Abstract
The genomes of influenza A viruses consist of eight negative-strand RNA segments. Recent studies suggest that influenza viruses are able to specifically package their segmented genomes into the progeny virions. Segment-specific packaging signals of influenza virus RNAs (vRNAs) are located in the 5′ and 3′ noncoding regions, as well as in the terminal regions, of the open reading frames. How these packaging signals function during genome packaging remains unclear. Previously, we generated a 7-segmented virus in which the hemagglutinin (HA) and neuraminidase (NA) segments of the influenza A/Puerto Rico/8/34 virus were replaced by a chimeric influenza C virus hemagglutinin/esterase/fusion (HEF) segment carrying the HA packaging sequences. The robust growth of the HEF virus suggested that the NA segment is not required for the packaging of other segments. In this study, in order to determine the roles of the other seven segments during influenza A virus genome assembly, we continued to use this HEF virus as a tool and analyzed the effects of replacing the packaging sequences of other segments with those of the NA segment. Our results showed that deleting the packaging signals of the PB1, HA, or NS segment had no effect on the growth of the HEF virus, while growth was greatly impaired when the packaging sequence of the PB2, PA, nucleoprotein (NP), or matrix (M) segment was removed. These results indicate that the PB2, PA, NP, and M segments play a more important role than the remaining four vRNAs during the genome-packaging process.
INTRODUCTION
Influenza viruses belong to the family Orthomyxoviridae. The viruses possess a host cell-derived envelope membrane carrying the hemagglutinin (HA) and the neuraminidase (NA) glycoproteins and the ion channel protein M2. Inside the membrane are a layer of matrix protein 1 (M1) and the core viral ribonucleoprotein (vRNP) complexes, which are composed of viral RNAs (vRNAs) and the binding proteins, including the heterotrimeric polymerase complex and multiple copies of nucleoprotein (NP) (28). Upon virus entry, the vRNPs are transported into the nucleus, where they undergo transcription and replication. At the late stage of infection, the newly synthesized vRNPs are exported from the nucleus through CRM1-dependent nuclear export machinery (28) and transported via Rab11- and human immunodeficiency virus rev-binding protein (HRB)-dependent trafficking pathways (1, 4, 5) to the plasma membrane, where the progeny virions are assembled and released.
Both the influenza A and B viruses possess eight vRNA segments, while the influenza C viruses have only seven (28). The mechanism utilized by influenza viruses to assemble their genomes is not well understood. Recent data from several groups suggested that influenza viruses do not incorporate their genomes randomly; instead, they use a rather specific mechanism to govern the genome-packaging process (3, 7–9, 17, 18, 20, 22–24, 26, 27, 34) (see Fig. 47.23 in reference 28). The cis-acting elements—the packaging sequences—that ensure each virion has a full complement of the vRNA genome have been identified for each influenza A virus segment (8, 9, 17, 18, 20, 22, 26, 27, 34). These segment-specific packaging signals include both the 3′ and 5′ noncoding regions (NCRs), as well as coding sequences, at the two ends of each open reading frame (ORF) (8, 9, 17, 18, 20, 22, 26, 27, 34). Precisely how these packaging signals function during the genome-packaging process has yet to be determined. Electron micrographs of budding viruses showed that the eight RNA segments are arranged in a specific configuration, with one in the center and seven in the surrounding positions (23). Interestingly, the architecture of vRNPs in the budding virions, elucidated by using electron tomography techniques, appeared to have multiple configurations (7, 24). Clearly, more work has to be done to further understand this phenomenon.
In the present study, we use a previously established 7-segmented virus system (10) to study the genome copackaging of influenza A virus. The recombinant virus was generated by replacing the HA and NA segments of the influenza A/Puerto Rico/8/34 (PR8) virus with a chimeric influenza C hemagglutinin/esterase/fusion (HEF) segment carrying the HA packaging sequences. Therefore, its genome is composed of only seven segments and it lacks the NA segment (10). Here, by utilizing reverse genetics and the “rewiring” (switching the packaging signals) approach (12), we examined the functional importance of all the influenza A virus segment-specific packaging sequences except the NA segment, which has been studied previously (10). Our results show that the packaging sequences from the PB2, PA, NP, and M segments are critical to the virus, while those of the remaining segments are not. Our results indicate that the PB2, PA, NP, and M segments play a more important role than the other segments during the genome-packaging process.
MATERIALS AND METHODS
Cells and viruses.
293T cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum. Viruses were grown in 8- or 10-day-old specific-pathogen-free chicken embryos at 33°C or 37°C (Charles River Laboratories; SPAFAS).
Plasmid construction.
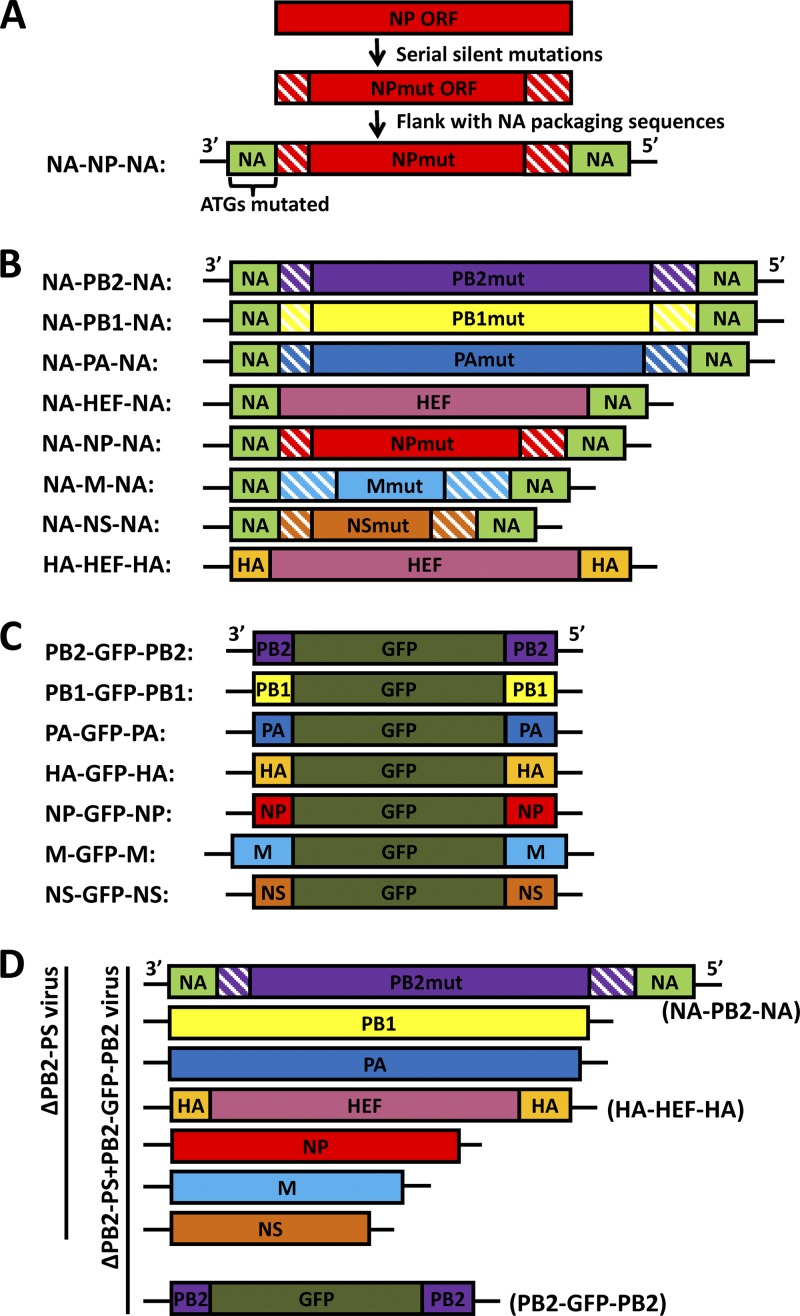
