Abstract
Listeria monocytogenes, a Gram-positive food-borne human pathogen, is able to grow at temperatures close to 0°C and is thus of great concern for the food industry. In this work, we investigated the physiological role of one DExD-box RNA helicase in Listeria monocytogenes. The RNA helicase Lmo1722 was required for optimal growth at low temperatures, whereas it was dispensable at 37°C. A Δlmo1722 strain was less motile due to downregulation of the major subunit of the flagellum, FlaA, caused by decreased flaA expression. By ribosomal fractionation experiments, it was observed that Lmo1722 was mainly associated with the 50S subunit of the ribosome. Absence of Lmo1722 decreased the fraction of 50S ribosomal subunits and mature 70S ribosomes and affected the processing of the 23S precursor rRNA. The ribosomal profile could be restored to wild-type levels in a Δlmo1722 strain expressing Lmo1722. Interestingly, the C-terminal part of Lmo1722 was redundant for low-temperature growth, motility, 23S rRNA processing, and appropriate ribosomal maturation. However, Lmo1722 lacking the C terminus showed a reduced affinity for the 50S and 70S fractions, suggesting that the C terminus is important for proper guidance of Lmo1722 to the 50S subunit. Taken together, our results show that the Listeria RNA helicase Lmo1722 is essential for growth at low temperatures, motility, and rRNA processing and is important for ribosomal maturation, being associated mainly with the 50S subunit of the ribosome.
INTRODUCTION
With reductions in temperature, most chemical reactions are slowed down. For bacteria to survive at low temperatures, they need to keep macromolecules active. Single-stranded RNA molecules form secondary structures that, through their physical properties, are more stable at low temperatures than at high temperatures. This raises the question of how psychrotolerant bacteria can survive and thrive at low temperatures. DExD-box RNA helicases (a subfamily of DEAD box RNA helicases) are enzymes that have been suggested to unwind occluding secondary RNA structures and are believed to be particularly important during growth at low temperatures (16). For instance, some RNA helicases have been shown to participate in the maturation and refolding of the 23S rRNA (DbpA and YxiN) or/and assembly of the 50S ribosomal subunit (SrmB and CsdA) (14, 15, 21, 30, 36). Several RNA helicases act together with additional partners, constituting an RNA degradosome (13, 35, 38, 52). Still other helicases can be associated with different targets and partners (16, 29, 30). Until now, most studies of RNA helicases in bacteria have been performed in Escherichia coli and Bacillus subtilis. However, neither of these bacteria can multiply at temperatures close to 0°C.
Listeria monocytogenes is a pathogenic Gram-positive bacterium causing listeriosis, manifested as gastrointestinal infection that can spread into the blood, the central nervous system, and, in the case of pregnant women, the fetus (17). As a soil bacterium, it can occasionally contaminate the food chain and thus can be found in nonpasteurized and ready-to-eat food (48). Listeria is a major concern for the food industry due to its ability to grow at very low temperatures, even below 0°C (32). Several factors have been identified that allow Listeria to grow at low temperatures (12). Among these are compatible solute transporters encoded by the opu and gbu operons; oligopeptide permease encoded by oppA; ferritin (Fri); and phosphohydrolase, encoded by pgpH and involved in ppGpp turnover (7, 9, 20, 23, 28, 41, 42, 45, 46). L. monocytogenes harbors 4 putative DExD-box RNA helicases, and in a previous study, the genes encoding three RNA helicases (lmo0866, lmo1450, and lmo1722) were found to be induced at low temperatures (11). A transposon insertion in a region directly upstream of a putative RNA helicase gene (homologous to lmo1722) in L. monocytogenes strain F2365 induced a cold-sensitive growth phenotype and reduced swarming motility on soft agar plates (4). However, the authors were unable to complement these phenotypes, suggesting additional mutations or downstream effects of the transposon mutant.
To our knowledge, only a few studies have investigated the roles of RNA helicases in organisms able to grow at very low temperatures. Most studies have focused on the roles of these RNA helicases in vitro and not in their natural context in vivo (8, 10, 40, 49). One exception is Bacillus cereus, where the physiological functions of five RNA helicase genes were examined with regard to their roles in cold growth and response to various stresses (50, 51). In this study, we investigated the function of an RNA helicase in a cold-growing bacterium, L. monocytogenes. Our data show that the RNA helicase Lmo1722 is required for growth at low temperatures, for motility, for accurate 23S rRNA processing, and for proper ribosome maturation. Intriguingly, a strain expressing a C-terminally deleted form of Lmo1722 displayed wild-type (WT) ribosomal maturation, 23S rRNA processing, and complete growth restoration in the cold, despite being dispersed in the cytoplasm (delocalized from ribosomal subunits).
MATERIALS AND METHODS
Strains and plasmid construction.
E. coli and L. monocytogenes strains are listed in Table 1. The plasmids used in the study are listed in Table 2 and the oligonucleotides in Table S1 in the supplemental material. E. coli was grown in LB and L. monocytogenes in brain heart infusion (BHI) unless otherwise noted. Where needed, antibiotics were included in the growth media at the following final concentrations: carbenicillin, 100 μg/ml; kanamycin, 50 μg/ml; nalidixic acid, 50 μg/ml; colistin sulfate, 10 μg/ml; and chloramphenicol, 25 μg/ml for E. coli and 7 μg/ml for L. monocytogenes. Cloning was performed using standard techniques (53). The flanking regions of lmo1722 were amplified with primer pairs lmo1722-A and lmo1722-B for the upstream region and lmo1722-C and lmo1722-D for the downstream region. The PCR products were ligated and digested with SalI and NcoI endonucleases and cloned in tandem into the SalI and NcoI sites of the pMAD vector (1). The deletion of lmo1722 was then performed as described previously (1). Complementation of lmo1722 deletion was done by cloning of the helicase gene into the E. coli-L. monocytogenes shuttle vector pMK4P harboring a constitutively active promoter (2). Oligonucleotide primers 1722 Dp+/p− and 1722 Up− were used for PCR amplification. The plasmid with an lmo1722 insert was transferred to L. monocytogenes by electroporation. For overexpression in E. coli, the gene lmo1722 was amplified with Phusion DNA polymerase (Finnzymes) from an EGDe chromosomal DNA template by PCR using lmo1722-F_64_KpnI and lmo1722-RH_66_XbaI primers. The appropriately cut DNA fragments purified from an agarose gel were ligated with pBAD18 vector, and E. coli DH5α was transformed with the ligation mixture. Selected clones were confirmed by sequencing the cloned region. Similarly, the primer pair lmo1722-F_64_BamHI and lmo1722-R_62_SalI was used to generate plasmid pKVA791 (as well as pKVA742 and pKVA789, which contain random mutations). The primer pair lmo1722-F_64_BamHI and lmo1722-R_noC_64_SalI was used to generate plasmid pKVA746. The resulting constructs were transferred to L. monocytogenes by conjugation with E. coli strain S17-1 carrying the plasmids (18). Transconjugants were selected by plating on BHI plates containing kanamycin, colistin sulfate, and nalidixic acid.
