Abstract
Francisella tularensis is the causative agent of tularemia. Due to its aerosolizable nature and low infectious dose, F. tularensis is classified as a category A select agent and, therefore, is a priority for vaccine development. Survival and replication in macrophages and other cell types are critical to F. tularensis pathogenesis, and impaired intracellular survival has been linked to a reduction in virulence. The F. tularensis genome is predicted to encode 31 major facilitator superfamily (MFS) transporters, and the nine-member Francisella phagosomal transporter (Fpt) subfamily possesses homology with virulence factors in other intracellular pathogens. We hypothesized that these MFS transporters may play an important role in F. tularensis pathogenesis and serve as good targets for attenuation and vaccine development. Here we show altered intracellular replication kinetics and attenuation of virulence in mice infected with three of the nine Fpt mutant strains compared with wild-type (WT) F. tularensis LVS. The vaccination of mice with these mutant strains was protective against a lethal intraperitoneal challenge. Additionally, we observed pronounced differences in cytokine profiles in the livers of mutant-infected mice, suggesting that alterations in in vivo cytokine responses are a major contributor to the attenuation observed for these mutant strains. These results confirm that this subset of MFS transporters plays an important role in the pathogenesis of F. tularensis and suggest that a focus on the development of attenuated Fpt subfamily MFS transporter mutants is a viable strategy toward the development of an efficacious vaccine.
INTRODUCTION
Francisella tularensis is a nonmotile, non-spore-forming, Gram-negative coccobacillus and the causative agent of tularemia. Of the four recognized subspecies, two cause the majority of disease in humans, the highly virulent F. tularensis subsp. tularensis (type A) and the less virulent F. tularensis subsp. holarctica (type B). Exposure to as few as 10 organisms of F. tularensis subsp. tularensis by the aerosol route can cause disease in humans, which can lead to a 30 to 50% mortality rate if left untreated (14, 40). Because of its highly infectious, aerosolizable nature and ability to cause severe morbidity and mortality, F. tularensis has long been considered a potential biological weapon. In fact, F. tularensis was developed and tested as a biological weapon by the Japanese during World War II and was developed and stockpiled for use as a biological weapon by both the United States and the Soviet Union during the Cold War (24). For these reasons, F. tularensis subsp. tularensis has been designated a category A select agent by the CDC.
Currently, there is no licensed vaccine against tularemia, but vaccine development has become a renewed priority since the 2001 anthrax attacks. A number of live-attenuated vaccine candidates were made from F. tularensis subsp. holarctica strains by the Soviet Union in the 1940s and 1950s. One of these live-attenuated derivatives, now known as F. tularensis live vaccine strain (LVS), was transferred to the United States in 1956 and was named as such after undergoing several passages to achieve further attenuation (40). However, LVS is not licensed in the United States due to the uncertainty of its history and basis of attenuation. In a number of volunteer studies, LVS was shown to provide partial protection against a type A challenge, with the level of protection being dependent on a combination of the dose and route of vaccination and the challenge dose (14, 23, 34). While LVS is not an optimal vaccine, its ability to confer partial protection against the most virulent strains demonstrates the potential for a live-attenuated strain to serve as an efficacious vaccine. LVS serves as a good model of study for F. tularensis subsp. tularensis since it causes a lethal disease in mice which resembles human tularemia and, unlike the type A strains, can be handled in a biosafety level 2 (BSL-2) laboratory.
The ability of F. tularensis to survive and replicate in macrophages is critical to its pathogenesis, and its intracellular life cycle has been well characterized (24). Francisella is phagocytosed by macrophages into a phagosome, from which it escapes into the cytosol and replicates. Strains that are unable to escape the phagosome, or that escape but are unable to replicate, have been shown to be attenuated for virulence (7, 32, 33). In animal models, F. tularensis has been shown to disseminate to a number of organs, where it survives and replicates in other cell types, such as hepatic cells. Dissemination has also been linked to virulence (28). Hence, for the development of an effective live vaccine, genes involved in intracellular survival and replication are ideal targets for attenuation.
The genome of type A strain Schu S4 is predicted to contain 31 genes that encode major facilitator superfamily (MFS) transporters (TransportDB [www.membranetransport.org]) (31). MFS transporters are ubiquitous across all classes of organisms, typically feature a 12-trans-membrane-domain architecture, and are involved in the transport of a wide variety of small-molecule substrates (19, 25). In Legionella pneumophila, another intracellular bacterial pathogen, a strain with a mutation of the phtA gene, a member of the phagosomal transporter (Pht) subfamily of MFS transporters, was unable to survive and replicate in murine macrophages (35). In that study, Sauer et al. highlighted some other bacterial pathogens that express homologs of this subfamily, including four genes in F. tularensis. Using BLAST-P analysis with PhtA as the query sequence, we identified nine MFS transporters that are phtA homologs in the Schu S4 genome, each of which has a virtually identical LVS homolog. These nine genes represent a specific subfamily analogous to the L. pneumophila Pht subfamily which is distinct from the other 22 F. tularensis MFS genes.
We hypothesized that one or more members of the F. tularensis Pht subfamily of MFS transporters, which we have named the Fpt (Francisella phagosomal transporter) subfamily, would be critical for intracellular replication and the overall pathogenesis of the bacteria. Using a suicide plasmid system developed in our laboratory, we created strains with deletions of all nine Fpt subfamily genes and characterized their abilities to survive and replicate intracellularly. In this study, we report that mutants with deletions of three Fpt subfamily genes showed altered intracellular replication phenotypes in at least one cell type. Each of these mutant strains demonstrated a reduced ability to colonize and replicate in mouse organs. Most importantly, each mutant strain was attenuated for virulence in vivo, and a single inoculation was protective against a lethal LVS challenge.
MATERIALS AND METHODS
Bacteria and growth conditions.
Bacterial strains used in this study are listed in Table S1 in the supplemental material. F. tularensis LVS (ATCC, Manassas, VA) was kindly provided by Karen Elkins (CBER/FDA, Rockville, MD) and was preserved at −80°C in Mueller-Hinton broth (MHB) (BD Microbiology Systems, Sparks, MD) supplemented with 1% Isovitalex (BD, Cockeysville, MD), 0.1% glucose, and 0.25% ferric pyrophosphate (Sigma, St. Louis, MO). MHB supplemented as outlined above was used for liquid cultures, and Mueller-Hinton agar (MHA) supplemented as outlined above and also containing 10% defibrinated sheep blood was used for solid medium when Francisella strains were grown. Liquid cultures were grown in a shaker at 37°C. Growth on solid medium was performed at 37°C in 5% CO2. When needed for selection, kanamycin (Km) was added to a final concentration of 10 μg/ml. Suicide plasmids used in this study were maintained in Escherichia coli DH5α cells and grown in LB broth supplemented with Km to a final concentration of 50 μg/ml.
Construction of suicide plasmids.
All plasmids used in this study are summarized in Table S1 in the supplemental material. The construction of suicide plasmids was accomplished by the cloning of a PCR fusion product consisting of the flanking regions surrounding a given target gene into pFT893, a suicide plasmid created in our laboratory for Francisella mutagenesis. To generate pFT893, pSacB (32) was modified by the removal of the ampicillin resistance marker, and a PgroEL-aphT fragment (aphT gene under the control of the F. tularensis PgroEL promoter) was cloned into the plasmid to serve as a kanamycin resistance marker. The F. tularensis PguaB promoter was then cloned upstream of the sacB gene to optimize the expression of the SacB protein in F. tularensis. Flanking regions of approximately 500 bp both upstream and downstream of a target gene were amplified from genomic DNA by PCR (primers are listed in Table S2 in the supplemental material). The individual amplicons were then fused in a second PCR using complementary sequences with cloning sites built into the internal primers and external primers. All PCRs for suicide plasmid construction were performed by using the Expand Long Template PCR system (Roche Diagnostics, Indianapolis, IN). The resulting 1-kb fusion product was then ligated into the BamHI site of pFT893. This plasmid features a ColE1 origin of replication that is not functional in Francisella, a kanamycin resistance cassette for the selection of cointegrant strains, and a sacB cassette under the control of Francisella PguaB for the counterselection of deletion mutants cured of the plasmid. Individual suicide plasmids were confirmed by both PCR and restriction digestion.
