of mitochondria (25).
Fig 1.
Them5 is a mitochondrial protein. (A) Mitochondrial localization of hThem5 in U2OS cells. TOM20 was used for mitochondria visualization. (B) Mitochondrial localization of hThem5 in HEK293 cells, transfected with full-length (FL) hThem5. (Left) The upper double band indicates FL Them5 and Them5 without a mitochondrial targeting sequence (ΔMTS). The lower band in the M fraction (*) suggests further Them5 processing in mitochondria. T, total; M, mitochondrial fraction; C, cytoplasmic fraction. (Right) Mitochondria were incubated in isolation buffer alone or treated with proteinase K (100 μg/ml) in the isolation buffer, in 20 mM HEPES, or in 1% Triton X-100. Portions (50 μg) of proteins were separated by SDS-PAGE and immunoblotted with the indicated antibodies. (C and D) Overexpressed in U2OS cells FL Them5 are localized in the mitochondria (C), whereas the ΔMTS Them5-HA mutant is excluded from the mitochondria (D). (E) Immunogold-EM staining of U2OS cells transfected with HA-tagged hThem5. To detect endogenous protein, sections of nontransfected cells were probed with anti-hThem5 antibodies (right). Staining was detected mainly in mitochondria; some background was present in other cellular compartments for both endogenous and transfected proteins.
Them4/5 are dimers of a Hotdog-fold.
Protein sequence analysis suggested that Them5 belongs to the Hotdog-fold thioesterase family of proteins. The Hotdog-fold enzymes are found in all organisms, with Them4 orthologs present in lower eukaryotes, including yeasts, but Them5 orthologs only in mammals (see Fig. S1B in the supplemental material). Them4 and Them5 genes are located on chromosome 1 in humans (chromosome 3 in mice) and share an exon-intron structure and protein domain organization (see Fig. S1A in the supplemental material).
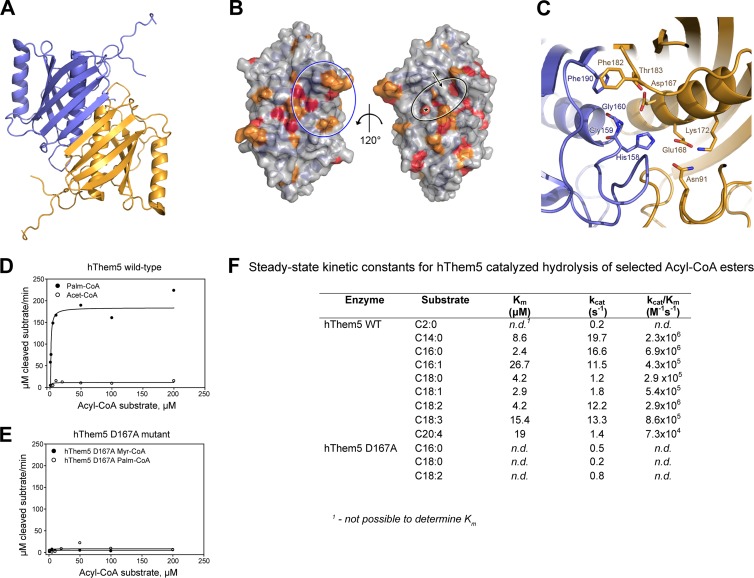
We determined the crystal structures of human Δ34Them5 and Δ36Them4 to resolutions of 1.45 and 1.6 Å, respectively (the data collection and refinement statistics are given in Table 1). The crystal structures (typical of thioesterases) confirmed both proteins as members of the Hotdog superfamily, namely, a core structure encompassing residues 119 to 231 and residues 125 to 237 of Δ36Them4 and Δ34Them5, respectively, and consisting of a long central α-helix surrounded by a six-stranded curved antiparallel β-sheet (Fig. 2A; see also Fig. S2C in the supplemental material). In both proteins, two of the core domains form a stable homodimer related by a 2-fold symmetry axis, thereby extending the two monomer β-sheets to a continuous antiparallel 12-stranded β-sheet. Furthermore, in both molecules, ∼120 N-terminal residues are mostly in a coiled conformation, with the exception of extended α-helixes formed by residues 55 to 68 (Δ36Them4) and residues 64 to 79 (Δ34Them5) that are tightly attached to the convex side of the curved β-sheet via hydrophobic interactions with the first and second strands. Them4 and Them5 proteins contain a highly variable sequence stretch (ca. residues 83 to 109 and ca. residues 91 to 114 for Them4 and Them5, respectively) that is partially disordered in both crystal structures.
Fig 2.
Them5 is a Hotdog-fold dimer. (A) Diagram of the Δ34Them5 crystal structure. (B) Surface representations of the Δ34Them5 crystal structure. Surface-exposed residues conserved in both Them4 and Them5 (see Fig. S2A in the supplemental material) are highlighted in red and orange. A black circle indicates the putative binding site of the CoA-linked acyl chain, an asterisk marks the catalytic center, and an arrow points to Them5 residues Glu168 and Lys172, which interrupt the long open channel present in the Them4 structure. A blue circle shows the conserved residues on the surface of the central β-sheet that are likely to be involved in binding the CoA phospho-ADP module. Residues 89 to 101 and residues 238 to 247 are not displayed to make the image clearer. (C) Stylized representation of the putative Δ34Them5 active site. Homodimer subunits are shown in blue and orange, and residues expected to be involved in catalysis and substrate recognition are displayed as sticks (atom colors). (D and E) Saturation curves of reaction rates for acyl-CoA substrates obtained for wild-type hThem5 (D) and the D167A hThem5 mutant (E) at 37°C. Lines indicate fitting of the experimental data to the Michaelis-Menten equation. (F) Them5 hydrolyzes long-chain acyl-CoA esters. Steady-state kinetic constants for hThem5-catalyzed hydrolysis of selected acyl-CoAs obtained at 37°C (pH 7.5).