Table 1.
Bacterial strains
| Strain | Relevant genotype/phenotype | Reference |
|---|---|---|
| E. coli | ||
| DH5α | Cloning host | 6 |
| LMG194 | Protein expression host for vectors with arabinose-inducible promoters | 27 |
| S17-1 | Strain used for conjugative plasmid transfer to L. monocytogenes | 55 |
| L. monocytogenes | ||
| EGDe | Wild-type L. monocytogenes | 24 |
| Δlmo1722 strain | EGDe with lmo1722 deleted | This study |
Table 2.
Plasmids
| Plasmid | Description | Source/reference |
|---|---|---|
| pMAD | Listeria allelic-replacement vector | 1 |
| pMK4P | E. coli-L. monocytogenes shuttle vector with constitutive Pprot promoter | 2 |
| plmo1722 | pMK4P with lmo1722 under constitutive promoter | This study |
| pBAD18 | Arabinose-inducible E. coli cloning vector; Cbr | 27 |
| pKVA429 | lmo1722 with C-terminal 6×His tag in pBAD18 | This study |
| pIMK3 | IPTG-inducible Listeria cloning vector; Kmr | P. Casey; 47 |
| pKVA791 | lmo1722 in pIMK3 | This study |
| pKVA746 | lmo1722 with a C-terminal deletion; clone in pIMK3 | This study |
| pKVA742 | lmo1722 in pIMK3; 1-bp ΔA deletion in the cloned region 5 nucleotides downstream of BamHI recognition site | This study |
| pKVA789 | lmo1722 in pIMK3; 19-bp deletion ΔG634-G652 of lmo1722 gene, resulting in a frameshift after Val211 of Lmo1722 | This study |
RNA isolation.
Bacterial cultures grown to a defined growth phase were mixed with 0.2 volume of 5% phenol in 95% ethanol (61), and bacteria were harvested by centrifugation. The bacterial pellets were frozen in liquid nitrogen and stored at −80°C. RNA from L. monocytogenes was isolated using a modification of guanidinium thiocyanate–phenol-chloroform extraction (56).
Northern blotting.
For Northern blotting, 20 μg of total RNA was separated on a formaldehyde agarose gel prior to blotting, as described previously (56). The Hybond-N membrane (GE Healthcare) was subsequently hybridized with [α-32P]dATP-labeled DNA fragments amplified with the corresponding primers using the Megaprime DNA-labeling system (GE Healthcare). Northern blots were developed, and band intensities were measured in the Storm 860 machine (Molecular Dynamics). The PCR primer pairs used to generate DNA probes for detection of flaA, degU, and transfer mRNA are listed in Table S1 in the supplemental material.
Primer extension.
A method of primer extension using a fluorescently labeled primer was described previously (43). Primer extension reactions were performed using RevertAid Premium reverse transcriptase (Thermo Scientific) according to the manufacturer's protocol. Each reaction mixture contained 1 μg of total RNA and 2.4 pmol 6-carboxyfluorescein (FAM)-labeled primer 23S-FAM (5′-CATATCGGTGTTAGTCCCG-3′). The primer was allowed to anneal to the template RNA by slowly cooling the reaction solution from 80°C to 30°C for 1 h. The rest of the reaction components were added to a final volume of 20 μl, and primer extension proceeded at 50°C for 1 h. The reaction products were ethanol precipitated and resolved on a 3130xl Genetic Analyzer using a GeneScan 500LIZ size standard (Applied Biosystems). The peaks of the fluorescent products (corresponding to transcripts of different lengths) were analyzed with GeneMapper 4.0 software (Applied Biosystems) (see Fig. S5 in the supplemental material). From this, the most prominent peak areas corresponding to mature 23S rRNA and immature 23S rRNA harboring a 160-nucleotide (nt) 5′ precursor sequence were quantified, and the immature/mature signal ratio was plotted.
Listeria chromosomal-DNA isolation.
Chromosomal DNA was extracted after lysis with mutanolysin (19). Briefly, bacteria were grown in BHI medium at 37°C in shaking culture. Listeria bacteria from 10 ml of overnight culture were harvested by centrifugation, suspended in 1 ml SET buffer (30 mM Tris-Cl, pH 8, 50 mM NaCl, and 5 mM EDTA), washed with 0.5 ml acetone, and incubated for 1 h at 37°C in 50 mM Tris-Cl, pH 6.5, 200 U/ml mutanolysin (Sigma-Aldrich), and 0.01 mg/ml RNase A. A 10× proteinase K incubation buffer was added to a final concentration of 50 mM Tris-Cl, pH 7.5, and 10 mM CaCl2, and treatment with 12,000 U of proteinase K (Fermentas) was continued for 30 min at 37°C. After phenol-chloroform extraction, chromosomal DNA was fished out from 75% ethanol and suspended in TE buffer (10 mM Tris-Cl, pH 8, 1 mM EDTA).
Lmo1722 purification and antibody production.
E. coli strain LMG194 carrying the pKVA429 (pBAD18:lmo1722H6) plasmid was grown at 37°C in 3 liters of LB medium supplemented with 100 μg/ml carbenicillin to an optical density at 500 nm (OD600) of 0.5. Lmo1722-H6 expression was induced with 0.02% arabinose and allowed to proceed at 30°C for 6 h. The bacteria were harvested by centrifugation at 6,000 × g for 30 min at 4°C. The bacteria were washed by suspending them in 1/10 culture volume of solution W (50 mM Tris-Cl, pH 8, 200 mM NaCl, 5 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM benzamidine) with 5 mM EDTA and 0.1 mg/ml lysozyme. After centrifugation, the pellet was again suspended in the same volume of solution W, and the bacteria were disrupted by sonication. Insoluble material was removed by centrifugation at 30,000 × g for 1 h. Polyethyleneimine, pH 8, was added dropwise to the stirred supernatant to a final concentration of 0.2% and allowed to precipitate at 0°C for 30 min, and the pellet was removed after centrifugation at 30,000 × g for 15 min (22, 59). Ammonium sulfate was added to 95% saturation and allowed to stand overnight in ice. The protein precipitate was collected by centrifugation at 30,000 × g for 2 h, washed in solution W saturated with ammonium sulfate, centrifuged again, suspended in solution A (20 mM Tris-Cl, pH 8, 500 mM NaCl, 50 mM imidazole, 10% glycerol, 5 mM 2-mercaptoethanol, 1 mM PMSF), and loaded onto a 1-ml HisTrap column (GE Healthcare). Lmo1722-H6 was then eluted with a linear imidazole gradient to a 300 mM concentration.
