Abstract
Virulence-related type III secretion systems are present in many Gram-negative bacterial pathogens. These complex devices translocate proteins, called effectors, from the bacterium into the eukaryotic host cell. Here, we identify the product of srfJ, a Salmonella enterica serovar Typhimurium gene regulated by SsrB, as a new substrate of the type III secretion system encoded by Salmonella pathogenicity island 2. The N-terminal 20-amino-acid segment of SrfJ was recognized as a functional secretion and translocation signal specific for this system. Transcription of srfJ was positively regulated by the PhoP/PhoQ system in an SsrB-dependent manner and was negatively regulated by the Rcs system in an SsrB-independent manner. A screen for regulators of an srfJ-lacZ transcriptional fusion using the T-POP transposon identified IolR, the regulator of genes involved in myo-inositol utilization, as an srfJ repressor. Our results suggest that SrfJ is synthesized both inside the host, in response to intracellular conditions, and outside the host, in myo-inositol-rich environments.
INTRODUCTION
Type III secretion systems (T3SS) are complex devices that are present in many Gram-negative bacteria, including the animal pathogens Yersinia spp., enteropathogenic Escherichia coli, Pseudomonas aeruginosa, Shigella flexneri, Chlamydia spp., and Salmonella enterica, and the plant pathogens or symbionts Pseudomonas syringae, Ralstonia solanacearum, Erwinia spp., Rhizobium spp., and Xanthomonas campestris (29). T3SS allows secretion and translocation of effector proteins from bacteria to eukaryotic cells. The injected proteins are often able to interfere with host signal transduction pathways to enable bacterial invasion and survival in subcellular niches.
Salmonella enterica is a facultative intracellular bacterium responsible for gastroenteritis and systemic infections in many animals, including humans (45). S. enterica virulence depends on two distinct T3SS, T3SS1 and T3SS2. T3SS1 translocates proteins through the plasma membrane of the host cell and is necessary for invasion, whereas T3SS2 is induced intracellularly, injects effector through the membrane of the Salmonella-containing vacuole (SCV), and is essential for survival and proliferation inside host cells. Both systems, however, depend on each other for efficient functioning (1). The genes encoding the structural components, many effectors, and some regulators of T3SS1 and T3SS2 are located in two Salmonella pathogenicity islands, SPI1 and SPI2, respectively (16, 17, 21, 41, 44, 54). Some effector proteins are encoded by genes outside SPI1 and SPI2.
SsrA (also named SpiR) and SsrB are the sensor and the response regulator, respectively, of a two-component regulatory system encoded by SPI2 that is necessary for the expression of T3SS2 (9, 28, 44). A genetic screen identified 12 genes (named srfA to srfM, for SsrB-regulated factors) outside SPI2 that were regulated by SsrB (62). These genes were located in horizontally acquired regions and, because of their pattern of expression, were suggested as candidates to encode T3SS2 effectors (61, 62). However, to date, the product of only one of these genes, srfH (also named sseI), has been demonstrated to be translocated into host cells through T3SS2 (39). One of the most interesting genes identified in this screen was srfJ. An srfJ null mutant has a slight virulence defect (49). More interestingly, the product of this gene, SrfJ, is similar to human lysosomal glucosylceramidase, an enzyme involved in regulating metabolism of ceramide, one of the most intensely studied classes of sphingolipids (4, 30).
The potential of srfJ prompted us to investigate in detail its pattern of expression and the possibility of secretion of its product, SrfJ, through a T3SS. Here, we show that SrfJ can be secreted in vitro and that the amino-terminal region of this protein directs translocation into macrophages specifically through T3SS2. srfJ is regulated not only by SsrB but also by PhoP, RcsB, and, surprisingly, by IolR, the main regulator of genes involved in myo-inositol utilization (33).
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and strain construction.
E. coli and S. enterica serovar Typhimurium strains used in this study are described in Table 1. Salmonella strains derive from the mouse-virulent strain ATCC 14028. Transductional crosses using phage P22 HT 105/1 int201 (53) were used for strain construction (37). To obtain phage-free isolates, transductants were purified by streaking on green plates (6). Phage sensitivity was tested by cross-streaking with the clear-plaque mutant P22 H5.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | supE44 ΔlacU169 (ϕ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 26 |
| TP610 | F− thi-1 thr-1 leuB6 lacY1 tonA21 supE44 hsdR hsdM recBC lop-11 lig+cya-610 | 27 |
| S. enterica serovar Typhimurium strains | ||
| 14028 | Wild type | ATCC |
| 55130 | 14028 pho-24 (PhoP constitutive) | E. A. Groisman |
| SV4699 | 14028 phoP7953::Tn10 | Laboratory stock |
| SV4758 | 14028 rcsC55 | 19 |
| SV5049 | 14028 ΔrcsB::Cmr | Laboratory stock |
| SV5093 | 14028 ΔrcsA::Cmr | 18 |
| SV5136 | 14028 ssaV::Cmr | Laboratory stock |
| SV5373 | 14028 ΔhilA | Laboratory stock |
| SV5379 | 14028 ΔprgH | Laboratory stock |
| SV5452 | 14028 ΔssrB::Cmr | 18 |
| SV5470 | 14028 ssaV::lacZ | 18 |
| SV5599a | 14028 srfJ::3×FLAG Kmr | This study |
| SV5600a | 14028 srfJ::lacZ | This study |
| SV5604 | 14028 ΔprgH ssaV::Cmr | Laboratory stock |
| SV6017 | 14028 ΔSPI2::Cmr | Laboratory stock |
| SV6055 | 14028 ΔSPI1::Kmr | Laboratory stock |
| SV6891 | 14028 ΔiolR::Cmr | This study |
| Plasmids | ||
| pBAD18 | bla PBAD promoter | 25 |
| pCE36 | aph FRT lacZY+ this oriR6K | 15 |
| pCP20 | bla cat cI857 λPR flp pSC101 oriTS | 7 |
| pKD3 | bla FRT cat FRT PS1 PS2 oriR6K | 11 |
| pKD4 | bla FRT aph FRT PS1 PS2 oriR6K | 11 |
| pKD46 | bla PBAD gam bet exo pSC101 oriTS | 11 |
| pIZ1673 | pSIF003-R1 ΔlacI | 5 |
| pIZ1843 | pIZ1673-SrfJ | This study |
| pIZ1844 | pIZ1673-SrfJ(1-42) | This study |
| pIZ1856 | pIZ1673-SrfJ(1-20) | This study |
| pIZ1897 | pIZ1673-SrfJ(1-10) | This study |
| pIZ1898 | pIZ1673-SrfJ(10-42) | This study |
| pIZ1899 | pIZ1673-SrfJ(20-42) | This study |
| pIZ1973 | pBAD18-iolR+ | This study |
| pIZ1974 | pBAD18-phoP+ | This study |
Derivatives of these strains were used as indicated in the text.
Bacterial culture.
The standard culture medium for S. enterica and E. coli was Luria-Bertani broth (LB). Solid LB contained 1.5% agar (final concentration). Antibiotics were used at the following concentrations: kanamycin (Km), 50 μg ml−1; chloramphenicol (Cm), 20 μg ml−1; ampicillin (Ap), 100 μg ml−1; and tetracycline (Tc), 20 μg ml−1. Subinhibitory concentrations of polymyxin B sulfate (Sigma) used in some experiments were 1 μg ml−1 in LB and 5 μg ml−1 in LPM. For some experiments, 40 μg ml−1 5-bromo-4-chloro-indolyl-β-d-galactopyranoside (X-Gal) or 55.5 mM myo-inositol was added. For SPI1-inducing conditions, Salmonella strains were grown overnight at 37°C in LB–0.3 M NaCl medium in static conditions. For SPI2-inducing conditions, cells from cultures in LB were washed and diluted 1:125 with minimal medium at pH 5.8 (LPM) containing 80 mM 2-(N-morpholino)ethanesulfonic acid (pH 5.8), 5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 0.1% Casamino Acids, 38 mM glycerol, 337.5 μM K2HPO4-KH2PO4 (pH 7.4), and 8 μM MgCl2, and then incubated overnight at 37°C with shaking. For some experiments, the concentration of MgCl2 or the pH of the medium was modified as indicated. Arabinose was used at a final concentration of 0.2%.
