Abstract
Phylogenetic relatedness and cocirculation of several major human pathogen flaviviruses are recognized as a possible cause of deleterious immune responses to mixed infection or immunization and call for a greater understanding of the inter-Flavivirus protein homologies. This study focused on the identification of human leukocyte antigen (HLA)-restricted West Nile virus (WNV) T-cell ligands and characterization of their distribution in reported sequence data of WNV and other flaviviruses. H-2-deficient mice transgenic for either A2, A24, B7, DR2, DR3, or DR4 HLA alleles were immunized with overlapping peptides of the WNV proteome, and peptide-specific T-cell activation was measured by gamma interferon (IFN-γ) enzyme-linked immunosorbent spot (ELISpot) assays. Approximately 30% (137) of the WNV proteome peptides were identified as HLA-restricted T-cell ligands. The majority of these ligands were conserved in ∼≥88% of analyzed WNV sequences. Notably, only 51 were WNV specific, and the remaining 86, chiefly of E, NS3, and NS5, shared an identity of nine or more consecutive amino acids with sequences of 64 other flaviviruses, including several major human pathogens. Many of the shared ligands had an incidence of >50% in the analyzed sequences of one or more of six major flaviviruses. The multitude of WNV sequences shared with other flaviviruses as interspecies variants highlights the possible hazard of defective T-cell activation by altered peptide ligands in the event of dual exposure to WNV and other flaviviruses, by either infection or immunization. The data suggest the possible preferred use of sequences that are pathogen specific with minimum interspecies sequence homology for the design of Flavivirus vaccines.
INTRODUCTION
West Nile virus (WNV), a member of the genus Flavivirus, is a mosquito-borne RNA pathogen closely related to numerous other flaviviruses of the Japanese encephalitis (JE) group, and to a lesser degree to Dengue virus (DENV), Yellow fever virus (YFV), and several other flaviviruses of about 70 different species (5, 18). The genome is a positive-sense, single-stranded RNA of approximately 10,293 nucleotides encoding a polyprotein of approximately 3,430 amino acids (aa) that is cleaved to produce three structural proteins, capsid (C), precursor membrane (prM), and envelope (E), and seven nonstructural (NS) proteins, NS1, 2a, 2b, 3, 4a, 4b, and 5 (24, 37). Originally isolated in Africa in 1937 (62), WNV has become an increasingly important human pathogen widely endemic in Africa, Asia, Europe, and North America and a significant agent of viral encephalitis (28, 38, 45). WNV and several of the major human pathogen flaviviruses are known to cocirculate (28, 71); for example, WNV and St. Louis encephalitis virus (LEV) in North America; WNV, DENV, and Japanese encephalitis virus (JEV) around the Indian subcontinent and portions of Southeast Asia (SEA); and WNV, DENV, JEV, and Murray Valley encephalitis virus (MVE) in neighboring pockets of SEA and Australasia.
The widespread colocalization of WNV with other flaviviruses calls for a greater understanding of the phylogenetic relatedness and possible pathophysiologic associations of flaviviruses. We previously reported a large-scale analysis of the conservation and variability of overlapping nonamers, the typical length of human leukocyte antigen (HLA) class I or class II binding cores of T-cell epitopes (47), of the 2,746 WNV protein sequences collected from the NCBI Entrez Protein Database (17). Notably, of the 88 completely conserved sequence fragments identified, representing 34% of the WNV proteome, 67 were also present in many other flaviviruses with identities of nine or more amino acids. These sequence homologies called attention to a broad inter-Flavivirus risk of altered peptide ligands (APL) (58), T-cell epitopes with one or more amino acid differences, that possibly would result in modified immune responses in the event of exposure to multiple Flavivirus pathogens.
This study focused on analyses of the diversity and presence in other flaviviruses of WNV peptide sequences that are HLA restricted and T-cell receptor (TCR) engaged (T-cell ligands). Several reports cite the role of CD8+ cytolytic T lymphocytes (CTL) and CD4+ helper T lymphocytes (HTL) in the immune response to WNV infection (3, 8, 42, 46); however, knowledge of the identities and properties of WNV T-cell epitopes in the human host is limited, and few have been reported to date (19–21, 32, 42, 61). Thus, for this study, large-scale identification of WNV T-cell ligands was performed by use of HLA transgenic (Tg) mice, a leading animal model system for analysis of HLA-restricted T-cell responses to human pathogens (4, 12, 25, 40, 41, 43, 49, 63, 65). Six mice transgenic for prototype HLA proteins, 3 class I and 3 class II, were immunized with 452 peptides covering the entire WNV proteome and peptide-specific T-cell responses were assayed by gamma interferon (IFN-γ) enzyme-linked immunosorbent spot (ELISpot) assay. Characterization of the identified sequences included their evolutionary conservation and variability in reported sequence data of WNV and other flaviviruses. The results provide details of protein sequence homologies of flaviviruses that are relevant to possible consequences of Flavivirus infection and immunization and to the design of a new generation of vaccines that avoid interspecies protein homologies.
MATERIALS AND METHODS
Ethics statement.
This study was performed in strict accordance to the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by The Johns Hopkins University (JHU) Institutional Animal Care & Use Committee (MO06M106).
HLA transgenic mice.
WNV T-cell ligand sequences were identified by use of HLA transgenic mice, a leading animal model system for analysis of HLA-restricted T-cell responses to human pathogens (4, 12, 25, 40, 41, 43, 49, 63, 65), as follows: H-2 class II deficient, HLA-DR2 (DRB1*1501) (68), HLA-DR3 (DRB1*0301) (29, 64), and HLA-DR4 (DRB1*0401)/human CD4 (huCD4) (6, 10); and H-2 class I deficient, HLA-A2.1 (A*0201) (HHD monochain) (44), HLA-A24 (A*2402)/huCD8α (F. Lemonnier unpublished data), and HLA-B7 (B*0702) (51). HLA-DR2 transgenic mice express chimeric molecules, with α1 and β1 domains encoded by the HLA-DRA1*0101 and -DRB1*1501 sequences and the other domains encoded by I-Eα and I-Eβ sequences from the I-E promoters (68). HLA-DR3 transgenic mice express the full-length HLA-DRA1*0101 and -DRB1*0301 sequences (29, 64). HLA-DR4/huCD4 transgenic mice express the full-length HLA-DRA1*0101 and -DRB1*0401 sequences from the I-Eα promoter and the human CD4 sequence from the murine CD3δ promoter (6, 10). HLA-DR2 and -DR3 mice have a homologous deletion of the murine H-2 class II region, and HLA-DR4/huCD4 mice are deficient for I-Aβ and I-Eα. HLA-DR2 mice have a predominant C57BL/6 background, and HLA-DR3 and -DR4/huCD4 mice have mixed backgrounds (B6, B10.H2b, and DBA/1J, 129/Sv, C57BL/6, respectively). HLA-A2.1 (HHD) transgenic mice express a chimeric monochain containing the HLA-A*0201 α1 and α2 domains and the murine H-2Db α3 domain linked to human β2-microglobulin (huβ2-m) from the HLA-A2.1 promoter and are deficient for H-2D and murine β2-m (mβ2-m) (44). HLA-A24/huCD8α mice express the full-length HLA-A*2402, huβ2-m, and huCD8α sequences and are deficient for H-2K, H-2D, and mβ2-m (Lemonnier et al., unpublished). HLA-B7 mice express a chimeric heavy chain with the HLA-B*0702 α1 and α2 domains and the H-2Kd α3 domain from the HLA-B7 promoter and are deficient for H-2K and H-2D (51). The three HLA class I transgenic strains have been backcrossed for 6 to 12 generations on the C57BL/6 genetic background (F. Lemonnier, Pasteur Institute). Animals were bred and maintained at the Johns Hopkins School of Medicine Research Animal Resources facilities. Specific-pathogen-free (SFP) transgenic mice were derived through iodine immersion of neonates (<1 day old) and transfer to outbred foster females (K. Thompson and J. Watson, The Johns Hopkins School of Medicine).