The protein was further purified by size exclusion on a Superdex 75pg (GE Healthcare) column using buffering solution S (50 mM Tris-Cl, pH 8, 500 mM NaCl, 5 mM 2-mercaptoethanol, 1 mM PMSF). For rabbit immunization, 2 mg of purified protein was separated on 10% SDS-PAGE, stained with Coomassie R250, cut out from the gel, and used for injection into a rabbit (Agrisera AB, Vännäs, Sweden).
Ribosome profiling.
The ribosomal profiling was essentially performed as previously described (25), with a modification of the lysis method adapted for Listeria.
L. monocytogenes strains were grown in BHI shaking culture at 16°C and 160 rpm to an OD600 of 0.5. Bacterial growth was stopped with 100 μg/ml chloramphenicol, and cells were harvested by centrifugation of 100 ml culture at 10,000 × g, and washed with half a culture volume of ice-cold solution RW (10 mM Tris-Cl, pH 7.5, 60 mM KCl, 10 mM MgCl2, 6 mM 2-mercaptoethanol, 1 mM PMSF, 100 μg/ml chloramphenicol). To the bacterial pellet, 0.5 ml of solution RM (10 mM Tris-Cl, pH 6.5, 60 mM KCl, 10 mM MgCl2, 6 mM 2-mercaptoethanol, 1 mM PMSF, 100 μg/ml chloramphenicol, 20% sucrose, 2,000 U/ml mutanolysin) was added. After incubation for 40 min at 37°C, samples were centrifuged for 10 min at 10,000 × g, and the supernatant was discarded. The bacteria were lysed by keeping them for 15 min in 0.5 ml ice-cold solution RL (10 mM Tris-Cl, pH 7.5, 60 mM KCl, 10 mM MgCl2, 6 mM 2-mercaptoethanol, 1 mM PMSF, 100 μg/ml chloramphenicol, 0.2% Triton X-100 Reduced [Sigma-Aldrich], 200 U/ml DNase I, RNase free [Roche]). After addition of 0.16% sodium deoxycholate, samples were centrifuged at 20,000 × g and 4°C for 1 h. The supernatant was either directly loaded for fractionation in a sucrose gradient or frozen in liquid nitrogen to be processed later. The sucrose gradients were prepared using the Gradient Master apparatus (Biocomp, Fredericton, NB, Canada). An amount of lysate corresponding to 7 A260 units was loaded on top of centrifugation tubes with a 10 to 40% sucrose gradient in solution R (10 mM Tris-Cl, pH 7.5, 60 mM KCl, 10 mM MgCl2). Samples were centrifuged for 3.5 h in an SW41Ti rotor (Beckman) at 35,000 rpm and 4°C. The ribosomal profiles were generated by UV absorbance (A254) measurements of the gradients using an ISCO sucrose gradient fractionator equipped with a UA-6 absorbance detector (Teledyne ISCO, Lincoln, NE). When necessary, 4-drop-volume fractions were collected and the absorbance of each was measured using Nanodrop (Thermo Scientific) and used for SDS-PAGE and Western blot analysis. Proteins for SDS-PAGE were concentrated by 6% trichloroacetic acid and sodium deoxycholate precipitation (5).
SDS-PAGE and Western blotting.
Listeria total-protein samples for electrophoresis were prepared by mutanolysin lysis (19) and analyzed by SDS-PAGE (37) and/or Western blotting on a polyvinylidene difluoride (PVDF) membrane (57). Briefly, the different cultures were grown in BHI before being disrupted. Protein samples were separated on 12% PAGE and either stained with Coomassie Brilliant Blue or transferred onto a PVDF membrane using a tank transfer apparatus (Bio-Rad). Development of the membrane essentially followed the protocol of the ECL+ Western blotting kit (Amersham) using anti-Lmo1722 as the primary antibody and horseradish peroxidase (HRP)-conjugated anti-rabbit as the secondary antibody (Bio-Rad). Measurement of the luminescence signal was carried out on a LAS4000 machine (Fuji). Quantification of the luminescence signal was done using Quantity One 4.6.3 1-D Analysis software (Bio-Rad).
RESULTS
Deletion of the DExD-box RNA helicase Lmo1722 abolishes L. monocytogenes growth at temperatures below 10°C.
To investigate whether DExD-box RNA helicases in L. monocytogenes are important for the overall bacterial physiology, the gene encoding one of them, Lmo1722, was deleted (Fig. 1). There were several reasons for choosing Lmo1722. It shows induced expression on the transcriptional level at low temperatures (11). Also, its B. subtilis orthologue, YfmL, is an RNA helicase that is not well characterized and is probably not part of the RNA degradosome, given that it was not able to interact with RNA degradosome components or other helicases in a bacterial two-hybrid system assay (38). The lmo1722 orthologue cshC in B. cereus was among three RNA helicase genes out of five that were required for cold growth and involved in adaptation to several stress conditions (50, 51). Lmo1722 has a core region similar to those of other DExD-box RNA helicases but also harbors a unique lysine-rich 72-amino-acid-long C terminus that is not present in the B. subtilis orthologue YfmL (see Fig. S1 in the supplemental material) (38, 44). Absence of Lmo1722 did not affect the growth of L. monocytogenes at 37°C (Table 3; see Fig. S2 in the supplemental material). However, the growth of the Δlmo1722 strain was severely impaired compared to the wild-type strain at lower temperatures (Fig. 1B and Table 3). For instance, the growth rate was almost 3-fold lower for the Δlmo1722 strain than for the wild type at 16°C, and the Δlmo1722 strain was unable to grow at 0°C. To rule out possible polar effects, the lmo1722 gene was introduced into plasmid pMK4P harboring a constitutive promoter (Pprot) (2). When the resulting plasmid, plmo1722, was introduced into the Δlmo1722 strain, growth was restored to wild-type levels at 26°C (Table 3). It also allowed the Δlmo1722 strain to grow at temperatures below 10°C (data not shown).
Fig 1.
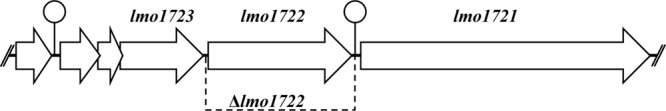
lmo1722 gene locus. The arrows indicate transcriptional directions, and the lollipops indicate terminators. The DNA between the double slashes was deleted in the Δlmo1722 strain.
Table 3.
