Abstract
Bacterial type III secretion systems (T3SS) are complex protein assemblies that mediate the secretion of protein substrates outside the cell. Type III secretion chaperones (T3SC) are always found associated with T3SS, and they serve in multiple roles to ensure that protein substrates are efficiently targeted for secretion. Bacterial pathogens with T3SS express T3SC proteins that bind effectors, a process important for effector protein delivery into eukaryotic cells during infection. In this minireview, we focus on multicargo and class 1B T3SC that associate with effectors within significant pathogens of animals and plants. As a primary role, multicargo and class 1B T3SC form homodimers and specifically bind different effectors within the cytoplasm, maintaining the effectors in a secretion-competent state. This role makes T3SC initial and central contributors to effector-mediated pathogenesis. Recent findings have greatly expanded our understanding of cellular events linked to multicargo T3SC function. New binding interactions with T3SS components have been reported in different systems, thereby implicating multicargo T3SC in critical roles beyond effector binding. Three notable interactions with the YscN, YscV, and YscQ family members are well represented in the literature. Similar T3SC interactions are reported in the putative related flagellar T3SS, suggesting that secretion mechanisms may be more similar than previously thought. The evidence implicates multicargo and class 1B T3SC in effector binding and stabilization, in addition to T3SS recruitment and docking events.
BACKGROUND
Bacterial type III secretion systems (T3SS) are complex molecular assemblies that mediate the secretion of proteins from the cytoplasm to the extracellular milieu or directly into eukaryotic cells. T3SS within pathogens have been linked to animal and plant diseases for a wide variety of species, including Yersinia (bubonic plague, enterocolitis) (12), Salmonella (typhoid fever) (33), Shigella (dysentery) (17, 66), Escherichia coli (hemolytic diarrhea) (49, 70), Chlamydia (trachoma) (29, 50), and Pseudomonas syringae and Xanthomonas campestris (tomato and pepper spot disease) (28, 76). The assembly of the T3SS and its subsequent function occur through a series of spatiotemporally regulated events. In this minireview, we will focus on type III secretion chaperones (T3SC), specifically multicargo and class 1B chaperones that are involved in coordinating interactions with effector proteins and critical components of the T3SS. The putative related flagellar T3SS will not be covered in detail (for reviews, see references 48 and 52), although discoveries for flagellar T3SS components and chaperones will be highlighted for contextual and putative similarities with the T3SS of pathogens.
DISCOVERY AND CLASSIFICATION OF T3SC
With the discovery of the T3SS and associated effectors (secreted substrates), supporting experimental observations implicated a number of T3SS ancillary proteins in substrate secretion. These proteins tend to be encoded by genes immediately adjacent to an effector gene (13, 31). Disruption of the adjacent gene often abolished or reduced effector secretion and in some cases reduced effector stability, resulting in degradation. Protein interaction studies revealed that these ancillary proteins were often stably and physically associated with effectors (1, 78), a feature in common with “traditional” molecular chaperones that bind and protect proteins within the cell (e.g., GroEL) (37), and hence the term “chaperone-like” was proposed (79). This name is controversial, as T3SC as a group of proteins do not exhibit the ATPase or protein folding activities that are hallmark features of traditional molecular chaperones. Certain T3SC display stabilizing interactions with effector proteins; therefore, the term chaperone is more akin to a factor involved in guiding or protecting partner proteins. In fact, cocrystallization and NMR studies have implicated T3SC in maintaining proteins in an extended conformation or a partially unfolded state, by providing a molecular scaffold (24, 65, 69).
T3SC are typically small (15- to 20-kDa) and acidic (pI 4 to 5) cytoplasmic proteins (Table 1) that remain within the bacterial cell. Three classes of T3SC have been proposed (13, 39, 62) and are briefly discussed here: class I T3SC bind effectors, class II T3SC bind translocon or pore-forming proteins of the T3SS, and class III T3SC bind needle and filament proteins. Class III T3SC are thought to prevent premature association or polymerization of monomeric needle and filament proteins within the bacterial cytoplasm. Class II T3SC bind their cargos within the bacterial cell and in many cases are required for translocon protein secretion. Class I T3SC has two subclasses: IA and IB. Class IA T3SC bind a single effector and are often encoded by a gene adjacent to an effector gene, whereas class IB T3SC bind to several effectors and are often encoded by a gene within an operon that carries genes encoding components of the secretion apparatus. Class IA and IB designations can be ambiguous, as new findings for some class IA T3SC showed binding to multiple effectors (e.g., CesT). For clarity, in this review, we will use the term multicargo chaperone, referring to a T3SC that binds several different effector substrates regardless of gene context. Within the last 10 years, a number of multicargo chaperones have been implicated in binding, recruitment, regulatory, and docking roles. The importance of these proteins is validated by findings where multicargo chaperone mutants exhibit significant virulence defects in animal and plant models of infection. The features of some of these chaperones are presented below and summarized in Table 1.
Table 1.
Summary of features and experimental observations for selected multicargo T3SC
| Multicargo chaperone | pI/103 Mw | Known binding interaction(s) with secreted substratesa | Mutant virulence defect | Interaction with conserved T3SS component | Relevant references |
|---|---|---|---|---|---|
| InvB | 4.5/14.9 | SspA(SipA), SopE/E2, SopA | Yes | 6, 19, 20, 43 | |
| Spa15 | 4.3/15.2 | IpaA, IpgB1, IpgB2, OspB, OspC1, OspC2, OspC3, OspD1, OspD2 | NDb | 60, 61, 67 | |
| CesT | 4.4/17.7 | Tir, Map, EspF, EspG, EspH, EspZ, NleA, NleG, NleH, NleH2 | Yes | EscN | 1, 16, 23, 73 |
| HpaB (162 aa) | 4.3/18.5 | AvrBs1, AvrBs3, XopF1 | Yes | HrcN, HrcU, HrcV | 3, 7, 8, 46 |
| Mcsc | 4.6/18.8 | Ct618, Cap1, Ct225 | ND | CdsQ | 68 |
| SrcA | 4.6/16.1 | SseL | Yes | SsaN | 11 |
Listed by direct interactions in experimental binding assays. Additional roles in effector secretion without demonstrated binding have been observed in some cases.
