Abstract
Mineralization potentials, rates, and kinetics of the three phenoxy acid (PA) herbicides, 2,4-dichlorophenoxyacetic acid (2,4-D), 4-chloro-2-methylphenoxyacetic acid (MCPA), and 2-(4-chloro-2-methylphenoxy)propanoic acid (MCPP), were investigated and compared in 15 soils collected from five continents. The mineralization patterns were fitted by zero/linear or exponential growth forms of the three-half-order models and by logarithmic (log), first-order, or zero-order kinetic models. Prior and subsequent to the mineralization event, tfdA genes were quantified using real-time PCR to estimate the genetic potential for degrading PA in the soils. In 25 of the 45 mineralization scenarios, ∼60% mineralization was observed within 118 days. Elevated concentrations of tfdA in the range 1 × 105 to 5 × 107 gene copies g−1 of soil were observed in soils where mineralization could be described by using growth-linked kinetic models. A clear trend was observed that the mineralization rates of the three PAs occurred in the order 2,4-D > MCPA > MCPP, and a correlation was observed between rapid mineralization and soils exposed to PA previously. Finally, for 2,4-D mineralization, all seven mineralization patterns which were best fitted by the exponential model yielded a higher tfdA gene potential after mineralization had occurred than the three mineralization patterns best fitted by the Lin model.
INTRODUCTION
In order to increase yields in agricultural practice and for weed control in general, the herbicidal group of phenoxy acids (PA) has been intensively used worldwide, resulting in numerous incidents of groundwater contaminations (21). In Denmark, the compounds 2,4-dichlorophenoxyacetic acid (2,4-D), 4-chloro-2-methylphenoxyacetic acid (MCPA), and 2-(4-chloro-2-methylphenoxy)propanoic acid (MCPP) have been the most frequently used. If we look at a comparison of data from more than 10,000 Danish groundwater wells, it is notable that although the compounds are used for the same purpose and in comparable amounts, 2,4-D is practically never found, while MCPA and MCPP are frequently found (37).
The fates of these compounds in many different environments have been studied intensively, and several mechanisms such as sorption, biodegradation, and chemical processes have been proved to be important (5, 20). Degradation performed by microorganisms is the most important process for complete transformation into inorganic metabolites, and biodegradation of PA is well known (3, 12). Especially for 2,4-D, an extensive amount of information is available on its degradation in soils, while MCPA and MCPP have been studied far less intensively. However, comparisons of the potentials to mineralize these three structurally related compounds are rather scarce.
Bacteria using 2,4-D as their sole source of carbon have been isolated, and genes and enzymes involved in the degradation have been characterized subsequently (35, 38). The first step in the degradation pathway is initiated by an α-ketoglutarate-dependent dioxygenase encoded by tfdA (14, 35, 36) or tfdA-like (22) genes. Pure-culture bacteria have been isolated, and they are able to proliferate utilizing PA as their only source of carbon. Furthermore, it has been demonstrated that the number of bacterial degraders in environmental samples in situ may increase as a result of PA degradation (1, 4, 16). Moreover, Bælum et al. (4) used an mRNA-based approach to directly link degradation of 2,4-D and MCPA to expression of tfdA genes in agricultural soil. Such an approach has never been applied to link MCPP degradation to the tfdA gene, but it has been shown that tfdA-harboring organisms proliferate as a result of MCPP degradation (19, 40).
Often, modeling is regarded as a powerful tool to obtain comprehensive information on the fates of organic contaminants in the environment. The models most commonly used in studies describing xenobiotic mineralization kinetics have been developed on the basis of Monod kinetics (33, 34). Especially for studies in liquid media, these models are ideal for describing the decomposition of carbon substrates, but due to bioavailability issues such as sorption/desorption processes and xenobiotic/microbial transport, degradation of substrates applied to soil is often much more complicated to describe (32). Brunner and Focht (6) developed the three-half-order models, the intention being to describe mineralization of carbon substrates in soil matrixes, taking zero/linear (Lin) or exponential (Exp) growth of the microbial degraders into account. These models have been successfully used to describe degradation of PA in natural soils (4) and in soils inoculated with a known degrader (19, 26). Even though the usefulness of the models to describe growth rates has been validated in soil experiments with inoculated degraders (12, 19), it has never been investigated quantitatively whether the models are able to distinguish between different levels of growth of indigenous microbial degraders.
Analyzing complex microbial communities, e.g., in environmental samples, in situ can be difficult; nevertheless, recently developed molecularly based methods have proved to be useful. Especially for quantification of indigenous microorganisms harboring specific functional genes, quantitative real-time PCR has been used successfully (1, 4, 16, 23). In our previous work (1, 4), we used quantitative real-time PCR to show increased numbers of tfdA genes as a result of mineralization events of 2,4-D and MCPA.
In the present study, we intend to elucidate the biodegradation of the three PA, 2,4-D, MCPA, and MCPP, in 15 soils sampled worldwide, representing soils pristine to PA as well as soils previously treated with PA herbicides. Levels of tfdA genes were quantified by molecularly based techniques and discussed in relation to mineralization data fitted to mathematical model describing mineralization kinetics influenced by zero, linear, or exponential growth of the degrading organisms.
MATERIALS AND METHODS
Soil.
Fifteen soils were collected from distinct locations on five continents. The 15 different sampling sites were chosen rather randomly but with a preference for a mixture of soils previously treated with PA and soils never directly exposed to PA. Six soils had previously been treated with PA, five soils had never been treated with PA, and the treatment history of four soils was unknown (Table 1). SjOreg is a regularly cultivated agricultural soil from Denmark, treated annually with a mixture of PA (mainly MCPA and MCPP for the most recent years). The sheep pen soil (United Kingdom) has been treated annually with MCPP for a period of at least 10 years. The pump ground soil (United Kingdom) has been set aside for a 5-year period but received MCPP prior to this. The Epoisses sample (France) is an agricultural soil regularly treated with many different types of herbicides, among these PA. The Suma-Paz sample (Columbia) is an agricultural soil irregularly treated with 2,4-D. KBSreg (1xR2+R2), KBSforest (TDF), and KBSorg (T4) are experimental soils from the Kellogg Biological Station, MI (names in parentheses are the specific plots named by KBS). KBSreg has been treated annually with 2,4-D for at least 20 years, KBSorg was sampled from the same area but never treated directly with any kind of herbicide, and KBSforest is a forest soil collected from a distinct location never treated with herbicides. Additionally, desert soil from the Negev desert (Israel) and two soils from the Yanchep National Park (Australia) (Yanchep, a bush land soil, and Crystal Cave, a sandy aquifer from an underground cave) are regarded as untreated soils. Even though it is a known fact that the soils from Quetigny (France), Sicily (Italy), and Kfar Vitkin (Israel) are agricultural soils treated with herbicides, the history of PA treatment remains unknown. Kings Park (Australia) is a city park, and again the history of PA treatment remains unknown.