Growth rates (minutes of doubling) at different temperatures of indicated strains
| Strain | Growth rate (min of doubling) |
||
|---|---|---|---|
| 16°C | 26°C | 37°C | |
| EGDe | 134 | 67 | 35 |
| Δlmo1722 strain | 345 | 80 | 37 |
| EGDe plus pMK4P | 231 | 110 | 56 |
| Δlmo1722 plus pMK4P | 393 | 147 | 61 |
| Δlmo1722 plus plmo1722 | 276 | 112 | 56 |
| EGDe plus pIMK3 | 176 | 69 | 45 |
| Δlmo1722 plus pIMK3 | 592 | 98 | 47 |
| Δlmo1722 plus pIMK3::mutlmo1722WT | 177 | 73 | 46 |
| Δlmo1722 plus pIMK3::lmo1722WT | 160 | 71 | 45 |
| Δlmo1722 plus pIMK3::lmo1722ΔCT | 167 | 70 | 45 |
Absence of Lmo1722 decreases L. monocytogenes motility by downregulating flaA expression.
L. monocytogenes is motile only at temperatures around and below 30°C (39). When plating the WT plus pMK4P, the Δlmo1722 plus pMK4P, and the Δlmo1722 plus plmo1722 strains on low-agar motility plates at 26°C, it was observed that the Δlmo1722 plus pMK4P strain showed reduced motility in comparison with the other strains (Fig. 2A). To analyze the reason for the motility deficiency, whole-cell protein preparations from the wild-type and Δlmo1722 strains grown at 37 and 16°C were separated on one-dimensional (1D) SDS-PAGE. This revealed a protein band more prominent in the wild-type than in the Δlmo1722 strain at 16°C (Fig. 2B). By trypsin digestion-mass spectrometry analysis, the protein was identified as FlaA, the major subunit of Listeria flagellin. To test if the regulation of FlaA expression was exerted on a transcriptional or posttranscriptional level, a Northern blot was performed with RNA from different strains. Compared to the wild type, the flaA mRNA level was decreased in the Δlmo1722 plus pMK4P strain but could be reestablished in the Δlmo1722 plus plmo1722 strain (Fig. 2C). These results are compatible with Lmo1722 affecting the expression of motility genes at the transcriptional level. The regulation of Listeria motility is complex (26, 33, 34). Both the gene encoding the repressor of motility (mogR) and the gene encoding the positive regulator of motility (gmaR) were downregulated in the Δlmo1722 strain compared to the wild-type strain (data not shown), indicating that Lmo1722 functions upstream of these regulators. One regulator stimulating motility and regulating gmaR expression is DegU. Expression of degU was decreased in the Δlmo1722 plus pMK4P strain but could be restored to wild-type levels in the Δlmo1722 plus plmo1722 strain (Fig. 2C). This indicates that Lmo1722 controls motility by regulating degU expression directly or by affecting the expression of regulators upstream of degU.
Fig 2.
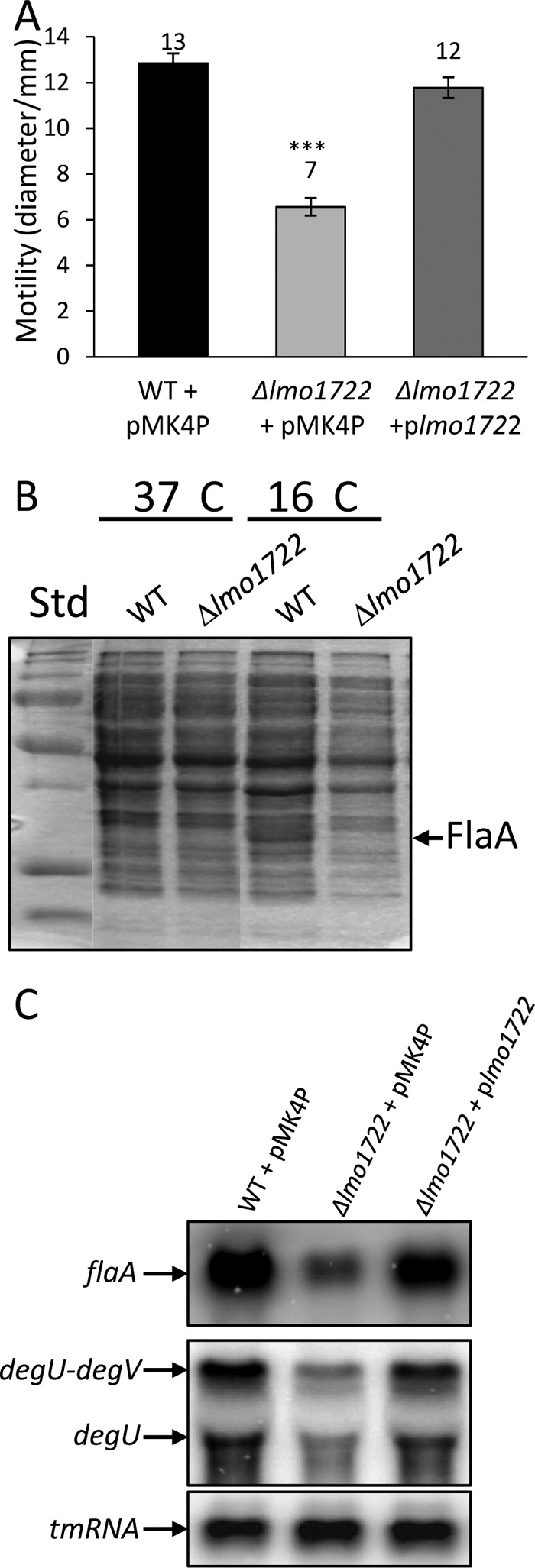
Motility and gene expression in different L. monocytogenes strains. (A) Motility of indicated strains on BHI soft agar plates at 26°C. Motility was scored after 48 h and measured as the diameter of the colonies originating from the center. The error bars indicate standard deviations (***, statistically significant difference [P < 0.001; Student t test] between the WT plus pMK4P and the Δlmo1722 plus pMK4P strains). (B) SDS-PAGE of total proteins from L. monocytogenes and its isogenic lmo1722 deletion strain grown at 37 or 16°C. Std, standard. (C) Northern blot analysis of flagellin flaA and transcriptional regulator degU and degV expression of bacteria grown at 26°C.
Absence of Lmo1722 decreases the number of mature 70S ribosomes at low temperatures.
Several studies in other bacterial species have shown that DEAD box RNA helicases are important for ribosomal maturation (14, 15, 21, 36). To test whether Lmo1722 was important for the formation of mature 70S ribosomes in L. monocytogenes, ribosomal fractions of the WT plus pMK4P, the Δlmo1722 plus pMK4P, and the Δlmo1722 plus plmo1722 strains grown at 16°C were separated on sucrose gradients. The results showed that the Δlmo1722 plus pMK4P strain formed fewer mature 70S ribosomes and instead contained more free 30S and 50S subunits, whereas the Δlmo1722 plus plmo1722 strain only marginally increased the number of mature 70S ribosomes (data not shown). This surprising result prompted us to measure the Lmo1722 levels in different strains. By Western blotting, we observed that the level of Lmo1722 protein was reduced in the Δlmo1722 plus plmo1722 strain compared to the WT plus pMK4P strain (see Fig. S3 in the supplemental material). Why the complemented strain displayed low levels of Lmo1722 protein is unclear, but it could be due to transcript or plasmid instability.