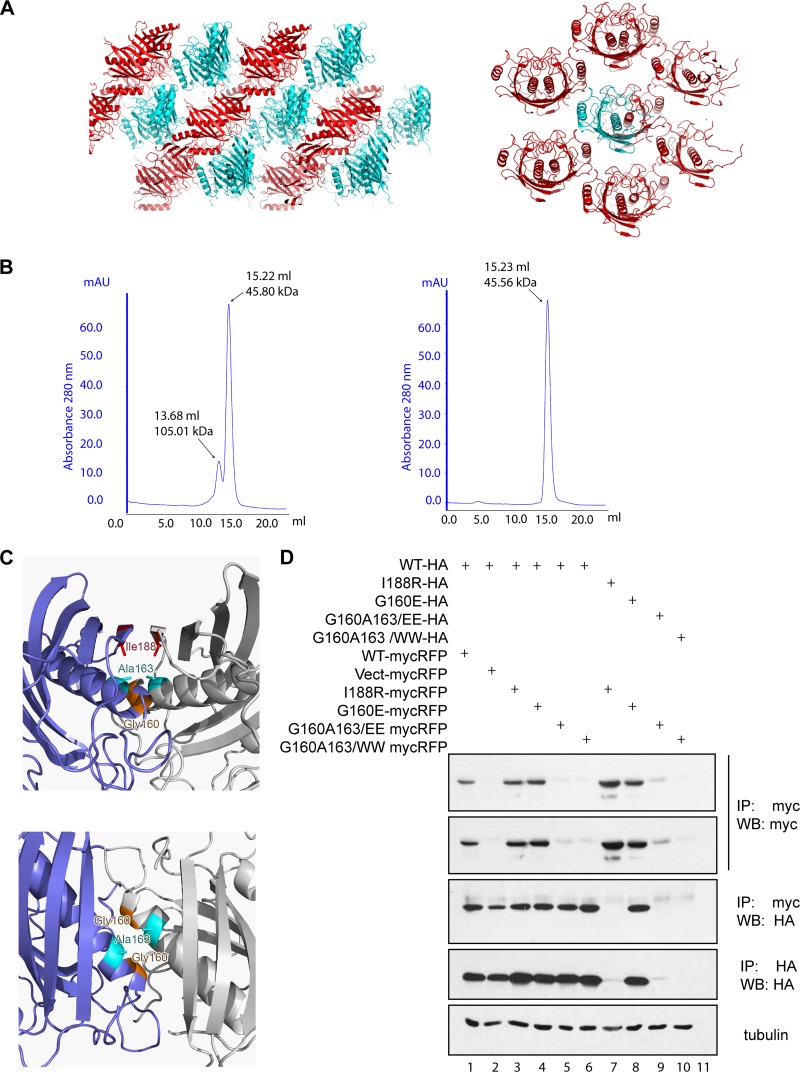
As in previous studies, we found that Δ36Them4 elutes as a dimer in size-exclusion chromatography, with a minor fraction present as a homotetramer (36). Δ34Them5, in contrast, elutes as a homodimer (Fig. 3B). A further strong argument for the dimeric state of Them4/5 in their native forms is the fact that they always appear as independent homodimers, irrespective of the crystal packing and the number of molecules in the asymmetric unit (a.u.) (Fig. 3A). However, it is still possible that Them4/5 can form higher-order oligomers in the specific environment in the mitochondria, interacting with membranes or proteins or binding substrates.
Fig 3.
Them4 and Them5 form homodimers. (A) Crystal packing of Δ36Them4 (left panel) and Δ34Them5 (right panel) molecules in the P1 and in the C2221 crystal lattice, respectively. Adjacent layers of symmetry-related molecules forming the crystal are displayed in cyan and red in the diagram. Δ34Them5 molecules are shown in the diagram, with the content of the asymmetric unit (a.u.) colored in cyan (1/2 of the Δ34Them5 homodimer) and the symmetry-related molecules displayed in red. The structure of Δ36Them4 was solved in space group P1 (two homodimers in the a.u.) and C2221 (one homodimer in the a.u.), while the structure of Δ34Them5 was solved in space group C2221 (1/2 homodimer in the a.u.), C2 (three homodimers in the a.u.), and P21 (one homodimer in the a.u.). In all of these cases, Δ36Them4 and Δ34Them5 are independent homodimers. (B) Analytical size-exclusion chromatography experiments were performed to determine the oligomeric states of Δ36Them4 (left panel) and Δ34Them5 (right panel) in solution. Δ36Them4 shows equilibrium between a homodimeric species (15.22 ml, 45.80 kDa, 48.50 kDa) and a tetrameric species (dimer of homodimers, 13.68 ml, 105.01 kDa, 97.01 kDa), whereas Δ34Them5 elutes as a homodimer only (15.23 ml, 45.56 kDa, 49.79 kDa). Elution volumes, approximate molecular masses, and expected molecular masses are given in parentheses, respectively. (C) Detailed view of Them5 mutation sites created to disrupt the homodimer. Sites are located at the homodimer interface and mapped onto the Δ34Them5 crystal structure (side view, left upper panel; top view, left bottom). The Them5 subunits are shown as diagrams (blue and gray); mutated residues are shown as sticks and colored in red (Ile188), orange (Gly160), and cyan (Ala163). The most C-terminal α-strand of both subunits is not shown in the bottom view for clarity reasons. (D) Coimmunoprecipitation experiments with ΔMTS-Them5 wild type and mutants with HA and myc-RFP tags. HEK293cells were transfected with the corresponding cDNAs 24 h after lysis, and the proteins were precipitated with anti-myc antibodies and then probed with the indicated antibodies. Mutations were introduced in the central helix, leading to instability of a dimer due to the electrostatic repulsion or steric hindrance (G160W/E, A163W/E); another mutation was made in order to disturb antiparallel β-sheet formation (I188R). Double 160E163E and 160W163W mutants have very low levels of expression but can still be coprecipitated with wild-type Them5 (lanes 5 and 6). However, cotransfection of two differently tagged mutants greatly diminishes their expression levels and abolishes interaction or coprecipitation (lanes 9 and 10). These results confirm that Them5 does indeed form dimers and cannot exist as a monomer in solution.