ND, not determined.
InvB.
The invABC genetic locus located within the Salmonella pathogenicity island 1 (SPI-1) was initially implicated in eukaryotic cell invasion, although invB mutants were still able to invade cells in vitro, making it difficult to assess the contribution of InvB to pathogenesis (22). invB mutants were shown to have a virulence defect in an avian model of Salmonella infection, yet the exact contribution or functional role of InvB in virulence remained unknown (63). At the sequence level, InvB shares 31% sequence similarity with Spa15 (SpaK), a protein encoded in the mxi-spa genetic locus associated with Shigella host cell invasion, and therefore a link to Salmonella invasion was suggested. The first demonstration for InvB being a chaperone was a study that used a yeast two-hybrid approach and in vitro infection experiments. Bronstein et al. (6) discovered that InvB interacts with SspA (SipA) and contributes to efficient translocation of SipA into target host cells. Two groups then answered a key question relating to Salmonella effectors encoded outside SPI-1 by demonstrating that InvB was required for SopE and SopE2 secretion and translocation into host cells (19, 43). Finally, using a polymutant effector Salmonella strain, InvB was shown to interact with SopA (20). Aside from SopE and SopE2, the InvB interacting effectors do not share extensive sequence similarity, which raised the important question of how InvB mediates binding specificity to effector partner proteins. As it turns out, intrinsic structural features within effectors are critical for InvB binding (45) (presented below).
Spa15.
The invasive enteric pathogen Shigella flexneri (and close relatives) contains a large 213-kb virulence plasmid that encodes components of a T3SS, effectors, chaperones, and transcriptional regulators. A screen for interacting proteins encoded by the virulence plasmid identified a number of potential chaperone-effector associations (58). Based on homology to InvB, which was previously known as a Salmonella chaperone for the effector SipA/SspA, a chaperone role for Spa15 was postulated based on an interaction with the effectors IpaA, IpgB1, and OspC3 (58, 60). This was the first evidence that T3SC could interact with multiple effectors and hence introduced the concept of “promiscuous” chaperones, i.e., chaperones that interact with multiple binding partners (35). In fact, a protein interaction platform assay has since revealed that Spa15 interacts with additional Shigella effectors, including IpgB2, OspB, OspC1, OspC2, OspD1, and OspD2 (for a total of nine) (67). Spa15 is involved in the efficient secretion of all nine of these effectors. Interestingly, the OspD1-Spa15 interaction has been implicated in an anti-activator role that negatively influences virulence gene transcription (61). Another observation is that to date Spa15 is the only example of a T3SC that is secreted (26). The relevance of Spa15 secretion remains to be determined.
CesT.
CesT of enteropathogenic E. coli (EPEC) is perhaps one of the most unusual multicargo chaperones, as it was first described as a class IA chaperone for the effector Tir (1, 23). CesT (chaperone E. coli secreted protein Tir) fits many of the criteria for assignment as a class IA chaperone; the cesT gene is immediately adjacent to tir within an operon in the locus of enterocyte effacement (LEE) pathogenicity island of attaching and effacing E. coli strains. This is similar to the cognate chaperone effector pairings for class IA chaperones seen in many pathogens (e.g., SycE/YopE, SigE/SigD, SicP/SptP). Additional interaction studies revealed that CesT binds to Map, another LEE-encoded effector (16). The discovery of many non-LEE-encoded effectors, accompanied by the paucity of chaperone candidate genes, raised the possibility that CesT could be involved in non-LEE-encoded effector secretion via the T3SS. Indeed this was the case, as CesT was shown to interact with all the effectors encoded within the LEE and many non-LEE-encoded effectors (73). CesT is known to interact with at least 10 effectors (Table 1) and possibly more given that some effectors are encoded by redundant genes with almost identical sequences (e.g., NleG and EspF protein families). Expectedly, cesT null mutants do not cause overt disease in a mouse model of infection (18).
HpaB.
Plant pathogens within the genus Xanthomonas use a T3SS to inject multiple effectors into plant cells during infection. The model organism for this group of bacteria is Xanthomonas campestris pv. vesicatoria, which causes bacterial spot disease in pepper and tomato. Studies have identified a 23-kb hypersensitive response and pathogenicity (hrp) gene cluster that contains six operons (hrpA-F) encoding components of a T3SS, effectors, regulators and chaperones (27). Genes without known functions or limited homology to known T3SS proteins in databases were initially termed hrp-associated genes (hpa). In the case of HpaB, it was shown to bind three effector proteins, AvrBs1, AvrBs3, and XopF1 using in vitro binding assays (7, 9). In vitro secretion assays demonstrated that five Xanthomonas effectors require HpaB for efficient secretion into culture supernatants. HpaB is required for Xanthomonas-mediated T3SS injection of two effectors into plant cells (XopJ, XopF1), implicating this protein in efficient effector translocation for a subset of effectors with which it interacts (8). Importantly, hpaB mutants display a complete loss of pathogenicity in susceptible plants and yet still produce a partial (reduced) hypersensitive response in resistant plants (7).
Mcsc.
The obligate intracellular pathogen Chlamydia trachomatis (and related species) uses a T3SS to inject effectors that localize to a membrane inclusion vacuole within the host cell. The study of specific Chlamydia T3SS components in situ is challenging due to the lack of an efficient genetic manipulation system. Researchers have relied on bioinformatic approaches and strategies to express T3SS components in heterologous systems to elucidate function. A comprehensive yeast two-hybrid protein interaction screen revealed that open reading frame (ORF) Ct260 of C. trachomatis interacts with three putative effectors (Ct618, Cap1, and Ct225) and CdsQ, a conserved component found in all T3SS (68) (discussed below). Immunolocalization and in vitro pulldown experiments validated the role of Ct260 as a multicargo T3SC, and it was named Mcsc (multiple cargo secretion chaperone). Mcsc was observed to form dimers, like other T3SC. While a crystal structure of Mcsc is not available, molecular modeling predicts common structural folds found in other T3SC.
SrcA.