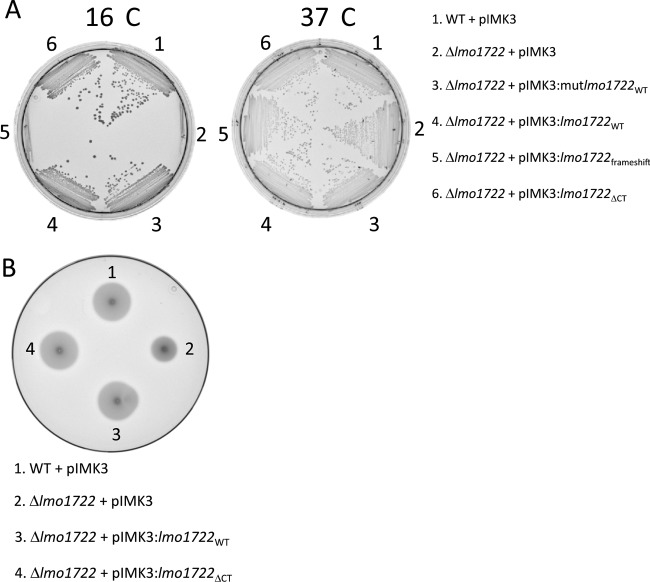
To analyze complementation phenotypes at wild-type Lmo1722 levels, lmo1722 alleles were placed under the control of an IPTG-inducible promoter in the chromosomally integrative pIMK3 vector (47). The resulting constructs were streaked on agar plates with or without 1 mM IPTG and incubated at 16 or 37°C (Fig. 3A and data not shown). Our results suggest that the Δlmo1722 plus pIMK3::lmo1722WT strain grew as fast as the WT plus pIMK3 strain on plates lacking IPTG (Fig. 3A), and the generation times of the strains were similar in bacterial cultures lacking IPTG (Table 3). During the cloning process, we obtained a base deletion in the promoter region of the construct (pIMK3::mutlmo1722WT), decreasing Lmo1722WT levels approximately 2-fold without IPTG induction compared to wild-type levels (see Fig. S3 in the supplemental material). However, the Δlmo1722 plus pIMK3::mutlmo1722WT strain still grew as fast as the WT plus pIMK3 strain at 16°C (Fig. 3A and Table 3). A construct harboring a deletion of 19 bp, creating a premature stop codon within the central core region, was also obtained during cloning. Unlike the other constructs, this strain (Δlmo1722 plus pIMK3::mutlmo1722Frameshift) was unable to complement growth at 16°C even in the presence of IPTG, suggesting that the Lmo1722 protein, but not the lmo1722 DNA or RNA, is sufficient for growth complementation (Fig. 3A and data not shown). In line with the above-mentioned results were strains expressing Lmo1722 (WT plus pIMK3, Δlmo1722 plus pIMK3::lmo1722WT, and Δlmo1722 plus pIMK3::mutlmo1722WT) able to grow at 0 and 5°C (see Fig. S4 in the supplemental material). This was in contrast to the Δlmo1722 plus pIMK3 strain, which was unable to grow at these lower temperatures. Some of the above-mentioned strains were examined for their motility on low-agar plates. Not surprisingly, expression of Lmo1722 reestablished motility to wild-type levels at 26°C (Fig. 3B).
Fig 3.
Analysis of low-temperature growth and motility of the indicated strains. (A) Growth of L. monocytogenes strains carrying different lmo1722 allele constructs on solid BHI plates lacking IPTG at 16°C or 37°C. (B) Motility of the indicated strains on BHI soft agar plates. The plates are shown after 48 h of incubation at 26°C.
Ribosomal maturation in L. monocytogenes does not require the C terminus of Lmo1722.
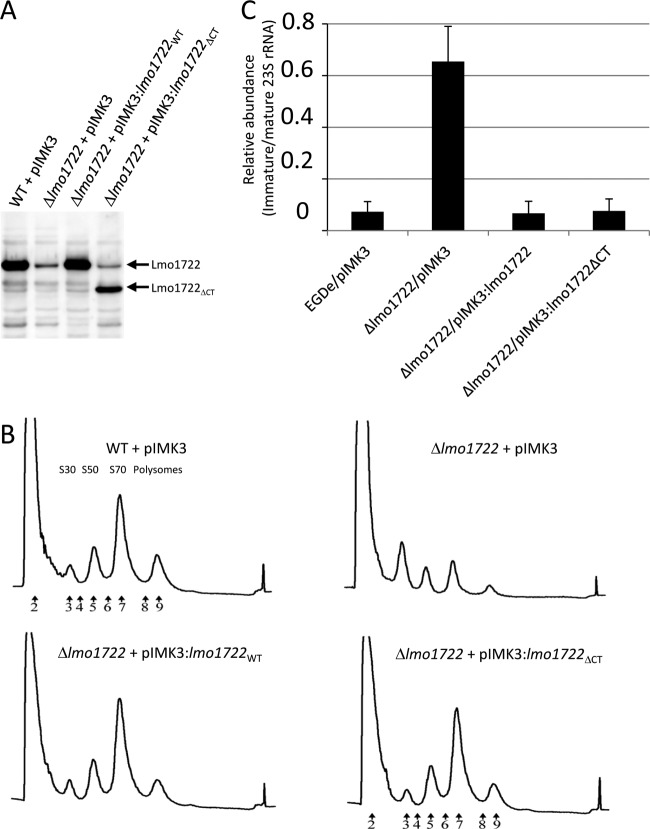
Since the Δlmo1722 strain harboring the pMK4P::lmo1722 construct did not restore ribosomal maturation to the wild-type profile, we were interested to investigate whether the pIMK3::lmo1722WT construct could reestablish the ribosomal profile. First, the expression levels of the Lmo1722 protein in various strains were determined. Our results showed that the expression of Lmo1722 followed the wild-type levels even in the absence of IPTG (Fig. 4A; see Fig. S3 in the supplemental material). We therefore performed experiments without IPTG induction. In contrast to the Δlmo1722 plus pIMK3 strain, a Δlmo1722 strain harboring pIMK3::lmo1722WT could restore ribosome maturation to a profile observed in the wild-type strain (Fig. 4B). A wild-type-like ribosomal maturation profile was also observed for the Δlmo1722 plus pIMK3::mutlmo1722WT strain, although this strain only express half of the wild-type Lmo1722 levels (data not shown). The Lmo1722 orthologue in B. subtilis, YfmL, lacks a major part of the C-terminal extension (see Fig. S1 in the supplemental material). To test if the C terminus of Lmo1722 was required for growth at low temperatures, motility, and ribosomal maturation, a C-terminally deleted form of Lmo1722 (Lmo1722ΔCT) was created. Expression of Lmo1722ΔCT reestablished growth, motility, and ribosome maturation as well as full-length Lmo1722 did (Fig. 3 and 4), indicating that the C-terminal part of Lmo1722 is redundant for the above deviations of the means from three experiments.
