Abstract
The genomes of the Tomato mosaic virus and many other plant and animal positive-strand RNA viruses of agronomic and medical importance encode superfamily 1 helicases. Although helicases play important roles in viral replication, the crystal structures of viral superfamily 1 helicases have not been determined. Here, we report the crystal structure of a fragment (S666 to Q1116) of the replication protein from Tomato mosaic virus. The structure reveals a novel N-terminal domain tightly associated with a helicase core. The helicase core contains two RecA-like α/β domains without any of the accessory domain insertions that are found in other superfamily 1 helicases. The N-terminal domain contains a flexible loop, a long α-helix, and an antiparallel six-stranded β-sheet. On the basis of the structure, we constructed deletion mutants of the S666-to-Q1116 fragment and performed split-ubiquitin-based interaction assays in Saccharomyces cerevisiae with TOM1 and ARL8, host proteins that are essential for tomato mosaic virus RNA replication. The results suggested that both TOM1 and ARL8 interact with the long α-helix in the N-terminal domain and that TOM1 also interacts with the helicase core. Prediction of secondary structures in other viral superfamily 1 helicases and comparison of those structures with the S666-to-Q1116 structure suggested that these helicases have a similar fold. Our results provide a structural basis of viral superfamily 1 helicases.
INTRODUCTION
Helicases are motor proteins that use the free energy of nucleoside triphosphate (NTP) hydrolysis to unwind nucleic acid duplexes during gene replication, transcription, translation, recombination, and repair (1). Helicases are encoded in the genomes of all living organisms and have been classified into six superfamilies (SF1 to SF6) according to their conserved motifs (11, 42).
Helicases are also encoded in the genomes of most viruses and play important roles in viral replication (20). Viral helicases belong to SF1, SF2, or SF3. The genomes of alpha-like viruses encode SF1 helicases. Those of potyviruses and flaviviruses encode SF2 helicases, and those of vaccinia virus and picornaviruses encode SF3 helicases (12). For the SF2 helicases of hepatitis C virus (21, 22, 29) and dengue virus (30, 51), three-dimensional structures have been determined. Viral SF3 helicase structures have also been determined (14). Conversely, a structure for an SF1 viral helicase has not been reported, although the genomes of many important positive-strand RNA viruses, e.g., severe acute respiratory syndrome (SARS) coronavirus and hepatitis E virus, encode SF1 helicases.
The tobamovirus group belongs to the alphavirus-like superfamily of positive-strand RNA viruses and includes Tobacco mosaic virus (TMV) and Tomato mosaic virus (ToMV). These viruses have long served as models of positive-strand RNA viruses to investigate the mechanisms of multiplication and interaction with host organisms (4). The genome of a tobamovirus encodes at least four proteins (10). After infection, the genomic RNA is translated in the cytoplasm of the host cell to produce a protein with a molecular mass of ∼130 kDa (130K protein) and a read-through product with a molecular mass of ∼180 kDa (180K protein). These two replication proteins are both involved in viral-RNA replication, and they harbor an SF1 helicase-like domain. The helicase-like domain interacts with itself (13) and with more than 10 host proteins (17) during viral-RNA replication and control of host defenses. For example, tobamovirus helicase domains interact with TOM1 and ARL8, which are host proteins required for tobamovirus multiplication (34, 52, 53), and with the protein products of the resistance genes N (46) and Tm-1 (16). The TMV helicase also interacts with auxin/indole-3-acetic acid proteins to avoid leaf age-related resistance (36, 37).
To fully characterize the functions of tobamovirus replication proteins, the structure must be solved. We recently reported that the fragment S666 to Q1116 (here denoted ToMV-Hel) from the ToMV replication proteins is stable and, notably, has NTPase activity (49). Here, we report the ToMV-Hel structure solved by X-ray crystallography to 1.9-Å resolution. We found that ToMV-Hel contains a core helicase domain composed of two RecA domains and an N-terminal accessory domain containing a long α-helix, which is structurally unlike the accessory domains found in nonviral SF1 helicases. A series of deletion mutagenesis studies based on the ToMV-Hel structure revealed that the long α-helix in the N-terminal domain is involved in the binding of the host factors TOM1 and ARL8 required for viral replication.
MATERIALS AND METHODS
Protein expression and purification.
Escherichia coli Rosetta(DE3) cells were transformed with the expression vector pDEST-trx-ToMV-Hel and cultured in Luria-Bertani medium. ToMV-Hel was then expressed and purified as described previously (49). To express selenomethionine-labeled ToMV-Hel, B834(DE3)pRARE cells were prepared by transferring the pRARE plasmid from Rosetta(DE3) cells (Novagen) into B834(DE3) competent cells (Novagen) and then transforming the cells with pDEST-trx-ToMV-Hel. The cells were cultured in LeMaster medium (28). SeMet-labeled ToMV-Hel was expressed and purified in the same manner as the unlabeled protein. MgCl2 (2 mM) and adenosine 5′-O-(3-thio)triphosphate (0.1 mM) were added to the purified ToMV-Hel preparations. The proteins were each concentrated to 2.0 mg ml−1 using an Amicon Ultra-30 (Millipore) with the buffer changed to 20 mM sodium phosphate, pH 7.5, 150 mM NaCl, 2 mM 2-mercaptoethanol, 0.4 mM ATPγS, and 0.4 mM MgCl2.
Crystallization and data collection.
The native and SeMet-labeled ToMV-Hels were crystallized using the hanging-drop vapor diffusion method at 285 K. The crystals were grown on siliconized coverslips by equilibrating a mixture containing 1.5 μl of a protein solution (2.0 mg ml−1 protein in 20 mM sodium phosphate, pH 7.5, 150 mM NaCl, 2 mM 2-mercaptoethanol, 0.4 mM ATPγS, and 0.4 mM MgCl2) and 1.5 μl of the reservoir solution (0.1 M sodium phosphate, pH 6.5, 1.7 M ammonium sulfate) with 500 μl of the reservoir solution.
