Abstract
Klebsiella pneumoniae M5a1 is capable of utilizing 3-hydroxybenzoate via gentisate, and the 6.3-kb gene cluster mhbRTDHIM conferred the ability to grow on 3-hydroxybenzoate to Escherichia coli and Pseudomonas putida PaW340. Four of the six genes (mhbDHIM) encode enzymes converting 3-hydroxybenzoate to pyruvate and fumarate via gentisate. MhbR is a gene activator, and MhbT is a hypothetical protein belonging to the transporter of the aromatic acid/H+ symporter family. Since a transporter for 3-hydrxybenzoate uptake has not been characterized to date, we investigated whether MhbT is responsible for the uptake of 3-hydroxybenzoate, its metabolic intermediate gentisate, or both. The MhbT-green fluorescent protein (GFP) fusion protein was located on the cytoplasmic membrane. P. putida PaW340 containing mhbRΔTDHIM could not grow on 3-hydroxybenzoate; however, supplying mhbT in trans allowed the bacterium to grow on the substrate. K. pneumoniae M5a1 and P. putida PaW340 containing recombinant MhbT transported 14C-labeled 3-hydroxybenzoate but not 14C-labeled gentisate and benzoate into the cells. Site-directed mutagenesis of two conserved amino acid residues (Asp-82 and Asp-314) and a less-conserved residue (Val-311) among the members of the symporter family in the hydrophilic cytoplasmic loops resulted in the loss of 3-hydroxybenzoate uptake by P. putida PaW340 carrying the mutant proteins. Hence, we demonstrated that MhbT is a specific 3-hydroxybenzoate transporter.
INTRODUCTION
A substantial number of microbes have been isolated for their capability to utilize aromatic acids as a sole source of carbon and energy. Their degradation pathways have been extensively investigated at both the biochemical and genetic levels in a number of cases (15, 17, 28, 40). The transport of the aromatic acids across the cytoplasmic membrane, an essential precursor to catabolism, has been found to be facilitated by members of the aromatic acid/H+ symporter (AAHS) family within the major facilitator superfamily (MFS) (34). Three members of the AAHS family involved in the catabolism of corresponding aromatic acids have been functionally identified by uptake assays using the respective 14C-labeled substrates: PcaK (4-hydroxybenzoate and protocatechuate transporter) from the 4-hydroxybenzoate and protocatechuate utilizer Pseudomonas putida PRS2000 (33), BenK (benzoate transporter) from the benzoate utilizer Acinetobacter sp. strain ADP1 (5), and TfdK (2,4-dichlorophenoxyacetate [2,4-D] transporter) from the 2,4-dichlorophenoxyacetate utilizer Ralstonia eutropha JMP134 (24). Nevertheless, the three identified transporters described above exhibit low amino acid sequence identities (28 to 33%) with each another, and no experiments have been conducted to show whether any of them is capable of transporting aromatic acids other than their respective primary substrates.
The microbial catabolism of 3-hydroxybenzoate has been shown to proceed via gentisate in several strains from phylogenetically diverse genera, including Corynebacterium glutamicum (40, 46), Rhodococcus sp. strain NCIMB 12038 (28), Polaromonas naphthalenivorans CJ2 (35), and Pseudomonas alcaligenes NCIMB 9867 (12), but no transporter for 3-hydroxybenzoate uptake has been reported so far. Notably, when 3-hydroxybenzoate catabolism in the Gram-positive organism Corynebacterium glutamicum was studied, a putative transporter gene within the catabolic cluster turned out to be involved in gentisate catabolism rather than 3-hydroxybenzoate catabolism (40).
Klebsiella pneumoniae M5a1 is a well-known nitrogen-fixing bacterium (10), which is able to metabolize 3-hydroxybenzoate via the gentisate pathway (20). An 8-kb DNA fragment from this strain conferred the ability to grow on 3-hydroxybenzoate to Escherichia coli (38). In this study, we report the identification of a specific transporter for 3-hydroxybenzoate uptake and its involvement in 3-hydroxybenzoate catabolism. This mhbT-encoded transporter shows 27 to 42% amino acid sequence identities with the three identified AAHS transporters described above and transports 14C-labeled 3-hydroxybenzoate but not 14C-labeled gentisate and benzoate into cells of Klebsiella pneumoniae M5a1 and Pseudomonas putida PaW340.
MATERIALS AND METHODS
Bacterial strains, plasmids, chemicals, media, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. Aromatic compounds were obtained from Sigma-Aldrich Co. (St. Louis, MO). The tracers [carboxyl-14C]3-hydroxybenzoate (55 mCi/mmol), [carboxyl-14C]gentisate (55 mCi/mmol), and [ring-UL-14C]benzoate (70 mCi/mmol) were purchased from American Radiolabeled Chemicals, Inc. (St. Louis, MO). Enzymes were purchased from TaKaRa Biotechnology Co. Ltd. (Dalian, China). Escherichia coli strains were grown in lysogeny broth (LB) medium at 37°C. Pseudomonas putida PaW340 (18, 43) and its variants were grown in LB or minimal medium (MM) (27) supplemented with 2 mM 3-hydroxybenzoate containing 0.015% yeast extract at 30°C. Klebsiella pneumoniae M5a1 (11) and its variants were grown in LB or MM supplemented with 2 mM 3-hydroxybenzoate at 30°C. Ampicillin, tetracycline, kanamycin, and chloramphenicol were used at final concentrations of 100, 25, 50, and 34 μg/ml, respectively, when necessary. In the expression experiments, cultures were grown to an optical density at 600 nm (OD600) of 0.6 and then induced by the addition of 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for E. coli and 1 mM IPTG for P. putida PaW340 and K. pneumoniae M5a1. A final concentration of 0.5 mM was used when 3-hydroxybenzoate was as an inducer.
General molecular biology methods.
