Abstract
Two novel phytases have been characterized from Bifidobacterium pseudocatenulatum and Bifidobacterium longum subsp. infantis. The enzymes belong to a new subclass within the histidine acid phytases, are highly specific for the hydrolysis of phytate, and render myo-inositol triphosphate as the final hydrolysis product. They represent the first phytases characterized from this group of probiotic microorganisms, opening the possibilities for their use in the processing of high-phytate-content foods.
TEXT
Phytases are a special class of phosphatases that catalyze the sequential hydrolysis of myo-inositol hexakis phosphate (InsP6) or phytate, the major form of phosphorous storage in plants, to lower myo-inositol phosphate derivatives. Based on biochemical properties and amino acid sequence, phytases can be categorized into two major classes, the histidine acid phytases and the alkaline phytases (9). The use of commercial phytases of fungal origin (usually Trichoderma and Aspergillus strains) is a common practice in animal feeding for improving the bioaccessibility of minerals (zinc, iron, calcium, and magnesium) that are otherwise chelated by InsP6 and InsP5 and insolubilized (4, 13). The presence of phytase activity has recently been reported to occur in the human intestinal Bifidobacterium pseudocatenulatum ATCC 27919 and Bifidobacterium longum subsp. infantis ATCC 15697 strains (5, 7). These bacteria were included in the production process of whole-wheat bread, showing their potential to reduce phytate content in fiber-rich products for human consumption (15, 16). However, no genetic information about the corresponding enzymes was available.
Identification and purification of bifidobacterial phytases.
When the genomes from B. pseudocatenulatum ATCC 27919 (GenBank accession no. NZ_ABXX02000003) and B. longum subsp. infantis ATCC 15697 (GenBank accession no. CP001095) were being searched for the presence of genes encoding putative phosphatases, two genes (BIFPSEUDO_03792 and BLON_0263, designated hereinafter PhypA and PhylA, respectively) attracted our attention. These genes encoded two proteins with 59% amino acid identity which possessed histidine acid phosphatase (HAP) domains (PF00328). Furthermore, PhypA and PhylA displayed N-terminal signal peptides for secretion and a predicted C-terminal sortase-dependent cell wall-anchoring domain. Initial characterization of the phytase activity in both bifidobacterial strains revealed that it was mainly located at the cell wall/membrane fractions (data not shown). Hence, the identified genes were good candidates for encoding the bifidobacterial phytases.
Sequence analysis additionally showed that the typical HAP signature sequence of the histidine-containing catalytic site (RHGXRXP) was replaced by RHGSRGL in PhypA and PhylA and that the aspartate of the C-terminal HD domain was replaced by alanine. Remarkably, the putative HAP homologues closer to PhylA and PhypA found in databases, belonging to the Actinobacteria, Betaproteobacteria, and Gammaproteobacteria, show the same variants in their sequences (N-terminal RHGSRXX and C-terminal HA-conserved motifs) (see Fig. S1 in the supplemental material). These results suggest the existence of a new class of bacterial HAPs within branch 2 of the histidine phosphatase superfamily, as the ones reported so far show the N-terminal RHGXRXP and C-terminal HD motifs. A phylogenetic analysis revealed that the putative HAPs with the RHGSRXX and HA motifs are in a cluster that is closer in the first instance to the plant, vertebrate, and fungal phytases than to a different bacterial HAP subgroup that includes the deeply characterized Escherichia coli (8) and Klebsiella sp. (1) phytases, among others (Fig. S2). In bifidobacteria, orthologues of PhypA and PhylA were found only in the genomes of B. longum, B. pseudocatenulatum, and Bifidobacterium dentium isolates (58 to 82% amino acid identity), which correlated with the results for previous enzymatic tests (6, 11, 12).
