Abstract
Fks1, with orthologs in nearly all fungi as well as plants and many protists, plays a central role in fungal cell wall formation as the putative catalytic component of β-1,3-glucan synthase. It is also the target for an important new antifungal group, the echinocandins, as evidenced by the localization of resistance-conferring mutations to Fks1 hot spots 1, 2, and 3 (residues 635 to 649, 1354 to 1361, and 690 to 700, respectively). Since Fks1 is an integral membrane protein and echinocandins are cyclic peptides with lipid tails, Fks1 topology is key to understanding its function and interaction with echinocandins. We used hemagglutinin (HA)-Suc2-His4C fusions to C-terminally truncated Saccharomyces cerevisiae Fks1 to experimentally define its topology and site-directed mutagenesis to test function of selected residues. Of the 15 to 18 transmembrane helices predicted in silico for Fks1 from evolutionarily diverse fungi, 13 were experimentally confirmed. The N terminus (residues 1 to 445) is cytosolic and the C terminus (residues 1823 to 1876) external; both are essential to Fks1 function. The cytosolic central domain (residues 715 to 1294) includes newly recognized homology to glycosyltransferases, and residues potentially involved in substrate UDP-glucose binding and catalysis are essential. All three hot spots are external, with hot spot 1 adjacent to and hot spot 3 largely embedded within the outer leaflet of the membrane. This topology suggests a model in which echinocandins interact through their lipid tails with hot spot 3 and through their cyclic peptides with hot spots 1 and 2.
INTRODUCTION
The predominant cell wall component in ascomycetous yeasts such as Saccharomyces cerevisiae and Candida albicans is β-1,3-glucan, and it remains essential even in fungi such as Aspergillus fumigatus and Cryptococcus neoformans, in which other polysaccharides predominate (8, 15, 25, 26, 44). In addition to this structural role, β-1,3-glucan has roles in innate immunity (12) and in fungal diagnostics (35) and as biofilm matrix (32). Moreover, β-1,3-glucan synthesis has emerged as a critical therapeutic target. Until recently, the antifungal agents available for treating invasive candidiasis or aspergillosis were limited to amphotericin B and azoles such as fluconazole. Amphotericin B is fungicidal but toxic; azoles are better tolerated, but their activity is fungistatic (6). Furthermore, resistance represents a serious limitation to azole use, particularly in Candida glabrata (37). Thus, the introduction of β-1,3-glucan synthesis-inhibiting echinocandins, beginning with caspofungin in 2001 followed by micafungin and anidulafungin, represented an important clinical development (6, 7). Echinocandins are well tolerated and exhibit potent fungicidal activity against most Candida species, including those that are azole resistant. Their activity against Aspergillus species is similarly potent, although fungistatic. For reasons that remain unclear, echinocandins exhibit little or no activity against other pathogenic fungi, including C. neoformans (8, 36, 44).
Echinocandins noncompetitively inhibit incorporation of substrate UDP-glucose into β-1,3-glucan (9). Since the substrate is cytosolic and the product is external, this glycosyltransferase reaction is topologically complex and, predictably, involves a membrane protein complex. Although it has resisted full purification, this complex appears to be limited to a small number of proteins, including Fks1, a large (ca. 1,900-residue) integral membrane protein and putative catalytic component, and Rho1, a small GTPase and putative regulatory component (9, 11, 28, 36, 38). C. albicans and related yeasts, including S. cerevisiae, encode Fks2 and Fks3 paralogs of Fks1; however, with few exceptions, these are minimally expressed under normal growth conditions (28, 30), and most molds, including Aspergillus species, encode only a single Fks1 ortholog. In all fungi examined, including intrinsically resistant C. neoformans, gene disruption studies have demonstrated that Fks1 (or Fks2 in fks1Δ strains of S. cerevisiae) is essential to viability (10, 15, 28, 30, 36, 44). In addition to enrichment of Fks1 in partially purified β-1,3-glucan synthase preparations, evidence for their equivalency derives from the fact that mutations conferring high-level echinocandin resistance map exclusively to this protein, specifically to a region known as hot spot 1 (residues 635 to 649 in S. cerevisiae Fks1) or, less commonly, hot spot 2 (residues 1354 to 1361) or the recently described hot spot 3 (residues 690 to 700) (9, 18, 36). Moreover, specific mutations within these hot spots confer differential echinocandin resistance (e.g., 16-fold-decreased caspofungin but unchanged micafungin susceptibilities), consistent with direct interaction between echinocandin and Fks1 (18).
Beyond this, however, our understanding of Fks1 structure, its role in β-1,3-glucan synthesis, and the mechanism by which echinocandins inhibit this activity is very limited. An initial attempt to directly map the echinocandin binding site on Candida albicans Fks1 by cross-linking with an azido derivative was unsuccessful (13, 39). A similar attempt to map the binding site for the substrate UDP-glucose using partially purified β-1,3-glucan synthase from Neurospora crassa also suffered from low specificity; however, among the cross-linked peptides were four clustered between residues equivalent to 1070 to 1273 in S. cerevisiae Fks1 (41). More recently, the characterization of temperature-sensitive S. cerevisiae mutants implicated specific Fks1 residues in β-1,3-glucan synthase activity, as well as other phenotypes, including polarized growth and endocytosis (33).
Here we explore one important aspect of Fks1 structure: its membrane topology. Initial in silico hydrophobicity analysis of the Fks1 sequence led to a model incorporating 16 transmembrane helices (TMH) divided into two clusters flanking a central region believed to represent the catalytic domain (11). While many of the predicted TMH appeared to be robust, others were questionable, and for all, their orientations (cytosolic versus external) were ambiguous. Of most concern with respect to understanding echinocandin action, the hot spots have inconsistent topologies in this initial model, localized either internally (hot spot 1) or externally (hot spots 2 and 3). This is difficult to reconcile with the apparent dominant effect of individual hot spot 1, 2, or 3 resistance-conferring mutations, which is most readily explained by a model in which these hot spots come together to form a single echinocandin binding site. It is similarly difficult to reconcile an internal hot spot with echinocandin structure: cyclic peptide coupled to a lipid tail essential to activity (2, 7). Analogous to the lipopeptide antibiotic daptomycin (1), the lipid tails most likely target echinocandins to the outer leaflet of the membrane.
A widely used approach to experimentally analyze membrane protein topology in S. cerevisiae involves construction of C-terminal fusions to hemagglutinin (HA)-Suc2-His4C, followed by complementary assays of His4C expression (requiring localization of that C terminus to the cytosol) and Suc2 glycosylation (requiring localization to the endoplasmic reticulum and subsequently to the outer leaflet in the case of plasma membrane proteins) (21, 42). This method has been used for topological analysis of numerous membrane proteins, including Pmt1 (43), Stt3 (22), Nce102 (27), and Arv1 (47). We combined this method with in silico analyses to generate a model of S. cerevisiae Fks1 topology. In this model, all three hot spots are external, adjacent to the membrane or partially embedded. The central domain, with newly identified homology to structurally defined glycosyltransferases, is cytosolic.