Salmonella makes use of two T3SS, encoded within the SPI-1 and SPI-2 pathogenicity islands, to subvert host cells during infection. The regulation of each pathogenicity island is complex, with master transcriptional regulators involved. For SPI-2, SsrB is involved in regulating gene expression, although SsrB also regulates other genes throughout the genome, including effector genes that encode protein substrates of the SPI-2 T3SS. Using a transcriptional profiling approach, ORF ST2138 was shown to be regulated by SsrB (11). ST2138 encodes a protein with features similar to T3SC (Table 1) and shares 59% amino acid sequence identity with T3SC CesT. Based on these properties and other findings, ST2138 was named srcA (SsrB-regulated chaperone A). Deletion of srcA from S. enterica serovar Typhimurium resulted in decreased fitness compared to that of wild-type bacteria in a mixed (competitive) oral infection of mice, thereby implicating SrcA as an important virulence determinant. SrcA was demonstrated to bind SseL, a SPI-2-encoded effector, and was further implicated as the chaperone for the PipB2 effector (11). Both of these respective effector genes are SsrB regulated, which correlates with the observed SsrB regulation for SrcA. Currently, it is debatable if SrcA is a bona fide multicargo chaperone, as it has only been shown to interact with SseL. The requirement of SrcA for PipB2 delivery implies a binding interaction, although additional work will be required to determine if SrcA binds to multiple effectors.
MECHANISMS OF EFFECTOR BINDING BY MULTICARGO CHAPERONES
In the majority of cases, structural studies have revealed that chaperone monomers form homodimers along a helical interface (reviewed in reference 59). One notable difference between class 1A and class 1B chaperones is that for class 1B, the helices contact each other at an angle of about 30°, as observed in Spa15 (74), whereas for class 1A chaperones (e.g., SycE, SicP), the helices run parallel. An impressive amount of surface-exposed hydrophobicity is available on chaperone homodimers, a feature that is thought to promote interactions with the amino-terminal region of effectors (47, 69). In support of that view, formation of chaperone-effector complexes can occur in the absence of other cellular proteins or ATP (1, 73, 79). Within the cell, a T3SS-associated ATPase has been implicated in dissociating chaperone-effector complexes (2) (discussed below).
A β-strand motif found in many (but not all) effectors has been implicated in binding to T3SC (45). Furthermore, alteration of the motif was shown to disrupt or weaken chaperone-effector binding interactions. Analysis of chaperone-effector cocrystals (e.g., SycE/YopE, SicP/SptP, InvB/SipA) reveals that β-strand motifs are positioned at the amino-terminal region of effectors in a “chaperone binding domain.” A conserved chaperone binding domain (CCBD) sequence [(LMIF)1XXX(IV)5XX(IV)8XN10] overlaps the β-motif (14). Furthermore, an interchangeability of InvB and Spa15 T3SC within Salmonella and Shigella was demonstrated, suggesting that the overall chaperone structure is sufficient to interact with diverse effectors. T3SC (class IA and IB) exhibit a contiguous conserved set of structural folds (α-β-β-β-α-β-β-α). In contrast, class II and class III T3SC exhibit multiple tetratricopeptide (TPR) folds (formed by pairs of antiparallel helices) (39). While structural differences are apparent between class I and class II or III T3SC, it is likely that subtle localized features for each T3SC class have evolved for optimal binding functions and interactions with the secretion apparatus (47, 73, 74).
MULTICARGO CHAPERONE INTERACTIONS WITH COMPONENTS OF THE T3SS
The role of T3SC in effector secretion has been controversial, since many early reports demonstrated that effector secretion still occurred in the absence of the respective binding chaperone (23, 79). Now multiple lines of experimental data from different systems support a role for T3SC in docking and recruitment events at the T3SS. Notably, three critical interactions with the conserved YscN (ATPase), YscQ (putative C-ring), and YscV (membrane protein) family members are observed.
Interactions with T3SS ATPases.
All T3SS have an associated ATPase (YscN protein family), which is believed to provide energy through ATP hydrolysis to dissociate effectors from T3SC. In vitro binding assays have demonstrated that purified CesT and SrcA interact with their respective T3SS ATPases (11, 34). These findings have been extended with coimmunoprecipitation of chaperone-ATPase from cell lysates in EPEC and Xanthomonas (46, 73). Therefore, observations from different bacterial pathogens provide evidence that multicargo chaperones interact with T3SS ATPases. This interaction can occur in the absence of effector cargo, although within the cell, we predict that chaperone-effector complexes interact with the T3SS ATPase.
A partially reconstituted system with purified proteins established that that the Salmonella T3SS ATPase InvC could bind to SicP (class IA chaperone) and to a SicP-SptP (chaperone-effector) complex (2). Additional experiments also provided evidence indicating that the InvC ATPase disassembles the SicP-SptP complex by ATP hydrolysis, an event that was required for SptP secretion into culture supernatants. Whether this interaction paradigm fits multicargo chaperones remains to be determined. The situation could be different for flagellar T3SS. The flagellar ATPase FliI is known to interact with FlgN and FliT chaperones (38, 71), although recent in vitro experiments suggest that FliI-mediated ATPase activity does not dissociate the filament capping protein FliD from a FliD/FliT complex (54). Interestingly, it is the proton motive force (PMF) that provides essential energy for flagellar protein secretion (55). Therefore, additional studies are required to critically address what role T3SS ATPases play in preparing chaperone-bound substrates for secretion.
The significance of a chaperone-ATPase interaction is paramount, since it appears to be a common feature in T3SS. T3SS ATPases have been observed and modeled as homohexamers, although cryoelectron microscopy studies of P. syringae HrcN indicate that a dodecamer, a double stack of hexameric rings, can be formed (57). The dodecameric form was shown to be the predominant form at the membrane and exhibited high ATPase activity. In support of those findings, crystallization studies of the ATPase EscN of EPEC revealed that EscN oligomerization is required for efficient ATPase activity (81). Consequently, the most likely site of ATP hydrolysis is at the cytoplasmic face of the inner membrane. Given that ATPase activity is likely required to dissociate chaperone-effector complexes (2), chaperones would seemingly need to exist in the cytoplasm to bind partner effectors and also at the membrane to mediate critical interactions with the T3SS ATPase. Indeed, we have shown that CesT partitions to cytoplasmic (soluble) and membrane (insoluble) fractions (73). This dual localization for CesT and presumably other chaperones implicates this group of proteins as recruitment factors for export substrates and further substantiates a role in secretion and trafficking events within the cell.