Deletion of target genes in F. tularensis.
The transformation of F. tularensis LVS was performed by electroporation. Competent cells were prepared by growing a 100-ml MHB culture of F. tularensis LVS at 37°C to the mid-log phase, followed by sequential pelleting and a 1/10-volume washing in 0.5 M sucrose. After the removal of the supernatant, the pellet was resuspended in 300 μl of 0.5 M sucrose for immediate electroporation or storage at −80°C. For electroporation, 150 μl of suicide plasmid DNA prepared from 300 ml of LB broth culture (using a Qiagen [Germantown, MD] Midi-Prep kit) was mixed with 300 μl of competent cells and pulsed 3 times at 1.75 kV, 25 μF, and 600 Ω. After electroporation, the suspension was recovered in MHB for 3 h at 37°C in 5% CO2. After recovery, the suspension was plated onto MHA containing kanamycin to select for cointegrant strains. Single colonies were confirmed by PCR. Once a cointegrant strain was confirmed, the plasmid was then cured from the strain by growth in MHB containing 10% sucrose, which allows for the selection of strains not expressing the sacB cassette and which have lost the plasmid. This culture was plated onto MHA, and individual colonies were screened by PCR to confirm that the target gene had been deleted.
Growth curves.
Growth curves were performed with MHB. Either LVS or Fpt mutant strains were grown in a starter culture overnight and were then subcultured in fresh MHB to an optical density at 600 nm (OD600) of 0.2. Cultures were incubated with shaking at 37°C. OD readings were taken approximately every 2 h over an 8-h period, and the growth profiles of Fpt mutant strains were compared to that of LVS.
Verification of Fpt gene expression.
The expression of Fpt genes both in broth culture and intracellularly in the J774.1 murine macrophage cell line (ATCC, Manassas, VA) was verified. To verify expression in broth culture, LVS was cultured in MHB, and RNA extraction was performed by using the RNeasy Protect minikit (Qiagen). To verify expression in J774.1 cells, a confluent T150 flask was infected with LVS at a multiplicity of infection (MOI) of 100. After a 2-h infection, the cells were treated with gentamicin to kill extracellular bacteria. Bacteria were then allowed to proliferate intracellularly for 20 h. The cells were then treated with TRIzol (Invitrogen, Carlsbad, CA) for 10 min, and RNA was extracted by using the PureLink RNA minikit (Invitrogen). In both cases, reverse transcription-PCR (RT-PCR) was performed by using the RevertAid first-strand cDNA synthesis kit (Fermentas, Glen Burnie, MD). Fpt gene-specific primers (see Table S2 in the supplemental material) were used to amplify each gene via PCR with a cDNA transcript to confirm expression.
Intracellular survival assays.
The abilities of Fpt mutant strains to survive and replicate intracellularly were evaluated with both the J774.1 murine macrophage cell line and the HepG2 human hepatic cell line (ATCC, Manassas, VA).
J774.1 cells were cultivated in Dulbecco's modified essential medium (DMEM) (Cellgro, Manassas, VA) supplemented with 10% fetal bovine serum (FBS) (Cellgro) and were maintained at 37°C in 5% CO2. To assess the intracellular survival abilities of Fpt mutant strains compared to that of parental LVS, J774.1 cells were seeded at a density of 2 × 105 cells per well in 12-well plates (Costar, Corning, NY) and infected at an MOI of 100 with either LVS or Fpt mutant strains for a period of 2 h. Following the 2-h infection, cells were washed twice with phosphate-buffered saline (PBS) and then incubated in DMEM containing 50 μg/ml gentamicin for 1 h. The cells were washed twice with PBS and then incubated in DMEM containing 2 μg/ml gentamicin for the duration of the experiment. Bacterial replication was assayed at 0-, 24-, 48-, and 72- h postinfection by lysing the cells with a 0.02% SDS solution followed by plating onto MHA.
HepG2 cells were cultivated in minimal essential medium (MEM) supplemented with 10% FBS and maintained at 37°C in 5% CO2. To assess the intracellular survival abilities of Fpt mutant strains compared to that of LVS, HepG2 cells were also seeded at a density of 2 × 105 cells per well in 12-well plates and infected with either LVS or Fpt mutant strains for 4 h. After the 4-h infection, cells were washed twice with PBS and then incubated in MEM containing 50 μg/ml gentamicin for 1 h. Next, cells were washed twice with PBS and incubated in antibiotic-free MEM for the duration of the experiment. Bacterial replication was assayed at 0, 24, 48, and 72 h postinfection by lysing the cells with 0.02% SDS and plating the cells onto MHA.
Intracellular doublings were calculated by using the formula [log10Tn − log10T(n − 1)] × 3.32, where Tn is the number of CFU at a given time point of interest and T(n − 1) is the number of CFU at the previous time point.
Complementation of mutants.
pFT906-FptB, pFT906-FptE, and pFT906-FptG were constructed to trans-complement their respective deletions in LVS. Each open reading frame (ORF) was amplified from LVS genomic DNA by using the Expand Long Template PCR system (Roche) and ligated into pFT906 (32) to be expressed under the control of the F. tularensis guaB promoter (F. tularensis PguaB). pFT906 features a multiple-cloning site directly downstream of F. tularensis PguaB and a kanamycin resistance cassette for selection. pFT906-FptB was created by the ligation of PCR-amplified ORF FTT_0056c into the SpeI site on pFT906. pFT906-FptE was created by the ligation of PCR-amplified ORF FTT_0129 into an SmaI-NdeI site on pFT906. pFT906-FptG was created by the ligation of PCR-amplified ORF FTT_1291 into an SpeI-KpnI site on pFT906. Once each of these plasmids was confirmed by restriction digestion, it was electroporated into its corresponding LVS mutant strain as described above.
Attenuation and protective capacity of MFS mutant strains in mice in vivo.
To determine whether any Fpt mutant strains were attenuated in vivo, a mouse infection model was used. Groups of four 6- to 8-week-old female BALB/c mice (Charles River Labs, MD) were injected intraperitoneally (i.p.) with either bacterial inoculum (the Fpt mutant strain or LVS in 0.1% gelatin–PBS) or a solution containing 0.1% gelatin–PBS and then monitored for survival and other clinical signs of infection (weight loss, ruffling of fur, and lethargy) for a period of 28 days postinfection. Any mice surviving the initial infection (or the control injection of 0.1% gelatin–PBS) were then challenged by the i.p. route with the parental LVS strain. Mice were monitored for survival and other clinical signs of infection for a period of 28 days postinfection. Any mouse showing two or more signs for a single day or one sign for two consecutive days was euthanized. Any mouse that maintained a loss of more than 20% of its starting weight was euthanized. All animal experiments were conducted with institutional approval.
Quantification of bacterial burden.
To determine whether any Fpt mutant strains were attenuated in vivo, a mouse infection model was used. Groups of three 6- to 8-week-old female BALB/c mice were injected by the i.p. route with either LVS or an Fpt mutant strain. Mice were euthanized, and organs were harvested to enumerate bacterial loads on days 1, 2, and 3 postinfection. The bacterial load was enumerated by serial plating. In a similar experiment to determine the time to complete bacterial clearance in the organs, mice were infected i.p. with either LVS or an Fpt mutant strain as described above. Mice were euthanized, and organs were harvested to enumerate bacterial loads on days 7, 14, 21, and 28 postinfection.
Real-time PCR.
Total RNA was isolated from infected murine livers by using TRIzol (Invitrogen) according to the manufacturer's protocol, and cDNA was generated by using the qScript cDNA synthesis kit (Quanta BioSciences, Gaithersburg, MD). Real-time PCR was performed with an ABI 7900HT (Applied Biosystems) sequence detection system and software as previously described (5). Relative mRNA levels for specific genes are reported as relative gene expression levels normalized to levels of mock-infected control samples. The primer sequences for tumor necrosis factor alpha (TNF-α), interleukin-1β (IL-1β), gamma interferon (IFN-γ), IFN-β, hypoxanthine phosphoribosyltransferase (HPRT), and inducible nitric oxide synthase (iNOS) were reported previously (5).