Synthetic WNV peptides.
A library of 452 overlapping peptides (see Table S6A to D in the supplemental material) covering the entire WNV proteome (NY99-flamingo 382-99 strain), each 12 to 20 amino acids in length with an overlap of 9 to 11 residues (mainly of 10) (>80% purity), was obtained as lyophilized powders from the Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH (BEI Resources). Each peptide was dissolved in 100% dimethyl sulfoxide (DMSO) and constituted to 20% with sterile filtered water. The final concentration of each peptide was 2 μg/μl. Dissolved peptides were stored at −20°C. Multiple studies have shown that peptides of these lengths are effective as immunogens (2, 9, 20).
Large-scale peptide pool immunization.
The 6 HLA transgenic mice were each immunized with the WNV peptides by use of a peptide pool protocol for large-scale T-cell ligand identification (50). The peptides were divided into 4 immunization pools containing 1 μg of each peptide in groups of about 100 peptides each, as follows: pool 1, 88 peptides spanning the prM and E proteins; pool 2, 107 peptides spanning the C, NS1, NS2a, and NS2b proteins; pool 3, 135 peptides spanning NS3, NS4a; and NS4b proteins; pool 4, 122 peptides spanning the NS5 protein (see Table S6A to D in the supplemental material). The criteria for identification of positive pools of peptides or individual peptides were based on the following combination of equations: (i) mean number of spots (peptides) − 2 · standard deviation (SD) > mean number of spots (background); (ii) mean number of spots (peptides) > mean number of spots (background) + 2 SD; (iii) mean number of spots (peptides) − mean number of spots (background) > 10 per million cells.
Day 1.
Each pool was mixed with 50 μl zymosan, 10 mg/ml (Sigma-Aldrich Co, St. Louis, MO) in phosphate-buffered saline (PBS) as the adjuvant and administered subcutaneously at the base of the tail to groups of 9 to 12 mice of each transgenic strain (7, 11). Mice immunized with zymosan without peptides served as negative controls.
Days 15 to 19.
After one immunization, two mice of each Tg strain on a given day were sacrificed, and their splenocytes were selectively depleted of CD8 or CD4 T cells for HLA class II and class I transgenic strains, respectively, and deconvolution analyses of the T-cell responses to the peptide pools (10 peptides, 1 μg peptide per pool) were performed by IFN-γ ELISpot assays. On the following day 2, for each Tg strain, additional mice, the number depending on the number of possible positive wells selected, were sacrificed, and the individual WNV peptide immunogens identified by the deconvolution analyses were individually tested by ELISpot assay. Experimental values for the validation assays reported herein (see Table S1 in the supplemental material) were obtained with a peptide concentration of 10 μg/ml and a minimum of 3 assays in duplicate with different immunized mice. Control mice were sacrificed on the same day.
Day 21.
The remaining mice were immunized a second time on day 21 with the original peptide pool without zymosan.
Day 35.
The mice immunized on day 21 and corresponding control mice were sacrificed, and splenocyte T-cell responses to individual WNV peptides were further assessed by ELISpot assay with peptide concentrations of 10, 1, and 0.1 μg/ml. Positive-control dengue virus peptides immunogenic in the relevant transgenic mouse were included in each immunization protocol to evaluate the responses of the individual immunized mouse.
DNA immunization.
The WNV strain NY99-flamingo 382-99 NS3 sequence (NCBI Entrez nucleotide database accession number AF196835) with NheI and KpnI sites was optimized using the Leto 1.0 software, synthesized by GeneArt Inc. (Toronto, Canada), and inserted into the p43 vector (15). A p43 vector encoding the dengue virus type 2 (strain 16681) prM and E antigens (p-DENV-prM-E) (J. Salmon, unpublished data) was used as a control. Four 6- to 8-week-old female HLA-DR2 transgenic mice were immunized subcutaneously at the base of the tail, twice at 3-week intervals, with 50 μg of the endotoxin-free DNA plasmid p-WNV-NS3, or p-DENV-prM-E. The mice were sacrificed on day 42, and the CD4 T-cell responses to the WNV NS3 peptides were assessed by IFN-γ ELISpot assays.
IFN-γ ELISpot assays.
Ex vivo IFN-γ ELISpot assays were performed using mouse IFN-γ ELISpot sets (BD Biosciences, San Jose, CA) following the manufacturer's recommendations. Briefly, 96-well ELISpot plates were coated with anti-IFN-γ antibody (5 μg/ml) by incubation at 4°C overnight and then blocked with RPMI 1640 medium containing 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, 100 μg/ml streptomycin, and 100 U penicillin, for 2 h at room temperature. Freshly isolated splenocytes from HLA class II and HLA class I transgenic mice were depleted of CD8 and CD4 T cells, respectively, using magnetic beads according to the manufacturer's protocol (Miltenyi Biotech, Auburn, CA). Flow cytometry analysis of the depleted cells indicated this method routinely achieved >95% depletion of the targeted cells. CD8- or CD4-depleted splenocytes (100 μl containing 0.5 × 106 to 1.0 × 106 cells/well) were plated together with WNV peptides. The final concentration of each peptide was 10 μg/ml in the peptide matrix pool and individual peptide validation assays, and 10, 1, and 0.1 μg/ml in the titration assays. Each peptide preparation was tested in duplicate wells. Cells plated without peptide (medium alone) served as negative controls, and concanavalin A (2.5 μg/ml; Sigma-Aldrich, St. Louis, MO) and known HLA-restricted peptides from dengue virus serotype 3 were included as positive controls. The cells were incubated at 37°C, 5% CO2 for 16 h. The plates were washed and incubated with biotinylated anti-IFN-γ antibody for 2 h at room temperature, followed by horseradish peroxidase (HRP)-conjugated streptavidin for 1 h at room temperature. Detection was performed with AEC substrate (Calbiochem, San Diego, CA) following the manufacturer's instructions. IFN-γ spot-forming cells (SFC) were counted using the Immunospot Series 3B Analyzer ELISpot reader and Immunospot software version 3.0 (Cellular Technologies, Shaker Heights, OH). Experimental values were expressed as the mean numbers of SFC/106 CD8- or CD4-depleted splenocytes ± SD, after subtraction of values from negative controls (background). Positive ELISpot responses were defined as values above 10 and above the background plus 2 SD. Each ELISpot-positive response was confirmed by three assays: by matrix screening, individually by the validation assay with the individual peptide, and by peptide titration. The apparent binding avidity of the WNV peptides to either the HLA or TCR molecules in the IFN-γ ELISpot assay was measured by titration of the peptides over a 100-fold range of concentrations (10, 1, and 0.1 μg/ml).