MATERIALS AND METHODS
Strains, media, and reagents.
S. cerevisiae strain STY50 (MATa his4-401 leu2-3,112 trp1-1 ura3-52 HOL1-1 suc2::LEU2) and plasmid pJK90 were obtained from J. Nickels (47). S. cerevisiae BY4742 fks1::KANMX was obtained from Open Biosystems (Huntsville, AL). Yeasts were cultured on YPD medium (1% yeast extract, 2% peptone, 2% dextrose) or, where indicated, synthetic defined SD-ura (DOB plus CSM-ura; Sunrise Science Products, San Diego, CA) or SDh-ura-his (DOB plus CSM-ura-his supplemented with 2 mM histidinol; Sunrise Science Products and Santa Cruz Biotechnology, Santa Cruz, CA). DNA primers (see Table S1 in the supplemental material) were obtained from IDT (Coralville, IA) and FK506 from Tecoland (Edison, NJ).
Protein sequences.
Fks1 sequences from Saccharomyces cerevisiae (NP_013446), Candida albicans (XP_721429), Aspergillus fumigatus (XP_751118), Cryptococcus neoformans (O93927), Arabidopsis thaliana (NM_123045), Phytophthora infestans (XM_002998508), Phaeodactylum tricornutum (XM_002177407), Chlamydomonas reinhardtii (XM_001691217), and Toxoplasma gondii (XM_002370473) were obtained from GenBank. Fks1 sequences from Rhizopus oryzae (RO3G_10349) and Allomyces macrogynus (AMAG_00576) were obtained from the Broad Institute Fungal Genome Initiative.
Construction of plasmids for topology analysis.
All constructs were generated by gap repair homologous recombination. PCR products were amplified with Phusion high-fidelity DNA polymerase (New England BioLabs, Ipswich, MA) as recommended by the manufacturer using S. cerevisiae genomic DNA as the template, with forward primer ScFks1-1TAGF and various reverse primers (e.g., ScFks1-300 TAGR) fusing Fks1 C termini as indicated below to the HA-Suc2-His4C tag. Both primers incorporated 33 bases at their 5′ ends complementary to pJK90 sequences flanking its unique SmaI restriction site. These products along with pJK90 that was SmaI linearized and alkaline phosphatase treated (New England Biolabs) were cotransformed into competent S. cerevisiae STY50 cells using the EZ Yeast Transformation II kit (Zymo Research, Irvine, CA). Transformants were selected on SD-ura. To confirm desired plasmid recombination, yeast lysates (19) were screened by PCR with Taq polymerase (New England BioLabs) and vector primer pJK90F or pJK90R (see Table S1 in the supplemental material) in conjunction with various FKS1 coding region primers (data not shown).
Plasmids were subsequently rescued, purified, sequenced with appropriate primers, and retransformed into STY50 before growth assays and protein analysis. For this, overnight SD-ura cultures (4 ml) were pelleted, resuspended in 2 ml SCE buffer (1 M sorbitol, 0.1 M sodium acetate, 60 mM EDTA [pH 8.0]) supplemented with 2.5 mg Zymolyase (ICN Biochemical, Aurora, OH) and 15 μl β-mercaptoethanol, and incubated at 37°C for 1 h. The resulting yeast spheroplasts were pelleted and processed using a plasmid miniprep kit (Denville Scientific, Metuchen, NJ). Plasmid products were transformed into Escherichia coli DH5α with selection on Luria broth agar containing 20 μg/ml ampicillin, purified from overnight cultures, and retransformed into STY50.
Growth assay of His4C expression.
Approximately 1,000 cells in 3 μl sterile water were replica plated on SD-ura and SDh-ura-his. Plates were incubated at 30°C for 72 h before being photographed.
Western blot assay of Suc2 glycosylation.
Cells from 5-ml SD-ura overnight cultures were pelleted, washed twice in 5 ml H2O, and resuspended in 150 μl sample buffer (10 mM Tris-HCl [pH 7.4], 140 mM NaCl) supplemented with 5% Triton X-114. Cells were mechanically lysed by addition of 100 μl cold glass beads and by vortexing 4 times in 2-min cycles at 4°C. Lysates were incubated on ice for 2 h and then centrifuged at 100 × g for 5 min at 4°C. Supernatants were collected and layered on top of 0.5 volume of 6% sucrose in sample buffer. These were incubated at 34°C for 20 min, followed by centrifugation at 2,000 × g in a swinging bucket rotor. The resulting crude membrane pellet was resuspended in 75 μl sample buffer. Aliquots of 18 μl each were supplemented with potassium acetate (pH 5.8) to a final concentration of 80 mM, followed by addition of 1 μl endoglycosidase H (Endo H; Roche, Indianapolis, IN) for treated samples or 1 μl H2O for mock controls (21 μl total volume), and incubated at 37°C for 1 h. Paired Endo H-treated and untreated samples were electrophoresed through a 5% SDS-polyacrylamide gel for 2 h, followed by overnight transfer to a nitrocellulose membrane. Membranes were blocked in 5% skim milk-TBST (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.1% Tween 20) and probed with a horseradish peroxidase-conjugated antihemagglutinin antibody (1:20,000 dilution; Santa Cruz Biotechnology).
Construction of plasmids for functional analysis.
For site-directed mutagenesis, gap repair with two partially overlapping PCR products was employed. One product was generated using ScFks1-1TAGF and a reverse primer encoding the desired mutation (e.g., ScFks1-D1102AR), and the second was generated using ScFks1-1876*TAGR (incorporating a stop codon before the tag) and a forward primer typically encoding the same mutation (e.g., ScFks1-D1102AF; for the R319A mutation, only the reverse primer encoded the mutation). Both products were cotransformed with SmaI-linearized and alkaline phosphatase-treated pJK90 into competent BY4742 fks1::KANMX with selection on SD-ura. PCR screens, plasmid rescue, purification, sequencing, and retransformation were performed as described above.
To construct C-terminal truncations, PCR products were generated with ScFks1-1TAGF and reverse primers (e.g., ScFks1-1830*TAGR) that incorporate a stop codon following the indicated Fks1 residue from 1830 to 1876. Products were cotransformed with SmaI-linearized and alkaline phosphatase-treated pJK90 into competent BY4742 fks1::KANMX, with selection and processing as described above.
To test the function of mutated or truncated Fks1, ∼1,000 cells were spotted on SD-ura with or without FK506 (1 μg/ml). Plates were incubated at 30°C for 72 h before being photographed.
