Abstract
Bacillus anthracis grows in chains of rod-shaped cells, a trait that contributes to its escape from phagocytic clearance in host tissues. Using a genetic approach to search for determinants of B. anthracis chain length, we identified mutants with insertional lesions in secA2. All isolated secA2 mutants exhibited an exaggerated chain length, whereas the dimensions of individual cells were not changed. Complementation studies revealed that slaP (S-layer assembly protein), a gene immediately downstream of secA2 on the B. anthracis chromosome, is also a determinant of chain length. Both secA2 and slaP are required for the efficient secretion of Sap and EA1 (Eag), the two S-layer proteins of B. anthracis, but not for the secretion of S-layer-associated proteins or of other secreted products. S-layer assembly via secA2 and slaP contributes to the proper positioning of BslO, the S-layer-associated protein, and murein hydrolase, which cleaves septal peptidoglycan to separate chains of bacilli. SlaP was found to be both soluble in the bacterial cytoplasm and associated with the membrane. The purification of soluble SlaP from B. anthracis-cleared lysates did not reveal a specific ligand, and the membrane association of SlaP was not dependent on SecA2, Sap, or EA1. We propose that SecA2 and SlaP promote the efficient secretion of S-layer proteins by modifying the general secretory pathway of B. anthracis to transport large amounts of Sap and EA1.
INTRODUCTION
The Gram-positive microbe Bacillus anthracis is the causative agent of anthrax and exists in two forms: vegetative, rod-shaped bacilli and small endospores (31). Following uptake into animal or human hosts, spores germinate into vegetative forms that replicate and disseminate into all organ tissues (31). Vegetative forms are thought to evade clearance by host phagocytes through the elaboration of a thick capsule, which is composed of poly-d-γ-glutamic acid (PDGA) tethered to the m-diaminopimelic acid cross bridge of peptidoglycan (7, 51), and through the ability to form elongated chains of bacilli tethered end-to-end at their septal peptidoglycan (47). Chains of bacilli present a physical obstacle for engulfment by immune cells (10, 53).
The envelope of B. anthracis is comprised of a plasma membrane and a peptidoglycan layer, which is decorated with the secondary cell wall polysaccharide (SCWP) [→6)-α-GlcNAc–(1→4)-β-ManNAc–(1→4)-β-GlcNAc–(1→]n, where α-GlcNAc is substituted with α-Gal and β-Gal at O3 and O4, respectively, and β-GlcNAc is substituted with α-Gal at O3 (8). The SCWP is tethered via GlcNAc-ManNAc linkage units to the C-6 position of N-acetylmuramic acid (MurNAc) in the repeating MurNAc-GlcNAc disaccharide structure of peptidoglycan (26). The B. anthracis S layer is comprised of the main S-layer proteins Sap and EA1 (14, 42, 43), which self-assemble into a paracrystalline layer of protein (41). The S layer also harbors B. anthracis S-layer-associated proteins (BSLs) (27), which provide for the uptake of nutrients across the envelope (63), adhesion to host tissues (28), and cell separation within chains of vegetative bacilli (1). The mature forms of S-layer and S-layer-associated proteins harbor SLH domains, which are responsible for the noncovalent association of these proteins with the SCWP (39, 40). SLH domains of S-layer and S-layer-associated proteins assemble into three-pronged spindle structures, where three interprong grooves are thought to capture the pyruvylated form of the SCWP, a modification catalyzed by the csaB gene product (16, 29).
Recently developed genetic tools have been applied to the study of B. anthracis chain formation and S-layer assembly (1, 62). By the use of flow cytometry analyses of bacilli, mutants with insertional lesions in csaB and bslO were isolated (1). BslO is an S-layer-associated protein that localizes to the septal portion of the B. anthracis S layer, where it cleaves peptidoglycan, thereby separating elongated chains of bacilli into shorter chains (1). Here we have expanded this search and report the isolation of chain length mutants with lesions in B. anthracis genes that are involved in the secretion of S-layer proteins across the bacterial envelope.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
B. anthracis Sterne 34F2 (60) and its mutants (Table 1) were cultured in brain heart infusion (BHI) broth supplemented with 0.8% NaHCO3 at 37°C or at 30°C when harboring the pLM4 vector (38). Escherichia coli strains DH5α (22) and K1077 (dcm dam) (18) were cultured in Luria-Bertani broth (LB) at 30°C. Media were supplemented with 20 μg/ml kanamycin to maintain plasmid selection in B. anthracis or with 50 μg/ml kanamycin in E. coli. B. anthracis strains were sporulated in modified G (modG) medium as described previously (30). Spore preparations were heat treated to kill any remaining vegetative bacilli and stored at 4°C. Spores were enumerated by CFU counts. Spores were germinated by inoculation into BHI broth and growth at 37°C.
Table 1.
Bacterial strains and plasmids used in this study
| B. anthracis strain or plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| Sterne 34F2 | Wild-type (pXO1+ pXO2−) genome sequence | 49 |
| SN1 | Deletion of secY2 (BAS2547) (nucleotides 2549507-2550730) in 34F2 | This study |
| SN2 | secA2::Sp transposon insertion at nucleotide 890561 in 34F2 | |
| SN3 | secA2::Sp transposon insertion at nucleotide 890732 in 34F2 | This study |
| SN4 | secA2::Sp transposon insertion at nucleotide 891556 in 34F2 | This study |
| SN5 | secA2::Sp transposon insertion at nucleotide 891568 in 34F2 | This study |
| SN6 | secA2::Sp transposon insertion at nucleotide 891758 in 34F2 | This study |
| SN7 | secA2::Sp transposon insertion at nucleotide 892627 in 34F2 | This study |
| SN8 | secA2::Sp transposon insertion at nucleotide 892786 in 34F2 | This study |
| SN9 | secA2::Sp transposon insertion at nucleotide 893012 in 34F2 | This study |
| SN10 | secA2::Sp transposon insertion at nucleotide 893039 in 34F2 | This study |
| SN11 | Deletion of sap (BAS0841) (nucleotides 896758-899063) in 34F2 | This study |
| SN12 | Deletion of eag (BAS0842) (nucleotides 899843-902414) in 34F2 | This study |
| SN13 | Deletion of sap and eag (BAS0841 and BAS0842, respectively) (nucleotides 896758-902414) in 34F2 | This study |
| SN14 | Deletion of slaP (BAS0837) (nucleotides 889657-890538) in 34F2 | This study |
| SN15 | bslO::Sp transposon insertion at nucleotide 1703151 in 34F2 | 16 |
| Plasmids | ||
| pLM4 | Temp-sensitive pE194 replicon; Kanr | 38 |
| pJK4 | Pspac; lacI regulator; Kanr | 26 |
| pSN1 | pJK4 harboring secA2 | This study |
| pSN2 | pJK4 harboring secA2 and slaP | This study |
| pSN3 | pJK4 harboring slaP | This study |
| pSN4 | pJK4 harboring slaP with a C-terminal Strep tag | This study |
| pSN5 | pET15b harboring full-length secA2 with an N-terminal His tag | This study |
| pSN6 | pET15b harboring full-length slaP with an N-terminal His tag | This study |
B. anthracis mutants and plasmids.
Plasmid DNA was purified from E. coli K1077 and was then used to transform B. anthracis. Deletion mutants were obtained by allelic replacement using temperature-sensitive plasmid pLM4 (Table 1) (38). Briefly, 1-kb upstream and downstream DNA sequences flanking the gene of interest were PCR amplified with specific primers (Table 2), cloned by restriction digestion into pLM4, and transformed into B. anthracis. Transformants were grown for 10 h at 42°C in the presence of 20 μg/μl kanamycin, diluted, and grown under the same conditions for four passages. Cultures were then diluted for another four passages into medium lacking antibiotics and grown for 10-h intervals at 30°C. Mutants were screened for growth on BHI agar and no growth on BHI-kanamycin agar. All complementation plasmids were derived from pJK4, and the expression of its Pspac promoter was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (26). Plasmid pSN1 was generated by amplifying the open reading frame of secA2 from genomic DNA using primers smn29 and smn30. Plasmid pSN3 was constructed by amplifying the open reading frame of slaP from genomic DNA using primers smn78 and smn79. Plasmid pSN2 was generated by amplifying both secA2 and slaP using primers smn29 and smn79. Strep-tagged SlaP (SlaPStrep) was constructed by amplifying slaP with primers smn96 and smn97, which contains a 24-bp insertion of the Strep tag coding sequence immediately adjacent to the stop codon (TGGTCTCATCCTCAATTTGAGAAG).
Table 2.