Prediction of putative minimal nonamer T-cell epitopes within the ELISpot-positive peptides.
The minimal T-cell epitopes within the 15 to 19 amino acids of the ELISpot-positive peptides (WNV T-cell ligands) were predicted by use of NetMHC 3.2 (http://www.cbs.dtu.dk/services/NetMHC-3.2/) (26) and NetMHCIIPan 2.0 (http://www.cbs.dtu.dk/services/NetMHCIIpan/) (39) immunoinformatics Web servers for HLA class I (A*0201, A*2402, and B*0702) and class II (DRB1*1501, DRB1*0301, and DRB1*0401) alleles, respectively. These servers were identified to be the best for the respective HLA class (22, 23). The ELISpot-positive peptides were analyzed by the servers as peptides of window size 9, overlapping by 8 aa for class I, and as peptides of window size 15, overlapping by 14 aa for class II, predicting the nonamer core. The predicted weak and strong nonamer binding epitopes of both classes were selected for further analysis.
WNV sequence data collection and processing for bioinformatics analyses.
Full-length and partial sequences of individual WNV proteins were retrieved from the NCBI Entrez Protein Database (1, 70) through the NCBI Taxonomy Browser application (Taxonomy ID 11082) as described by Koo et al. (17). These included over 100 aligned sequences of each protein (C, 264; prM, 417; E, 927; NS1, 164; NS2a, 143; NS2b, 146; NS3, 146; NS4a, 142; NS4b, 141; and NS5, 256).
Entropy analysis of WNV T-cell ligand sequences.
The evolutionary conservation and variability of the identified T-cell ligand regions in the alignments of the recorded WNV sequences were measured by use of a modified Shannon's entropy computation function (16, 56) in the Antigenic Variability Analyzer (AVANA) software (http://sourceforge.net/projects/avana/) (33). The entropy of a ligand sequence (15 to 19 aa) position in the alignment was measured as the average of the entropies of the (7 to 11) nonamer positions within the ligand sequence position. AVANA was also used to study the incidence of the individual ligand sequence and its intraspecies variants in the corresponding WNV protein alignments. For a given WNV T-cell ligand sequence present in the alignment, the intraspecies variants are defined as peptides from that region of the alignment that differed by at least 1 amino acid from the ligand.
T-cell ligand sequence homologies with other flaviviruses.
Homologs of the WNV (Taxonomy ID 11082) T-cell ligand sequences were identified by performing a BLASTP (31) search against all protein sequences of all flaviviruses (Taxonomy ID 11051) present in the NCBI “nr” database (as of January 2009). The parameters set for the search by use of the NCBI BLAST Web server (http://blast.ncbi.nlm.nih.gov) were as follows: set limit by Entrez query to “txid11051[Organism:exp] NOT txid11082[Organism:exp] NOT txid81077[ORGN],” disabled the option “automatically adjust parameters for short sequences,” disabled “low-complexity” filter, and set maximum number of aligned sequences to be displayed to 20,000, expect threshold to 200,000, word size to 2, matrix to “PAM30,” gap costs to “Existence: 9, Extension: 1,” and compositional adjustments to “no adjustment.” Artificial sequence hits were removed by the “NOT txid81077[ORGN]” keyword.
The incidence of each shared WNV ligand was analyzed in reported sequences of the following six major flaviviruses that had sufficient published data (20 or more sequences corresponding to the ligand): LEV, JEV, Tick-borne encephalitis virus (TBEV), Powassan encephalitis virus (PV), YFV, and DENV (one or more of the four dengue virus serotypes). The incidence was determined by use of a BLASTP search performed at the NCBI BLAST Web server with a similar setting as above, except that the Entrez query was in the form of “<txid of virus>[Organism:exp] NOT txid81077[ORGN]”, where “<txid of virus>” was “txid11080” for LEV, “txid11072” for JEV, “txid11084” for TBEV, “txid11083” for PV, “txid11089” for YFV, and “txid12637” for DENV. The incidence of a shared ligand sequence in these other flaviviruses was determined for both full-length and partial identity matches. This was done for partial matches by dividing the number of BLAST hits that had nine or more, but not full-length, consecutive amino acid matches to the ligand sequence by the total number of hits from the species for the ligand. Similarly, the incidence of full-length matches was computed by determining the number of hits with nine or more matches that were full length.
RESULTS
Identification of HLA-restricted T-cell ligand sequences of the WNV proteome.
Six HLA transgenic (Tg), H-2-deficient mice, each expressing one of the predominant HLA class I (A2, A24, or B7) or class II (DR2, DR3, or DR4) alleles, were each immunized with 4 peptide pools (88 to 135 peptides per pool) that contained a total of 452 overlapping peptides, 12 to 20 amino acids each, comprising the entire WNV proteome. Each pool was mixed with 50 μl zymosan, 10 mg/ml, as the adjuvant. Splenocytes of the immunized mice, selectively depleted of murine CD4 or CD8 T cells for HLA class I or class II alleles, respectively, were analyzed by IFN-γ ELISpot assays and a peptide matrix deconvolution approach (see Materials and Methods). A total of 137 peptides, ∼30% of the 452 total, were found to activate T cells of one or more of the immunized Tg mice (Table 1; also see Table S1 in the supplemental material), the majority with high apparent avidity in the ELISpot assays at peptide concentrations of 1.0 or 0.1 μg/ml, except for splenocytes of the B7 Tg mice (see Table S2). The efficacy of the peptide immunization approach was further established by comparison with naked DNA immunization of HLA-DR2 Tg mice with a plasmid expression vector encoding the WNV NS3 protein. All but two (14/16; ∼88%) of the peptide-specific T-cell responses following peptide immunization were also detected after DNA immunization (see Fig. S1 in the supplemental material).
Table 1.