Plasmid DNA was isolated by using the alkaline lysis method (39), except in the case of plasmid pVLT33 (7) and its derivatives, which were isolated by using the boiling lysis method (41). Restriction endonuclease digestion and ligation with T4 DNA ligase were conducted in accordance with the manufacturer's instructions. E. coli strains were transformed according to standard procedures (39). All the pRK415 (22) and pVLT33 constructs were transferred into P. putida PaW340 or K. pneumoniae M5a1 cells by triparental mating as described previously (45). Nucleotide sequences of pBSI containing the 8-kb fragment were determined by MWG-Biotech Ltd. (Ebersberg, Germany). Nucleotide sequences of the constructs were verified by Invitrogen Life Technologies Co. Ltd. (Shanghai, China). Analyses and comparison of amino acid sequences (or nucleotide sequences) were performed with BLAST programs at the National Center for Biotechnology Information website.
Site-directed mutagenesis.
The desired mutants of MhbT were obtained by overlap extension PCR (36). The outer primers for amplification were mhbT forward and mhbT reverse (see Table S2 in the supplemental material). The inner primers were designed to incorporate one codon change. The overlap extension PCR products were digested before ligation into similarly digested pVLT33, resulting in the expression constructs (see Table S1 in the supplemental material). The sequences of all mutated mhbT gene constructs were verified by DNA sequencing to ensure that only the desired mutations had occurred.
Cloning and expression of mhbDHIM genes in E. coli.
Entire individual genes, including their Shine-Dalgarno sequences, were amplified from pBSI by PCR, and the primers used are listed in Table S2 in the supplemental material. The purified PCR products were digested with EcoRI and ligated into similarly treated pUC18 to generate the desired constructs (see Table S1 in the supplemental material). The inserts in these clones were sequenced to ensure that no mutation had been incorporated during the PCR. Fragments from each of these clones were excised by using EcoRI and NdeI and individually religated into the expression vector pET5a to produce the corresponding constructs (see Table S1 in the supplemental material). The Mhb proteins encoded on the pET5a recombinants were individually expressed in E. coli Rosetta(DE3)(pLysS).
Preparation of cell extracts and enzyme assays.
E. coli cells were lysed by ultrasonic treatment, and the extracts were prepared as described previously (50). All assays were performed with 50 mM phosphate buffer (pH 7.4) at room temperature. The activity of 3-hydroxybenzoate 6-hydroxylase was determined by measuring the decrease in the absorbance at 340 nm due to the substrate-dependent oxidation of NADH, the molar extinction coefficient of which was taken to be 6,220 M−1 cm−1 (42). The activities of gentisate 1,2-dioxygenase, maleylpyruvate isomerase, and fumarylpyruvate hydrolase were all assayed according to a methods reported previously (50). Protein concentrations were determined by the Bradford method (2), with bovine serum albumin as the standard. One unit of enzyme activity was defined as the amount required for the disappearance (or production) of 1 μmol of substrate (or product) per minute at room temperature. Specific activities are expressed as units per milligram of protein. Analysis of fumarate and pyruvate was performed by using high-performance liquid chromatography (HPLC), as described previously (50).
Cellular localization of MhbT-green fluorescent protein (GFP) fusion proteins.
Cultures of E. coli BL21(DE3), K. pneumoniae M5a1, and P. putida PaW340 carrying the appropriate constructs (see Table S1 in the supplemental material) were induced with IPTG for 4 h before cells were harvested. Cells were washed and maintained in 50 mM phosphate buffer (pH 7.4), which was mixed with agarose (0.3%) to immobilize cells and for imaging under a confocal microscope with a 490-nm excitation filter and a 520-nm emission filter (45). The imaging experiments were performed by using a Leica TCS SP2 laser scanning spectral confocal microscope equipped with a cooled charge-coupled-device (CCD) camera (Leica Microsystems, Mannheim, Germany). The background cell fluorescence was subtracted.
mhbT disruption.
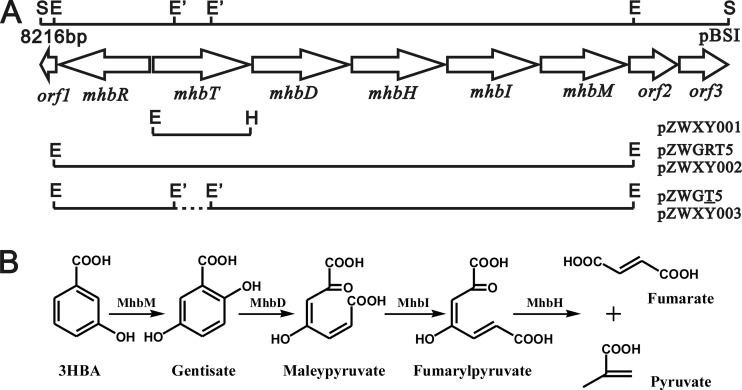
Plasmid pZWGRT5 was digested with EcoNI within mhbT and then religated to produce pZWGT5, resulting in the loss of a 639-bp EcoNI fragment. The complete mhb gene cluster of pZWGRT5 and the mhb gene cluster with truncated mhbT from pZWGT5 were both digested with EcoRI and inserted into pRK415 to produce pZWXY002 and pZWXY003, respectively (Fig. 1A; see also Table S1 in the supplemental material).
Fig 1.
(A) Physical map of the mhb gene cluster from pBSI with the 8,216-bp SphI fragment of K. pneumoniae M5a1 inserted into pUC18 and inserts of recombinant plasmids constructed from it. Open reading frames are marked by open arrows, with the directions of the arrowheads indicating the transcriptional directions. The restriction sites are designated follows: S, SphI; E, EcoRI; E′, EcoNI; H, HindIII. Both pZWGT5 and pZWXY003 contain a 5.6-kb fragment carrying the mhb gene cluster with a 639-bp EcoNI fragment deletion in mhbT. (B) Proposed pathway for 3-hydroxybenzoate catabolism in strain M5a1, together with the catabolic reactions catalyzed by mhb gene products in vivo.
Uptake assays.