Interactions with the structural C-ring protein.
Another conserved component for all flagellar and T3SS is the YscQ/FliM-N family. YscQ/FliM-N members (PscQ, Spa33, SpaO, EscQ) are believed to form a cytoplasmic ring structure (C-ring) at the inner membrane (25, 56, 82). This membrane-associated ring structure is conceivably amenable to localized interactions for soluble chaperone-effector complexes. Pulldown and yeast hybrid protein interaction experiments suggest that a YscQ-YscL-YscK-YscN complex may exist in vivo (40, 64). Pulldown experiments in EPEC and Shigella identified similar interactions (4, 41). Therefore, a high-order protein complex composed of YscNLQ, and probably YscK, likely exists within the cell. A recent report indicated that SpaO (Salmonella YscQ homologue) forms a sorting platform for chaperone-effector complexes (42). This sorting platform also contained the ATPase InvC, an observation in agreement with the existence of a high-order YscNLQ complex. SicA (class IA chaperone) interactions with the sorting platform were detected, although no evidence of class 1B or multicargo chaperones was presented. Data for multicargo chaperone binding to the YscQ family was recently demonstrated for the chlamydial Mcsc chaperone. Mcsc was shown to interact with the CdsQ, the C. trachomatis homologue of YscQ (68). Hence T3SC interactions with the YscQ family are likely important binding events that support efficient targeting of effector proteins to the membrane-embedded T3SS.
Interactions with the YscV/FlhA family.
YscV/FlhA family members are large proteins (>600 amino acids) that contain six to eight predicted transmembrane domains and a large cytoplasmic extension spanning over 300 amino acids. Recent findings suggest that this family of proteins forms oligomers at the membrane (44). A yeast two-hybrid screen using bait proteins encoded from the hrp locus of Xanthomonas axonopodis pv. citri identified an interaction between HpaB and HrcV (325 to 646 amino acids [aa]) (3). In support of that finding, purified GST-HrcV and HpaB were shown to interact with each other in a pulldown assay (8). These protein interaction studies suggest that in Xanthomonas, HpaB may interact with HrcV, presumably at the inner membrane. Such an interaction has yet to be observed within Xanthomonas, although it is reasonable to speculate that an HpaB-HrcV interaction occurs near the cytoplasmic face of the T3SS given that HrcV (325 to 646 aa) is predicted to be within a large cytoplasmic domain. Whether other multicargo chaperones interact with their corresponding YscV family member remains to be determined.
An interesting discovery relating to FlhA interaction with the flagellar multicargo chaperone FlgN has been reported by Minamino and colleagues (53). FlgN is the chaperone for hook filament junction proteins FlgK and FlgL (30). Previous studies identified a Salmonella ΔfliH-I flhB(P28T) strain that unexpectedly exhibits reduced motility on soft agar plates (55). This polymutant strain still produces some flagella, which indicates that flagellar filament assembly can occur to some extent in the absence of FliH and FliI and in the presence of a FlhB variant. Deletion of flgN from this polymutant background resulted in a further reduction in motility described as a “very weak motility phenotype” (53). Interestingly, multiple pseudorevertants that modestly improved motility for the ΔfliH-I flhB(P28T) ΔflgN strain were found to have suppressor mutations exclusively within flhA. Possibly even more intriguing was the fact that within these isolated pseudorevertants, the mutations were found in only two sites, FlhA(D456A) and FlhA(T490M), both within the FlhA cytoplasmic domain. Correspondingly, a direct FlgN-FlhA interaction was observed in follow-up protein interaction studies.
From the aforementioned findings, YscV/FlhA family members are central to chaperone-effector complex interactions at the T3SS. The observations of multicargo T3SC and flagellar chaperones binding to the YscV/FlhA large cytoplasmic domain strongly suggest that this is a conserved mechanism for T3SS.
ENGAGEMENT OF PROTEIN SUBSTRATES WITH THE T3SS—A CHANNEL AWAITS
While evidence links chaperones to substrate binding and recruitment events prior to substrate secretion, it is difficult to reconcile published observations where substrates are secreted in the absence of chaperones. In some cases, short N-terminal signal sequences of effectors are sufficient in mediating translocation of reporter proteins into target cells (5, 15). It should be noted that the translocation of artificial reporters is not necessarily equivalent to naturally translocated effectors. In fact, in many cases, translocation efficiencies are typically higher when the N-terminal sequence and chaperone binding domain are intact within effectors and when translocation is assayed in the presence of the associated chaperone (10, 15, 51).
If T3SC have recruitment functions, one might anticipate T3SS recruitment deficiencies for relevant mutants. This has been observed in Salmonella with studies evaluating secretion of SopE, which is chaperoned by InvB. SopE variants that were unable to bind InvB due to alteration of the chaperone binding domain or deletion of invB were found to be secreted by the flagellar export pathway, a situation that did not occur when InvB and wild-type SopE were within the cell (21, 43). Unfortunately, these studies did not establish an actual recruitment of an InvB-SopE complex to the T3SS, although the results clearly demonstrate that in the absence of InvB binding, SopE is no longer exclusively recruited to the T3SS.
Perhaps one of the most intriguing findings relating to engagement of substrates at a T3SS is found in the flagellar system, where dominant negative FlgN variants were shown to block export of multiple flagellar substrates (71). FlgN-FlgK/L complexes were demonstrated to interact with FliI (ATPase) independently of other flagellar components, although within the cell, FlgN variants blocked flagellar protein export, presumably by inappropriately engaging the export channel and preventing entry for other substrates. In such a model, one could envision a shuttling or adaptor mechanism where the FlgN-FlgK/L complexes initially interact with FliI followed by localized FliI ATPase activity to dissociate FlgN complexes in close proximity to FlhA as described above (53), allowing for efficient and productive engagement of the flagellar export apparatus. This hypothetical model requires experimental validation and will be an area of considerable interest. Moreover, it is unclear whether such a model applies to the T3SS of pathogens, although there appear to be enough similarities to warrant further investigation.