Distribution of HLA-restricted ELISpot responses in WNV proteinsa
| WNV protein | Protein size (aa)b | No. of overlapping peptides | No. (∼%) of ELISpot-positive peptides | HLA-restricted T-cell activation |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A2 | A24 | B7 | DR2 | DR3 | DR4 | Total | ||||
| C | 123 | 15 | 1 (7) | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| prM | 167 | 21 | 6 (29) | 1 | 2 | 1 | 6 | 3 | 4 | 17 |
| E | 501 | 67 | 25 (37) | 8 | 6 | 2 | 8 | 12 | 6 | 42 |
| NS1 | 352 | 46 | 10 (22) | 2 | 1 | 0 | 1 | 3 | 5 | 12 |
| NS2a | 231 | 30 | 10 (33) | 0 | 3 | 0 | 8 | 3 | 3 | 17 |
| NS2b | 131 | 16 | 6 (38) | 1 | 3 | 1 | 2 | 0 | 2 | 9 |
| NS3 | 619 | 84 | 46 (55) | 26 | 0 | 1 | 16 | 11 | 4 | 58 |
| NS4a | 149 | 18 | 5 (28) | 0 | 3 | 0 | 1 | 1 | 2 | 7 |
| NS4b | 255 | 33 | 9 (27) | 0 | 1 | 0 | 3 | 3 | 5 | 12 |
| NS5 | 905 | 122 | 19 (16) | 2 | 5 | 4 | 5 | 2 | 7 | 25 |
| Total | 3,433 | 452 | 137 (30) | 40 | 24 | 10 | 50 | 38 | 38 | 200 |
| Percentage of positive ligand sequences per complete proteome array | ∼9% | ∼5% | ∼2% | ∼11% | ∼8% | ∼8% | ||||
As identified by IFN-γ ELISpot assay in HLA Tg mice immunized with WNV overlapping peptides.
Corresponding to the WNV flamingo strain (NCBI Entrez Protein Database accession number AAF20092.2).
Nearly one-third of the 137 WNV T-cell ligands were immunogenic in 2 or more Tg strains, both class I and class II, resulting in a total of 200 HLA-restricted T-cell ligand responses, 74 class I (40 A2, 24 A24, and 10 B7) and 126 class II (50 DR2, 38 DR3, and 38 DR4). T-cell responses to the same peptide by 2 or more Tg mice can be attributed to overlapping epitopes in the same peptide and/or to promiscuous epitope presentation by multiple HLA alleles (55). The allele-specific T-cell responses were lowest for HLA-B7 (∼2%) and from ∼5 to 11% for the remaining alleles. NS3 (∼55%) and E (∼37%) had the greatest fractions of HLA class I and class II ligand sequences, while C (∼7%) had the least. About half of the ligand sequences were in clustered localizations of 3 or more overlapping ligand peptides each of the peptides overlapped with their adjacent peptides by 9 or more amino acids (see Table S1 in the supplemental material). This included all of the 6 prM ligand peptides forming a single cluster, 15 E peptides of 4 clusters, 3 NS1 peptides of 1 cluster, 21 NS3 peptides of 5 clusters, and 6 NS5 peptides of 2 clusters.
A correlation of the 137 HLA-restricted T-cell ligand sequences with reports of human T-cell epitopes (19–21, 32, 42, 61) showed that 22 of the ligand sequences matched 9 or more consecutive amino acids of 28 reported T-cell epitope peptides, both class I and class II (see Table S1 in the supplemental material). Additionally, the 137 ligand sequences were analyzed for candidate minimal HLA class I and II binding motifs within the 15 to 19 amino acids by use of NetMHC 3.2 and NetMHCIIPan 2.0 algorithms (26, 39). A total of 76 (38%) of the 200 positive HLA-restricted ELISpot responses corresponded to at least one predicted nonamer epitope with a concordant predicted HLA restriction (see Table S1). This relatively low extent of correspondence among the discovered HLA-restricted immunogenic peptides and predicted HLA binding motifs is comparable to those reported for other pathogens (27) and can be attributed, at least in part, to the fact that HLA binding prediction does not account for the pivotal role of the TCR in the immunogenicity of a peptide (67).
Evolutionary conservation and variants of the WNV T-cell ligand sequences.
The identification of ligand sequences was conducted with the peptides of the NY99-flamingo 382-99 WNV strain. The incidence (% occurrence) of these ligands in the aligned sequences of reported WNV was analyzed. Further, entropy, a measure of overall diversity (see Materials and Methods), was determined for the positions in the alignment that contained the ligands. Five of the 137 T-cell ligands, of NS3 and NS5, were completely conserved in all of the analyzed WNV sequences, with a 100% incidence and zero entropy (see Table S3 in the supplemental material). The other 132 ligands had an incidence of 69 to 99% of the analyzed sequences (alignment entropies of <0.1 to 1.6), with the remaining 1 to 31% of the analyzed sequences containing one or more amino acids that differed from the ligands. Only seven of the ligands, with alignment entropies of 0.7 to 1.6, had an individual variant of incidence of >10% (∼11 to 23%) of the analyzed WNV sequences (Table 2). Most of the variants of the ligand sequences were minor with negligible to low individual incidences (<1 to 9%) in the analyzed sequences (see Table S3). For example, about ∼6% of the WNV sequences at the alignment position E 62 to 77 (entropy of 0.5) contained 16 intraspecies variants, each with a negligible incidence of ∼1% or less (see Table S4).
Table 2.
WNV T-cell ligands with individual variants of high incidencea
| Protein and position | Peptide sequenceb | Incidence (∼%)c | Entropy |
|---|---|---|---|
| E 119–136 | FACSTKAIGRTILKENIK | 81 | 0.9 |
| .......T.......... | 16 | ||
| ...T...T.WI.Q..... | 2 | ||
| 11 variants | <1 each | ||
| NS2a 168–183 | CLNLDVYRILLLMVGI | 78 | 1.0 |
| ...............V | 18 | ||
| .........V...I.. | 1 | ||
| 4 variants | <1 each | ||
| NS2a 211–229 | GLFNPMILAAGLIACDPNR | 75 | 1.3 |
| .............T..... | 12 | ||
| .V..........M...... | 7 | ||
| .F..........V...... | 3 | ||
| .M.S.LV............ | 1 | ||
| 3 variants | <1 each | ||
| NS3 317–333 | ATPPGTSDPFPESNSPI | 87 | 0.7 |
| ..............A.. | 11 | ||
| 4 variants | <1 each | ||
| NS3 343–360 | RAWNSGYEWITEYTGKTV | 69 | 1.3 |
| .............I.... | 23 | ||
| ....T........V.... | 6 | ||
| .............V.... | 1 | ||
| 1 variant | <1 | ||
| NS4a 54–71 | IALIALLSVMTMGVFFLL | 80 | 1.1 |
| ....T............. | 11 | ||
| ..........SL...... | 4 | ||
| .V........SL...... | 3 | ||
| 4 variants | <1 each | ||
| NS5 863–879 | TWAENIQVAINQVRAII | 71 | 1.6 |
| ..............S.. | 12 | ||
| ......H.......SV. | 9 | ||
| ....D.........S.. | 2 | ||
| ......H.......SL. | 2 | ||
| 10 variants | <1 each |
Seven ligand sequence sites in the protein alignment, with average nonamer entropy of greater than 0.7, had at least one of the variants present in more than 10% of the analyzed WNV protein sequences.