Fig 4.
Absence of Lmo1722 alters rRNA processing and ribosomal maturation. (A) Western blot analysis of Lmo1722 expression levels in the indicated strains grown at 16°C. The arrows indicate Lmo1722 and Lmo1722ΔCT. The background signal in the Δlmo1722 plus pIMK3 strain probably reflects expression of Lmo1450, an RNA helicase 32% identical to Lmo1722 and with a similar calculated molecular mass of 50 kDa. (B) Ribosomal profile analysis. Samples from the indicated strains grown at 16°C were prepared and separated by sucrose gradient. The gradients were normalized by the addition of equivalent A260 units, as described in Materials and Methods. The numbered arrows indicate samples used in Fig. 5. The different ribosomal subunits are indicated above the peaks for the wild-type plus pIMK3 profile. (C) Relative processing of immature versus mature 23S rRNA transcripts in the indicated strains grown at 16°C (see the text for details). The error bars indicate standard deviations of the means from three experiments.
Absence of Lmo1722 alters 23S rRNA precursor processing.
A commonly observed phenotype of bacterial strains showing an immature ribosomal profile is the inability to properly process the 23S rRNA precursor transcript. The E. coli RNA helicases SrmB and CsdA affect 23S rRNA maturation, as reviewed in reference 54. To analyze whether a Δlmo1722 strain displayed altered 23S rRNA processing, a primer extension analysis was performed. Our results suggest that Lmo1722 is indeed important for proper 23S rRNA precursor processing, since a strain lacking Lmo1722 showed a large fraction of an unprocessed precursor (Fig. 4C; see Fig. S5 in the supplemental material). This immature 23S rRNA harbors a 160-nucleotide-long 5′ precursor not present in the mature 23S rRNA. The phenotype was restored in the Δlmo1722 plus pIMK3::lmo1722WT strain (Fig. 4C). Interestingly, a C-terminally truncated Lmo1722 could fully restore 23S rRNA processing to a wild-type appearance (Fig. 4C).
Lmo1722 is associated with free 50S subunits and 70S ribosomes.
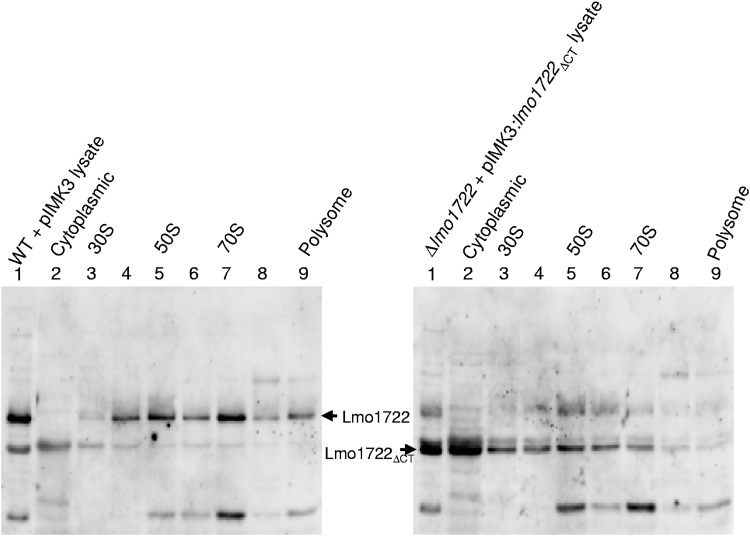
To further investigate the role of Lmo1722 during ribosomal maturation, the association of Lmo1722 with the ribosomal subunits was examined. Sucrose gradient-fractionated extracts from the Lmo1722- or Lmo1722ΔCT-expressing strains were used for Western blotting. The results suggested that Lmo1722 was associated with the 50S subunit, but also with mature 70S ribosomes. Almost no full-length Lmo1722 could be detected in cytoplasmic or 30S fractions (Fig. 5). In contrast, Lmo1722ΔCT was observed in all isolated fractions (also cytoplasmic and 30S fractions), indicating that the C-terminal part guides Lmo1722 to the 50S subunit and 70S ribosomes (Fig. 5).
Fig 5.
Localization of Lmo1722 in different fractions of ribosomal profiles from bacteria grown at 16°C. Equal volumes of fractions with either the highest or the lowest A260 value (indicated in Fig. 4B) were pooled, and the proteins were concentrated by trichloroacetic acid (TCA) precipitation and analyzed by Western blotting. The arrows indicate Lmo1722 and Lmo1722ΔCT.
DISCUSSION
In this work, we show that the L. monocytogenes DExD-box RNA helicase Lmo1722 is essential for growth at low temperatures, a condition L. monocytogenes encounters in nature, but also when present in refrigerated food. In contrast, Lmo1722 is dispensable for growth at 37°C (Fig. 1 and Table 3). DExD-box RNA helicases in bacteria have mostly been studied in E. coli and B. subtilis (16, 30). However, neither of these strains is able to grow at extremely low temperatures like L. monocytogenes. Considering that expression of lmo1722 is induced during cold growth (11), it is not surprising that the presence of Lmo1722 is needed for growth at low temperatures (Fig. 1 and 3 and Table 3; see Fig. S4 in the supplemental material). In support of the above-mentioned results, we observed an increase in Lmo1722 protein levels at 16°C compared to 37°C (see Fig. S3 in the supplemental material). Absence of Lmo1722 decreased L. monocytogenes motility, due to downregulated expression of flaA, encoding the main subunit of the flagellin FlaA (Fig. 2). The exact mechanism by which Lmo1722 controls flagellin expression remains to be elucidated, but it most probably lies somewhere upstream of the most prominent regulators (MogR, GmaR, and DegU).
Absence of Lmo1722 decreases the number of polysomes, mature 70S ribosomes, and 50S subunits but increases the level of 30S subunits. Similar ribosomal profiles have been detected in other bacteria lacking certain RNA helicases (31). However, we did not observe any ribosomal subunit intermediates (i.e., 40S and 45S) for the Δlmo1722 mutant strain, as found in E. coli helicase mutant strains. Whether this is due to differences in the ribosomal maturation patterns between Gram-positive and Gram-negative strains remains to be investigated. A Δlmo1722 mutant strain supplemented with wild-type levels of Lmo1722 reestablished the ribosomal profile to wild-type appearance (Fig. 4B).
