Abstract
Glutathione- and mycothiol-dependent maleylpyruvate isomerases are known to be involved, respectively, in gentisate catabolism in Gram-negative and high G+C Gram-positive strains. In the present study, a low-G+C Gram-positive Paenibacillus sp. strain, NyZ101, was isolated and shown to degrade 3-hydroxybenzoate via gentisate. A 6.5-kb fragment containing a conserved region of gentisate 1,2-dioxygenase genes was cloned and sequenced, and four genes (bagKLIX) were shown to encode the enzymes involved in the catabolism to central metabolites of 3-hydroxybenzoate via gentisate. The Bag proteins share moderate identities with the reported enzymes in the 3-hydroxybenzoate catabolism, except BagL that had no obvious homology with any functionally characterized proteins. Recombinant BagL was purified to homogeneity as a His-tagged protein and likely a dimer by gel filtration. BagL was demonstrated to be a novel thiol-dependent maleylpyruvate isomerase catalyzing the isomerization of maleylpyruvate to fumarylpyruvate with l-cysteine, cysteinylglycine, or glutathione, as its cofactor. The Km values of these three thiols for BagL were 15.5, 8.4, and 552 μM, respectively. Since cysteine and coenzyme A were reported to be abundant in low-G+C Gram-positive strains, BagL should utilize l-cysteine as its physiological cofactor in vivo. The addition of Ni2+ increased BagL activity, and site-directed mutagenesis experiments indicated that three conserved histidines in BagL were associated with binding to Ni2+ ion and were necessary for its enzyme activity. BagL is the first characterized l-cysteine-dependent catabolic enzyme in microbial metabolism and is likely a new and distinct member of DinB family, with a four-helix-bundle topology, as deduced by sequence analysis and homology modeling.
INTRODUCTION
The gentisate (2,5-dihydroxybenzoate) pathway is one of the important ring-cleavage pathways in the bacterial degradation of various aromatic compounds, such as naphthalene in Ralstonia sp. strain U2 (17, 45) and Rhodococcus sp. strain NCIMB 12038 (26, 28), 3-hydroxybenzoate in Salmonella enterica serovar Typhimurium (18) and Klebsiella pneumoniae M5a1 (22), and xylenol in Pseudomonas alcaligenes NCIMB 9867 (23). This pathway starts with the ring cleavage oxidation of gentisate, catalyzed by gentisate 1,2-dioxygenase (GDO; EC 1.13.11.4) (25). The ring fission product maleylpyruvate is further degraded by either direct hydrolysis to maleate and pyruvate (2, 20) or isomerization to fumarylpyruvate before hydrolysis to fumarate and pyruvate (12, 25, 36, 45). Two phylogenetically unrelated maleylpyruvate isomerases have been identified thus far, and both are known to be low-molecular-weight (LMW) thiol dependent. One is the glutathione (GSH)-dependent enzyme found only in Gram-negative strains including Ralstonia sp. strain U2 (45) and Klebsiella pneumoniae M5a1 (35). The other is the mycothiol (MSH)-dependent enzyme exclusively found in the high-G+C Gram-positive bacteria (Actinobacteria), as demonstrated in Corynebacterium glutamicum ATCC 13032 (16) and Rhodococcus sp. strain NCIMB 12038 (28).
Previous studies have revealed that the richness of LMW thiols in phylogenetically divergent species varied significantly, showing that GSH is abundant in Gram-negative strains, while MSH is abundant in the high-G+C Gram-positive bacteria (14, 31). In low-G+C Gram-positive bacteria Bacillus spp., however, l-cysteine and coenzyme A were the two abundant thiols, whereas both GSH and MSH were at low or undetectable levels (15, 31). Studies on the gentisate catabolism by Bacillus species were previously reported from 1970 to 1980 (8, 10, 11, 13, 19), but their molecular and biochemical basis remains unidentified. This has stimulated interest in investigating whether there is a novel type of thiol-dependent maleylpyruvate isomerase involved in the gentisate pathway in Bacillus spp. or its phylogenetically closely related genera. We report here the identification and characterization of a novel thiol-dependent maleylpyruvate isomerase, distinct from two previous thiol-dependent isomerases, from a newly isolated 3-hydroxybenzoate/gentisate utilizer, Paenibacillus sp. strain NyZ101. This study presents a novel l-cysteine-dependent catabolic enzyme and also provides us with a better understanding of the genetic and biochemical diversity of gentisate catabolic pathways in bacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains, primer sequences for PCR, and plasmids used in the present study are listed in Table 1. Escherichia coli strains were grown in lysogeny broth (LB) at 37°C with 100 μg of ampicillin/ml or 50 μg of kanamycin/ml, as necessary. Paenibacillus sp. strain NyZ101 was grown at 30°C in LB or minimal medium (MM) (27) with 5 mM 3-hydroxybenzoate or 2 mM gentisate as the sole carbon source.
Table 1.
Bacterial strains, plasmids, and primers in this study
| Strain, plasmid, or primer | Characteristic(s)a or sequence (5′–3′) | Reference, source, and/or function |
|---|---|---|
| Strains | ||
| Paenibacillus sp. strain NyZ101 | Wild type; 3-hydoxybenzoate+ gentisate+ | This study |
| E. coli DH5α | supE44 lacU169 (ϕ80lacZΔM15) recA1 endA1 hsdR17 Δthi gyrA96 relA1 | 42 |
| E. coli BL21(DE3) | F− ompT hsdS (rB− mB−) gal dcm lacY1(DE3) | Novagen |
| Plasmids | ||
| pGEM-T | Apr; cloning vector | Promega |
| pET-28a | Kmr; expression vector | Novagen |
| pZWT50 | NdeI-BamHI fragment containing bagI inserted into pET28a | This study |
| pZWT51 | NdeI-EcoRI fragment containing bagL inserted into pET28a | This study |
| pZWT52 | NdeI-EcoRI fragment containing bagK inserted into pET28a | This study |
| pZWT53 | SacI-XhoI fragment containing bagX inserted into pET28a | This study |
| pZWTH46 | pZWT51, in which CAT code for His46 was changed to GCT (alanine) | This study |
| pZWTH143 | pZWT51, in which CAT code for His143 was changed to GCT (alanine) | This study |
| pZWTH147 | pZWT51, in which CAT code for His147 was changed to GCT (alanine) | This study |
| Primers | ||
| g1 | GACTGGATCCATATGGCAGAAAAAATGGAAT | To amplify the bagI gene |
| g2 | ACATGGATCCTAGAATAGGATGGATGAAAG | |
| m1 | GGAGAATTCATATGAGTATTCAGGAAGCGA | To amplify the bagL gene |
| m2 | AGGTGAATTCTGGTACTTCTCGATTTCCT | |
| f1 | CGACGAATTCATATGAAACTAGTGAGCTTTCG | To amplify the bagK gene |
| f2 | TAGAGAATTCGGATCGCTTCCTGAATACTCA | |
| h1 | ACTGAGCTCGCATTGCTTTCATCCATCCT | To amplify the bagX gene |
| h2 | CTGCTCGAGATGTTTTCGTATAGCCGGTT | |
| RTFM | GGCGACATAGTCCGTATTGAA | To amplify 219 bp of the bagK-bagL spanning region |
| CATGGCACAGCACCTCGATAA | ||
| RTMG | CCATCTGCTGGTCGAGCATCT | To amplify 219 bp of the bagK-bagL spanning region |
| CAGGGATGGCATTCCAGAGAG | ||
| RTGH | TCTTCTCTGTCAGCGATCTG | To amplify 207 bp of bagL-bagI spanning region |
| CCAGCACTTGAACGGATTTG | ||
| RT16S | GCTTGAGTGCAGGAGAGGAAA | To amplify a 110-bp fragment of 16S rDNA |
| TCAGCGTCAGTTACAGTCCAG |
+, The ability to grow on the indicated substrates. Apr, ampicillin resistance; Kmr, kanamycin resistance.