Evolutionary variants of the WNV T-cell ligand (in boldface) are shown. The sequences of the ligand and its variants are compared, with “.” denoting identical amino acids. Individual variants with less than 1% incidence are not shown; only the sum of the number of such variants is indicated.
Incidence (to the nearest whole number) of the T-cell ligand sequence and its variants at the alignment position. Those with <1% incidence are more than 0%.
WNV-specific and Flavivirus-shared T-cell ligand sequences.
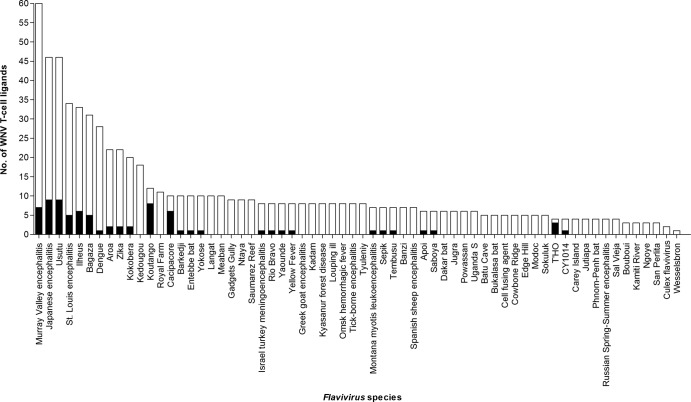
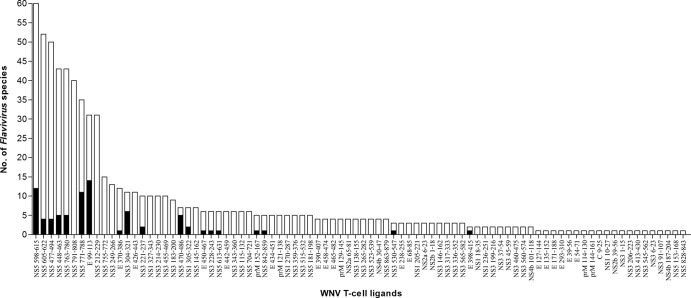
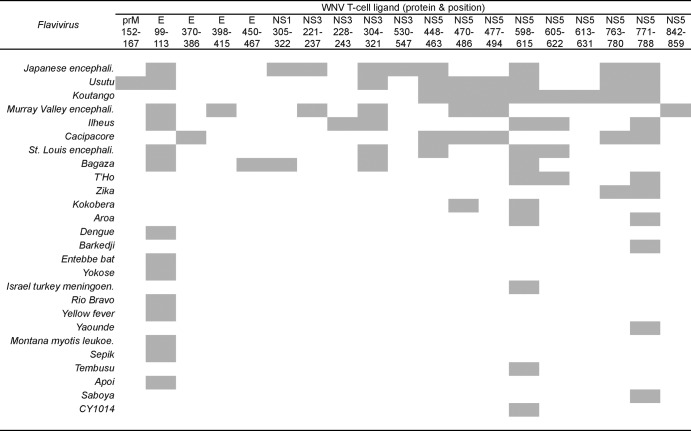
Notably, only 51 (∼37%) of the 137 HLA-restricted WNV ligand sequences were specific for WNV; the remaining 86 (∼63%) shared nine or more consecutive amino acid identity with sequences of one or more of other flaviviruses (see Table S1 in the supplemental material). The distinction between the WNV-specific and shared ligand sequences was striking. WNV-specific sequences were concentrated in NS2a, NS2b, NS4a, and NS4b, while the shared sequences were concentrated in E, NS3, and NS5. None of the 5 NS4a ligands was shared, and none of the 19 NS5 ligand sequences was WNV specific. Remarkably, the 86 shared sequences were present in 64 other flaviviruses, mainly of the JE group (JEV, MVE, LEV, and Koutango and Usutu viruses), and also DENV and YFV (Fig. 1 and 2). These 86 matches of nine or more consecutive amino acids to sequences of other flaviviruses were mostly partial in relation to the WNV ligand sequence length. Only 19 of the 86 shared ligands had full-length identity among sequences of 26 other flaviviruses, most commonly E 99 to 113, NS5 598 to 615, and NS5 771 to 788, which individually were present in 11 to 14 other flaviviruses (Fig. 3). The majority of the shared ligands were in six major reported flaviviruses: the Japanese encephalitis group viruses (LEV and JEV), tick-borne encephalitis group viruses (TBEV and PV), YFV, and the four dengue virus serotypes (DENV1 to 4) (see Table S5 in the supplemental material). LEV and JEV, as expected, had the greatest number of partial and full-length identity matches, followed by DENV with mostly partial matches. Many of the full-length and partial matches among the flaviviruses were of high incidence, present in ∼90 to 100% of the analyzed sequences, with low incidence matches mostly for DENV. Each shared ligand (either of full-length or partial identity) with an incidence less than 100% of the analyzed viral sequences had the remaining fraction, which lacked nine or more consecutive amino acid identity, highly similar to the shared ligand, with mostly one to two amino acid differences (data not shown). Both of the shared and highly similar sequences of the other flaviviruses are potential interspecies TCR altered peptide ligands of WNV, with one or more amino acid differences in the region corresponding to or flanking the WNV T-cell ligand sequence.
Fig 1.
Number of WNV T-cell ligand sequences shared by other flaviviruses. The black bars indicate shared sequences with full-length identity to the WNV ligand; the transparent bars indicate shared sequences with identity of 9 or more consecutive amino acids, but not full-length identity, to the WNV ligand.
Fig 2.
Number of other flaviviruses that shared WNV T-cell ligand sequences. The black bars indicate shared sequences with full-length identity to the WNV ligand; the transparent bars indicate shared sequences with identity of 9 or more consecutive amino acids, but not full-length identity, to the WNV ligand.
Fig 3.
Distribution of WNV T-cell ligands shared with full-length identity to sequences of other flaviviruses. Abbreviations: encephali., encephalitis; meningoen., meningoencephalomyelitis; leukoe., leukoencephalitis.