An Lmo1722 protein lacking its C-terminal part was not affected in growth or motility, and the ribosomal profile and rRNA processing in that strain overlapped with the profile of a wild-type strain. In this respect, the Lmo1722 helicase is comparable to several other well-characterized bacterial helicases. E. coli SrmB helicase with its 60-amino-acid lysine-rich C-teminus replaced with an affinity tag or deleted was able to completely restore the ribosomal profile of a ΔsrmB deletion mutant (58). Yet another E. coli helicase, CsdA, involved in ribosomal maturation, could not fully complement bacterial growth at 15°C when the C terminus was truncated (3, 60). Here, complementation with physiological levels of Lmo1722ΔCT expression completely restored the analyzed phenotypes. However, in contrast to a wild-type strain in which full-length Lmo1722 protein was detected almost exclusively with 50S subunits and mature ribosomes, C-terminally deleted Lmo1722 could also be found in cytoplasmic and 30S subunit fractions (Fig. 4C). This was not due to overexpression of Lmo1722ΔCT compared to the wild-type expression of Lmo1722 (Fig. 4A). The results suggest that the function of the C-terminal part of Lmo1722 involves appropriate guidance of Lmo1722 to the 50S subunit of the ribosome. The presence of Lmo1722 at polysomes indicates that it might be important for active translation (Fig. 4C). Such a mechanism has been suggested previously to be most important at the initiation step of translation for DEAD box RNA helicases (16, 29).
Supplementary Material
ACKNOWLEDGMENTS
We thank Siv Sääf for assistance in primer extension DNA fragment analysis.
J.J. was supported by Umeå University, Swedish Research Council grants K2011-56X-15144-08-6 and 621-2009-5677, and ERC starting grant no. 260764-RNAntibiotics.
Footnotes
Published ahead of print 15 June 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Arnaud M, Chastanet A, Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70: 6887–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aucher W, et al. 2004. Differences in mesentericin secretion systems from two Leuconostoc strains. FEMS Microbiol. Lett. 232: 15–22 [DOI] [PubMed] [Google Scholar]
- 3. Awano N, et al. 2007. Complementation analysis of the cold-sensitive phenotype of the Escherichia coli csdA deletion strain. J. Bacteriol. 189: 5808–5815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Azizoglu RO, Kathariou S. 2010. Inactivation of a cold-induced putative RNA helicase gene of Listeria monocytogenes is accompanied by failure to grow at low temperatures but does not affect freeze-thaw tolerance. J. Food Prot. 73: 1474–1479 [DOI] [PubMed] [Google Scholar]
- 5. Bensadoun A, Weinstein D. 1976. Assay of proteins in the presence of interfering materials. Anal. Biochem. 70: 241–250 [DOI] [PubMed] [Google Scholar]
- 6. Bethesda Research Laboratories 1986. BRL pUC host: E. coli DH5α competent cells. Focus 8: 9 [Google Scholar]
- 7. Borezee E, Pellegrini E, Berche P. 2000. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect. Immun. 68: 7069–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Broussolle V, et al. 2010. Insertional mutagenesis reveals genes involved in Bacillus cereus ATCC 14579 growth at low temperature. FEMS Microbiol. Lett. 306: 177–183 [DOI] [PubMed] [Google Scholar]
- 9. Burall LS, Laksanalamai P, Datta AR. 2012. Listeria monocytogenes mutants with altered growth phenotypes at refrigeration temperature and high salt concentrations. Appl. Environ. Microbiol. 78: 1265–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cartier G, Lorieux F, Allemand F, Dreyfus M, Bizebard T. 2010. Cold adaptation in DEAD-box proteins. Biochemistry 49: 2636–2646 [DOI] [PubMed] [Google Scholar]
- 11. Chan YC, Raengpradub S, Boor KJ, Wiedmann M. 2007. Microarray-based characterization of the Listeria monocytogenes cold regulon in log- and stationary-phase cells. Appl. Environ. Microbiol. 73: 6484–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chan YC, Wiedmann M. 2009. Physiology and genetics of Listeria monocytogenes survival and growth at cold temperatures. Crit. Rev. Food Sci. Nutr. 49: 237–253 [DOI] [PubMed] [Google Scholar]
- 13. Chandran V, et al. 2007. Recognition and cooperation between the ATP-dependent RNA helicase RhlB and ribonuclease RNase E. J. Mol. Biol. 367: 113–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Charollais J, Dreyfus M, Iost I. 2004. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 32: 2751–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. 2003. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 48: 1253–1265 [DOI] [PubMed] [Google Scholar]
- 16. Cordin O, Banroques J, Tanner NK, Linder P. 2006. The DEAD-box protein family of RNA helicases. Gene 367: 17–37 [DOI] [PubMed] [Google Scholar]
- 17. Drevets DA, Bronze MS. 2008. Listeria monocytogenes: epidemiology, human disease, and mechanisms of brain invasion. FEMS Immunol. Med. Microbiol. 53: 151–165 [DOI] [PubMed] [Google Scholar]
- 18. Flamm RK, Hinrichs DJ, Thomashow MF. 1984. Introduction of pAM beta 1 into Listeria monocytogenes by conjugation and homology between native L. monocytogenes plasmids. Infect. Immun. 44: 157–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fliss I, Emond E, Simard RE, Pandian S. 1991. A rapid and efficient method of lysis of Listeria and other gram-positive bacteria using mutanolysin. Biotechniques 11: 453, 456–457. [PubMed] [Google Scholar]
- 20. Fraser KR, Harvie D, Coote PJ, O'Byrne CP. 2000. Identification and characterization of an ATP binding cassette L-carnitine transporter in Listeria monocytogenes. Appl. Environ. Microbiol. 66: 4696–4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fuller-Pace FV, Nicol SM, Reid AD, Lane DP. 1993. DbpA: a DEAD box protein specifically activated by 23s rRNA. EMBO J. 12: 3619–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gegenheimer P. 1990. Preparation of extracts from plants. Methods Enzymol. 182: 174–193 [DOI] [PubMed] [Google Scholar]
- 23. Gerhardt PN, Tombras Smith L, Smith GM. 2000. Osmotic and chill activation of glycine betaine porter II in Listeria monocytogenes membrane vesicles. J. Bacteriol. 182: 2544–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Glaser P, et al. 2001. Comparative genomics of Listeria species. Science 294: 849–852 [DOI] [PubMed] [Google Scholar]
- 25. Gould H, Herbert BN, Loviny T. 1969. Polysomes from Bacillus subtilis and Bacillus thuringiensis. Nature 223: 855–857 [DOI] [PubMed] [Google Scholar]
- 26. Gueriri I, et al. 2008. The DegU orphan response regulator of Listeria monocytogenes autorepresses its own synthesis and is required for bacterial motility, virulence and biofilm formation. Microbiology 154: 2251–2264 [DOI] [PubMed] [Google Scholar]