Computational methods.
In silico TMH predictions were generated using TMHMM (http://www.cbs.dtu.dk/services/TMHMM) (23) and PRO-TMHMM (http://topcons.cbr.su.se) (46). Sequences were aligned using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2). Homology detection and structure prediction were performed using HHpred (http://hhpred.tuebingen.mpg.de/hhpred) (17).
RESULTS
In silico modeling of Fks1 topology.
Significant advances in transmembrane helix (TMH) prediction have been made since the original S. cerevisiae Fks1 model was proposed in 1994 (5, 46). Also, many additional Fks1 sequences are now available from evolutionarily diverse fungi, including representatives of all 4 fungal phyla, e.g., the ascomycetous yeast C. albicans and mold A. fumigatus, basidiomycete C. neoformans, zygomycete Rhizopus oryzae, and chytrid Allomyces macrogynus. Alignment of these sequences using ClustalW2 (see Fig. S1 in the supplemental material) confirms previous observations (9) that conservation is concentrated in the central domain (S. cerevisiae Fks1 residues ∼715 to 1300), with 43% identity among the 6 diverse species. In contrast, the N- and C-terminal domains (residues ∼1 to 715 and ∼1300 to 1876) are relatively divergent, with 13% and 11% identities, respectively.
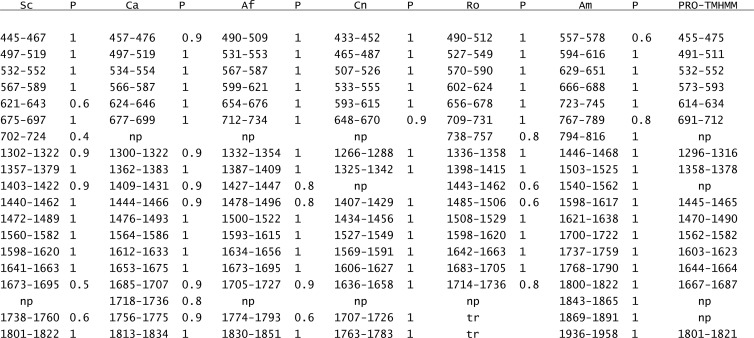
TMHMM is the most widely used and validated single sequence TMH prediction algorithm (5, 23, 29, 31). For S. cerevisiae Fks1, TMHMM predicts up to 18 TMH (Fig. 1; see also Fig. S2 in the supplemental material), with probabilities ranging from high (1.0) to low (0.4 to 0.6). Seven of these TMH localize to the N-terminal domain between residues 445 to 724, and 11 localize in the C-terminal domain between 1302 to 1822. As an initial test of these predictions, Fks1 sequences from the 5 additional diverse fungi listed above were similarly analyzed by TMHMM (Fig. S2). Predicted TMH locations and their probabilities are listed in Fig. 1. Comparison of these locations (allowing for minor shifts) with the ClustalW2 alignment (Fig. S1) indicated that 16 of 18 TMH predicted for S. cerevisiae Fks1 are similarly predicted for the 5 other fungal Fks1. The exceptions involve the final N-terminal domain TMH and the third C-terminal domain TMH (TMH 702–724 and 1403–1422, respectively) (Fig. 1).
Fig 1.
TMHMM-predicted TMH locations (residue number) and probabilities (P) for Fks1 from S. cerevisiae (Sc), C. albicans (Ca), A. fumigatus (Af), C. neoformans (Cn), R. oryzae (Ro), and A. macrogynus (Am). The PRO-TMHMM prediction was generated using S. cerevisiae Fks1 as a query. TMH on the same line are considered to be equivalent based on overlap in the Fks1 ClustalW2 alignment (see Fig. S1 in the supplemental material). np, not predicted; tr, truncated C terminus.
To complement the single sequence-based TMHMM, Fks1 topology predictions were also generated using PRO-TMHMM (46), using S. cerevisiae Fks1 as a query. Analogous to our approach above, PRO-TMHMM incorporates evolutionary analysis, but it does so more comprehensively by generating a consensus sequence. A total of 15 TMH were predicted: 6 in the N-terminal domain between residues 445 and 712 and 9 in the C-terminal domain between 1296 and 1821 (Fig. 1). The PRO-TMHMM predictions coincide with those described above, with the following exceptions: N-terminal domain TMH 675–697 and 702–724 predicted by TMHMM are replaced by TMH 691–712, and C-terminal domain TMH 1402–1422 and 1738–1760 predicted by TMHMM are not predicted by PRO-TMHMM.
Topology of Fks1 N-terminal domain.
To directly test these in silico predictions, we employed Fks1 fragment C-terminal fusions to the HA-Suc2-Hic4C topology reporter (21). The C termini were selected to fall within loop regions before or after predicted TMH (Fig. 1). Constructs were generated by gap repair between FKS1 PCR products and SmaI- and alkaline phosphatase-treated plasmid vector pJK90 which were cotransformed into S. cerevisiae STY50 with selection on SD-ura medium. In this his4 strain, cytosolic localization of the C-terminal fusion permits His4C-mediated growth on SDh-ura-his medium (SD-ura-his medium supplemented with histidinol). Alternatively, C-terminal fusions destined for external localization undergo N-linked glycosylation within the endoplasmic reticulum on the Suc2 moiety; this is detected by a band shift following endoglycosidase H (Endo H) treatment in a Western blot with anti-HA antibody. Together, these complementary assays provide a robust system for topological analysis.
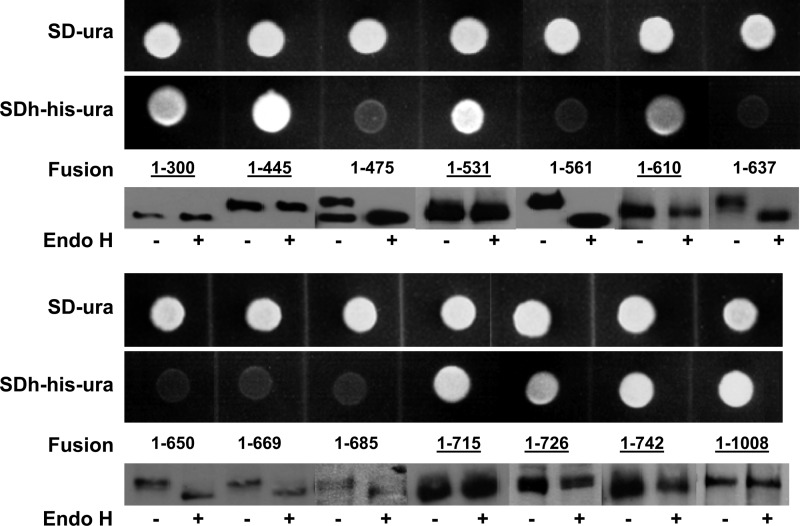
S. cerevisiae STY50 expressing Fks1 residues 1 to 300 fused at its C terminus to HA-Suc2-Hic4C (designated Fks1-300 fusion) demonstrated robust growth on SDh-ura-his medium and no detectable band shift upon Endo H digestion, both indicating cytosolic localization (Fig. 2). This was also predicted by TMHMM (see Fig. S2 in the supplemental material), although the localization predictions provided by these algorithms are less reliable than their TMH predictions, as evidenced by external predictions for the N termini of other Fks1 proteins (Fig. S2). For an Fks1-445 fusion, a similar cytosolic localization was determined (Fig. 2), confirming the predicted lack of TMH within this N-terminal region. In contrast, an Fks1-475 fusion grew minimally on SDh-ura-his and on a Western blot exhibited a clear Endo H-mediated band shift, indicating external localization (Fig. 2). This supports formation of predicted TMH 445–467 (Fig. 1).