The PhypA and PhylA genes were amplified by PCR excluding the sequences encoding the N-terminal signal peptides (48 amino acids for PhypA and 31 amino acids for PhylA) and the C-terminal hydrophobic domains (deletion of the 27 and 24 C-terminal amino acids in PhypA and PhylA, respectively) and cloned into the expression vector pQE80 (Qiagen) for their purification as 6×His-tagged proteins (see Fig. S3 and Table S1 in the supplemental material). The enzymes were purified from Escherichia coli M15-induced cultures grown in 1 liter of LB by chromatography on Ni-nitrilotriacetic acid (NTA) agarose columns (1-ml bed volume) according to the supplier's instructions (Qiagen). The fractions containing the expressed proteins were dialyzed against 100 mM Tris-HCl, pH 7.4, 1 mM EDTA, 10% glycerol, 50 mM NaCl before being applied to a Resource Q 1-ml column (GE Healthcare) installed in an Äktapurifier fast-performance liquid chromatography (FPLC) system (GE Healthcare). The elution profile consisted of a linear gradient of NaCl from 0 to 1 M in 20 mM Tris-HCl, pH 6.0, and the purified recombinant proteins were stored at −80°C.
Characterization of the phytase activity of the purified enzymes.
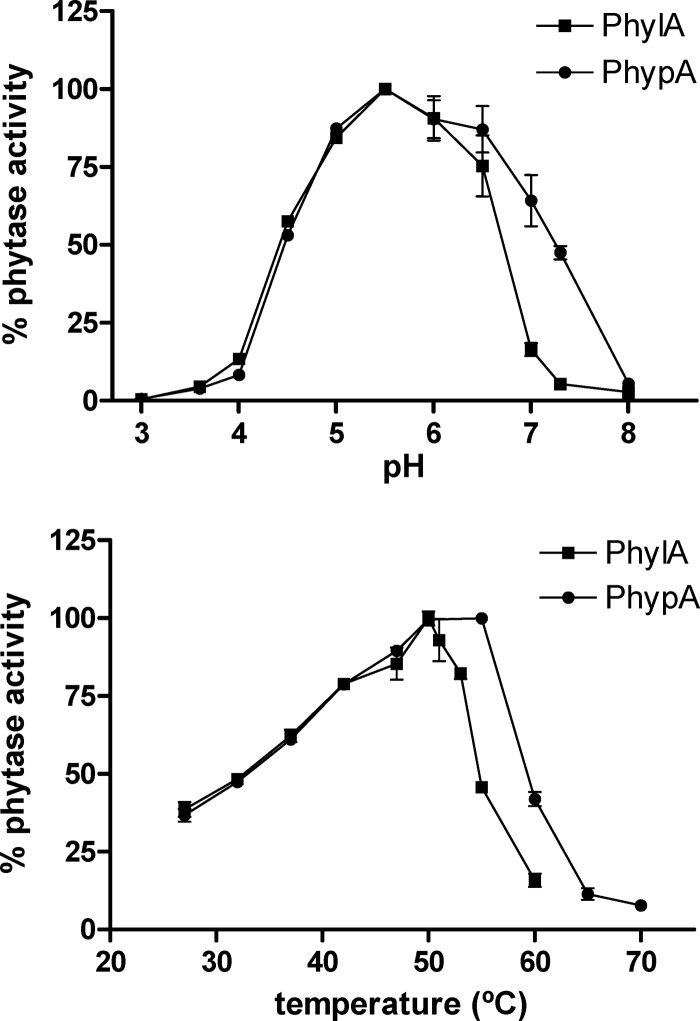
Both enzymes displayed phytase activity that could be evidenced by measuring the release of inorganic phosphate from phytate as previously described (6). The activity was determined with a range of buffers (sodium citrate-NaOH buffer for pH 3.0, sodium acetate/acetic acid buffer for the pH range of 3.6 to 5.5, bis-Tris-HCl buffer for the pH range of 6.0 to 7.3, and Tris-HCl for pH 8 at a final concentration of 100 mM) containing 0.2 to 0.3 μg/ml of the purified enzymes and 1.2 mM potassium phytate (Sigma). While both enzymes showed their optimum activities at pH 5.5 and 50°C, PhypA displayed a broader activity range, conserving more than 90% activity at pH 6.5 and 55°C (Fig. 1). The two enzymes showed a remarkable thermal stability at 80°C, preserving 44% (PhylA) and 73% (PhypA) of their activities after 15 min. The activities were negatively affected by all the assayed compounds to a low extent (less than 40% inhibition) (Table 1), and both enzymes were very specific for phytate (Table 2). Thus, PhylA and PhypA were closer to the E. coli phytase (3) than to other fungal HAP phytases, which are able to hydrolyze a wide range of phosphorylated compounds (9).