Histopathology.
Organs to be examined for tissue pathology were fixed in 4% paraformaldehyde. After fixation, the tissues were trimmed, dehydrated in graded ethanols, and embedded in paraffin. Tissue blocks were sectioned at 5 to 6 μm and stained with hematoxylin and eosin by the University of Maryland School of Medicine Histology Core Laboratory.
Statistical analysis.
Regression analysis was used to determine statistical significance in intracellular survival assays. The 0-, 24-, 48-, and 72-h time points were analyzed separately, with LVS serving as the reference strain. Results were considered statistically significant at a P value of <0.05. A two-sample t test was used to determine statistical significance in organ bacterial burden assays. Results were considered statistically significant at a P value of <0.05. Dunnett's two-sided multiple-comparison test was used to determine statistical significance in cytokine gene expression assays. Results were considered statistically significant at a P value of <0.05.
RESULTS
Identification of Francisella MFS genes.
There are 31 genes predicted to encode MFS transporters in the F. tularensis genome (Transport DB [www.membranetransport.org]) (31). We identified nine of these as having homology to the L. pneumophila phtA gene and named them fptA to fptI, ranking them based on protein similarity to L. pneumophila PhtA, as determined by BLAST scores (Table 1). We first determined if each gene was expressed during growth in both broth and intracellular environments. RNA was isolated from LVS cells grown overnight in a broth culture or from the J774.1 macrophage cell line that had been infected with LVS at an MOI of 100 for 18 h. After cDNA was generated, PCR was performed with gene-specific primers to confirm the presence of transcripts. The expression of all nine Fpt subfamily genes was confirmed both intracellularly (Fig. 1) and in broth culture (data not shown).
Table 1.
Gene designations and similarity of F. tularensis Schu S4 Fpt proteins to L. pneumophila PhtA
| Gene | Schu S4 IDa | LVS IDb | Schu S4 vs LVS aa identity (%)c | BLAST scored |
|---|---|---|---|---|
| fptA | FTT_0104c | FTL_1673 | 99 | 9e−39 |
| fptB | FTT_0056c | FTL_1803 | 99 | 1e−36 |
| fptC | FTT_0671 | FTL_0946 | 99 | 4e−32 |
| fptD | FTT_0053 | FTL_1806 | 100 | 1e−30 |
| fptE | FTT_0129 | FTL_1645 | 100 | 6e−27 |
| fptF | FTT_0127c | FTL_1647 | 99 | 3e−22 |
| fptG | FTT_1291 | FTL_0443 | 99 | 4e−13 |
| fptH | FTT_0488c | FTL_1573 | 98 | 1e−10 |
| fptI | FTT_0708 | FTL_1528 | 99 | 1e−4 |
Schu S4 GenBank accession number NC_006570.2.
LVS GenBank accession number NC_007880.1.
aa, amino acid.
Comparison of each Fpt protein sequence from Schu S4 to that of PhtA by using BLAST-P.
Fig 1.
fpt genes are expressed during intracellular growth. J774.1 cells were infected with LVS for a period of 20 h. Following overnight incubation, RNA was isolated, cDNA was generated, and the presence of fpt transcripts was confirmed by PCR with Fpt gene-specific primers. For each labeled gene, lane 1 represents the gene-specific PCR product amplified from cDNA, lane 2 is a non-RT negative control for genomic DNA contamination, and lane 3 is a positive control where the specified gene was amplified from LVS genomic DNA.
Replication kinetics are altered in multiple cell types in a subset of Fpt mutants.
LVS strains with a complete deletion of each individual fpt gene were constructed. The ability of each mutant strain to grow in broth culture was compared to that of parental LVS. Standardized growth curves showed no growth defects for any Fpt mutant strain in comparison with parental LVS (data not shown).
The ability to survive and replicate in macrophages is central to the pathogenesis of F. tularensis. LVS is typically phagocytosed by J774.1 macrophages in high numbers and proceeds to undergo 6 to 7 doublings within the first 24 h following infection (our unpublished observations). Each Fpt mutant was assessed for survival and growth within macrophages. While the majority of Fpt mutants had survival and replication kinetics similar to those of LVS, two mutant strains, the LVS ΔfptB and LVS ΔfptG strains, exhibited altered intracellular replication profiles compared to that of LVS. Specifically, each of these two strains exhibited a significant defect in replication (P < 0.01) during the first 24 h after infection (Fig. 2). LVS underwent approximately 7 doublings during the first 24 h after infection, whereas the LVS ΔfptB and LVS ΔfptG strains underwent 1.5 and 3 doublings, respectively. By 48 h postinfection, both mutants reached intracellular levels closer to those of LVS. By 72 h postinfection, the LVS ΔfptB strain had reached intracellular levels equivalent to those of LVS, while both LVS and the LVS ΔfptG strain ceased replication. The trans-complementation of both mutants restored intracellular replication to wild-type (WT) levels (Fig. 2).
Fig 2.
Two Fpt mutant strains exhibit early altered replication kinetics in J774.1 cells. The LVS ΔfptB (A) and LVS ΔfptG (B) strains exhibited a significant defect in replication in J774.1 cells at 24 h postinfection. J774.1 cells were infected with either LVS or the specified mutant strain at an MOI of 100 for 2 h. Cells were then washed twice with PBS and incubated with 50 μg/ml gentamicin for 1 h. Following the incubation with gentamicin, cells were again washed twice with PBS and then maintained in medium containing 2 μg/ml gentamicin for the duration of the experiment. Intracellular bacteria were counted at 0, 24, 48, and 72 h postinfection. Data are plotted is the arithmetic mean CFU counts ± standard deviations and are from a single representative experiment of two experiments with similar designs and outcomes. *, P < 0.01 by regression analysis.
F. tularensis is also capable of survival and replication in the liver (8). Therefore, we also performed intracellular survival assays with HepG2 cells, a hepatic cell line. The majority of Fpt mutants showed replication kinetics similar to those of LVS. The two mutant strains that showed altered replication kinetics in macrophages (the LVS ΔfptB and LVS ΔfptG strains) and a third mutant strain, the LVS ΔfptE strain, also demonstrated delayed replication kinetics in HepG2 cells (P < 0.05) at 24 h postinfection but recovered to levels at or near those of LVS by 72 h postinfection (Fig. 3). The trans-complementation of each mutant restored intracellular replication to WT levels. These results demonstrate that three members of the Fpt subfamily are important for the intracellular replication of F. tularensis and, therefore, may play a role in pathogenesis.
Fig 3.
Three Fpt mutant strains exhibit early altered replication kinetics in HepG2 cells. The LVS ΔfptB (A), LVS ΔfptE (B), and LVS ΔfptG (C) strains exhibited significant defects in replication in HepG2 cells at 24 h postinfection. HepG2 cells were infected with either LVS or the specified mutant strain at an MOI of 100 for 4 h. Cells were then washed twice with PBS and incubated with 50 μg/ml gentamicin for 1 h. Following the incubation with gentamicin, cells were again washed twice and then maintained in antibiotic-free medium for the duration of the experiment. Intracellular bacteria were counted at 0, 24, 48, and 72 h postinfection. Data are plotted is the arithmetic mean CFU counts ± standard deviations and are from a single representative experiment of two experiments with similar designs and outcomes. *, P < 0.05; **, P < 0.01 (by regression analysis).
Fpt mutants with altered intracellular replication kinetics are attenuated in mice and protective against WT challenge.