DNA amplification with PCR.
Amplification reactions were carried out in a Perkin Elmer Gene-Amp PCR system 2400 (Perkin Elmer Cetus). The final volume of reactions was 50 μl, and the final concentration of MgCl2 was 1.5 mM. Reagents were used at the following concentrations: deoxynucleoside triphosphates (dNTPs), 300 μM; primers, 0.3 μM; and Taq polymerase (KAPA HiFi DNA Polymerase; Kapa Biosystems), 1 U per reaction. The thermal program included the following steps: (i) initial denaturation for 5 min at 95°C; (ii) 25 cycles of denaturation (98°C, 20 s), annealing (57°C, 15 s), and extension (72°C, 30 s); and (iii) final incubation at 72°C for 5 min to complete the extension. Primers are listed in Table 2. PCR constructs were sequenced with an automated DNA sequencer (Stab Vida, Oeiras, Portugal) to confirm that the sequence was correct.
Table 2.
Oligonucleotides used in this study
| Oligonucleotide and use | Sequence (5′–3′) |
|---|---|
| Insertion in srfJ | |
| srfJP1 | CAGATCGACTCCTGCCGCCATAGCAACGTACTGGCGCCTGGTGTAGGCTGGAGCTGCTTC |
| srfJP2 | ATGAAAGGCAGACTCATCTCTTCCGATCCGTATCGTCAGCCATATGAATATCCTCCTTAG |
| Insertion in iolR | |
| iolRP1 | TGGTTATTTACGAAATTTTCGTTCTATTAGAGTATCATGCGTGTAGGCTGGAGCTGCTTC |
| iolRP2 | AATGCCGATGATCGCTAAATACGATCATCGGCTTGTTTTTCATATGAATATCCTCCTTAG |
| Epitope tagging of srfJ | |
| srfJP1flag | GTCAGGCGCCAGTACGTTGCTATGGCGGCAGGAGTCGATCGACTACAAAGACCATGACGG |
| srfJP2flag | TTCGCATACAGGTCGCTTCATTAAATCCCAGCTTCATCATCATATGAATATCCTCCTTAG |
| Construction of pIZ1843 | |
| srfJbampsif5′ | GAATGGATCCAGGAGGTTCCCTATGAAAGGCAGACTCATCTC |
| srfJbampsif3′ | CAAGGGATCCAGATCGACTCCTGCCGCCATAG |
| Construction of pIZ1844 | |
| srfJbampsif5′ | |
| srfJ42bampsif3′ | CAAGGGATCCGTAACGCGTGGCGCGGTAAGAC |
| Construction of pIZ1856 | |
| srfJbampsif5′ | |
| srfJ20bampsif3′ | CAAGGGATCCCCGCACGCTCAACAAGGAATTG |
| Construction of pIZ1897 | |
| srfJbampsif5′ | |
| srfJ10bampsif3′ | CAAGGGATCCACGGATCGGAAGAGATGAGTC |
| Construction of pIZ1898 | |
| srfJ10bampsif5′ | GAATGGATCCAGGAGGTTCCCTATGCCGTATCGTCAGCAATTCCTTG |
| srfJ42bampsif3′ | |
| Construction of pIZ1899 | |
| srfJ20bampsif5′ | GAATGGATCCAGGAGGTTCCCTATGGCGGTCTCTTTTTCGCATCG |
| srfJ42bampsif3′ | |
| Construction of pIZ1973 | |
| iolReco5′ | CTAGGAATTCAGGAGGTCATGCATGTCTAAACATCAAAC |
| iolRhind3′ | CATGAAGCTTTTACTCCGTCGCCAGCGCCAG |
| Construction of pIZ1974 | |
| phoPeco5′ | CTAGGAATTCAGGAGGAAGAGATGATGCGCGTACTG |
| phoPhind3′ | CATGAAGCTTTTAGCGCAATTCAAAAAGATATC |
| ST-PCR | |
| tpop1 | GCCTTCTTATTCGGCCTTGAATTGATCATATGCGG |
| tpop2 | CTTTTTCCGTGATGGTAACC |
| st1 | GGCCACGCGTCGACTAGTAC |
| stACGCC | GGCCACGCGTCGACTAGTACNNNNNNNNNNACGCC |
| stGATAT | GGCCACGCGTCGACTAGTACNNNNNNNNNNGATAT |
Plasmids.
Plasmids used in this study are listed in Table 1. Plasmids expressing CyaA′ fusions were derivatives of pIZ1673, a modification of pSIF003-R1 (58) (a gift from I. Rosenshine, The Hebrew University of Jerusalem) that was constructed by deletion of the lacI gene using enzymes HpaI and SphI. Therefore, expression of CyaA fusions from derivatives of pIZ1673 is no longer dependent on isopropyl-β-d-thiogalactopyranoside (IPTG). To construct these plasmids, DNA from strain 14028 was used as a template for PCR amplification with the primers listed in Table 2. The amplified fragments were digested with BamHI and ligated with BamHI-digested and dephosphorylated pIZ1673. To detect successful constructs (plasmid with insert in the correct orientation), the ligation mixture was transformed into E. coli TP610 and transformants were selected in MacConkey agar supplemented with Ap and 1% maltose. TP610 is a cya mutant strain (22) that can be complemented with the basal adenylate cyclase activity that possesses CyaA from Bordetella pertussis in the absence of calmodulin. Transformants with empty plasmids or with inserts in the wrong orientation give white colonies in MacConkey-maltose, whereas plasmids with inserts in the correct orientation give red colonies in the same medium. Derivatives of plasmid pBAD18 were used for complementation studies. To construct pIZ1973 and pIZ1974, iolR and phoP, respectively, were PCR amplified with the primers indicated in Table 2, which introduce EcoRI and HindIII sites. Digestions with these enzymes were used for oriented cloning in pBAD18.
Mammalian cell culture.
RAW264.7 cells (murine macrophages; ECACC no. 91062702) were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum; 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin were included in the culture media. Cells were maintained in a 5% CO2 humidified atmosphere at 37°C.
Construction of mutants and of a chromosomal lacZ fusion.
Disruption and replacement of srfJ with a Km resistance gene and of iolR with a Cm resistance gene were performed as described previously (11). Briefly, the Km resistance gene from plasmid pKD4 or the Cm resistance gene from plasmid pKD3 was PCR amplified with primers srfJP1 and srfJP2 or with primers iolRP1 and iolRP2, respectively. The sequences of the primers used are shown in Table 2. The PCR products were used to transform the wild-type strain carrying the Red recombinase expression plasmid pKD46. The antibiotic resistance cassette introduced by the gene-targeting procedure was eliminated by recombination using the FLP helper plasmid pCP20 (11). The FRT site (FLP recombinase recognition target) generated by excision of the antibiotic resistance cassette in srfJ was used to integrate plasmid pCE36 to generate a transcriptional lacZ fusion (15).
Chromosomal gene epitope tagging.
Addition of a DNA fragment encoding the 3× FLAG epitope tag at the 3′ end of srfJ (designated SrfJ-3×FLAG) was carried out as described previously (57) using primers srfJP1flag and srfJP2flag.
β-Galactosidase assays.
Levels of β-galactosidase activity were assayed for stationary-phase cultures in LB or in minimal medium as described previously (40) using the CHCl3/SDS permeabilization procedure.
Western blotting and antibodies.
Salmonella strains were grown under different conditions. Usually, cultures in LB medium were diluted and grown in different media. The bacteria were then pelleted by centrifugation and resuspended in SDS-PAGE sample buffer. Proteins from the same numbers of bacteria were separated by gradient SDS-PAGE (Mini-Protean TGX precast gels; 4 to 15%; Bio-Rad) and electrophoretically transferred to nitrocellulose filters for Western blot analysis using anti-Flag M2 monoclonal antibodies (1:5,000; Sigma), anti-GroEL polyclonal antibodies (1:20,000; Sigma), or anti-DnaK (8E2/2) monoclonal antibodies (1:5,000; Assay Designs). Goat anti-mouse horseradish peroxidase (HRP)-conjugated antibodies (1:5,000; Bio-Rad) and goat anti-rabbit HRP-conjugated antibodies (1:10,000; GE Healthcare) were used as secondary antibodies.