Oligonucleotides used in this study
| Primer | Sequencea | Use(s) |
|---|---|---|
| smn11 F | TTTCCCGGGTGGTGATTGGACAGCTGAAA | sap |
| smn12 R | TTTGGTACCGCTAGCCAGGAACGTCTGGGAATGTT | sap |
| smn13 F | TTTGGTACCGCTAGCGAAGCAAAACCTGCAACAAA | sap |
| smn14 R | TTTGAGCTCTACAGCTGCAGAAGCACGAT | sap |
| smn17 F | TTTCCCGGGCTGCGACTACAGCAAAAGCA | eag |
| smn18 R | TTTGGTACCGCTAGCTTGCTGCTGTCATTGTACCTG | eag |
| smn19 F | TTTGGTACCGCTAGCTGAAAAATCAGTGGGGATTC | eag |
| smn20 R | TTTGAGCTCGACCAATCCATGCCTGCTAT | eag |
| smn23 F | TTTCCCGGGTTGGCACAAATAAATCAACCTTT | secY2 |
| smn24 R | TTTGGTACCGCTAGCTGTCGCCTCCTATATTTTTATCA | secY2 |
| smn25 F | TTTGGTACCGCTAGCTGTGGCACTTGAAACAATGAA | secY2 |
| smn26 R | TTTGAGCTCCAATTGTGAATTTGCACGTGACTGGT | secY2 |
| smn29 F | TTTTCTAGAATGCTGAATTCGGTAAAAAAGC | pSN1, pSN2 |
| smn30 R | TTTGGTACCTTATTGTACGTTTTCAGGAACACC | pSN1 |
| smn78 F | TTTTCTAGAATGTTATCATTCCTAAAAAAAGCTAAGAA | pSN3 |
| smn79 R | TTTTGGTACCTTAGTCTTGTGGTACAAGTGC | pSN2, pSN3 |
| smn86 F | TTTTCCCGGGGGAGCAAGAAGCTGATTTAATCG | slaP |
| smn87 R | CGAGCCTCTCTATGACTCGAGACTAGTAATCTTTCACCTCGTTATTGTACGTTTTC | slaP |
| smn90 F | ACGAGGTGAAAGATTACTAGTCTCGAGTCATAGAGAGGCTCGCCTC | slaP |
| smn91 R | TTTTGAATTCGTGCAAAGTGTAGGTGGTGAT | slaP |
| smn96 F | TTTTCCATGGCAGCAGCATGTTATCATTCCTAAAAAAAGCTAAGAAAAACG | pSN4 |
| smn97 R | TTTTGGTCCTCAGATTACTTCTCAAATTGAGGATGAGACCAGTCTTGTGGTACAAGTGCTTTC | pSN4 |
| smn137 | TTTTCTCGAGATGCTGAATTCGGTAAAAAAGC | pSN5 |
| smn138 | TTTTGGATCCTTATTGTACGTTTTCAGGAACACC | pSN5 |
| smn122 | TTTTCATATGTTATCATTCCTAAAAAAAGCTAAGAAAAACG | pSN6 |
| smn95 | TTTTGGATCCTCAGATTAGTCTTGTGGTACAAGTGCTTTC | pSN6 |
Primer sequences encompassing restriction enzyme cleavage sites are underlined.
Antibody production.
Recombinant SecA2 and SlaP were produced in E. coli BL21(DE3) cells harboring plasmids pSN5 and pSN6, respectively. Cultures grown overnight were diluted 1:50 in LB supplemented with 100 μg/ml ampicillin at 30°C. IPTG (1 mM) was added to the cultures at an A600 of 0.5, and the expressions of SecA2 and SlaP were induced for 3 h. Cells were collected by centrifugation, suspended in 50 mM Tris-HCl and 150 mM NaCl, and lysed in a French press. Cell lysates were subjected to affinity purification over a Ni-nitrilotriacetic acid (NTA) column, and affinity-tagged proteins were eluted with 250 mM imidazole. Eluates were pooled and dialyzed in 1× phosphate-buffered saline (PBS). Five hundred micrograms of purified protein was emulsified in complete Freund's adjuvant (Difco) and injected subcutaneously into female New Zealand White rabbits. Antibody production was stimulated in 21-day intervals with two booster injections of antigen emulsified in incomplete Freund's adjuvant.
Flow cytometry analysis of Bacillus anthracis vegetative forms.
B. anthracis spores were germinated in 3 ml BHI broth at 37°C for 3 h. Cells were fixed with 4% paraformaldehyde and subjected to flow cytometry using an LSR Fortessa instrument (BD Biosciences). Forward-scatter A (FSC-A) data were collected for 10,000 events. Files were analyzed by using FlowJo software.
Light microscopy of bacilli.
Cells were fixed with 4% paraformaldehyde and imaged. Images were captured with a charge-coupled-device (CCD) camera on an Olympus IX81 microscope using a 100× or 40× objective. The chain lengths of bacilli were measured from acquired differential interference contrast (DIC) images by using ImageJ and converted to lengths in micrometers by using reference images of an objective micrometer.
Immunofluorescence microscopy of bacilli.
B. anthracis strain Sterne and its mutants were germinated in 3 ml of BHI broth at 37°C for 3 h. Cells were centrifuged and fixed with 4% paraformaldehyde. Samples were treated with diluted rabbit antiserum raised against purified recombinant Sap or BslO and labeled with a secondary antibody conjugated to a fluorophore (Thermoscientific). Cells were counterstained with boron dipyrromethene (BODIPY)-vancomycin (Invitrogen). A Leica SP5 Tandem Scanner Spectral 2-Photon confocal microscope with a 63× objective was used to observe cells. Images were captured at various zooms to include the entire chain. The scale bar reflects the actual length in micrometers regardless of the zoom function.
S-layer fractionation.
B. anthracis cultures grown overnight were diluted 1:100 into fresh medium and grown to an A600 of 2. One milliliter of culture was centrifuged at 16,000 × g and separated into medium (supernatant) and pellet fractions. Proteins in the medium were precipitated with 10% (vol/vol) trichloroacetic acid (TCA) for 30 min on ice and centrifuged at 16,000 × g for 10 min. The bacterial sediments were washed twice with 1× PBS and boiled at 95°C for 10 min in 3 M urea. Cells were sedimented by centrifugation at 16,000 × g, and the supernatant (S-layer fraction) was removed. The pellet was washed twice with PBS and mechanically lysed by silica bead beating for 3 min (Fastprep-24; MP Biomedical). After the sedimentation of the beads, proteins in the cell lysates were precipitated with TCA. All TCA precipitates were washed with ice-cold acetone and centrifuged at 16,000 × g for 10 min. Acetone was removed, and protein precipitates were dried. Samples were suspended in 50 μl of 0.5 M Tris-HCl (pH 7.5)–4% SDS and mixed with an equal volume of sample buffer (4% SDS, 1% β-mercaptoethanol, 10% glycerol, 50 mM Tris-HCl [pH 7.5], bromophenol blue). Proteins were separated on 10% SDS-PAGE gels and analyzed by Coomassie staining or electrotransferred onto a polyvinylidene difluoride (PVDF) membrane for immunoblot analysis. Proteins were detected with rabbit antisera raised against purified antigens. Strep-tagged proteins were detected by using monoclonal antibody StrepMAB (IBA). Immunoreactive products were revealed by chemiluminescent detection after incubation with a horseradish peroxidase (HRP)-conjugated secondary antibody (Cell Signaling Technology). The percentage of protein in subcellular fractions was calculated by averaging the ratio of immunoblot signals from one fraction to the sum of the signal in all fractions of three trials. Protective antigen (PA) (a protein secreted into the medium), PrsA (a membrane lipoprotein), and L6 (a ribosomal protein in the cytoplasm) were used as internal controls for the proper fractionation of bacilli.
Affinity purification of SlaPStrep.
B. anthracis vegetative forms were diluted 1:50 from a culture grown overnight in BHI broth supplemented with 0.8% sodium carbonate and 20 μg/ml kanamycin, induced with 1 mM IPTG, and grown to an A600 of 1.5. Cells were sedimented by centrifugation, suspended in column buffer (100 mM Tris-HCl [pH 8.0], 150 mM NaCl), and lysed by bead beating. Lysates were centrifuged for 5 min at 1,000 × g to remove beads and unbroken cells. Lysates were then subjected to ultracentrifugation at 100,000 × g for 30 min. Cleared lysates were loaded onto Strep-tactin Sepharose preequilibrated with column buffer, and the column was washed extensively with column buffer (IBA). Bound proteins were eluted with 2.5 mM desthiobiotin in column buffer, separated by 10% SDS-PAGE, and analyzed by Coomassie staining or immunoblotting.
Extraction of proteins from B. anthracis membranes with sodium carbonate.
B. anthracis cultures grown overnight were diluted 1:50 into fresh medium, the expression of SlaPStrep was induced with 1 mM IPTG, and bacilli were grown to an A600 of 1.5. Vegetative bacilli were sedimented by centrifugation, suspended in cytosol buffer (50 mM HEPES [pH 7.5], 10 mM magnesium acetate (MgOAc), 66 mM potassium acetate [KOAc]), and lysed by bead beating. Lysates were centrifuged for 5 min at 1,000 × g to remove beads and unbroken cells, and the supernatant was subjected to ultracentrifugation at 100,000 × g for 30 min. Proteins in the supernatant were precipitated with 10% TCA. Membrane sediments were suspended in 0.1 M sodium carbonate (pH 11.4) for 30 min on ice and then subjected to ultracentrifugation at 100,000 × g for 30 min. Proteins in the supernatant and pellet were precipitated with TCA, separated on 10% SDS-PAGE gels, and analyzed by Coomassie staining or immunoblotting.
RESULTS
Isolation of B. anthracis secA2 mutants.