Fig 2.
His4C expression and Suc2 glycosylation assays of Fks1 N-terminal and central domain fusions. His4C expression was assayed by comparing growth on SD-ura and SDh-ura-his media. N-linked glycosylation was assayed by Western blot detection of HA-tagged fusions either treated (+) or untreated (−) with Endo H; band shift indicates glycosylation. Fusions localized to the cytosol are underlined.
The growth and band shift assays similarly indicated cytosolic, external, and cytosolic localizations, respectively, for Fks1-531, Fks1-561, and Fks1-610 fusions (Fig. 2). These results support predicted TMH 497–519, 532–552, and 567–589, respectively (Fig. 1).
Importantly, with respect to the region representing echinocandin resistance hot spot 1 (residues 635 to 649), both growth and band shift assays of the Fks1-650 and Fks1-669 fusions indicate external localization (Fig. 2). TMH 621–643, responsible for this localization, was predicted with only low probability in S. cerevisiae Fks1 but with high probability in all other fungi (Fig. 1). Unexpectedly, an Fks1-637 fusion also showed external localization (Fig. 2), suggesting that the actual TMH may be shifted upstream of the predicted TMH. This alignment is supported by the alternative algorithm PRO-TMHMM, which predicts a TMH from residues 614 to 634 (Fig. 1).
The cytosolic localization of the Fks1-715 fusion (Fig. 2) provides support for either TMH 675–697 (predicted by TMHMM) or TMH 692–712 (predicted by PRO-TMHMM). To resolve this, an additional Fks1-685 fusion was constructed; its external localization supports TMH 692–712. Thus, as with hot spot 1, hot spot 3 (residues 690 to 700) is externally located, albeit largely membrane embedded. The final, inconsistently predicted TMH 702–724 of the N-terminal domain (Fig. 1) was not supported, since an Fks1-726 fusion remained in the cytosol (Fig. 2).
Topology of Fks1 central domain.
The Fks1-715 and Fks1-726 fusions described above tentatively localized the central domain to the cytosol. This was confirmed with the Fks1-742 and Fks1-1008 fusions, which similarly exhibited robust growth on SDh-ura-his and no band shift following Endo H treatment (Fig. 2).
For Fks1 regions beyond residue 1008, Western blot analysis using fusions initiated at residue 1 was complicated by inadequate resolution of Endo H band shift due to the large size of the resulting protein. Consequently, additional fusions were constructed that initiated at Met residue 1204, located ∼100 residues upstream of the first predicted TMH of the C-terminal domain. Growth on SDh-ura-his, however, was determined for fusions initiated at both residues 1 and 1204.
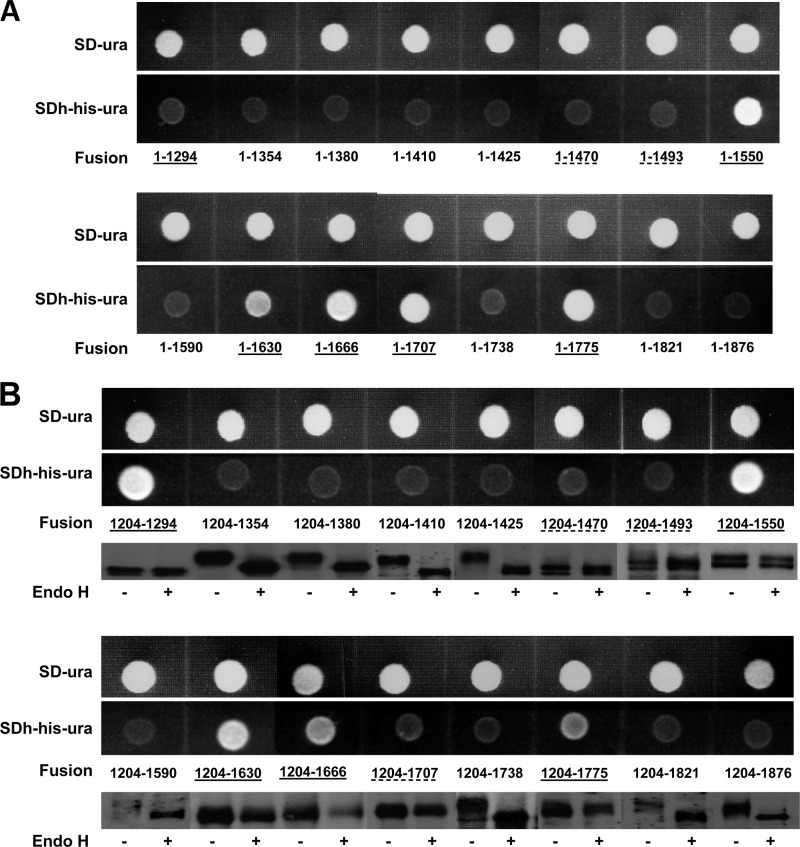
As expected, an Fks1204–1294 fusion (i.e., initiated at residue 1204 and fused at 1294 to HA-Suc2-His4C) was cytosolically localized as indicated by growth on SDh-ura-his and lack of band shift (Fig. 3), consistent with its inclusion in the central domain. Conversely, an Fks1204–1354 fusion was external (Fig. 3), in support of predicted TMH 1302–1322 (Fig. 1). Combined with fusion Fks1-715, these results define the limits of the cytosolic central domain as residues 715 to 1294.
Fig 3.
(A) His4C expression of Fks1 central and C-terminal domain fusions initiated at amino acid 1; (B) His4C expression and Suc2 glycosylation assays of Fks1 central and C-terminal domain fusions initiated at amino acid 1204. See the legend to Fig. 2 for details. Ambiguous results (lack of band shift but weak growth on SDh-ura-his) are indicated with a dashed underline.