Protein secretion analysis.
Salmonella strain SV5599 (14028 expressing SrfJ-3×FLAG) and prgH or ssaV derivatives were grown overnight in LPM at pH 5.8, followed by 1 h of incubation at pH 7.2 or overnight incubation in LB supplemented with 55.5 mM myo-inositol. Whole cells and culture supernatants were separated by centrifugation at 13,000 × g for 10 min. The supernatants were filtered (0.22-μm pore size) and incubated for 30 min on ice with 0.02% deoxycholate. Proteins were then precipitated by adding trichloroacetic acid at a final concentration of 10% (vol/vol), followed by incubation at −20°C for 5 min, placement on ice for 15 min, and then centrifugation (13,000 × g, 4°C, 15 min). The supernatant was discarded and the pellet was incubated with 300 μl of cold acetone for 15 min on ice. After a new centrifugation step, the pellet and the whole cells were processed for electrophoresis and Western blotting.
Bacterial infection of cultured cells.
RAW 264.7 cells were plated in 24-well plates at 1.5 × 105 cells per well and incubated for 24 h at 37°C with 5% CO2. Bacteria were grown in LB medium for 24 h at 37°C with shaking and were added at a multiplicity of infection (MOI) of 250. Bacteria were centrifuged onto the cell monolayer at 200 × g for 5 min and then incubated at 37°C with 5% CO2. The cell culture was washed twice with phosphate-buffered saline (PBS) 1 h postinfection, overlaid with DMEM containing 100 μg ml−1 gentamicin, and incubated for another hour. The culture was then washed twice with PBS, covered with DMEM with gentamicin (16 μg ml−1), and incubated for 14 h.
Protein translocation assays.
Following the infections described above, the translocation of SrfJ-CyaA fusions into the eukaryotic cells was monitored by measuring the levels of cyclic AMP (cAMP). The infected cells were lysed and the levels of cAMP in the lysates were determined using a colorimetric direct cAMP enzyme immunoassay kit (Arbor Assays) according to the manufacturer's instructions.
Mutagenesis with T-POP.
Strain 14028 of S. enterica serovar Typhimurium was mutagenized with T-POP, a derivative of Tn10dTc (48). Pools of 5,000 colonies, each carrying an independent T-POP insertion, were then prepared and lysed with phage P22 HT. The lysates were used to transduce strain SV5600 (14028 srfJ::lacZ), with selection of Tc-resistant transductants on LB plates supplemented with X-Gal, Tc, and Km.
Molecular characterization of T-POP insertions.
A semirandom, two-step PCR protocol, ST-PCR (8), was used to amplify genomic regions adjacent to the T-POP insertions. The first reaction was carried out on bacterial colonies with primers tpop1 and stACGCC or stGATAT (Table 2) in a final volume of 25 μl using a thermal program with the following steps: (i) initial denaturation for 2 min at 94°C; (ii) 6 cycles of denaturation (94°C, 30 s), annealing (42°C, 30 s, −1°C for each cycle), and extension (72°C, 3 min); (iii) 25 cycles of denaturation (94°C, 30 s), annealing (65°C, 30 s), and extension (72°C, 3 min); and (iv) final incubation at 72°C for 7 min to complete extension. The second reaction was carried out with primers tpop2 and st1 using 1 μl of a 5-fold dilution of the product of the first reaction. The thermal program for the second reaction was (i) initial denaturation for 30 s at 94°C; (ii) 30 cycles of denaturation (94°C, 30 s), annealing (56°C, 30 s), and extension (72°C, 2 min); and (iii) final incubation at 72°C for 7 min to complete extension. The final product was sequenced using primers tpop2 and st1. Sequence analysis was performed with molecular biology algorithms from the National Center for Biotechnology Information at www.ncbi.nlm.nih.gov.
RESULTS
srfJ is expressed under SPI2-inducing conditions.
In the course of an infection, S. enterica receives different environmental cues that lead to expression of T3SS1 or T3SS2 and translocation into the host cells of their cognate effectors. Different host environments can be partially mimicked in vitro using different culture conditions: overnight incubation with low aeration in LB medium supplemented with 0.3 M NaCl favors expression of T3SS1, and culture in LPM medium at pH 5.8 with good aeration induces expression of T3SS2. A lacZ transcriptional fusion was used to study the pattern of expression of srfJ. Data shown in Fig. 1A indicated that srfJ was transcribed specifically under T3SS2-inducing conditions. Increases in pH or Mg2+ concentration dramatically reduced transcription of srfJ, indicating that these were relevant factors in the regulation of the expression of this gene. The results at the transcriptional level were confirmed at the protein level using an SrfJ-3×FLAG fusion and immunoblotting with an anti-FLAG antibody (Fig. 1B).
Fig 1.
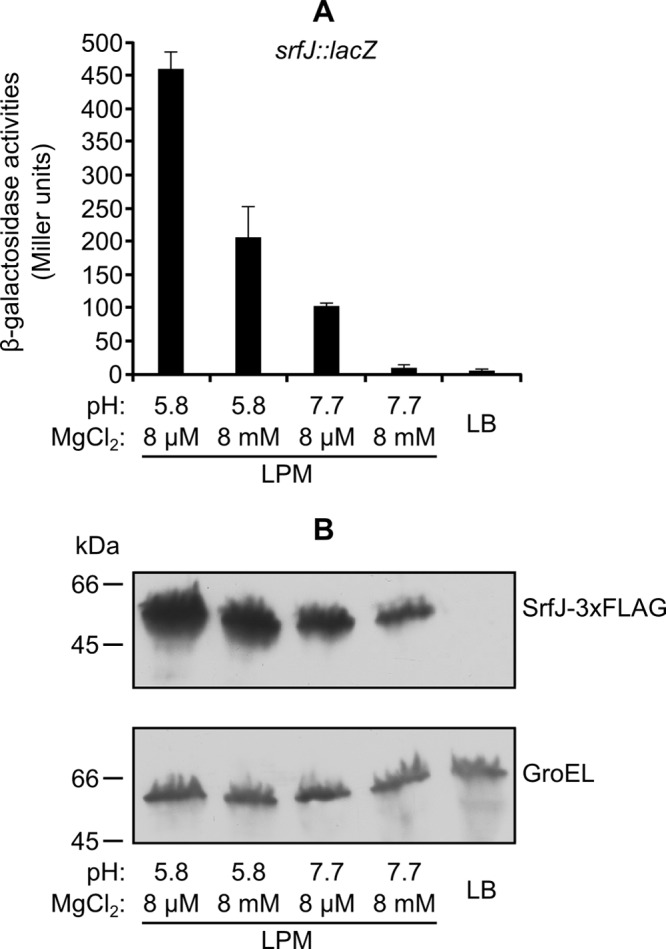
Expression of srfJ in different media. Bacteria were incubated overnight at 37°C with shaking in LPM or without shaking in LB with 0.3 M NaCl (LB). Different pH and Mg2+ concentrations were used as indicated. (A) β-Galactosidase activities were measured from cultures of an S. enterica strain carrying a chromosomal srfJ::lacZ transcriptional fusion. Means and standard deviations from duplicate experiments are represented. (B) Expression at the protein level was studied by Western blotting using a strain expressing SrfJ-3×FLAG. Extracts from this strain were resolved by SDS-PAGE (4 to 15% gradient), and a monoclonal anti-FLAG antibody was used for immunoblotting (upper panel). Polyclonal anti-GroEL antibody was used to get a loading control (lower panel).
SrfJ is secreted into the culture media under SPI2-inducing conditions.