Random mutagenesis with the bursa aurealis transposon was used to generate mutants of B. anthracis Sterne (62), an attenuated strain that harbors virulence plasmid pXO1, which contains the genes for anthrax toxins (protective antigen [PA], lethal factor [LF], and edema factor [EF]) but lacks pXO2 and the PDGA capsule genes (45, 60). Flow cytometry forward-scatter (FSC) and side-scatter (SSC) parameters were used to assess B. anthracis chain length and refractivity (1). By comparing mean FSC-A and SSC-A signals (n = 10,000 events per strain), we identified nine mutants that produced increased forward scattering relative to that of an age-matched wild-type (WT) control (Fig. 1B). Inverse PCR mapped the transposon insertions to BAS0838, a gene specifying a 788-residue polypeptide with homology (43% sequence identity) to the SecA ATPase of E. coli (46) (Fig. 1A). The genome of B. anthracis contains a second gene, BAS5038, whose 835-amino-acid product displays 51% sequence identity to E. coli SecA (901 amino acids). We assigned BAS5038 to be the secA homologue of B. anthracis and BAS0838 to be secA2, an accessory secretion gene whose function is required for the efficient secretion of the S-layer proteins of this microbe (see below).
Fig 1.
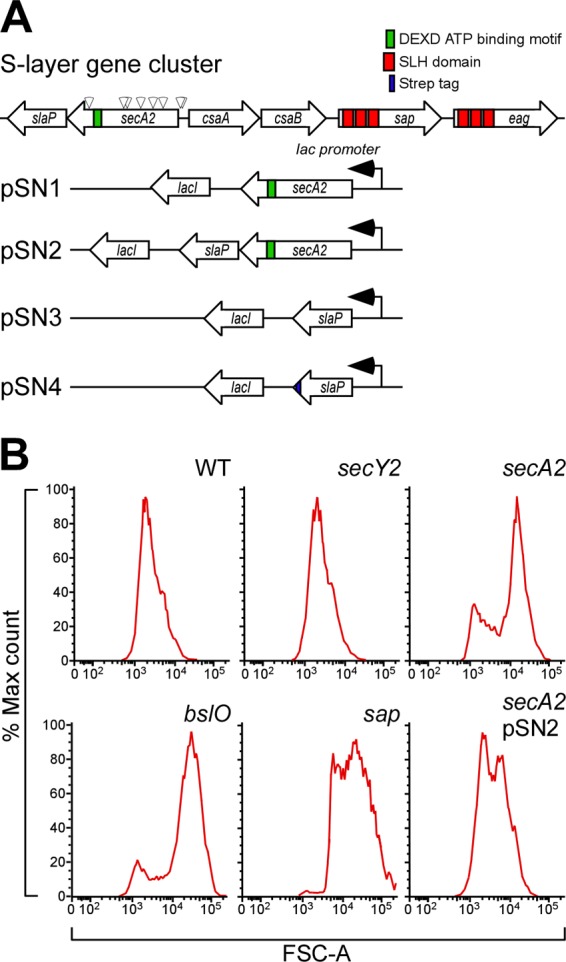
Isolation of Bacillus anthracis mutants that display chain length phenotypes. (A) Diagram illustrating the B. anthracis chromosomal gene cluster for the expression of S-layer proteins (sap and eag encoding signal peptide-bearing precursors with three SLH domains), the pyruvylation of the B. anthracis secondary cell wall polysaccharide (csaB), as well as the secretion of S-layer proteins, including the secA2 gene product, a predicted ATPase with a DEXD binding motif, and the slaP gene product. Arrowheads positioned above the secA2 gene identify nine different bursa aurealis insertional lesions that confer a chain length phenotype to mutant strains. Plasmids used in this study harbor wild-type secA2 (pSN1), wild-type secA2 and slaP (pSN2), wild-type slaP (pSN3), or slaP with a 3′ extension of its open reading frame specifying a Strep tag peptide for affinity chromatography of the encoded product (pSN4). All plasmids are recombinants of pJK4 and express inserted secretion gene products via the lacI-controlled, IPTG-inducible Pspac promoter. (B) The chain length of B. anthracis strain Sterne (WT) or its mutants with mutational lesions in the secY2, secA2, bslO, and sap genes was assessed by flow cytometry. B. anthracis secA2 mutants were transformed with pSN2 (harboring secA2 and slaP) to analyze the complementation of the chain length phenotype of the secA2 insertional lesion.
B. anthracis secA2 variants display a chain length phenotype.
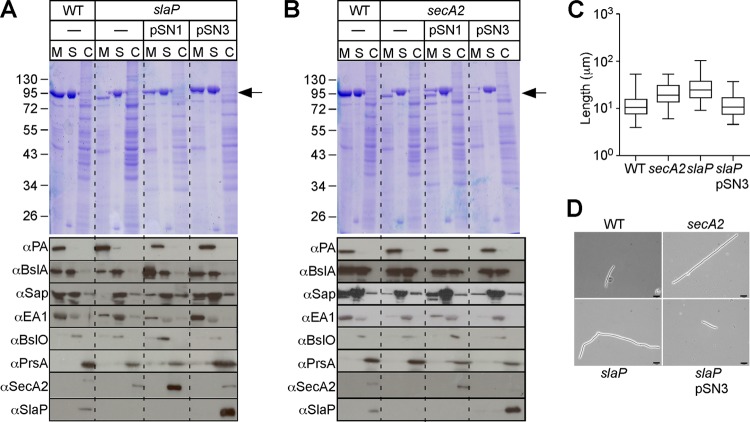
B. anthracis spores were suspended in fresh medium and monitored for changes in optical density (A600) over time. The growth curves of B. anthracis Sterne and its secA2 mutants were superimposable, indicating that secA2 is not required for growth (data not shown). Three hours after the inoculation, i.e., during the early exponential phase, culture aliquots were analyzed by light microscopy (Fig. 2). B. anthracis Sterne formed chains that were on average 34 (±22.68) μm in length (Fig. 2A). As previously reported, the chain lengths of B. anthracis Sterne or its mutants are variable when many different chains are analyzed (n = 100) (1) (Fig. 2A). The average chain length of bacilli increases during the early exponential phase and gradually decreases as cultures enter the late exponential phase (1). At a 3-h growth interval, the average chain length of the secA2 mutant was increased to 83 (±62.05) μm (P < 0.001 for the WT versus the secA2 mutant). Similar increases in chain lengths (secA2 mutants versus the WT) were observed when cultures were sampled at later time intervals (data not shown). The chain length phenotype of the secA2 mutant was reduced when secA2 bacilli were transformed with plasmid pSN2, carrying wild-type secA2 and slaP [51 (±31.66) μm; P < 0.001 for the secA2 versus the secA2(pSN2) mutant]. As a control, a mutant with a bursa aurealis insertion in the bslO gene formed chains with a greatly exaggerated length of 298 (±290.46) μm (Fig. 2) (1). Furthermore, a mutant lacking sap, the gene encoding the surface array protein, also formed chains that were increased in length (79 [±47.52] μm; P < 0.001 for the WT versus the sap mutant). The chain length phenotype of the sap mutant appears to be due to its inability to properly position BslO in the vicinity of septal peptidoglycan (see below); BslO cleaves septal peptidoglycan and controls the chain length of B. anthracis (1).
Fig 2.
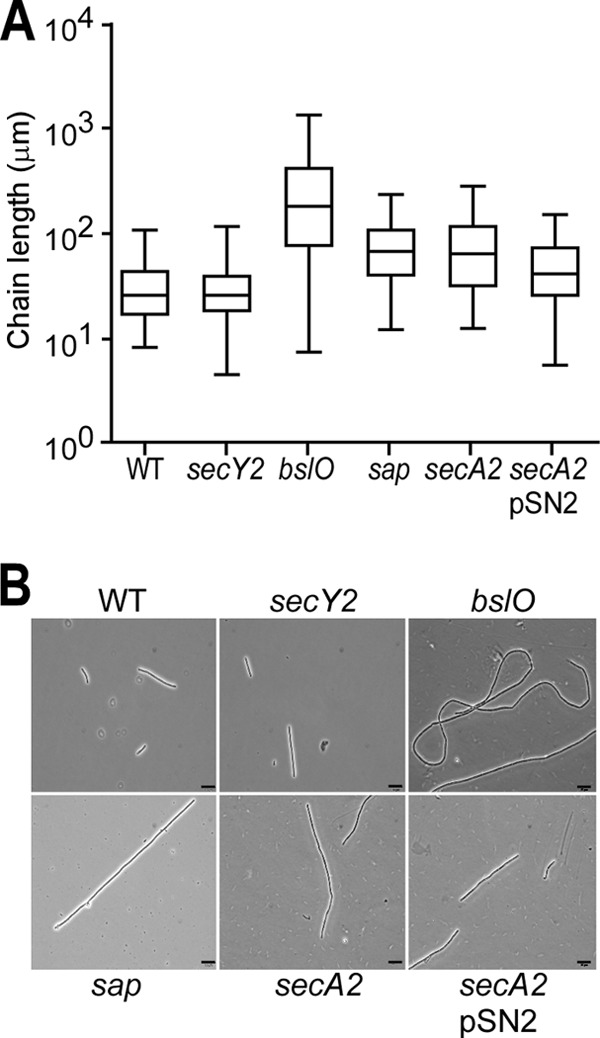
Chain lengths of B. anthracis secA2 mutants. (A) Box-and-whisker plot of the chain length of B. anthracis Sterne (WT) and its mutants with mutational lesions in secY2, bslO, sap, or secA2 or a secA2 mutant harboring pSN2 at 3 h postgermination in BHI medium. (B) Chain lengths were measured from DIC micrographs of vegetative bacilli (n = 100). Scale bars represent 10 μm. The statistical significance of differences in B. anthracis chain lengths were examined with the two-tailed Student t test [P > 0.5 for the WT versus the secY2 mutant, P < 0.001 for the WT versus the bslO mutant, P < 0.001 for the WT versus the sap mutant, P < 0.001 for the WT versus the secA2 mutant, and P < 0.001 for the secA2 versus the secA2(pSN2) mutant].