The determination of the uptake rates of aromatic acids, measured by liquid scintillation counting of 14C-labeled substrates, was performed as described previously (33), with minor modifications. P. putida PaW340, K. pneumoniae M5a1, and their transconjugants were harvested during the exponential phase by centrifugation. After being washed twice with 50 mM Tris-HCl buffer (pH 8.5), cells were resuspended in the same buffer to an OD600 of 5 to 10 and kept on ice. Before the uptake assay, cells were incubated for 3 min at 30°C with 10 mM glucose for energy generation (48). The assays were initiated by mixing 600 μl of 50 mM Tris-HCl buffer (pH 8.5) containing 40 μM 14C-labeled substrates and 400 μl of the cell suspension. Samples (100 μl) were taken at timed intervals and filtered through Nuclepore polycarbonate membranes (0.22-μm pore size; Xinya, Shanghai, China), which were previously soaked with 50 mM Tris-HCl buffer (pH 8.5) containing 400 μM unlabeled substrates. The filters were then immediately washed with 2 ml of cold 0.1 M LiCl. For uptake assays performed at different pH values, cells in 50 mM phosphate buffer (pH 6.5, 7.5, and 8.0) and 50 mM Tris-HCl buffer (pH 7.5 and 8.5) were assayed at 1 min in triplicate. In competition and inhibition experiments, uptake measurements were performed in the presence of saturating substrate concentrations (40 μM 3-hydroxybenzoate) and other possible substrates in a 20-fold excess. The amount of substrate accumulated in the cells on the filters was determined with a scintillation counter (1450 MicroBeta TriLux; Perkin-Elmer Life Sciences, Boston, MA). The uptake activity was expressed as nanomoles of substrates taken up per milligram of protein. Cell protein contents were determined according to a protocol described previously (33). Apparent Km and Vmax values were obtained by measuring the uptake of 14C-labeled 3-hydroxybenzoate with nine substrate concentrations at 1 min in triplicate. Data were fitted according to the Michaelis-Menten equation using the least-squares method (4).
Quantitative PCR.
cDNA was obtained as previously described (44), and quantitative PCR was conducted according to methods described in a previous study (47). The 2−ΔΔCT method was used to calculate the relative changes in gene expression levels (29). The 16S rRNA gene (GenBank accession number FJ482128) from strain M5a1 was used as the internal reference gene.
Nucleotide sequence accession number.
The DNA sequence of 8,216 bp of the mhb genes has been submitted to GenBank under accession number AY648560.
RESULTS
Mhb proteins are involved in 3-hydroxybenzoate catabolism and its regulation.
The 8-kb DNA fragment of K. pneumoniae (38) was sequenced, and a gene cluster (orf1-mhbRTDHIM-orf2-orf3) was identified (Fig. 1). In our previous studies, the mhb structural genes were demonstrated to be transcribed as an operon, and their expression was regulated by MhbR of the LysR family (25). MhbM and MhbD were shown previously to be 3-hydroxybenzoate 6-hydroxylase (26) and gentisate 1,2-dioxygenase (30), respectively. In this study, cell extracts containing MhbI were found to contain glutathione (GSH)-dependent maleylpyruvate isomerase with a specific activity of 4.99 U/mg. Cell extracts containing MhbH were found to contain fumarylpyruvate hydrolase with a specific activity of 1.45 U/mg. The products from the latter reaction were confirmed to be fumarate and pyruvate by HPLC. Sequentially, MhbM, MhbD, MhbI, and MhbH converted 3-hydroxybenzoate to yield pyruvate and fumarate in vitro. It is reasonable to propose that this is also the case for the 3-hydroxybenzoate catabolic pathway in vivo (Fig. 1B). Additionally, plasmids pZWGRT5 and pZWXY002, carrying the 6.3-kb EcoRI fragment with the complete mhbRTDHIM cluster from the 8-kb fragment in pBSI (Fig. 1A), also conferred the ability to grow on 3-hydroxybenzoate to E. coli DH5α and P. putida PaW340, respectively.
MhbT is an AAHS family transmembrane protein.
As shown in Fig. 1A, mhbT was present in the gene cluster between the regulatory-protein-encoding gene mhbR and the enzyme genes mhbDHIM, encoding a 452-amino-acid polypeptide. Three topology prediction methods, TMHMM (http://www.cbs.dtu.dk/services/TMHMM/), MEMSAT (http://saier-144-37.ucsd.edu/cgi-bin/memsat.cgi), and HMMTOP (http://www.enzim.hu/hmmtop/), indicated that MhbT contained 12 α-helix transmembrane spanners, a typical feature of MFS members (34). By sequence comparison, MhbT exhibited 42% identity with the 4-hydroxybenzoate and protocatechuate transporter PcaK from P. putida (33), 27% identity with the benzoate transporter BenK from Acinetobacter sp. ADP1 (5), and 30% identity with the 2,4-dichlorophenoxyacetate (2,4-D) transporter TfdK from Ralstonia eutropha JMP134 (24). With this degree of identity, the function and substrate specificity of MhbT cannot be convincingly deduced.
Localization of MhbT-GFP to the periphery of cells.
To confirm the prediction that MhbT is associated with the membrane, mhbT was tagged with gfp in plasmids pZWXY004 and pZWXY006 (see Table S1 in the supplemental material) to facilitate the detection of its localization in bacterial cells. K. pneumoniae M5a1[pZWXY004], P. putida PaW340[pZWXY004], and E. coli BL21(DE3)[pZWXY006] (all carrying mhbT-gfp) cells were observed by using confocal microscopy. The results clearly showed that MhbT-GFP fusion proteins in these strains from three different genera were located at the periphery of cells (Fig. 2), consistent with the cytoplasmic membrane location of MhbT predicted by bioinformatics. In the control strains carrying gfp only, GFP was distributed in the cytoplasm (Fig. 2).
Fig 2.