Chemicals.
3-Hydroxybenzoate, gentisate, cysteinylglycine (Cys-Gly), and γ-glutamylcysteine (γ-Glu-Cys) were purchased from Sigma Chemical Co. (St. Louis, MO). GSH, l-cysteine, and coenzyme A (CoA) were from Bio Basic, Inc. (Ontario, Canada). Maleylpyruvate was prepared by the oxidation of gentisate with purified nagI-encoded GDO from Ralstonia sp. strain U2 as described previously (16). Fumarylpyruvate was prepared via sequential reaction by GDO and nagL-encoded GSH-dependent maleylpyruvate isomerase from Ralstonia sp. strain U2 (45). MSH was kindly supplied by Shuang-Jiang Liu of Institute of Microbiology, Chinese Academy of Sciences.
Strain isolation.
Soil samples were collected from garden soil in Wuhan, China. In order to isolate bacterial strains with heat-resistant spores, soil suspensions in 5 ml of sterile water was incubated at 80°C for 25 min before it was enriched with 3-hydroxybenzoate as the sole source of carbon and energy in MM at 30°C. After several cycles, 50 μl of sample was spread onto MM agar containing 5 mM 3-hydroxybenzoate to screen for colonies capable of growing on 3-hydroxybenzoate.
Cloning of 3-hydroxybenzoate/gentisate degradation genes and sequence analyses.
Primers (GDO-BSF, 5′-TGAAGTCGCTCCATCCCATCGACATAT-3′; GDO-BSR, 5′-GGATTGATGTAATCTACAGCGTATCC-3′) were designed based on a conserved region of GDO genes from Bacillus spp. (accession nos. BAB05721, EDX59360, and EEN92715) to amplify potential DNA fragment encoding GDO from strain NyZ101. A 403-bp fragment was PCR amplified from the genomic DNA of strain NyZ101 and ligated into pGEM-T vector (Promega Corp., Madison, WI) for sequencing. The flanking regions of the fragment obtained above were cloned by Genome walking strategy (37). DNA sequences were determined by Invitrogen Technologies Co. (Shanghai, China). Analyses of open reading frames (ORFs) and an amino acid identity search were performed using ORF finder programs and BLASTX on NCBI website (43). The model three-dimensional (3D) structure of BagL was generated by Swiss-Model (1) using the chain A of YfiT (PDB code 1rxq) from Bacillus subtilis as the template structure. The structure figures were performed using PyMOL (http://www.pymol.org/). The phylogenetic tree was generated by MEGA5 software (39).
Cloning and expression of bag genes.
Individual bag genes were amplified by PCR from genomic DNA of strain NyZ101 with Pyrobest polymerase (TaKaRa, Dalian, China). The products were then cloned into pET28a to produce plasmids listed in Table 1, resulting in N-terminal His6-tagged fusion proteins. E. coli BL21(DE3) strains carrying the recombinant plasmids were grown in LB, supplemented with 50 μg of kanamycin/ml, at 37°C to an optical density at 600 nm of 0.6 and then induced with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 6 h at 16°C.
Preparation of cell extracts.
Washed cells were suspended in ice-cold phosphate buffer (50 mM, pH 8.0) and then disrupted by sonification at 4°C (3-s sonication, followed by a 7-s break). After centrifugation at 13,000 × g for 40 min at 4°C, the cell supernatant was collected for enzyme assay or protein purification.
Protein purification.
Cell extracts containing His-tagged BagL protein (H6-BagL) were applied to nickel-nitrilotriacetic acid (Ni2+-NTA) agarose (Merck Biosciences, Darmstadt, Germany) according to the supplier's recommendations. The column containing the extracts was washed with binding buffer (50 mM sodium phosphate buffer, 300 mM NaCl and 20 mM imidazole [pH 8.0]), and the protein was released with elution buffer (50 mM sodium phosphate buffer, 300 mM NaCl, 120 mM imidazole [pH 8.0]) at a flow rate 1.0 ml/min. The purified protein was dialyzed against 50 mM sodium phosphate buffer (pH 8.0) in dialysis bags and concentrated by ultrafiltration through an Ultra-15 filter unit with a molecular mass cutoff value of 10 kDa (Millipore, Bedford, MA) and then stored at 4°C.
Molecular weight determination.
The molecular weight was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Gel filtration chromatography of purified H6-BagL was performed to determine the native molecular mass of the purified protein by using a Superdex 200 10/300 GL column (Amersham Pharmacia Biotech) on AKTÄ fast-performance liquid chromatography system (Amersham Pharmacia Biotech) (44). The column was equilibrated and eluted with 50 mM phosphate buffer (pH 7.2) that contained 0.15 M NaCl at a flow rate 0.5 ml/min. The native molecular mass was estimated from a calibration curve plotted with carbonic anhydrase (Mr, 29,000), bovine serum albumin (Mr, 66,000), alcohol dehydrogenase (Mr, 150,000), β-amylase (Mr, 200,000), apoferritin (Mr, 443,000), and thyroglobulin (Mr, 669,000) (Sigma Chemical Co.).
Enzyme assays.