Table 1.
Soils and soil properties
| Soil | Location | Soil application |
Kda |
Soil texture (%) |
pHb | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2,4-D | MCPA | MCPP | C | Clay | Silt | Sand | ||||
| SjOreg | Denmark | Arable | 0.26 ± 0.05 | 0.19 ± 0.02 | 0.20 ± 0.02 | 1.2 | 19 | 18 | 62 | 7.2 |
| Sheep pens | UK | Arable | 0.14 ± 0.05 | 0.13 ± 0.01 | 0.18 ± 0.02 | 1.0 | 16 | 7 | 77 | 7.0 |
| Pump ground | UK | Set aside | 0.20 ± 0.01 | 0.15 ± 0.02 | 0.18 ± 0.0 | 1.1 | 18 | 9 | 73 | 6.8 |
| Epoisses | France | Arable | 0.30 ± 0.10 | 0.20 ± 0.03 | 0.21 ± 0.02 | 1.0 | 27 | 41 | 31 | 6.6 |
| Quetiyly | France | Arable | 0.29 ± 0.02 | 0.20 ± 0.08 | 0.18 ± 0.01 | 1.8 | 34 | 27 | 40 | 7.7 |
| Sicily | Italy | Arable | 0.47 ± 0.0 | 0.32 ± 0.02 | 0.30 ± 0.01 | 7.4 | 37 | 31 | 32 | 7.9 |
| Kfar Vitkin | Israel | Arable | 2.12 ± 0.13 | 1.93 ± 0.19 | 1.49 ± 0.06 | 1.1 | 32 | 59 | 9 | 5.5 |
| Negev | Israel | Desert | 1.17 ± 0.23 | 1,14 ± 0.07 | 0.62 ± 0.24 | 0.1 | 18 | 75 | 7 | 6.1 |
| Kings Park | Australia | City park | 0.72 ± 0.11 | 0.63 ± 0.02 | 0.65 ± 0.15 | 2.6 | 3 | 0 | 97 | 5.0 |
| Crystal Cave | Australia | Cave sand | <0.01 ± 0.0 | <0.01 ± 0.0 | 0.02 ± 0.0 | 0.02 | 1 | 0 | 99 | 8.3 |
| Yanchep | Australia | Bushland peat | 4.33 ± 0.38 | 2.50 ± 0.14 | 1.62 ± 0.03 | 4.9 | 4 | 6 | 90 | 8.2 |
| Suma-Paz | Columbia | Arable | 3.18 ± 0.12 | 1.91 ± 0.05 | 1.57 ± 0.07 | 8.8 | 18 | 22 | 60 | 5.9 |
| KBSreg | MI/USA | Arable | 0.28 ± 0.02 | 0.18 ± 0.01 | 0.21 ± 0.01 | 0.7 | 12 | 27 | 61 | 5.8 |
| KBSforest | MI/USA | Forest | 0.46 ± 0.02 | 0.32 ± 0.01 | 0.37 ± 0.01 | 1.5 | 7 | 20 | 73 | 5.0 |
| KBSorg | MI/USA | Arable/organic | 0.31 ± 0.01 | 0.20 ± 0.01 | 0.25 ± 0.0 | 1.0 | 15 | 30 | 55 | 6.2 |
Kd values were measured in H2O and at a only one concentration, 0.09 mmol · kg−1.
pH values were measured in H2O.
Soils were collected as composite samples from the upper 10 cm, i.e., as small subsamples taken within an area of a couple of square meters and mixed thoroughly. Subsequently, they passed through a 4-mm-pore-size sieve and were stored in the dark at 4°C until experiment setup but not longer than 2 to 3 months.
The Kd (dissociation constant) value was determined in soil pH for the three phenoxy acids at concentrations of 0.09 mmol kg−1 of soil for all of the soils. Soil texture was determined by the combined hydrometer and sieving method (9), and soil carbon was determined by dry combustion at 1,250°C in oxygen (11) (Table 1).
Mineralization assay.
Microcosms were set up as 5 g (dry weight) of soil inside 20-ml glass flasks with air-tight lids. Individual stock solutions of 2,4-D, MCPA, and MCPP (>97% purity; Ehrenstorfer, Augsburg, Germany) were prepared in sterile MilliQ water (Millipore, Birkerød, Denmark) as 1.8 mM solutions. Additionally, U-ring 14C-labeled 2,4-D, MCPA, or MCPP (radiochemical purity of > 95%; Izotop, Budapest, Hungary) was added in trace amounts. An initial 250 μl of stock was added to each microcosm, giving final phenoxyacetic acid concentrations of 0.09 mmol kg−1 of soil and radiochemical activity of ∼50,000 dpm in each microcosm. To ensure that moisture in the soils was ∼80% of the water holding capacity, additional MilliQ was added. A small glass vial containing 0.8 ml of 0.5 M NaOH was placed inside the glass flask to trap CO2 as HCO3− evolved from the PA decomposition. The NaOH traps were replaced at intervals of 3 to 5 days for the first 30 days, followed by intervals of 7 to 10 days for the remaining period, and radioactivity was quantified as described elsewhere (1). Microcosms were set up in triplicates, which, including control samples without phenoxyacetic acid, resulted in 180 microcosms in total. They were incubated in the dark at 10°C. A composite sampling technique was used to sample 0.25 g of soil for DNA extraction after mineralization of ∼60% had occurred. All soil samples for nucleic acid extractions were frozen immediately in liquid nitrogen and stored at −80°C until extraction.
Nucleic acid extraction.
DNA extractions were performed in triplicates on 0.25-g soil samples from all of the soil scenarios after mineralization had occurred and on soil samples sampled prior to pesticide application. For soils exhibiting low mineralization potentials (<20%), only one soil sample was extracted. Extractions were performed using a PowerSoil DNA isolation kit (MoBio Laboratories, Carlsbad, CA) according to the manufacturer's instructions. Consistency of the general nucleic acid extraction efficiency was checked by running 4-μl aliquots on standard 1.5% agarose gels stained with ethidium bromide (EtBr). The intensity of the bands was quantified using Quantity One software, version 4.6.3 (Bio-Rad, Hercules, CA), and Lambda DNA-BstE II digest (New England BioLabs, Ipswich, MA) as the standard ladder. Furthermore, the DNA concentrations in the extracts were quantified, and the A260/280 and A260/230 ratios were determined using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA).