Them5 is an acyl-CoA thioesterase with unusual substrate specificity.
We analyzed the protein sequences of the Them4 and Them5 remote homologs in a search for conserved sequence motifs (see Fig. S2A in the supplemental material). Despite the low conservation of amino acid sequence between Hotdog thioesterases, their folding is conserved, as is the catalytic machinery (37). We identified a highly conserved HGG…D motif that was reported earlier to be an important consensus sequence motif in bacterial and mammalian Hotdog thioesterases (26). Both Δ36Them4 and Δ34Them5 crystal structures have two active sites per homodimer, located at the end of each Hotdog helix, with residues from both subunits contributing to catalysis. The main catalytic residues for Δ36Them4 and Δ34Them5, respectively, are indeed the HGG motif from residues 152 to 154 and from residues 158 to 160 on one subunit and Asp161/Thr177 and Asp167/Thr183 on the other subunit (Fig. 2C and see Fig. S2D in the supplemental material). These residues are proposed to bind and hydrolyze the thioester moiety of the acyl-CoA substrate, as described for a number of other Hotdog thioesterases (8, 26). They are located in the middle of a long L-shaped channel that is limited in length by a stretch of residues from the N-terminal part (residues 74 to 79 in Δ36Them4 and residues 83 to 88 in Δ34Them5), which likely defines the length of the CoA-coupled fatty acids accepted for turnover. This channel is less deep and less open in Them5 than in Them4, where the Them4 residues Ala162 and Met166 are positionally exchanged with Glu168 and Lys172, which together with Asn91 form an extended hydrogen bond network.
Mapping of conserved residues onto the surface of the Them5 crystal structure (Fig. 2B) showed full conservation of the catalytic machinery; a comparison with related structures revealed Asp161 (Them4) and Asp167 (Them5) to be critically important (16).
We next confirmed that recombinant Them5 is an active thioesterase in vitro (Fig. 2D to F). Like Them4 (12), Them5 was particularly active on medium-chain (e.g., C14:0) and long-chain acyl-CoA esters (Fig. 2F). However, Them5 displayed a distinctive specificity for linoleyl-CoA among the long-chain fatty acid esters (Fig. 2F). This difference in substrate affinities may be related to the structural differences we observed in the substrate-binding channels of Them4 and Them5. In Them5, the side chains of Glu168 and Lys172 form a polar, barrier-like elevation in the middle of this channel, restricting binding to the conserved residues (Ser87, Thr117, Arg118, and Tyr129) that form a cavity (Fig. 2B and C).
According to our crystallographic model of the active centers of Them4 and Them5 (Fig. 2C and see Fig. S2D in the supplemental material) and previously published structures of Hotdog thioesterases, we propose that D167 in Them5 is the key catalytic residue. Consistent with this hypothesis, an alanine-substituted Them5 mutant (D167A) no longer displayed catalytic activity (Fig. 2E and F). Additional mutations of the predicted active-site residues further confirmed their critical role in catalysis (see Fig. S2E in the supplemental material).
Them5 ablation modifies the metabolism of cardiolipin.
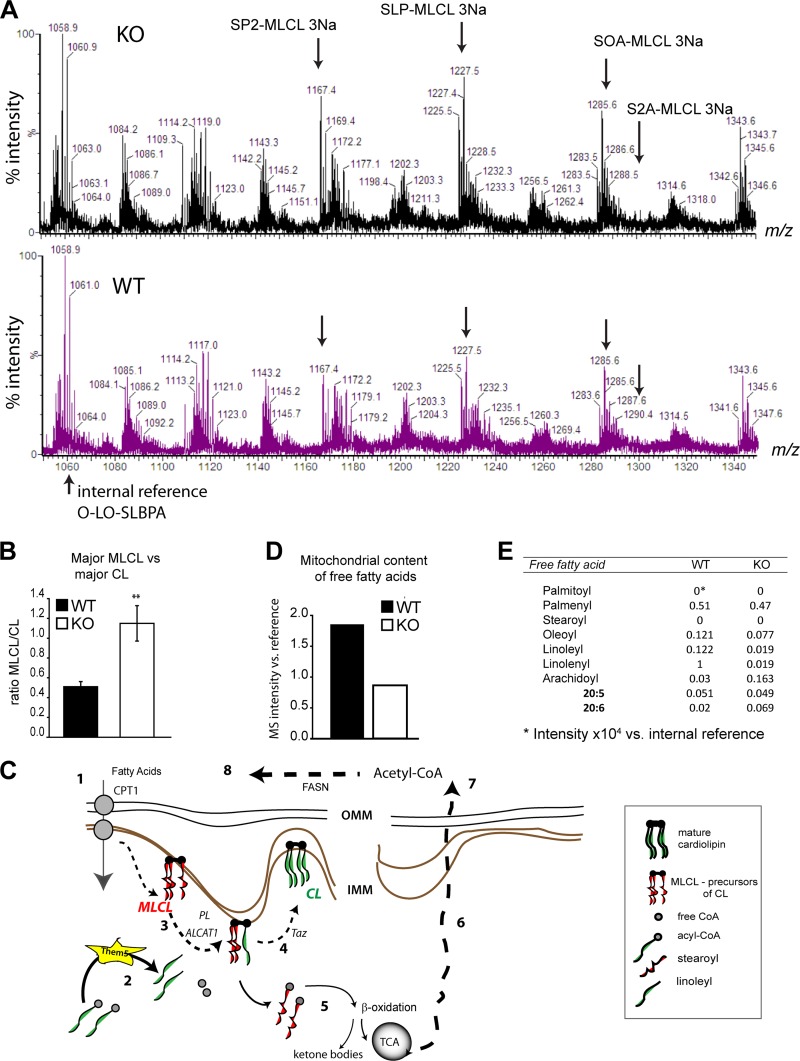
To investigate the biological role of Them5, we generated mice deficient in the Them5 protein (see Fig. S3 in the supplemental material). In view of the mitochondrial localization of Them5 and its acyl-CoA thioesterase activity, we carried out a detailed MS analysis of mitochondrial lipids from Them5−/− and control mice. There were very specific changes in the lipid profile of cardiolipin (CL) and its metabolites in the Them5 knockout mice, but no changes in phospholipids synthesized outside mitochondria (Fig. 4A; see also Fig. S4 in the supplemental material). Loss of Them5 led to a 2-fold increase in major species of monolysocardiolipin (MLCL) (Fig. 4B), which act as upstream metabolites in the remodeling cycle of the inner mitochondrial membrane CL (15). Intriguingly, loss of only one allele of Them5 (in heterozygous mice [HET]) produced a stronger increase in these MLCL species and a more significant alteration in the profile of CL species than complete loss of the Them5 gene (see Fig. S4 in the supplemental material). However, in both KO and HET mitochondria, the common effect of Them5 on cardiolipin was altered ratios of major species of CL and MLCL (see Fig. S4C in the supplemental material), analogous to the phenotypic changes in Barth syndrome (8). Subsequently, we focused on characterization of the complete Them5 knockout.