EVIDENCE FOR MULTICARGO CHAPERONES IN HIERARCHY OF EFFECTOR TRANSLOCATION?
An unresolved issue is whether there is a hierarchical order to effector secretion. Do some substrates get secreted before others? If so, does the putative hierarchical mechanism involve multicargo chaperones? Examples from the literature provide evidence that an effector translocation hierarchy does exist; however, limited data are available to establish a role for multicargo chaperones in hierarchy.
In Salmonella, T3SS-delivered SopE/E2, SspA, and SopB(SigD) act in concert to promote actin polymerization at sites of bacterial invasion (83). SopE/E2 act as guanine nucleotide exchange factors (GEF) to activate Cdc42, which triggers membrane ruffling and eventual bacterial uptake (36). In contrast, T3SS-delivered SptP acts as a GTPase activating protein (GAP), antagonizing Cdc42 (32). These effector-mediated activities on Cdc42 would conceivably be canceled out if the effectors were delivered concurrently. As it turns out, translocation of SspA (SipA) and SopE2 has been experimentally observed to occur before SptP translocation (75, 80). The chaperone for SspA and SopE2 is InvB, whereas the chaperone for SptP is SicP. It is tempting to speculate that InvB-effector complexes engage the T3SS export apparatus first or perhaps with higher affinity than SicP-SptP complexes.
In EPEC, Tir delivery results in intimate attachment to the host cell. Interestingly, Tir secretion has been shown to be important for the secretion of other effectors (72). Real-time quantitative infection assays revealed that Tir delivery into host cells precedes that of other effectors (51). Moreover, during EPEC infection, Tir-mediated actin remodeling promotes efficient effector translocation (77). Collectively, these findings suggest a hierarchy for Tir over other effectors. At least 10 EPEC effectors interact with the T3SC CesT, including Tir, which implies that a mechanism to ensure Tir hierarchy likely involves CesT. The tir and cesT genes are cotranscribed, which may favor immediate CesT-Tir complex formation before other CesT-effector complexes can be formed. In support of this view, coexpressing NleA or NleH with CesT in a tir mutant increased NleA and NleH secretion levels (72). Therefore, temporal coexpression of effectors and chaperones could be a simple mechanism that promotes hierarchical effector translocation.
FUTURE DIRECTIONS AND CONCLUDING REMARKS
Multicargo chaperones are critical players in protein trafficking events with the cell. Through multiple interactions with different effectors, these T3SC coordinate binding and recruitment actions leading to efficient effector secretion. Although T3SC were once thought to serve only as scaffolds and bodyguards, recent findings clearly implicate T3SC in recruitment and docking roles with the YscN, YscQ, and YscV components of the T3SS (Fig. 1). A considerable amount of primary evidence has been learned from the related flagellar export pathway, where motility screens and efficient genetic approaches have proven very useful in elucidating T3SC function. A major challenge will be to determine if multicargo chaperones of pathogens behave in a similar manner or whether novel interactions or trafficking roles are required for effector secretion. Another challenging issue is whether effectors exhibit different binding affinities for multicargo chaperones. If so, does it contribute to a hierarchy of effector translocation that has been observed in some infection models?
Fig 1.
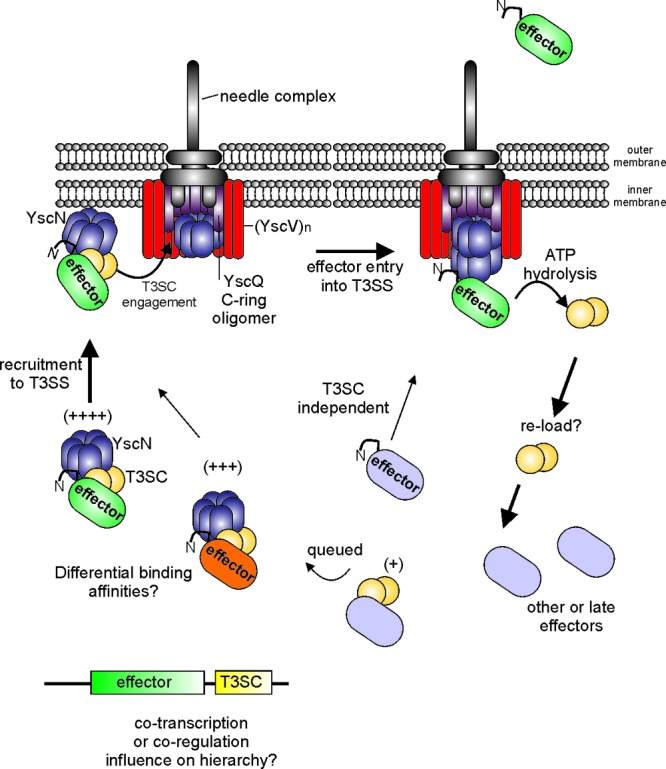
Hypothetical model depicting three steps involved in trafficking a T3SC-effector complex to the T3SS: (i) recruitment, (ii) engagement, and (iii) entry into the T3SS. The model incorporates multiple findings from the literature as described in the text. The secretion of a putative hierarchical effector (green) is shown over other effectors based on experimental observations where some effectors are secreted before others. Note that in this case, the dimeric T3SC (yellow) is depicted to interact with YscN (blue), YscV (red), and YscQ (purple). The sequence of these binding interactions is not known and could be different than depicted. Whether this occurs in all systems remains to be determined. YscN-mediated ATP hydrolysis is shown and presumably favors T3SC dissociation, allowing for effector entry into the T3SS. The uncomplexed T3SC could potentially reload with other effectors in the cytoplasm. A T3SC-independent pathway is shown to indicate that such events have been observed in different experimental systems. The plus signs indicate the possibility of binding affinities or favorable formation of tripartite T3SC-effector-YscN complexes. Other possible mechanisms of effector hierarchy such as cotranscription or coregulation are presented. YscN (ATPase) is shown as a ring-like homohexamer in the cytoplasm and a dodecamer at the membrane based on experimental observations. A YscQ oligomer forming a C-ring at the base of the T3SS is in close proximity to a (YscV)n oligomer. The exact stoichiometry of these oligomers is not known, although evidence exists to suggest oligomer formation. The N attached to effector ovals represents an amino-terminal signal sequence. Some components of the T3SS are not depicted for clarity and/or lack of experimental data. The thickness of the arrows denotes the putative efficiency of the process, as in the majority of cases effector secretion is more efficient with T3SC function.