Quantitative real-time PCR.
For quantitative real-time PCR, a DyNAmo SYBR Green qPCR kit (FinnZymes, Helsinki, Finland) was used as Master Mix with 0.4 μM each primer for the tfdA genes (forward, 5′-GAGCACTACGCRCTGAAYTCCCG-3′; reverse, 5′-GTCGCGTGCTCGAGAAG-3′) (1) and the rpoB gene (forward, 5′-AACATCGGTTTGATCAAC-3′; reverse, 5′-CGTTGCATGTTGGTACCCAT-3′) (8) and 1 μg/μl bovine serum albumin (BSA). Subsequently, 1 μl of soil DNA extract (∼10 ng) was added as the template in 20-μl reaction mixtures. The reaction conditions were as follows: 15 min at 95°C for enzyme activation, followed by 50 cycles of 60 s at 95°C, 30 s at 64°C, and 60 s at 72°C, with a final step of 7 min at 72°C in an iCycler iQ5. All quantitative PCRs (qPCRs) were run in triplicates, producing a total of 312 (28 × 3 × 3 + 20 × 1 × 3) individual real-time PCRs. For qPCR on the tfdA gene, seven 96-well plates were used, each with individual positive-control standards with plasmids containing the target gene of interest (for details, see below) in the range 103 to 106 plasmids per reaction mixture, covering the range of all samples with a positive signal. Reaction efficiencies for all plates were in the range 94.7% to 97.9% with R2 values of >0.989. For qPCR on the rpoB gene, positive plasmid standards with a concentration range of 104 to 107 genes per reaction mixture were used to cover the signal from all samples. The efficiency for this assay was in the range of 91.3 to 96.7% with R2 values of >0.993.
Standards for the qPCR assay were prepared by cloning the entire tfdA gene from the pure culture 2,4-D degrader Cupriavidus necator JMP134 (29) into a pCR 2.1-TOPO cloning vector (Invitrogen, Carlsbad, CA) as described in Bælum and Jacobsen (2). Plasmids were obtained and purified using an UltraClean 6-min Mini Plasmid Prep Kit (MoBio Laboratories, Carlsbad, CA), and the DNA concentration in the crude extract was measured using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA). The tfdA gene copy number was calculated as described elsewhere (27). Ten-fold serial dilutions were made and used as a template in qPCRs. qPCR of the standards was performed in triplicates, and at least four dilutions were included in each single run.
PCR inhibition assay.
Because of the many different soils investigated in the present study, there was a chance that the level of PCR inhibitors varied in the soil DNA extracts, giving rise to biases in the qPCR. Therefore, we made a test in order to reveal any possible effect of inhibitors in the extracts. An extra qPCR was completed on all extracts targeting pCR 2.1-TOPO plasmids (Invitrogen, Paisley, United Kingdom), which were added at a concentration of 106 per reaction mixture. qPCR was done with the M13 primers (0.4 μM each) and 1 μl of the soil DNA extract. The PCR conditions were as follows: 6 min at 95°C, followed by 50 cycles of 45 s at 94°C, 30 s at 60°C, and 2 min at 72°C, with a final step of 6 min at 72°C. A subsequent temperature ramping was performed to analyze melting curve profiles of the PCR products. The conditions were as follows: 80 cycles of 30 s starting at 58°C with an increase in temperature of 0.5°C for every cycle to a temperature of 98°C at the final cycle. Inhibition levels of the qPCRs were determined by comparing the mean threshold cycle (CT) values obtained from the soil extracts and cDNA dilutions with those of the plasmid controls. Three replicate qPCRs were performed for each soil DNA extract.
Data analysis.
All microcosm incubations, sorption experiments, DNA extractions, and qPCRs were performed in three replicates, and mean values and standard errors were calculated. Mineralization data were corrected for background radioactivity, and a nonlinear regression analysis was performed on the cumulative mineralization curves using the curve-fitting program in SigmaPlot (version 10; Systat Software Inc., San Jose, CA). All of the mineralization curves exceeding a threshold value of 5% mineralization were fitted by the zero/linear (Lin) and exponential (Exp) growth forms of the 3/2-order models (6) and by logarithmic (log), first-order, and zero-order kinetic models (19). The best fit was primarily chosen based on F tests (P < 0.05) (31), and additionally R2 values and a visual evaluation were taken into account. Subsequently, the maximum mineralization rate for each mineralization curve was estimated by optimizing the differentiated form of the equation of the best-fitting model.
To assess direct relationships between variables, Spearman rank correlations were computed using the statistical program package R (R, version 2.14.1 [r-project.org]). Then, in order to perform a multidimensional investigation of the whole data set, nonmetric multidimensional scaling analysis (NMDS) was conducted using the Vegan R library (version 2.0.2). As the data were heterogeneous, square root transformation and standardization were performed prior to NMDS analysis. In order to maintain an assumption-free framework, the distances between each pair of variables were computed according to the Euclidean formula.
RESULTS
Sorption/soil characteristics.
The soil texture and sorption abilities varied between the soils used in this study (Table 1). Kd values were in general low, varying between 0 in the Crystal Cave soil and 4.33 for 2,4-D in the Yanchep soil. In general, the Kd values for 2,4-D were slightly higher than those for MCPA and MCPP, while Kd values for MCPA and MCPP were more or less equal (Table 1).
Mineralization assay.