Topology of Fks1 C-terminal domain.
As with Fks1204–1354 above, an Fks1204–1380 fusion was similarly external (Fig. 3). This was unexpected, since TMH 1357–1379 was consistently predicted (Fig. 1), but was confirmed by an Fks1204–1410 fusion which also indicated external location (Fig. 3). Thus, these results demonstrate an external location for echinocandin resistance hot spot 2 (residues 1354 to 1361).
TMH 1403–1422 was also not supported, since an Fks1204–1425 fusion remained external as indicated by both absence of growth on SDh-ura-his and band shift following Endo H treatment (Fig. 3). Indeed, cytosolic localization appears to require extension to residue 1470. This was demonstrated by an Fks1204–1470 fusion which exhibited a clear lack of band shift, although growth on SDh-ura-his was minimal for unclear reasons (Fig. 3). The same pattern was observed with an Fks1204–1493 fusion; however, an Fks1204–1550 fusion provided both strong growth on SDh-ura-his and lack of band shift indicating cytosolic localization (Fig. 3). We interpret these data as supporting TMH 1440–1462 or, in its absence, TMH 1472–1489.
Subsequent TMH 1560–1582 and 1598–1620 are supported by both growth and band shift assays with Fks1204–1590 (external) and Fks1204–1630 (cytosol) fusions (Fig. 3); both are predicted with high probability in diverse fungi (Fig. 1). Predicted TMH 1641–1663, however, was not supported, since an Fks1204–1666 fusion remained cytosolic. This was confirmed by the band shift and growth assays for the Fks1204–1707 and Fks1-1707 fusions, respectively; the growth assay for the Fks1204–1707 fusion was negative for unclear reasons (Fig. 3).
Interestingly, a TMH predicted only in C. albicans involving residues 1718 to 1736, equivalent to S. cerevisiae residues 1706 to 1724 (see Fig. S1 in the supplemental material), appears to be supported by the external location of an Fks1204–1738 fusion. The final two predicted TMH, TMH 1738–1760 and 1801–1822, are supported by fusions Fks1204–1775 and Fks1204–1821, respectively (Fig. 3). Thus, the C terminus of S. cerevisiae Fks1 is external, as confirmed by fusion Fks1204–1876 (Fig. 3).
Functional analysis of selected N- and C-terminal residues.
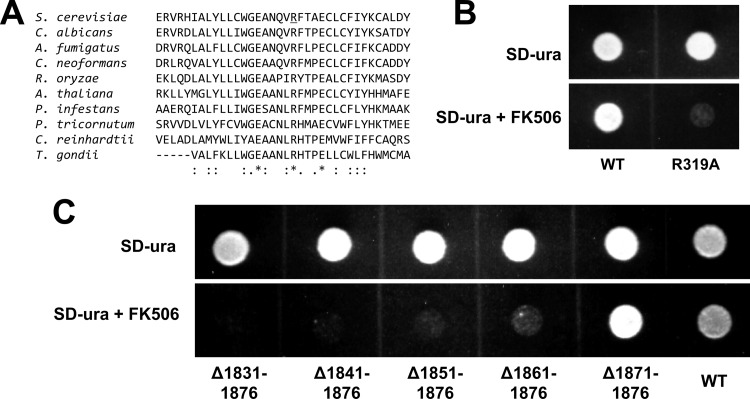
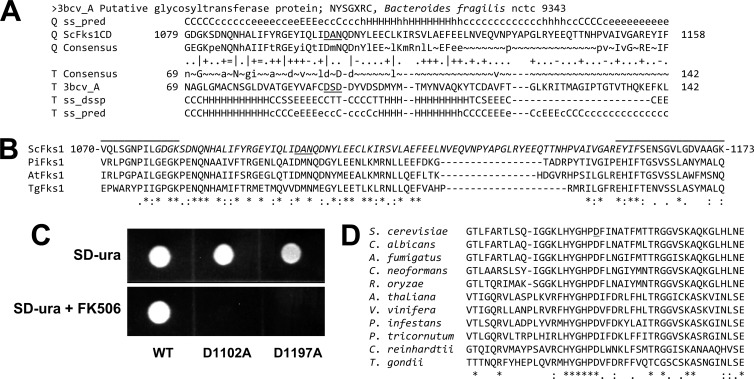
As demonstrated above, both the N-terminal (residues 1 to 445) and central domain regions of Fks1 are cytosolic; however, unlike the latter, the former is evolutionarily divergent, with the exception of an ∼25-residue segment which is conserved in Fks1 homologs from species representing three kingdoms: fungi, plants, and protozoa (Fig. 4A). In particular, several residues are universally conserved, implying functional importance. To test this, we mutated one of these, R319, to A and examined the effect on Fks1 function. This was accomplished by plasmid-based expression of the mutated gene in an fks1::KANMX strain and then blocking of expression of the “backup” gene FKS2 with calcineurin inhibitor FK506. Cells expressing wild-type Fks1-R319 were unaffected by FK506, while those expressing Fks1-R319A exhibited clearly reduced growth in the presence of FK506 (Fig. 4B).
Fig 4.
Mutational analysis of a conserved residue within the Fks1 cytosolic N-terminal domain and truncation analysis of the external C terminus. (A) ClustalW2 alignment of S. cerevisiae Fks1 residues 300 to 335 and homologous sequences from divergent species: ascomycetes S. cerevisiae, C. albicans, and A. fumigatus; basidiomycete C. neoformans; zygomycete R. oryzae; plant Arabidopsis thaliana; oomycete Phytophthora infestans; diatom Phaeodactylum tricornutum; green alga Chlamydomonas reinhardtii; and apicomplexan Toxoplasma gondii. Asterisk, conserved in all sequences; colon, highly conservative substitution; period, less conservative substitution. (B) Functional analysis of plasmid-expressed wild-type (WT) versus R319A-mutated Fks1. Function was tested by comparing growth in an fks1::KANMX strain background with and without calcineurin inhibitor FK506 (1 μg/ml), which blocks FKS2 expression. (C) Functional analysis of full-length versus C-terminally truncated Fks1; see description of panel B for details.
The external C terminus of S. cerevisiae Fks1 (residues 1823 to 1876) is enriched in N, S, and T residues (13 within the final 30 residues; see Fig. S1 in the supplemental material) that could serve as glycosylation sites. This is true as well for the C termini of C. albicans and A. fumigatus Fks1, although their sequences are otherwise divergent (Fig. S1). To test the functional importance of the C terminus, truncations were constructed. In the fks1::KANMX strain background, plasmids expressing wild-type Fks1 or an Fks1Δ1871–1876 truncation were unaffected by FK506, indicating full function (Fig. 4C). However, an Fks1Δ1861–1876 truncation led to reduced growth in the presence of FK506, and further truncation eliminated growth (Fig. 4C).