For such small proteins, multicargo chaperones play a significant role in effector biology. Preventing multicargo chaperone function could be an efficient strategy to limit effector-mediated pathogenesis. Therefore, future searches for inhibitors that impede multicargo chaperone function may prove useful in disease prevention.
ACKNOWLEDGMENTS
While we have tried to be complete, we apologize to those researchers whose findings are not presented in this minireview due to space limitations.
This work was supported by the Canadian Institutes of Health Research (CIHR, MOP84472) and the Dalhousie Medical Research Foundation. N.A.T is the recipient of a CIHR/Nova Scotia Health Research Foundation New Investigator Award.
Footnotes
Published ahead of print 25 May 2012
REFERENCES
- 1. Abe A, et al. 1999. Enteropathogenic Escherichia coli translocated intimin receptor, Tir, requires a specific chaperone for stable secretion. Mol. Microbiol. 33:1162–1175 [DOI] [PubMed] [Google Scholar]
- 2. Akeda Y, Galan JE. 2005. Chaperone release and unfolding of substrates in type III secretion. Nature 437:911–915 [DOI] [PubMed] [Google Scholar]
- 3. Alegria MC, et al. 2004. New protein-protein interactions identified for the regulatory and structural components and substrates of the type III secretion system of the phytopathogen Xanthomonas axonopodis pathovar citri. J. Bacteriol. 186:6186–6197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Biemans-Oldehinkel E, Sal-Man N, Deng W, Foster LJ, Finlay BB. 2011. Quantitative proteomic analysis reveals formation of an EscL-EscQ-EscN type III complex in enteropathogenic Escherichia coli. J. Bacteriol. 193:5514–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boyd AP, Lambermont I, Cornelis GR. 2000. Competition between the Yops of Yersinia enterocolitica for delivery into eukaryotic cells: role of the SycE chaperone binding domain of YopE. J. Bacteriol. 182:4811–4821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bronstein PA, Miao EA, Miller SI. 2000. InvB is a type III secretion chaperone specific for SspA. J. Bacteriol. 182:6638–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buttner D, Gurlebeck D, Noel LD, Bonas U. 2004. HpaB from Xanthomonas campestris pv. vesicatoria acts as an exit control protein in type III-dependent protein secretion. Mol. Microbiol. 54:755–768 [DOI] [PubMed] [Google Scholar]
- 8. Buttner D, Lorenz C, Weber E, Bonas U. 2006. Targeting of two effector protein classes to the type III secretion system by a HpaC- and HpaB-dependent protein complex from Xanthomonas campestris pv. vesicatoria. Mol. Microbiol. 59:513–527 [DOI] [PubMed] [Google Scholar]
- 9. Buttner D, Noel L, Stuttmann J, Bonas U. 2007. Characterization of the nonconserved hpaB-hrpF region in the hrp pathogenicity island from Xanthomonas campestris pv. vesicatoria. Mol. Plant Microbe Interact. 20:1063–1074 [DOI] [PubMed] [Google Scholar]
- 10. Charpentier X, Oswald E. 2004. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J. Bacteriol. 186:5486–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cooper CA, et al. 2010. Structural and biochemical characterization of SrcA, a multi-cargo type III secretion chaperone in Salmonella required for pathogenic association with a host. PLoS Pathog. 6:e1000751 doi:10.1371/journal.ppat.1000751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cornelis GR. 2002. The Yersinia Ysc-Yop ‘type III’ weaponry. Nat. Rev. Mol. Cell Biol. 3:742–752 [DOI] [PubMed] [Google Scholar]
- 13. Cornelis GR, Van Gijsegem F. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735–774 [DOI] [PubMed] [Google Scholar]
- 14. Costa SC, et al. 2012. A new means to identify type 3 secreted effectors: functionally interchangeable class IB chaperones recognize a conserved sequence. mBio 3:e00243–11 doi:10.1128/mBio.00243-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crawford JA, Kaper JB. 2002. The N-terminus of enteropathogenic Escherichia coli (EPEC) Tir mediates transport across bacterial and eukaryotic cell membranes. Mol. Microbiol. 46:855–868 [DOI] [PubMed] [Google Scholar]
- 16. Creasey EA, et al. 2003. CesT is a bivalent enteropathogenic Escherichia coli chaperone required for translocation of both Tir and Map. Mol. Microbiol. 47:209–221 [DOI] [PubMed] [Google Scholar]
- 17. Demers B, Sansonetti PJ, Parsot C. 1998. Induction of type III secretion in Shigella flexneri is associated with differential control of transcription of genes encoding secreted proteins. EMBO J. 17:2894–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deng W, et al. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. U. S. A. 101:3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ehrbar K, Friebel A, Miller SI, Hardt WD. 2003. Role of the Salmonella pathogenicity island 1 (SPI-1) protein InvB in type III secretion of SopE and SopE2, two Salmonella effector proteins encoded outside of SPI-1. J. Bacteriol. 185:6950–6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ehrbar K, Hapfelmeier S, Stecher B, Hardt WD. 2004. InvB is required for type III-dependent secretion of SopA in Salmonella enterica serovar Typhimurium. J. Bacteriol. 186:1215–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ehrbar K, Winnen B, Hardt WD. 2006. The chaperone binding domain of SopE inhibits transport via flagellar and SPI-1 TTSS in the absence of InvB. Mol. Microbiol. 59:248–264 [DOI] [PubMed] [Google Scholar]
- 22. Eichelberg K, Ginocchio CC, Galan JE. 1994. Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. J. Bacteriol. 176:4501–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Elliott SJ, et al. 1999. Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Mol. Microbiol. 33:1176–1189 [DOI] [PubMed] [Google Scholar]
- 24. Evdokimov AG, et al. 2003. Similar modes of polypeptide recognition by export chaperones in flagellar biosynthesis and type III secretion. Nat. Struct. Biol. 10:789–793 [DOI] [PubMed] [Google Scholar]