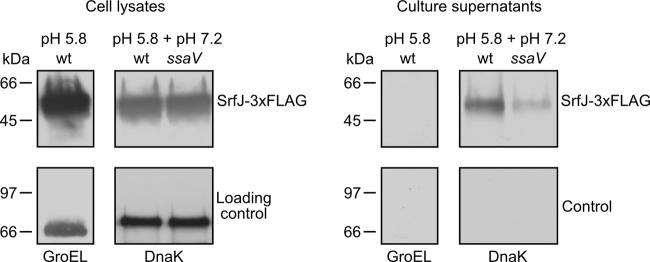
Although coexpression of srfJ with T3SS2 (shown in the previous section) and regulation by SsrB (62) are consistent with a role of SrfJ as a substrate of T3SS2, secretion of this protein had not been previously shown. A derivative of strain 14028 of S. enterica serovar Typhimurium expressing a SrfJ-3×FLAG fusion was used to test secretion into the culture medium. Initial attempts to detect the protein in the supernatants of overnight cultures in LPM (pH 5.8) were unsuccessful. Secretion of many T3SS2 effectors into the culture medium can be triggered by shifting the pH of the culture medium from 5.8 to 7.2 during the last hour of incubation (65). Indeed, these conditions allowed detection of SrfJ in the culture supernatant. DnaK, a cytosolic protein used as a control, was not detected, as expected (Fig. 2). Interestingly, secretion of SrfJ was significantly reduced in an ssaV mutant (SsaV is an essential component of T3SS2), suggesting that secretion of SrfJ was, at least partially, dependent on T3SS2 under these in vitro conditions.
Fig 2.
SrfJ is secreted into the culture medium through T3SS2. A derivative of strain 14028 of S. enterica serovar Typhimurium expressing SrfJ-3×FLAG either was grown overnight in LPM at pH 5.8 or was grown overnight in LPM at pH 5.8, the pH shifted to 7.2, and the incubation continued for an additional hour. After centrifugation, the supernatants were filtered and precipitated. The supernatants and aliquots of the pellets (cell lysates) were submitted to SDS-PAGE and immunoblotting with anti-FLAG antibodies (upper panels) and with anti-GroEL or anti-DnaK antibodies (lower panels). The same experiment was carried out in an ssaV mutant (lacking T3SS2), as indicated. wt, wild type.
The N-terminal portion of SrfJ directs translocation into macrophages through T3SS2.
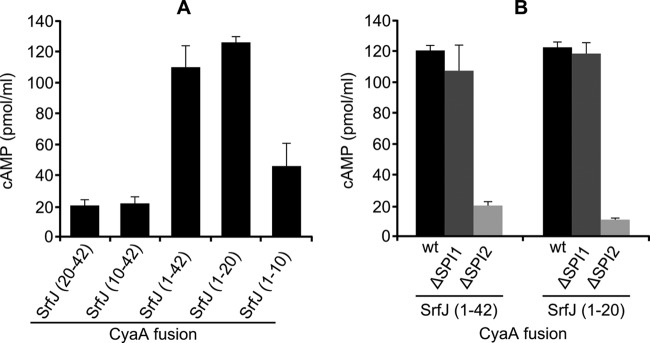
We decided to test the translocation of SrfJ into host cells. To this end, we used a sensitive method based on the generation of fusions with the catalytic domain of CyaA from Bordetella pertussis, a calmodulin-dependent adenylate cyclase. Calmodulin is not produced in bacteria but is present in eukaryotic host cells. After infection of eukaryotic cell cultures with bacteria expressing a CyaA′ fusion, translocation is detected as an increase in the concentration of cAMP, whose production is catalyzed by adenylate cyclase. Analysis of the sequence of SrfJ, a protein of 447 amino acids, revealed the presence of a C-terminal domain similar to glucosylceramidases starting at amino acid 43. Since translocation signals of T3SS effectors are usually located in the N-terminal portion of the protein, we generated a CyaA fusion with amino acids 1 to 42 of SrfJ expressed from a constitutive promoter in a plasmid. A derivative of S. enterica serovar Typhimurium 14028 carrying this plasmid was used to infect cultures of RAW264.7 macrophages. An increase in the level of cAMP in the culture 16 h postinfection indicated translocation of the fusion into the cytosol of the host cells (Fig. 3A). Fragment 1-20 of SrfJ, but not fragment 1-10, 10-42, or 20-42, was also able to direct translocation of CyaA into macrophages. Fusions with fragments 1-42 and 1-20 were also translocated in a strain lacking T3SS1 (ΔSPI1), but they were no longer translocated in a strain lacking T3SS2 (ΔSPI2) (Fig. 3B). These results suggest that the N-terminal 20 amino acids of SrfJ are enough to direct translocation into macrophages specifically through T3SS2.
Fig 3.
The 20 N-terminal amino acids of SrfJ are able to direct translocation into macrophages through T3SS2. (A) Different fragments of SrfJ were expressed in the wild-type strain of S. enterica serovar Typhimurium in fusion with the catalytic domain of CyaA from B. pertussis. RAW264.7 cells were infected with bacteria grown for 24 h in LB at 37°C with shaking, and the concentrations of cAMP were measured 16 h postinfection. Numbers in parentheses indicate the amino acids of SrfJ included in the fusion. The results are represented as the means from four independent experiments. Error bars represent standard deviations. (B) Fragments containing amino acids 1 to 42 and 1 to 20 from SrfJ fused to the catalytic domain of CyaA from B. pertussis were expressed in the wild-type (wt) strain of S. enterica serovar Typhimurium, in a strain lacking T3SS1 (ΔSPI1), and in a strain lacking T3SS2 (ΔSPI2). Cultures from these bacteria were used to infect RAW264.7 cells, and the concentrations of cAMP were measured 16 h postinfection. Means and standard deviations from two independent experiments are represented.
Expression of srfJ is regulated positively by SsrB and PhoP and negatively by RcsB.
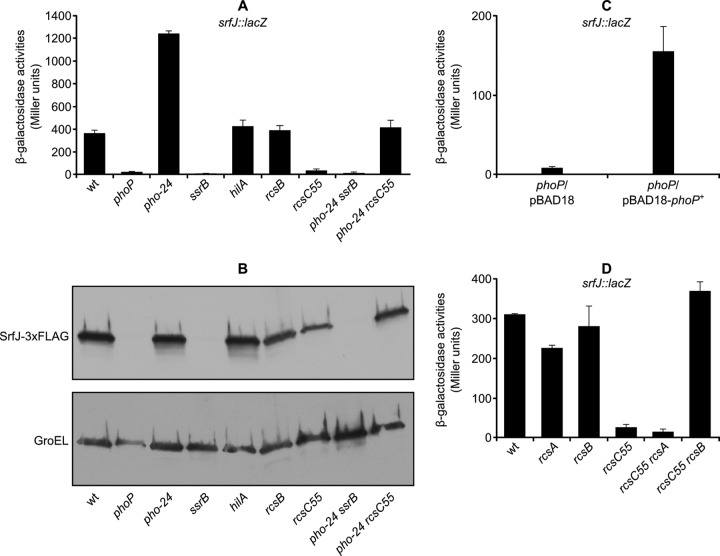
The srfJ::lacZ transcriptional fusion and the SrfJ-3×FLAG fusion were used to search for regulators of this gene. The expression of the fusions was analyzed in LPM using Salmonella strains with mutations in genes encoding known virulence regulators: PhoP, SsrB, HilA, and RcsB. In addition to null mutations, the activating mutations pho-24 and rcsC55 were also used. As seen in Fig. 4A and B, PhoP and SsrB were necessary for expression of srfJ in LPM. Expression of srfJ was increased in a pho-24 background, confirming that PhoP was a positive regulator of this gene. Additional confirmation was provided by complementation analysis. Plasmid pIZ1974, carrying the phoP gene under the control of the arabinose-inducible PBAD promoter, was constructed for this purpose. Arabinose-induced phoP expression, but not the empty vector (pBAD18), complemented the effect of phoP disruption and activated srfJ expression (Fig. 4C). The phoP mutant used in our experiments was the product of a Tn10 insertion that is expected to have a polar effect on phoQ. Partial complementation due to expression of phoP alone is explained by overexpression of the transcriptional regulator PhoP, which can activate its target genes in a PhoQ-independent manner (34). In addition, overactivation of the Rcs system by the rcsC55 mutation prevented srfJ transcription, suggesting that RcsB is a negative regulator of srfJ. This conclusion was confirmed using a ΔrcsB::Cmr mutation: introduction of this null mutation in the rcsC55 mutant restored wild-type levels of srfJ::lacZ transcription (Fig. 4D). In contrast, a ΔrcsA::Cmr mutation was unable to suppress the effect of the rcsC55 mutation on the expression of srfJ::lacZ, indicating that the coregulator RcsA does not participate in the regulation of srfJ transcription. pho-24 ssrB and pho-24 rcsC55 double mutants were constructed to carry out epistasis analysis. The results shown in Fig. 4 suggested that PhoP activates srfJ through SsrB, whereas RcsB acts independently. Since both the Rcs and the PhoP/PhoQ systems are activated by cationic antimicrobial peptides, we decided to study the effect of subinhibitory concentrations of polymyxin B on srfJ expression. No significant modifications in the expression of srfJ::lacZ were detected after addition of polymyxin B to LB–0.3 M NaCl or to LPM (data not shown). This result is consistent with the opposite effects that these regulatory systems have on srfJ.