B. anthracis secY2 variants do not display a chain length phenotype.
The genome of B. anthracis harbors two copies of the secY gene, which in E. coli encodes one of the three components of preprotein translocase (SecYEG) (20) and represents the site for SecA-mediated protein secretion (12, 13). The predicted product of BAS0130 (433 residues) displays 43% sequence identity to E. coli SecY (443 residues) (24), whereas BAS2547 (434 residues) is 42% identical. As BAS0130 is located in the B. anthracis operon for ribosomal protein synthesis, similar to the E. coli operon expressing the secY gene product (23), the gene was designated secY. To test whether BAS2547 functions as an accessory secretion gene in the same pathway as B. anthracis secA2, we deleted its open reading frame. Mutants lacking BAS2547 were viable and without general defects in protein secretion (see below). We therefore designated BAS2547 secY2. The secY2 mutant did not display a B. anthracis chain length phenotype (32 [±20.44] μm; P > 0.5), suggesting that secY2 and secA2 may not function in the same accessory secretion pathway (Fig. 1B and 2).
B. anthracis secA2 variants display reduced secretion of the S-layer proteins Sap and EA1.
Cultures of B. anthracis Sterne or its secA2 and secY2 mutants were grown to an A600 of 2 and centrifuged to separate the extracellular medium (M) from the bacterial sediment. Bacilli were suspended in 3 M urea and boiled, thereby extracting S-layer proteins from B. anthracis cells. Extracts were again centrifuged to separate extracted S-layer proteins in the supernatant from B. anthracis cells in the sediment. The peptidoglycan of bacilli was broken with glass beads to release cellular proteins. Proteins in the medium and cellular fractions were precipitated with TCA, washed in acetone, and solubilized in sample buffer prior to SDS-PAGE and immunoblot analysis (Fig. 3). Similar amounts of Sap were detected in the medium (47% ± 8.48%) and in the S layer (42% [±13.48%]) of B. anthracis Sterne cultures (Fig. 3); however, only small amounts of Sap (11% [±5.11%]) were found within bacilli. PrsA, a membrane-associated lipoprotein involved in the folding of secreted polypeptides (67), was detected predominantly in the cellular fraction (Fig. 3B). PA, a secreted toxin component (64, 68), was found in the medium (Fig. 3). Most of the EA1 protein was detected in the medium of B. anthracis Sterne cultures (53% [±15.51%]), while the remainder was found in the S layer (Fig. 3). The S-layer-associated protein BslO was detected only in the S layer (1); BslA was found in the extracellular medium (42.62%) and in the S layer (57.38%) (27). These results suggest that B. anthracis secretes significant amounts of Sap and EA1 into the medium and that not all of these S-layer proteins are permanently retained in the bacterial envelope through their binding to the SCWP (26).
Fig 3.
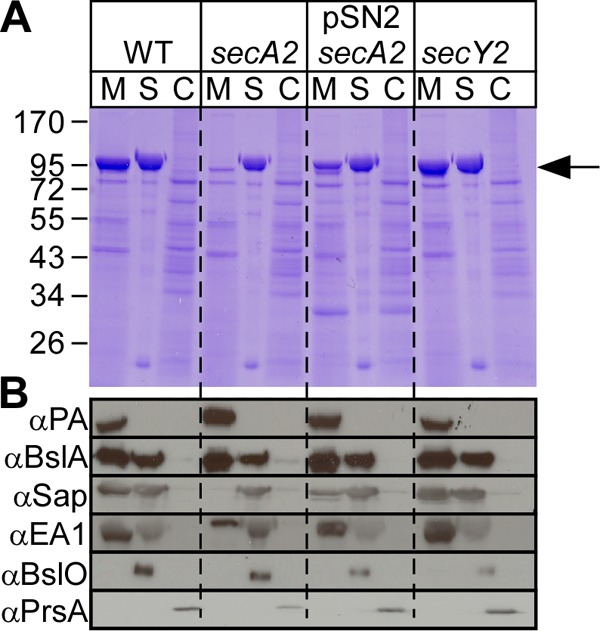
B. anthracis secA2 mutants are defective for the secretion of the S-layer proteins Sap and EA1. (A) Vegetative forms of B. anthracis Sterne (WT), its secA2 or secY2 mutant, or a secA2 mutant harboring pSN2 were grown to the mid-log phase. Cultures were centrifuged to separate the extracellular medium (M) from the bacterial sediment. The S layer (S) of bacilli was extracted by boiling in 3 M urea. Extracted cells (C) were broken in a bead beater. Proteins in all fractions were precipitated with TCA, washed in acetone, and analyzed with Coomassie-stained 10% SDS-PAGE gels. The position of Sap/EA1 is indicated by a black arrow. (B) B. anthracis cultures fractionated as described above for panel A were subjected to immunoblotting with rabbit antisera raised against purified Sap, EA1, PA, BslA, BslO, PrsA, SecA2, or SlaP.
Mutants with an insertional lesion in the secA2 gene secreted reduced amounts of Sap (7% [±4.50%]) and EA1 (13% [±12.52%]) into the culture medium; however, significant amounts of these polypeptides were retained in the S layer (Fig. 3). The defect in Sap and EA1 secretion could be complemented by plasmid pSN2 (33% [±0.41%] secreted Sap and 48% [±14.32%] secreted EA1), which harbors the secA2 and slaP genes (see below) from the IPTG-inducible Pspac promoter (Fig. 1A and 3). The subcellular locations of PA, BslA, BslO, and PrsA were not affected in the B. anthracis secA2 mutant (Fig. 3B). The deletion of the secY2 gene did not have an effect on S-layer protein secretion, as 48% (±14.62%) Sap and 57% (±23.80%) EA1 were detected in the medium of B. anthracis secY2 mutant cultures (Fig. 3B).
B. anthracis slaP mutants display a chain length phenotype and a partial defect in Sap and EA1 secretion.
Plasmid pSN1 harbors wild-type secA2 under the control of an IPTG-inducible promoter; the transformation of pSN1 into B. anthracis secA2 mutants did not completely restore Sap and EA1 secretion to wild-type levels (Fig. 4B), suggesting that the insertional lesion resulted in a polar effect on the neighboring gene BAS0837 (Fig. 4B). To explore this further, we generated a mutant with a deletion in the 294-codon open reading frame of BAS0837 (Fig. 1A). Compared to B. anthracis Sterne, the mutant was defective for the efficient secretion of Sap (10% [±3.00%]) and EA1 (15% [±15.28%]), whereas the secretion of PA and BslA was not altered (Fig. 4A). We therefore designated BAS0837 slaP (B. anthracis S-layer assembly protein). Similar to B. anthracis secA2 mutants, the variant lacking slaP displayed an increased chain length (76 [±46.97] μm; P < 0.001 for the WT versus the slaP mutant) (Fig. 4C). The transformation of the slaP mutant with pSN3, a plasmid carrying wild-type slaP under the control of the IPTG-inducible Pspac promoter, restored the efficient secretion of Sap (32% [±9.91%]) and EA1 (55% [±8.64%]) (Fig. 4A) and reduced the increased-chain-length phenotype of the slaP mutant (Fig. 4C and D). In contrast, the transformation of the slaP mutant with pSN1 (secA2) did not lead to a complete restoration of S-layer protein secretion (Fig. 4A) or to a reduction in chain length (data not shown).
Fig 4.
B. anthracis slaP mutants are defective for the secretion of S-layer proteins Sap and EA1. (A) Vegetative forms of B. anthracis Sterne (WT) and its slaP mutant without a plasmid or with pSN1 or pSN3 were grown to the mid-log phase. Cultures were centrifuged to separate the extracellular medium (M) from the bacterial sediment. Bacilli were extracted with hot urea to remove the S layer (S) and again centrifuged to sediment cell extracts (C). (Top) Proteins were precipitated with TCA, washed in acetone, and analyzed with Coomassie-stained 10% SDS-PAGE gels. The position of Sap/EA1 is indicated by an arrow. (Bottom) Samples from the top panel were subjected to immunoblotting with rabbit antisera raised against purified Sap, EA1, PA, BslA, BslO, PrsA, SecA2, and SlaP. (B) Vegetative forms of B. anthracis Sterne (WT) and its secA2 mutant without a plasmid or with pSN1 or pSN3 were grown to the mid-log phase and analyzed as described above for panel A. (C) Box-and-whisker plot of the chain lengths of B. anthracis Sterne (WT), its mutant with mutational lesions in secA2 or slaP, or a slaP mutant harboring pSN3 at 3 h postgermination. (D) Chain lengths were measured from DIC micrographs of vegetative bacilli (n = 100). Scale bars represent 10 μm. The statistical significances of differences in B. anthracis chain lengths were examined with the two-tailed Student t test [P < 0.001 for the WT versus the slaP mutant and P < 0.001 for the slaP versus the slaP(pSN3) mutant].