Fig 4.
Them5 is a new player in cardiolipin remodeling. (A) MS profiles of liver mitochondrial lipids from Them5KO and WT in the MLCL region. P, palmitoyl; S, stearoyl; O, oleoyl; L, lineaoyl; A, arachidoyl. (B) Increase in major monolysocardiolipin (MLCL) over cardiolipin (CL) levels upon Them5 ablation. (C) Fatty acids are imported into mitochondria from the cytosol via CPT transporters (step 1). Acyl-CoAs are hydrolyzed by Them5 (step 2), releasing acyl chains used for reacylation of the CL precursor, SP2-MLCL, by ALCAT1 or MLCLAT1 (20) (step 3). The other steps of CL remodeling are mediated by transacylase Taz (step 4). Normally, strearoyl and palmitoyl acyls are re-esterified and can be used in β-oxidation and, subsequently, the TCA cycle (step 5). The final product acetyl-CoA is exported to the cytosol (step 6) and is used in fatty acid synthesis (steps 7 and 8). CL, cardiolipin; LPC, lysophosphatidylcholine; MLCL, monolysocardiolipin; CPT, carnitine-palmitoyl transferase; CoA, coenzyme A; ALCAT1, monolyso-acyl-CoA:lysocardiolipin acyltransferase 1; TCA cycle, tricarboxylic acid cycle. (D) Quantification of total mitochondrial content of FFA. (E) Detailed composition of FFA from Them5 WT and KO liver mitochondria. Note the strong decrease in linolenic acid in Them5−/− samples compared to the WT. This fatty acid has the fastest turnover in mitochondria but does not contribute much to CL remodeling.
We propose that Them5 specifically regulates upstream remodeling of mitochondrial CL by maintaining, in particular, the pool of linoleyl groups used to reacetylate one metabolic intermediate of cardiolipin, namely, SP2-MLCL (stearoyl-dipalmitoyl-monolysocardiolipin) (Fig. 4). Consequently, it is likely that the mitochondrial accumulation of SP2-MLCL and other palmitoyl-containing MLCL species reflects an early impairment in the remodeling cycle of CL, which progressively adds linoleyl (and, to a lesser extent, other unsaturated fatty acids) to the saturated precursors, producing mature forms enriched in linoleyl (30). In addition, Them5 loss resulted in an ∼2-fold decrease in free fatty acids detected by MS in mitochondria, with a particularly marked decrease in linoleic and linolenic acids in the KO samples (Fig. 4D and E). The substrate preference of Them5 shown by a 4- to 5-fold higher kcat for linoleyl-CoA (C18:2) over either C18:3 or C18:1 explains the observed changes in free fatty acids and the remodeling of mitochondrial CL metabolites after Them5 ablation (Fig. 2F). Thus, combination of the MS analysis and the data on enzyme properties strongly suggests that Them5 is a thioesterase with a distinct specificity for C18 polyunsaturated fatty acids, which are essential metabolites for mammalian CL remodeling via acyl-CoA-dependent transferases (6).
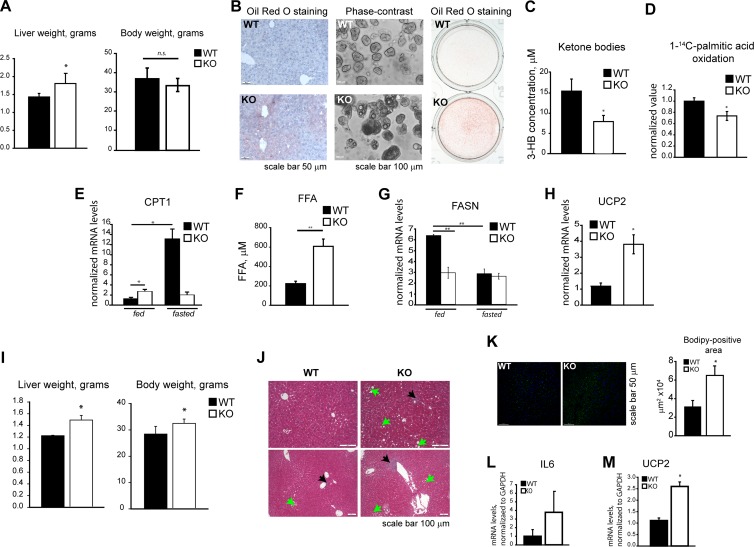
Them5−/− mice develop fatty liver.
The decrease in free fatty acids inside mitochondria after Them5 ablation (Fig. 4D) interferes with the formation and recycling of acyl-CoA molecules, particularly those essential for CL remodeling with linoleyl-CoA, and leads to age-dependent fatty liver disease (Fig. 5A and B; see also Fig. S5A and B in the supplemental material) and the inhibition of β-oxidation and ketone body formation (Fig. 5C and D). Free fatty acids are exported from Them5−/− mitochondria by the carnitine transport system (Fig. 5E) and accumulate in the cytosol, leading to the development of fatty liver.
Fig 5.