The crystals were flash frozen under a cryogenic stream of nitrogen. The X-ray diffraction data set for the native crystal was collected at the KEK Photon Factory BL17A beamline, Japan (wavelength, 1.0 Å) at 100 K. The crystal-to-detector distance was set to 299.6 mm, and diffraction images were recorded using an ADSC/Quantum 315 charge-coupled device (CCD) detector. Data were collected in 1°-oscillation steps for a total rotation of 180° and an exposure time of 20 s per frame. The X-ray diffraction data set for the SeMet-labeled ToMV-Hel crystal was collected at the Spring-8 BL38B1 beamline, Japan (wavelength, 0.97914 Å), at 100 K. The crystal-to-detector distance was set to 300 mm, and diffraction images were recorded using an ADSC/Quantum 315 CCD detector. Data were collected in 1°-oscillation steps for a total rotation of 225° and an exposure time of 20 s per frame. The detailed crystallization conditions and data collection have been described elsewhere (50).
Structure determination.
The diffraction data sets were processed using the HKL2000 suite of programs (35). The data collection statistics are summarized in Table 1. Of the 12 possible selenium atoms in an asymmetric unit, seven were found by SHELXD (41) using the anomalous signal in the peak data set of SeMet-labeled ToMV-Hel. Initial phases for SeMet-labeled ToMV-Hel were calculated and refined using SHELXE (41) with the graphical interface HKL2MAP (39). ARP/wARP (33) was used for autotracing. Then, manual model building was carried out using COOT (7). The crystallographic model was refined to 1.9-Å resolution using CNS (2). More than 90% of the residues in the model were found in the most favored regions of the Ramachandran plot by PROCHECK (26). Phasing and refinement statistics are given in Table 1.
Table 1.
Data collection and refinement statistics for ToMV-Hel
| Parameter | Value |
|
|---|---|---|
| Native | SeMet | |
| Data collection | ||
| Wavelength (Å) | 1.00 | 0.9780 |
| Temp (K) | 100 | 100 |
| Space group | P212121 | P212121 |
| Unit cell parameters (Å) | a = 85.44 | a = 85.52 |
| b = 128.82 | b = 128.54 | |
| c = 40.74 | c = 40.55 | |
| Resolution (Å)a | 50.0–1.9 (1.93–1.9) | 50.0–1.75 (1.81–1.75) |
| Reflections | ||
| Unique (no.)a | 36,315 (1,776) | 43,116 (3,045) |
| Completeness (%)a | 99.8 (97.8) | 93.6 (67.4) |
| Redundancy (no.)a | 9.1 (6.1) | 3.9 (2.0) |
| I/σ(I)a | 8.5 (1.3) | 12.5 (1.8) |
| Rsym (I)a | 0.080 (35.3) | 0.069 (38.6) |
| Phasing | ||
| Heavy-atom sites (no.) | 7 | |
| Correlation coefficientsc | ||
| SHELXD CC/CCweak | 37.62/22.69 | |
| SHELXE CCoverall | 33.04 | |
| CCfree left/right hand | 67.7/59.36 | |
| FOMd | 0.55 | |
| Refinement | ||
| Resolution (Å) | 40–1.90 | |
| Reflections (work/test) (no.) | 35,277/1,752 | |
| Rwork/Rfree (%) | 18.0/22.3 | |
| No. of water molecules | 568 | |
| No. of SO4 ions | 7 | |
| Avg B factors | 19.6 | |
| RmsD | ||
| Bond length (Å) | 0.005 | |
| Bond angle (Å) | 1.6 | |
| Ramachandran (no./% of residues) | ||
| Allowed | 29/7.2 | |
| Favored | 372/92.5 | |
| Disallowed | 1/0.2 | |
Data for the highest-resolution shell are given in parentheses.
Rsym(I) = ΣhklΣi|Ii(hkl) − <I(hkl)>|/ΣhklΣi|Ii(hkl)| for n independent reflections and i observations of a given reflection; <I(hkl)>, average intensity of the i observations.
CC = [ΣwEoEcΣw − ΣwEoΣwEc]/{[ΣwEo2Σw − (ΣwEo)2] [ΣwEc2Σw − (ΣwEc)2]}½; w, weight (see http://shelx.uni-ac.gwdg.de/SHELX/shelx_de.pdf for full definitions).
FOM (figure of merit) = |F(hkl)best|/|F(hkl)|; F(hkl)best = ΣαP(α)Fhkl(α)/ΣαP(α).
Split-ubiquitin-based interaction assays in yeast.
The Saccharomyces cerevisiae reporter strain L40 and plasmids for NubG-ALG5, TOM1-Cub-PLV, and ARL8-Cub-PLV expression have been described previously (34, 43, 45). DNA fragments corresponding to the ToMV helicase and upstream regions were PCR amplified from pTLW3 (24) with primer sets 1 and 4 (S666 to Q1116), 2 and 4 (V802 to Q1116), 1 and 3 (S666 to V801), and 5 and 6 (G699 to A732) (see Table S2 in the supplemental material). The fragments were each digested with NcoI and PstI. To create an expression plasmid for each ToMV-Hel fragment as an N-terminal NubG-tagged fusion protein, the TOM2A fragment bordered by the NcoI and PstI sites in pTN-NubG-TOM2A (45) was replaced with one of the ToMV-Hel fragments. The DNA fragment encoding G699 to A732 from the N-terminal domain of ToMV-Hel that also encoded an N-terminal methionine [Met-(G699-A732)] was PCR amplified from pTLW3 using primers 7 and 8 (see Table S2 in the supplemental material). The fragment was digested with SpeI and SalI and used to replace the mating factor coding region bordered by the SpeI and SalI sites in pTEF-MF (Dualsystems Biotech) to obtain an expression plasmid for Met-(G699-A732). For the three-hybrid experiment, the reporter strain NMY51 [MATa his3delta200 trp1-901 leu2-3,112 ade2 LYS2::(lexAop)4-HIS3 ura3::(lexAop)8-lacZ (lexAop)8-ADE2 GAL4; Dualsystems Biotech] was transformed with a ClaI-linearized pTN-TOM1-Cub-PLV plasmid (45) that was then integrated into the leu2 locus of NMY51. A transformant containing a single copy of the integrated plasmid was selected using Southern blot hybridization (3). Then, the strain was transformed with the pMN-ARL8-NubG (34) and pMN-TEF-699-732 plasmids. The β-galactosidase assay was performed as described previously (45).
Accession number.
The coordinates and structure factors for the native ToMV-Hel structure have been deposited in the Protein Data Bank (PDB) under accession number 3VKW.
RESULTS AND DISCUSSION
Overall structure of ToMV-Hel.