The assay of maleylpyruvate isomerase was initiated by addition of maleylpyruvate. The reaction mixtures (1.0 ml) contained 70 μM substrate, 20 μM single thiol, and 1 to 5 μg of purified protein in 50 mM Tris-HCl buffer (pH 8.0). The rate of change was measured at A330, the molar extinction coefficient was taken as 2,400 M−1 cm−1 (19, 36). For the determination of specific activity in kinetic measurement, the rate of change of A330 was measured in the presence of excess fumarylpyruvate hydrolase as described previously (19), the molar extinction coefficient for maleylpyruvate was taken as 13,000 M−1 cm−1 (25). The fumarylpyruvate hydrolase used here was purified NagK from strain U2 (45) or BagK from strain NyZ101 in the present study. The 3-hydroxybenzoate 6-monooxygenase activity was determined by measuring the decrease in the absorbance at 340 nm due to the substrate-dependent oxidation of NADH, the molar extinction coefficient of which was taken as 6,200 M−1 cm−1 (40). GDO was assayed by measuring the increase in absorbance at 330 nm due to conversion of gentisate to maleylpyruvate; the molar extinction coefficient was taken as 13,000 M−1 cm−1 (25). Fumarylpyruvate hydrolase was assayed by measuring the decrease at 345 nm due to fumarylpyruvate disappearance; the molar extinction coefficient of fumarylpyruvate was taken as 12,000 M−1 cm−1 (19). All of the assays were conducted in 50 mM Tris-HCl buffer at pH 8.0. Protein concentration was determined according to the Bradford method (6), with bovine serum albumin as the standard. One unit of enzyme activity was defined as the amount required for the disappearance (or production) of 1 μmol of substrate (or product) per min at 30°C. Specific activities are expressed as units per milligram of protein.
Kinetic measurements of BagL.
Michaelis-Menten kinetics of the maleylpyruvate isomerization catalyzed with BagL were identified by plotting reaction rates against seven maleylpyruvate concentrations ranging from 2 to 200 μM from three independent sets of experiments, with the concentration of l-cysteine fixed at 400 μM. For thiol compounds, the concentration of maleylpyruvate was fixed at 100 μM, whereas the concentration of l-cysteine varied from 1 to 400 μM, the concentration of Cys-Gly varied from 0.5 to 400 μM, and the concentration of GSH varied from 20 to 2,000 μM. The Km and Vmax values were determined by nonlinear regression analysis of the plots of the initial reaction rates and substrate concentrations by using Grafit 7.0 software (R. J. Leatherbarrow, Erithicus Software, Ltd., Horley, United Kingdom).
RT-PCR and real-time PCR.
The total RNA was isolated from strain NyZ101 with an RNAprep pure bacterial kit (Tiangen Biotech, Beijing, China). DNase digestion was carried out to remove any DNA contamination. Reverse transcription-PCR (RT-PCR) was performed by using Moloney murine leukemia virus reverse transcriptase (Promega Corp., Madison, WI) and then amplifying the resulting cDNAs with gene-specific primers for each bag gene. cDNAs obtained from the RT-PCR were used as the template by RT-Cycle (CapitalBio Corp., Beijing, China). The primers used for the RT-PCR are listed in Table 1. A 110-bp fragment of the 16S rRNA gene of strain NyZ101 was used as the reference to evaluate the relative differences in the integrity of individual RNA samples. Real-time PCR was conducted with a commercial kit (RealMasterMix, SYBR green; Tiangen Biotech). The amplification conditions were as follows: denaturation at 95°C for 300 s, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 62°C for 30 s, and extension at 68°C for 30 s. Post-PCR melting curves confirmed the specificity of single-target amplification. The 2−ΔΔCT method was used to calculate relative changes in gene expression (29).
HPLC analysis.
High-pressure liquid chromatography (HPLC) analysis of catabolic products was performed on an Agilent series 1200 system (Agilent Technologies, Palo Alto, CA) equipped with a C18 reversed-phase column (5 μm, 4.6 by 250 mm; Agilent Technologies). For analysis of 3-hydroxybenzoate and gentisate, the mobile phase was methanol-water (20:80 [vol/vol]) containing 1% acetic acid at a flow rate of 1 ml/min. Detection was performed by UV at 254 nm. The retention times for 3-hydroxybenzoate and gentisate were 15.9 and 10.0 min, respectively. The protocol for analysis of fumarate and pyruvate were as described previously (45), and the retention times were 15.1 and 9.1 min for fumarate and pyruvate, respectively.
Site-directed mutagenesis.
Site-directed mutagenesis of BagL was carried out by overlapped PCR. Three conserved histidines (His46, His143, and His147) were replaced by alanine, respectively. The PCR fragments with mutagenesis were then ligated into pET28a to produce plasmids pZWTH46, pZWTH143, and pZWTH147, respectively. The recombinant plasmids were sequenced to ensure that the mutations occurred as desired and that no unintended mutation had been incorporated during the PCR.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the nucleotide sequence reported here are HQ897157 for the incomplete 16S rRNA gene of strain NyZ101 and HQ897158 for the 3-hydroxybenzoate/gentisate degradation gene cluster.
RESULTS
Isolation and characterization of the 3-hydroxybenzoate/gentisate-degrading strain NyZ101.
After several cycles of enrichment, a 3-hydroxybenzoate-degrading bacterial strain, designated NyZ101, was isolated with 3-hydroxybenzoate as the sole source of carbon and energy. It was identified as a Paenibacillus sp. based on sequence analysis of its 16S rRNA gene (sharing more than 97% identity with several Paenibacillus spp.). It was also able to grow on gentisate but not on salicylate, 4-hydroxybenzoate, or catechol. Gentisate was identified as the product of 3-hydroxybenzoate catabolism by HPLC analysis. Addition of the cell extracts of 3-hydroxybenzoate-grown culture to a cuvette containing maleylpyruvate caused an obvious shift in absorbance from 330 to 345 nm, indicating isomerization from maleylpyruvate (λmax, 330 nm) to fumarylpyruvate (λmax, 345 nm) at pH 8.0 (19). This suggested that a maleylpyruvate isomerase was involved in the gentisate catabolism of this newly isolated strain.
Cloning and sequence analyses of the 3-hydroxybenzoate/gentisate degradation gene cluster.
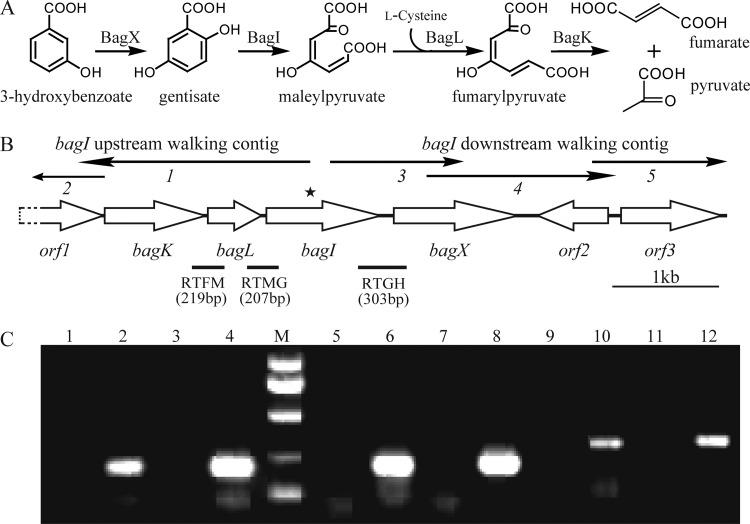
A pair of primers based on a conserved region of the GDO genes was used to amplify a PCR product with an anticipated size of 403 bp. A genome-walking strategy for both directions was used to obtain the flanking sequence of this fragment, as shown in Fig. 1B. After five cycles of genome walking, a 6,537-bp DNA fragment around the GDO gene was obtained, and six complete ORFs and one truncated ORF (orf1) were deduced (Fig. 1B). These genes have been designated the bag genes (Bacillus gentisate pathway). Among the products encoded by these genes, BagX, BagI, and BagK exhibit moderate identities with the enzymes involved in the gentisate pathway from different bacterial genera. Interestingly, although no obvious homology was found between BagL and any functionally identified proteins, it is similar to a group of hypothetical proteins with 39 to 48% identities in Bacillus and Geobacillus spp. On the other hand, all three gentisate catabolism-encoding genes were usually found to be clustered together in previous studies (21, 28, 35, 36, 45). Given the fact that only the gene encoding the maleylpyruvate isomerase of known functions is “missing” in the bag cluster, bagL is likely to encode a novel type of maleylpyruvate isomerase.