- 25. Fadouloglou VE, et al. 2004. Structure of HrcQB-C, a conserved component of the bacterial type III secretion systems. Proc. Natl. Acad. Sci. U. S. A. 101:70–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Faherty CS, Maurelli AT. 2009. Spa15 of Shigella flexneri is secreted through the type III secretion system and prevents staurosporine-induced apoptosis. Infect. Immun. 77:5281–5290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fenselau S, Balbo I, Bonas U. 1992. Determinants of pathogenicity in Xanthomonas campestris pv. vesicatoria are related to proteins involved in secretion in bacterial pathogens of animals. Mol. Plant Microbe Interact. 5:390–396 [DOI] [PubMed] [Google Scholar]
- 28. Fenselau S, Bonas U. 1995. Sequence and expression analysis of the hrpB pathogenicity operon of Xanthomonas campestris pv. vesicatoria which encodes eight proteins with similarity to components of the Hrp, Ysc, Spa, and Fli secretion systems. Mol. Plant Microbe Interact. 8:845–854 [DOI] [PubMed] [Google Scholar]
- 29. Fields KA, Mead DJ, Dooley CA, Hackstadt T. 2003. Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol. Microbiol. 48:671–683 [DOI] [PubMed] [Google Scholar]
- 30. Fraser GM, Bennett JC, Hughes C. 1999. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol. Microbiol. 32:569–580 [DOI] [PubMed] [Google Scholar]
- 31. Frithz-Lindsten E, Rosqvist R, Johansson L, Forsberg A. 1995. The chaperone-like protein YerA of Yersinia pseudotuberculosis stabilizes YopE in the cytoplasm but is dispensible for targeting to the secretion loci. Mol. Microbiol. 16:635–647 [DOI] [PubMed] [Google Scholar]
- 32. Fu Y, Galan JE. 1999. A Salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature 401:293–297 [DOI] [PubMed] [Google Scholar]
- 33. Galan JE. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53–86 [DOI] [PubMed] [Google Scholar]
- 34. Gauthier A, Finlay BB. 2003. Translocated intimin receptor and its chaperone interact with ATPase of the type III secretion apparatus of enteropathogenic Escherichia coli. J. Bacteriol. 185:6747–6755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ghosh P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68:771–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hardt WD, Chen LM, Schuebel KE, Bustelo XR, Galan JE. 1998. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93:815–826 [DOI] [PubMed] [Google Scholar]
- 37. Hartl FU, Hayer-Hartl M. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852–1858 [DOI] [PubMed] [Google Scholar]
- 38. Imada K, Minamino T, Kinoshita M, Furukawa Y, Namba K. 2010. Structural insight into the regulatory mechanisms of interactions of the flagellar type III chaperone FliT with its binding partners. Proc. Natl. Acad. Sci. U. S. A. 107:8812–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Izore T, Job V, Dessen A. 2011. Biogenesis, regulation, and targeting of the type III secretion system. Structure 19:603–612 [DOI] [PubMed] [Google Scholar]
- 40. Jackson MW, Plano GV. 2000. Interactions between type III secretion apparatus components from Yersinia pestis detected using the yeast two-hybrid system. FEMS Microbiol. Lett. 186:85–90 [DOI] [PubMed] [Google Scholar]
- 41. Jouihri N, et al. 2003. MxiK and MxiN interact with the Spa47 ATPase and are required for transit of the needle components MxiH and MxiI, but not of Ipa proteins, through the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 49:755–767 [DOI] [PubMed] [Google Scholar]
- 42. Lara-Tejero M, Kato J, Wagner S, Liu X, Galan JE. 2011. A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331:1188–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee SH, Galan JE. 2003. InvB is a type III secretion-associated chaperone for the Salmonella enterica effector protein SopE. J. Bacteriol. 185:7279–7284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li H, Sourjik V. 2011. Assembly and stability of flagellar motor in Escherichia coli. Mol. Microbiol. 80:886–899 [DOI] [PubMed] [Google Scholar]
- 45. Lilic M, Vujanac M, Stebbins CE. 2006. A common structural motif in the binding of virulence factors to bacterial secretion chaperones. Mol. Cell 21:653–664 [DOI] [PubMed] [Google Scholar]
- 46. Lorenz C, Buttner D. 2009. Functional characterization of the type III secretion ATPase HrcN from the plant pathogen Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 191:1414–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Luo Y, et al. 2001. Structural and biochemical characterization of the type III secretion chaperones CesT and SigE. Nat. Struct. Biol. 8:1031–1036 [DOI] [PubMed] [Google Scholar]
- 48. Macnab RM. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77–100 [DOI] [PubMed] [Google Scholar]
- 49. Marches O, et al. 2000. Role of tir and intimin in the virulence of rabbit enteropathogenic Escherichia coli serotype O103:H2. Infect. Immun. 68:2171–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Matsumoto A. 1982. Electron microscopic observations of surface projections on Chlamydia psittaci reticulate bodies. J. Bacteriol. 150:358–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mills E, Baruch K, Charpentier X, Kobi S, Rosenshine I. 2008. Real-time analysis of effector translocation by the type III secretion system of enteropathogenic Escherichia coli. Cell Host Microbe 3:104–113 [DOI] [PubMed] [Google Scholar]
- 52. Minamino T, Imada K, Namba K. 2008. Mechanisms of type III protein export for bacterial flagellar assembly. Mol. Biosyst. 4:1105–1115 [DOI] [PubMed] [Google Scholar]
- 53. Minamino T, et al. 2012. Interaction of a bacterial flagellar chaperone FlgN with FlhA is required for efficient export of its cognate substrates. Mol. Microbiol. 83:775–788 [DOI] [PubMed] [Google Scholar]
- 54. Minamino T, Kinoshita M, Imada K, Namba K. 2012. Interaction between FliI ATPase and a flagellar chaperone FliT during bacterial flagellar protein export. Mol. Microbiol. 83:168–178 [DOI] [PubMed] [Google Scholar]