Fig 4.
srfJ is regulated by PhoP, SsrB, and RcsB. (A) β-Galactosidase activities were measured from cultures in LPM of several S. enterica strains: 14028 (wild type [wt]), null mutants (phoP, ssrB, hilA, and rcsB), mutants with constitutive activation of the PhoP/PhoQ or the Rcs system (pho-24 and rcsC55, respectively), and double mutants (pho-24 ssrB and pho-24 rcsC55), all of them carrying a chromosomal srfJ::lacZ transcriptional fusion. Means and standard deviations from duplicate experiments are represented. (B) Expression at the protein level was studied by Western blotting using strains expressing SrfJ-3×FLAG. Extracts from these strains were resolved by SDS-PAGE (4 to 15% gradient), and a monoclonal anti-FLAG antibody was used for immunoblotting (upper panel). A polyclonal anti-GroEL antibody was used to get a loading control (lower panel). (C) A plasmid expressing wild-type phoP (phoP+) under a PBAD promoter was used to complement the effect of a null phoP mutation on srfJ transcription. The empty vector (pBAD18) was used as a control. Means and standard deviations of β-galactosidase activities obtained from strains carrying a chromosomal srfJ::lacZ transcriptional fusion grown in LPM supplemented with 0.2% arabinose are represented. (D) To study the role of RcsB and RcsA in the regulation of srfJ, β-galactosidase activities were measured from cultures in LPM of wild-type (wt) S. enterica or mutants, as indicated, all of them carrying a chromosomal srfJ::lacZ transcriptional fusion. Means and standard deviations from duplicate experiments are represented.
A T-POP-based screen identifies IolR as a negative regulator of srfJ transcription.
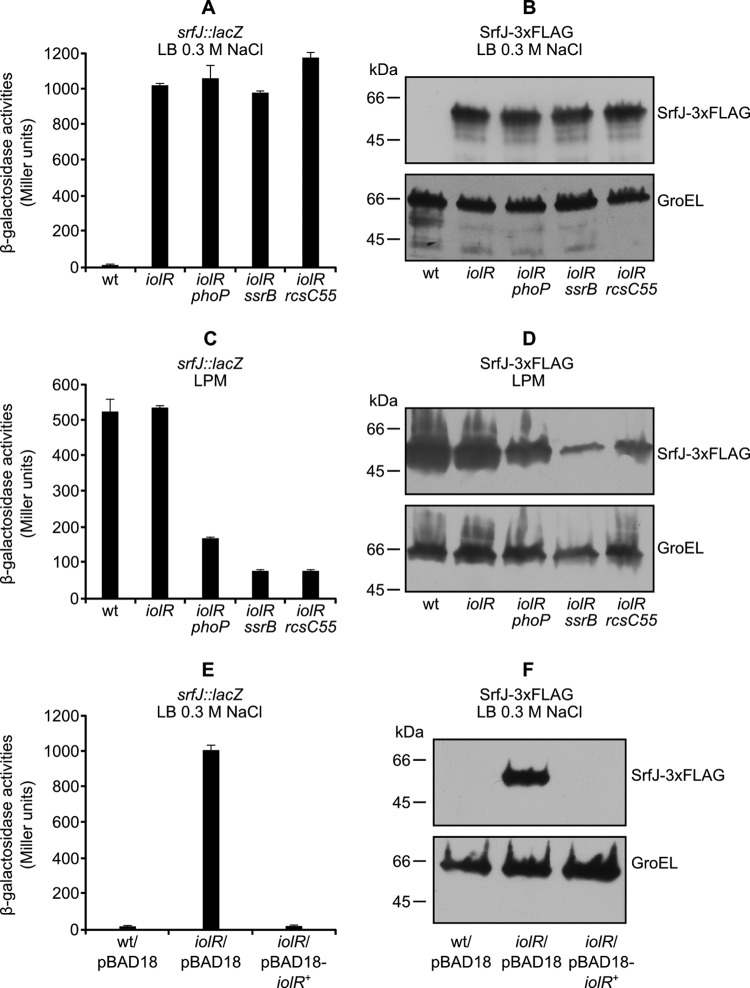
We took advantage of the srfJ::lacZ fusion and the T-POP transposon to look for new regulators of srfJ. A pool of random T-POP insertions was generated in strain SV5600, a derivative of strain 14028 of S. enterica serovar Typhimurium carrying an srfJ::lacZ fusion. Tc-resistant isolates were selected on LB plates supplemented with Tc and the chromogenic indicator X-Gal. Most colonies were white, as expected, since srfJ shows a very low level of expression in this medium. However, 15 colonies out of 20,000 screened were blue. All were also blue in the absence of tetracycline, suggesting that their phenotypes were due to disruption of a regulatory gene by T-POP and not to the Tc-dependent overexpression of an adjacent gene(s) from a T-POP promoter (48). To identify the location of the T-POP insertion, a semirandom, two-step PCR protocol (described in Materials and Methods) was used to amplify the DNA adjacent to the transposon for sequencing. All of the isolates carried the T-POP transposon in the same gene: STM14_5307 (STM4417 in strain LT2), also known as iolR (33). To confirm the results, an iolR null mutant was constructed using λ Red recombineering, and the expression of srfJ::lacZ was measured in wild-type and iolR backgrounds. The iolR mutation caused a >200-fold increase in β-galactosidase activity of LB–0.3 M NaCl cultures (Fig. 5A). This increase was confirmed by immunoblotting using the SrfJ-3×FLAG fusion (Fig. 5B). Epistasis analysis showed that this regulation was independent of SsrB and PhoP (Fig. 5A and B). Similar experiments carried out in LPM (pH 5.8) showed that the iolR mutation was unable to increase expression of srfJ in this medium (Fig. 5C and D). Interestingly, this mutation increased expression of srfJ in a phoP or an ssrB background (compare data in Fig. 4 and 5). Additional support for the conclusion that IolR was a repressor of srfJ transcription was provided by complementation analysis. As seen in Fig. 5E, derepression of srfJ::lacZ transcription in LB–0.3 M NaCl caused by disruption of iolR was abolished by a derivative of plasmid pBAD18 expressing wild-type iolR from a PBAD promoter (plasmid pIZ1973) but not by the empty vector. Complementation was confirmed at the protein level in the strains expressing the SrfJ-3×FLAG fusion (Fig. 5F).
Fig 5.
IolR represses srfJ. Expression of srfJ at the transcriptional level was monitored using a chromosomal srfJ::lacZ fusion. β-Galactosidase activities were measured in LB–0.3 M NaCl (A) or LPM (pH 5.8) (C) cultures from strains with different genetic backgrounds: the wild type (wt) and iolR, iolR phoP, iolR ssrB, and iolR rcsC55 mutants. Means and standard deviations from duplicate experiments are represented. Expression of srfJ at the protein level was monitored using an SrfJ-3×FLAG fusion. An anti-FLAG antibody was used to detect the fusion protein by immunoblotting in extracts from cultures in LB–0.3 M NaCl (B) or LPM (pH 5.8) (D) of S. enterica serovar Typhimurium strains with the indicated genetic backgrounds (upper panels). An anti-GroEL antibody was used as a loading control (lower panels). Complementation of the effect of the iolR mutation on srfJ transcription observed in LB–0.3 M NaCl cultures expressing wild-type iolR (iolR+) from a PBAD promoter was demonstrated at the transcriptional level (E) and the protein level (F).
myo-inositol induces transcription of srfJ but not secretion into the medium.