SlaP is located in the cytoplasm of B. anthracis.
When examined with the Kyte-Doolittle hydrophobicity plot (33) or a signal peptide algorithm (65), the in silico-translated product of slaP, a polypeptide of 293 amino acids, was not predicted to harbor an N-terminal signal peptide or a hydrophobic transmembrane domain. Homologues of SlaP were identified in the genome sequences of B. anthracis, Bacillus cereus, and Bacillus thuringiensis strains but not in the genome of Bacillus subtilis or Clostridium difficile (data not shown). To detect a slaP gene product, we generated specific rabbit antisera against full-length recombinant SlaP. Immunoblotting with rabbit antisera detected SlaP in the cellular fraction of B. anthracis Sterne (WT) but not in the cellular fraction of the slaP mutant (Fig. 4A). Increased amounts of SlaP were detected in the cytoplasm of the slaP mutant harboring plasmid pSN3 but not in slaP(pSN1) cells (Fig. 4A). To identify SlaP binding partners, a plasmid harboring a slaP variant with a 3′ extension of its open reading frame for eight codons (WSHPQFEK) specifying a Strep tag (pSN4) (Fig. 1) was constructed. Similar to pSN3, the transformation of the slaP mutant with pSN4 restored the efficient secretion of the S-layer proteins Sap and EA1 (Fig. 5A). Cleared lysates derived from the vegetative forms of B. anthracis slaP(pSN3) and B. anthracis slaP(pSN4) cells were subjected to affinity chromatography on Strep-tactin Sepharose, eluted with desthiobiotin, and analyzed with Coomassie-stained SDS-PAGE gels (55). The data in Fig. 5B reveal the affinity purification of a 32-kDa polypeptide (white arrowhead) from the cleared lysate of B. anthracis slaP(pSN4) cells but not from B. anthracis slaP(pSN3) cells. Mass spectrometry experiments with tryptic peptides derived from the 32-kDa polypeptide confirmed its identity as SlaPStrep (data not shown). The eluates of affinity chromatography samples were subjected to immunoblotting with antibodies specific for the Strep tag as well as Sap and EA1. As expected, only the eluate of B. anthracis slaP(pSN4) cells, and not the eluate of B. anthracis slaP(pSN3) cells, harbored SlaPStrep (Fig. 5B). The S-layer proteins Sap and EA1 were not detected in the eluate of B. anthracis slaP(pSN4) cells (Fig. 5B). As a control, the medium fraction of B. anthracis Sterne cultures was subjected to SDS-PAGE in order to calibrate immunoblotting experiments for Sap and EA1 (Fig. 5B). Thus, the soluble form of SlaPStrep does not copurify with Sap or EA1 precursors, i.e., the substrates for an accessory secretion pathway that is defined by its unique requirement for slaP and secA2 functions.
Fig 5.
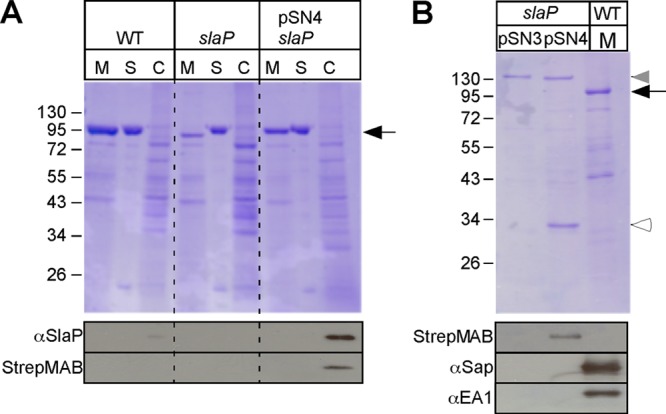
B. anthracis slaP mutants can be complemented by affinity-tagged slaP, which encodes a cytoplasmic protein. (A) Vegetative forms of B. anthracis Sterne (WT) and its slaP mutant without a plasmid or with pSN4, harboring slaP with a 3′ extension of its open reading frame specifying a Strep-tagged peptide, were grown to the mid-log phase. Cultures were centrifuged to separate the extracellular medium (M) from the bacterial sediment. Bacilli were extracted with hot urea to remove the S layer (S) and again centrifuged to sediment cells, which were subsequently broken in a bead beater (C). (Top) Proteins in all fractions were precipitated with TCA, washed in acetone, and analyzed with Coomassie-stained 10% SDS-PAGE gels. The position of Sap/EA1 is indicated by a black arrow. (Bottom) Samples were subjected to immunoblotting with a Strep-tag-specific monoclonal antibody, which identified SlaPStrep in B. anthracis slaP(pSN4) cells. (B) B. anthracis slaP(pSN3) and slaP(pSN4) cells were broken in a bead beater instrument, and cleared lysates were subjected to affinity chromatography on Strep-tactin Sepharose, eluted with desthiobiotin, and analyzed with Coomassie-stained SDS-PAGE gels. A medium (M) sample from B. anthracis Sterne cultures was added as a control. The black arrow identifies the position of Sap/EA1, and the white arrowhead identifies that of SlaPStrep. The gray arrowhead identifies the position of a B. anthracis protein in slaP(pSN3) and slaP(pSN4) cells that copurified during chromatography on Strep-tactin Sepharose but that did not react with the Strep-tag-specific monoclonal antibody (see below). Shown at the bottom are samples that were subjected to immunoblotting with a Strep-tag-specific monoclonal antibody or with rabbit antisera raised against purified Sap or EA1.
SlaP associates with the plasma membrane of B. anthracis.
To examine the membrane association of SlaP, a lysate derived from the vegetative forms of B. anthracis (obtained by mechanically lysing bacilli with glass beads) was subjected to slow-speed centrifugation to remove unbroken cells. The lysate was then subjected to ultracentrifugation to separate soluble cytoplasmic components in the supernatant from integral membrane proteins and membrane-associated proteins in the sediment (pellet). Membrane samples were extracted with 0.1 M sodium carbonate (Na2CO3) (pH >11), a perturbant that displaces proteins peripherally associated with membranes (59), and again centrifuged to separate extracted proteins in the supernatant from integral membrane proteins in the sediment (pellet). Proteins in all fractions were precipitated with trichloroacetic acid (TCA) and analyzed with Coomassie-stained SDS-PAGE gels and by immunoblotting (Fig. 6). Following fractionation, most of SlaP sedimented with the membranes of B. anthracis cells; however, a small portion of SlaP was extracted from the membranes by treatment with sodium carbonate. These data suggest a peripheral membrane association for SlaP (Fig. 6).
Fig 6.
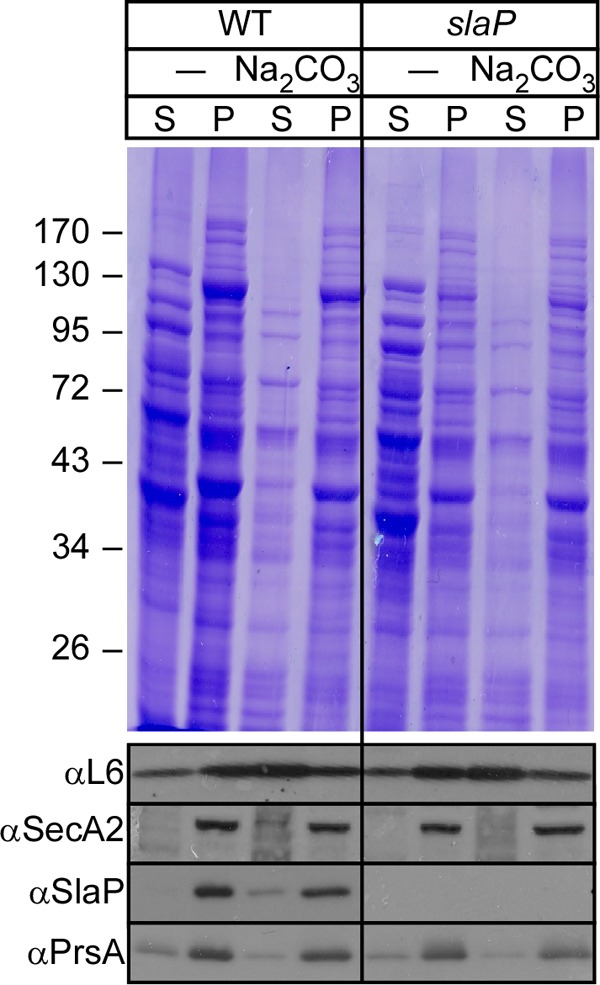
SlaP occurs as soluble and membrane-associated species in B. anthracis. Vegetative forms of B. anthracis Sterne and the slaP mutant were grown to the mid-log phase. Cultures were centrifuged to sediment vegetative forms, which were subsequently lysed in a bead beater. Beads and unbroken cells were removed by slow-speed centrifugation, and crude extracts were subjected to ultracentrifugation at 100,000 × g, separating soluble cytoplasmic proteins in the supernatant (S) from membrane proteins in the pellet (P). Membranes were extracted on ice with 0.1 M Na2CO3 and again subjected to ultracentrifugation at 100,000 × g, separating peripheral membrane proteins in the supernatant from integral membrane proteins in the pellet. All samples were analyzed with Coomassie-stained 10% SDS-PAGE gels and by immunoblotting with SlaP-specific rabbit antiserum.