Fig 1.
Determination of optimal pH and temperature for 6×(His)PhylA and 6×(His)PhypA enzymes by use of phytate as the substrate. Different pHs were assayed at 50°C, and different temperatures were assayed at pH 5.5. Experiments were run in triplicate.
Table 1.
Effects of different compounds on phytase activity
| Compound (5 mM) | % phytase activity (mean ± SD)a |
|
|---|---|---|
| PhypA | PhylA | |
| None | 100 ± 0.90 | 100 ± 1.04 |
| CoCl2 | 79.8 ± 3.14 | 84 ± 1.04 |
| MnCl2 | 71 ± 0.27 | 76 ± 2.5 |
| EDTA | 85.3 ± 0.1 | 69.2 ± 3.8 |
| Phenylmethylsulfonyl fluoride | 92.1 ± 0.1 | 65.1 ± 3.4 |
| β-Mercaptoethanol | 61 ± 0.2 | 64.1 ± 0.26 |
| Iodoacetic acid | 100 ± 0.3 | 99.9 ± 1.43 |
| Potassium fluoride | 70.3 ± 0.63 | 79.3 ± 1.16 |
The reactions were carried out in triplicate in 100 mM acetate buffer, pH 5.5, with 1 mM phytate at 50°C.
Table 2.
Activity of bifidobacterial phytases on different phosphorylated substrates
| Substrate (1 mM) | % phytase activity (mean ± SD)a |
|
|---|---|---|
| PhypA | PhylA | |
| Phytate | 100 ± 0.25 | 100 ± 0.34 |
| p-Nitrophenyl-phosphate | 0.9 ± 0.42 | 1.05 ± 0.11 |
| Phosphoenolpyruvate | ND | ND |
| Acetyl-phosphate | 6.7 ± 1.8 | 2.3 ± 0.45 |
| AMP | ND | 0.4 ± 0.11 |
| ADP | ND | ND |
| ATP | ND | ND |
| Glyceraldehyde-3-phosphate | ND | ND |
| Glucose 1-phosphate | ND | ND |
| Glucose 6-phosphate | ND | ND |
| Fructose 1-phosphate | ND | ND |
| Fructose 1,6-bisphosphate | 4.9 ± 0.1 | 1.45 ± 0.45 |
The reactions were carried out in triplicate in 100 mM acetate buffer, pH 5.5, at 50°C. ND, no activity detected.