The Fpt mutants with altered in vitro intracellular replication kinetics were assessed for virulence in vivo. Groups of BALB/c mice were infected i.p. with LVS or the Fpt mutant strains and monitored for survival and clinical signs of infection over a period of 28 days. LVS has been shown to cause a lethal infection by this route in mice with a dose of fewer than 100 CFU (8, 10). We have observed a 100% mortality rate by day 7 postinfection in mice infected with 500 CFU of LVS. The attenuation of the Fpt mutant strains was tested in a series of studies in which mice were infected with each mutant strain at dose levels ranging from 102 to 105 CFU and monitored over 28 days (Table 2 and Fig. 4). Mice infected with the LVS ΔfptB strain had a 75% survival rate at doses of both 102 and 103 CFU and a 50% survival rate at doses of 3 × 104 and 3 × 105 CFU (Fig. 4A). At both doses, both survivors lost a notable amount of weight and showed obvious signs of infection but recovered. Mice infected with the LVS ΔfptE strain had a 100% survival rate at doses ranging from 200 to 3.5 × 103 CFU, but only 50% and 25% of mice survived doses of 3.5 × 104 and 3.5 × 105 CFU, respectively (Fig. 4B). The LVS ΔfptG strain was the most attenuated mutant strain, and 100% of mice survived all doses up to 2.7 × 106 CFU (Fig. 4C). At this high dose, the mice did not exhibit weight loss and showed only minor, transient signs of illness.
Table 2.
Summary of survival and protective capacities of Fpt mutant strains
| Treatment or strain | Vaccination dose (CFU) | Survival rate after vaccination (no. of surviving mice/total no. of mice tested) | Time to challenge (days) | Challenge dose (CFU) | Survival rate after challenge (no. of surviving mice/total no. of mice tested) |
|---|---|---|---|---|---|
| 0.1% gelatin–PBS | 4/4 | 28 | 5.1 × 103 | 0/4 | |
| 0.1% gelatin–PBS | 4/4 | 28 | 1.2 × 105 | 0/4 | |
| 0.1% gelatin–PBS | 5/5 | 28 | 2 × 106 | 0/5 | |
| LVS | 5 × 102 | 0/4 | |||
| LVS ΔfptB mutant | 5 × 102 | 3/4 | 28 | 6.6 × 103 | 3/3 |
| LVS ΔfptB mutant | 5 × 103 | 3/4 | 28 | 6.6 × 103 | 3/3 |
| LVS ΔfptB mutant | 3 × 103 | 3/4 | 28 | 1.2 × 105 | 3/3 |
| LVS ΔfptB mutant | 3 × 104 | 2/4 | 28 | 1.2 × 105 | 2/2 |
| LVS ΔfptB mutant | 3 × 105 | 2/4 | 28 | 1.2 × 105 | 2/2 |
| LVS ΔfptE mutant | 2 × 102 | 4/4 | 28 | 5.1 × 103 | 4/4 |
| LVS ΔfptE mutant | 2 × 103 | 4/4 | 28 | 5.1 × 103 | 4/4 |
| LVS ΔfptE mutant | 3.5 × 103 | 4/4 | 28 | 1.2 × 105 | 4/4 |
| LVS ΔfptE mutant | 3.5 × 104 | 2/4 | 28 | 1.2 × 105 | 2/2 |
| LVS ΔfptE mutant | 3.5 × 105 | 1/4 | 28 | 1.2 × 105 | 1/1 |
| LVS ΔfptG mutant | 5 × 102 | 4/4 | 28 | 5.1 × 103 | 4/4 |
| LVS ΔfptG mutant | 5 × 103 | 4/4 | 28 | 5.1 × 103 | 4/4 |
| LVS ΔfptG mutant | 3.4 × 103 | 4/4 | 28 | 1.2 × 105 | 4/4 |
| LVS ΔfptG mutant | 3.4 × 104 | 4/4 | 28 | 1.2 × 105 | 4/4 |
| LVS ΔfptG mutant | 3.4 × 105 | 4/4 | 28 | 1.2 × 105 | 4/4 |
| LVS ΔfptG mutant | 2.7 × 106 | 5/5 | 28 | 2 × 106 | 5/5 |
| 0.1% gelatin–PBS | 4/4 | 90 | 2.5 × 105 | 0/4 | |
| LVS ΔfptB mutant | 1.5 × 103 | 4/4 | 90 | 2.5 × 105 | 4/4 |
| LVS ΔfptE mutant | 2.5 × 103 | 4/4 | 90 | 2.5 × 105 | 4/4 |
| LVS ΔfptG mutant | 4 × 105 | 4/4 | 90 | 2.5 × 105 | 4/4 |
Fig 4.
LVS Fpt mutant strains are attenuated in the mouse model. The survival rate of BALB/c mice following infection with in vitro-attenuated Fpt mutant strains was measured. Groups of 4 BALB/c mice were inoculated i.p. with either LVS (3.4 × 103 CFU); the LVS ΔfptB (3 × 103, 3 × 104, or 3 × 105 CFU) (A), LVS ΔfptE (3.5 × 103, 3.5 × 104, or 3.5 × 105 CFU) (B), or LVS ΔfptG (3.4 × 103, 3.4 × 104, or 3.4 × 105 CFU) (C) strain; or a 0.1% PBS–gelatin solution and monitored for survival and clinical signs of disease for 28 days.
Surviving mice were tested for protection against lethal challenge. Following a challenge dose of 5 × 103 to 6 × 103 CFU of LVS, all immunized mice, regardless of the vaccination strain, survived, with no clinical signs of disease (Table 2). At this challenge dose, there was a 100% mortality rate for the unimmunized control group. When the challenge dose was increased to 1.2 × 105 CFU of LVS (approximately 1,000 times the lethal dose), all vaccinated mice were fully protected and displayed no signs of disease. Mice vaccinated with 2.7 × 106 CFU of the LVS ΔfptG strain, the most attenuated mutant, were fully protected against an LVS challenge of 2 × 106 CFU (Table 2).
We next sought to determine how long mice vaccinated with Fpt mutant strains would remain protected. Groups of 4 BALB/c mice were inoculated with each Fpt mutant at a dose previously determined to be well tolerated (i.e., 1.5 × 103 CFU of the LVS ΔfptB strain, 2.5 × 103 CFU of the LVS ΔfptE strain, and 4 × 105 CFU of the LVS ΔfptG strain). After 90 days, all mice were challenged with 2.5 × 105 CFU of LVS and monitored for survival and clinical signs of infection for 28 days. All mice were protected against challenge and showed no signs of illness for the duration of the experiment (Table 2). These results demonstrate that members of the Fpt subfamily are critical for the in vivo virulence of F. tularensis and suggest that these genes represent potential targets for the development of an effective vaccine.