Membrane association of SlaP does not require S-layer proteins or SecA2.
We wondered whether the membrane association of SlaP is caused by an engagement of the precursors of the Sap and EA1 S-layer proteins with the SecA2 secretory pathway. If so, mutants that are unable to express the secretion substrates (Sap or EA1) or the accessory secretion component (SecA2) would be expected to release SlaPStrep from the bacterial membrane (Fig. 7). This was tested; however, mutants lacking secY2, secA2, sap, eag, or both S-layer protein genes (sap eag) displayed similar patterns of SlaPStrep associations with the membranes of bacilli; i.e., the protein was found to be associated with bacterial membranes in a manner that could be perturbed by treatment with sodium carbonate (Fig. 7). Thus, neither the accessory secretion genes (secA2 and secY2) nor the genes for the S-layer protein substrates of SlaP- and SecA2-mediated secretion are required for the association of SlaPStrep with the bacterial membrane.
Fig 7.
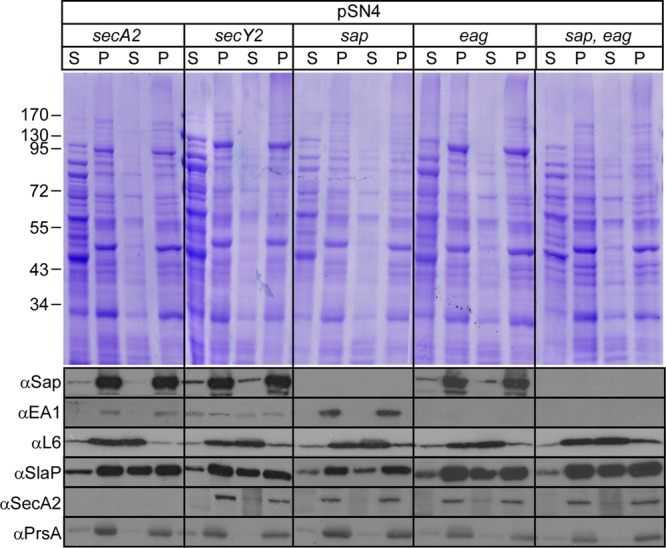
SlaPStrep association with the membranes of B. anthracis does not require S-layer proteins (Sap/EA1) or SecA2. B. anthracis mutants with mutational lesions in secY2, secA2, sap, eag, or sap and eag were transformed with pSN4. Lysates of vegetative forms that had been broken in a bead beater instrument were subjected to membrane cosedimentation analysis of SlaPStrep as described in the legend to Fig. 6. All samples were analyzed with Coomassie-stained 10% SDS-PAGE gels and by immunoblotting with rabbit antiserum against Sap, EA1, L6, SecA2, SlaP, and PrsA.
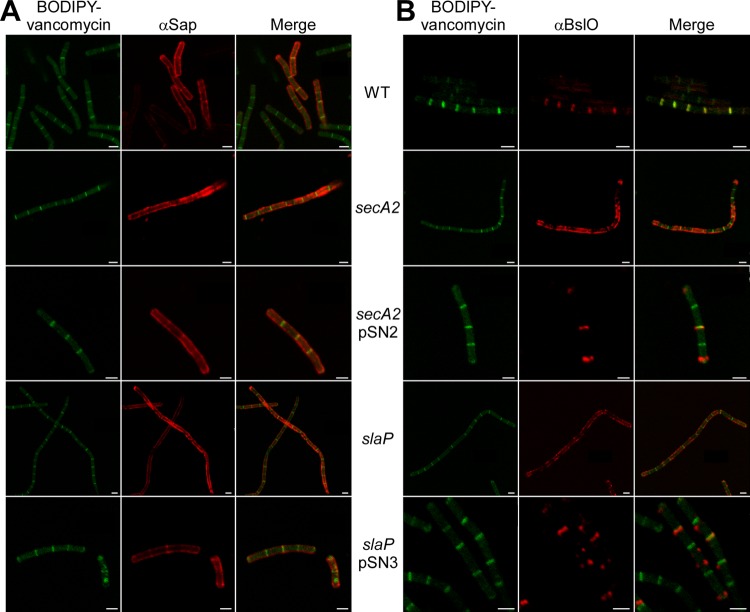
BslO is mislocalized in B. anthracis secA2 and slaP mutants.
Spores of B. anthracis strain Sterne and its secA2 and slaP mutants were germinated for 3 h. Cells were fixed in 4% paraformaldehyde and stained for Sap and BslO by using specific rabbit antisera and a secondary antibody conjugated to a fluorophore. BODIPY-vancomycin was used to stain peptidoglycan lipid II, which accumulates in the septum between adjacent vegetative cells (1). Bacilli were imaged with a Leica SP5 Tandem Scanner Spectral 2-Photon confocal microscope. As noted in the accompanying article by Kern et al. (29a), the S-layer protein Sap was found to be distributed throughout the envelope of B. anthracis Sterne vegetative cells (Fig. 8A). BslO localized to the septal portion of the B. anthracis envelope, as demonstrated by the superimposable fluorescence signals for BslO and BODIPY-vancomycin (Fig. 8B) (1). The distribution of Sap in the envelope of secA2 and slaP mutant cells appeared uneven, with patches of strong fluorescence intensity (Fig. 8A). Unlike its physiological septal localization in wild-type bacilli, BslO was found to be deposited in patches throughout the envelope of B. anthracis secA2 and slaP mutant vegetative forms. The localization of BslO in the secA2 and slaP mutants was restored to the septal position when the wild-type secA2 or slaP gene products were expressed from complementing plasmids (Fig. 8B). These data suggest that the secretion defect of the secA2 and slaP mutants for the S-layer proteins (Sap and EA1) perturbs the distribution of the S-layer-associated protein BslO, whose localization to the septal portion of the envelope is required for the physiological control of B. anthracis chain length (1).
Fig 8.
Localization of Sap and BslO in the envelope of B. anthracis secA2 and slaP mutants. B. anthracis strain Sterne and its mutants with mutational lesions in secA2 and slaP were fixed in 4% buffered formalin at 3 h postgermination. The localizations of Sap (A) and BslO (B) were observed with specific rabbit antisera as well as secondary antibody conjugates and counterstained with BODIPY-vancomycin to reveal the septal peptidoglycan. Images were obtained with a Leica SP5 Tandem Scanner Spectral 2-Photon confocal microscope (100× objective). Scale bars represent 2 μm.
DISCUSSION
The secretion of signal peptide-bearing precursor proteins in B. anthracis has not yet been studied. Assuming that the protein secretory pathway, as defined for E. coli, is operational in B. anthracis, signal peptide-bearing precursors would likely use the ATPase SecA and the membrane translocon SecY/SecE/SecG complex as well as the SecD/SecF/YajC complex for their travels across the plasma membrane (54). The removal of the signal peptide by signal peptidase (9) would release mature proteins into the extracellular medium of B. anthracis, unless polypeptides are endowed with S-layer homology domains (39), which retain S-layer proteins and S-layer-associated proteins in the envelope by binding to the SCWP (26, 27, 29), or with sorting signals for sortase-mediated anchoring to peptidoglycan (19, 36, 37). E. coli also employs the signal recognition particle (SRP), a ribonucleoprotein complex comprised of an Ffh polypeptide and 4.5S RNA, which interacts with the nascent precursors of membrane proteins to regulate translation and deliver the ribosome to the SRP receptor (FtsY) and eventually to the SecY/SecE/SecG translocon for cotranslational secretion (21, 44). Several chaperones, including SecB (48), heat shock proteins (DnaK/DnaJ/GrpE) (66), as well as trigger factor, a peptidyl-prolyl isomerase (61), contribute to secretion by maintaining specific substrate proteins of E. coli in a secretion-competent state (3, 11). The genome of B. anthracis harbors the secA, secD, secE, secF, secG, secY, ffh, ftsY, and yajC genes (50). A notable difference from E. coli is that B. anthracis lacks the secB gene but harbors three homologues of prsA (67), a lipoprotein peptidyl-prolyl isomerase involved in the folding of secreted polypeptides (32, 57).