Fig 1.
(A) Proposed pathway for the 3-hydroxybenzoate catabolism via gentisate in Paenibacillus sp. strain NyZ101, together with the catabolic reactions catalyzed by the bag gene products in vivo. (B) Organization of the bag gene cluster of Paenibacillus sp. strain NyZ101. The arrows indicate the size and direction of transcription of each gene or ORF. The star above bagI indicates the region where the genome walking started in both directions. The thin arrows above the gene cluster represent the fragments obtained by genome-walking reactions. The locations of the primer sets RTFM, RTMG, and RTGH and the amplified DNA fragments for RT-PCR are indicated below. (C) Analysis of bagILKX transcription by RT-PCR. Reactions performed without RT were used as negative controls, as shown in lanes 1, 3, 5, 7, 9, and 11. Lane M, molecular size markers at 1,000, 750, 500, 250, and 100 bp. Lanes 2, 6, and 10 (template from succinate-grown strain NyZ101) and lanes 4, 8, and 12 (template from succinate-grown strain NyZ101 induced by 3HBA) show products amplified using the RTFM, RTMG, and RTGH primer sets, respectively, with products of RT.
Overexpression and purification of H6-BagL.
Recombinant BagL was overexpressed in E. coli BL21(DE3) as an N-terminal His6-tagged fusion protein for easy purification. A substantial amount of soluble and active H6-BagL was synthesized and purified to apparent homogeneity by Ni2+-NTA affinity chromatography. After 49 mg of cell extract protein with a specific activity of 0.15 U/mg was applied to 5 ml of Ni2+-NTA agarose beads, 2.9 mg of H6-BagL was purified with a specific activity of 18.9 U/mg. A single polypeptide with an apparent molecular mass of 19 kDa was detected by SDS-PAGE (Fig. 2), as expected from the deduced amino acid composition. The result of gel filtration chromatography for H6-BagL showed a peak value corresponding to 33 kDa, indicating that the native BagL is likely a dimer.
Fig 2.
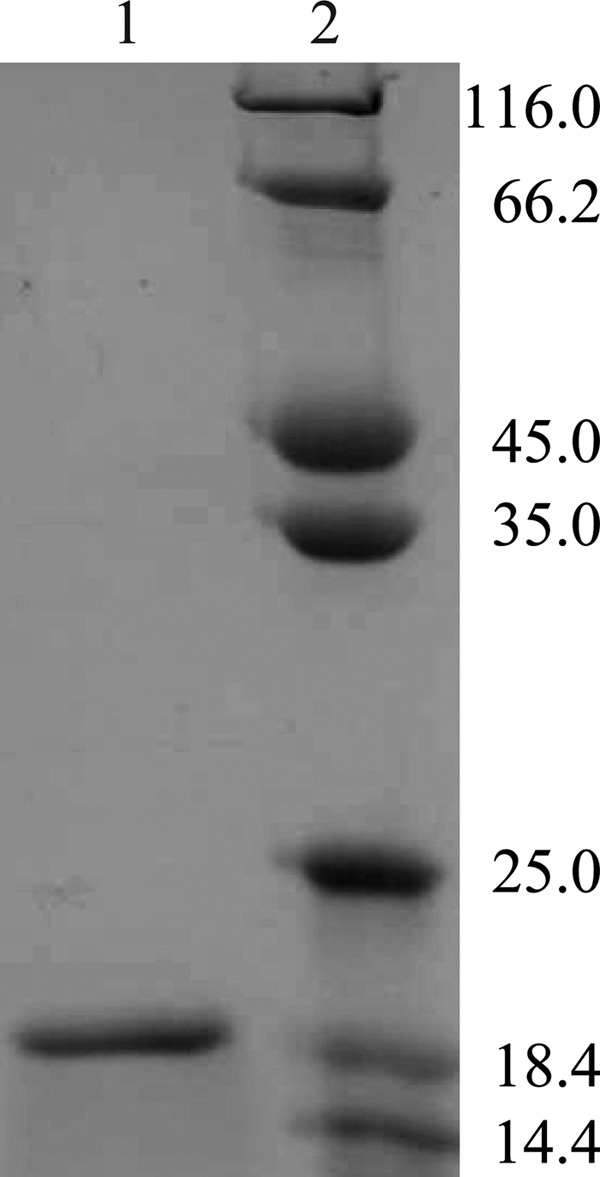
SDS-PAGE of H6-BagL. Lane 1, purified H6-BagL; lane 2, molecular mass standards. Molecular masses are indicated in kilodaltons on the right.
BagL is a novel l-cysteine-dependent maleylpyruvate isomerase.
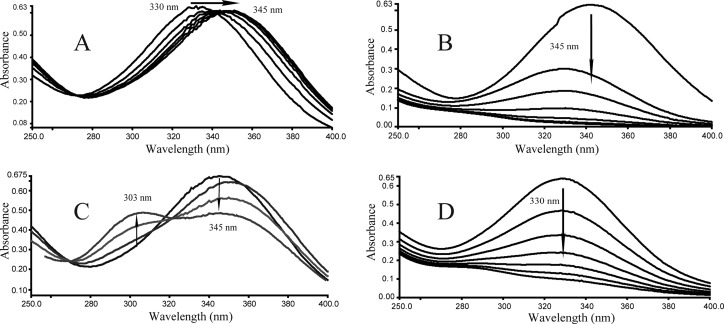
When purified H6-BagL was incubated with maleylpyruvate, no change in the absorption spectrum between 250 to 400 nm took place. Since l-cysteine and CoA are known to be the abundant thiol compounds in low G+C Gram-positive strains, they were added individually into the reaction to test whether they are the cofactors for this isomerase. No activity for BagL was observed when CoA was introduced into the reaction. In the presence of l-cysteine, however, rapid transformation of maleylpyruvate (λmax, 330 nm) occurred to form a new peak with a λmax of 345 nm and with a lower extinction coefficient (Fig. 3A), a finding in line with the spectral properties of fumarylpyruvate at pH 8.0 (19). The formed fumarylpyruvate was then able to be degraded by nagK-encoded fumarylpyruvate hydrolase from strain U2 (45) to yield fumarate and pyruvate (Fig. 3B), which were confirmed by HPLC analysis. Noticeably, the presence of excess l-cysteine (20 μM) in the isomerization resulted in the formation of a product with a λmax of ∼303 nm (Fig. 3C). This is similar to the phenomenon described by Lack (24) and Zhou (45) during the GSH-dependent isomerization in the presence of excess of GSH, which was assumed to be the result of a spontaneous nonenzymatic reaction between GSH and fumarylpyruvate. In this case, it is likely the reaction between the l-cysteine and fumarylpyruvate. However, synergistic effects of BagL and NagK resulted in maleylpyruvate disappearance without observing the formation of the product with a λmax of ∼303 nm, in the presence of excess l-cysteine, as shown in Fig. 3D. This also suggested the product of a λmax of ∼303 nm was formed between the l-cysteine and fumarylpyruvate.