Loss of Them5 deregulates lipid metabolism. (A) Increased liver weight in Them5−/− animals but no changes in body weight (n = 8 to 11). (B) Age-dependent accumulation of lipids in liver. Oil Red O staining of liver sections from 6-month-old mice (left). Phase-contrast images (middle) and ORO staining (right) of primary hepatocytes from 9-month-old mice are shown. (C and D) Decreased serum levels of 3-hydroxybutyrate in Them5−/− (fasted, 12 months old, n = 5 to 6) (C) and lower rates of [1-14C]palmitic acid oxidation by primary hepatocytes (D). (E) Them5−/− mice show increases in basal levels of liver CPT1 but fail to induce it upon fasting (6 to 7 months old, n = 3 to 4). (F) Increase in total levels of plasma FFA in Them5−/− mice (fasted, 6 to 7 months old, n = 5). (G) Absence of fatty acid synthase induction in liver of Them5−/− mice after refeeding (6 to 7 months old, n = 3 to 4). (H) Increase in UCP2 mRNA in Them5−/− liver (6 to 7 months old, n = 4). (I to M) Mice (males, n = 6 to 8) were kept on a high-fat diet for 15 weeks and analyzed afterward. (I) Increase in liver and body weights. (J) Increased lipid accumulation (green arrows) and lymphocytic infiltration (black arrows) in Them5−/− liver sections compared to Them5+/+ after 15 weeks of a high-fat diet. (K) BODIPY 493/503 staining of liver sections. Quantification of lipid-positive staining (right side) shows an ∼2-fold increase in Them5−/− samples. (L and M) RNA was extracted from the livers of randomly fed Them5−/− and Them5+/+ mice after 15 weeks of high-fat diet, and the interleukin-6 mRNA (L) and UCP2 (M) expression levels were measured by real-time PCR.
The levels of FFA in plasma also increased significantly in Them5−/− mice upon fasting (Fig. 5F). Whereas the fatty acid synthase (FASN) mRNA levels were similar in fasted mutant and control mice, these levels were not stimulated in Them5−/− animals by refeeding (Fig. 5G). Since the induction of FASN normally occurs in response to elevated levels of acetyl-CoA, a β-oxidation product, and is negatively regulated by FA, the impaired β-oxidation, i.e., reduced production of acetyl-CoA, and steatosis may be explained by the failure of FASN induction. In line with hypoketosis and reduced oxidation of [14C]palmitic acid, the levels of CPT1 did not respond to fasting (Fig. 5E). In addition, UCP2 expression was considerably increased in Them5−/− mice (Fig. 5H), most probably as a result of lipid accumulation. Indeed, a high-fat diet exacerbated fatty liver development. The reduced ability of Them5−/− animals to cope with an increased FA load resulted in increased body weight, pronounced lipid accumulation, and inflammatory infiltration (Fig. 5I to M).
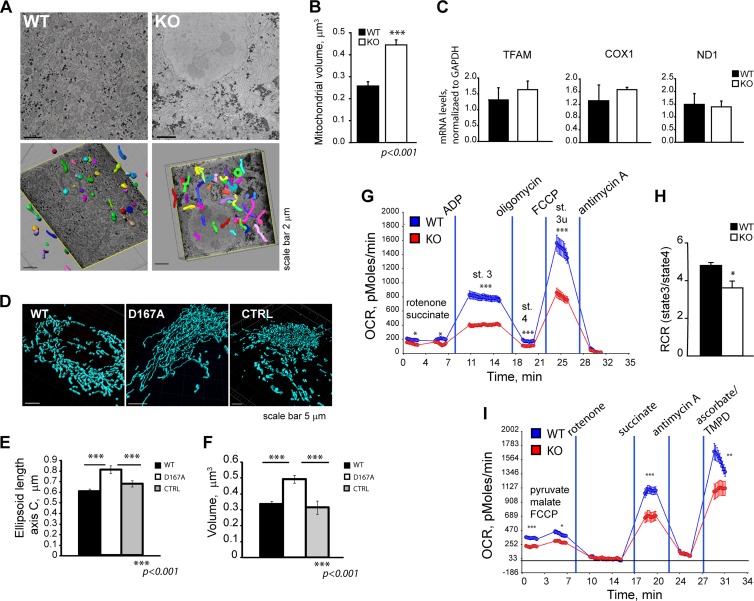
Loss of thioesterase activity of Them5 leads to changes in mitochondrial morphology and function.
Loss of mitochondrial proteins often affects function and, as a consequence, may alter the morphology of these organelles. To assess possible changes in mitochondrial morphology upon Them5 loss, we performed serial block-face scanning EM analysis of hepatocytes. 2D-EM images showed elongated, filamentous mitochondria in Them5 mutant mice (Fig. 6A, upper). Subsequent 3D reconstruction indicated a 2-fold increase in mitochondria volume and the formation of a highly interconnected network in Them5−/− mice, which was dramatically different from that in WT mitochondria (Fig. 6A, lower, and B; see also Fig. S5C and D in the supplemental material).
Fig 6.
Loss of the enzymatic activity of Them5 protein affects mitochondrial morphology and functions. (A) Electron micrographs of liver cells (upper) and subsequent 3D reconstruction of mitochondria (lower) (3 months old, fed). (B) Increased mitochondrial volume in Them5−/− hepatocytes from fed mice compared to WT controls. (C) mtDNA copy number as assessed by qPCR of single-copy mitochondrial genes ND1 and COX1, and mitochondrial transcription factor TFAM (n = 3). (D) Changes in morphology of mitochondria upon transfection of the D167A-Them5 mutant compared to WT Them5 or vector control animals; mitochondria were visualized with anti-TOM20 staining. (E and F) Increase in the length (E) and volume (F) of 3D reconstructed mitochondria in cells expressing D167A-Them5 compared to WT Them5 or vector control (a minimum of 10 cells per experiment were analyzed, n = 3). (G) Decreased oxygen consumption rates in Them5−/− liver mitochondria, as assayed using Seahorse XF analyzer. (n = 10). For a similar analysis in MEFs, see Fig 7. (H) Decrease in the respiratory control ratio (RCR) of Them5−/− liver mitochondria compared to WT controls (n = 8 to 10). (I) Reduced electron transfer abilities in isolated Them5−/− liver mitochondria (n = 10; TMPD, N,N,N′,N′-tetramethyl-p-phenylenediamine).