The pDZ plasmids expressing NA-PB2-NA, NA-PB1-NA, NA-PA-NA, or HA-HEF-HA were described previously (10, 11). The pDZ plasmid expressing NA-HEF-NA vRNA was generated by replacing the green fluorescent protein (GFP) gene of the pDZ-NA-GFP-NA construct with the HEF ORF (10). The pDZ plasmids expressing NA-NP-NA, NA-M-NA, and NA-NS-NA were generated in this study by using a method described previously (10, 11). Briefly, serial mutations were introduced into the terminal regions of the ORFs of the PR8 virus NP, M, and NS segments to remove the ORF region packaging signals, and then the mutated ORFs were each flanked with the previously described NA packaging sequences in a pDZ vector (10). Figure 1A shows one example of the method. The pDZ plasmids expressing PB2-GFP-PB2, PB1-GFP-PB1, and PA-GFP-PA were described previously (11). The pDZ plasmids expressing HA-GFP-HA and NS-GFP-NS were generated from pDZ plasmids HA-NSmut-HA and NS-HAmut-NS (12) by replacing the ORFs with a GFP gene (Clontech). The pDZ plasmids expressing NP-GFP-NP and M-GFP-M were generated in this study using a method similar to that described for PB2-GFP-PB2 (11). The lengths of the PR8 virus NP packaging sequences are 171 nucleotides (nt) in the 3′ end and 145 nt in the 5′ end (26); the lengths of the PR8 virus M packaging sequences are 281 nt in the 3′ end and 236 nt in the 5′ end (27).
Fig 1.
Diagrams of chimeric constructs and recombinant HEF viruses. (A) Construction of the NA-NP-NA chimera. This construct contains the PR8 NA packaging sequences in the flanking regions and the mutated PR8 NP ORF (NPmut) in the middle. The hatched boxes represent serial silent mutations introduced into the terminal ORF regions. The ATGs in the 3′ NA ORF packaging sequence region were all mutated to TTGs (12). (B) Rewired RNA segments used for generating recombinant viruses. All these constructs carry the NA or HA packaging sequences in the flanking regions and the mutated ORFs in the center. Only the HEF ORF of the NA-HEF-NA and HA-HEF-HA constructs does not carry serial silent mutations at the two ends. In these constructs, the 3′-end NA and HA packaging sequences are not translated due to the removal of ATGs. The hatched boxes also represent serial silent mutations introduced into the terminal ORF regions for each construct. (C) GFP constructs carrying segment-specific influenza A virus packaging signals. All of the constructs carry a GFP gene in the middle and influenza PR8 virus vRNA packaging sequences at the two ends. The ATGs in the 3′-end packaging sequences were removed by silent mutations. (D) Diagrams of a 7-segmented HEF virus (ΔPB2-PS) and an 8-segmented HEF GFP virus (ΔPB2-PS+PB2-GFP-PB2). ΔPB2-PS includes two chimeric segments (NA-PB2-NA and HA-HEF-HA), shown in panel B, and five wild-type PR8 virus segments (PB1, PA, NP, M, and NS). The ΔPB2-PS+PB2-GFP-PB2 virus contains an eighth GFP segment (PB2-GFP-PB2), shown in panel C. The remaining 7-segmented and 8-segmented viruses listed in Table 1 were generated in a similar fashion.
Reverse genetics for recombinant viruses.
The method for generating recombinant influenza viruses was as described previously (6, 10, 29).
Viral growth in eggs.
Eight- or 10-day-old embryonated chicken eggs were inoculated with influenza viruses (100 50% tissue culture infective doses [TCID50]/egg) and incubated at 33°C or 37°C. At 72 h postinoculation, the allantoic fluids were harvested, and the titers of the viruses were determined by measuring the TCID50 in 293T cells. Three eggs were analyzed for each virus.
Immunofluorescence of virus-infected cells.
The 293T cells in a poly-d-lysine-coated plate were infected with HEF viruses. One day later, the cells were fixed with 4% formaldehyde and then permeabilized with 0.5% Triton X-100. The cells were incubated with mouse anti-PR8 NP monoclonal antibody (HT103; 5 μg/ml) (25) and then incubated with Alexa Fluor 594-labeled donkey anti-mouse IgG (Invitrogen). Red and green images of the cells were taken using a fluorescence microscope.
Acrylamide gel electrophoresis of purified vRNA.
The viruses were grown in 8- to 10-day-old embryonated chicken eggs at either 33°C for HEF viruses or 37°C for PR8 virus. Two to 3 days later, the embryos were killed by transferring them to 4°C and left overnight. The allantoic fluids were harvested and clarified by centrifugation at 6,000 rpm at 4°C for 15 min using a Beckman SW28 rotor. The clarified supernatant was then layered on a 30% sucrose cushion and further centrifuged at 25,000 rpm for 2.5 h. The pelleted virus was resuspended in 1× phosphate-buffered saline (PBS) buffer, and vRNA was extracted by using TRIzol reagent (Invitrogen). Precipitated vRNA was resuspended in a final volume of 15 μl DNase- and RNase-free H2O and stored at −80°C; 0.5 to 1 μg of RNA was separated on a 2.8% denaturing polyacrylamide gel containing 7 M urea, and the gel was stained with a SilverXpress silver-staining kit (Invitrogen) to visualize the RNA bands. The bands of the gel were quantified using ImageQuant software (GE Healthcare).
qPCR analysis of packaged vRNAs.
The viruses were grown in embryonated chicken eggs and purified using a sucrose cushion method. Extracted vRNA (approximately 200 ng) was reverse transcribed using random hexamers and Superscript II reverse transcriptase (RT) (Invitrogen). The RT product was then diluted 10,000-fold and used as a template for quantitative PCR (qPCR). Separate qPCRs were then carried out with segment-specific primers for the NP and GFP genes using a SYBR green-based method. Relative concentrations of vRNA were determined on the basis of an analysis of cycle threshold values. The results are presented as the ratios of the GFP vRNA versus NP vRNA. The standard deviations were calculated based on triplicate analysis of the samples.
Single virus particle immobilization and fluorescence in situ hybridization.