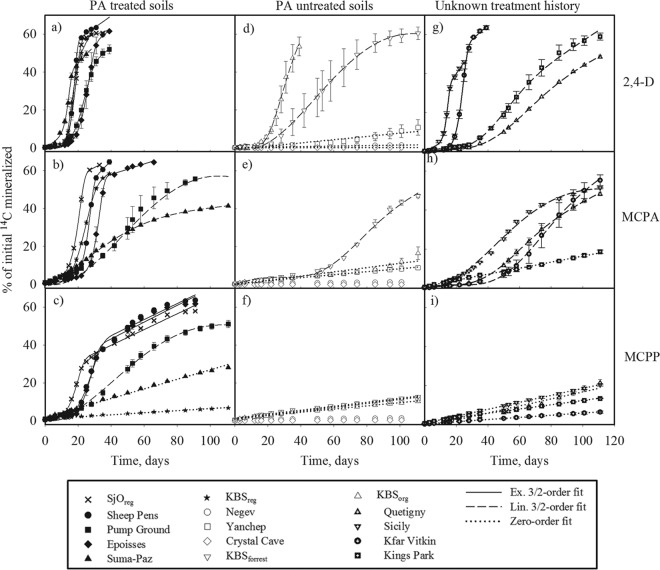
The mineralization curves showing accumulated amounts of 14CO2 released from mineralization of [14C]2,4-D, [14C]MCPA, and [14C]MCPP were obtained from 45 scenarios, i.e., from the three phenoxy acids in 15 different soils (see Fig. S1 in the supplemental material). Of these, 27 mineralization curves clearly had a sigmoid curvature, indicating growth of microbial degraders during mineralization. The remaining 18 mineralization curves had a linear curvature with a constant mineralization rate.
For 11 of the 15 soils, the PA treatment history could be documented (6 soils had been more or less extensively treated with PA, and 5 soils had never been directly treated with PA), and by comparing these, we observed a correlation between PA pretreatment of the soils and rapid mineralization (Fig. 1). For the soils Quetigny, Sicily, Kfar Vitkin, and Kings Park, no treatment history was available; however, they still exhibited the potential to mineralize 2,4-D and MCPA, while MCPP mineralization followed slower, zero-order mineralization kinetics (Fig. 1g and h). All curves showing mineralization of 2,4-D in previously treated soils (SjOreg, sheep pens, pump ground, Epoisses, Suma-Paz, and KBSreg soils) were best fitted by the Exp model (Fig. 1a), exhibiting rapid mineralization, while the mineralization curves for MCPA in pump ground and Suma-Paz soils (Fig. 1b) and for MCPP in pump ground soil (Fig. 1c) were significantly slower and best described by the Lin model. Furthermore, for MCPP in Suma-Paz and in KBSreg, the mineralization pattern followed zero-order kinetics (Fig. 1d). In the soils not previously treated with PA (Negev, Crystal Cave, Yanchep, KBSforest, and KBSorg), however, a notably different pattern was observed (Fig. 1d to f). None of these soils exhibited a mineralization pattern which could be fitted by the Exp model; the mineralization of 2,4-D in only two soils (KBSforest and KBSorg) and of MCPA in one soil (KBSforest) (Fig. 1e) could be fitted by the Lin model. Mineralizations in the remaining soils (Negev, Crystal Cave, and Yanchep for 2,4-D, MCPA, and MCPP, KBSorg soil for MCPA and MCPP, and the KBSforest soil for MCPP) were best fitted by 0-order kinetics or did not exceed a threshold value of 5% mineralization.
Fig 1.
Mineralization curves fitted to Exp, Lin, and zero-order kinetics. Panels show mineralization curves for 2,4-D, MCPA, and MCPP in PA-treated or -untreated soils and in soils with unknown treatment histories, as indicated on the figure. See Table 1 for further information on soils. Error bars represent standard errors of the means for soil triplicates.
The lag phase was passed and exceeded its maximum mineralization rate within 30 days for nine soils applied with 2,4-D, while this was observed in only four and three soils for MCPA and MCPP, respectively (see Fig. S1 in the supplemental material). The parameters Vmax and time for Vmax (tmax) derived by differentiation of the fitted equations (Table 2) clearly suggest that the potential for mineralization of these three PA in the studied soils was in the order 2,4-D > MCPA > MCPP. Maximum mineralization rates (υmax) varied between 10.6 and 0.8% day−1 for the 2,4-D experiments, while they varied between 7.2 and 0.1% day−1 for MCPA and between 3.3 and 0.1% day−1 for MCPP. The ratios of the best fits of the entire set of mineralization curves according to the order Exp/Lin/zero-order/not fitted (curves were not fitted when mineralization did not exceed 5%) were 9:4:0:3, 5:6:1:4, and 3:1:9:3 for 2,4-D, MCPA, and MCPP, respectively. Furthermore, we observed a trend that the order of the best model fit was consistent in a way that the MCPP fit was never of a higher growth order than MCPA and the MCPA was never of a higher growth order than 2,4-D. Actually, none of the MCPA mineralization data could be fitted by a model describing faster growth than the model describing mineralization of 2,4-D in the same soil. The same trend occurred for models describing MCPP and MCPA and for models describing MCPP and 2,4-D. In general, the mineralization curves with high mineralization rates, >∼3, were best fitted by the Exp model, while the ones with lower rates were best fitted by the Lin model.
Table 2.
Estimates derived from mathematical fits of the mineralization rates
| Soil | 2,4-D |
MCPA |
MCPP |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Model of best fit (R2)a | Vmaxb | tmaxc | Model of best fit (R2)a | Vmaxb | tmaxc | Model of best fit (R2)a | Vmaxb | tmaxc | |
| SjOreg | Exp (0.99) | 7.8 | 18.9 | Exp (0.99) | 6.4 | 21.1 | Exp (0.99) | 3.0 | 19.8 |
| Sheep pens | Exp (0.99) | 10.6 | 17.7 | Exp (0.99) | 7.2 | 27.3 | Exp (0.99) | 2.6 | 28.8 |
| Pump ground | Exp (0.98) | 3.4 | 24.4 | Lin (0.94 | 1.1 | 42.9 | Lin (0.99) | 0.8 | 42.2 |
| Epoisses | Exp (0.96) | 5.3 | 26.5 | Exp (0.99) | 6.4 | 33.1 | Exp (0.99) | 3.3 | 28.3 |
| Quetigny | Lin (0.99) | 0.8 | 65.9 | Lin (0.99) | 0.8 | 60.7 | 0-order (0.97) | 0.3 | |
| Sicily | Exp (0,99) | 7.0 | 14.7 | Exp (0.99) | 1.0 | 51.8 | 0-order (0.99) | 0.2 | |
| Kfar Vitkin | Exp (0.99) | 6.8 | 24.5 | Lin (0.95) | 1.0 | 74.5 | 0-order (0.95) | 0.1 | |
| Negev | NA | NA | NA | ||||||
| Kings Park | Lin (0.98) | 1.0 | 52.1 | 0-Order (0.99) | 0.2 | 0-Order (0.99) | 0.1 | ||
| Crystal Cave | NA | NA | NA | ||||||
| Yanchep | NA | NA | NA | ||||||
| Suma-Paz | Exp (0.99) | 6.8 | 14.6 | Lin (0.99) | 0.6 | 25.7 | 0-Order (0.99) | 0.3 | |
| KBSreg | Exp (0.99) | 6.8 | 16.7 | Exp (0.99) | 3.6 | 25.0 | 0-Order (0.96) | 0.1 | |
| KBSforest | Lin (0.88) | 1.0 | 48.7 | Lin (0.99) | 0.9 | 76.8 | 0-Order (0.98) | 0.1 | |
| KBSorg | Lin (0.91) | 3.0 | 28.6 | NA | 0-Order (0.99) | 0.1 | |||
Exp, exponential growth form of the 3/2-order model; Lin, linear or zero growth form of the 3/2-order model; NA, not applicable since mineralization did not exceed 5% and the curve was not fitted. Values in brackets are R2-values.