In silico and functional analyses of the putative catalytic central domain.
On the primary sequence level, homology between the Fks1 cytosolic central domain (defined by the topological analysis described above as residues 715 to 1294) and known glycosyltransferases cannot be discerned by conventional search tools such as BLASTP (data not shown). As a more sensitive alternative, HHpred employs profile-hidden Markov model searches at the primary and secondary structure levels to identify distant homologs to proteins with experimentally defined structures (17). Indeed, using the central domain as a query, the top 2 matches returned by HHpred are glycosyltransferases that employ UDP-sugar substrates, i.e., have the same basic catalytic activity (and presumably catalytic mechanism) as β-1,3-glucan synthase. With all other regions of Fks1 as queries, matches were minimal and random (data not shown). The central domain match to glycosyltransferases encompasses two sections of Fks1, residues 825 to 881 and 1079 to 1158 (Fig. 5A). Related glycosyltransferase matches were similarly identified by HHpred using Fks1 central domain regions from other fungi, as well as from distantly related orthologs found in plants and oomycetes (data not shown). The glycosyltransferase region homologous to S. cerevisiae Fks1 residues 1079 to 1158 includes the DXD motif which coordinates Mn2+ interaction with the diphosphate of UDP-glucose substrate, and it is catalytically essential (3, 14). In Fks1, the corresponding sequence is DAN (residues 1102 to 1104 [Fig. 5B; see also Fig. S1 in the supplemental material]). To directly test its role in Fks1 function, a D1102A mutation was generated using the plasmid-based approach described above, with expression in the fks1::KANMX strain. This mutation led to a modest decrease in growth rate relative to the wild-type R319 control and to a complete loss of function in the presence of FK506 (Fig. 5C).
Fig 5.
In silico and mutational analyses of Fks1 central domain. (A) Representative example of HHpred output using the S. cerevisiae Fks1 central domain as a query, showing apparent homology to a structurally defined bacterial glycosyltransferase. The DXD motif which coordinates Mn2+ interaction with UDP-glucose is underlined. (B) Alignment of Fks1 central domain regions from S. cerevisiae (Sc), the oomycete Phytophthora infestans (Pi), the plant Arabidopsis thaliana (At), and the protozoan Toxoplasma gondii (Tg). Sequences corresponding to the Neurospora crassa Fks1 peptides cross-linked to azido-UDP-glucose are overlined. Glycosyltransferase homology region within ScFks1 is in italics; residues that align with the DXD motif are underlined. (C) Functional analysis of Fks1 mutants D1102A and D1197A; see the legend to Fig. 4B for details. (D) ClustalW2 alignment of putative Fks1 catalytic regions equivalent to S. cerevisiae residues 1177 to 1221 from divergent species representing fungi, plants, and protists. Mutated S. cerevisiae residue D1197 is underlined.
Fks1 belongs to the GT-A superfamily of glycosyltransferases (20) in which the catalytic residue is a D (less commonly E) located ∼100 residues downstream of the substrate-binding DXD motif (4, 40). ClustalW2 alignment of Fks1 orthologs from highly divergent species identified a conserved stretch of six residues from 1192 to 1197 terminating in D (Fig. 5D). Generation of a D1197A mutation at this putative catalytic residue completely blocked Fks1 function, as indicated by a loss of growth on FK506 medium (Fig. 5C).
DISCUSSION
By combining analysis of sequences from evolutionarily diverse fungi with HA-Suc2-His4C fusion technology in S. cerevisiae, we have defined the membrane topology of Fks1, the putative catalytic component of β-1,3-glucan synthase. This topology (Fig. 6) includes 6 TMH in the N-terminal domain, 7 TMH in the C-terminal domain, a cytosolic N terminus, a cytosolic central domain, and an external C terminus. All 6 TMH consistently predicted for the N-terminal domain were experimentally confirmed, as were 7 of the predicted TMH for the C-terminal domain. On the other hand, the remaining 4 TMH predicted for the C-terminal domain were not confirmed. This emphasizes the importance of experimental verification; similar discrepancies between in silico and experimental topology results have been reported by others (22, 27, 43, 47).
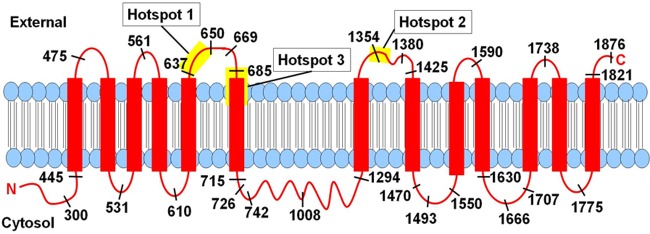
Fig 6.
Model of S. cerevisiae Fks1 membrane topology (red bars, TMH) and correlation with echinocandin resistance hot spots (yellow bars). Drawing is not to scale and does not attempt to show how the three hot spots might come together to form a single echinocandin binding pocket. Numbers represent Fks1 residue locations of C-terminal HA-Suc2-His4C fusions tested in this study (fusion 1410 omitted due to space constraints).
Fks1 has emerged as an important target for antifungals, notably the clinically approved echinocandins (24), as well as investigational agents, including the enfumafungins (16, 34) and recently described piperazinyl-pyridazinone derivatives (48). Since numerous mutations conferring resistance (or in some cases hypersensitivity) to echinocandins have been mapped to three distinct hot spots within Fks1 (residues 635 to 649, 690 to 700, and 1354 to 1361), we reasoned that correlating these mutation sites with Fks1 topology would shed light on the mechanism behind this resistance. Furthermore, since some mutations confer differential resistance (e.g., to micafungin but not caspofungin), implying a direct interaction of an echinocandin-specific side chain with the mutated residue (18), Fks1 topology should also shed light on the echinocandin mechanism of action.
In this regard, a major finding of this work is the observation that all three hot spots are externally located (Fig. 6). This is reassuringly consistent with the echinocandin structure, which includes a common cyclic hexapeptide and a lipid tail essential to activity (2, 7). Specifically, we propose that the lipid tail anchors the drug in the outer leaflet, where it interacts with membrane-embedded regions of Fks1 within hot spot 3 or immediately upstream of hot spot 1. (With regard to hot spot 2, predicted TMH 1357–1379, which would have anchored hot spot 2 in the membrane, was not experimentally supported; however, it is quite possible that these residues associate with the membrane without spanning it.) This positions the peptide moiety for interaction with the externally located hot spot residues. We further speculate that the three hot spots are juxtaposed in the three-dimensional structure of Fks1 to form a single echinocandin binding pocket. While there is no direct evidence for this, it is consistent with the apparent dominant effect (i.e., high-level resistance) of specific mutations at any one hot spot.