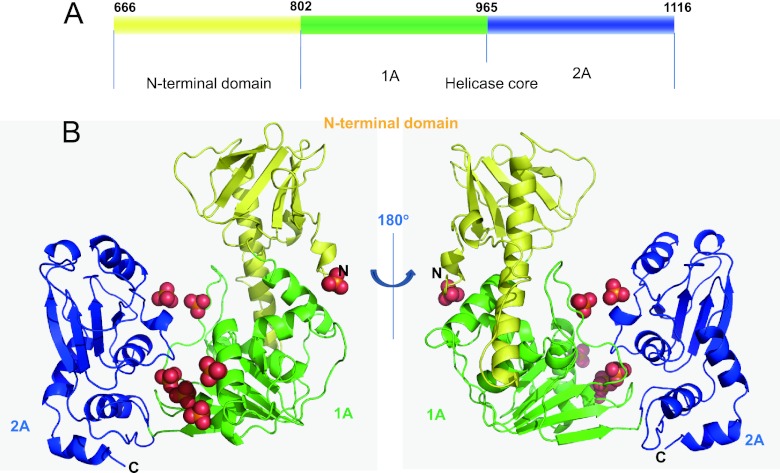
The asymmetric unit of the crystal contains one ToMV-Hel molecule. The final model contains 440 of the 451 expected residues (E672 to V1111), 7 sulfate molecules, and 568 water molecules. Two RecA-like α/β domains (1A and 2A) (42) are present and form the helicase core (Fig. 1A). Domain 1A (residues V802 to R964) contains a parallel six-stranded β-sheet surrounded by four helices on one side and two helices on the other (Fig. 1 and 2A). Domain 2A (residues T965 to Q1116) contains a parallel four-stranded β-sheet sandwiched between two helices on each of its sides (Fig. 1 and 2A). Three sulfate ions are found in a deep cleft that separates domains 1A and 2A (Fig. 1B). The N-terminal region (residues S666 to V801) forms a domain that tightly binds domain 1A (Fig. 1B; see Fig. S1 in the supplemental material). The N-terminal domain contains a flexible loop (L688 to P700), a long α-helix (L701 to D730), and an antiparallel, six-stranded β-sheet (F742 to E798) (Fig. 1B). The long α-helix in the N-terminal domain interacts with the groove formed by the domain 1A α-helices D805 to R814 and H911 to F918 (see Fig. S1 in the supplemental material). E674, K702, Q704, and K707 in the N-terminal domain interact electrostatically with R814, E956, K954, and E820 in domain 1A, respectively. Nonpolar residues, e.g., V703, F709, and W763 in the N-terminal domain and V823, F918, and Y803 in domain 1A also interact and help stabilize the association of the two domains (see Fig. S1 in the supplemental material). Consistently, two fragments (residues K549 to R868 and R814 to S1128) from the TMV replication protein interact with each other in a yeast two-hybrid assay (13).
Fig 1.
Structure of ToMV-Hel. (A) ToMV-Hel domain structure. (B) Ribbon diagrams of the ToMV-Hel crystal structure before and after a 180° rotation around a y axis. Domains 1A (V802 to R964) and 2A (T965 to Q1116) and the N-terminal domain (S666 to V801) are colored green, blue, and yellow, respectively. Sulfates are represented as space-filling models, with the oxygen and sulfate atoms colored red and yellow, respectively.
Fig 2.
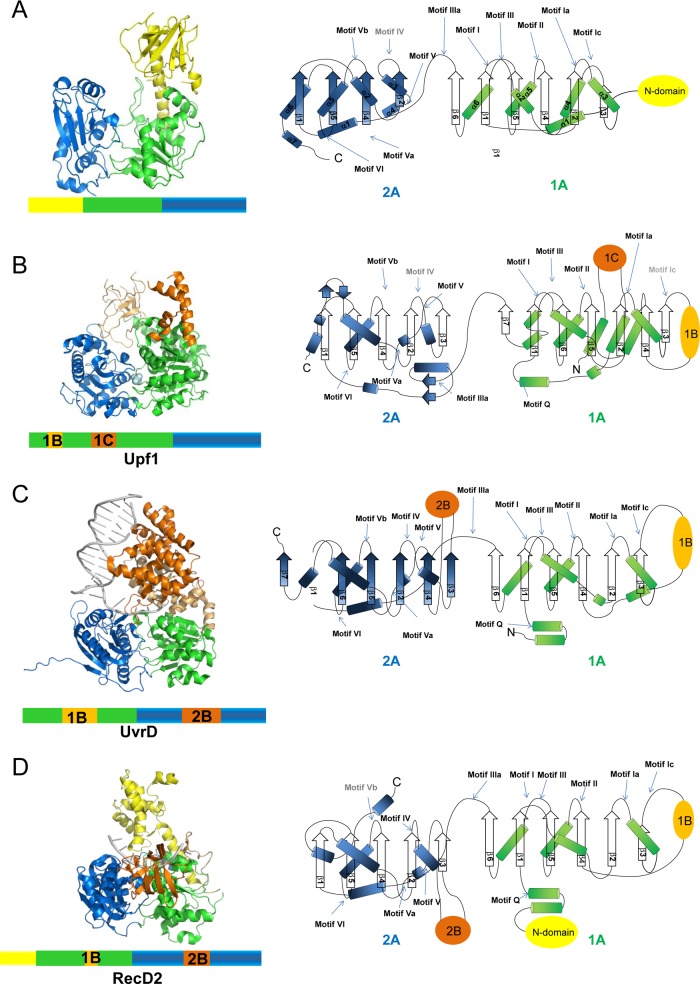
Comparison of the ToMV-Hel structure with those of nonviral SF1 helicases. (Left) The three-dimensional structures shown as ribbon diagrams. Domains 1A and 2A, which form the helicase cores, are colored green and blue, respectively. N-terminal domains are shown in yellow. Accessory domains are shown in light orange and orange. (Right) Topology diagrams of the helicase core domains. DNAs are shown in gray. The positions of the 12 conserved motifs defined by Fairman-Williams and colleagues (9) are marked. The sequences for the motifs labeled in gray are absent in the corresponding proteins. α-helices and β-sheets are shown as cylinders and arrows, respectively. The representative structures of the three nonviral SF1 helicases that have the highest Z scores against ToMV-Hel core domain in the DALI (15) conservation list (see Table S3 in the supplemental material) are shown in panels B, C, and D. (A) ToMV-Hel. (B) Upf1 (PDB code 2GK7; Z score, 21.7; root mean square deviation [RMSD], 3.3 Å). (C) UvrD (PDB code 2IS1; Z score, 18.4; RMSD, 3.1 Å). (D) RecD2 (PDB code 3GPL; Z score, 15.2; RMSD, 3.7 Å). The diagrams were created using PyMol (http://pymol.org/pymol).