A similar phenotype was observed upon overexpression of the D167A Them5 mutant. Mitochondria showed a significant increase in volume (45%) and length (35%) compared to WT protein transfected cells (Fig. 6D), indicating the importance of Them5 enzymatic activity in the maintenance of mitochondrial morphology (Fig. 5E and F). The plasticity and dynamics of mitochondria are both dependent on the content of CL and other lipids and on the balance between CL and MLCL (1, 3, 13). Thus, the apparent importance of the thioesterase activity of Them5 for mitochondrial morphology may be explained by the observed >2-fold increase in major MLCL relative to CL species (Fig. 4 and 6; see also Fig. S4C in the supplemental material).
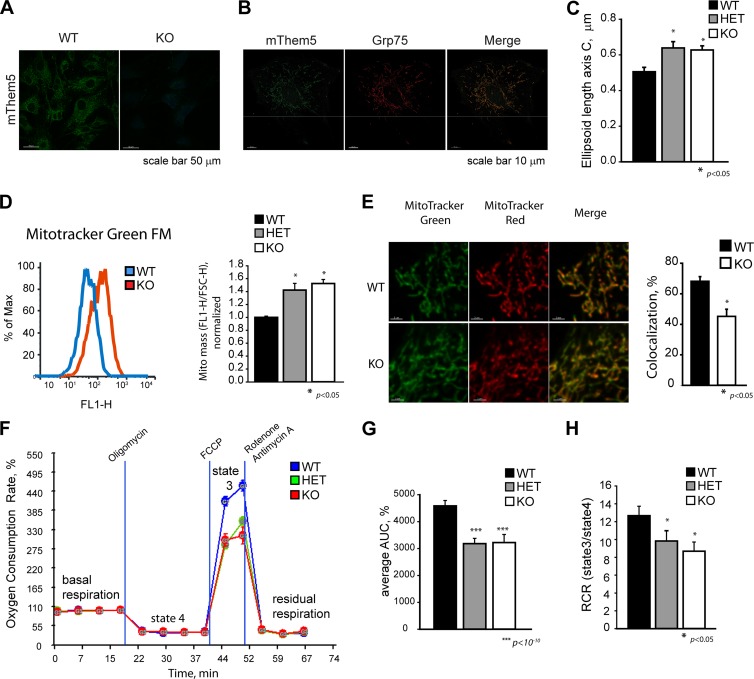
There were no significant changes in the mRNA levels of ND1, COX1, or TFAM, suggesting that the mtDNA copy number was not affected (Fig. 6C). However, fluorescence-activated cell sorting (FACS) analysis of MitoTracker green-labeled cells showed a 30% increase in the mitochondrial mass in Them5 mutant cells (Fig. 7D), possibly due to a compensatory mechanism for the reduced respiratory capacity of mitochondria. Indeed, we observed a reduction in the Them5−/− cells of the colocalization of MitoTracker Green and MitoTracker Red, which responds to the membrane potential of mitochondria (Fig. 7E).
Fig 7.
Them5 KO and HET liver cells and MEFs show changed mitochondrial morphology and decreased respiratory capacity. (A) Primary MEFs from Them5−/− and wild-type controls (gender-matched siblings) were stained with anti-mouse Them5 antibodies and visualized with LSM700 microscope. (B) Primary MEFs were costained with anti-mouse Them5 and anti-Grp75 (mortallin) antibodies. Images were taken with a LSM700 microscope. The staining pattern shows colocalization of Them5 and Grp75. (C) Primary MEFs from Them5−/−, Them5+/−, and wild-type control animals were fixed and stained with anti-TOM20 antibodies. Confocal images were taken with the LSM700 microscope, and mitochondria were reconstructed and their lengths were quantified using Imaris software (Bitplane). A minimum of three experiments were performed, with at least 10 cells in each analyzed. (D) Increased mitochondrial mass in Them5 KO and HET MEFs, as analyzed by FACS. Live cells were stained with MitoTracker Green FM, and the ratio between fluorescence intensity (FL1-H) and forward scatter (FSC-H, corresponding to the cell size) was determined and normalized to WT values. A total of five to six independent MEF lines (gender matched) for each genotype, using two to three independent sets of experiments, with 10,000 events for each sample, were analyzed. (E) Representative confocal images of live MEFs stained with MitoTracker Red and MitoTracker Green FM (left panel). Confocal images with 200-nm steps in z-stacks were taken using piezo motor-driven stage. Images were deconvolved and analyzed (quantification of colocalization, right side) with Imaris software. (F) Oxygen consumption rates were measured for Them5−/−, Them5+/−, and Them5+/+ primary MEFs using a Seahorse XF24 analyzer. (n = 10 for each genotype). (G) Quantification of the integral maximum respiration rate presented as the area under curve (AUC) shows a 35% decrease for both HET and KO samples compared to WT. (H) RCR, as determined by the ratio of OCR between state 3 and state 4, shows 25 and 30% decrease in Them5−/+ and Them5−/− MEFs compared to the wild-type control, respectively.
Analysis of Them5−/− mitochondria revealed a significant decrease in state 3 and state 4 respiration (Fig. 6G). The respiratory control ratio was lower in both Them5−/− liver mitochondria and MEFs (Fig. 6H and Fig. 7F and G), indicating increased uncoupling in knockout mitochondria, potentially due to elevated UCP2 expression (Fig. 5H). In studying the respiratory capacity of mitochondria functionally dissected with specific inhibitors of complex I and III, we found a decrease in the coupling efficiency at both site I (pyruvate + malate) and site II (succinate) (Fig. 6I). These results are consistent with the MitoTracker Red staining pattern (Fig. 7E). The rates of electron transfer were also reduced in Them5−/− mice (Fig. 6I). The previously documented impairment in β-oxidation and tricarboxylic acid (TCA) cycle could be partially responsible for the observed decrease (almost 50%) in maximal respiratory rates with endogenous substrates, which mainly derives from intramitochondrial metabolites and imported pyruvate (Fig. 5C and D and Fig. 6G and I).