A second important finding is the cytosolic location of the conserved central domain (residues 715 to 1294), consistent with its presumed role as the catalytic core of β-1,3-glucan synthase. Until recently, evidence in support of this role has been limited. Four Neurospora crassa Fks1 peptides mapping to this domain, equivalent to S. cerevisiae Fks1 residues 1070 to 1082, 1156 to 2273, 1249 to 1261, and 1262 to 1273, were cross-linked in vitro to an azido derivative of UDP-glucose, the substrate for β-1,3-glucan synthase (41). However, specificity in this cross-linking study was apparently limited, since an additional peptide mapped to the equivalent of residues 650 to 665, demonstrated above to be external, and other peptides mapped to proteins not known to bind UDP-glucose. By characterizing in vitro glucan synthase activity of S. cerevisiae temperature-sensitive Fks1 mutants, Okada et al. (33) implicated multiple central domain residues between 853 and 1047 in catalysis. Most recently, two mutations conferring resistance to a novel, nonechinocandin inhibitor of β-1,3-glucan synthase have been mapped to residues 1175 and 1297 (48).
In light of this mounting evidence, we were encouraged to reexamine the cytosolic central domain for homology to structurally defined glycosyltransferases using HHpred. The most significant Fks1 region identified in this analysis, residues 1079 to 1158, overlaps with sequences corresponding to two of the UDP-glucose cross-linked peptides and includes a variant of the DXD motif involved in binding UDP-glucose substrate in known glycosyltransferases. Mutation of the D1102 residue within this motif abolished Fks1 function. Mutation of a candidate catalytic site residue, D1197, identified on the basis of its location within a highly conserved region ∼100 residues downstream of the DXD motif similarly abolished Fks1 function. Needless to say, this initial mutational analysis is far from exhaustive, but it does lead to hypotheses regarding residues involved in Fks1 catalysis that can be more definitively tested in future studies.
Two additional implications of this work relate to the Fks1 N terminus (residues 1 to 445) and C terminus (1823 to 1876). Although the N terminus is minimally conserved, it has one or more essential functions, as indicated by the effects of R319A mutation described here and the temperature-sensitive mutations (residues 146, 302, 329, and 335) described previously (33). Various roles can be envisioned for the N terminus that are consistent with its cytosolic localization as determined here; for example, it may interact with Rho1. With respect to the C terminus, four different Fks1 fusions (Fks1-1821, Fks1-1876, Fks1204-1821, and Fks1204-1876) demonstrated clear external phenotypes, consistent with previously reported results for Fks2 using the same methodology (15). (This appears to contradict the results obtained with a strain expressing Fks1 with C-terminally fused green fluorescent protein [GFP] [45]; however, the fluorescence associated with GFP fusions to other integral membrane proteins with external C termini such as Stt3 [22; http://yeastgfp.yeastgenome.org] implies that this is not a reliable marker of cytosolic versus external topology.) As with the N terminus, the C terminus has an essential role as indicated by truncation analysis. Within ascomycetes, at least, this C-terminal region is enriched in N, S, and T residues (see Fig. S1 in the supplemental material), which may serve as N- or O-linked glycosylation sites required, for example, for appropriate Fks1 trafficking to the membrane.
In conclusion, the model for Fks1 membrane topology presented here forms a framework for future studies of Fks1 structure-activity relationships. These studies will include detailed analysis of the central domain, since the evidence for its role as the catalytic component of β-1,3-glucan synthase is now compelling. Also, further modeling of the interaction between echinocandin, Fks1, and the plasma membrane can now be pursued, with the goal of designing expanded-spectrum agents with improved antifungal activity.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Villasmil, J. Nickels, T. Daly, and J. Alaro for providing reagents and invaluable advice.
This work was supported by National Institute of Allergy and Infectious Disease grant AI075272.
Footnotes
Published ahead of print 11 May 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Baltz R. 2009. Daptomycin: mechanisms of action and resistance, and biosynthetic engineering. Curr. Opin. Chem. Biol. 13:144–151 [DOI] [PubMed] [Google Scholar]
- 2. Boeck LD, Fukuda DS, Abbott BJ, Debono M. 1989. Deacylation of echinocandin B by Actinoplanes utahensis. J. Antibiot. 42:382–388 [DOI] [PubMed] [Google Scholar]
- 3. Breton C, Bettler E, Joziasse DH, Geremia RA, Imberty A. 1998. Sequence-function relationships of prokaryotic and eukaryotic galactosyltransferases. J. Biochem. 123:1000–1009 [DOI] [PubMed] [Google Scholar]
- 4. Charnock SJ, Davies GJ. 1999. Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry 38:6380–6385 [DOI] [PubMed] [Google Scholar]
- 5. Chen CP, Kernytsky A, Rost B. 2002. Transmembrane helix predictions revisited. Protein Sci. 11:2774–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen SCA, Sorrell TC. 2007. Antifungal agents. Med. J. Aust. 187:404–409 [DOI] [PubMed] [Google Scholar]
- 7. Denning DW. 2003. Echinocandin antifungal drugs. Lancet 362:1142–1151 [DOI] [PubMed] [Google Scholar]
- 8. Doering TL. 2009. How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans. Annu. Rev. Microbiol. 63:223–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Douglas CM. 2001. Fungal beta(1,3)-D-glucan synthesis. Med. Mycol. 39(Suppl 1):55–66 [DOI] [PubMed] [Google Scholar]
- 10. Douglas CM, et al. 1997. Identification of the FKS1 gene of Candida albicans as the essential target of 1,3-beta-D-glucan synthase inhibitors. Antimicrob. Agents Chemother. 41:2471–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Douglas CM, et al. 1994. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-beta-D-glucan synthase. Proc. Natl. Acad. Sci. U. S. A. 91:12907–12911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Drummond RA, Brown GD. 2011. The role of dectin-1 in the host defence against fungal infections. Curr. Opin. Microbiol. 14:392–399 [DOI] [PubMed] [Google Scholar]
- 13. Edlind TD, Katiyar SK. 2004. The echinocandin “target” identified by cross-linking is a homolog of Pil1 and Lsp1, sphingolipid-dependent regulators of cell wall integrity signaling. Antimicrob. Agents Chemother. 48:4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fulton Z, et al. 2008. Crystal structure of a UDP-glucose-specific glycosyltransferase from a Mycobacterium species. J. Biol. Chem. 283:27881–27890 [DOI] [PubMed] [Google Scholar]