DISCUSSION
Sequence homologies among proteins of flaviviruses are expected because of the genetic relationships that established the phylogeny of these viruses. Nevertheless, these homologies have exceptional relevance when viewed in the context of the immunity of T-cell epitopes. WNV has particular pertinence because of its remarkable evolutionary conservation (17). Eighty-eight sequences of nine or more amino acids, representing 34% of the WNV proteome, were completely conserved in recorded WNV, and the remaining regions of the proteome generally had less than 10% of the sequences as variants of the predominant sequence at the position, hence the conserved nature of the majority of the 137 HLA-restricted WNV ligands and the low incidence of variants that differed from the ligand sequences. The high conservation of WNV is favorable for a satisfactory host immune response following natural infection or vaccination, because the low cumulative and negligible individual incidences of variant sequences provide little chance of immune escape by the proliferation of variants of the founder virus.
However, a negative aspect of the WNV evolutionary conservation is the extensive protein sequence homology with as many as 64 other viruses of the genus Flavivirus. Only 51 of the 137 ligand sequences were specific to WNV; the remaining 86 were shared with other flaviviruses, as sequences of either full-length or partial identity. The most frequently shared were the highly conserved sequences of E, NS1, NS3, and NS5, present in several major flaviviruses, including LEV, JEV, TBEV, PV, YFV, and DENV. Many of the shared sequences had high incidences, >50% of the analyzed sequences of one or more of the six major flaviviruses, particularly the closely related LEV and JEV. Further, shared sequences that did not occur in all (100%) of the analyzed sequences of the other flaviviruses had a highly similar remaining fraction, with mostly one to two amino acid differences. Both the shared and highly similar sequences of the other flaviviruses, with one or more amino acid differences corresponding to the WNV T-cell ligand and/or flanking sequences, represent potential interspecies TCR altered peptide ligands of WNV. Others (19, 72) have shown that interspecies variants of WNV T-cell ligands can be epitopes with the same HLA restriction, and Moran et al. (36) reported that T-cell clones from an acute dengue patient responded to epitope variants from other flaviviruses and also suggested that this may be of pathological significance, particularly in geographical regions where related viruses cocirculate.
This demonstration that a large number of WNV HLA-restricted T-cell ligand sequences shared 9 or more consecutive amino acid identities with multiple other flaviviruses serves to display the full extent of potential immune cross-reactivity among Flavivirus proteins. It is clear that exposure to multiple flaviviruses by infection or immunization may risk immune responses to a large number of cross-reactive T cell epitopes that, as altered peptide ligands (APL), may significantly affect the immune responses to the pathogens. The concept of “original antigenic sin” in the role of cross-reactive immune responses was initially applied to serum antibody responses (39, 40) and later extended to T-cell responses (41, 42). It now has been extensively studied with respect to a variety of immune responses, including thymic education, T-cell memory, pathogen virulence, and autoimmune disease (43, 44, 45, 46, 47). In the context of T-cell responses, this concept relates to the role of cross-reactive T-cell epitopes resulting from dual exposure to viruses with sequence homology and epitope sequences that contain one or more amino acid substitutions at TCR contact residues as compared to the epitope of a previous pathogen or immunogen. Memory T cells selectively engaged by a variant epitope sequence may exhibit an impaired immune response, depending on the positions and types of amino acid substitutions surrounding or within T-cell epitope cores and the effect of these changes on the affinity of the interaction (48, 49). Protein structural analysis has provided evidence for extensive epitope interresidue interactions whereby an amino acid substitution at an HLA anchor site of epitope binding to a memory T cell can diminish or modify the TCR response to this epitope and block amplification of the immune response (13, 48, 66). T-cell anergy may also result from partial activation, such as from impaired costimulation of T cells by antigen-presenting cells (APC) (57, 60). The effect in humans of APL inhibition or modification of T-cell immune responses has been widely recognized (14, 53, 58, 59). A recognized example is the immunopathology observed during secondary DENV infection, where misdirected immune responses of cross-reactive T cells are suggested to contribute to the severe dengue hemorrhagic fever (30, 34, 35, 52, 54, 69).
The selection of evolutionarily conserved protein sequences has widely been considered important to vaccine design in order to limit the selective loss of immunity resulting from mutation and protein modifications. However, as shown herein, sequences conserved in the evolution of viruses can be present in many different forms in viruses of related species. In this case, such conserved sequences could be recognized by the TCR of cells previously activated through infection or vaccination by a homologous sequence of another virus. Thus, we propose that the selection of conserved sequences that are also virus specific should have precedence in vaccine design.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Lars Fugger (Weatherall Institute of Molecular Medicine, Oxford, United Kingdom) and Arthur Vanderbark (Oregon Health and Science University, Portland, OR), Chella S. David (Mayo Clinic, Rochester, MN), and Grete Sønderstrup (Stanford University School of Medicine) for providing HLA-DR2, -DR3, and -DR4 transgenic mice, respectively. Overlapping WNV peptide arrays were obtained through the NIH Biodefense and Emerging Infectious Disease Research Resources Repository, NIAID, NIH. We thank Paul Nordstrom August, Natascha May Thevasagayam, and Rashmi Sukumaran for their help with the manuscript.
No competing interests exist.