DISCUSSION
We show here that Them5 is a novel mitochondrial acyl-CoA thioesterase most likely involved in the metabolism of cardiolipin (CL), specifically in remodeling steps requiring linoleyl-CoA species. This new evidence provides a chemical and metabolic explanation for the mitochondrial phenotype observed after the genetic ablation of Them5 and also throws light on the biological function of Them4. Them5 seems to be a result of gene duplication, and thus Them4 and 5 may be paralogs. We show here that human Them4 and Them5 are both homodimers of the Hotdog-fold, supporting the hypothesis that these proteins are a distinct group of mammalian thioesterases (5). They seem to be most related to the homodimer subclass of the PAAI family, members of which are present as homodimers only (hypothetical proteins with PDB code 1IXl in P. horikoshii or PDB code 2OV9 in Rhodococcus sp.). The catalytic machinery of Them4/5 is conserved and comprises the HGG motif and Asp/Thr residues. The main sequence differences map onto the partially disordered loop covering the active-site cleft and onto the bottom of the cleft, which is the putative binding site for the CoA-linked fatty acid alkyl chain. This may provide a structural explanation for the different substrate preferences of Them4 and Them5. Considering the late appearance of Them4 in evolution and the even later appearance of Them5, these proteins may have evolved specific and nonoverlapping activities in mitochondria.
Indeed, our data indicate that Them5 loss induces the accumulation of major species of MLCL. Together with the kinetics analysis, these results indicate that Them5 is a linoleyl-CoA-specific thioesterase. We propose that Them5 thioesterase activity regulates upstream remodeling of CL by maintaining the pool of acyl groups, specifically linoleyl, which in particular reacylate stearoyl- and palmitoyl-containing MLCL species lying upstream in the CL remodeling cycle (Fig. 4 and see Fig. S4 in the supplemental material) (15, 30). Several enzymes involved in CL biosynthesis are known, but no specific thioesterases providing acyls for CL remodeling have been identified (23). To our knowledge, Them5 is the first C18 polyunsaturated specific thioesterase to have been defined and, given its location and substrate preference, it may firmly be considered to be involved in CL metabolism. Remodeling is essential for CL functions, namely, in the regulation of mitochondrial bioenergetics and dynamics. This has been clearly shown in the example of ALCAT1, an acyltransferase involved in the remodeling of CL (6, 20). Upregulation of ALCAT1 on a high-fat diet, aggravated by its lack of acyl-chain substrate specificity, leads to alteration of the “normal” CL acyl profile due to changes in the substrates provided. This results further in the exaggeration of oxidative stress. Loss of tafazzin, another CL remodeling enzyme, is associated with the Barth syndrome and involves changes in mitochondrial morphology that are much more dramatic than those observed here in Them5−/− mice (11, 31). Clearly, interference with either chain selection during CL remodeling or CL synthesis itself leads to a number of mitochondrion-associated metabolic conditions (10). Hence, our work, together with the recent identification of PTPMT1 as a novel CL biosynthetic enzyme, prompts further studies on CL involvement in human diseases (34, 35).
We show that loss of Them5 decreases the pool of FFA in mitochondria, resulting in age-dependent progression of fatty liver in Them5−/− mice, compounded by hypoketosis and decreased β-oxidation. This resembles results from a previous study, where loss of the acyl-CoA binding protein altered the supply of acyl-CoA metabolites and thus modified CL remodeling in yeasts (28). CoA is then trapped in long acyl-CoA molecules (stearoyl- and palmitoyl-CoA) that are not normally used in CL remodeling. The absence of Them5 increases the MLCL/CL ratio and causes an imbalance in CL homeostasis. This results in the reduction of respiratory rates and impairment of electron transfer between the respiratory complexes. OXPHOS complexes are known to require normal levels of CL and its metabolites for their function; however, the documented respiratory defects have not been linked previously to thioesterases (10, 12). The observed patterns of phospholipid composition in Them5+/− and Them5−/− mice suggest that further enzymes are involved in the rewiring of CL synthesis/remodeling, which warrants further analysis. Given that Them5−/− animals develop even stronger lipid accumulation than Them5+/− animals, it is possible that Them5 interferes with other signaling pathways. For example, the observed increase in C20:n levels could affect BDNF/TrkB-dependent signaling in a context-dependent manner (27). Given the unusual specificity of Them5 enzymatic activity, it would be reasonable to investigate possible mutations and/or polymorphisms involving catalytic residues or dimerization motifs present in humans that may affect its substrate preference.
Considering the importance of cardiolipin synthesis and remodeling pathways in mitochondrial function and generally in physiology, our results indicate the possible involvement of Them5 in human pathologies, such as fatty liver disease, obesity, and related disorders.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael Rebhan for help with the initial bioinformatics analysis, Ahmad Bechara for discussion of the EM data, David Knight for MS analysis, and Gregor Lotz for respirometry analysis. We also thank Susanne Schenk for Them5 antibodies generation and the staff of beamlines X10SA and X06DA at the Swiss Light Source (Villigen, Switzerland) for their excellent support in X-ray data collection.
E.Z. was supported by a Swiss Bridge fellowship. Work in Manchester was undertaken with the FLS Biomolecular Analysis Facility. The Friedrich Miescher Institute for Biomedical Research is part of the Novartis Research Foundation.