Fig 3.
(A) Isomerization of maleylpyruvate (λmax, 330 nm) to fumarylpyruvate (λmax, 345 nm) catalyzed by H6-BagL. Sample and reference cuvettes contained 10 μM l-cysteine, 50 mM Tris-HCl buffer (pH 8.0), and 4.8 μg of purified H6-BagL in a 1.0-ml mixture. The reaction was initiated by the addition of 50 μM maleylpyruvate. The spectra were recorded every 30 s after the addition of maleylpyruvate. (B) Hydrolysis of fumarylpyruvate by nagK-encoded fumarylpyruvate hydrolase. After the isomerization of maleylpyruvate to fumarylpyruvate, 30 μg of purified H6-NagK was added to both sample and reference cuvettes from the reaction depicted in panel A, and the spectra were recorded every 30 s. (C) Formation of a product with a λmax of ∼303 nm in the presence of excess l-cysteine. The experiment was performed as described for panel A, except the l-cysteine was used at a concentration of 20 μM. (D) Spectrophotometric changes during the transformation of maleylpyruvate catalyzed by both H6-BagL and H6-NagK in the presence of excess l-cysteine. The experiment was performed as described for panel C, except that fumarylpyruvate hydrolase was also present.
Cofactor specificity of BagL.
Several thiol compounds were also tested to determine whether BagL exhibited extended cofactor substrate specificity. Interestingly, H6-BagL also showed activity with GSH and Cys-Gly, but no activities were observed with γ-Glu-Cys, MSH, β-mercaptoethanol, or dithiothreitol as cofactors. H6-BagL exhibited approximately one-eighth and 2-fold of the activity when GSH and Cys-Gly replacing l-cysteine, respectively. When GSH was preincubated with H6-BagL for 1 min before the reaction started, the enzyme activity was increased by 30%. This phenomenon, however, was not observed when l-cysteine or Cys-Gly was used as the cofactor. In addition, H6-BagL exhibited much higher affinity with l-cysteine and Cys-Gly than GSH. The kinetic parameters of the thiol substrate of BagL are shown in Table 2.
Table 2.
Kinetic parameters of the thiol substrate of BagL
| Thiol | Mean parameter for thiol ± SDa |
||
|---|---|---|---|
| Km (μM) | kcat (min−1) | kcat/Km (μM−1 min−1) | |
| Cysteine | 15.4 ± 2.1 | 4,286 ± 142 | 277 ± 28 |
| Cysteinyl glycine | 8.4 ± 1.3 | 4,489 ± 163 | 536 ± 60 |
| GSH | 552 ± 78 | 6,129 ± 309 | 11.1 ± 3.9 |
The kinetic constants were calculated by nonlinear regression analysis. kcat values were calculated on the basis of a subunit Mr of 19,000. The values are expressed as means calculated from triplicate assays.
Biochemical properties of BagL.
The Km value of H6-BagL for maleylpyruvate was 19.5 ± 5.6 μM. Maximum activity was exhibited at pH 8.3 in the range of pH 7.0 to 8.9 in 50 mM Tris-HCl buffer. The purified H6-BagL was fairly stable in the elution buffer on ice. Long-term storage of the purified enzyme seemed possible, since 85% of the activity remained after 1 month when the enzyme was kept at 4°C. When l-cysteine was used as its cofactor, H6-BagL activity was increased by 20% after 0.2 mM metal ion Ni2+ was incubated with the enzyme for 15 min at room temperature. However, the activity was increased by 90% with Cys-Gly as its cofactor. Other divalent metal ions, including 0.2 mM Mg2+, Ca2+ Cu2+, and Zn2+, inhibited the activity by 20 to 53%.
Residues in histidine triads are necessary for BagL activity.
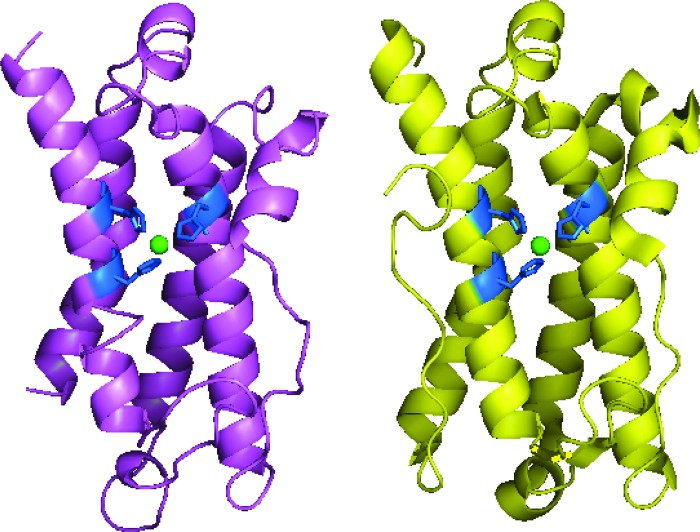
With DinB superfamily member YfiT's 3D structure (PDB code 1rxq) as the template, the 3D structure model of BagL was constructed (see Fig. 5). The same histidine triad (His46-His143-His147) was also identified to be probably associated with the bound of metal ion Ni2+ (see Fig. 5) as in YfiT in the DinB superfamily (34). After changing the histidines to alanines, all three mutants (H46A, H143A, and H147A) were devoid of enzyme activity.
Fig 5.
(Left) Diagram of BagL, shown in purple. (Right) Diagram of YfiT (PDB code 1rxq), chain A, shown in yellow. The Ni2+ ion contained in the structure is shown as green sphere. The Ni2+-coordinating histidine triads are represented as sticks in blue.
bagX, bagI, and bagK encode functional enzymes involved in 3-hydroxybenzoate catabolism via gentisate.