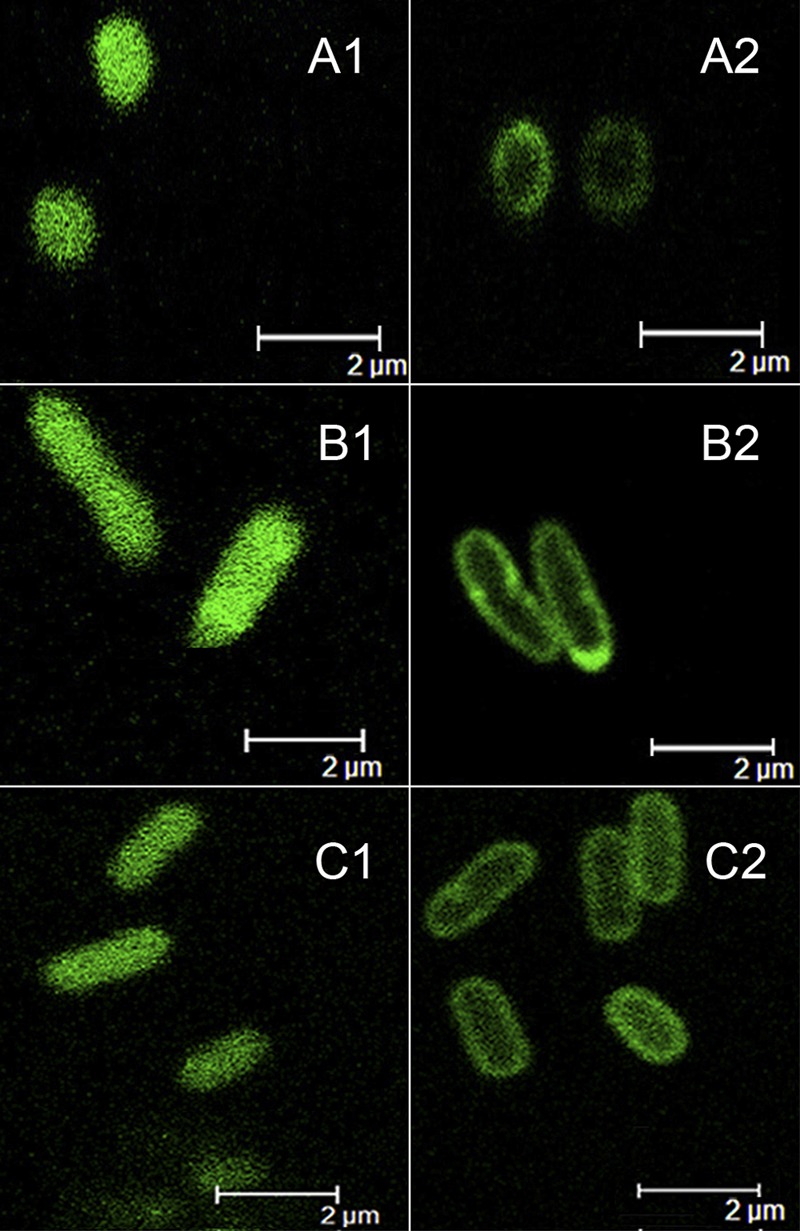
Localization of green fluorescent proteins by confocal microscopy. (A1, B1, and C1) GFP expressed in K. pneumoniae M5a1[pZWXY005] (A1), P. putida PaW340[pZWXY005] (B1), and E. coli BL21(DE3) harboring plasmid pGFPe (37) (C1) (controls), showing that GFP was distributed throughout the cytoplasm. (A2, B2, and C2) MhbT-GFP expressed in K. pneumoniae M5a1[pZWXY004] (A2), P. putida PaW340[pZWXY004] (B2), and E. coli BL21(DE3)[pZWXY006] (C2), showing that the fusion proteins were all located at the periphery of the cells.
MhbT is essential for 3-hydroxybenzoate catabolism.
In a previous study involving strain M5a1, all four 3-hydroxybenzoate catabolic enzymes exhibited high activities in 3-hydroxybenzoate-grown cells but very low and even undetectable activities in extracts from glycerol-grown cells (20). In this study, the quantitative PCR results indicated that there was a 6.2-fold increase in the transcription level of mhbT in the presence of 3-hydroxybenzoate. To identify the possible involvement of mhbT in 3-hydroxybenzoate catabolism, unsuccessful attempts to delete mhbT in strain M5a1 were conducted with a suicide vector, pZH5 (49), and a temperature-sensitive vector, pKD46 (6). Because of its ability to grow on 3-hydoxybenzoate when carrying the 6.3-kb EcoRI fragment of the mhb gene cluster, P. putida PaW340 was chosen as a substitute for the determination of the involvement of mhbT in the 3-hydoxybenzoate catabolic pathway by the introduction of pZWXY002 (carrying mhbRTDHIM) or pZWXY003 (carrying mhbRΔTDHIM with a truncated mhbT gene). Strain PaW340[pZWXY002] was able to utilize 3-hydroxybenzoate, and its OD600 reached 0.4 after a 48-h incubation, whereas strain PaW340[pZWXY003] virtually lost the ability to utilize 3-hydroxybenzoate. However, after pVLT33-based pZWXY001 (carrying mhbT) was introduced into strain PaW340[pZWXY003], its capability of growing on 3-hydroxybenzoate was restored. Such experiments were repeated three times, and similar results were obtained in all cases. This clearly indicated that mhbT in the mhb cluster of K. pneumoniae M5a1 was essential for 3-hydroxybenzoate assimilation in P. putida PaW340, which is presumably the case for K. pneumoniae M5a1.
MhbT transports 3-hydroxybenzoate.