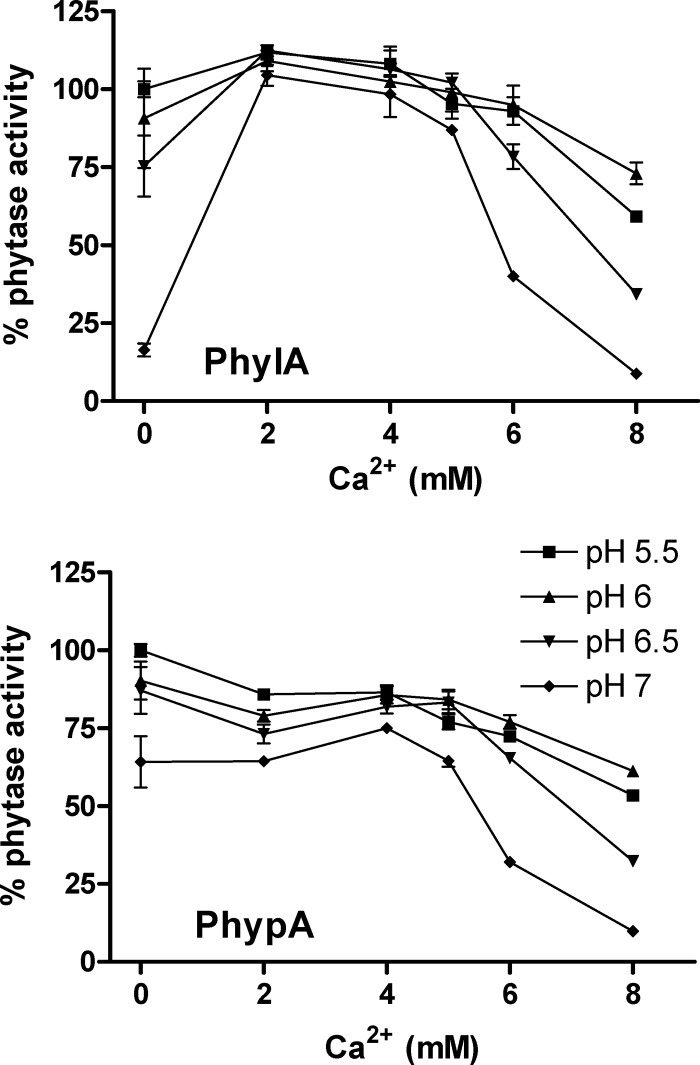
Strict dependence for Ca2+ is typical for alkaline phytases, where the phytate-Ca2+ complex is the molecule which is accommodated at the active center (10). In contrast, a negative effect of Ca2+ on phytase activity is the general rule for phytases belonging to the HAP family, which act on metal-free phytate (9). PhypA and PhylA were equally inhibited by Ca2+ at concentrations above 4 mM. However, at low concentrations (2 mM), PhylA displayed activation by Ca2+, which was more pronounced at pH 7 (Fig. 2). In PhypA, such Ca2+-dependent activation was not produced. Chelating agents such as EDTA generally have a positive effect ranging from a 10 to a 50% increase in activity (18, 19). This was not the case for bifidobacterial phytases, where a slight negative effect of EDTA on activity was monitored (Table 1). Calcium activation in HAP phytases is not unprecedented. As an example, similar to PhylA, the phytase from the pollen grains of Lilium longiflorum is activated by Ca2+ at 2 mM and inhibited by EDTA (17).
Fig 2.
Phytase activity of 6×(His)PhylA and 6×(His)PhypA in the presence of different concentrations of Ca2+ (as CaCl2) at different pHs. Values are means of results from at least three replicates.