Structure comparison of ToMV-Hel by the DALI server (15) showed that the ToMV-Hel helicase core is most similar to those of Upf1-like, UvrD/Rep, and Pif1-like helicases. These helicases are members of three different nonviral SF1 helicase families as defined by their structures and sequential placements of their accessory domains (32) (Fig. 2; see Table S3 in the supplemental material). All previously characterized SF1 helicases have accessory domains, which are inserted in domains 1A and/or 2A (Fig. 2). Upf1, which is a member of the Upf1-like family, contains two accessory domains, 1B and 1C, situated in 1A. Domain 1B is a six-stranded antiparallel β-barrel, and domain 1C is α-helical (5). In UvrD, which is a member of the Uvr/Rep family, there are two large insertions, one found in 1A (domain 1B) and the second in 2A (domain 2B). Domains 1B and 2B are composed of α-helices and act as a wedge to destabilize double-stranded nucleic acid (27). RecD2, which is a member of the Pif1-like family, contains three accessory domains, an N-terminal domain, and domains 1B and 2B. Domain 1B is a rigid β-hairpin turn, and domain 2B has an SH3-type fold (40). These accessory domains are spatially located over the nucleic-acid-binding sites of the helicase cores (19), and this arrangement encloses the bound nucleic acid strand. In contrast, no insertions are found in the helicase core of ToMV-Hel. Instead, ToMV-Hel has an N-terminal domain. The N-terminal domain of the ToMV-Hel forms a novel fold not represented by the aforementioned families.
SF1 helicases contain at least 12 characteristic sequence motifs at defined positions in their helicase cores (9). However, it was difficult to identify all the helicase motifs in viral SF1 helicases only through comparison of amino acid sequences because of low sequence conservation. The three-dimensional structure of ToMV-Hel allowed easy identification of helicase motifs by comparing it with the previously determined structure of SF1 helicases. The positions and sequences of these motifs in ToMV and other SF1 helicases are summarized in Table 2. Motifs I, II, and VI, which mediate binding to and hydrolysis of NTPs, are well conserved. Motif Q forms part of the α-helix in the N-terminal region of domain 1A, and the glutamine side chain in the motif interacts with the adenine base of the ATP analog in nonviral SF1 helicase-ATP analog complexes (Fig. 2 and 3) (5, 47). However, ToMV-Hel does not contain a Q motif or an α-helix at the corresponding position in the crystal structure (Fig. 2 and Table 2). The tyrosine residue in motif IIIa of nonviral SF1 helicases provides a stacking platform for the adenine base of the ATP analog, but this tyrosine residue is not found in the ToMV-Hel sequence or structure (Table 2 and Fig. 3). Motifs III and Va, which contribute to the binding of NTP and nucleic acids, are conserved in ToMV-Hel (Table 2). Motifs Ia, Ic, IV, V, and Vb are located on the face opposite the ATP-binding and nucleic acid-binding sites in other helicases (Fig. 2 and 4). However, except motif V, the ToMV-Hel sequences at the positions of those motifs show low similarity (Table 2).
Table 2.
Conserved helicase motifs in SF1 helicases


Helicase motifs as defined by Fairman-Williams et al. (9).
Conserved and conservatively replaced residues are highlighted in green and yellow, respectively. Gray shading indicates that there are no consensus amino acids, but they are at the same positions as other proteins.
Fig 3.
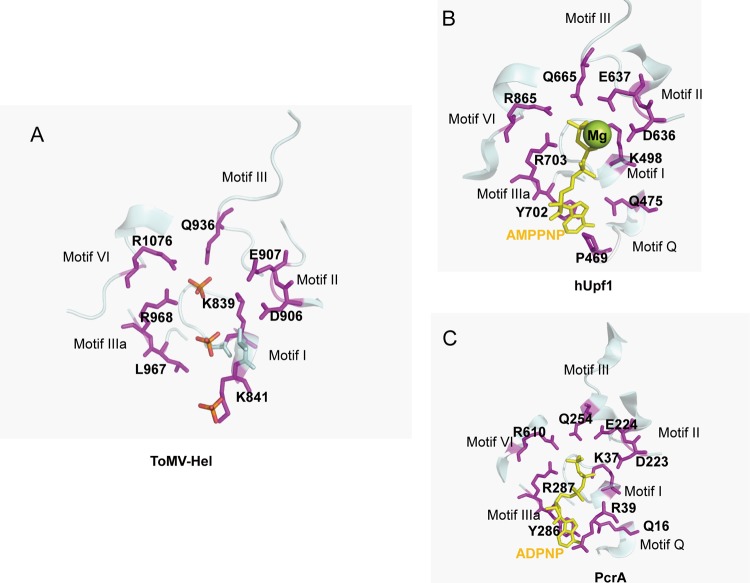
NTP-binding sites. (A) The putative amino acid residues comprising the NTPase active site in the ToMV-Hel–sulfate complex structure. (B) The AMPPNP interaction site in hUpf1 (PDB code 2GJK) (5). (C) The ADPNP interaction site in PcrA (PDB code 3PJR) (47). The structures are shown as ribbon diagrams. The residues that interact with the sulfates in ToMV-Hel, with AMPPNP in hUpf1, and with ADPNP in PcrA are shown as magenta stick models and are labeled by residue number. AMPPNP and ADPNP are shown as a yellow stick models. The diagrams were created using PyMol.
Fig 4.