Footnotes
Published ahead of print 14 May 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Acehan D, et al. 2011. Cardiac and skeletal muscle defects in a mouse model of human Barth syndrome. J. Biol. Chem. 286:899–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ban T, Heymann JA, Song Z, Hinshaw JE, Chan DC. 2010. OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation. Hum. Mol. Genet. 19:2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berrow NS, et al. 2007. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 35:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brocker C, Carpenter C, Nebert DW, Vasiliou V. 2010. Evolutionary divergence and functions of the human acyl-CoA thioesterase gene (ACOT) family. Hum. Genomics 4:411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cao J, Liu Y, Lockwood J, Burn P, Shi Y. 2004. A novel cardiolipin-remodeling pathway revealed by a gene encoding an endoplasmic reticulum-associated acyl-CoA:lysocardiolipin acyltransferase (ALCAT1) in mouse. J. Biol. Chem. 279:31727–31734 [DOI] [PubMed] [Google Scholar]
- 7. Cao J, Shen W, Chang Z, Shi Y. 2009. ALCAT1 is a polyglycerophospholipid acyltransferase potently regulated by adenine nucleotide and thyroid status. Am. J. Physiol. Endocrinol. Metab. 296:E647–E653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cao J, Xu H, Zhao H, Gong W, Dunaway-Mariano D. 2009. The mechanisms of human hotdog-fold thioesterase 2 (hTHEM2) substrate recognition and catalysis illuminated by a structure and function based analysis. Biochemistry 48:1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cerqua C, et al. 2010. Trichoplein/mitostatin regulates endoplasmic reticulum-mitochondria juxtaposition. EMBO Rep. 11:854–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chicco AJ, Sparagna GC. 2007. Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am. J. Physiol. Cell Physiol. 292:C33–C44 [DOI] [PubMed] [Google Scholar]
- 11. Claypool SM, Boontheung P, McCaffery JM, Loo JA, Koehler CM. 2008. The cardiolipin transacylase, tafazzin, associates with two distinct respiratory components providing insight into Barth syndrome. Mol. Biol. Cell 19:5143–5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Claypool SM, Koehler CM. 2011. The complexity of cardiolipin in health and disease. Trends Biochem. Sci. 37:32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Claypool SM, Whited K, Srijumnong S, Han X, Koehler CM. 2011. Barth syndrome mutations that cause tafazzin complex lability. J. Cell Biol. 192:447–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 15. Esposti MD, Cristea IM, Gaskell SJ, Nakao Y, Dive C. 2003. Proapoptotic Bid binds to monolysocardiolipin, a new molecular connection between mitochondrial membranes and cell death. Cell Death Differ. 10:1300–1309 [DOI] [PubMed] [Google Scholar]
- 16. Forwood JK, et al. 2007. Structural basis for recruitment of tandem hotdog domains in acyl-CoA thioesterase 7 and its role in inflammation. Proc. Natl. Acad. Sci. U. S. A. 104:10382–10387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hunt MC, Alexson SE. 2002. The role Acyl-CoA thioesterases play in mediating intracellular lipid metabolism. Prog. Lipid Res. 41:99–130 [DOI] [PubMed] [Google Scholar]
- 18. Kang HW, Niepel MW, Han S, Kawano Y, Cohen DE. 2012. Thioesterase superfamily member 2/acyl-CoA thioesterase 13 (Them2/Acot13) regulates hepatic lipid and glucose metabolism. FASEB J. 26:2209–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kirkby B, Roman N, Kobe B, Kellie S, Forwood JK. 2010. Functional and structural properties of mammalian acyl-coenzyme A thioesterases. Prog. Lipid Res. 49:366–377 [DOI] [PubMed] [Google Scholar]
- 20. Li J, et al. 2010. Cardiolipin remodeling by ALCAT1 links oxidative stress and mitochondrial dysfunction to obesity. Cell Metab. 12:154–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61:458–464 [DOI] [PubMed] [Google Scholar]
- 22. Moisoi N, et al. 2009. Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brain-specific transcriptional stress response. Cell Death Differ. 16:449–464 [DOI] [PubMed] [Google Scholar]
- 23. Osman C, Voelker DR, Langer T. 2011. Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 192:7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pagliarini DJ, et al. 2008. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134:112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Parcellier A, et al. 2009. Carboxy-terminal modulator protein (CTMP) is a mitochondrial protein that sensitizes cells to apoptosis. Cell Signal. 21:639–650 [DOI] [PubMed] [Google Scholar]
- 26. Pidugu LS, Maity K, Ramaswamy K, Surolia N, Suguna K. 2009. Analysis of proteins with the ‘hot dog’ fold: prediction of function and identification of catalytic residues of hypothetical proteins. BMC Struct. Biol. 9:37 doi:10.1186/1472-6807-9-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rapoport SI, Rao JS, Igarashi M. 2007. Brain metabolism of nutritionally essential polyunsaturated fatty acids depends on both the diet and the liver. Prostaglandins Leukot. Essent. Fatty Acids 77:251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rijken PJ, et al. 2009. Cardiolipin molecular species with shorter acyl chains accumulate in Saccharomyces cerevisiae mutants lacking the acyl coenzyme A-binding protein Acb1p: new insights into acyl chain remodeling of cardiolipin. J. Biol. Chem. 284:27609–27619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sali A, Blundell TL. 1993. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815 [DOI] [PubMed] [Google Scholar]
- 30. Schlame M. 2008. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J. Lipid Res. 49:1607–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schlame M, Ren M. 2006. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS Lett. 580:5450–5455 [DOI] [PubMed] [Google Scholar]
- 32. Slot JW, Geuze HJ. 2007. Cryosectioning and immunolabeling. Nat. Protoc. 2:2480–2491 [DOI] [PubMed] [Google Scholar]
- 33. vom Brocke J, Schmeiser HH, Reinbold M, Hollstein M. 2006. MEF immortalization to investigate the ins and outs of mutagenesis. Carcinogenesis 27:2141–2147 [DOI] [PubMed] [Google Scholar]
- 34. Xiao J, et al. 2011. Structural and functional analysis of PTPMT1, a phosphatase required for cardiolipin synthesis. Proc. Natl. Acad. Sci. U. S. A. 108:11860–11865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang J, et al. 2011. Mitochondrial phosphatase PTPMT1 is essential for cardiolipin biosynthesis. Cell Metab. 13:690–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao H, Martin BM, Bisoffi M, Dunaway-Mariano D. 2009. The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-fold family. Biochemistry 48:5507–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhuang Z, et al. 2008. Divergence of function in the Hotdog-fold enzyme superfamily: the bacterial thioesterase YciA. Biochemistry 47:2789–2796 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.