To identify the functions of other genes in the bag cluster, bagX, bagI, and bagK, were cloned into pET28a to produce pZWT53, pZWT50, and pZWT52, respectively. SDS-PAGE analyses of cell extracts of E. coli strain BL21(DE3) containing these constructs showed elevated levels of polypeptides of ∼45, ∼43, and ∼35 kDa, as expected from the deduced amino acid compositions of H6-BagX, H6-BagI, and H6-BagK, respectively. The same extracts were used for the identifications of their functions. BagX acted as a 3-hydroxybenzoate 6-monooxygenase converting 3-hydroxybenzoate to gentisate in the presence of NADH with a specific activity of 0.16 U/mg. The activity was increased by 70% when 10 μM FAD was added. Gentisate was confirmed as the product of 3-hydroxybenzoate monooxygenation by HPLC analysis. BagI is a GDO that catalyzes the ring cleavage of gentisate to maleylpyruvate, with a specific activity of 5.6 U/mg. BagK acted as a fumarylpyruvate hydrolase with a specific activity of 0.45 U/mg. Together or sequentially, the products from four bag genes with added l-cysteine were able to convert 3-hydroxybenzoate to pyruvate and fumarate via gentisate in vitro, and it is reasonable to propose that this is also true in vivo (Fig. 1A).
The bag cluster is highly transcribed in 3-hydroxybenzoate-induced cells of strain NyZ101.
RT-PCR was performed with RNA derived from the succinate-grown cultures of strain NyZ101, with or without the induction of 3-hydroxybenzoate. The results showed that the transcription of bag cluster was initiated under both situations, and the bagXILK genes were suggested to be a cluster transcribed together, as shown in Fig. 1C. Real-time quantitative PCR analysis showed a 57-fold change in gene transcription of the bag cluster from the 3-hydroxybenzoate-induced samples compared to the data from the noninduced samples. This suggested that although the induction was not stringent, the transcription efficiency of the 3-hydroxybenzoate-gentisate catabolic cluster was significantly enhanced in the presence of 3-hydroxybenzoate.
DISCUSSION
It was generally thought that the maleylpyruvate isomerization in the gentisate catabolism took place either in a GSH-dependent (12, 25, 45) or GSH-independent (12, 28, 36) manner, and NagL was studied from Gram-negative Ralstonia sp. strain U2 as a representative of GSH-dependent maleylpyruvate isomerase (30, 45). This was the view that had been upheld until recent identification of the MSH-dependent maleylpyruvate isomerase NCgl2918 in Corynebacterium glutamicum (16, 41). In the present study, a novel l-cysteine-dependent maleylpyruvate isomerase, BagL, was identified from a newly isolated Paenibacillus strain. This particular finding provides yet another example leading to a better understanding of the genetic and biochemical diversity of gentisate pathway in bacteria. Previous reports have demonstrated that the content of LMW thiols among bacterial genera is significantly different (15, 31). In Gram-negative species, GSH is considered to play a key role against oxygen toxicity. MSH, which functions in a manner similar to that of GSH, is a dominant thiol in high-G+C Gram-positive actinomycetes, whereas l-cysteine and coenzyme A are the major thiols in bacillus, the representative of low G+C group (31). It seems that the availability and richness of the thiol types present in the phylogenetically divergent species have been playing key roles in the evolution of the thiol-dependent enzymes, as seen in the maleylpyruvate isomerase from three phylogenetically divergent bacterial genera. The only similar case reported is that a fosfomycin resistance protein (FosA) from Gram-negative strains is dependent on GSH (3, 4), but its isoenzyme FosB in B. subtilis uses l-cysteine as the cofactor (7). However, this enzyme, unlike BagL in present study, is not involved in a metabolic pathway. Notably, Cys-Gly was able to function as a cofactor of BagL that was as efficient as l-cysteine, although its availability in bacteria was not documented. Given the fact that Cys-Gly was reported to be produced during the process of GSH metabolism (38) and GSH is undetectable or at a low level in low-G+C Gram-positive strains (15, 31), Cys-Gly is unlikely the physiological thiol for BagL instead of cysteine in this case.
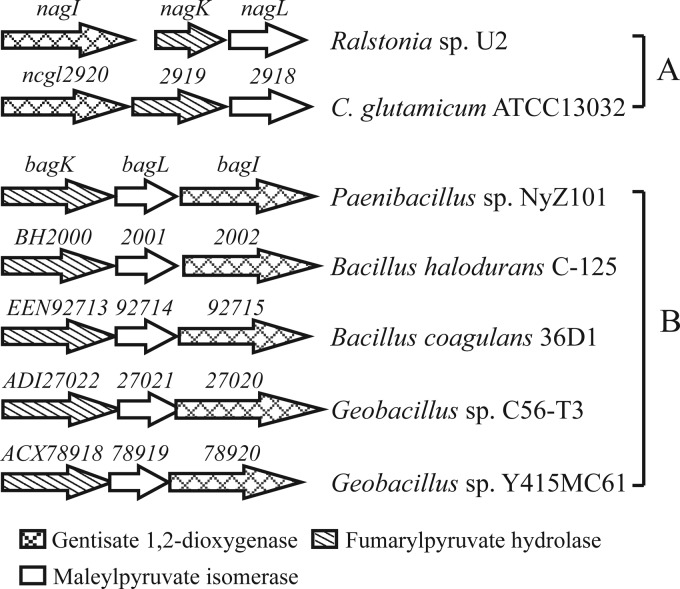
Despite the fact that the three thiol-dependent maleylpyruvate isomerases involved in the gentisate pathway are phylogenetically unrelated proteins, the other two enzymes (GDO and fumarylpyruvate hydrolase) are conserved. The three gentisate catabolism-encoding genes are prone to be clustered together in either the clusters involving GSH- and MSH-dependent isomerization pathways (Fig. 4A) or the bag cluster and its homologs from the genome sequences of Bacillus and its phylogenetically related genera (Fig. 4B). Indeed, in Bacillus halodurans C-125, BH1999, located immediately next to genes BH2000 to BH2002 (Fig. 4B), has been identified to code for a gentisyl-CoA thioesterase catalyzing the hydrolysis of gentisyl-coenzyme A to gentisate (46). Thus, it is likely that the products from BH2000 to BH2002 are also involved in gentisate catabolism, with BH2001 being an l-cysteine-dependent maleylpyruvate isomerase.
Fig 4.
Comparison of genetic organization of gentisate catabolic gene clusters from different strains. (A) Cluster containing GSH-dependent maleylpyruvate isomerase-encoding nagL in Ralstonia sp. strain U2 (45) and cluster containing MSH-dependent maleylpyruvate isomerase-encoding gene ncgl2918 in Corynebacterium glutamicum (36). (B) Cluster containing l-cysteine-dependent maleylpyruvate isomerase-encoding gene bagL in Paenibacillus sp. strain NyZ101 with its homologous clusters in Bacillus halodurans C-125 (accession no. NC_002570.2), Bacillus coagulans 36D1 (NC_016023.1), Geobacillus sp. strain C56-T3 (NC_014206.1), and Geobacillus sp. stain Y415MC61 (NC_013411.1).