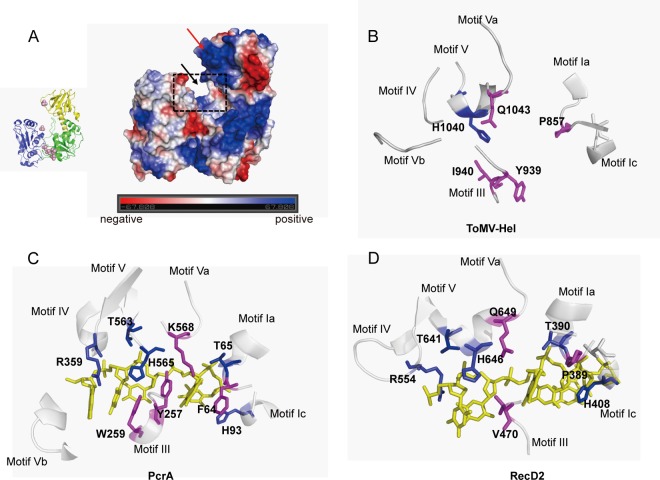
Putative RNA-binding sites. (A) Electrostatic surface of the ToMV-Hel structure. Positive, negative, and uncharged residues are shown in blue, red, and white, respectively. The putative RNA-binding site is a shallow groove indicated by the black arrow. The N-terminal-domain β-sheet, which is positively charged, is indicated by the red arrow. (B) The positions of the ToMV amino acid residues comprising the putative RNA-binding site are structurally homologous to those found in nonviral SF1 helicase motifs that interact with DNA. (C) Interactions between PcrA and DNA in the PcrA binary complex (PDB code 2PJR). (D) Interactions between RecD2 and DNA in the RecD2 binary complex (PDB code 3GP8) (40). Conserved and nonconserved residues that recognize DNA or RNA are shown as blue and magenta stick models, respectively. DNA is shown as a yellow stick model in panels C and D. The diagrams were created using PyMol.
The NTP-binding site of ToMV-Hel.
As the structure of the ToMV-Hel core domain is very similar to those of nonviral SF1 helicases, recognition of NTPs by the 130K replication protein likely involves a mechanism similar to those of nonviral SF1 helicases. Although we crystallized ToMV-Hel in the presence of ATPγS, ATPγS was not observed in the crystal structure. Instead, sulfates, which were included in the crystallization solution as a precipitant, are found in the deep cleft formed by domains 1A and 2A (Fig. 3A). ToMV-Hel residues Q936 in motif III, R968 in motif IIIa, and R1076 in motif VI directly contact one of these sulfates (Fig. 3A). The position of this sulfate corresponds to those of the 5′-adenylyl-β,γ-imidodiphosphate (ADPNP) and adenylyl imidodiphosphate (AMPPNP) γ-phosphates in the PcrA-ADPNP and hUPf1-AMPPNP complexes, respectively (Fig. 3B and C). The ToMV-Hel K839 in motif I directly contacts a second sulfate also positioned deep within the cleft (Fig. 3A). This sulfate position corresponds to those of the ADPNP and AMPPNP β-phosphates in the PcrA-ADPNP and in the hUPf1-AMPPNP complexes, respectively (Fig. 3B and C). Notably, Wang and colleagues reported that amino acid substitutions in the TMV helicase at positions equivalent to K839 in motif I and R1076 in motif VI in the ToMV 130K replication protein disrupt ATPase activity (48).
The aspartic and glutamic acid residues in motif II are highly conserved among SF1 helicase sequences (Fig. 3B and C). The ToMV sequence also contains the aspartic and glutamic acid residues (D906 and E907), and they are positioned in a spatial arrangement similar to those in nonviral SF1 helicases (Fig. 3A). These residues coordinate an Mg2+ that is essential for NTP hydrolysis. As noted above, ToMV-Hel does not have a Q motif or the tyrosine residue in motif IIIa, which are important for the binding of the ATP adenine base. The absence of substrate specificity found for the hydrolysis of NTPs by ToMV-Hel (49) may be a consequence of the lack of a Q motif and conserved tyrosine residue.
Possible RNA-binding site in ToMV-Hel.
A fragment of TMV replication protein containing the helicase domain has been shown to bind RNA (48), and it is therefore reasonable to assume that the ToMV-Hel binds RNA. The β-sheet of the N-terminal domain of ToMV-Hel is located over the putative nucleic-acid-binding site (Fig. 4), which is topologically similar to the positions of accessory domains in nonviral SF1 helicases. Furthermore, the net charge on the β-sheet surface is positive (Fig. 4A), which has been found for the RNA-binding surfaces of nonviral SF1 helicases. Therefore, the surface of this β-sheet might interact with RNA.
Saikrishnan and colleagues reported that the modes of interaction between DNA and different SF1 helicases are not conserved, as those for Rep and PcrA mainly involve aromatic side chains stacked against the DNA bases whereas those for RecD2 mainly involve ionic pairs formed by cationic side chains in the protein and the phosphates of the DNA backbone (Fig. 4C and D) (40). It was interesting, therefore, to compare residues in these regions in the SF1 helicases studied by Saikrishnan and colleagues with those of ToMV-Hel. For PcrA, F64 (motif Ia) regulates access to the pocket by the translocating base in response to ATP binding and hydrolysis. In RecD2 and ToMV replication protein, the corresponding positions contain a proline residue (P389 in RecD2 and P857 in ToMV replication protein). Furthermore, the residues that correspond to W259 in PcrA (motif III), which binds the bases of single-stranded DNA (ssDNA), are nonpolar (V470 in RecD2 and I940 in ToMV replication protein) (Fig. 4B). The backbone positions are similar for Y939 in ToMV-Hel and Y257 in PcrA (motif III), which stacks against two of the bound ssDNA bases, even though their side chain conformations are different. Between PcrA and RecD2, other DNA-recognizing residues, e.g., the threonine in motif Ia, the histidine in motif Ic, the arginine in motif IV, and the threonine and histidine in motif V, are well conserved (Fig. 4C and D). However, these residues are not conserved in the sequence of ToMV-Hel, with the exception of the histidine in motif V. Taken together, the similarities and differences in the motif sequences between the nonviral SF1 helicases and ToMV-Hel suggest that ToMV-Hel might recognize the bases and backbone phosphates of RNA via a mechanism(s) that is different from those of nonviral SF1 helicases. To clarify how ToMV-Hel recognizes RNA, the structure of a ToMV-Hel-RNA complex should be solved.
Locations of previously characterized mutations in the helicase domains of tobamovirus replication proteins.
The helicase domains of tobamovirus replication proteins interact with many host proteins (17). Some residues involved in these interactions have been identified (16, 36, 46). In Fig. 5, we have highlighted the residues that correspond to the residues known to be involved in the interactions with host proteins.
Fig 5.