Virus particle immobilization and single-molecule imaging were performed as previously described (2, 16) with several modifications. In brief, passivated flow chambers were prepared, and after incubating the chamber with NeutrAvidin (Thermo), the recombinant HEF virus particles were specifically immobilized on the surface using biotinylated mouse monoclonal anti-HEF antibody (8D6D3) (10). The virus particles were diluted in T50 buffer (10 mM Tris-HCl, pH 8.0, 50 mM NaCl, 1 mg/ml bovine serum albumin [BSA]) and incubated for 30 min at room temperature over the surface with immobilized antibody. The unbound antibodies and virus particles were washed away with T50 buffer. The virus was fixed with 4% paraformaldehyde and permeabilized with 0.25% Triton X-100 in the presence of 2 mM vanadyl ribonucleoside complex (VRC) to expose the viral RNPs. The flow chambers were then washed twice with wash buffer (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 10% formamide, and 2 mM VRC), followed by fluorescence in situ hybridization against the NP and GFP viral RNAs. Probes designed against the viral RNAs were purchased from Biosearch Technologies, Novato, CA. Fifteen probes against different regions of the NP viral RNA were labeled with Cy3 fluorophore, and 24 probes against the GFP were labeled with Cy5 fluorophore, and they were high-performance liquid chromatography (HPLC) purified according to a published protocol (30). After hybridization, the surfaces on which virus was immobilized were washed with wash buffer (2× SSC, 10% formamide, and 2 mM VRC) for 30 min and then equilibrated in 2× SSC buffer before imaging. Single-molecule imaging was performed using a prism-type total internal reflection fluorescence (TIRF) microscope, and the single-molecule analysis was performed as described previously (16, 32).
Colocalization analysis for the single-molecule fluorescence in situ hybridization (smFISH) experiment.
Colocalization between Cy3 and Cy5 images was analyzed similarly to what was previously described (16, 33). The colocalization efficiency was calculated as a percentage (the number of Cy3 and Cy5 colocalized spots over the total number of spots of either Cy3 or Cy5 signal detected).
Nucleotide sequence accession numbers.
The sequences of the constructs used in this study were submitted to GenBank under the following accession numbers: NA-PB2-NA (JQ733034), NA-PB1-NA (JQ733035), NA-PA-NA (JQ733036), NA-HEF-NA (JQ733037), NA-NP-NA (JQ733038), NA-M-NA (JQ733039), NA-NS-NA (JQ733040), HA-HEF-HA (JQ733041), PB2-GFP-PB2 (JQ733043), PB1-GFP-PB1 (JQ733033), PA-GFP-PA (JQ733044), HA-GFP-HA (JQ733042), NP-GFP-NP (JQ733045), M-GFP-M (JQ733046), NS-GFP-NS (JQ733047).
RESULTS
Rewiring the vRNAs of the 7-segmented HEF virus.
The packaging sequences of influenza virus A vRNA segments include the NCRs and terminal ORF regions at both the 3′ and 5′ ends. Previously, we established a “rewiring” approach to switch the packaging signals of the influenza virus A vRNAs (11, 12). In Fig. 1A, the NP segment was used as an example to show how the packaging sequences were substituted in this study. Briefly, the NP ORF region was amplified using PCR to introduce multiple serial silent mutations into the terminal ORF region packaging sequences identified previously (26). The mutated ORF was flanked by the NA packaging sequences (11), and then the ATGs (translation start codons) in the 3′ NA packaging sequences were removed, generating the NA-NP-NA construct (Fig. 1A). Based on previous observations (8, 12–14, 18–20), this rewired NA-NP-NA segment utilizes the flanking NA packaging signals during genome assembly. Using a similar method, the NA-PB2-NA (11); NA-PB1-NA (11); NA-PA-NA (11); and NA-HEF-NA, NA-M-NA, NA-NS-NA, and HA-HEF-HA (10) constructs were generated either in this study (Fig. 1B) or previously (10, 11). During mutagenesis to knock out the terminal ORF region packaging signals, transitions were preferred over transversions, and every possible silent mutation was introduced in order to completely disable the packaging signals. The lengths of the mutated regions for each ORF were decided based upon previous observations (8, 9, 17, 18, 20, 22, 26, 27, 34). For the NA-M-NA construct (GenBank accession number JQ733039), more mutations were introduced into both ends, considering the fact that the 5′-end sequences of the M ORF were insensitive to mutation (27). In addition, for the NA-M-NA and NA-NS-NA constructs, the sequences surrounding the 5′ mRNA splice sites in the M and NS ORF regions were kept intact. It should be noted that the NA-HEF-NA construct was constructed by replacing the HA packaging sequences of the HA-HEF-HA construct with those of NA and that the HEF ORF region did not carry silent mutations (Fig. 1B). The GFP genes flanked by PB2, PB1, PA, HA, NP, M, or NS packaging signals (Fig. 1C) were either generated in this study or described previously (11) (see Materials and Methods for details). These constructs and the wild-type PR8 virus ambisense rescue plasmids (10) were used to generate recombinant HEF viruses carrying rewired segments.
Generation of multiple 7-segmented HEF viruses carrying rewired segments and 8-segmented HEF viruses carrying an extra GFP segment.
The successful generation and efficient growth of the 7-segmented HEF virus suggested that the NA segment is not required for the packaging of other segments during the genome-packaging process (10). In order to determine the significance of the other packaging sequences during genome incorporation, we generated multiple HEF viruses carrying rewired segments (Table 1). For example, to analyze the role of the PB2 packaging sequences, we generated two viruses using the reverse-genetics technique: a 7-segmented ΔPB2-PS virus (“PS” represents packaging sequences) (Fig. 1D and Table 1) carrying two rewired segments (NA-PB2-NA and HA-HEF-HA) (Fig. 1B) and five PR8 wild-type segments (PB1, PA, NP, M, and NS) (Fig. 1D and Table 1) and an 8-segmented ΔPB2-PS+PB2-GFP-PB2 virus carrying all the segments of the ΔPB2-PS virus (Fig. 1D) and an eighth PB2-GFP-PB2 segment (Table 1 and Fig. 1D). The ΔPB2-PS virus possessed all the packaging signals of the PR8 virus except those of the PB2 segment; the ΔPB2-PS+PB2-GFP-PB2 virus carried all eight packaging sequences of the PR8 virus. Following a similar strategy, we generated five additional 7-segmented viruses: ΔPA-PS, ΔNP-PS, ΔM-PS, ΔPB1-PS, and ΔNS-PS, all of which carried two rewired segments and five wild-type PR8 segments (Table 1 and Fig. 1B). We also generated five more 8-segmented viruses, ΔPA-PS+PA-GFP-PA, ΔNP-PS+NP-GFP-NP, ΔM-PS+M-GFP-M, ΔPB1-PS+PB1-GFP-PB1, and ΔNS-PS+NS-GFP-NS, all of which carried an eighth GFP segment flanked by the packaging sequences that were mutated in the corresponding 7-segmented HEF viruses (Table 1 and Fig. 1C). These viruses were utilized to assess the roles of the PB2, PA, NP, M, PB1, and NS packaging signals. The 7-segmented virus lacking the HA packaging sequences, ΔHA-PS, carried only one rewired segment (NA-HEF-NA) (Fig. 1B) and six PR8 wild-type segments (PB2, PB1, PA, NP, M, and NS) (Table 1), and the ΔHA-PS+HA-GFP-HA virus carried an eighth HA-GFP-HA segment (Fig. 1C and Table 1). These two viruses were generated to study the roles of the HA packaging signals. The ΔNA-PS and ΔNA-PS+NA-GFP-NA viruses were described previously (10) (Table 1).
Table 1.