- 55. Minamino T, Namba K. 2008. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 451:485–488 [DOI] [PubMed] [Google Scholar]
- 56. Morita-Ishihara T, et al. 2006. Shigella Spa33 is an essential C-ring component of type III secretion machinery. J. Biol. Chem. 281:599–607 [DOI] [PubMed] [Google Scholar]
- 57. Muller SA, et al. 2006. Double hexameric ring assembly of the type III protein translocase ATPase HrcN. Mol. Microbiol. 61:119–125 [DOI] [PubMed] [Google Scholar]
- 58. Page AL, Fromont-Racine M, Sansonetti P, Legrain P, Parsot C. 2001. Characterization of the interaction partners of secreted proteins and chaperones of Shigella flexneri. Mol. Microbiol. 42:1133–1145 [DOI] [PubMed] [Google Scholar]
- 59. Page AL, Parsot C. 2002. Chaperones of the type III secretion pathway: jacks of all trades. Mol. Microbiol. 46:1–11 [DOI] [PubMed] [Google Scholar]
- 60. Page AL, Sansonetti P, Parsot C. 2002. Spa15 of Shigella flexneri, a third type of chaperone in the type III secretion pathway. Mol. Microbiol. 43:1533–1542 [DOI] [PubMed] [Google Scholar]
- 61. Parsot C, et al. 2005. A secreted anti-activator, OspD1, and its chaperone, Spa15, are involved in the control of transcription by the type III secretion apparatus activity in Shigella flexneri. Mol. Microbiol. 56:1627–1635 [DOI] [PubMed] [Google Scholar]
- 62. Parsot C, Hamiaux C, Page AL. 2003. The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 6:7–14 [DOI] [PubMed] [Google Scholar]
- 63. Porter SB, Curtiss R., III 1997. Effect of inv mutations on Salmonella virulence and colonization in 1-day-old White Leghorn chicks. Avian Dis. 41:45–57 [PubMed] [Google Scholar]
- 64. Riordan KE, Schneewind O. 2008. YscU cleavage and the assembly of Yersinia type III secretion machine complexes. Mol. Microbiol. 68:1485–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rodgers L, Mukerjea R, Birtalan S, Friedberg D, Ghosh P. 2010. A solvent-exposed patch in chaperone-bound YopE is required for translocation by the type III secretion system. J. Bacteriol. 192:3114–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sansonetti PJ. 2001. Rupture, invasion and inflammatory destruction of the intestinal barrier by Shigella, making sense of prokaryote-eukaryote cross-talks. FEMS Microbiol. Rev. 25:3–14 [DOI] [PubMed] [Google Scholar]
- 67. Schmitz AM, Morrison MF, Agunwamba AO, Nibert ML, Lesser CF. 2009. Protein interaction platforms: visualization of interacting proteins in yeast. Nat. Methods 6:500–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Spaeth KE, Chen YS, Valdivia RH. 2009. The Chlamydia type III secretion system C-ring engages a chaperone-effector protein complex. PLoS Pathog. 5:e1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Stebbins CE, Galan JE. 2001. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 414:77–81 [DOI] [PubMed] [Google Scholar]
- 70. Tacket CO, et al. 2000. Role of EspB in experimental human enteropathogenic Escherichia coli infection. Infect. Immun. 68:3689–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Thomas J, Stafford GP, Hughes C. 2004. Docking of cytosolic chaperone-substrate complexes at the membrane ATPase during flagellar type III protein export. Proc. Natl. Acad. Sci. U. S. A. 101:3945–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Thomas NA, Deng W, Baker N, Puente J, Finlay BB. 2007. Hierarchical delivery of an essential host colonization factor in enteropathogenic Escherichia coli. J. Biol. Chem. 282:29634–29645 [DOI] [PubMed] [Google Scholar]
- 73. Thomas NA, et al. 2005. CesT is a multi-effector chaperone and recruitment factor required for the efficient type III secretion of both LEE- and non-LEE-encoded effectors of enteropathogenic Escherichia coli. Mol. Microbiol. 57:1762–1779 [DOI] [PubMed] [Google Scholar]
- 74. van Eerde A, Hamiaux C, Perez J, Parsot C, Dijkstra BW. 2004. Structure of Spa15, a type III secretion chaperone from Shigella flexneri with broad specificity. EMBO Rep. 5:477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Van Engelenburg SB, Palmer AE. 2008. Quantification of real-time Salmonella effector type III secretion kinetics reveals differential secretion rates for SopE2 and SptP. Chem. Biol. 15:619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Van Gijsegem F, et al. 1995. The hrp gene locus of Pseudomonas solanacearum, which controls the production of a type III secretion system, encodes eight proteins related to components of the bacterial flagellar biogenesis complex. Mol. Microbiol. 15:1095–1114 [DOI] [PubMed] [Google Scholar]
- 77. Vingadassalom D, et al. 2010. Enterohemorrhagic E. coli requires N-WASP for efficient type III translocation but not for EspFU-mediated actin pedestal formation. PLoS Pathog. 6:e1001056 doi:10.1371/journal.ppat.1001056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wattiau P, Bernier B, Deslee P, Michiels T, Cornelis GR. 1994. Individual chaperones required for Yop secretion by Yersinia. Proc. Natl. Acad. Sci. U. S. A. 91:10493–10497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wattiau P, Cornelis GR. 1993. SycE, a chaperone-like protein of Yersinia enterocolitica involved in Ohe secretion of YopE. Mol. Microbiol. 8:123–131 [DOI] [PubMed] [Google Scholar]
- 80. Winnen B, et al. 2008. Hierarchical effector protein transport by the Salmonella Typhimurium SPI-1 type III secretion system. PLoS One 3:e2178 doi:10.1371/journal.pone.0002178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zarivach R, Vuckovic M, Deng W, Finlay BB, Strynadka NC. 2007. Structural analysis of a prototypical ATPase from the type III secretion system. Nat. Struct. Mol. Biol. 14:131–137 [DOI] [PubMed] [Google Scholar]
- 82. Zhao R, Pathak N, Jaffe H, Reese TS, Khan S. 1996. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J. Mol. Biol. 261:195–208 [DOI] [PubMed] [Google Scholar]
- 83. Zhou D, Galan J. 2001. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 3:1293–1298 [DOI] [PubMed] [Google Scholar]