Footnotes
Published ahead of print 9 May 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Berman HM, et al. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Betts MR, et al. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4(+) and CD8(+) T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 75:11983–11991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brien JD, Uhrlaub JL, Nikolich-Zugich J. 2007. Protective capacity and epitope specificity of CD8(+) T cells responding to lethal West Nile virus infection. Eur. J. Immunol. 37:1855–1863 [DOI] [PubMed] [Google Scholar]
- 4. Cheuk E, et al. 2002. Human MHC class I transgenic mice deficient for H2 class I expression facilitate identification and characterization of new HLA class I-restricted viral T cell epitopes. J. Immunol. 169:5571–5580 [DOI] [PubMed] [Google Scholar]
- 5. Cook S, Holmes EC. 2006. A multigene analysis of the phylogenetic relationships among the flaviviruses (Family: Flaviviridae) and the evolution of vector transmission. Arch. Virol. 151:309–325 [DOI] [PubMed] [Google Scholar]
- 6. Cope AP, et al. 1999. T cell responses to a human cartilage autoantigen in the context of rheumatoid arthritis-associated and nonassociated HLA-DR4 alleles. Arthritis Rheum. 42:1497–1507 [DOI] [PubMed] [Google Scholar]
- 7. de la Rosa G, et al. 2005. Regulated recruitment of DC-SIGN to cell-cell contact regions during zymosan-induced human dendritic cell aggregation. J. Leukoc. Biol. 77:699–709 [DOI] [PubMed] [Google Scholar]
- 8. Diamond MS, Mehlhop E, Oliphant T, Samuel MA. 2009. The host immunologic response to West Nile encephalitis virus. Front. Biosci. 14:3024–3034 [DOI] [PubMed] [Google Scholar]
- 9. Draenert R, et al. 2003. Comparison of overlapping peptide sets for detection of antiviral CD8 and CD4 T cell responses. J. Immunol. Methods 275:19–29 [DOI] [PubMed] [Google Scholar]
- 10. Fugger L, Michie SA, Rulifson I, Lock CB, McDevitt GS. 1994. Expression of HLA-DR4 and human CD4 transgenes in mice determines the variable region beta-chain T-cell repertoire and mediates an HLA-DR-restricted immune response. Proc. Natl. Acad. Sci. U. S. A. 91:6151–6155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goodridge HS, Simmons RM, Underhill DM. 2007. Dectin-1 stimulation by Candida albicans yeast or zymosan triggers NFAT activation in macrophages and dendritic cells. J. Immunol. 178:3107–3115 [DOI] [PubMed] [Google Scholar]
- 12. Hu N, et al. 2005. Highly conserved pattern of recognition of influenza A wild-type and variant CD8+ CTL epitopes in HLA-A2+ humans and transgenic HLA-A2+/H2 class I-deficient mice. Vaccine 23:5231–5244 [DOI] [PubMed] [Google Scholar]
- 13. Kalergis AM, Nathenson SG. 2000. Altered peptide ligand-mediated TCR antagonism can be modulated by a change in a single amino acid residue within the CDR3 beta of an MHC class I-restricted TCR. J. Immunol. 165:280–285 [DOI] [PubMed] [Google Scholar]
- 14. Katsara M, Minigo G, Plebanski M, Apostolopoulos V. 2008. The good, the bad and the ugly: how altered peptide ligands modulate immunity. Expert Opin. Biol. Ther. 8:1873–1884 [DOI] [PubMed] [Google Scholar]
- 15. Kessler PD, et al. 1996. Gene delivery to skeletal muscle results in sustained expression and systemic delivery of a therapeutic protein. Proc. Natl. Acad. Sci. U. S. A. 93:14082–14087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khan AM, et al. 2008. Conservation and variability of dengue virus proteins: implications for vaccine design. PLoS Negl. Trop. Dis. 2:e272 doi:10.1371/journal.pntd.0000272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koo QY, et al. 2009. Conservation and variability of West Nile virus proteins. PLoS One 4:e5352 doi:10.1371/journal.pone.0005352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. 1998. Phylogeny of the genus Flavivirus. J. Virol. 72:73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kurane I, et al. 1995. Flavivirus-cross-reactive, HLA-DR15-restricted epitope on NS3 recognized by human CD4+ CD8− cytotoxic T lymphocyte clones. J. Gen. Virol. 76:2243–2249 [DOI] [PubMed] [Google Scholar]
- 20. Lanteri MC, et al. 2008. Comprehensive analysis of West Nile virus-specific T cell responses in humans. J. Infect. Dis. 197:1296–1306 [DOI] [PubMed] [Google Scholar]
- 21. Larsen MV, et al. 2010. Identification of CD8+ T cell epitopes in the West Nile virus polyprotein by reverse-immunology using NetCTL. PLoS One 5:e12697 doi:10.1371/journal.pone.0012697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin HH, Ray S, Tongchusak S, Reinherz EL, Brusic V. 2008. Evaluation of MHC class I peptide binding prediction servers: applications for vaccine research. BMC Immunol. 9:8 doi:10.1186/1471-2172-9-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin HH, Zhang GL, Tongchusak S, Reinherz EL, Brusic V. 2008. Evaluation of MHC-II peptide binding prediction servers: applications for vaccine research. BMC Bioinformatics 9(Suppl. 12):S22 doi:10.1186/1471-2105-9-S12-S22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lindenbach BD, Rice CM. 2003. Molecular biology of flaviviruses. Adv. Virus Res. 59:23–61 [DOI] [PubMed] [Google Scholar]
- 25. Loirat D, Lemonnier FA, Michel ML. 2000. Multiepitopic HLA-A*0201-restricted immune response against hepatitis B surface antigen after DNA-based immunization. J. Immunol. 165:4748–4755 [DOI] [PubMed] [Google Scholar]
- 26. Lundegaard C, et al. 2008. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res. 36:W509–W512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lundegaard C, Lund O, Buus S, Nielsen M. 2010. Major histocompatibility complex class I binding predictions as a tool in epitope discovery. Immunology 130:309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mackenzie JS, Gubler DJ, Petersen LR. 2004. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 10:S98–S109 [DOI] [PubMed] [Google Scholar]
- 29. Madsen L, et al. 1999. Mice lacking all conventional MHC class II genes. Proc. Natl. Acad. Sci. U. S. A. 96:10338–10343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mathew A, Rothman AL. 2008. Understanding the contribution of cellular immunity to dengue disease pathogenesis. Immunol. Rev. 225:300–313 [DOI] [PubMed] [Google Scholar]
- 31. McGinnis S, Madden TL. 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32:W20–W25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McMurtrey CP, et al. 2008. Epitope discovery in West Nile virus infection: identification and immune recognition of viral epitopes. Proc. Natl. Acad. Sci. U. S. A. 105:2981–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miotto O, Heiny A, Tan TW, August JT, Brusic V. 2008. Identification of human-to-human transmissibility factors in PB2 proteins of influenza A by large-scale mutual information analysis. BMC Bioinformatics 9(Suppl. 1):S18 doi:10.1186/1471-2105-9-S1-S18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mongkolsapaya J, et al. 2003. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat. Med. 9:921–927 [DOI] [PubMed] [Google Scholar]
- 35. Mongkolsapaya J, et al. 2006. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal? J. Immunol. 176:3821–3829 [DOI] [PubMed] [Google Scholar]