Of these three different maleylpyruvate isomerases, BagL (18.2 kDa with 162 residues) is significantly smaller in molecular size than both NagL (23.5 kDa with 212 residues) and NCgl2918 (26.4 kDa with 241 residues) and does not show homology to either of the two known isomerases NagL and NCgl2918. NagL acts as a dimer and is a member of the glutathione S-transferase zeta family, including a motif of S(X)4RXRIAL as the distinctive feature (5, 30). No metal ion was reported to participate in the catalysis by NagL. Searches conducted in the InterProScan database (33) suggested that BagL is possibly a member of DinB superfamily. They are a group of proteins containing a single domain with four-helix-bundle topology, but few of them have identified functions. The MSH-dependent maleylpyruvate isomerase NCgl2918 was thought to be the only functionally identified protein in this family (see below for this expression). NCgl2918 is a monomer and its N-terminal domain is a Zn2+-binding domain with the motif His52-Glu144-His148, while its C-terminal domain is necessary for its enzyme activity and stability (41). Based upon its crystal structure, NCgl2918 contains a DinB-like moiety as its N-terminal domain (42). Based on alignment of the 3D structure between modeling BagL and NCgl2918 (PDB code 2nsf), a 65.6% structure overlap was deduced by click server (http://mspc.bii.a-star.edu.sg/minhn/click.html). However, truncated NCgl2918 without a C-terminal domain did not show any activities against maleylpyruvate (34). As deduced from these DinB superfamily structures, a histidine triad has been suggested to coordinate with the metal ion Ni2+ (9). Indeed, the same histidine triad—His46-His143-His147—is also present in BagL when its 3D structure model was constructed (Fig. 5) using DinB superfamily member YfiT as the template. YfiT is a probable metal-dependent hydrolase with four-helix bundle topology in which three histidines were located around the center metal ion Ni2+ (34). In the present study, the increased activity of H6-BagL by the addition of Ni2+ suggested that Ni2+ was likely involved in the catalytic process. The complete loss of enzyme activity in three BagL mutants indicated that the three histidines are necessary for its activity and probably function in binding with Ni2+.
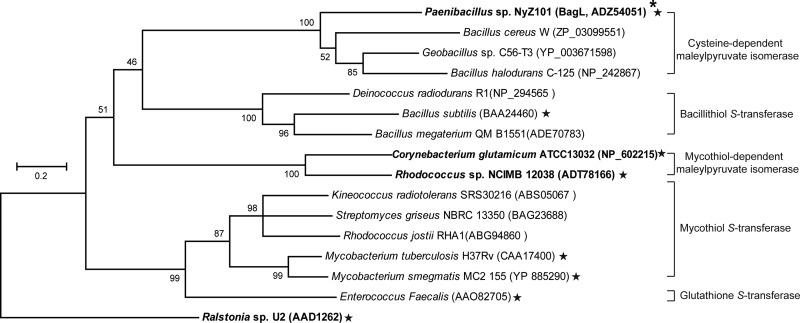
During the preparation of the present study the characterization of three additional enzymes in DinB superfamily was reported to suggest that thiol-dependent S-transferase activity is dominant in this family, including MSH, bacillithiol (BSH), and GSH S-transferases (32). In particular, B. subtilis YfiT, mentioned above, was shown to have BSH S-transferase activity (32). As shown in the phylogenetic tree of the DinB superfamily in Fig. 6, the l-cysteine-dependent maleylpyruvate isomerase represented by BagL forms an independent clade with a closer relationship to BSH S-transferases or MSH-dependent maleylpyruvate isomerase than to MSH S-transferases. Therefore, BagL, the first characterized metabolic enzyme with l-cysteine as its thiol cofactor, is likely a new, distinct member in the DinB superfamily.
Fig 6.
Phylogenetic relationship between l-cysteine-dependent maleylpyruvate isomerase BagL and selected members of the DinB superfamily. The phylogenetic tree was constructed by using the neighbor-joining method with MEGA5 software. The bootstrap confidence limits are indicated at the nodes, and the scale on the left indicates sequence divergence. BagL is marked by a an asterisk (*). The proteins of known function were indicated by a star (★). The GSH-dependent maleylpyruvate isomerase NagL from Ralstonia sp. strain U2 is used as the outgroup. The three types of maleylpyruvate isomerase are indicated in boldface.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (grant 30730002) and by the National Key Basic Research Program of China (973 Program, grant 2012CB725202).
Footnotes
Published ahead of print 25 May 2012
REFERENCES
- 1. Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22:195–201 [DOI] [PubMed] [Google Scholar]
- 2. Bayly RC, Chapman PJ, Dagley S, Diberardino D. 1980. Purification and some properties of maleylpyruvate hydrolase and fumarylpyruvate hydrolase from Pseudomonas alcaligenes. J. Bacteriol. 143:70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beharry Z, Palzkill T. 2005. Functional analysis of active site residues of the fosfomycin resistance enzyme FosA from Pseudomonas aeruginosa. J. Biol. Chem. 280:17786–17791 [DOI] [PubMed] [Google Scholar]
- 4. Bernat BA, Laughlin LT, Armstrong RN. 1997. Fosfomycin resistance protein (FosA) is a manganese metalloglutathione transferase related to glyoxalase I and the extradiol dioxygenases. Biochemistry 36:3050–3055 [DOI] [PubMed] [Google Scholar]
- 5. Board PG, Baker RT, Chelvanayagam G, Jermiin LS. 1997. Zeta, a novel class of glutathione transferases in a range of species from plants to humans. Biochem. J. 328(Pt 3):929–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 7. Cao M, Bernat BA, Wang Z, Armstrong RN, Helmann JD. 2001. FosB, a cysteine-dependent fosfomycin resistance protein under the control of sigma(W), an extracytoplasmic-function sigma factor in Bacillus subtilis. J. Bacteriol. 183:2380–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clark JS, Buswell JA. 1979. Catabolism of gentisic acid by two strains of Bacillus stearothermophilus. J. Gen. Microbiol. 112:191–195 [Google Scholar]
- 9. Cooper DR, Grelewska K, Kim CY, Joachimiak A, Derewenda ZS. 2010. The structure of DinB from Geobacillus stearothermophilus: a representative of a unique four-helix-bundle superfamily. Acta Crystallogr. Sect F Struct. Biol. Crystallogr. Commun. 66:219–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Crawford RL. 1975. Degradation of 3-hydroxybenzoate by bacteria of the genus Bacillus. Appl. Microbiol. 30:439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crawford RL. 1976. Pathways of 4-hydroxybenzoate degradation among species of Bacillus. J. Bacteriol. 127:204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Crawford RL, Frick TD. 1977. Rapid spectrophotometric differentiation between glutathione-dependent and glutathione-independent gentisate and homogentisate pathways. Appl. Environ. Microbiol. 34:170–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Crawford RL, Olson PE. 1979. Catabolism of 2-hydroxybenzoate by Bacillus species. FEMS Microbiol. Lett. 5:193–195 [Google Scholar]