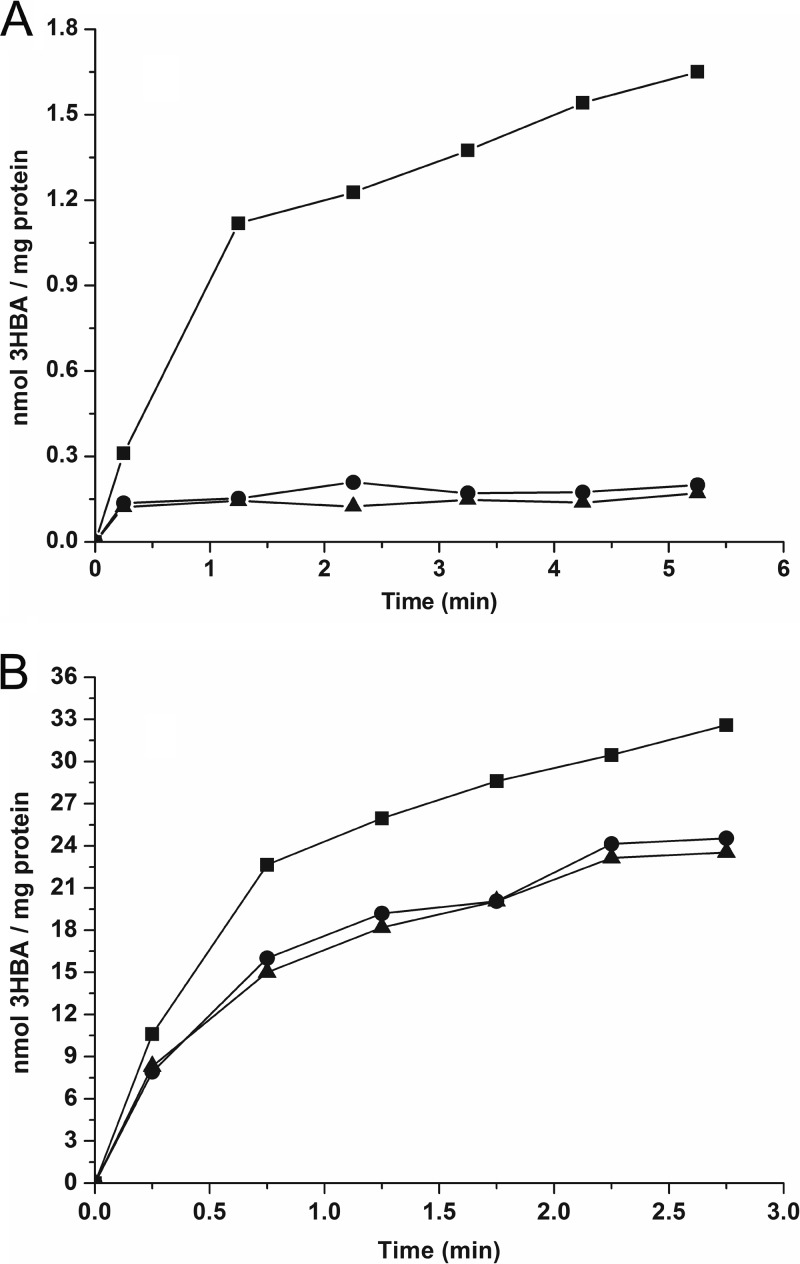
To detect the ability for 3-hydroxybenzoate transport by MhbT, the radiolabeled accumulation and the kinetics of 3-hydroxybenzoate transport were measured in resting cells. In a homogenous expression system, when K. pneumoniae M5a1 and its variants were grown in LB with IPTG induction, time courses of 3-hydroxybenzoate transport demonstrated that only cells of strain M5a1[pZWXY001] (expressing recombinant MhbT) were able to accumulate 14C-labeled 3-hydroxybenzoate (Fig. 3A), with a Vmax of 0.92 ± 0.06 nmol/min/mg of protein and a Km of 4.36 ± 0.12 μM. Wild-type strain M5a1 exhibited no transport activity, apparently because of a lack of mhbT transcription in the absence of 3-hydroxybenzoate. However, when they were grown on 3-hydroxybenzoate in MM with IPTG induction, all strains tested had the ability to transport 14C-labeled 3-hydroxybenzoate (Fig. 3B). Under these conditions, the Vmax was 18.35 ± 0.18 nmol/min/mg of protein and the Km was 5.57 ± 0.11 μM for 3-hydroxybenzoate-grown strain M5a1, in which all four enzymes for 3-hydroxybenzoate catabolism were active.
Fig 3.
MhbT transports [carboxyl-14C]3-hydroxybenzoate in the homogenous expression system. (A) K. pneumoniae M5a1 and its transconjugants grown in LB with IPTG induction at 30°C. (B) K. pneumoniae M5a1 and its transconjugants grown on 3-hydroxybenzoate in MM with IPTG induction at 30°C. The same experiments were repeated three times, with the same trends. 3HBA, 3-hydroxybenzoate; ■, K. pneumoniae M5a1[pZWXY001] (containing overexpressed MhbT); ●, K. pneumoniae M5a1[pVLT33]; ▲, K. pneumoniae M5a1.
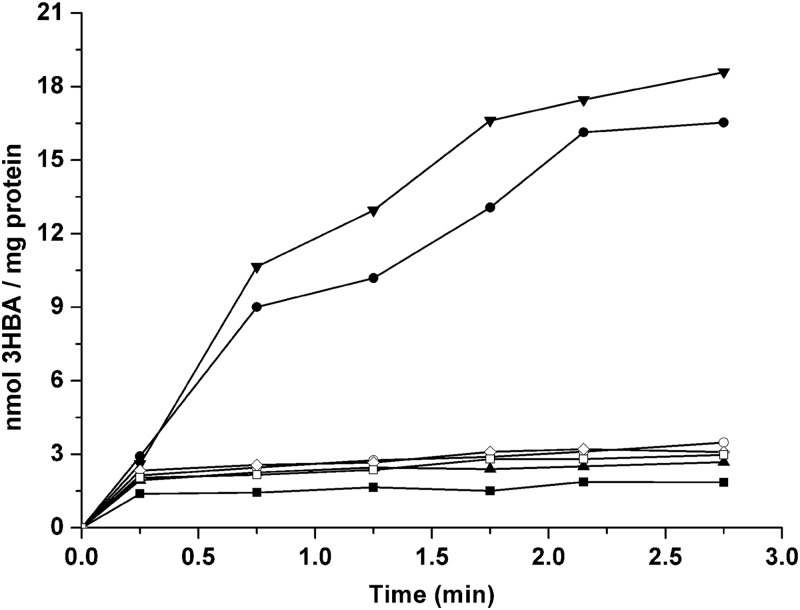
Additionally, a heterologous expression system was performed in P. putida PaW340, which was shown to be able to grow on 3-hydroxybenzoate after the introduction of the 6.3-kb EcoRI fragment with the complete mhbRTDHIM operon. Strain PaW340 and its transconjugants were grown in LB with 3-hydroxybenzoate and IPTG induction. As shown in Fig. 4, it was evident that strain PaW340[pZWXY002] (carrying mhbRTDHIM) accumulated 14C-labeled 3-hydroxybenzoate with a Vmax of 12.05 ± 0.18 nmol/min/mg of protein and a Km of 4.92 ± 0.08 μM. However, strain PaW340[pZWXY003] (carrying mhb genes with a truncated mhbT gene) showed virtually no ability to transport 3-hydroxybenzoate, similar to plasmid-free strain PaW340. This ability was restored in complemented strain PaW340[pZWXY001,pZWXY003].