Summary of recombinant HEF viruses
| Virus | Rewired segments | WTa PR8 segments | GFP segment | Virus growthb | Reference |
|---|---|---|---|---|---|
| ΔPB2-PS | NA-PB2-NA, HA-HEF-HA | PB1, PA, NP, M, NS | ± | This study | |
| ΔPB2-PS+PB2-GFP-PB2 | NA-PB2-NA, HA-HEF-HA | PB1, PA, NP, M, NS | PB2-GFP-PB2 | ++++ | This study |
| ΔPA-PS | NA-PA-NA, HA-HEF-HA | PB2, PB1, NP, M, NS | ++ | This study | |
| ΔPA-PS+PA-GFP-PA | NA-PA-NA, HA-HEF-HA | PB2, PB1, NP, M, NS | PA-GFP-PA | +++++ | This study |
| ΔNP-PS | NA-NP-NA, HA-HEF-HA | PB2, PB1, PA, M, NS | ± | This study | |
| ΔNP-PS+NP-GFP-NP | NA-NP-NA, HA-HEF-HA | PB2, PB1, PA, M, NS | NP-GFP-NP | +++++ | This study |
| ΔM-PS | NA-M-NA, HA-HEF-HA | PB2, PB1, PA, NP, NS | ± | This study | |
| ΔM-PS+M-GFP-M | NA-M-NA, HA-HEF-HA | PB2, PB1, PA, NP, NS | M-GFP-M | +++++ | This study |
| ΔPB1-PS | NA-PB1-NA, HA-HEF-HA | PB2, PA, NP, M, NS | +++++ | This study | |
| ΔPB1-PS+PB1-GFP-PB1 | NA-PB1-NA, HA-HEF-HA | PB2, PA, NP, M, NS | PB1-GFP-PB1 | +++++ | This study |
| ΔHA-PS | NA-HEF-NA | PB2, PB1, PA, NP, M, NS | +++++ | This study | |
| ΔHA-PS+HA-GFP-HA | NA-HEF-NA | PB2, PB1, PA, NP, M, NS | HA-GFP-HA | +++++ | This study |
| ΔNA-PS | HA-HEF-HA | PB2, PB1, PA, NP, M, NS | +++++ | 10 | |
| ΔNA-PS+NA-GFP-NA | HA-HEF-HA | PB2, PB1, PA, NP, M, NS | NA-GFP-NA | +++++ | 10 |
| ΔNS-PS | NA-NS-NA, HA-HEF-HA | PB2, PB1, PA, NP, M | +++++ | This study | |
| ΔNS-PS+NS-GFP-NS | NA-NS-NA, HA-HEF-HA | PB2, PB1, PA, NP, M | NS-GFP-NS | +++++ | This study |
WT, wild type.
Virus growth are determined by measuring the TCID50/ml in embryonated chicken eggs 3 days postinoculation at 33°C. ±, virus that grows poorly and can be lost during passages; ++, titer around 105 TCID50/ml; ++++, titer around 107 TCID50/ml; +++++, titer around 108 TCID50/ml.
PB2, PA, NP, and M are required for efficient packaging of a full complement of the RNA genome.
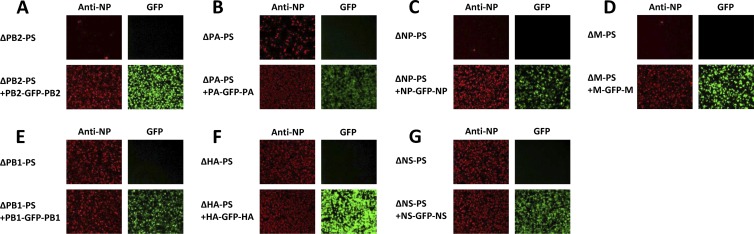
After rescue, all the recombinant 7-segmented and 8-segmented viruses listed in Table 1 were further passaged in embryonated chicken eggs at 33°C two or three times to reach the highest infectious titer. The undiluted viruses were then used to infect 293T cells in a 24-well plate coated with poly-d-lysine. One day postinfection, the infected cells were fixed with formaldehyde and incubated with mouse monoclonal antibody against the PR8 virus NP (HT103). The expression of NP (shown in red) and GFP by infected cells was then visualized by using a fluorescence microscope (Fig. 2). For the ΔPB2-PS (Fig. 2A), ΔNP-PS (Fig. 2C), and ΔM-PS (Fig. 2D) viruses, only a few cells expressing NP could be seen, suggesting the titers of these viruses were very low; however, for the viruses carrying an eighth GFP segment flanked by the missed packaging sequences, ΔPB2-PS+PB2-GFP-PB2 (Fig. 2A), ΔNP-PS+NP-GFP-NP (Fig. 2C), and ΔM-PS+M-GFP-M (Fig. 2D), high numbers of NP-expressing cells were observed, suggesting the infectious titers of these viruses were much higher than those of the corresponding 7-segmented viruses. Cells infected with these GFP viruses showed GFP expression, indicating that the GFP viruses packaged the eighth GFP segment (Fig. 2A, C, and D). These results showed that the existence of an eighth GFP segment carrying the additional packaging sequences greatly increased the growth of these GFP viruses. Even though more NP-expressing cells were observed among cells infected with the ΔPA-PS virus (Fig. 2B) than among those infected with the ΔPB2-PS (Fig. 2A), ΔNP-PS (Fig. 2C), and ΔM-PS (Fig. 2D) viruses, the GFP virus carrying an eighth PA-GFP-PA segment had an increase in the number of NP-expressing cells (approximately 10-fold) (Fig. 2B), indicating that the PA-GFP-PA segment is also required for efficient growth of the virus. These results indicated that the packaging sequences of the PB2, PA, NP, and M segments are required for efficient virus growth.
Fig 2.
Immunofluorescence of 293T cells infected with 7-segmented and 8-segmented HEF viruses. The rescued HEF viruses were passaged in embryonated eggs two or three times to reach peak titers. 293T cells in 24-well plates treated with poly-d-lysine were then infected with undiluted 7- or 8-segmented HEF viruses—ΔPB2-PS and ΔPB2-PS+PB2-GFP-PB2 (A), ΔPA-PS and ΔPA-PS+PA-GFP-PA (B), ΔNP-PS and ΔNP-PS+NP-GFP-NP (C), ΔM-PS and ΔM-PS+M-GFP-M (D), ΔPB1-PS and ΔPB1-PS+PB1-GFP-PB1 (E), ΔHA-PS and ΔHA-PS+HA-GFP-HA (F), and ΔNS-PS and ΔNS-PS+NS-GFP-NS (G)—and incubated in a 37°C incubator overnight. The cells were then visualized for NP (red) and GFP expression by using an immunofluorescence assay (see Materials and Methods). All the GFP viruses (A to G) and the three 7-segmented viruses ΔPB1-PS (E), ΔHA-PS (F), and ΔNS-PS (G) were normalized based on the hemagglutination assay titers measured by using chicken red blood cells, and the multiplicity of infection for these viruses was around 5 to 10; for the remaining 7-segmented viruses, those with the highest TCID50/ml after passages in eggs were used to infect 293T cells.