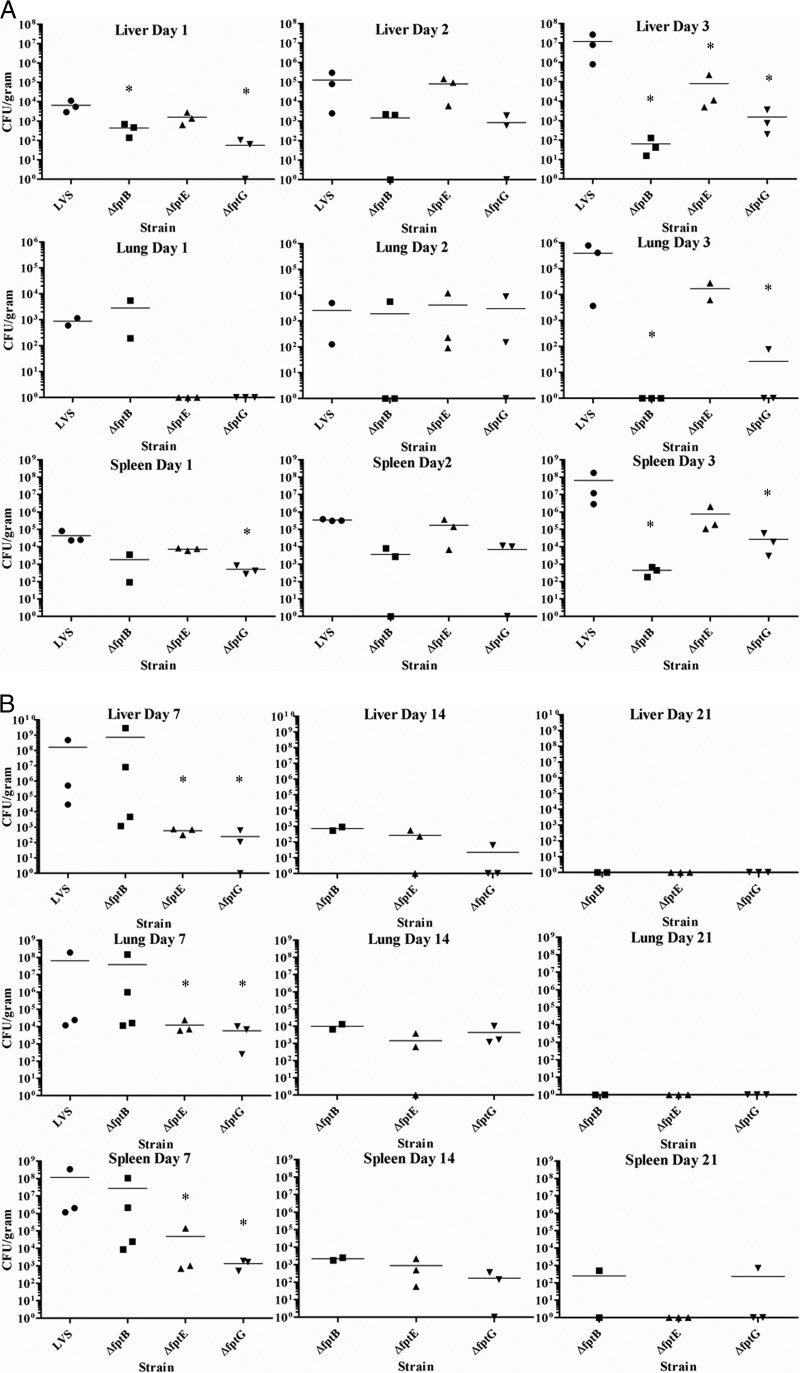
Fpt mutant strains have reduced bacterial burdens in the liver, lung, and spleen.
Following infection, F. tularensis colonizes and replicates in a number of organs, including the liver, lung, and spleen (8). The ability of each of the attenuated mutant strains to spread to and colonize organs was characterized. In initial studies, mice were infected with a low dose of each mutant strain (3 × 102 to 7 × 102 CFU) i.p. and euthanized at days 1, 2, and 3 postinfection, at which times livers, lungs, and spleens were harvested, homogenized, and assayed for bacterial burdens. Organs from mice infected with LVS were colonized on day 1 postinfection, and the bacterial burdens increased through days 2 and 3 in all organs. The spleen was most heavily colonized, followed by the liver and lung. The high bacterial burdens observed by day 3 correlated with the onset of clinical signs of infection in LVS-infected mice. While all Fpt mutant strains colonized each organ, some mutants had a delay in colonization, and the level of colonization was often reduced compared to that of LVS (Fig. 5A).
Fig 5.
(A) The bacterial burden of Fpt mutant strains is less severe than that of LVS in mouse organs. BALB/c mice were infected with a low dose of either an Fpt mutant strain or LVS, and organs were harvested and homogenized for bacterial enumeration on days 1, 2, and 3 postinfection. Bars indicate arithmetic means of bacterial counts. *, P < 0.05 by a two-sample t test. The statistical significance represented is between a given Fpt mutant strain and LVS at the indicated time point. (B) LVS Fpt mutant strains are cleared from mouse organs by day 28 after infection. BALB/c mice were infected with either a well-tolerated dose of an Fpt mutant strain or a lethal dose of LVS, and organs were harvested and homogenized. Organs from LVS-infected mice were harvested and homogenized on day 5, and organs from Fpt mutant-infected mice were harvested and homogenized on days 7, 14, 21, and 28 postinfection. Bars indicate arithmetic means of bacterial counts. *, P < 0.05 by a two-sample t test. The statistical significance represented is between a given Fpt mutant strain and LVS at the indicated time point.
In mice infected with the LVS ΔfptB strain, the lung, liver, and spleen were all colonized on day 1. The bacterial burden in the lung was similar to that of LVS but was reduced by more than a log in both the liver and the spleen. By day 2, the bacterial burden in mice infected with LVS was beginning to increase in all organs, but the burden in LVS ΔfptB strain-infected mice was still similar to that observed on day 1, and some individual mice showed bacterial clearance. By day 3, the bacterial burden in LVS-infected mice had increased sharply, but the burden in LVS ΔfptB strain-infected mice had begun to decline in all organs, with the lung being totally free of bacteria. In mice infected with the LVS ΔfptE strain, the liver and spleen were colonized at levels similar to those of LVS on day 1, but the lungs were not colonized. The bacterial burden in LVS ΔfptB mutant-infected mice was observed to increase in all organs on days 2 and 3. The burden observed for all organs on day 2 was similar to that of LVS but was 10 to 100 times lower in every organ than with LVS on day 3. Mice infected with the LVS ΔfptG strain were colonized at lower levels than with LVS in the liver and spleen on day 1 and were free of bacteria in the lung. On day 2, the levels of colonization in the liver and spleen were still lower than those of LVS and were similar in the lung. On day 3, the level of colonization in each organ was 1,000 to 10,000 times lower than that of LVS, and two of three mice were actually free of bacteria in the lung (Fig. 5A).
To extend this study, groups of mice were infected by the i.p. route with either LVS at a dose of 103 CFU or an Fpt mutant strain at the highest dose that was well tolerated in the previous experiments (103 CFU of the LVS ΔfptB and LVS ΔfptE strains and 105 CFU of the LVS ΔfptG strains) and were euthanized at days 7, 14, 21, and 28 postinfection to determine the time to completely clear each strain. All LVS-infected mice either had succumbed to infection or were euthanized at day 5, and all organs were infected with large numbers of bacteria, up to 109 CFU per gram of tissue in some organs (Fig. 5B). Mice infected with Fpt mutant strains generally exhibited a lower overall bacterial burden in the organs at day 7 postinfection. The LVS ΔfptB strain was present at slightly reduced levels in 2 of 4 mice at day 7 compared to LVS, and the two mice with higher organ burdens displayed clinical signs of illness, while the two with low burdens did not. Bacterial burdens for the LVS ΔfptE and LVS ΔfptG strains were 4 to 5 logs lower than those for LVS. Bacterial burdens steadily decreased through the course of the experiment, and bacteria were cleared from some individual organs from mice infected with the LVS ΔfptE or LVS ΔfptG strain by day 14 and were completely cleared in all but the spleens of 2 mice by day 21 (one spleen was minimally infected by the LVS ΔfptB mutant, and another was minimally infected by the LVS ΔfptG mutant). All organs showed bacterial clearance by day 28.
The severity of tissue pathology observed for the livers of the infected mice was closely correlated with the bacterial burden (Fig. 6). The LVS-infected mice that were euthanized or had succumbed at day 5 exhibited extensive inflammation and granuloma formation. Numerous necrotic and some apoptotic lesions were also observed (Fig. 6A). Inflammation and granuloma formation were also observed in livers of mice infected with Fpt mutants (observed at day 7), but the pathology was generally less severe than that of LVS-infected livers (Fig. 6C and D). Notably, there were fewer instances of necrotic or apoptotic lesions observed for the livers of LVS ΔfptE mutant-infected mice, and there were few to no such lesions observed for mice infected with the LVS ΔfptG mutant. The two LVS ΔfptB mutant-infected mice that had higher bacterial burdens in their livers showed pathology more similar to that of the LVS-infected mice (Fig. 6E), whereas the two with lower bacterial burdens exhibited reduced pathology similar to that of the LVS ΔfptG mutant-infected mice. At day 14, livers from Fpt mutant-infected mice showed decreased pathology with minor inflammation and granuloma formation and no evidence of necrotic or apoptotic lesions regardless of the infecting strain. By day 21, only very minor signs of inflammation were present, with no granulomas or lesions being observed regardless of the infecting strain (Fig. 6F).
Fig 6.