Fig 4.
MhbT transports [carboxyl-14C]3-hydroxybenzoate in the heterologous expression system. P. putida PaW340 and its transconjugants were grown in LB with 3-hydroxybenzoate and IPTG induction at 30°C. The same experiments were repeated three times, with the same trends. 3HBA, 3-hydroxybenzoate; ■, P. putida PaW340; ●, P. putida PaW340[pZWXY002] (carrying the mhb gene cluster); ▲, P. putida PaW340[pZWXY003] (carrying the mhb gene cluster with truncated mhbT); ▼, P. putida PaW340[pZWXY001,pZWXY003] (carrying the mhb gene cluster with truncated mhbT and recombinant MhbT); ○, P. putida PaW340[pZWXY003,pZWXY007] (carrying the mhb gene cluster with truncated mhbT and the recombinant MhbT D82A mutant); ♢, P. putida PaW340[pZWXY003,pZWXY008] (carrying the mhb gene cluster with truncated mhbT and the recombinant MhbT V311W mutant); □, P. putida PaW340[pZWXY003,pZWXY009] (carrying the mhb gene cluster with truncated mhbT and the recombinant MhbT D314A mutant).
As described in the introduction, it is not known whether PcaK, BenK, or TfdK had extended transport activity, in addition to their respective primary substrates. In this study, measurements of uptake by MhbT were also performed with 14C-labeled benzoate and gentisate (a dihydroxylated benzoate) in the above-described systems, but no uptake of these two 3-hydroxybenzoate analogues by MhbT was detected.
Effect of pH values on uptake activity.
Strains PaW340[pZWXY002] (carrying mhbRTDHIM) and PaW340[pZWXY003] (carrying mhb genes with a truncated mhbT gene) were grown in LB with 3-hydroxybenzoate and IPTG induction, and their 3-hydroxybenzoate transport activities were detected at different pH values, as described in Materials and Methods. Strain PaW340[pZWXY002] accumulated 14C-labeled 3-hydroxybenzoate with similar activities of 10.37 ± 0.48, 12.89 ± 0.30, and 13.57 ± 0.59 nmol/min/mg of protein at pH 6.5, 7.5, and 8.5, respectively. In contrast, while strain PaW340[pZWXY003] exhibited a 3-hydroxybenzoate transport activity of 6.59 ± 0.18 nmol/min/mg of protein at pH 6.5, its activity was almost suppressed at pH 7.5 and 8.5 (1.89 ± 0.43 nmol/min/mg of protein and 2.01 ± 0.28 nmol/min/mg of protein, respectively). This finding suggested that active transport by MhbT played a particular role in substrate uptake under basic conditions.
Identification of residues in MhbT critical for 3-hydroxybenzoate transport.
Among the AAHS family members, two conserved stretches of amino acids in the cytoplasmic hydrophilic loops (the 2-3 and 8-9 loops) are known to be required for substrate transport (8, 9). To detect the involvement of the conserved motifs in MhbT in 3-hydroxybenzoate transport, two conserved charged amino acids, Asp-82 and Asp-314, in the 2-3 and 8-9 loops, respectively, were changed to an alanine. Val-311, which was specific in the 8-9 loop of MhbT, was replaced with tryptophan (V311W). The three mutated mhbT genes were inserted into pVLT33 to produce pZWXY007, pZWXY008, and pZWXY009, respectively (see Table S1 in the supplemental material). Uptake assays showed that all three mutants lost the ability to transport 14C-labeled 3-hydroxybenzoate (Fig. 4). This indicated that the conserved motifs of MhbT in the cytoplasmic hydrophilic loops were important for 3-hydroxybenzoate transport.
Overexpression of MhbT increases the specific activities of 3-hydroxybenzoate catabolic enzymes.
3-Hydroxybenzoate 6-hydroxylase (MhbM) and gentisate 1,2-dioxygenase (MhbD) catalyze the initial reactions of 3-hydroxybenzoate catabolism in strain M5a1. To investigate whether the overexpression of MhbT would affect the activities of these two enzymes, the specific activities of cell extracts from strain M5a1 and strain M5a1[pZWXY001] (containing overexpressed MhbT) were determined. Both cultures were grown in LB with 3-hydroxybenzoate, with strain M5a1[pZWXY001] being induced with IPTG. The overexpression of MhbT in strain M5a1[pZWXY001] resulted in a 2.5-fold increase of the 3-hydroxybenzoate 6-hydroxylase activity (0.262 U/mg) and a 3.3-fold increase of the gentisate 1,2-dioxygenase activity (0.695 U/mg) in comparison with those measured for a 3-hydroxybenzoate-grown wild-type culture. This observation is similar to that reported in a previous study, in which the overexpression of the styrene transporter, StyE, in P. putida CA-3 resulted in a 4.2-fold increase of the styrene monooxygenase activity (32).
Competition and inhibition experiments.