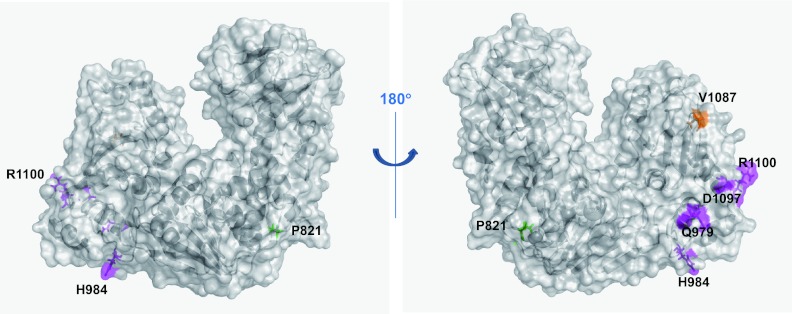
Locations of previously characterized mutations in tobamovirus helicase domains. The residues that are important for interaction with the N protein (46), with PAP1 (36), and with Tm-1 (31, 44) are shown as green, orange, and magenta stick and surface models and are labeled by residue name and number.
The helicase domain polypeptide of TMV (residues R814 to G1211) interacts with the Aux/IAA protein PAP1 (IAA26) from Arabidopsis thaliana and tomato, a putative regulator of auxin response genes involved in plant development. The TMV V1087I helicase mutant has a decreased affinity for PAP1 (36). The solvent-accessible V1087 is positioned in the loop that connects β5 and α6 in domain 2A, which suggests that the surface region near or surrounding V1087 interacts with PAP1.
The tomato Tm-1 encodes a protein that inhibits ToMV RNA replication by binding to the replication proteins (16). ToMV mutants capble of multiplying in Tm-1-containing tomato plants have mutations in their replication proteins, e.g., Q979E and H984Y (31) or D1097V and R1100Q (44). Both sets of mutations abolish the interaction between the replication proteins and Tm-1 (16) (K. Ishibashi and M. Ishikawa, unpublished results). Q979 is positioned in α1 of domain 2A, H984 is positioned in the loop between α1 and β1 of domain 2A, and D1097 and R1100 are positioned in α6 of domain 2A. Despite the relatively large separations between these two sets of residues in the replication protein amino acid sequence, the four residues are in close spatial proximity on the surface of domain 2A in ToMV-Hel, suggesting that Tm-1 targets this surface region.
Tobacco plants harboring the N gene elicit hypersensitive responses (HR) when infected with TMV. The HR is also induced by the expression of p50 (residues E672 to Q1116), which is a helicase domain-containing fragment from the TMV replication proteins; however, deletion derivatives of p50 (residues A733 to Q1116 or M692 to K1082) are ineffective (8). Ueda and colleagues reported that p50 directly interacts with the N protein, whereas a p50 mutant (P821L) does not (46). P821 is positioned in the domain 1A loop between α1 and β1 and is at the interface formed by the N-terminal domain and domain 1A (Fig. 5). P821 is surrounded by the hydrophobic residues V703, M706, and I710 of the long α-helix in the N-terminal domain and L815, L816, and V823 in the loop between α1 and β1 in domain 1A. We speculate that the N protein recognizes a TMV-Hel region containing the N-terminal domain and that the structure of this region cannot be maintained in the P821L mutant and the P50 deletion derivatives.
The N-terminal domain of ToMV-Hel interacts with host proteins.
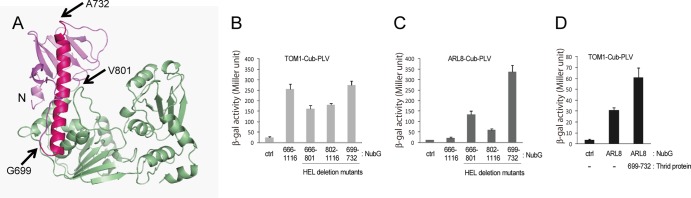
We previously demonstrated, using a split-ubiquitin-based yeast two-hybrid assay, that the host seven-pass transmembrane protein TOM1 and small GTP-binding protein ARL8, which are essential for ToMV RNA replication, interact with a fragment (residues M648 to Q1116) of the ToMV replication protein that contains the helicase domain (34, 43). Because replication protein mutants that cannot bind TOM1 or ARL8 are not available, we designed deletion mutants of ToMV-Hel, considering its three-dimensional structure, and used them to determine which region(s) of ToMV-Hel binds these host proteins by the split-ubiquitin system. The results suggested that TOM1 interacts with both the N-terminal domain (S666 to V801) and the helicase core (V802 to Q1116) and that ARL8 strongly interacts with the N-terminal domain (Fig. 6A to C). However, the signal arising from the interaction of ARL8 and ToMV-Hel was weak, suggesting that the helicase core sterically hinders the binding of ARL8 to the N-terminal domain. Because in vivo ARL8 strongly binds the full-length 130K replication protein (34), conformational changes in ToMV-Hel that ameliorate steric hindrance may be responsible for the in vivo association and/or additional factors might be involved in ToMV replication protein-ARL8 association in infected plant cells. Additional assays suggested that the fragment (G699 to A732) that contains the long α-helix could interact with TOM1 and ARL8 (Fig. 6B and C) and that the interaction between TOM1 and ARL8 was facilitated by the presence of the long α-helix (Fig. 6D). Therefore, the α-helix probably binds TOM1 and ARL8 simultaneously, which also suggests that the two host proteins interact with distinct sites on the α-helix. These results are consistent with our previous finding that TOM1 copurifies with ARL8 more efficiently from ToMV-infected tobacco cells than from uninfected tobacco cells (34).
Fig 6.
Interactions between the ToMV helicase domain and host proteins. (A) Start and stop positions of the ToMV-Hel deletion mutants used in the split-ubiquitin-based yeast two-hybrid assay. (B) Interactions between TOM1 and ToMV-Hel deletion mutants assessed by the split-ubiquitin-based yeast two-hybrid assay. (C) Interaction between ARL8 and ToMV-Hel deletion mutants assessed by the split-ubiquitin-based yeast two-hybrid assay. (D) Effect of the long α-helix from the ToMV-Hel N-terminal domain on the interaction of TOM1 with ARL8 as assayed by the split-ubiquitin-based yeast three-hybrid system. Averages and standard deviations of β-galactosidase activity (Miller units) for three or four independent yeast transformants are shown in panels B to D.
Comparison of the ToMV-Hel structure with those of other viral helicases.