- 14. Fahey RC. 2001. Novel thiols of prokaryotes. Annu. Rev. Microbiol. 55:333–356 [DOI] [PubMed] [Google Scholar]
- 15. Fahey RC, Brown WC, Adams WB, Worsham MB. 1978. Occurrence of glutathione in bacteria. J. Bacteriol. 133:1126–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feng J, et al. 2006. The gene ncgl2918 encodes a novel maleylpyruvate isomerase that needs mycothiol as cofactor and links mycothiol biosynthesis and gentisate assimilation in Corynebacterium glutamicum. J. Biol. Chem. 281:10778–10785 [DOI] [PubMed] [Google Scholar]
- 17. Fuenmayor SL, Wild M, Boyes AL, Williams PA. 1998. A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J. Bacteriol. 180:2522–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goetz FE, Harmuth LJ. 1992. Gentisate pathway in Salmonella typhimurium: metabolism of m-hydroxybenzoate and gentisate. FEMS Microbiol. Lett. 76:45–49 [DOI] [PubMed] [Google Scholar]
- 19. Hagedorn SR, Bradley G, Chapman PJ. 1985. Glutathione-independent isomerization of maleylpyruvate by Bacillus megaterium and other gram-positive bacteria. J. Bacteriol. 163:640–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hopper DJ, Chapman PJ, Dagley S. 1968. Enzymic formation of d-malate. Biochem. J. 110:798–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jeon CO, Park M, Ro HS, Park W, Madsen EL. 2006. The naphthalene catabolic (nag) genes of Polaromonas naphthalenivorans CJ2: evolutionary implications for two gene clusters and novel regulatory control. Appl. Environ. Microbiol. 72:1086–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones DC, Cooper RA. 1990. Catabolism of 3-hydroxybenzoate by the gentisate pathway in Klebsiella pneumoniae M5a1. Arch. Microbiol. 154:489–495 [DOI] [PubMed] [Google Scholar]
- 23. Laapoh C, Bayly RC. 1980. Evidence for isofunctional enzymes used in meta-cresol and 2,5-xylenol degradation via the gentisate pathway in Pseudomonas alcaligenes. J. Bacteriol. 143:59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lack L. 1961. Enzymic cis-trans isomerization of maleylpyruvic acid. J. Biol. Chem. 236:2835–2840 [PubMed] [Google Scholar]
- 25. Lack L. 1959. The enzymic oxidation of gentisic acid. Biochim, Biophys, Acta 34:117–123 [DOI] [PubMed] [Google Scholar]
- 26. Larkin MJ, Day MJ. 1986. The metabolism of carbaryl by three bacterial isolates, Pseudomonas spp. (NCIB 12042 and 12043) and Rhodococcus sp. (NCIB 12038) from garden soil. J. Appl. Bacteriol. 60:233–242 [DOI] [PubMed] [Google Scholar]
- 27. Liu DQ, Liu H, Gao XL, Leak DJ, Zhou NY. 2005. Arg169 is essential for catalytic activity of 3-hydroxybenzoate 6-hydroxylase from Klebsiella pneumoniae M5a1. Microbiol. Res. 160:53–59 [DOI] [PubMed] [Google Scholar]
- 28. Liu TT, et al. 2010. Functional characterization of a gene cluster involved in gentisate catabolism in Rhodococcus sp. strain NCIMB 12038. Appl. Microbiol. Biotechnol. [DOI] [PubMed] [Google Scholar]
- 29. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 30. Marsh M, et al. 2008. Structure of bacterial glutathione S-transferase maleyl pyruvate isomerase and implications for mechanism of isomerization. J. Mol. Biol. 384:165–177 [DOI] [PubMed] [Google Scholar]
- 31. Newton GL, et al. 1996. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 178:1990–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Newton GL, Leung SS, Wakabayashi JI, Rawat M, Fahey RC. 2011. The DinB superfamily includes novel mycothiol, bacillithiol, and glutathione S-transferases. Biochemistry 50:10751–10760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quevillon E, et al. 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33:W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rajan SS, Yang XJ, Shuvalova L, Collart F, Anderson WF. 2004. YfiT from Bacillus subtilis is a probable metal-dependent hydrolase with an unusual four-helix bundle topology. Biochemistry 43:15472–15479 [DOI] [PubMed] [Google Scholar]
- 35. Robson ND, Parrott S, Cooper RA. 1996. In vitro formation of a catabolic plasmid carrying Klebsiella pneumoniae DNA that allows growth of Escherichia coli K-12 on 3-hydroxybenzoate. Microbiology 142:2115–2120 [DOI] [PubMed] [Google Scholar]
- 36. Shen XH, Jiang CY, Huang Y, Liu ZP, Liu SJ. 2005. Functional identification of novel genes involved in the glutathione-independent gentisate pathway in Corynebacterium glutamicum. App. Environ. Microbiol. 71:3442–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA. 1995. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23:1087–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Suzuki H, Hashimoto W, Kumagai H. 1999. Glutathione metabolism in Escherichia coli. J. Mol. Catal. B Enzymol. 6:175–184 [Google Scholar]
- 39. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang LH, Hamzah RY, Yu Y, Tu SC. 1987. Pseudomonas cepacia 3-hydroxybenzoate 6-hydroxylase: induction, purification, and characterization. Biochemistry 26:1099–1104 [DOI] [PubMed] [Google Scholar]
- 41. Wang R, et al. 2007. Crystal structures and site-directed mutagenesis of a mycothiol-dependent enzyme reveal a novel folding and molecular basis for mycothiol-mediated maleylpyruvate isomerization. J. Biol. Chem. 282:16288–16294 [DOI] [PubMed] [Google Scholar]
- 42. Woodcock DM, et al. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ye J, McGinnis S, Madden TL. 2006. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 34:W6–W9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang JJ, Liu H, Xiao Y, Zhang XE, Zhou NY. 2009. Identification and characterization of catabolic para-nitrophenol 4-monooxygenase and para-benzoquinone reductase from Pseudomonas sp. strain WBC-3. J. Bacteriol. 191:2703–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhou NY, Fuenmayor SL, Williams PA. 2001. nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J. Bacteriol. 183:700–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhuang Z, Song F, Takami H, Dunaway-Mariano D. 2003. The BH1999 protein of Bacillus halodurans C-125 is gentisyl-coenzyme A thioesterase. J. Bacteriol. 186:393–399 [DOI] [PMC free article] [PubMed] [Google Scholar]