Pathology is more severe in the livers of mice infected with LVS than in the livers of mice infected with Fpt mutants. (A) Liver section taken from a mock-infected mouse on day 1. (B) Liver section taken from an LVS-infected mouse at day 5 postinfection. (C and D) Liver sections taken from LVS ΔfptB (C) and LVS ΔfptG (D) mutant-infected mice at day 7 postinfection. (E) Liver section taken from an LVS ΔfptB mutant-infected mouse with a high bacterial burden at day 7 postinfection. (F) Liver section taken from an LVS ΔfptG mutant-infected mouse at day 21 postinfection. Arrows denote areas of inflammation and granuloma formation containing necrotic lesions. All images were taken at a ×100 magnification.
Altered cytokine responses to Fpt mutants in livers of infected mice.
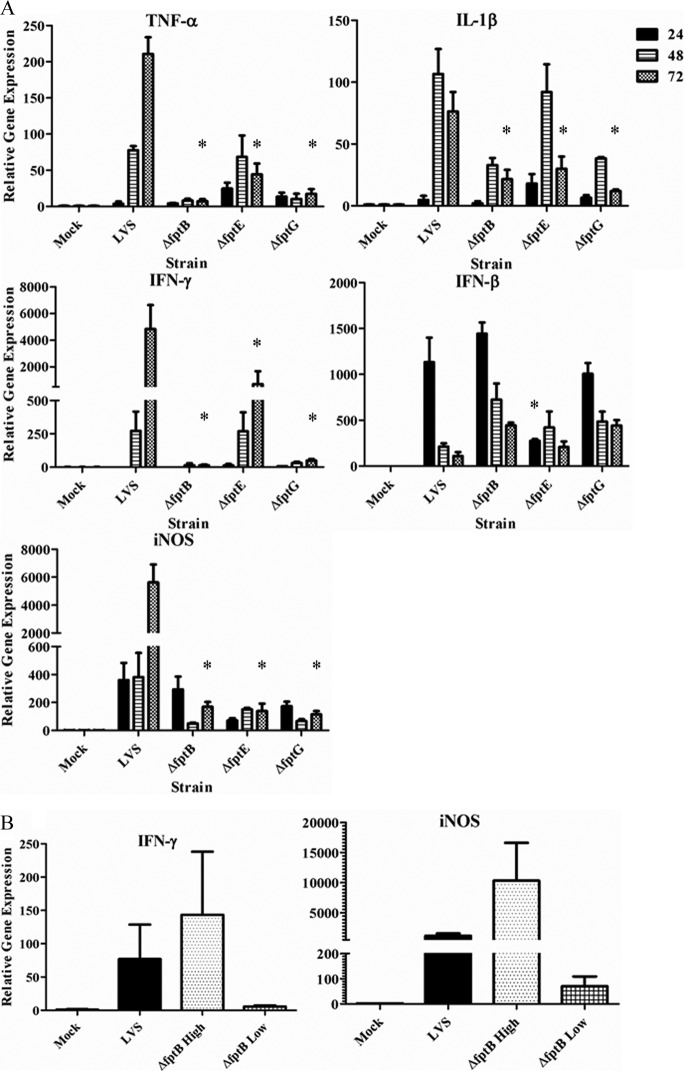
Since the pathology induced by LVS has been attributed to the ability of this organism to produce an early cytokine storm (5, 21, 22), we hypothesized that the attenuated Fpt mutants would also exhibit a diminished early proinflammatory response. Livers from LVS-infected mice showed significantly increased levels of cytokine and iNOS mRNAs over a 24- to 72-h period, as previously reported (5, 6). In contrast, the two more attenuated strains, the LVS ΔfptB and LVS ΔfptG strains, exhibited markedly decreased cytokine (TNF-α, IL-1β, and IFN-γ) and iNOS gene expression levels (Fig. 7A). The LVS ΔfptE mutant, which showed intermediate attenuation (Fig. 4), also showed intermediate expression levels of TNF-α, IL-1β, and IFN-γ over the 72-h time period. Interestingly, only the LVS ΔfptE strain induced diminished IFN-β gene expression levels compared to the levels observed for LVS-infected mice or mice infected with the other two Fpt mutants (Fig. 7A).
Fig 7.
Cytokine gene expression in murine livers in response to infection with Fpt mutant strains is altered compared to that in response to infection with LVS. Groups of 3 BALB/c mice were infected with either PBS (mock infected), 500 CFU of LVS, 400 CFU of the LVS ΔfptB strain, 300 CFU of the LVS ΔfptE strain, or 700 CFU of the LVS ΔfptG strain. (A) Livers were harvested and homogenized at 24, 48, and 72 h after infection. (B) Difference in IFN-γ and iNOS expression levels in LVS ΔfptB mutant-infected mice (1.5 × 103 CFU) with high bacterial burdens versus those with low bacterial burdens at day 7. High bacterial burden versus low bacterial burden refers to Fig. 5B, where two mice had bacterial burdens similar to those for LVS infection (approximately 108 CFU/gram tissue) and are considered “high” and two mice had bacterial burdens similar to those for infection with the other two mutants (approximately 103 CFU/gram tissue) and are considered low. Relative cytokine gene expression levels were measured by using quantitative RT-PCR. *, P < 0.05 by Dunnett's two-sided multiple-comparison test. The statistical significance represented is between a given Fpt mutant strain and LVS at the indicated time point.
We also observed that increased expression levels of a subset of these inflammatory cytokines were correlated directly with an increased bacterial burden in organs and the onset of clinical signs of illness. At day 7 after infection, two of four mice infected with the LVS ΔfptB strain had a very high bacterial burden in the liver that was similar to that observed for LVS-infected mice (approximately 108 CFU/gram tissue) (Fig. 5B) and exhibited obvious signs of illness. Both IFN-γ and iNOS mRNA levels in these two mice were considerably higher than those in livers of mock-infected mice and not statistically different from those in the LVS-infected mice (Fig. 7B). In contrast, the other two mice infected with the LVS ΔfptB strain had considerably lower bacterial burdens in the liver (approximately 103 CFU/gram tissue) (Fig. 5B), had no obvious signs of illness, and showed a level of induction of IFN-γ and iNOS mRNAs that was considerably lower than that observed for the LVS-infected mice or their LVS ΔfptB strain-infected counterparts, which had much higher bacterial burdens in the liver (Fig. 7B).
DISCUSSION
There has been a surge of research interest in Francisella tularensis due in large part to concern for its use in a bioterror attack. Due to the lack of a licensed vaccine, much effort has been dedicated to the development of vaccines against tularemia. One common strategy used for the development of vaccine candidates has been the construction of live-attenuated strains by the targeted deletion of genes suspected to encode virulence factors. Advantages to this approach include the ability to engineer strains with a clearly defined molecular basis of attenuation with multiple distinct genetic deletions, greatly reducing the risk of reversion. While the highly virulent type A strains remain the target for a protective vaccine, type B LVS provides a good model system to use for the engineering of prototype attenuated strains. LVS retains the ability to mimic the lethal disease caused by type A strains (such as Schu S4) in humans in animal models and can be studied in BSL-2 containment. Mutations of various metabolic genes and virulence factors in the LVS background, such as mutations of purMCD, guaB and guaA, sodB, and tolC, have conferred various degrees of attenuation and protective capacity against the parental strain in mouse models (2, 11, 26, 27, 32). Three mutants have been shown to provide significant protection against Schu S4 challenge in mice, an LVS sodB mutant (1); a Schu S4 FTT_1103 mutant, which encodes a DsbA-like protein (29); and a Schu S4 clpB mutant (9, 39). The creation of attenuated strains with defined genetic deletions toward the goal of creating a safe and highly efficacious vaccine continues.