- 15. Ha Y-S, Covert SF, Momany M. 2006. FsFKS1, the 1,3-β-glucan synthase from the caspofungin-resistant fungus Fusarium solani. Eukaryot. Cell 5:1036–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hector RF, Bierer DE. 2011. New β-glucan inhibitors as antifungal drugs. Expert Opin. Ther. Pat. 21:1597–1610 [DOI] [PubMed] [Google Scholar]
- 17. Hildebrand A, Remmert M, Biegert A, Söding J. 2009. Fast and accurate automatic structure prediction with HHpred. Protein Struct. Func. Bioinform. 77:128–132 [DOI] [PubMed] [Google Scholar]
- 18. Johnson ME, Katiyar SK, Edlind TD. 2011. New Fks hot spot for acquired echinocandin resistance in Saccharomyces cerevisiae and its contribution to intrinsic resistance of Scedosporium species. Antimicrob. Agents Chemother. 55:3774–3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katiyar SK, Edlind TD. 2009. Role for Fks1 in the intrinsic echinocandin resistance of Fusarium solani as evidenced by hybrid expression in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 53:1772–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kikuchi N, Kwon Y-D, Gotoh M, Narimatsu H. 2003. Comparison of glycosyltransferase families using the profile hidden Markov model. Biochem. Biophys. Res. Commun. 310:574–579 [DOI] [PubMed] [Google Scholar]
- 21. Kim H, Melén K, von Heijne G. 2003. Topology models for 37 Saccharomyces cerevisiae membrane proteins based on C-terminal reporter fusions and predictions. J. Biol. Chem. 278:10208–10213 [DOI] [PubMed] [Google Scholar]
- 22. Kim H, von Heijne G, Nilsson I. 2005. Membrane topology of the STT3 subunit of the oligosaccharyl transferase complex. J. Biol. Chem. 280:20261–20267 [DOI] [PubMed] [Google Scholar]
- 23. Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 24. Kurtz MB, Douglas CM. 1997. Lipopeptide inhibitors of fungal glucan synthase. J. Med. Vet. Mycol. 35:79–86 [DOI] [PubMed] [Google Scholar]
- 25. Latgé J-P. 2007. The cell wall: a carbohydrate armour for the fungal cell. Mol. Microbiol. 66:279–290 [DOI] [PubMed] [Google Scholar]
- 26. Lesage G, Bussey H. 2006. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70:317–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loibl M, et al. 2010. C terminus of Nce102 determines the structure and function of microdomains in the Saccharomyces cerevisiae plasma membrane. Eukaryot. Cell 9:1184–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mazur P, et al. 1995. Differential expression and function of two homologous subunits of yeast 1,3-beta-D-glucan synthase. Mol. Cell. Biol. 15:5671–5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Melén K, Krogh A, von Heijne G. 2003. Reliability measures for membrane protein topology prediction algorithms. J. Mol. Biol. 327:735–744 [DOI] [PubMed] [Google Scholar]
- 30. Mio T, et al. 1997. Cloning of the Candida albicans homolog of Saccharomyces cerevisiae GSC1/FKS1 and its involvement in beta-1,3-glucan synthesis. J. Bacteriol. 179:4096–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Möller S, Croning MDR, Apweiler R. 2001. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17:646–653 [DOI] [PubMed] [Google Scholar]
- 32. Nett JE, Crawford K, Marchillo K, Andes DR. 2010. Role of Fks1p and matrix glucan in Candida albicans biofilm resistance to an echinocandin, pyrimidine, and polyene. Antimicrob. Agents Chemother. 54:3505–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Okada H, et al. 2010. Multiple functional domains of the yeast 1,3-β-glucan synthase subunit Fks1p revealed by quantitative phenotypic analysis of temperature-sensitive mutants. Genetics 184:1013–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Onishi J, et al. 2000. Discovery of novel antifungal (1,3)-β-d-glucan synthase inhibitors. Antimicrob. Agents Chemother. 44:368–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ostrosky-Zeichner L. 2012. Invasive mycoses: diagnostic challenges. Am. J. Med. 125:S14–S24 [DOI] [PubMed] [Google Scholar]
- 36. Perlin DS. 2007. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 10:121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pfaller MA, Diekema DJ. 2004. Twelve years of fluconazole in clinical practice: global trends in species distribution and fluconazole susceptibility of bloodstream isolates of Candida. Clin. Microbiol. Infect. 10:11–23 [DOI] [PubMed] [Google Scholar]
- 38. Qadota H, et al. 1996. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-β-glucan synthase. Science 272:279–281 [DOI] [PubMed] [Google Scholar]
- 39. Radding JA, Heidler SA, Turner WW. 1998. Photoaffinity analog of the semisynthetic echinocandin LY303366: identification of echinocandin targets in Candida albicans. Antimicrob. Agents Chemother. 42:1187–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saxena IM, Brown RM, Jr, Dandekar T. 2001. Structure-function characterization of cellulose synthase: relationship to other glycosyltransferases. Phytochemistry 57:1135–1148 [DOI] [PubMed] [Google Scholar]
- 41. Schimoler-O'Rourke R, Renault S, Mo W, Selitrennikoff CP. 2003. Neurospora crassa FKS protein binds to the (1,3)-β-glucan synthase substrate, UDP-glucose. Curr. Microbiol. 46:408–412 [DOI] [PubMed] [Google Scholar]
- 42. Sengstag C, Stirling C, Schekman R, Rine J. 1990. Genetic and biochemical evaluation of eukaryotic membrane protein topology: multiple transmembrane domains of Saccharomyces cerevisiae 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mol. Cell. Biol. 10:672–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Strahl-Bolsinger S, Scheinost A. 1999. Transmembrane topology of Pmt1p, a member of an evolutionarily conserved family of protein O-mannosyltransferases. J. Biol. Chem. 274:9068–9075 [DOI] [PubMed] [Google Scholar]
- 44. Thompson JR, et al. 1999. A glucan synthase FKS1 homolog in Cryptococcus neoformans is single copy and encodes an essential function. J. Bacteriol. 181:444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Utsugi T, et al. 2002. Movement of yeast 1,3-β-glucan synthase is essential for uniform cell wall synthesis. Genes Cells 7:1–9 [DOI] [PubMed] [Google Scholar]
- 46. Viklund H, Elofsson A. 2004. Best α-helical transmembrane protein topology predictions are achieved using hidden Markov models and evolutionary information. Protein Sci. 13:1908–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Villasmil ML, Nickels JJT. 2011. Determination of the membrane topology of Arv1 and the requirement of the ER luminal region for Arv1 function in Saccharomyces cerevisiae. FEMS Yeast Res. 11:524–527 [DOI] [PubMed] [Google Scholar]
- 48. Walker SS, et al. 2011. Discovery of a novel class of orally active antifungal β-1,3-d-glucan synthase inhibitors. Antimicrob. Agents Chemother. 55:5099–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.