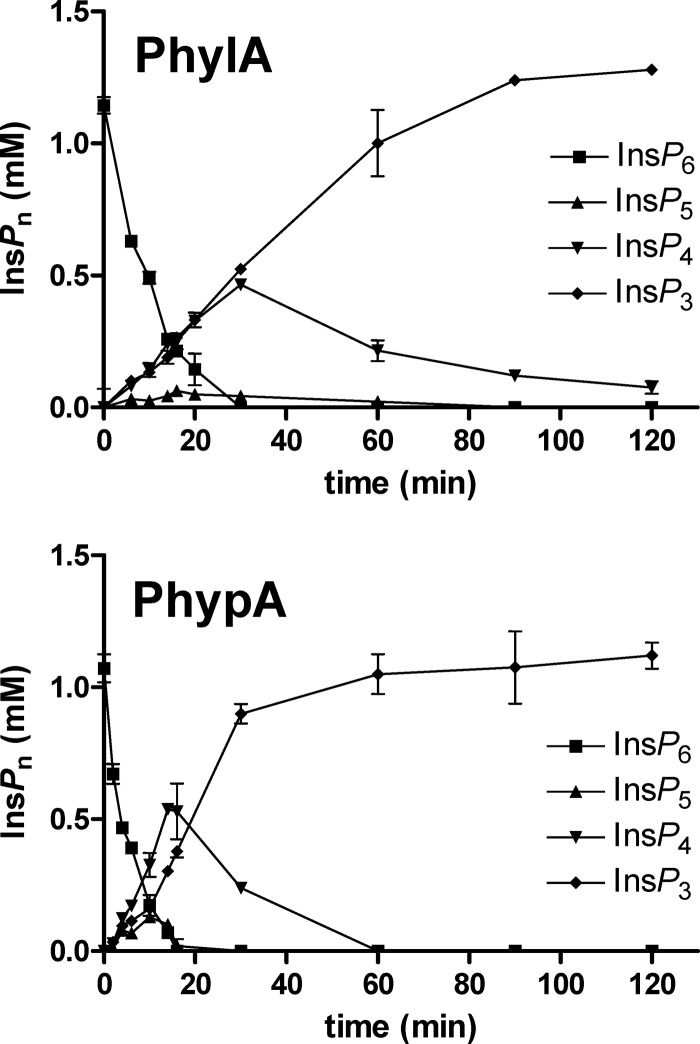
A time course evaluation of phytate hydrolysis was carried out, analyzing the different products by HPLC as previously described (14). As can be seen in Fig. 3, both phytases produced a transient accumulation of InsP5 and InsP4 that were rapidly hydrolyzed to InsP3, which remained over the experimental period. These results are in agreement with the myo-inositol phosphate products generated from phytate by B. longum subsp. infantis ATCC 15697 (5) and B. pseudocatenulatum ATCC 27919 (7) and suggest that, in contrast to other histidine acid phytases (e.g., fungal phytases [9]), the bifidobacterial enzymes do not completely hydrolyze phytate to myo-inositol. This could be of particular interest, because lower myo-inositol phosphate derivatives generate soluble and absorbable chelates with minerals and crucial biological functions have been postulated for specific isomers of InsP4 and InsP3 (2).
Fig 3.
Time course analysis of phytate (InsP6) hydrolysis by 6×(His)PhylA and 6×(His)PhypA at pH 5.5 and 50°C. Reactions were carried out in triplicate.
These are the first phytases identified in Bifidobacterium. They are probably surface anchored in these bacteria and may play a role in phosphate scavenging from phytate in the gastrointestinal habitat. The qualified presumption of safety/generally regarded as safe (QPS/GRAS) status and the probiotic characteristics of this group of microorganisms enable their phytases to be used to reduce phytate contents in products possibly fit for human consumption.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by grants AGL2006-09613/ALI and Consolider Fun-C-Food CSD2007-00063 from the Ministry of Science and Innovation, Spain (MICINN). The contract of Juan Antonio Tamayo-Ramos and the fellowship to Juan Mario Sanz-Penella from MICINN are greatly acknowledged.