Toward the goal of defining improved targets for mutation in generating live-attenuated vaccine strains, we investigated a family of genes encoding MFS transporters as vaccine candidates. MFS transporters are ubiquitous across bacteria and are involved in the transport of a wide variety of substrates, including but not limited to amino acids, carbohydrates, and Krebs cycle metabolites (25). F. tularensis is predicted to encode 31 total MFS transporters. We studied a subset of nine of these genes with homology to the phtA gene of L. pneumophila. phtA is a member of the Pht (phagosomal transporter) subfamily of MFS transporters and was shown previously to be important for the intramacrophage survival of L. pneumophila (35). Including phtA, a total of 12 Pht subfamily MFS transporters have been identified in L. pneumophila (4). Of the other members, phtJ has been defined as a valine transporter, and phtC and phtD are suspected to be involved in nucleoside assimilation (4). Based on their homology to phtA and the other Pht genes in L. pneumophila, we named these nine genes the Fpt (Francisella phagosomal transporter) subfamily. In this study, we created genetic deletions of each member of the Fpt subfamily and characterized their effects on pathogenesis.
Three mutants, the LVS ΔfptB, LVS ΔfptE, and LVS ΔfptG mutants, showed altered replication kinetics in one or more cell types, including macrophages and HepG2 cells. The defect in all cases was a decreased level of replication within the first 24-h period. Two Schu S4 transposon mutations of Fpt genes, FTT_0056c and FTT_0129 (corresponding to fptB and fptG), were previously identified as being defective for replication in HepG2 cells, and FTT_0056c was identified as being defective for replication in J774.1 cells (28), suggesting that the phenotypes which we have observed for LVS are conserved in Schu S4. This is not surprising given the high (98 to 100%) degree of amino acid identity between proteins encoded by LVS and those encoded by Schu S4. There are several possible reasons for the observed lag in intracellular replication, including a metabolic defect in the mutant strains that forces the use of an alternate metabolite or metabolic pathway for an essential cellular function upon reaching the cytosol, resulting in the observed lag in replication. Studies are under way to identify the function of each Fpt protein and its mechanism in the infection process of Francisella.
These three mutant strains were highly attenuated in BALB/c mice following i.p. inoculation, where at least partial survival was observed for mice inoculated with each of the three mutants at doses ranging up to 3.5 × 105 CFU, a dose at least 1,000 times higher than the 100% lethal dose for LVS. Our data demonstrate that the LVS ΔfptG strain was the most attenuated of these three strains, with a 100% survival rate for mice infected with a dose as high as 2.7 × 106 CFU. The levels of attenuation of the other two mutant strains were similar.
The three mutant strains all conferred protection against lethal challenge following a single immunizing dose. Regardless of the immunizing strain or dose, all mice were protected against a challenge dose of 1.2 × 105 CFU of LVS, which is at least 1,000 times higher than the lethal dose. The protection from challenge elicited from these strains after a single dose was also shown to be long lasting, as mice that were challenged 3 months after vaccination were also fully protected. We also observed that vaccination doses as low as 200 to 500 CFU were completely protective against a challenge dose of 5.1 × 103 CFU. This is important to note, since many of the strains previously reported to be attenuated and protective either were not challenged with such a high dose, required a higher vaccination dose, or required multiple vaccinations to achieve the same level of protection (15, 16, 18, 20, 26, 30, 32, 36). There has been one other LVS mutant reported to provide full protection against an LVS challenge of 105 CFU with a vaccination dose of only 103 CFU (37). The ability of the Fpt mutants to confer protection with a small vaccination dose may be related to the fact that they are able to replicate in macrophages, unlike other attenuated strains, such as the ΔguaA, ΔguaB, and ΔpurMCD mutant strains, which are unable to replicate in macrophages.
During infection in mice, F. tularensis colonizes target organs, including the lungs, liver, and spleen. While Fpt mutant strains colonize the lungs, liver, and spleen in mice, they fail to replicate to the same levels as those of parental LVS during the first 3 days after infection. These mutants were almost completely cleared from the organs by day 21 after inoculation and were totally cleared by day 28. This is significant, because even if attenuated and protective, a strain that causes a persistent bacterial burden in target organs which fails to clear over time would likely not be a viable vaccine candidate.
The cytokine responses in the livers of mice infected with the Fpt mutant strains were altered in comparison to that of LVS. We focused on the livers because the cytokine response to Francisella infection is much more muted in the lungs and spleen following i.p. infection (5, 12, 13). Consistent with previous reports, we observed a robust inflammatory response, particularly at 48 and 72 h postinfection in the LVS-infected mice (3, 5, 12, 13, 21, 22). In contrast, the Fpt mutant strains induced significantly less proinflammatory gene expression, and this reduction correlated with lower bacterial burdens and reduced pathology. It was shown previously that a cytokine storm resulting in severe tissue damage is a factor contributing to F. tularensis virulence in mice (3, 21, 22, 38). The reduced induction of gene expression of the proinflammatory cytokines TNF-α, IFN-γ, and iNOS in the livers of mice infected with Fpt mutant strains corresponded to decreased liver pathology compared with that of mice infected with LVS, where more inflammatory foci, apoptosis, and necrosis were observed.
Also of note is the elevation of IFN-β gene expression levels in the livers of mice infected with Fpt mutants compared with the level in mice infected with LVS. Cole et al. previously showed the importance of IFN-β in controlling F. tularensis replication (6). While the inducible levels of IFN-β mRNA did not reach the level of statistical significance, we believe that this finding suggests that this cytokine may contribute to the control of the replication of the Fpt mutant strains. Interestingly, despite the observed elevation of IFN-β gene expression levels in response to Fpt mutant infection at 72 h compared with levels in response to LVS infection, iNOS expression levels at 72 h in response to infection with these mutants were significantly reduced. While LVS-induced IFN-β was previously shown to be required for iNOS expression in macrophages (6), mice that lack the IFN-α/β receptor have been found to be more resistant to infection (17). Thus, the role of IFN-β in F. tularensis infection remains controversial, and its possible relationship to the phenotypes of the attenuated strains will be examined in future studies.
Additional evidence that a reduced proinflammatory response contributes to attenuation and eventual survival was seen in the pathology of mice infected with the LVS ΔfptB mutant. When we examined cytokine expression levels in livers from mice infected with this mutant at day 7 after infection, we observed that two mice with a low bacterial burden which exhibited no signs of illness had levels of IFN-γ and iNOS expression similar to those of mock-infected mice, whereas two mice with a high bacterial burden which exhibited severe signs of illness had levels of IFN-γ and iNOS expression that were dramatically elevated compared to those of mock-infected mice. Tissue pathology such as inflammation, granuloma formation, and necrotic cell death, which correlated with the high bacterial burdens in the two mice, was also observed to be more severe in the two mice with higher bacterial burdens.
During our analysis of the LVS and Schu S4 strains for the presence of the Fpt genes, we discovered that each of the LVS Fpt genes has a virtually identical homolog in Schu S4 (Table 1). We have previously shown that our suicide plasmid system is amenable to the creation of deletions in the Schu S4 strain, and the development of live-attenuated Schu S4 Fpt mutants is under way.
This is the first report to link MFS transporters to pathogenesis and virulence in F. tularensis, making it the second intracellular pathogen shown to rely on MFS transporters for virulence. Three genes in this family, fptB, fptE, and fptG, are critical for normal survival and replication in one or more cell types that are important for Francisella pathogenesis. Furthermore, we have shown that these Fpt mutant strains are severely attenuated for virulence in mice and protective against high-level challenge with parental LVS and that the protection is long lasting. There have been limited reports of live vaccine strains conferring protection against a type A challenge (1, 27). Future studies will determine if these LVS Fpt mutants or the corresponding mutants in the Schu S4 background can protect against a type A challenge. While further investigation of the role of the Fpt transporters in pathogenesis and the mechanism underlying the attenuation and protection of these strains is required, we have shown that members of the Fpt subfamily of MFS transporters are critical virulence factors in F. tularensis and may represent promising targets for the development of a live-attenuated vaccine.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH NIAID U01 AI077909 (E.M.B.) and U54 AI057168 (E.M.B. and S.N.V.; principal investigator, M. Levine).
We acknowledge William Blackwelder and Yukun Wu for performing statistical analyses and Christina Faherty for critical review of the manuscript.