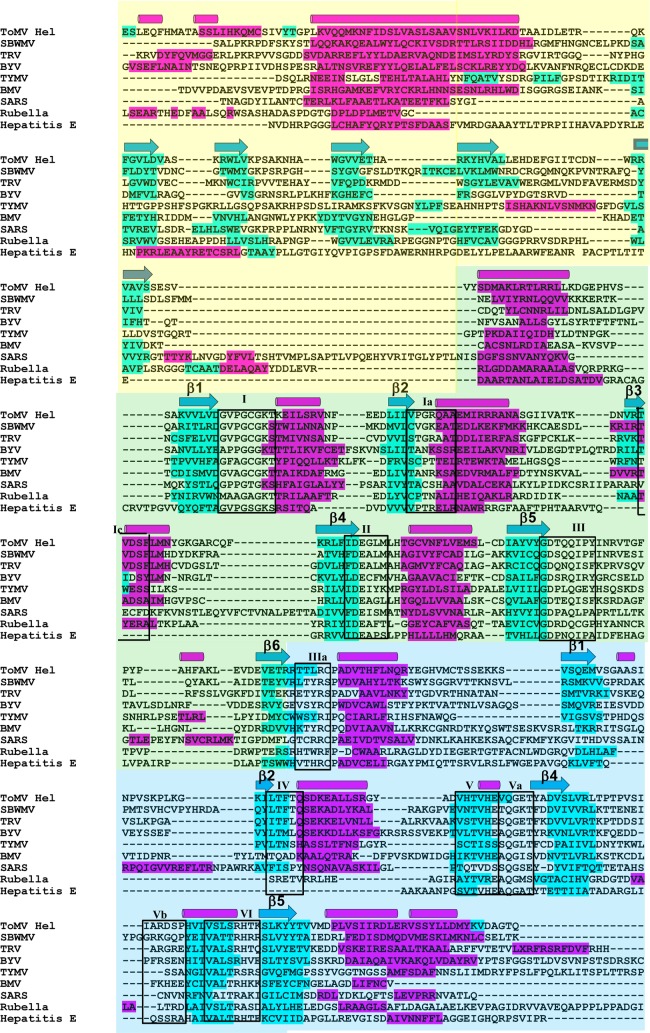
We have determined the first viral SF1 helicase structure and, by doing so, have found an α-helix that is important for the association of the helicase and host factors. Therefore, it is interesting to compare the secondary structural elements of ToMV-Hel with those predicted for other viral SF1 helicases. To investigate this, the secondary structures of SF1 helicases of eight positive-strand RNA viruses were predicted using Jpred (6), and then the sequences were aligned according to their secondary structures. For ToMV-Hel, the secondary structures predicted by Jpred agree with those in the crystal structure, with the exceptions of the sequences for the 4th and 5th α-helices in domain 2A, which were predicted by Jpred to be β-strands. The alignment of their sequences indicates that none of the viral helicases contain inserted accessory domains, which allows easy identification of SF1-type helicase motifs (Fig. 7). For the N-terminal domain sequences examined, with a few exceptions, the long α-helix and β-sheet sequences were predicted by Jpred in positions similar to those in ToMV-Hel, even though the sequences of the N-terminal domains are not similar. Because the host factors involved in viral-RNA replication vary from one virus to another (23, 25, 38), and because TOM1 and ARL8 are specifically required for RNA replication of ToMV and related viruses belonging to the genus Tobamovirus (18, 34, 53), the N-terminal accessory domains, and especially their conserved long α-helices, may act as customized scaffolds for the different sets of host factors.
Fig 7.
Sequence alignment of the helicase N-terminal and core domains from the replication proteins of positive-strand RNA viruses. Secondary-structure predictions were made by Jpred (6) for the viral SF1 helicases, and then the nine sequences were manually aligned according to their secondary structures and sequence similarities. The sequences that form β-strands and α-helices in the ToMV-Hel crystal structure are shown above the sequences as blue arrows and magenta cylinders, respectively. The sequences predicted by Jpred to form β-strands and α-helices are highlighted in blue and magenta, respectively. The 11 helicase motifs that are conserved in all of the sequences are boxed. SBWMV, Soilborne wheat mosaic virus (Furovirus, Virgaviridae); TRV, Tobacco rattle virus (Tobravirus, Virgaviridae); BYV, Beet yellows virus (Closterovirus, Closteroviridae); TYMV, Turnip yellow mosaic virus (Tymovirus, Tymoviridae); BMV, Brome mosaic virus (Bromovirus, Bromoviridae); SARS, SARS coronavirus (Coronavirus, Coronaviridae); Rubella, Rubella virus (Rubivirus, Togaviridae); Hepatitis E, Hepatitis E virus (Hepevirus, Hepeviridae).
Supplementary Material
ACKNOWLEDGMENTS
This work was performed at the Spring-8 BL38B1 and BL41XU beamlines with the approval of the Japan Synchrotron Radiation Research Institute (proposals 2011A2031 and 2011A6640) and at the KEK Photon Factory BL17A and BL5A beamlines with the approval of the Photon Factory Advisory Committee (proposals 2011P005 and 2009G129).
This work was supported by grants from the Program for Promotion of Basic Research Activities for Innovative Biosciences of Japan to M.I. and E.K. and by Core Research for Evolution Science and Technology (CREST) grants to S.S., T.I., and H.M.