- 36. Moran E, et al. 2008. Preservation of a critical epitope core region is associated with the high degree of flaviviral cross-reactivity exhibited by a dengue-specific CD4(+) T cell clone. Eur. J. Immunol. 38:1050–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mukhopadhyay S, Kuhn RJ, Rossmann MG. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3:13–22 [DOI] [PubMed] [Google Scholar]
- 38. Murray KO, Walker C, Gould E. 2011. The virology, epidemiology, and clinical impact of West Nile virus: a decade of advancements in research since its introduction into the Western Hemisphere. Epidemiol. Infect. 139:807–817 [DOI] [PubMed] [Google Scholar]
- 39. Nielsen M, et al. 2008. Quantitative predictions of peptide binding to any HLA-DR molecule of known sequence: NetMHCIIpan. PLoS Comput. Biol. 4:e1000107 doi:10.1371/journal.pcbi.1000107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pajot A, et al. 2004. A mouse model of human adaptive immune functions: HLA-A2.1-/HLA-DR1-transgenic H-2 class I-/class II-knockout mice. Eur. J. Immunol. 34:3060–3069 [DOI] [PubMed] [Google Scholar]
- 41. Pajot A, et al. 2006. Identification of novel HLA-DR1-restricted epitopes from the hepatitis B virus envelope protein in mice expressing HLA-DR1 and vaccinated human subjects. Microbes Infect. 8:2783–2790 [DOI] [PubMed] [Google Scholar]
- 42. Parsons R, et al. 2008. The memory T cell response to West Nile virus in symptomatic humans following natural infection is not influenced by age and is dominated by a restricted set of CD8+ T cell epitopes. J. Immunol. 181:1563–1572 [DOI] [PubMed] [Google Scholar]
- 43. Pascolo S. 2005. HLA class I transgenic mice: development, utilisation and improvement. Expert Opin. Biol. Ther. 5:919–938 [DOI] [PubMed] [Google Scholar]
- 44. Pascolo S, et al. 1997. HLA-A2.1-restricted education and cytolytic activity of CD8(+) T lymphocytes from beta2 microglobulin (beta2m) HLA-A2.1 monochain transgenic H-2Db beta2m double knockout mice. J. Exp. Med. 185:2043–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Petersen LR, Roehrig JT. 2001. West Nile virus: a reemerging global pathogen. Emerg. Infect. Dis. 7:611–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Purtha WE, et al. 2007. Antigen-specific cytotoxic T lymphocytes protect against lethal West Nile virus encephalitis. Eur. J. Immunol. 37:1845–1854 [DOI] [PubMed] [Google Scholar]
- 47. Rammensee HG. 1995. Chemistry of peptides associated with MHC class I and class II molecules. Curr. Opin. Immunol. 7:85–96 [DOI] [PubMed] [Google Scholar]
- 48. Regner M, Lobigs M, Blanden RV, Milburn P, Mullbacher A. 2001. Antiviral cytotoxic T cells cross-reactively recognize disparate peptide determinants from related viruses but ignore more similar self- and foreign determinants. J. Immunol. 166:3820–3828 [DOI] [PubMed] [Google Scholar]
- 49. Richards KA, et al. 2007. Direct ex vivo analyses of HLA-DR1 transgenic mice reveal an exceptionally broad pattern of immunodominance in the primary HLA-DR1-restricted CD4 T-cell response to influenza virus hemagglutinin. J. Virol. 81:7608–7619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Roederer M, Koup RA. 2003. Optimized determination of T cell epitope responses. J. Immunol. Methods 274:221–228 [DOI] [PubMed] [Google Scholar]
- 51. Rohrlich PS, et al. 2003. HLA-B*0702 transgenic, H-2KbDb double-knockout mice: phenotypical and functional characterization in response to influenza virus. Int. Immunol. 15:765–772 [DOI] [PubMed] [Google Scholar]
- 52. Rothman AL. 2010. Cellular immunology of sequential dengue virus infection and its role in disease pathogenesis. Curr. Top. Microbiol. Immunol. 338:83–98 [DOI] [PubMed] [Google Scholar]
- 53. Rothman AL. 2004. Dengue: defining protective versus pathologic immunity. J. Clin. Invest. 113:946–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Screaton G, Mongkolsapaya J. 2006. T cell responses and dengue haemorrhagic fever. Novartis Found. Symp. 277:164–171; discussion 171–176, 251–253 [PubMed] [Google Scholar]
- 55. Sette A, Sidney J. 1999. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics 50:201–212 [DOI] [PubMed] [Google Scholar]
- 56. Shannon CE. 1948. A mathematical theory of communication. Bell System Tech. J. 27:379–423 and 623–656 [Google Scholar]
- 57. Sharpe AH. 2009. Mechanisms of costimulation. Immunol. Rev. 229:5–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sloan-Lancaster J, Allen PM. 1996. Altered peptide ligand-induced partial T cell activation: molecular mechanisms and role in T cell biology. Annu. Rev. Immunol. 14:1–27 [DOI] [PubMed] [Google Scholar]
- 59. Sloan-Lancaster J, Allen PM. 1995. Significance of T-cell stimulation by altered peptide ligands in T cell biology. Curr. Opin. Immunol. 7:103–109 [DOI] [PubMed] [Google Scholar]
- 60. Sloan-Lancaster J, Evavold BD, Allen PM. 1994. Th2 cell clonal anergy as a consequence of partial activation. J. Exp. Med. 180:1195–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Smith HL, et al. 2011. Development of antigen-specific memory CD8+ T cells following live-attenuated chimeric West Nile virus vaccination. J. Infect. Dis. 203:513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Smithburn KC, Hughes TP, Burke AW, Paul JH. 1940. A neurotropic virus isolated from the blood of a native of Uganda. Am. J. Trop. Med. 20:471–492 [Google Scholar]
- 63. Sonderstrup G, et al. 1999. HLA class II transgenic mice: models of the human CD4+ T-cell immune response. Immunol. Rev. 172:335–343 [DOI] [PubMed] [Google Scholar]
- 64. Strauss G, Vignali DA, Schonrich G, Hammerling GJ. 1994. Negative and positive selection by HLA-DR3(DRw17) molecules in transgenic mice. Immunogenetics 40:104–108 [PubMed] [Google Scholar]
- 65. Taneja V, David CS. 1999. HLA class II transgenic mice as models of human diseases. Immunol. Rev. 169:67–79 [DOI] [PubMed] [Google Scholar]
- 66. Theodossis A, et al. 2010. Constraints within major histocompatibility complex class I restricted peptides: presentation and consequences for T-cell recognition. Proc. Natl. Acad. Sci. U. S. A. 107:5534–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tung CW, Ziehm M, Kamper A, Kohlbacher O, Ho SY. 2011. POPISK: T-cell reactivity prediction using support vector machines and string kernels. BMC Bioinformatics 12:446 doi:10.1186/1471-2105-12-446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vandenbark AA, et al. 2003. Recombinant TCR ligand induces tolerance to myelin oligodendrocyte glycoprotein 35-55 peptide and reverses clinical and histological signs of chronic experimental autoimmune encephalomyelitis in HLA-DR2 transgenic mice. J. Immunol. 171:127–133 [DOI] [PubMed] [Google Scholar]
- 69. Welsh RM, Rothman AL. 2003. Dengue immune response: low affinity, high febrility. Nat. Med. 9:820–822 [DOI] [PubMed] [Google Scholar]
- 70. Wheeler DL, et al. 2006. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 34:D173–D180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Whitehead SS, Blaney JE, Durbin AP, Murphy BR. 2007. Prospects for a dengue virus vaccine. Nat. Rev. Microbiol. 5:518–528 [DOI] [PubMed] [Google Scholar]
- 72. Zeng L, Kurane I, Okamoto Y, Ennis FA, Brinton MA. 1996. Identification of amino acids involved in recognition by dengue virus NS3-specific, HLA-DR15-restricted cytotoxic CD4+ T-cell clones. J. Virol. 70:3108–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.