Vmax, maximum rate of mineralization (percent mineralized day−1).
tmax, time for maximum rate of mineralization (days).
qPCR data.
The A260/280 ratio was 1.7 to 2.0 in all DNA extracts, while the A260/230 ratio was 1.8 or greater in all samples except in the Sicily and Suma-Paz soils, in which it was ∼1.6, probably due to coextracted nucleic acids.
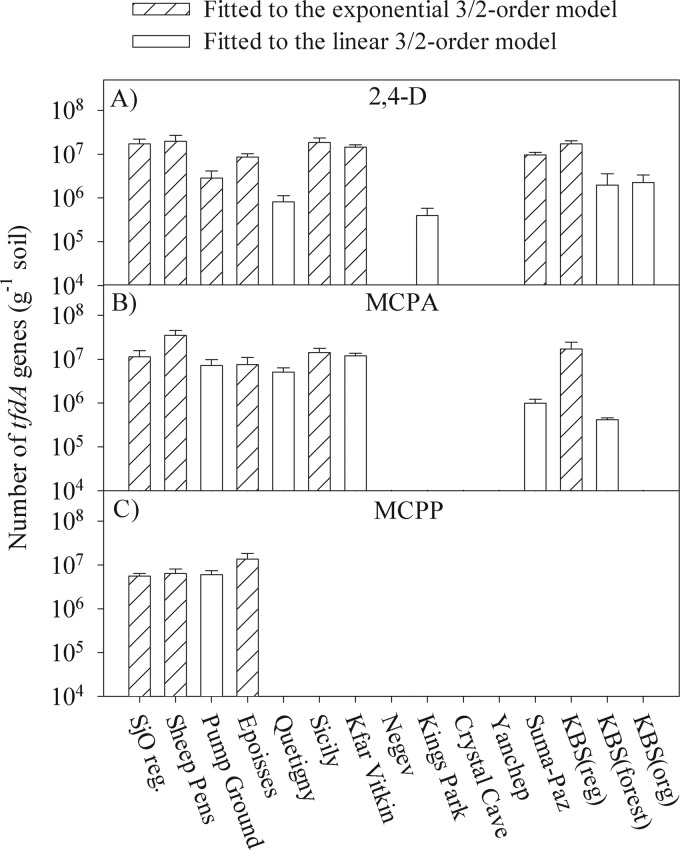
Occurrence of tfdA genes in the indigenous soils prior to PA mineralization could be detected only in the soils Suma-Paz, KBSreg, and SjOreg. In these soils ∼105, ∼104, and ∼104 tfdA genes g−1 of soil were detected, respectively (data not shown). After PA mineralization, however, elevated abundances of the functional gene tfdA were observed in many of the soils (Fig. 2). In all of the soils where the mineralization pattern was best fitted by the Exp or Lin models, elevated abundance of tfdA genes was observed, while none of the soils exhibiting zero-order mineralization kinetics showed any sign of elevated tfdA gene abundance (Fig. 2).
Fig 2.
Growth yield of tfdA gene copies in the soils after ∼60% mineralization of 2,4-D, MCPA, and MCPP. Error bars represent standard errors of the means for soil triplicates.
A general trend in the copy number of tfdA genes after mineralization was that higher levels were observed in soils where mineralization was best fitted by the Exp model. In the majority of the soils, the tfdA copy number was ∼107 genes g−1 of soil, but levels as low as 105 genes g−1 and as high as 5 × 107 genes g−1 were also observed. In the control soils without PA addition, we did not detect tfdA proliferation in any of the soils.
The number of bacteria in the different soils was quantified before and after mineralization using the rpoB gene as a biomarker (see Fig. S2 in the supplemental material). In all of the soils except Negev and Crystal Cave, the number of rpoB genes varied between 108 and 8 × 108 g−1 of soil, and no clear correlation between increase/decrease in rpoB gene content as a result of PA addition was found. In the Negev soil and in the Crystal Cave soil, fewer than 107 rpoB genes g−1 of soil were found.
In order to avoid the occurrence of any possible PCR-inhibiting compounds in the DNA extracts, an inhibition assay was performed. We found inhibitory compounds only in the soil from Sicily and Suma-Paz, in which a high content of organic carbon was also found. After a 10-fold dilution of the DNA extract, we were unable to detect any influence of the inhibiting compounds, and consequently for these soils the real-time PCR quantifications were made on these diluted extracts.
Statistical analysis.
Spearman rank correlations were estimated (Table 3) and significant (P < 0.05) correlations were identified. A positive correlation was observed between the Kd value and carbon content (percent) in the soil, between tmax and Vmax, and finally between the increase in tfdA gene abundance and both Vmax and tmax.
Table 3.
Soil specification variables according to Spearman's ρ test result
| Parameter |
P value for the indicated parametera |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Kd value | C (%) | Clay (%) | Silt (%) | Sand (%) | pHsoil | Vmax | tmax | No. of tfdA genes | |
| Kd | 1.29E−04 | 6.87E−01 | 6.69E−02 | (−) 5.21E−02 | (−) 5.72E−03 | (−) 2.12E−01 | (−) 3.68E−01 | (−) 1.51E−01 | |
| C (%) | 1.22E−01 | (−) 3.00E−01 | 6.60E−01 | (−) 9.44E−01 | 5.42E−01 | 4.67E−01 | (−) 9.49E−01 | ||
| Clay (%) | 1.63E−07 | (−) 1.92E−10 | 3.85E−01 | 4.35E−03 | 2.44E−02 | 4.42E−03 | |||
| Silt (%) | (−) 0.00E+00 | (−) 9.31E−02 | 4.80E−01 | 5.34E−01 | 3.34E−01 | ||||
| Sand (%) | 2.01E−01 | (−) 3.73E−01 | (−) 4.37E−01 | (−) 2.83E−01 | |||||
| pHsoil | (−) 4.41E−01 | (−) 3.83E−01 | (−) 9.86E−01 | ||||||
| Vmax | 8.38E−06 | 0.00E+00 | |||||||
| tmax | 3.74E−07 | ||||||||
| No. of tfdA genes | |||||||||
Spearman's ρ correlation between each pair of soil specification variables was computed and tested against 0. This table shows the resulting P values. Values in boldface mean that the link measurement is considered significant according to a risk α of 5%. The minus sign (−) before some of the values means that the link between the two variables is negative (ρ < 0); i.e., when one variable increases, the other decreases.