Footnotes
Published ahead of print 9 May 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Abdelhaleem M. 2010. Helicases: an overview. Methods Mol. Biol. 587:1–12 [DOI] [PubMed] [Google Scholar]
- 2. Adams PD, Pannu NS, Read RJ, Brunger AT. 1997. Cross-validated maximum likelihood enhances crystallographic simulated annealing refinement. Proc. Natl. Acad. Sci. U. S. A. 94:5018–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amberg DC, Burke DJ, Strathern JN. 2005. Methods in yeast genetics, p 161–164 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 4. Buck KW. 1999. Replication of tobacco mosaic virus RNA. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354:613–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheng Z, Muhlrad D, Lim MK, Parker R, Song H. 2007. Structural and functional insights into the human Upf1 helicase core. EMBO J. 26:253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cole C, Barber JD, Barton GJ. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36:W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 8. Erickson FL, et al. 1999. The helicase domain of the TMV replicase proteins induces the N-mediated defence response in tobacco. Plant J. 18:67–75 [DOI] [PubMed] [Google Scholar]
- 9. Fairman-Williams ME, Guenther UP, Jankowsky E. 2010. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 20:313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goelet P, et al. 1982. Nucleotide sequence of tobacco mosaic virus RNA. Proc. Natl. Acad. Sci. U. S. A. 79:5818–5822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gorbalenya AE, Koonin EV. 1993. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3:419–429 [Google Scholar]
- 12. Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. 1989. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 17:4713–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goregaoker SP, Lewandowski DJ, Culver JN. 2001. Identification and functional analysis of an interaction between domains of the 126/183-kDa replicase-associated proteins of tobacco mosaic virus. Virology 282:320–328 [DOI] [PubMed] [Google Scholar]
- 14. Hickman AB, Dyda F. 2005. Binding and unwinding: SF3 viral helicases. Curr. Opin. Struct. Biol. 15:77–85 [DOI] [PubMed] [Google Scholar]
- 15. Holm L, Rosenstrom P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38:W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ishibashi K, Masuda K, Naito S, Meshi T, Ishikawa M. 2007. An inhibitor of viral RNA replication is encoded by a plant resistance gene. Proc. Natl. Acad. Sci. U. S. A. 104:13833–13838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ishibashi K, Nishikiori M, Ishikawa M. 2010. Interactions between tobamovirus replication proteins and cellular factors: their impacts on virus multiplication. Mol. Plant Microbe Interact. 23:1413–1419 [DOI] [PubMed] [Google Scholar]
- 18. Ishikawa M, Obata F, Kumagai T, Ohno T. 1991. Isolation of mutants of Arabidopsis thaliana in which accumulation of tobacco mosaic virus coat protein is reduced to low levels. Mol. Gen. Genet. 230:33–38 [DOI] [PubMed] [Google Scholar]
- 19. Jankowsky E. 2011. RNA helicases at work: binding and rearranging. Trends Biochem. Sci. 36:19–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kadare G, Haenni AL. 1997. Virus-encoded RNA helicases. J. Virol. 71:2583–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim JL, et al. 1998. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6:89–100 [DOI] [PubMed] [Google Scholar]
- 22. Kim JL, et al. 1996. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87:343–355 [DOI] [PubMed] [Google Scholar]
- 23. Krishnan MN, et al. 2008. RNA interference screen for human genes associated with West Nile virus infection. Nature 455:242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kubota K, Tsuda S, Tamai A, Meshi T. 2003. Tomato mosaic virus replication protein suppresses virus-targeted posttranscriptional gene silencing. J. Virol. 77:11016–11026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kushner DB, et al. 2003. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc. Natl. Acad. Sci. U. S. A. 100:15764–15769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. Procheck—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291 [Google Scholar]
- 27. Lee JY, Yang W. 2006. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 127:1349–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. LeMaster DM, Richards FM. 1985. 1H–15N heteronuclear NMR studies of Escherichia coli thioredoxin in samples isotopically labeled by residue type. Biochemistry 24:7263–7268 [DOI] [PubMed] [Google Scholar]
- 29. Love RA, et al. 1996. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell 87:331–342 [DOI] [PubMed] [Google Scholar]
- 30. Luo D, et al. 2008. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 27:3209–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meshi T, et al. 1988. Two concomitant base substitutions in the putative replicase genes of tobacco mosaic virus confer the ability to overcome the effects of a tomato resistance gene, Tm-1. EMBO J. 7:1575–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Montpetit B, et al. 2011. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature 472:238–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morris RJ, Perrakis A, Lamzin VS. 2003. ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 374:229–244 [DOI] [PubMed] [Google Scholar]
- 34. Nishikiori M, et al. 2011. A host small GTP-binding protein ARL8 plays crucial roles in Tobamovirus RNA replication. PLoS Pathog. 7:e1002409 doi:10.1371/journal.ppat.1002409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. A 276:307–326 [DOI] [PubMed] [Google Scholar]
- 36. Padmanabhan MS, Goregaoker SP, Golem S, Shiferaw H, Culver JN. 2005. Interaction of the tobacco mosaic virus replicase protein with the Aux/IAA protein PAP1/IAA26 is associated with disease development. J. Virol. 79:2549–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Padmanabhan MS, Kramer SR, Wang X, Culver JN. 2008. Tobacco mosaic virus replicase-auxin/indole acetic acid protein interactions: reprogramming the auxin response pathway to enhance virus infection. J. Virol. 82:2477–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Panavas T, Serviene E, Brasher J, Nagy PD. 2005. Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 102:7326–7331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pape T, Schneider TR. 2004. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr. 37:843–844 [Google Scholar]
- 40. Saikrishnan K, Powell B, Cook NJ, Webb MR, Wigley DB. 2009. Mechanistic basis of 5′-3′ translocation in SF1B helicases. Cell 137:849–859 [DOI] [PubMed] [Google Scholar]
- 41. Sheldrick GM. 2008. A short history of SHELX. Acta Crystallogr. A 64:112–122 [DOI] [PubMed] [Google Scholar]
- 42. Singleton MR, Dillingham MS, Wigley DB. 2007. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76:23–50 [DOI] [PubMed] [Google Scholar]
- 43. Stagljar I, Korostensky C, Johnsson N, te Heesen S. 1998. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. U. S. A. 95:5187–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Strasser M, Pfitzner AJ. 2007. The double-resistance-breaking Tomato mosaic virus strain ToMV1-2 contains two independent single resistance-breaking domains. Arch. Virol. 152:903–914 [DOI] [PubMed] [Google Scholar]
- 45. Tsujimoto Y, et al. 2003. Arabidopsis TOBAMOVIRUS MULTIPLICATION (TOM) 2 locus encodes a transmembrane protein that interacts with TOM1. EMBO J. 22:335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ueda H, Yamaguchi Y, Sano H. 2006. Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Mol. Biol. 61:31–45 [DOI] [PubMed] [Google Scholar]
- 47. Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. 1999. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97:75–84 [DOI] [PubMed] [Google Scholar]
- 48. Wang X, Kelman Z, Culver JN. 2010. Helicase ATPase activity of the Tobacco mosaic virus 126-kDa protein modulates replicase complex assembly. Virology 402:292–302 [DOI] [PubMed] [Google Scholar]
- 49. Xiang H, et al. 2012. Expression, purification, and functional characterization of a stable helicase domain from a tomato mosaic virus replication protein. Protein Expr. Purif. 81:89–95 [DOI] [PubMed] [Google Scholar]
- 50. Xiang H, et al. 2011. Crystallization and preliminary X-ray crystallographic analysis of a helicase-like domain from a tomato mosaic virus replication protein. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 67:1649–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu T, et al. 2005. Structure of the Dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 A. J. Virol. 79:10278–10288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamanaka T, et al. 2002. Complete inhibition of tobamovirus multiplication by simultaneous mutations in two homologous host genes. J. Virol. 76:2491–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yamanaka T, et al. 2000. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc. Natl. Acad. Sci. U. S. A. 97:10107–10112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.