Some Gram-positive bacteria express accessory secretion genes designated secA2 or secY2 (52). In Streptococcus gordonii and Staphylococcus aureus, the secA2 and secY2 genes are essential for the secretion of the large glycoprotein GspB and SraP, respectively (4). Due to posttranslational glycosylation, GspB cannot be secreted via the canonical SecA pathway (4, 58). A 90-residue N-terminal signal peptide as well as the first 20 amino acids of mature GspB are required for its initiation into an accessory secretion pathway requiring SecA2 and SecY2 as well as the accessory secretion proteins Asp1 to Asp5 (5, 56). Several Gram-positive microbes also use the accessory secretion genes secA2 and secY2 (either alone or together) for the selective transport of specific substrates (52). For example, Listeria monocytogenes employs a SecA2-dependent pathway for the secretion of the p60 murein hydrolase, a protein whose gene is located immediately adjacent to the secA2 gene (34, 35). In contrast to the narrow substrate specificity of SecA2/SecY2 in streptococci and staphylococci, both Listeria and mycobacteria, i.e., Mycobacterium smegmatis and Mycobacterium tuberculosis, appear to transport a broader spectrum of proteins via SecA2; this includes some proteins that lack an N-terminal signal peptide (2, 6). The biochemical details for SecA2 function in any one of these bacteria, i.e., the selection of a specific substrate by SecA2 or its presumed interaction with other components of the secretory pathway, are still unknown (52).
Several spore-forming Firmicutes, including B. cereus, B. anthracis, B. thuringiensis, C. difficile, as well as other clostridial species, elaborate a proteinaceous S layer from precursor proteins harboring an N-terminal signal peptide (15, 17). C. difficile, an anaerobic microbe, contains two secA genes, of which the noncanonical secretion gene secA2 is required for the assembly of its S-layer proteins and its cell wall protein CwpV (15). In contrast to C. difficile, members of the B. cereus sensu lato group (25), which includes B. cereus, B. anthracis, and B. thuringiensis, harbor the slaP gene and require SLH domains within S-layer proteins for their envelope deposition via a noncovalent association with the SCWP (26, 29). Here we demonstrate that the secA2 and slaP genes of B. anthracis are both required for the efficient secretion of the S-layer proteins Sap and EA1 and for the physiological control of the chain length of the bacterium's vegetative forms. The latter phenotype is explained as a result of the reduced deposition of S-layer proteins in the B. anthracis envelope. S-layer proteins, for example, Sap, are required for the proper positioning of the BslO murein hydrolase in the bacterial envelope (1). Furthermore, we show that SlaP can be isolated as a soluble protein from the bacterial cytoplasm; this species does not appear to interact with other proteins of B. anthracis. SlaP is associated with bacterial membranes in a manner that can be perturbed with sodium carbonate, which is indicative of a peripheral membrane association. We surmise, but do not know, that SlaP interacts with components of the secretion pathway to promote the efficient secretion of Sap and EA1. This interaction does not seem to involve SecA2, SecY2, Sap, or EA1, as mutants lacking any one of these components still retain SlaP in the bacterial membrane. To characterize the SecA2/SlaP-mediated secretion of Sap and EA1 in greater detail, future work must focus on characterizing the interaction of these proteins with components of the B. anthracis secretion pathway.
Why does B. anthracis require an alternative secretion pathway for the assembly of its S-layer and S-layer-associated functions? Currently available data cannot provide a conclusive answer; however, we have entertained two possibilities. First, the S-layer proteins Sap and EA1 are perhaps the most abundantly synthesized proteins of B. anthracis (27). Because protein translocation across the plasma membrane is a catalytic process, the considerable abundance of S-layer precursors may affect the secretion of other substrates. To prevent a traffic jam at the plasma membrane, B. anthracis may have evolved another catalyst (SecA2/SlaP). Second, S-layer protein secretion and assembly may be confined to discrete locations in the B. anthracis envelope, of which SecA2/SlaP may represent the secretion machinery components. Such a dedicated S-layer secretion/assembly pathway could further involve folding catalysts enabling the transfer of proteins into the envelope and the formation of the paracrystalline lattice. The latter model implies that S-layer proteins are transferred directly (without prior secretion into the medium) to their final destination; the former model implies a pathway whereby S-layer proteins are first secreted into the medium and subsequently seated by binding to the SCWP in the bacterial envelope. Although a mechanism of direct transfer seems favorable to us, we also appreciate that the experimental tools presented here cannot distinguish between the above-mentioned models for S-layer protein secretion and assembly.
ACKNOWLEDGMENTS
We thank members of our laboratory, specifically Bill Blaylock, Matthew Frankel, and Justin Mark Lunderberg, for discussion and comments on the manuscript.
This work has been supported by National Institute of Allergy and Infectious Diseases (NIAID) Infectious Disease Branch grant AI069227 to O.S. and D.M.M. S.-M.N.-M. and V.J.K. received support from NIH training grants GM007183 (molecular cell biology) and AI065382 (host-pathogen interactions). We acknowledge membership within and support from the Region V Great Lakes Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (GLRCE) (National Institute of Allergy and Infectious Diseases award 1-U54-AI-057153).
Footnotes
Published ahead of print 18 May 2012
REFERENCES
- 1. Anderson VJ, Kern JW, McCool JW, Schneewind O, Missiakas DM. 2011. The SLH domain protein BslO is a determinant of Bacillus anthracis chain length. Mol. Microbiol. 81:192–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Archambaud C, Nahori MA, Pizarro-Cerda J, Cossart P, Dussurget O. 2006. Control of Listeria superoxide dismutase by phosphorylation. J. Biol. Chem. 281:31812–31822 [DOI] [PubMed] [Google Scholar]
- 3. Beck K, Wu L-F, Brunner J, Müller M. 2000. Discrimination of SRP- and SecA/SecB-dependent substrates involves selective recognition of nascent chains by SRP and trigger factor. EMBO J. 19:134–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bensing BA, Sullam PM. 2002. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol. Microbiol. 44:1081–1094 [DOI] [PubMed] [Google Scholar]
- 5. Bensing BA, Sullam PM. 2010. Transport of preproteins by the accessory Sec system requires a specific domain adjacent to the signal peptide. J. Bacteriol. 192:4223–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Braunstein M, Espinosa B, Chan J, Beisle JT, Jacobs WR., Jr 2003. SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 48:453–464 [DOI] [PubMed] [Google Scholar]
- 7. Bruckner V, Kovacs J, Denes G. 1953. Structure of poly-D-glutamic acid isolated from capsulated strains of B. anthracis. Nature 172:508. [DOI] [PubMed] [Google Scholar]
- 8. Choudhury B, et al. 2006. The structure of the major cell wall polysaccharide of Bacillus anthracis is species specific. J. Biol. Chem. 281:27932–27941 [DOI] [PubMed] [Google Scholar]
- 9. Dalbey RE, Wickner W. 1985. Leader peptidase catalyzes the release of exported proteins from the outer surface of the Escherichia coli plasma membrane. J. Biol. Chem. 260:15925–15931 [PubMed] [Google Scholar]
- 10. Dixon TC, Fadl AA, Koehler TM, Swanson JA, Hanna PC. 2000. Early Bacillus anthracis-macrophage interactions: intracellular survival and escape. Cell. Microbiol. 2:453–463 [DOI] [PubMed] [Google Scholar]
- 11. Duong F, Eichler J, Price A, Leonard MR, Wickner W. 1997. Biogenesis of the gram-negative bacterial envelope. Cell 91:567–573 [DOI] [PubMed] [Google Scholar]
- 12. Economou A, Pogliano JA, Beckwith J, Oliver DB, Wickner W. 1995. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell 83:1171–1181 [DOI] [PubMed] [Google Scholar]
- 13. Economou A, Wickner W. 1994. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell 78:835–843 [DOI] [PubMed] [Google Scholar]
- 14. Etienne-Toumelin I, Sirard JC, Duflot E, Mock M, Fouet A. 1995. Characterization of the Bacillus anthracis S-layer: cloning and sequencing of the structural gene. J. Bacteriol. 177:614–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fagan RP, Fairweather NF. 2011. Clostridium difficile has two parallel and essential Sec secretion systems. J. Biol. Chem. 286:27483–27493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Forsberg LS, et al. 3 May 2012. Localization and structural analysis of a conserved pyruvylated epitope in Bacillus anthracis secondary cell wall polysaccharides and characterization of the galactose deficient wall polysaccharide from avirulent B. anthracis CDC 684. Glycobiology. [Epub ahead of print.] doi:10.1093/glycob/cws080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fouet A, Mesnage S. 2002. Bacillus anthracis cell envelope components. Curr. Top. Microbiol. Immunol. 271:87–113 [DOI] [PubMed] [Google Scholar]
- 18. Fulford W, Model P. 1984. Specificity of translational regulation by two DNA-binding proteins. J. Mol. Biol. 173:211–226 [DOI] [PubMed] [Google Scholar]