The NMDS shows a link between tfdA gene copy numbers and Vmax and tmax (Fig. 3a and b). Also, the Kd value and Vmax were in opposite directions, suggesting that substrate availability is important for the degradation of PA (Fig. 3a). The compound type, on the other hand, did not explain the dispersion of the data (Fig. 3b).
Fig 3.
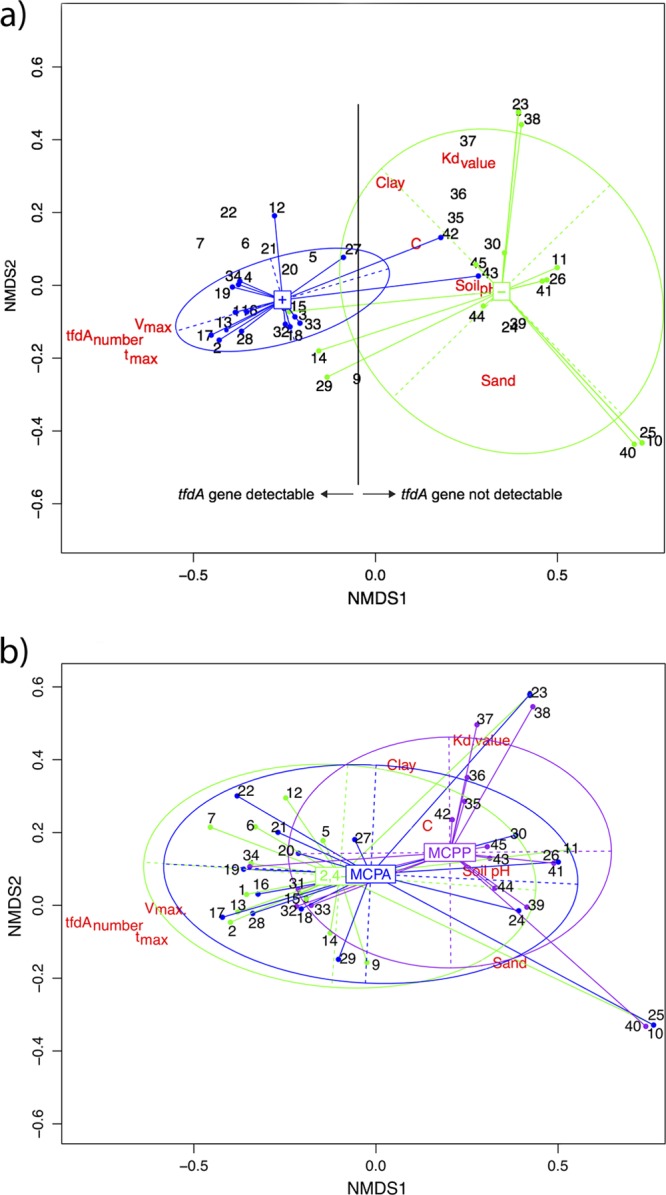
Bidimensional NMDS plots. Panel a focuses on the effect of the presence (blue)/absence (green) of tfdA genes. Panel b focuses on the effect of the different PA 2,4-D (green), MCPA (blue), and MCPP (purple). If a sample is known to belong to a class, it is linked to the inertia center of its class. The horizontal and vertical widths of the class ellipses are equal to the variance of the NMDS coordinates, and the slope is equal to their covariance.
DISCUSSION
In the present study, a large data set comparing mineralization data of 2,4-D, MCPA, and MCPP in 15 different soils from five continents was obtained. For all soils exhibiting a potential for mineralization above 50%, 2,4-D was degraded faster than MCPA and MCPP, and MCPA was degraded faster than MCPP. Previous investigations reported the influence of single parameters such as bioavailability (20), adaptation (4, 10, 13), PA concentration (17, 30), moisture level (18), or temperature (17) on degradation rates, but this study is the first to compare the degradation of all the three compounds, 2,4-D, MCPA, and MCPP, in different soils incubated under the same controlled laboratory conditions. Crespin et al. (7) found almost the same degradation rates of 2,4-D and MCPA under field conditions, whereas other studies of the three PA individually showed mineralization within 5 to 50 days (1, 4, 17, 18, 28). This is in accordance with our results for soil previously exposed to PA and in contrast to our PA-pristine soils. Growth of degraders utilizing the carbon in PA generates the general sigmoid mineralization pattern observed here and reported in almost all other studies of PA mineralization as well (18, 25). However, the zero-order mineralization kinetics and very low mineralization potentials observed here for a number of the soils not previously exposed to PA have only rarely been reported for PA mineralization in top soils under realistic conditions (15). The linear pattern may be due to a cometabolic process in which PA is mineralized along with the general metabolic pathways without initiation of any growth of the degraders. Anther, and more likely, explanation is that the incubation period was too short and that it was only the initial lag phase for these soils that was elucidated. Such a slow mineralization pattern may be further extended because of the relatively low incubation temperature chosen for the study reported here. It is well known that temperature is a highly determinative factor for the speed of biodegradation of PA and xenobiotic compounds in general (17). Also, it has to be noted that changes might have occurred in the soil microbial communities during sampling, transport, and storage. Our experience is, however, that the genomic potential to perform specific functions, such as PA mineralization, does not change much over time, even at varying temperatures (25).