The iolR gene product was described as a negative regulator of genes involved in myo-inositol utilization in S. enterica serovar Typhimurium (33). The presence of myo-inositol is expected to prevent binding of IolR to promoters. Therefore, we tested the effect of myo-inositol on the expression of srfJ at both the transcriptional and protein level. As seen in Fig. 6A and B, addition of 55.5 mM myo-inositol to LB–0.3 M NaCl induced expression of srfJ to the same level as that obtained for the iolR mutant in LB–0.3 M NaCl in a PhoP-, SsrB-, and RcsB-independent manner. SrfJ-3×FLAG was not detected, however, in the supernatant of cultures in LB–0.3 M NaCl with myo-inositol (Fig. 6B), indicating that myo-inositol induces expression but not secretion under these conditions. The effect of myo-inositol on srfJ expression seems to be specific, since transcription of ssaV, another SsrB-regulated gene used for comparison, was not affected by myo-inositol (Fig. 6A). Finally, we compared the growth of the wild-type strain to that of a strain lacking srfJ in minimal medium with myo-inositol as the sole carbon source. No differences were observed (data not shown), indicating that SrfJ is not involved in the utilization of myo-inositol.
Fig 6.
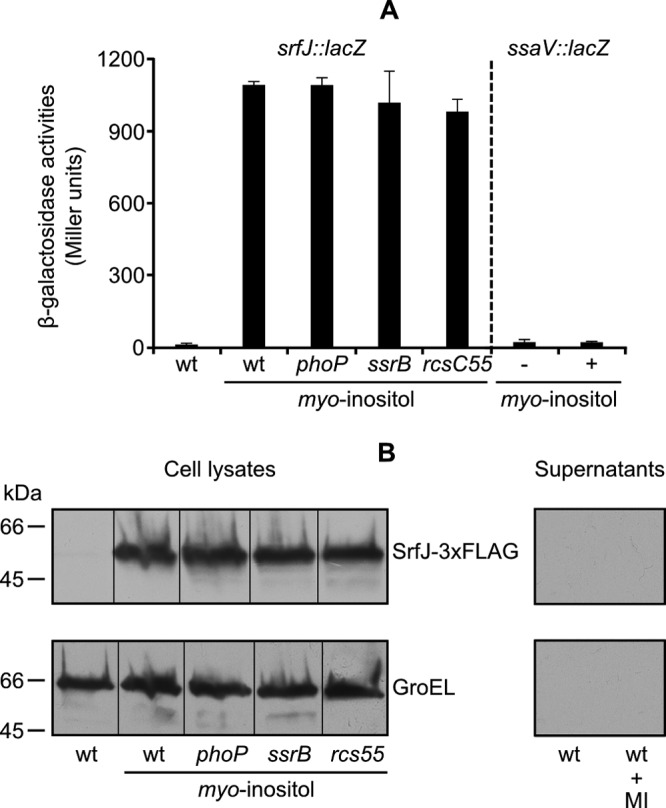
myo-inositol induces expression, but not secretion, of srfJ in LB–0.3 M NaCl. (A) Expression of srfJ at the transcriptional level was monitored using a chromosomal srfJ::lacZ fusion. Strains carrying the indicated mutations were cultured in LB–0.3 M NaCl medium or LB–0.3 M NaCl medium supplemented with myo-inositol. Expression of ssaV in LB–0.3 M NaCl medium (−) or the same medium supplemented with myo-inositol (+) was monitored using a chromosomal ssaV::lacZ transcriptional fusion. Means and standard deviations from duplicate experiments are represented. (B) Expression and secretion of srfJ at the protein level was monitored using an SrfJ-3×FLAG fusion. An anti-FLAG antibody was used to detect the fusion protein by immunoblotting in cell lysates and in concentrated supernatants from cultures in LB–0.3 M NaCl medium of S. enterica serovar Typhimurium strains with the indicated mutations (upper panel). An anti-GroEL antibody was used as the loading control (lower panel). wt, wild type; MI, myo-inositol.
DISCUSSION
Characterization of the whole catalogue of effector proteins is an important step in the understanding of the pathogenesis of bacteria that rely on T3SS for their interaction with the host. Although the components of the secretion apparatus are well conserved between distantly related bacteria, the secreted proteins are usually very different. This is a hurdle in the identification of effector proteins, in spite of the increasing availability of whole-genome sequences. Proteomic and genetic screens have led to the identification of many effectors in different bacteria but the catalogue of effectors is still growing, indicating that it is not completed yet (12, 20, 43, 46, 51, 52, 55).
In this work, we provide evidence suggesting that SrfJ, the product of an SsrB-regulated gene, is a new substrate of Salmonella T3SS2. Regulation by SsrB is a feature shared by most T3SS2 effectors encoded inside or outside SPI2. Therefore, all srf genes (62) were, in principle, candidates to encode such an effector. For instance, SrfH, also called SseI, was shown to be translocated through T3SS2 into the host cytosol (39), where it binds the actin-cross-linking protein filamin and colocalizes with polymerizing actin in the cytoskeleton and with TRIP6 (38, 63). There was no evidence of translocation for the other srf gene products. Previous work in our laboratory showed that the srfABC operon is not expressed in coordination with SPI2; therefore, its products, SrfA, SrfB, and SrfC, are not expected to be secreted through T3SS2 (18). In addition, no translocation was detected for SrfK using the CyaA fusion system (our unpublished results).
Translocation of SrfJ into macrophages was dependent on T3SS2 and independent of T3SS1. In addition, secretion of SrfJ into the culture medium occurred only under SPI2-inducing conditions, although a shift from pH 5.8 to 7.2 was essential to detect the protein in the supernatant of the cell cultures. Experiments by Yu et al. (65) suggest that the SPI2-encoded proteins SsaL, SsaM, and SpiC form a complex that is required for secretion of translocon proteins and suppress secretion of effectors at pH 5.0. Exposure to pH 7.2 after growth at pH 5.0 causes dissociation and degradation of the complex and allows effector secretion. The same requirement was previously observed for other T3SS2, like SseF and SseJ (65), but not for other effectors, like SteA, that can also be secreted through T3SS1 (5).
In most T3SS effectors studied, a small N-terminal domain is enough to direct translocation, although a conserved sequence has been identified in only a few cases (39). As a consequence, computational methods used to detect secretion signals are only partially accurate (2, 36, 50, 60, 64). Our experiments with SrfJ fragments fused to CyaA showed that the minimal signal sequence necessary for translocation is contained in the N-terminal 20 amino acids of this protein. This short sequence was enough to direct translocation of CyaA specifically through T3SS2, suggesting that in this case interaction with a chaperone, as has been described for other effectors, was dispensable.
As expected for a T3SS2 effector, expression of srfJ is coordinated with expression of SPI2. In fact, high expression is observed in LPM (pH 5.8), a medium that mimics some of the conditions found inside the SCV and activates some regulators that are activated within the phagosome. Our experiments reveal that both acidic pH and Mg2+ limitation are important factors in the regulation of srfJ. Transcription of srfJ is positively regulated by the two-component systems PhoP/PhoQ and SsrA/SsrB and negatively regulated by the RcsC/RcsD/RcsB phosphorelay system. Our epistasis experiments suggest that, whereas PhoP regulation is SsrB dependent, RcsB acts independently from the other two regulators. Positive regulation by PhoP is consistent with expression in LPM (pH 5.8), since the PhoQ sensor is activated in response to low Mg2+ concentrations and acidic pH (31, 47). Regulation of SPI2 genes by the two-component system PhoP/PhoQ occurs because PhoP controls expression of ssrB and ssrA at transcriptional and posttranscriptional levels, respectively (3). Here, we show that the same regulatory cascade affects srfJ, a T3SS2-related gene located outside SPI2. The Rcs system is also involved in the control of Salmonella virulence (13, 14, 19, 42) through the regulation of many genes, including SPI1 and SPI2 genes (35, 59). A general regulatory overlap between Rcs and PhoP/PhoQ in the control of Salmonella virulence was already shown (18). The regulation of srfJ by both systems is a new element contributing to this overlap.