- 27. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177: 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hebraud M, Guzzo J. 2000. The main cold shock protein of Listeria monocytogenes belongs to the family of ferritin-like proteins. FEMS Microbiol. Lett. 190: 29–34 [DOI] [PubMed] [Google Scholar]
- 29. Hunger K, Beckering CL, Wiegeshoff F, Graumann PL, Marahiel MA. 2006. Cold-induced putative DEAD box RNA helicases CshA and CshB are essential for cold adaptation and interact with cold shock protein B in Bacillus subtilis. J. Bacteriol. 188: 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iost I, Dreyfus M. 2006. DEAD-box RNA helicases in Escherichia coli. Nucleic Acids Res. 34: 4189–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jagessar KL, Jain C. 2010. Functional and molecular analysis of Escherichia coli strains lacking multiple DEAD-box helicases. RNA 16: 1386–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Junttila JR, Niemela SI, Hirn J. 1988. Minimum growth temperatures of Listeria monocytogenes and non-haemolytic Listeria. J. Appl. Bacteriol. 65: 321–327 [DOI] [PubMed] [Google Scholar]
- 33. Kamp HD, Higgins DE. 2011. A protein thermometer controls temperature-dependent transcription of flagellar motility genes in Listeria monocytogenes. PLoS Pathog. 7: e1002153 doi:10.1371/journal.ppat.1002153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kamp HD, Higgins DE. 2009. Transcriptional and post-transcriptional regulation of the GmaR antirepressor governs temperature-dependent control of flagellar motility in Listeria monocytogenes. Mol. Microbiol. 74: 421–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Khemici V, Toesca I, Poljak L, Vanzo NF, Carpousis AJ. 2004. The RNase E of Escherichia coli has at least two binding sites for DEAD-box RNA helicases: functional replacement of RhlB by RhlE. Mol. Microbiol. 54: 1422–1430 [DOI] [PubMed] [Google Scholar]
- 36. Kossen K, Uhlenbeck OC. 1999. Cloning and biochemical characterization of Bacillus subtilis YxiN, a DEAD protein specifically activated by 23S rRNA: delineation of a novel sub-family of bacterial DEAD proteins. Nucleic Acids Res. 27: 3811–3820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- 38. Lehnik-Habrink M, et al. 2010. The RNA degradosome in Bacillus subtilis: identification of CshA as the major RNA helicase in the multiprotein complex. Mol. Microbiol. 77: 958–971 [DOI] [PubMed] [Google Scholar]
- 39. Leifson E, Palen MI. 1955. Variations and spontaneous mutations in the genus Listeria in respect to flagellation and motility. J. Bacteriol. 70: 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lim J, Thomas T, Cavicchioli R. 2000. Low temperature regulated DEAD-box RNA helicase from the Antarctic archaeon, Methanococcoides burtonii. J. Mol. Biol. 297: 553–567 [DOI] [PubMed] [Google Scholar]
- 41. Liu S, Bayles DO, Mason TM, Wilkinson BJ. 2006. A cold-sensitive Listeria monocytogenes mutant has a transposon insertion in a gene encoding a putative membrane protein and shows altered (p)ppGpp levels. Appl. Environ. Microbiol. 72: 3955–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu S, Graham JE, Bigelow L, Morse PD, 2nd, Wilkinson BJ. 2002. Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl. Environ. Microbiol. 68: 1697–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lloyd AL, Marshall BJ, Mee BJ. 2005. Identifying cloned Helicobacter pylori promoters by primer extension using a FAM-labelled primer and GeneScan analysis. J. Microbiol. Methods 60: 291–298 [DOI] [PubMed] [Google Scholar]
- 44. Lopez-Ramirez V, Alcaraz LD, Moreno-Hagelsieb G, Olmedo-Alvarez G. 2011. Phylogenetic distribution and evolutionary history of bacterial DEAD-Box proteins. J. Mol. Evol. 72: 413–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mattila M, Lindstrom M, Somervuo P, Markkula A, Korkeala H. 2011. Role of flhA and motA in growth of Listeria monocytogenes at low temperatures. Int. J. Food Microbiol. 148: 177–183 [DOI] [PubMed] [Google Scholar]
- 46. Michel E, Stephan R, Tasara T. 2011. The lmo0501 gene coding for a putative transcription activator protein in Listeria monocytogenes promotes growth under cold, osmotic and acid stress conditions. Food Microbiol. 28: 1261–1265 [DOI] [PubMed] [Google Scholar]
- 47. Monk IR, Gahan CG, Hill C. 2008. Tools for functional postgenomic analysis of Listeria monocytogenes. Appl. Environ. Microbiol. 74: 3921–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Oliver HF, Wiedmann M, Boor KJ. 2007. Environmental reservoir and transmission into the mammalian host, p 111–137 in Goldfine H, Shen H. (ed), Listeria monocytogenes: pathogenesis and host response. Springer Science andBusiness Media, LLC, New York, NY [Google Scholar]
- 49. Palonen E, et al. 2012. Requirement for RNA helicase CsdA for growth of Yersinia pseudotuberculosis IP32953 at low temperatures. Appl. Environ. Microbiol. 78: 1298–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pandiani F, et al. 2010. Differential involvement of the five RNA helicases in adaptation of Bacillus cereus ATCC 14579 to low growth temperatures. Appl. Environ. Microbiol. 76: 6692–6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pandiani F, Chamot S, Brillard J, Carlin F, Nguyen-The C, Broussolle V. 2011. Role of the five RNA helicases in the adaptive response of Bacillus cereus ATCC 14579 cells to temperature, pH, and oxidative stresses. Appl. Environ. Microbiol. 77: 5604–5609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Py B, Higgins CF, Krisch HM, Carpousis AJ. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381: 169–172 [DOI] [PubMed] [Google Scholar]
- 53. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 54. Shajani Z, Sykes MT, Williamson JR. 2011. Assembly of bacterial ribosomes. Annu. Rev. Biochem. 80: 501–526 [DOI] [PubMed] [Google Scholar]
- 55. Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotech. 1: 784–791 [Google Scholar]
- 56. Toledo-Arana A, et al. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459: 950–956 [DOI] [PubMed] [Google Scholar]
- 57. Towbin H, Staehelin T, Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Trubetskoy D, Proux F, Allemand F, Dreyfus M, Iost I. 2009. SrmB, a DEAD-box helicase involved in Escherichia coli ribosome assembly, is specifically targeted to 23S rRNA in vivo. Nucleic Acids Res. 37: 6540–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tsu CA, Uhlenbeck OC. 1998. Kinetic analysis of the RNA-dependent adenosinetriphosphatase activity of DbpA, an Escherichia coli DEAD protein specific for 23S ribosomal RNA. Biochemistry 37: 16989–16996 [DOI] [PubMed] [Google Scholar]
- 60. Turner AM, Love CF, Alexander RW, Jones PG. 2007. Mutational analysis of the Escherichia coli DEAD box protein CsdA. J. Bacteriol. 189: 2769–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Udekwu KI, et al. 2005. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 19: 2355–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.