On the other hand, for the three 7-segmented HEF viruses ΔPB1-PS (Fig. 2E), ΔHA-PS (Fig. 2F), and ΔNS-PS (Fig. 2G), the majority of infected 293T cells were observed to express NP, suggesting these viruses could reach high titers in the absence of the PB1, HA, or NS packaging signals; for cells infected with the 8-segmented viruses carrying an eighth GFP segment (ΔPB1-PS+PB1-GFP-PB1 [Fig. 2E], ΔHA-PS+HA-GFP-HA [Fig. 2F], and ΔNS-PS+NS-GFP-NS [Fig. 2G]), the numbers of NP-expressing cells were similar to those infected with the corresponding 7-segmented HEF viruses, and they all expressed GFP. These results indicated that the packaging sequences of the PB1, HA, and NS segments are not required for efficient replication of the virus.
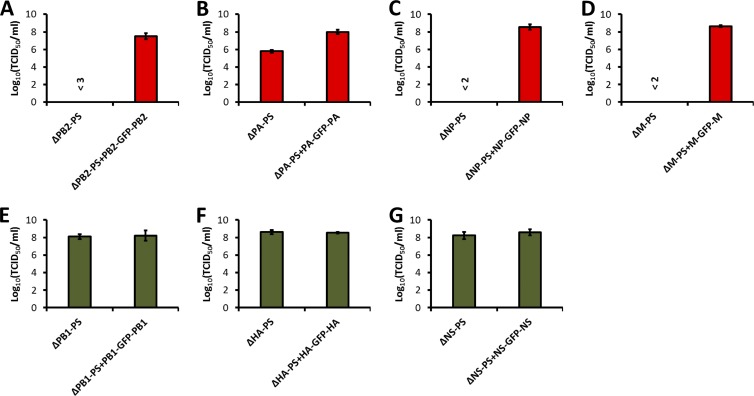
To further confirm the virus growth property observed in Fig. 2, we inoculated 100 TCID50 of each virus into embryonated chicken eggs and determined the titers of these viruses in the eggs 3 days postinoculation (Fig. 3). For three viruses, ΔPB2-PS (Fig. 3A), ΔNP-PS (Fig. 3C), and ΔM-PS (Fig. 3D), we were unable to determine an accurate titer because of the low growth of the viruses in the eggs. We estimated that the titer of the ΔPB2-PS virus were lower than 103 TCID50/ml (Fig. 3A) and the titers of the ΔNP-PS (Fig. 3C) and ΔM-PS (Fig. 3D) viruses were lower than 102 TCID50/ml. In fact, these viruses were often lost during passages in eggs. When an eighth GFP segment was added, all the viruses reached titers on the order of 107 to 108 TCID50/ml (Fig. 3A, C, and D). The ΔPA-PS virus grew to a titer of less than 106 TCID50/ml, and the ΔPA-PS+PA-GFP-PA virus carrying an eighth GFP segment reached a titer of around 108 TCID50/ml (Fig. 3B). The titers of the three 7-segmented viruses, ΔPB1-PS (Fig. 3E), ΔHA-PS (Fig. 3F), and ΔNS-PS (Fig. 3G), were around 108 TCID50/ml, similar to those of the 8-segmented viruses carrying a GFP segment, ΔPB1-PS+PB1-GFP-PB1 (Fig. 3E), ΔHA-PS+HA-GFP-HA (Fig. 3F), and ΔNS-PS+NS-GFP-NS (Fig. 3G). These results further confirmed the results in Fig. 2, indicating that the packaging signals of the PB2, PA, NP, and M segments are required for efficient virus growth, while those of the PB1, HA, and NS segments are not. Since segment-specific packaging signals were shown to affect the incorporation of the corresponding parental segments into virus particles and the genomes of all the rewired 7-segmented viruses encode the same viral proteins required for viral replication, it is possible that the PB2, PA, NP, and M packaging signals are involved in the packaging of not only themselves, but also other viral segments into virions.
Fig 3.
Growth of HEF viruses in embryonated chicken eggs. Eight-day-old chicken eggs were inoculated with 100 TCID50 of ΔPB2-PS and ΔPB2-PS+PB2-GFP-PB2 (A), ΔPA-PS and ΔPA-PS+PA-GFP-PA (B), ΔNP-PS and ΔNP-PS+NP-GFP-NP (C), ΔM-PS and ΔM-PS+M-GFP-M (D), ΔPB1-PS and ΔPB1-PS+PB1-GFP-PB1 (E), ΔHA-PS and ΔHA-PS+HA-GFP-HA (F), and ΔNS-PS and ΔNS-PS+NS-GFP-NS (G) viruses; incubated at 33°C for 3 days; and transferred to 4°C overnight. The allantoic fluids were then harvested, and the virus titers (TCID50/ml) were determined in 96-well plates using an immunofluorescence method with anti-PR8 NP mouse monoclonal antibody HT103 (25) and Alexa Fluor 594-labeled donkey anti-mouse IgG (Invitrogen) (Fig. 2). Three viruses, ΔPB2-PS (A), ΔNP-PS (C), and ΔM-PS (D), grew very poorly and were easily lost during the passages. Therefore, we could not determine accurate titers for them. We estimated their titers at less than 103, 102, and 102 TCID50/ml, respectively. The mean titer ± standard deviation calculated from three eggs is shown for each virus.
vRNA packaging of the recombinant HEF viruses.
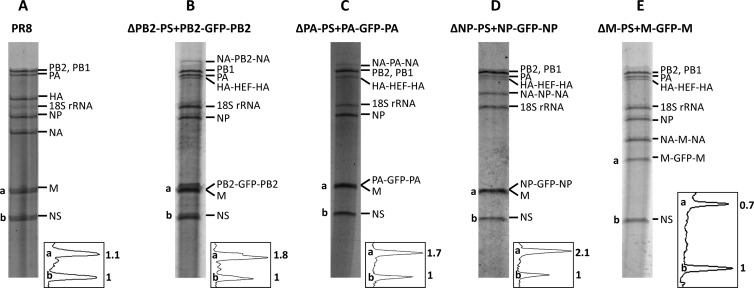
The packaging of vRNAs into the recombinant HEF viruses was assessed by examination of the RNA compositions of concentrated egg-grown viruses. The HEF viruses were grown in eggs and concentrated through a 30% sucrose cushion. RNA was isolated from purified virus and resolved on a 2.8% acrylamide gel to visualize the virus genome composition by silver staining. We first examined the four HEF viruses in which the eighth GFP segment was essential for the virus to reach high growth titers: ΔPB2-PS+PB2-GFP-PB2 (Fig. 4B), ΔPA-PS+PA-GFP-PA (Fig. 4C), ΔNP-PS+NP-GFP-NP (Fig. 4D), and ΔM-PS+M-GFP-M (Fig. 4E). Because of the low growth of the four 7-segmented HEF viruses—ΔPB2-PS, ΔPA-PS, ΔNP-PS, and ΔM-PS—we were unable to analyze their genomic RNA compositions by using this method. The silver-staining results show that these four GFP viruses lacked the HA and NA segments of the PR8 virus shown in Fig. 4A. Instead, they all contained a chimeric HA-HEF-HA segment (Fig. 4B to E). Furthermore, they all carried a rewired segment, namely, NA-PB2-NA for the ΔPB2-PS+PB2-GFP-PB2 virus (Fig. 4B), NA-PA-NA for the ΔPA-PS+PA-GFP-PA virus (Fig. 4C), NA-NP-NA for the ΔNP-PS+NP-GFP-NP virus (Fig. 4D), and NA-M-NA for the ΔM-PS+M-GFP-M virus (Fig. 4E). The three GFP segments (PB2-GFP-PB2, PA-GFP-PA, and NP-GFP-NP) comigrated with the M segment due to their length similarity to the M segment (Fig. 4B to D); therefore, we were unable to differentiate them from the M segment. However, after the intensities of the bands were normalized to that of the NS segment (labeled “b” in Fig. 4A to D), comigration of the GFP segment resulted in a band with approximately 2-fold-higher intensity at the M vRNA position (labeled “a” in Fig. 4B to D) than the M segment of the PR8 virus (Fig. 4A). This suggested that the GFP segments were efficiently incorporated into these viruses. For the ΔM-PS+M-GFP-M virus (Fig. 4E), the positions of the rewired M segment, NA-M-NA, and the eighth GFP segment, M-GFP-M, were all visible in the gel, with the intensity of the M-GFP-M segment similar to that of NA-M-NA, indicating that the GFP segment was also incorporated well (Fig. 4E).