Using a T-POP-based screen, we have identified IolR as an unexpected regulator of srfJ. IolR is the main regulator of five operons, encompassing the genes iolA-iolB, iolE-iolG1, iolC1-iolC2, iolD1-iolD2-iolG2, and iolI2-iolH, which are involved in the utilization of myo-inositol as a carbon source (33). Transcription of these operons is repressed in minimal medium with glucose and induced in minimal medium with myo-inositol. Deletion of iolR resulted in stimulation of the transcription of the iol operons in minimal medium with glucose, suggesting that the IolR protein was a repressor. In fact, IolR is similar to proteins belonging to the RpiR family of transcriptional regulators. The iol operons and the iolR gene are located in a 22.6-kb genomic island. Sequence comparisons using BLAST identified this island in the genomes of S. enterica serovar Typhimurium strains LT2, 14028, D23580, UK-1, T000240, ST4/74, and SL1344; S. enterica serovar Paratyphi B strain SPB7; S. enterica serovar Agona strain SL483; S. enterica serovar Weltevreden strain 2007-60-3289-1; and E. coli ED1a. Most genes in the island are absent from the genomes of Salmonella bongori and certain S. enterica strains from serovars Typhi, Paratyphi, Gallinarum, Enteritidis, Dublin, and Diarizonae. Interestingly, srfJ is present, downstream of iolG1, in the strains that contain the island and is absent from the genomes of the strains that lack the island.
The IolR repressor is expected to be inactivated in the presence of myo-inositol. Consistent with this notion, addition of this sugar to LB–0.3 M NaCl, a medium where srfJ exhibits a very low level of expression, had the same effect as the iolR null mutation and resulted in a more than 200-fold increase in srfJ transcription. The iolR mutation, or the addition of myo-inositol, failed to induce expression of srfJ in LPM. This result suggests that IolR can repress srfJ when SsrB is inactive but not when SsrB is active (LB–0.3 M NaCl versus LPM). In fact, the iolR mutation significantly increased transcription of srfJ in LPM when tested in an ssrB background.
Our results show that srfJ can be expressed in two different physiological conditions: (i) intracellularly inside the SCV (conditions mimicked by LPM) in coordination with T3SS2 and (ii) in the presence of myo-inositol, a carbohydrate abundant in soil and plants. Whereas intracellular expression is consistent with SrfJ being a T3SS2 effector, the significance of myo-inositol-dependent expression is unclear. In fact, SrfJ is not secreted in LB–0.3 M NaCl supplemented with myo-inositol in spite of its high level of expression, probably owing to the lack of T3SS2 under these conditions. One possibility is that accumulation of SrfJ before ingestion by the host animal (i.e., in environments where myo-inositol is present) is advantageous for early secretion as soon as bacteria enter the host cells and the T3SS2 is mounted. However, an independent role for SrfJ in environments where myo-inositol is present cannot be excluded. In support of this idea is the fact that srfJ and the myo-inositol utilization island are present in E. coli ED1a (10, 56), a human host-specific commensal strain belonging to genetic group B2 that is avirulent in a mouse model of extraintestinal infection and does not possess an identified virulence-related T3SS.
Finally, the identification of a new T3SS effector opens the way to physiological studies. SrfJ is especially interesting because it contains a C-terminal domain that is similar to human lysosomal glucosylceramidase (also known as acid β-glucocerebrosidase or acid β-glucosidase). Both proteins share 30% amino acid sequence identity over 447 residues and show structural similarities in the overall fold and the active-site environment (32). The human enzyme catalyzes the hydrolysis of glucosylceramide to β-glucose and ceramide (23). Ceramide is a lipid molecule composed of sphingosine and a fatty acid. The formation of ceramide within the cell membrane alters biophysical characteristics of membranes and leads to the generation of ceramide-enriched membrane domains. These domains have a general function in signal transduction through amplification of receptor- and stress-mediated signaling events (66). Well-known events modulated by ceramide include differentiation, proliferation, and apoptosis. In addition, ceramide and ceramide-enriched membrane domains have been involved in host responses to many pathogens (24).
ACKNOWLEDGMENTS
This work was supported by grant SAF2010-15015, from the Spanish Ministry of Science and Innovation and the European Regional Development Fund, and grant P08-CVI-03487, from the Consejería de Economía, Innovación y Ciencia, Junta de Andalucía, Spain.
We are grateful to I. Rosenshine (The Hebrew University, Israel) for kindly providing plasmid pSIF003-R1, to E. Krin and A. Danchin (Institut Pasteur, France) for the generous gift of E. coli TP610, and to J. Casadesús (Universidad de Sevilla, Spain) for critical reading of the manuscript.
Footnotes
Published ahead of print 1 June 2012
REFERENCES
- 1. Agbor TA, McCormick BA. 2011. Salmonella effectors: important players modulating host cell function during infection. Cell. Microbiol. 13: 1858–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arnold R, et al. 2009. Sequence-based prediction of type III secreted proteins. PLoS Pathog. 5: e1000376 doi:10.1371/journal.ppat.1000376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bijlsma JJ, Groisman EA. 2005. The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol. Microbiol. 57: 85–96 [DOI] [PubMed] [Google Scholar]
- 4. Canals D, Perry DM, Jenkins RW, Hannun YA. 2011. Drug targeting of sphingolipid metabolism: sphingomyelinases and ceramidases. Br. J. Pharmacol. 163: 694–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cardenal-Muñoz E, Ramos-Morales F. 2011. Analysis of the expression, secretion and translocation of the Salmonella enterica type III secretion system effector SteA. PLoS One 6: e26930 doi:10.1371/journal.pone.0026930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan RK, Botstein D, Watanabe T, Ogata Y. 1972. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high-frequency-transducing lysate. Virology 50: 883–898 [DOI] [PubMed] [Google Scholar]
- 7. Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158: 9–14 [DOI] [PubMed] [Google Scholar]
- 8. Chun KT, Edenberg HJ, Kelley MR, Goebl MG. 1997. Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast 13: 233–240 [DOI] [PubMed] [Google Scholar]
- 9. Cirillo DM, Valdivia RH, Monack DM, Falkow S. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30: 175–188 [DOI] [PubMed] [Google Scholar]
- 10. Clermont O, et al. 2008. Evidence for a human-specific Escherichia coli clone. Environ. Microbiol. 10: 1000–1006 [DOI] [PubMed] [Google Scholar]
- 11. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deng W, et al. 2010. A comprehensive proteomic analysis of the type III secretome of Citrobacter rodentium. J. Biol. Chem. 285: 6790–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Detweiler CS, Monack DM, Brodsky IE, Mathew H, Falkow S. 2003. virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol. Microbiol. 48: 385–400 [DOI] [PubMed] [Google Scholar]
- 14. Domínguez-Bernal G, et al. 2004. Repression of the RcsC-YojN-RcsB phosphorelay by the IgaA protein is a requisite for Salmonella virulence. Mol. Microbiol. 53: 1437–1449 [DOI] [PubMed] [Google Scholar]
- 15. Ellermeier CD, Janakiraman A, Slauch JM. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290: 153–161 [DOI] [PubMed] [Google Scholar]
- 16. Galan JE, Curtiss R., III 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. U. S. A. 86: 6383–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garai P, Marathe SA, Chakravortty D. 2011. Effectors of Salmonella pathogenicity island 2: an island crucial to the life of Salmonella. Virulence 2: 177–180 [DOI] [PubMed] [Google Scholar]
- 18. García-Calderón CB, Casadesús J, Ramos-Morales F. 2007. Rcs and PhoPQ regulatory overlap in the control of Salmonella enterica virulence. J. Bacteriol. 189: 6635–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. García-Calderón CB, García-Quintanilla M, Casadesús J, Ramos-Morales F. 2005. Virulence attenuation in Salmonella enterica rcsC mutants with constitutive activation of the Rcs system. Microbiology 151: 579–588 [DOI] [PubMed] [Google Scholar]