Adaptation of the soil microbial community is regarded as an important parameter for mineralization of PA in soils. Increasing the genetic potential, either by horizontal gene transfer of mobile genetic elements between indigenous cells (10, 39) or by growth of organisms possessing the ability to mineralize PA (24), is considered the most important adaptation mechanism. In Bælum et al. (4), we observed that elevated numbers of tfdA-harboring organisms, following an event of 2,4-D or MCPA mineralization, significantly reduced the initial lag period of mineralization of a second application of PA to the same system. In accordance with this, we observed a clear correlation between higher Vmax and shorter tmax in soils previously treated with PA in the present study. Also, we found that the three soils, SjOreg, Suma-Paz, and KBSreg, were excellent examples of the adaptation theory, as tfdA genes could be detected in the soils prior to PA mineralization, which corresponds to the relatively short lag phases exhibited in these three soils especially for 2,4-D and MCPA mineralization.
As stated above, elevated numbers of tfdA genes as a consequence of PA mineralization have been reported in several studies, and the numbers reported here correspond to a great extent to what has been reported previously (1, 4, 16). The number of tfdA genes in the soil following ∼60% mineralization was higher in the mineralization scenarios best fitted to the Exp model than in those best fitted to the Lin model. Our statistical model also suggests a strong correlation between the increase in tfdA genes and the mineralization of PA, and according to our data, in some soils 60% mineralization can be reached with an increase of only 105 copies of the gene. As the tfdA gene content in the soils prior to PA mineralization was below our qPCR detection limit, we are not able to determine a potential correlation between natural tfdA gene abundance and PA mineralization. Unlike tfdA gene abundance, no effect of PA addition on the abundance of general bacteria could be detected. In some of the soils, the increase in tfdA gene abundance was equivalent to 10% of the total bacterial abundance (based on rpoB genes). Hence, other conditions (e.g., water content) may affect the general bacterial number more than the PA mineralization.
Even though it has previously been shown that sorption has an impact on degradation of PA (20), we did not observe any correlation between the potential for mineralizing PA and the Kd value or other soil parameters. In fact, we observed slightly higher Kd values for 2,4-D than for MCPA and MCPP, but despite this, mineralization of 2,4-D was significantly faster. This is strong evidence that the slower mineralization of MCPA and MCPP is not due to higher sorption but, rather, to lower affinity of the soil microbial degraders to MCPA and MCPP than to 2,4-D. This trend was also observed in a previous study (3) although this was based on a pure culture collection most likely biased in favor of 2,4-D-degrading organisms by the isolation technique. This higher microbial affinity toward 2,4-D might also explain the elevated presence of MCPA and MCPP in Danish groundwater compared to 2,4-D levels.
Conclusion.
In the present study, we were able to link an increase in the presence of the functional phenoxy acid-degrading gene tfdA to growth-linked mineralization kinetics. A large increase in tfdA genes correlated with mineralization kinetics taking exponential growth of the degraders into account, while a lower tfdA content correlated with mineralization kinetics taking linear growth into account. We found that the degradability of the three most commonly used phenoxy acid herbicides was in the order 2,4-D > MCPA > MCPP, and a clear tendency was also revealed that the mineralization occurred significantly faster in soils previously treated with phenoxy acid herbicides than in soils never treated.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Danish Research Council for Technology and Production Sciences for its financial support of the project Agricultural Practice, Microbial Activity and Pesticide Leaching (FTP no 274-05-0199). We thank Ziv Arbelli, Shai Arnon, Gary Bending, and Fabrice Martin-Laurent for kindly providing soils. We thank Pia Bach Jakobsen and Szymon Kopalski for skillful technical assistance and Kirsa Demant and Michael Belt at Ad Hoc Translation service for proofreading the manuscript.
Footnotes
Published ahead of print 25 May 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Bælum J, Henriksen T, Hansen HCB, Jacobsen CS. 2006. Degradation of 2-methyl-4-chlorophenoxyacetic acid in top- and subsoil is quantitatively linked to the class III tfdA gene. Appl. Environ. Microbiol. 72:1476–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bælum J, Jacobsen CS. 2009. TaqMan probe-based real-time PCR assay for detection and discrimination of class I, II, and III tfdA genes in soils treated with phenoxy acid herbicides. Appl. Environ. Microbiol. 75:2969–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bælum J, Jacobsen CS, Holben WE. 2010. Comparison of 16S rRNA gene phylogeny and functional tfdA gene distribution in thirty-one different 2,4-dichlorophenoxyacetic acid and 4-chloro-2-methylphenoxyacetic acid degraders. Syst. Appl. Microbiol. 33:67–70 [DOI] [PubMed] [Google Scholar]
- 4. Bælum J, et al. 2008. Direct analysis of tfdA gene expression by indigenous bacteria in phenoxy acid amended agricultural soil. ISME J. 2:677–687 [DOI] [PubMed] [Google Scholar]
- 5. Bollag JM, Helling CS, Alexander M. 1967. Metabolism of 4-chloro-2-methylphenoxyacetic acid by soil bacteria. Appl. Microbiol. 15:1393–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brunner W, Focht DD. 1984. Deterministic 3-half-order kinetic-model for microbial-degradation of added carbon substrates in soil. Appl. Environ. Microbiol. 47:167–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crespin MA, Gallego M, Valcarcel M, Gonzalez JL. 2001. Study of the degradation of the herbicides 2,4-D and MCPA at different depths in contaminated agricultural soil. Environ. Sci. Technol. 35:4265–4270 [DOI] [PubMed] [Google Scholar]
- 8. Dahllof I, Baillie H, Kjelleberg S. 2000. rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl. Environ. Microbiol. 66:3376–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Day PR. 1965. Particle fractionation and particle-size analysis, p 545–567 In Black CA, Evans DD, White JL, Ensminger LE, Clark FE. (ed), Methods of soil analysis. American Society of Agronomy, Madison, WI [Google Scholar]