Fig 4.
Genome compositions of four HEF GFP viruses. (A) The PR8 virus was grown in 10-day-old chicken eggs at 37°C for 2 days. (B to E) The HEF viruses ΔPB2-PS+PB2-GFP-PB2 (B), ΔPA-PS+PA-GFP-PA (C), ΔNP-PS+NP-GFP-NP (D), and ΔM-PS+M-GFP-M (E) were grown in 8-day-old chicken eggs at 33°C for 3 days. The viruses were then purified by passing them through a sucrose cushion, and RNA was isolated using TRIzol (Invitrogen). The purified RNA was separated on a 2.8% denaturing polyacrylamide gel containing 7 M urea. The gel was stained with a SilverXpress silver-staining kit (Invitrogen) to visualize the RNA bands. The bands representing the genomic segments of the virus were labeled for each gel. The two bands corresponding to the M (or rewired M [E]) segment and the NS segment (labeled “a” and “b,” respectively) were quantified for intensity using ImageQuant software. The graphs used to quantify the two peaks (a and b) are shown at the lower right of each gel. The numbers represent the relative peak volumes (with peak b set to 1). Note that the gels shown were run at different times.
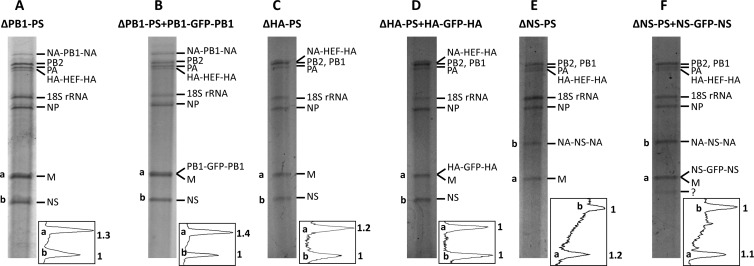
We also examined the genomic compositions of the three 7-segmented viruses, ΔPB1-PS (Fig. 5A), ΔHA-PS (Fig. 5C), and ΔNS-PS (Fig. 5E), and the corresponding GFP viruses, ΔPB1-PS+PB1-GFP-PB1 (Fig. 5B), ΔHA-PS+HA-GFP-HA (Fig. 5D), and ΔNS-PS+NS-GFP-NS (Fig. 5F). Both the ΔPB1-PS (Fig. 5A) and ΔNS-PS (Fig. 5E) viruses contained an HA-HEF-HA segment and a rewired segment: NA-PB1-NA for the ΔPB1-PS virus (Fig. 5A) and NA-NS-NA for the ΔNS-PS virus (Fig. 5E). The ΔHA-PS virus contained only one chimeric segment, NA-HEF-NA, which comigrated with PB2 and PB1 because of length similarity (Fig. 5C). For three GFP viruses (ΔPB1-PS+PB1-GFP-PB1 [Fig. 5B], ΔHA-PS+HA-GFP-HA [Fig. 5D], and ΔNS-PS+NS-GFP-NS [Fig. 5F]), due to the length similarity, this method could not differentiate the M segment from the eighth GFP segment the viruses carried. Unlike the three GFP viruses examined previously (ΔPB2-PS+PB2-GFP-PB2 [Fig. 4B], ΔPA-PS+PA-GFP-PA [Fig. 4C], and ΔNP-PS+NP-GFP-NP [Fig. 4D], the comigration of the GFP segments with the M segment in these three GFP viruses did not result in a heavier band at the M vRNA position, suggesting the GFP segments were not incorporated efficiently (Fig. 5B, D, and F). However, the expression of GFP by cells infected with these viruses did indicate their presence in the particles (Fig. 2E to G).
Fig 5.
Genome compositions of recombinant 7- and 8-segmented HEF viruses carrying a rewired PB1, HA, or NS segment. Using the same method as in Fig. 4, six HEF viruses, ΔPB1-PS (A), ΔPB1-PS+PB1-GFP-PB1 (B), ΔHA-PS (C), ΔHA-PS+HA-GFP-HA (D), ΔNS-PS (E), and ΔNS-PS+NS-GFP-NS (F), were analyzed for genome composition using an RNA gel and silver-staining method. The quantifications of the bands at the bottom of each gel are also shown. ?, unknown band.
Taken together, the vRNA compositions of these HEF viruses showed that they indeed contained the rewired segments we designed (Fig. 4 and 5). More importantly, the GFP segments carrying the PB2, PA, NP, or M packaging signals were incorporated more efficiently into the viruses than those carrying the PB1, HA, or NS packaging signals (Fig. 4B to E and 5B, D, and F).
qRT-PCR results confirmed the high packaging efficiency of GFP segments flanked by the PB2, PA, NP, or M packaging sequences.
To further evaluate the effects of packaging sequences on the packaging efficiencies of the engineered GFP segments and to overcome the issue of the GFP segments comigrating with the M segment due to length similarity (Fig. 4 and 5), qRT-PCR was performed to quantify the GFP vRNA levels relative to the NP vRNA levels in these HEF GFP viruses (Fig. 6). Prior to the experiment, the viruses were further passaged three times (to passage 6) in embryonated chicken eggs, harvested, and purified by passing them through a sucrose cushion. The purified RNA was reverse transcribed, and the GFP/NP vRNA ratio was determined (Fig. 6A). The GFP/NP vRNA ratio for the ΔNP-PS+NP-GFP-NP virus was arbitrarily set at 1 (Fig. 6B). The results showed that for the four HEF GFP viruses (ΔPB2-PS+PB2-GFP-PB2, ΔPA-PS+PA-GFP-PA, ΔNP-PS+NP-GFP-NP, and ΔM-PS+M-GFP-M), the GFP/NP vRNA ratios were all higher than 0.6; for the other three GFP viruses (ΔPB1-PS+PB1-GFP-PB1, ΔHA-PS+HA-GFP-HA, and ΔNS-PS+NS-GFP-NS), the GFP/NP vRNA ratios were all lower than 0.2 (Fig. 6B). Combining these data and the RNA gel silver-staining results (Fig. 4 and 5), we conclude that the GFP segments flanked by the PB2, PA, NP, or M packaging sequence are packaged much more efficiently into the viruses, suggesting they play a more important role during the genome assembly process.