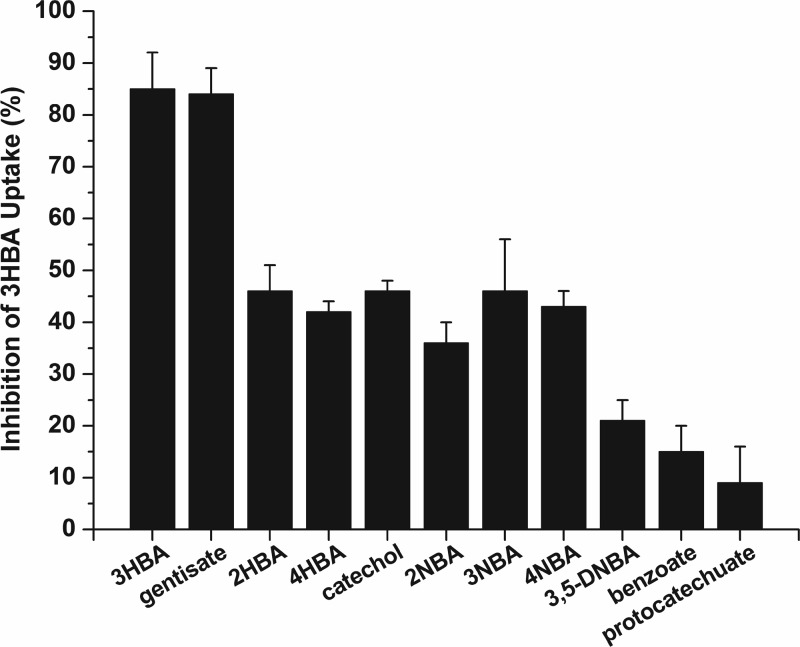
In order to detect whether 3-hydroxybenzoate transport by MhbT could be inhibited by other aromatic compounds, 14C-labeled 3-hydroxybenzoate uptake was determined in the presence of the structural analogues of 3-hydroxybenzoate by P. putida PaW340[pZWXY002] (carrying mhbRTDHIM). The uptake assays for transport were performed in the presence of 20-fold excesses of 3-hydroxybenzoate structural analogues. The impact on 3-hydroxybenzoate transport is shown in Fig. 5. In a control experiment, a 20-fold excess of unlabeled 3-hydroxybenzoate inhibited the rate of transport of radiolabeled 3-hydroxybenzoate by 85%. Gentisate inhibited the transport of 3-hydroxybenzoate by 84%. Catechol, the two monohydroxybenzoate isomers (2-hydroxybenzoate and 4-hydroxybenzoate), and the three nitro-substituted benzoates (2-nitrobenzoate, 3-nitrobenzoate, and 4-nitrobenzoate) inhibited approximately 40% of the uptake activity. The addition of benzoate, protocatechuate, and 3,5-dinitrobenzoate had a moderate effect on uptake, with about 20% inhibition.
Fig 5.
Substrate inhibition of MhbT-mediated 3-hydroxybenzoate uptake in P. putida PaW340[pZWXY002]. The concentrations of 3-hydroxybenzoate and the competing substrate were 40 and 800 μM, respectively. The rate of accumulation of 3-hydroxybenzoate (without competing substrates) was 11.86 ± 0.54 nmol/min/mg of protein, which was set as 100% activity. Inhibition was determined by comparing the rates of 3-hydroxybenzoate uptake in the presence and absence of competing substrates. Values are the averages of data from three experiments ± standard deviations. 2HBA, 2-hydroxybenzoate; 3HBA, 3-hydroxybenzoate; 4HBA, 4-hydroxybenzoate; 2NBA, 2-nitrobenzoate; 3NBA, 3-nitrobenzoate; 4NBA, 4-nitrobenzoate; 3,5-DNBA, 3,5-dinitrobenzoate.
DISCUSSION
Although 3-hydroxybenzoate catabolism has been studied thoroughly at both the genetic and biochemical levels in phylogenetically diverse strains (28, 35, 40, 46), no report on its transport was documented until MhbT was identified as an active 3-hydroxybenzoate transporter in the current study. MhbT transported 14C-labeled 3-hydroxybenzoate but not 14C-labeled gentisate and benzoate into the cells of two Gram-negative strains. Of the identified 3-hydroxybenzoate catabolic clusters in Rhodococcus sp. strain NCIMB 12038 (28) and Polaromonas naphthalenivorans CJ2 (35), no putative transporter-encoding genes were found in the vicinity of the catabolic clusters. However, a gentisate transporter gene was found in Corynebacterium glutamicum in the 3-hydroxybenzoate catabolic cluster (40). Extensive searching for MhbT homologues in GenBank resulted in the identification of its putative homologues in Rhodococcus jostii RHA1 (accession number NC_008268) and Burkholderia sp. strain NCIMB 10467 (accession number EU165545), where they are located near putative 3-hydroxybenzoate catabolic clusters. Transport was perhaps theoretically unnecessary, because aromatic acids could diffuse across biological membranes (21), but accumulating evidence indicates that the active transport of aromatic acids is widespread among bacteria (1, 5, 15, 24, 31, 33). The aromatic acids have pKa values of around 4.0 to 4.5, depending on the substituents on the ring, and the pKa of 3-hydroxybenzoate is 4.202 (ACD/Labs Software). It is generally accepted that these acids are present in an undissociated form that diffuses across membranes by passive diffusion under acidic conditions. Moreover, under neutral or alkaline conditions, less than 0.1% aromatic acids would be present in the undissociated form, and active transport is necessary for their transport. Therefore, the pH of the medium is a major factor in the decision of whether microbes require certain transporters to grow on aromatic acids. In this study, strain PaW340[pZWXY003] (carrying mhb genes and truncated mhbT) was unable to grow on 3-hydroxybenzoate at pH 6.5. However, it still retained 64% 3-hydroxybenzoate transport activity compared with that of strain PaW340[pZWXY002] (carrying all mhb genes) at this pH value. This is likely because the undissociated form of 3-hydroxybenzoate was capable of entering cells by passive diffusion under acidic conditions. In contrast, the transport activity of strain PaW340[pZWXY003] was almost suppressed at pH 7.5 and 8.5, with a loss of more than 85% of the activity. Although aromatic acids can enter cells by passive diffusion, active transport is thought to increase the efficiency and rate of substrate absorption and may impart a growth advantage in natural environments, where these compounds are present at low concentrations (33).
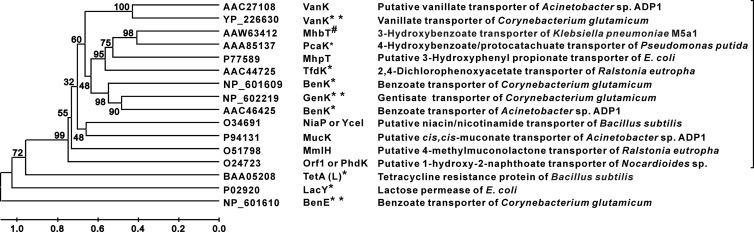
MhbT showed relatively low amino acid sequence identities (27 to 42%) with three functionally identified AAHS transporters. A phylogenetic tree was generated to further elucidate the relationship among AAHS family members (Fig. 6). Phylogenetic analysis indicated that the AAHS family of transporters (including both putative and functionally identified members) loosely formed one cluster, which was distant from the outgroup. However, the relatively low amino acid sequence identities among the identified AAHS transporters suggest that they may have undergone adaptive evolution to result in the specific transport of the respective aromatic acids in the process of their catabolism. Therefore, the functions and substrate specificities of putative transporters cannot be convincingly deduced without functional analyses using radiolabeled compounds, such as in the current study.