Footnotes
Published ahead of print 11 May 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bohm K, Herter T, Muller JJ, Borriss R, Heinemann U. 2010. Crystal structure of Klebsiella sp. ASR1 phytase suggests substrate binding to a preformed active site that meets the requirements of a plant rhizosphere enzyme. FEBS J. 277: 1284–1296 [DOI] [PubMed] [Google Scholar]
- 2.Greiner R, Konietzny U. 2006. Phytase for food application. Food Technol. Biotechnol. 44: 125–140 [Google Scholar]
- 3.Greiner R, Konietzny U, Jany KD. 1993. Purification and characterization of two phytases from Escherichia coli. Arch. Biochem. Biophys. 303: 107–113 [DOI] [PubMed] [Google Scholar]
- 4.Haefner S, et al. 2005. Biotechnological production and applications of phytases. Appl. Microbiol. Biotechnol. 68: 588–597 [DOI] [PubMed] [Google Scholar]
- 5.Haros M, Bielecka M, Honke J, Sanz Y. 2007. Myo-inositol hexakisphosphate degradation by Bifidobacterium infantis ATCC 15697. Int. J. Food Microbiol. 117: 76–84 [DOI] [PubMed] [Google Scholar]
- 6.Haros M, Bielecka M, Sanz Y. 2005. Phytase activity as a novel metabolic feature in Bifidobacterium. FEMS Microbiol. Lett. 247: 231–239 [DOI] [PubMed] [Google Scholar]
- 7.Haros M, et al. 2009. Phytate degradation by human gut isolated Bifidobacterium pseudocatenulatum ATCC27919 and its probiotic potential. Int. J. Food Microbiol. 135: 7–14 [DOI] [PubMed] [Google Scholar]
- 8.Lim D, Golovan S, Forsberg CW, Jia Z. 2000. Crystal structures of Escherichia coli phytase and its complex with phytate. Nat. Struct. Biol. 7: 108–113 [DOI] [PubMed] [Google Scholar]
- 9.Oh BC, Choi WC, Park S, Kim YO, Oh TK. 2004. Biochemical properties and substrate specificities of alkaline and histidine acid phytases. Appl. Microbiol. Biotechnol. 63: 362–372 [DOI] [PubMed] [Google Scholar]
- 10.Oh BC, et al. 2006. Ca(2+)-inositol phosphate chelation mediates the substrate specificity of beta-propeller phytase. Biochemistry 45: 9531–9539 [DOI] [PubMed] [Google Scholar]
- 11.Palacios MC, Haros M, Rosell CM, Sanz Y. 2008. Selection of phytate-degrading human bifidobacteria and application in whole wheat dough fermentation. Food Microbiol. 25: 169–176 [DOI] [PubMed] [Google Scholar]
- 12.Palacios MC, Haros M, Sanz Y, Rosell CM. 2008. Selection of lactic acid bacteria with high phytate degrading activity for application in whole wheat breadmaking. LWT Food Sci. Technol. 41: 82–92 [Google Scholar]
- 13.Rao DE, Rao KV, Reddy TP, Reddy VD. 2009. Molecular characterization, physicochemical properties, known and potential applications of phytases: an overview. Crit. Rev. Biotechnol. 29: 182–198 [DOI] [PubMed] [Google Scholar]
- 14.Sandberg AS, Ahderinne R. 1986. HPLC method for determination of inositol tri-, tetra-, penta-, and hexaphosphates in foods and intestinal contents. J. Food Sci. 51: 547–550 [Google Scholar]
- 15.Sanz-Penella JM, Tamayo-Ramos JA, Haros M. 2011. Application of bifidobacteria as strater culture in whole wheat sourdough breadmaking. Food Bioprocess. Technol. doi:10.1007/s11947-011-0547-1 [Google Scholar]
- 16.Sanz-Penella JM, Tamayo-Ramos JA, Sanz Y, Haros M. 2009. Phytate reduction in bran-enriched bread by phytase-producing bifidobacteria. J. Agric. Food Chem. 57: 10239–10244 [DOI] [PubMed] [Google Scholar]
- 17.Scott JJ, Loewus FA. 1986. A calcium-activated phytase from pollen of Lilium longiflorum. Plant Physiol. 82: 333–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vats P, Banerjee UC. 2006. Catalytic characterization of phytase (myoinositolhexakisphosphate phosphohydrolase) from Aspergillus niger van Teighem: glycosylation pattern, kinetics and molecular properties. Enzyme Microb. Technol. 39: 596–600 [Google Scholar]
- 19.Zhang GQ, et al. 2010. Purification, characterization, and cloning of a novel phytase with low pH optimum and strong proteolysis resistance from Aspergillus ficuum NTG-23. Biores. Technol. 101: 4125–4131 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.