- 20. Geddes K, Worley M, Niemann G, Heffron F. 2005. Identification of new secreted effectors in Salmonella enterica serovar Typhimurium. Infect. Immun. 73: 6260–6271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ginocchio CC, Olmsted SB, Wells CL, Galan JE. 1994. Contact with epithelial cells induces the formation of surface appendages on Salmonella typhimurium. Cell 76: 717–724 [DOI] [PubMed] [Google Scholar]
- 22. Glaser P, Danchin A, Ladant D, Barzu O, Ullmann A. 1988. Bordetella pertussis adenylate cyclase: the gene and the protein. Tokai J. Exp. Clin. Med. 13 (Suppl.): 239–252 [PubMed] [Google Scholar]
- 23. Grabowski GA, Gatt S, Horowitz M. 1990. Acid beta-glucosidase: enzymology and molecular biology of Gaucher disease. Crit. Rev. Biochem. Mol. Biol. 25: 385–414 [DOI] [PubMed] [Google Scholar]
- 24. Grassme H, Becker KA, Zhang Y, Gulbins E. 2008. Ceramide in bacterial infections and cystic fibrosis. Biol. Chem. 389: 1371–1379 [DOI] [PubMed] [Google Scholar]
- 25. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177: 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166: 557–580 [DOI] [PubMed] [Google Scholar]
- 27. Hedegaard L, Danchin A. 1985. The cya gene region of Erwinia chrysanthemi B374: organisation and gene products. Mol. Gen. Genet. 201: 38–42 [Google Scholar]
- 28. Hensel M, et al. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30: 163–174 [DOI] [PubMed] [Google Scholar]
- 29. Hueck CJ. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62: 379–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jenkins RW, Canals D, Hannun YA. 2009. Roles and regulation of secretory and lysosomal acid sphingomyelinase. Cell. Signal. 21: 836–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kato A, Groisman EA. 2008. The PhoQ/PhoP regulatory network of Salmonella enterica. Adv. Exp. Med. Biol. 631: 7–21 [DOI] [PubMed] [Google Scholar]
- 32. Kim YG, Kim JH, Kim KJ. 2009. Crystal structure of the Salmonella enterica serovar Typhimurium virulence factor SrfJ, a glycoside hydrolase family enzyme. J. Bacteriol. 191: 6550–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kröger C, Fuchs TM. 2009. Characterization of the myo-inositol utilization island of Salmonella enterica serovar Typhimurium. J. Bacteriol. 191: 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lejona S, et al. 2004. PhoP can activate its target genes in a PhoQ-independent manner. J. Bacteriol. 186: 2476–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin D, Rao CV, Slauch JM. 2008. The Salmonella SPI1 type three secretion system responds to periplasmic disulfide bond status via the flagellar apparatus and the RcsCDB system. J. Bacteriol. 190: 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lower M, Schneider G. 2009. Prediction of type III secretion signals in genomes of gram-negative bacteria. PLoS One 4: e5917 doi:10.1371/journal.pone.0005917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maloy SR. 1990. Experimental techniques in bacterial genetics. Jones & Barlett, Boston, MA [Google Scholar]
- 38. Miao EA, et al. 2003. Salmonella effectors translocated across the vacuolar membrane interact with the actin cytoskeleton. Mol. Microbiol. 48: 401–415 [DOI] [PubMed] [Google Scholar]
- 39. Miao EA, Miller SI. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. U. S. A. 97: 7539–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 41. Mills DM, Bajaj V, Lee CA. 1995. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol. Microbiol. 15: 749–759 [DOI] [PubMed] [Google Scholar]
- 42. Mouslim C, Delgado M, Groisman EA. 2004. Activation of the RcsC/YojN/RcsB phosphorelay system attenuates Salmonella virulence. Mol. Microbiol. 54: 386–395 [DOI] [PubMed] [Google Scholar]
- 43. Niemann GS, et al. 2011. Discovery of novel secreted virulence factors from Salmonella enterica serovar Typhimurium by proteomic analysis of culture supernatants. Infect. Immun. 79: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ochman H, Soncini FC, Solomon F, Groisman EA. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc. Natl. Acad. Sci. U. S. A. 93: 7800–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ohl ME, Miller SI. 2001. Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 52: 259–274 [DOI] [PubMed] [Google Scholar]
- 46. Petnicki-Ocwieja T, et al. 2002. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 99: 7652–7657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Prost LR, Miller SI. 2008. The salmonellae PhoQ sensor: mechanisms of detection of phagosome signals. Cell. Microbiol. 10: 576–582 [DOI] [PubMed] [Google Scholar]
- 48. Rappleye CA, Roth JR. 1997. A Tn10 derivative (T-POP) for isolation of insertions with conditional (tetracycline-dependent) phenotypes. J. Bacteriol. 179: 5827–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ruiz-Albert J, et al. 2002. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol. Microbiol. 44: 645–661 [DOI] [PubMed] [Google Scholar]
- 50. Samudrala R, Heffron F, McDermott JE. 2009. Accurate prediction of secreted substrates and identification of a conserved putative secretion signal for type III secretion systems. PLoS Pathog. 5: e1000375 doi:10.1371/journal.ppat.1000375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sato Y, Takaya A, Yamamoto T. 2011. Meta-analytic approach to the accurate prediction of secreted virulence effectors in gram-negative bacteria. BMC Bioinformatics 12: 442 doi:10.1186/1471-2105-12-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schechter LM, et al. 2006. Multiple approaches to a complete inventory of Pseudomonas syringae pv. tomato DC3000 type III secretion system effector proteins. Mol. Plant Microbe Interact. 19: 1180–1192 [DOI] [PubMed] [Google Scholar]
- 53. Schmieger H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119: 75–88 [DOI] [PubMed] [Google Scholar]
- 54. Shea JE, Hensel M, Gleeson C, Holden DW. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. U. S. A. 93: 2593–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tobe T, et al. 2006. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc. Natl. Acad. Sci. U. S. A. 103: 14941–14946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Touchon M, et al. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5: e1000344 doi:10.1371/journal.pgen.1000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. U. S. A. 98: 15264–15269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Valinsky L, et al. 2002. A host-specific virulence protein of Erwinia herbicola pv. gypsophilae is translocated into human epithelial cells by the type III secretion system of enteropathogenic Escherichia coli. Mol. Plant Pathol. 3: 97–101 [DOI] [PubMed] [Google Scholar]
- 59. Wang Q, Zhao Y, McClelland M, Harshey RM. 2007. The RcsCDB signaling system and swarming motility in Salmonella enterica serovar Typhimurium: dual regulation of flagellar and SPI-2 virulence genes. J. Bacteriol. 189: 8447–8457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang Y, Zhang Q, Sun MA, Guo D. 2011. High-accuracy prediction of bacterial type III secreted effectors based on position-specific amino acid composition profiles. Bioinformatics 27: 777–784 [DOI] [PubMed] [Google Scholar]
- 61. Waterman SR, Holden DW. 2003. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell. Microbiol. 5: 501–511 [DOI] [PubMed] [Google Scholar]
- 62. Worley MJ, Ching KH, Heffron F. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36: 749–761 [DOI] [PubMed] [Google Scholar]
- 63. Worley MJ, Nieman GS, Geddes K, Heffron F. 2006. Salmonella typhimurium disseminates within its host by manipulating the motility of infected cells. Proc. Natl. Acad. Sci. U. S. A. 103: 17915–17920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang Y, Zhao J, Morgan RL, Ma W, Jiang T. 2010. Computational prediction of type III secreted proteins from gram-negative bacteria. BMC Bioinformatics 11 (Suppl. 1): S47 doi:10.1186/1471-2105-11-S1-S47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yu XJ, McGourty K, Liu M, Unsworth KE, Holden DW. 2010. pH sensing by intracellular Salmonella induces effector translocation. Science 328: 1040–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang Y, Li X, Becker KA, Gulbins E. 2009. Ceramide-enriched membrane domains–structure and function. Biochim. Biophys. Acta 1788: 178–183 [DOI] [PubMed] [Google Scholar]