- 19. Gaspar AH, et al. 2005. Bacillus anthracis sortase A (SrtA) anchors LPXTG motif-containing surface proteins to the cell wall envelope. J. Bacteriol. 187:4646–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Görlich D, Rapoport TA. 1993. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 75:615–630 [DOI] [PubMed] [Google Scholar]
- 21. Halic M, et al. 2006. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature 444:507–511 [DOI] [PubMed] [Google Scholar]
- 22. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–572 [DOI] [PubMed] [Google Scholar]
- 23. Ito E, Strominger JL. 1964. Enzymatic synthesis of the peptide in bacterial uridine nucleotides. III. Purification and properties of L-lysine adding enzyme. J. Biol. Chem. 239:210–214 [PubMed] [Google Scholar]
- 24. Ito K, Bassford PJJ, Beckwith J. 1981. Protein localization in E. coli: is there a common step in the secretion of periplasmic and outer membrane proteins? Cell 24:707–717 [DOI] [PubMed] [Google Scholar]
- 25. Jensen GB, Hensen BM, Eilenberg J, Mahillon J. 2003. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 5:631–640 [DOI] [PubMed] [Google Scholar]
- 26. Kern J, Ryan C, Faull K, Schneewind O. 2010. Bacillus anthracis surface-layer proteins assemble by binding to the secondary cell wall polysaccharide in a manner that requires csaB and tagO. J. Mol. Biol. 401:757–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kern JW, Schneewind O. 2008. BslA, a pXO1-encoded adhesin of Bacillus anthracis. Mol. Microbiol. 68:504–515 [DOI] [PubMed] [Google Scholar]
- 28. Kern JW, Schneewind O. 2009. BslA, the S-layer adhesin of Bacillus anthracis, is a virulence factor for anthrax pathogenesis. Mol. Microbiol. 75:324–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kern JW, et al. 2011. Structure of the SLH domains from Bacillus anthracis surface array protein. J. Biol. Chem. 286:26042–26049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a. Kern VJ, Kern JW, Theriot JA, Schneewind O, Missiakas D. 2012. Surface-layer (S-layer) proteins Sap and EA1 govern the binding of the S-layer-associated protein BslO at the cell septa of Bacillus anthracis. J. Bacteriol. 194:3833–3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim HU, Goepfert JM. 1974. A sporulation medium for Bacillus anthracis. J. Appl. Bacteriol. 37:265–267 [DOI] [PubMed] [Google Scholar]
- 31. Koch R. 1876. Die Ätiologie der Milzbrand-Krankheit, begründet auf die Entwicklungsgeschichte des Bacillus anthracis. Beitr. Biol. Pflanzen 2:277–310 [Google Scholar]
- 32. Kontinen VP, Saris P, Sarvas M. 1991. A gene (prsA) of Bacillus subtilis involved in a novel, late stage of protein export. Mol. Microbiol. 5:1273–1283 [DOI] [PubMed] [Google Scholar]
- 33. Kyte J, Doolittle RF. 1982. A simple method for displaying the hydrophobic character of a protein. J. Mol. Biol. 157:105–132 [DOI] [PubMed] [Google Scholar]
- 34. Lenz LL, Mohammadi S, Geissler A, Portnoy DA. 2003. SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 100:12432–12437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lenz LL, Portnoy DA. 2002. Identification of a second Listeria secA gene associated with protein secretion and the rough phenotype. Mol. Microbiol. 45:1043–1056 [DOI] [PubMed] [Google Scholar]
- 36. Maresso AW, Chapa TJ, Schneewind O. 2006. Surface protein IsdC and sortase B are required for heme-iron scavenging of Bacillus anthracis. J. Bacteriol. 188:8145–8152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marraffini LA, Schneewind O. 2007. Sortase C-mediated anchoring of BasI to the cell wall envelope of Bacillus anthracis. J. Bacteriol. 189:6425–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marraffini LA, Schneewind O. 2006. Targeting proteins to the cell wall of sporulating Bacillus anthracis. Mol. Microbiol. 62:1402–1417 [DOI] [PubMed] [Google Scholar]
- 39. Mesnage S, et al. 2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19:4473–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mesnage S, Tosi-Couture E, Fouet A. 1999. Production and cell surface anchoring of functional fusions between the SLH motifs of the Bacillus anthracis S-layer proteins and the Bacillus subtilis levansucrase. Mol. Microbiol. 31:927–936 [DOI] [PubMed] [Google Scholar]
- 41. Mesnage S, Tosi-Couture E, Gounon P, Mock M, Fouet A. 1998. The capsule and S-layer: two independent and yet compatible macromolecular structures in Bacillus anthracis. J. Bacteriol. 180:52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mesnage S, Tosi-Couture E, Mock M, Gounon P, Fouet A. 1997. Molecular characterization of the Bacillus anthracis main S-layer component: evidence that it is the major cell-associated antigen. Mol. Microbiol. 23:1147–1155 [DOI] [PubMed] [Google Scholar]
- 43. Mignot T, Mesnage S, Couture-Tosi E, Mock M, Fouet A. 2002. Developmental switch of S-layer protein synthesis in Bacillus anthracis. Mol. Microbiol. 43:1615–1627 [DOI] [PubMed] [Google Scholar]
- 44. Miller JD, Bernstein HD, Walter P. 1994. Interaction of E. coli Ffh/4.5S ribonucleoprotein and FtsY mimics that of mammalian signal recognition particle and its receptor. Nature 367:657–659 [DOI] [PubMed] [Google Scholar]
- 45. Okinaka R, et al. 1999. Sequence, assembly and analysis of pX01 and pX02. J. Appl. Microbiol. 87:261–262 [DOI] [PubMed] [Google Scholar]
- 46. Oliver DB, Beckwith J. 1981. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell 25:765–772 [DOI] [PubMed] [Google Scholar]
- 47. Preisz H. 1909. Experimentelle Studien über Virulenz, Empfänglichkeit und Immunität beim Milzbrand. Z. Immunitatsforsch. 5:341–452 [Google Scholar]
- 48. Randall LL. 1992. Peptide binding by chaperone SecB: implications for recognition of non-native structure. Science 257:241–245 [DOI] [PubMed] [Google Scholar]
- 49. Ravel J, et al. 2009. The complete genome sequence of Bacillus anthracis Ames “ancestor.” J. Bacteriol. 191:445–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Read TD, et al. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81–86 [DOI] [PubMed] [Google Scholar]
- 51. Richter GS, et al. 2009. Capsule anchoring in Bacillus anthracis occurs by a transpeptidation mechanism that is inhibited by capsidin. Mol. Microbiol. 71:404–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rigel NW, Braunstein M. 2008. A new twist on an old pathway—accessory Sec systems. Mol. Microbiol. 69:291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ruthel G, Ribot WJ, Bavari S, Hoover T. 2004. Time-lapse confocal imaging of development of Bacillus anthracis in macrophages. J. Infect. Dis. 189:1313–1316 [DOI] [PubMed] [Google Scholar]
- 54. Sardis MF, Economou A. 2010. SecA: a tale of two protomers. Mol. Microbiol. 76:1070–1081 [DOI] [PubMed] [Google Scholar]
- 55. Schmidt TG, Skerra A. 2007. The Strep-tag system for one-step purification and high affinity detection or capturing of proteins. Nat. Protoc. 2:1528–1535 [DOI] [PubMed] [Google Scholar]
- 56. Seepersaud R, Bensing BA, Yen YT, Sullam PM. 2010. Asp3 mediates multiple protein-protein interactions within the accessory Sec system of Streptococcus gordonii. Mol. Microbiol. 78:490–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sibbald MJ, et al. 2006. Mapping the pathways to staphylococcal pathogenesis by comparative secretomics. Microbiol. Mol. Biol. Rev. 70:755–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Siboo IR, Chaffin DO, Rubens CE, Sullam PM. 2008. Characterization of the accessory Sec system of Staphylococcus aureus. J. Bacteriol. 190:6188–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Steck TL, Yu J. 1973. Selective solubilization of proteins from red blood cell membranes by protein perturbants. J. Supramol. Struct. 1:220–232 [DOI] [PubMed] [Google Scholar]
- 60. Sterne M. 1937. Avirulent anthrax vaccine. Onderstepoort J. Vet. Sci. Anim. Ind. 21:41–43 [PubMed] [Google Scholar]
- 61. Stoller G, et al. 1995. A ribosome-associated peptidyl-prolyl cis/trans isomerase identified as the trigger factor. EMBO J. 14:4939–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tam C, Glass EM, Anderson DM, Missiakas D. 2006. Transposon mutagenesis of Bacillus anthracis strain Sterne using bursa aurealis. Plasmid 56:74–77 [DOI] [PubMed] [Google Scholar]
- 63. Tarlovsky Y, et al. 2010. A Bacillus anthracis S-layer homology protein that binds heme and mediates heme delivery to IsdC. J. Bacteriol. 192:3503–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vodkin MH, Leppla SH. 1983. Cloning of the protective antigen gene of Bacillus anthracis. Cell 34:693–697 [DOI] [PubMed] [Google Scholar]
- 65. von Heijne G. 1992. Membrane protein structure prediction: hydrophobic analysis and the positive inside rule. J. Mol. Biol. 225:487–494 [DOI] [PubMed] [Google Scholar]
- 66. Wild J, Rossmeissl P, Walter WA, Gross CA. 1996. Involvement of the DnaK-DnaJ-GrpE chaperone team in protein secretion in Escherichia coli. J. Bacteriol. 178:3608–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Williams RC, et al. 2003. Production of Bacillus anthracis protective antigen is dependent on extracellular chaperone, PrsA. J. Biol. Chem. 278:18056–18062 [DOI] [PubMed] [Google Scholar]
- 68. Young JA, Collier JR. 2007. Anthrax toxin: receptor binding, internalization, pore formation and translocation. Annu. Rev. Biochem. 76:243–265 [DOI] [PubMed] [Google Scholar]