Fig 6.
qRT-PCR analysis of recombinant HEF GFP viruses. (A) Procedure for the experiment. (B) GFP/NP vRNA ratios for each virus. In total, seven viruses were analyzed: ΔPB2-PS+PB2-GFP-PB2, ΔPA-PS+PA-GFP-PA, ΔNP-PS+NP-GFP-NP, ΔM-PS+M-GFP-M, ΔPB1-PS+PB1-GFP-PB1, ΔHA-PS+HA-GFP-HA, and ΔNS-PS+NS-GFP-NS. The GFP/NP vRNA ratio for the ΔNP-PS+NP-GFP-NP virus was arbitrarily set at 1. The experiment was performed in triplicate, and the mean value ± standard deviation is shown for each virus.
smFISH analysis showed high copackaging efficiency of the GFP and NP segments in ΔPA-PS+PA-GFP-PA virus.
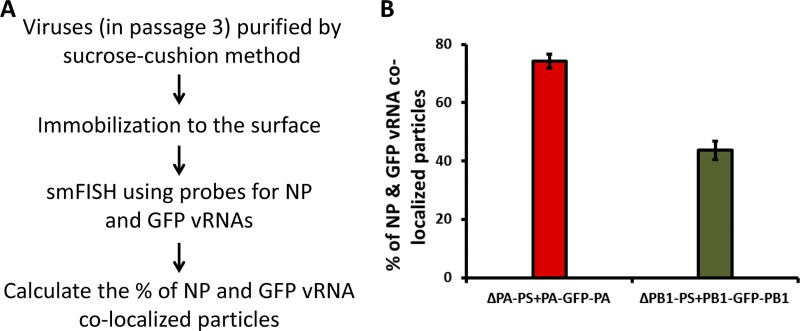
Since both RNA gel silver staining and qRT-PCR techniques measured the efficiency of vRNA incorporation in a virus population, we conducted smFISH experiments (2, 31) to analyze the incorporated viral RNAs in individual virus particles and measured the efficiencies of two viral RNAs being copackaged. The efficiencies of the GFP and NP segments incorporated into the same virus particles were analyzed for the ΔPA-PS+PA-GFP-PA and ΔPB1-PS+PB1-GFP-PB1 viruses. The two viruses showed similar growth characteristics, while the corresponding 7-segmented ΔPA-PS virus grew much less efficiently than the 7-segmented ΔPB1-PS virus (Fig. 3B and E). To do smFISH analysis, a slide surface was passivated with an inert polymer polyethylene glycol (PEG) doped with biotinylated PEG. After incubating the surface with NeutrAvidin (Thermo), the purified egg-grown viruses were immobilized on the imaging surface through biotinylated antibody against the HEF protein captured by the NeutrAvidin. Fifteen Cy5-labeled probes against the NP vRNA and 24 Cy3-labeled probes against the GFP vRNA were used to detect viral RNAs in the fixed and permeabilized virus particles (Fig. 7A). The percentages of particles detected with both NP and GFP probes among the sum of particles detected with either NP or GFP probes were calculated. For the ΔPA-PS+PA-GFP-PA virus, the percentage of NP and GFP being copackaged into the same virus particle was around 75%, while for the ΔPB1-PS+PB1-GFP-PB1 virus, the ratio was around 40% and was statistically lower than that for the ΔPA-PS+PA-GFP-PA virus (Fig. 7B). This result showed that the GFP segment flanked by the PA packaging sequences was copackaged more efficiently with the NP segment than the one flanked by the PB1 packaging sequences.
Fig 7.
smFISH analysis of single virus particles. The smFISH experiment (see Materials and Methods) was performed to measure the percentages of GFP and NP vRNA colocalized particles. (A) Experimental procedure. (B) Percentages of GFP and NP colocalized particles. Two viruses, ΔPA-PS+PA-GFP-PA and ΔPB1-PS+PB1-GFP-PB1, were analyzed. For each virus, at least 10 images were analyzed, and the mean values ± standard deviations are shown. On average, 500 to 700 particles were counted for each image.
DISCUSSION
Segmented RNA viruses face a challenging task of assembling their genomes during the late stage of infection. Based on the recent identification of influenza A virus segment-specific packaging signals and the electron microscopy data showing that one particle incorporates eight RNA segments in budding virions (8, 9, 17, 18, 20, 22, 23, 26, 27, 34) (see Fig. 47.23 in reference 28), the packaging of the influenza A virus vRNAs appears to be a specific process. The FISH analysis of vRNAs using fluorophore-labeled probes also showed that influenza A virus assembles its genome in a highly specific fashion by incorporating only one copy of each segment into the majority of virus particles (2). Nevertheless, due to the technical difficulties in studying large RNP complexes inside infected cells, how the eight segments interact to form the vRNP complex inside virions has yet to be determined. Recent electron tomography analyses of vRNP structure within the budding virions showed that the eight segments are arranged in multiple configurations (7, 24), indicating that there might be a random element in this seemingly very specific process. The question we wanted to address is whether each segment of the influenza A virus has a similar role during genome packaging. Previous findings suggested some segments might play unequal roles. Muramoto et al. showed that when reverse genetics was applied to generate virus-like particles carrying influenza A vRNAs, the omission of the PB2 segment had a greater impact on the production of vRNA-containing particles than removing the PB1 or PA vRNA, suggesting PB2 might play a more important role during the packaging process (22). Furthermore, Marsh et al. showed that mutations in the conserved region of the PB1 segment have much less of an effect on the packaging of other segments than mutations in the PB2 and PA segments, indicating PB1 is less important than PB2 and PA (20). Moreover, an NA-lacking 7-segmented HEF virus we generated grew to a high titer, demonstrating that the NA segment is not crucial for influenza A virus genome assembly (10). All these experiments were performed using quite different approaches; therefore, it is difficult to compare the relative contributions of individual RNA segments during genome packaging from these studies alone.
In the present study, to overcome this issue, we used a 7-segmented HEF virus as a tool and applied the “rewiring” method to determine the roles of individual segment-specific packaging signals in the genome assembly process. This unique approach allowed us to compare the functions of all eight influenza A virus genome-packaging signals under one condition. We rewired the segments of the 7-segmented HEF virus by replacing the packaging signals with those of the NA segment and assessed its effect on viral growth and genome packaging (Fig. 2 to 6). Our results showed that the 7-segmented HEF viruses lacking the PB2, M, or NP segment grew poorly and the viruses were easily lost during passages in eggs (Fig. 3); however, when an eighth GFP segment carrying the missing PB2, M, or NP packaging sequence was included, the resulting 8-segmented HEF virus reached a high titer (Fig. 2 and 3). Although the 7-segmented HEF virus lacking the PA packaging sequences can grow to a titer around 105 TCID50/ml, adding an eighth GFP segment increased the titer by about 3 log units (Fig. 2 and 3). These results suggested that the efficient growth of the virus requires the presence of the packaging signals of the PB2, PA, NP, and M segments. Previously, Hutchinson et al. showed that the silent mutations in the M segment packaging signals can result in a packaging defect in all seven other segments (14), and silent mutations in the NP segment can result in inefficient incorporation of the PA segment (15). These observations also indicate that the M and NP segments play a more important role during genome packaging. Taken together, these segments (PB2, PA, NP, and M) may function to recruit other segments during the genome-packaging process. For the PB1, HA, NA, and NS segments, the rewired HEF viruses grew to high titers in both the presence and absence of the eighth GFP segment carrying the missing packaging signals (Fig. 2 and 3) (10), suggesting the four segments are not required for the packaging of the other vRNA segments. In fact, for the three HEF GFP viruses (ΔPB1-PS+PB1-GFP-PB1, ΔHA-PS+HA-GFP-HA, and ΔNS-PS+NS-GFP-NS), the levels of GFP vRNA relative to NP in purified particles fluctuated during passages in eggs (data not shown). It should be noted that all the flanking PB1, HA, NA, and NS packaging sequences used in this study and a previous study (10) have been shown to be efficient in mediating the packaging of a GFP reporter vRNA (8, 9, 22, 34). These data suggested that the PB1, HA, NA, and NS segments play nonessential roles during genome assembly, and as a result, they might be more permissive toward reassortment.