Footnotes
Published ahead of print 16 April 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Bakshi CS, et al. 2008. An improved vaccine for prevention of respiratory tularemia caused by Francisella tularensis SchuS4 strain. Vaccine 26:5276–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bakshi CS, et al. 2006. Superoxide dismutase B gene (sodB)-deficient mutants of Francisella tularensis demonstrate hypersensitivity to oxidative stress and attenuated virulence. J. Bacteriol. 188:6443–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bosio CM, Bielefeldt-Ohmann H, Belisle JT. 2007. Active suppression of the pulmonary immune response by Francisella tularensis Schu4. J. Immunol. 178:4538–4547 [DOI] [PubMed] [Google Scholar]
- 4. Chen DE, Podell S, Sauer JD, Swanson MS, Saier MH., Jr 2008. The phagosomal nutrient transporter (Pht) family. Microbiology 154:42–53 [DOI] [PubMed] [Google Scholar]
- 5. Cole LE, et al. 2006. Immunologic consequences of Francisella tularensis live vaccine strain infection: role of the innate immune response in infection and immunity. J. Immunol. 176:6888–6899 [DOI] [PubMed] [Google Scholar]
- 6. Cole LE, et al. 2008. Macrophage proinflammatory response to Francisella tularensis live vaccine strain requires coordination of multiple signaling pathways. J. Immunol. 180:6885–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cole LE, et al. 2007. Toll-like receptor 2-mediated signaling requirements for Francisella tularensis live vaccine strain infection of murine macrophages. Infect. Immun. 75:4127–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Conlan JW, Chen W, Shen H, Webb A, KuoLee R. 2003. Experimental tularemia in mice challenged by aerosol or intradermally with virulent strains of Francisella tularensis: bacteriologic and histopathologic studies. Microb. Pathog. 34:239–248 [DOI] [PubMed] [Google Scholar]
- 9. Conlan JW, et al. 2010. Differential ability of novel attenuated targeted deletion mutants of Francisella tularensis subspecies tularensis strain SCHU S4 to protect mice against aerosol challenge with virulent bacteria: effects of host background and route of immunization. Vaccine 28:1824–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fortier AH, Slayter MV, Ziemba R, Meltzer MS, Nacy CA. 1991. Live vaccine strain of Francisella tularensis: infection and immunity in mice. Infect. Immun. 59:2922–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gil H, et al. 2006. Deletion of TolC orthologs in Francisella tularensis identifies roles in multidrug resistance and virulence. Proc. Natl. Acad. Sci. U. S. A. 103:12897–12902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Golovliov I, Kuoppa K, Sjostedt A, Tarnvik A, Sandstrom G. 1996. Cytokine expression in the liver of mice infected with a highly virulent strain of Francisella tularensis. FEMS Immunol. Med. Microbiol. 13:239–244 [DOI] [PubMed] [Google Scholar]
- 13. Golovliov I, Sandstrom G, Ericsson M, Sjostedt A, Tarnvik A. 1995. Cytokine expression in the liver during the early phase of murine tularemia. Infect. Immun. 63:534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hornick RB, Eigelsbach HT. 1966. Aerogenic immunization of man with live tularemia vaccine. Bacteriol. Rev. 30:532–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jayakar HR, et al. 2011. A galU mutant of Francisella tularensis is attenuated for virulence in a murine pulmonary model of tularemia. BMC Microbiol. 11:179 doi:10.1186/1471-2180-11-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jia Q, et al. 2010. A Francisella tularensis live vaccine strain (LVS) mutant with a deletion in capB, encoding a putative capsular biosynthesis protein, is significantly more attenuated than LVS yet induces potent protective immunity in mice against F. tularensis challenge. Infect. Immun. 78:4341–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jones JW, Broz P, Monack DM. 2011. Innate immune recognition of Francisella tularensis: activation of type-I interferons and the inflammasome. Front. Microbiol. 2:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim TH, Pinkham JT, Heninger SJ, Chalabaev S, Kasper DL. 2012. Genetic modification of the O-polysaccharide of Francisella tularensis results in an avirulent live attenuated vaccine. J. Infect. Dis. 205:1056–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Law CJ, Maloney PC, Wang DN. 2008. Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 62:289–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li J, et al. 2007. Attenuation and protective efficacy of an O-antigen-deficient mutant of Francisella tularensis LVS. Microbiology 153:3141–3153 [DOI] [PubMed] [Google Scholar]
- 21. Mares CA, Ojeda SS, Morris EG, Li Q, Teale JM. 2008. Initial delay in the immune response to Francisella tularensis is followed by hypercytokinemia characteristic of severe sepsis and correlating with upregulation and release of damage-associated molecular patterns. Infect. Immun. 76:3001–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mares CA, et al. 2010. Attenuated response of aged mice to respiratory Francisella novicida is characterized by reduced cell death and absence of subsequent hypercytokinemia. PLoS One 5:e14088 doi:10.1371/journal.pone.0014088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McCrumb FR. 1961. Aerosol infection of man with Pasteurella tularensis. Bacteriol. Rev. 25:262–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oyston PC, Sjostedt A, Titball RW. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2:967–978 [DOI] [PubMed] [Google Scholar]
- 25. Pao SS, Paulsen IT, Saier MH., Jr 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pechous R, et al. 2006. Construction and characterization of an attenuated purine auxotroph in a Francisella tularensis live vaccine strain. Infect. Immun. 74:4452–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pechous RD, et al. 2008. A Francisella tularensis Schu S4 purine auxotroph is highly attenuated in mice but offers limited protection against homologous intranasal challenge. PLoS One 3:e2487 doi:10.1371/journal.pone.0002487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qin A, Mann BJ. 2006. Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 6:69 doi:10.1186/1471-2180-6-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qin A, Scott DW, Thompson JA, Mann BJ. 2009. Identification of an essential Francisella tularensis subsp. tularensis virulence factor. Infect. Immun. 77:152–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Raynaud C, et al. 2007. Role of the wbt locus of Francisella tularensis in lipopolysaccharide O-antigen biogenesis and pathogenicity. Infect. Immun. 75:536–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ren Q, Kang KH, Paulsen IT. 2004. TransportDB: a relational database of cellular membrane transport systems. Nucleic Acids Res. 32:D284–D288 doi:10.1093/nar/gkh016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Santiago AE, et al. 2009. Characterization of rationally attenuated Francisella tularensis vaccine strains that harbor deletions in the guaA and guaB genes. Vaccine 27:2426–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Santic M, Molmeret M, Klose KE, Jones S, Kwaik YA. 2005. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell. Microbiol. 7:969–979 [DOI] [PubMed] [Google Scholar]
- 34. Saslaw S, Eigelsbach HT, Prior JA, Wilson HE, Carhart S. 1961. Tularemia vaccine study. II. Respiratory challenge. Arch. Intern. Med. 107:702–714 [DOI] [PubMed] [Google Scholar]
- 35. Sauer JD, Bachman MA, Swanson MS. 2005. The phagosomal transporter A couples threonine acquisition to differentiation and replication of Legionella pneumophila in macrophages. Proc. Natl. Acad. Sci. U. S. A. 102:9924–9929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sebastian S, et al. 2007. A defined O-antigen polysaccharide mutant of Francisella tularensis live vaccine strain has attenuated virulence while retaining its protective capacity. Infect. Immun. 75:2591–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sen B, Meeker A, Ramakrishnan G. 2010. The fslE homolog, FTL_0439 (fupA/B), mediates siderophore-dependent iron uptake in Francisella tularensis LVS. Infect. Immun. 78:4276–4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sharma J, Li Q, Mishra BB, Georges MJ, Teale JM. 2009. Vaccination with an attenuated strain of Francisella novicida prevents T-cell depletion and protects mice infected with the wild-type strain from severe sepsis. Infect. Immun. 77:4314–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shen H, et al. 2010. Molecular immune responses to aerosol challenge with Francisella tularensis in mice inoculated with live vaccine candidates of varying efficacy. PLoS One 5:e13349 doi:10.1371/journal.pone.0013349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sjostedt A. 2007. Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann. N. Y. Acad. Sci. 1105:1–29 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.