- 10. de Lipthay JR, Barkay T, Sorensen SJ. 2001. Enhanced degradation of phenoxyacetic acid in soil by horizontal transfer of the tfdA gene encoding a 2,4-dichlorophenoxyacetic acid dioxygenase. FEMS Microbiol. Ecol. 35:75–84 [DOI] [PubMed] [Google Scholar]
- 11. Eltra 1995. CS500 Simultaneous carbon/sulfur determinator. ELTRA GmbH, Neuss, Germany [Google Scholar]
- 12. Focht DD, Brunner W. 1985. Kinetics of biphenyl and polychlorinated biphenyl metabolism in soil. Appl. Environ. Microbiol. 50:1058–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fournier JC, Codaccioni P, Soulas G. 1981. Soil adaptation to 2,4-D degradation in relation to the application rates and the metabolic behavior of the degrading microflora. Chemosphere 10:977–984 [Google Scholar]
- 14. Fukumori F, Hausinger RP. 1993. Purification and characterization of 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase. J. Biol. Chem. 268:24311–24317 [PubMed] [Google Scholar]
- 15. Fulthorpe RR, Rhodes AN, Tiedje JM. 1996. Pristine soils mineralize 3-chlorobenzoate and 2,4-dichlorophenoxyacetate via different microbial populations. Appl. Environ. Microbiol. 62:1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gonod LV, Martin-Laurent F, Chenu C. 2006. 2,4-D impact on bacterial communities, and the activity and genetic potential of 2,4-D degrading communities in soil. FEMS Microbiol. Ecol. 58:529–537 [DOI] [PubMed] [Google Scholar]
- 17. Helweg A. 1987. Degradation and adsorption of C-14 MCPA in soil—influence of concentration, temperature and moisture content on degradation. Weed Res. 27:287–296 [Google Scholar]
- 18. Helweg A. 1993. Degradation and adsorption of 14C mecoprop (MCPP) in surface soils and in subsoil: influence of temperature, moisture content, sterilization and concentration on degradation. Sci. Total Environ. 132:229–241 [Google Scholar]
- 19. Jacobsen CS, Pedersen JC. 1992. Mineralization of 2,4-D in soil inoculated with DBO1 (pRO101), AEO106 (pRO101) and JMP134 (pJP4): effects of inoculation level and substrate concentration. Biodegradation 2:253–263 [DOI] [PubMed] [Google Scholar]
- 20. Jensen PH, Hansen HCB, Rasmussen J, Jacobsen OS. 2004. Sorption-controlled degradation kinetics of MCPA in soil. Environ. Sci. Technol. 38:6662–6668 [DOI] [PubMed] [Google Scholar]
- 21. Jørgensen LF, Stockmarr J. 2009. Groundwater monitoring in Denmark: characteristics, perspectives and comparison with other countries Hydrogeol. J. 17:827–842 [Google Scholar]
- 22. Kitagawa W, et al. 2002. Novel 2,4-dichlorophenoxyacetic acid degradation genes from oligotrophic Bradyrhizobium sp. strain HW13 isolated from a pristine environment. J. Bacteriol. 184:509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee TH, Kurata S, Nakatsu CH, Kamagata Y. 2005. Molecular analysis of bacterial community based on 16S rDNA and functional genes in activated sludge enriched with 2,4-dichlorophenoxyacetic acid (2,4-D) under different cultural conditions. Microb. Ecol. 49:151–162 [DOI] [PubMed] [Google Scholar]
- 24. Macur RE, Wheeler JT, Burr MD, Inskeep WP. 2007. Impacts of 2,4-D application on soil microbial community structure and on populations associated with 2,4-D degradation. Microbiol. Res. 162:37–45 [DOI] [PubMed] [Google Scholar]
- 25. Mortensen SK, Jacobsen CS. 2004. Influence of frozen storage on herbicide degradation capacity in surface and subsurface sandy soils. Environ. Sci. Technol. 38:6625–6632 [DOI] [PubMed] [Google Scholar]
- 26. Nicolaisen MH, Bælum J, Jacobsen CS, Sorensen J. 2008. Transcription dynamics of the functional tfdA gene during MCPA herbicide degradation by Cupriavidus necator AEO106 (pRO101) in agricultural soil. Environ. Microbiol. 10:571–579 [DOI] [PubMed] [Google Scholar]
- 27. Park JW, Crowley DE. 2005. Normalization of soil DNA extraction for accurate quantification real-time PCR and of target genes by DGGE. Biotechniques 38:579–586 [DOI] [PubMed] [Google Scholar]
- 28. Parker LW, Doxtader KG. 1982. Kinetics of microbial decomposition of 2,4-D in soil: effects of herbicide concentration. J. Environ. Qual. 11:679–684 [Google Scholar]
- 29. Pemberton JM, Corney B, Don RH. 1979. Evolution and spread of pesticide degrading ability among soil microorganisms, p 287–299 In Timmis KN, PÅhler A. (ed), Plasmids of medical, environmental and commercial importance. Elsevier, Amsterdam, the Netherlands [Google Scholar]
- 30. Reffstrup TK, Sorensen H, Helweg A. 1998. Degradation of mecoprop at different concentrations in surface and sub-surface soil. Pestic. Sci. 52:126–132 [Google Scholar]
- 31. Robinson JA. 1985. Determining microbial kinetic-parameters using nonlinear-regression analysis: advantages and limitations in microbial ecology. Adv. Microb. Ecol. 8:61–114 [Google Scholar]
- 32. Scow KM, Simkins S, Alexander M. 1986. Kinetics of mineralization of organic compounds at low concentration in soil. Appl. Environ. Microbiol. 51:1028–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Simkins S, Alexander M. 1984. Models for mineralization kinetics with the variables of substrate concentration and population density. Appl. Environ. Microbiol. 47:1299–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Simkins S, Alexander M. 1985. Nonlinear estimation of the parameters of Monod kinetics that best describe mineralization of several substrate concentrations by dissimilar bacterial densities. Appl. Environ. Microbiol. 50:816–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Streber WR, Timmis KN, Zenk MH. 1987. Analysis, cloning, and high-level expression of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J. Bacteriol. 169:2950–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Suwa Y, et al. 1996. Characterization of a chromosomally encoded 2,4-dichlorophenoxyacetic acid α-ketoglutarate dioxygenase from Burkholderia sp. strain RASC. Appl. Environ. Microbiol. 62:2464–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thorling L, et al. 2011. Groundwater. Status and development, 1989–2010. Technical report. Geological Survey of Denmark and Greenland, Copenhagen, Denmark: (In Danish.) [Google Scholar]
- 38. Tonso NL, Matheson VG, Holben WE. 1995. Polyphasic characterization of a suite of bacterial isolates capable of degrading 2,4-D. Microb. Ecol. 30:3–24 [DOI] [PubMed] [Google Scholar]
- 39. Top EM, Van Daele P, De Saeyer N, Forney LJ. 1998. Enhancement of 2,4-dichlorophenoxyacetic acid (2,4-D) degradation in soil by dissemination of catabolic plasmids. Antonie Van Leeuwenhoek 73:87–94 [DOI] [PubMed] [Google Scholar]
- 40. Zakaria D, Lappin-Scott H, Burton S, Whitby C. 2007. Bacterial diversity in soil enrichment cultures amended with 2 (2-methyl-4-chlorophenoxy) propionic acid (mecoprop). Environ. Microbiol. 9:2575–2587 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.