Finally, we propose a model for influenza A virus genome packaging. Because of its multisegmented nature, it may be unlikely that influenza A virus assembles all its genomic segments together at the same time. For example, another segmented RNA virus, the double-stranded RNA (dsRNA) bacteriophage Φ6, specifically packages its RNA genome (S, M, and L) in a sequential manner, first S, then M, and finally the L segment (21). Considering the findings in this study and previous observations (7–10, 17, 18, 20, 22–24, 26, 27, 34), we propose that during influenza A virus genome packaging, the four segments PB2, PA, NP, and M interact in the packaging signal regions, bringing the PB1, HA, NA, and NS segments to join and eventually form the “7 + 1” vRNP complex observed by electron microscopy (7, 23, 24).
ACKNOWLEDGMENTS
We thank John Steel for providing the NA-HEF-NA plasmid.
This work was partially supported by NIH grants HHSN2662000700010C (Center for Research on Influenza Pathogenesis to P.P.) and U54 AI057158-04 (Northeast Biodefense Center to P.P.), National Science Foundation grants (0646550 and 0822613 to T.H.), and a National Institutes of Health grant (U19AI083025 to T.H.). T.H. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Published ahead of print 24 April 2012
REFERENCES
- 1. Amorim MJ, et al. 2011. A Rab11- and microtubule-dependent mechanism for cytoplasmic transport of influenza A virus viral RNA. J. Virol. 85:4143–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chou Y-Y, et al. 30 April 2012. One influenza virus particle packages eight unique viral RNAs as shown by FISH analysis. Proc. Natl. Acad. Sci. U. S. A. doi:10.1073/pnas.1206069109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Duhaut SD, McCauley JW. 1996. Defective RNAs inhibit the assembly of influenza virus genome segments in a segment-specific manner. Virology 216:326–337 [DOI] [PubMed] [Google Scholar]
- 4. Eisfeld AJ, Kawakami E, Watanabe T, Neumann G, Kawaoka Y. 2011. RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J. Virol. 85:6117–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eisfeld AJ, Neumann G, Kawaoka Y. 2011. Human immunodeficiency virus rev-binding protein is essential for influenza A virus replication and promotes genome trafficking in late-stage infection. J. Virol. 85:9588–9598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fodor E, et al. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679–9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fournier E, et al. 2012. A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic Acids Res. 40:2197–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fujii K, et al. 2005. Importance of both the coding and the segment-specific noncoding regions of the influenza A virus NS segment for its efficient incorporation into virions. J. Virol. 79:3766–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fujii Y, Goto H, Watanabe T, Yoshida T, Kawaoka Y. 2003. Selective incorporation of influenza virus RNA segments into virions. Proc. Natl. Acad. Sci. U. S. A. 100:2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao Q, Brydon EW, Palese P. 2008. A seven-segmented influenza A virus expressing the influenza C virus glycoprotein HEF. J. Virol. 82:6419–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao Q, Lowen AC, Wang TT, Palese P. 2010. A nine-segment influenza a virus carrying subtype H1 and H3 hemagglutinins. J. Virol. 84:8062–8071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao Q, Palese P. 2009. Rewiring the RNAs of influenza virus to prevent reassortment. Proc. Natl. Acad. Sci. U. S. A. 106:15891–15896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gog JR, et al. 2007. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic Acids Res. 35:1897–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hutchinson EC, Curran MD, Read EK, Gog JR, Digard P. 2008. Mutational analysis of cis-acting RNA signals in segment 7 of influenza A virus. J. Virol. 82:11869–11879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hutchinson EC, Wise HM, Kudryavtseva K, Curran MD, Digard P. 2009. Characterisation of influenza A viruses with mutations in segment 5 packaging signals. Vaccine 27:6270–6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jain A, et al. 2011. Probing cellular protein complexes using single-molecule pull-down. Nature 473:484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liang Y, Hong Y, Parslow TG. 2005. cis-Acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments. J. Virol. 79:10348–10355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liang Y, Huang T, Ly H, Parslow TG, Liang Y. 2008. Mutational analyses of packaging signals in influenza virus PA, PB1, and PB2 genomic RNA segments. J. Virol. 82:229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marsh GA, Hatami R, Palese P. 2007. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J. Virol. 81:9727–9736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marsh GA, Rabadan R, Levine AJ, Palese P. 2008. Highly conserved regions of influenza A virus polymerase gene segments are critical for efficient viral RNA packaging. J. Virol. 82:2295–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mindich L. 1999. Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage phi6. Microbiol. Mol. Biol. Rev. 63:149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Muramoto Y, et al. 2006. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J. Virol. 80:2318–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Noda T, et al. 2006. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 439:490–492 [DOI] [PubMed] [Google Scholar]
- 24. Noda T, et al. 2012. Three-dimensional analysis of ribonucleoprotein complexes in influenza A virus. Nat. Commun. 3:639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. O'Neill RE, Talon J, Palese P. 1998. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 17:288–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ozawa M, et al. 2007. Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J. Virol. 81:30–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ozawa M, et al. 2009. Nucleotide sequence requirements at the 5′ end of the influenza A virus M RNA segment for efficient virus replication. J. Virol. 83:3384–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Palese P, Shaw ML. 2007. Orthomyxoviridae: the viruses and their replication, p 1647–1689 In Knipe DM, Howley PM. (ed), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 29. Quinlivan M, et al. 2005. Attenuation of equine influenza viruses through truncations of the NS1 protein. J. Virol. 79:8431–8439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Raj A, Tyagi S. 2010. Detection of individual endogenous RNA transcripts in situ using multiple singly labeled probes. Methods Enzymol. 472:365–386 [DOI] [PubMed] [Google Scholar]
- 31. Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. 2008. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5:877–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Roy R, Hohng S, Ha T. 2008. A practical guide to single-molecule FRET. Nat. Methods 5:507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ulbrich MH, Isacoff EY. 2007. Subunit counting in membrane-bound proteins. Nat. Methods 4:319–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Watanabe T, Watanabe S, Noda T, Fujii Y, Kawaoka Y. 2003. Exploitation of nucleic acid packaging signals to generate a novel influenza virus-based vector stably expressing two foreign genes. J. Virol. 77:10575–10583 [DOI] [PMC free article] [PubMed] [Google Scholar]