Fig 6.
Phylogenetic relationship of the AAHS family transporters. The construction of the phylogenetic tree (using the neighbor-joining method) and multiple-sequence alignments were performed with Mega 3.1 software. The bootstrap confidence limits are indicated at the nodes, and the scale at the bottom indicates sequence divergence. “∗” indicates that the function of transporters was examined by genetic disruption/complementation and uptake assays with radioisotopes. “∗∗” indicates that the function of transporters was examined by genetic disruption/complementation. “#” indicates the 3-hydroxybenzoate transporter identified in this study. LacY (13), TetA(L) (14), and BenE (NCgl2326) (3) were used as the outgroups.
Unlike the broad substrate ranges for many catabolic enzymes in aromatic degradation, the transporters for these substrates exhibited a relatively narrow substrate range. Among the transporters studied in the AAHS family, BenK (5) and TfdK (24) were shown previously to transport benzoate and 2,4-dichlorophenoxyacetate, respectively. PcaK was able to transport both 4-hydroxybenzoate and protocatechuate (33). MhbT specifically transported 3-hydroxybenzoate in the current study. It was thought that the 12 transmembrane α-helices in the MFS transporters packed to form the perimeter of a pore through which the substrate crosses the cell membrane (13, 16, 23). The changing of a single amino acid residue made conformational changes of the transporters, affecting the ability of the substrate to cross the cell membrane (19). The two conserved domains [GXXXD(R/K)XGR(R/K)] in two identified cytoplasmic hydrophilic loops (the 2-3 and 8-9 loops) were known to be required for substrate transportation in the AAHS family (8, 9). This was also the case for MhbT, in which two strictly conserved residues, Asp-82 and Asp-314 in the 2-3 and 8-9 loops, respectively, were important for 3-hydroxybenzoate transport activity (Fig. 4). The less-conserved Val-311 of MhbT in the 8-9 loop was different from the corresponding residues in PcaK (W-309), BenK (W-305), and TfdK (L-314) but was also necessary for activity. This specific residue in MhbT possibly has an important role in the substrate specificity of this 3-hydroxybenzoate transport.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant 30730002) and the National Key Basic Research Program of China (973 Program) (grant 2012CB725202).
We thank R. A. Cooper for providing pBSI, Ji-Lun Li for Klebsiella pneumoniae M5a1, Ping Zheng for pZH5, and Hai-Hong Wang for pKD46.
Footnotes
Published ahead of print 22 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Allende JL, Gibello A, Martin M, Garrido-Pertierra A. 1992. Transport of 4-hydroxyphenylacetic acid in Klebsiella pneumoniae. Arch. Biochem. Biophys. 292: 583–588 [DOI] [PubMed] [Google Scholar]
- 2. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- 3. Chaudhry MT, et al. 2007. Genome-wide investigation of aromatic acid transporters in Corynebacterium glutamicum. Microbiology 153: 857–865 [DOI] [PubMed] [Google Scholar]
- 4. Cleland WW. 1967. The statistical analysis of enzyme kinetic data. Adv. Enzymol. Relat. Areas Mol. Biol. 29: 1–32 [DOI] [PubMed] [Google Scholar]
- 5. Collier LS, Nichols NN, Neidle EL. 1997. benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobacter sp. strain ADP1. J. Bacteriol. 179: 5943–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Lorenzo V, Eltis L, Kessler B, Timmis KN. 1993. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123: 17–24 [DOI] [PubMed] [Google Scholar]
- 8. Ditty JL, Harwood CS. 2002. Charged amino acids conserved in the aromatic acid/H+ symporter family of permeases are required for 4-hydroxybenzoate transport by PcaK from Pseudomonas putida. J. Bacteriol. 184: 1444–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ditty JL, Harwood CS. 1999. Conserved cytoplasmic loops are important for both the transport and chemotaxis functions of PcaK, a protein from Pseudomonas putida with 12 membrane-spanning regions. J. Bacteriol. 181: 5068–5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dixon RA. 1984. The genetic complexity of nitrogen fixation. The ninth Fleming lecture. J. Gen. Microbiol. 130: 2745–2755 [DOI] [PubMed] [Google Scholar]
- 11. Dixon RA, Postgate JR. 1971. Transfer of nitrogen-fixation genes by conjugation in Klebsiella pneumoniae. Nature 234: 47–48 [DOI] [PubMed] [Google Scholar]
- 12. Gao X, Tan CL, Yeo CC, Poh CL. 2005. Molecular and biochemical characterization of the xlnD-encoded 3-hydroxybenzoate 6-hydroxylase involved in the degradation of 2,5-xylenol via the gentisate pathway in Pseudomonas alcaligenes NCIMB 9867. J. Bacteriol. 187: 7696–7702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guan L, Kaback HR. 2006. Lessons from lactose permease. Annu. Rev. Biophys. Biomol. Struct. 35: 67–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guffanti AA, Krulwich TA. 1995. Tetracycline/H+ antiport and Na+/H+ antiport catalyzed by the Bacillus subtilis TetA(L) transporter expressed in Escherichia coli. J. Bacteriol. 177: 4557–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harwood CS, Nichols NN, Kim MK, Ditty JL, Parales RE. 1994. Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J. Bacteriol. 176: 6479–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hirai T, Heymann JA, Maloney PC, Subramaniam S. 2003. Structural model for 12-helix transporters belonging to the major facilitator superfamily. J. Bacteriol. 185: 1712–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ishiyama D, Vujaklija D, Davies J. 2004. Novel pathway of salicylate degradation by Streptomyces sp. strain WA46. Appl. Environ. Microbiol. 70: 1297–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jeenes DJ, Williams PA. 1982. Excision and integration of degradative pathway genes from TOL plasmid pWW0. J. Bacteriol. 150: 188–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jessen-Marshall AE, Parker NJ, Brooker RJ. 1997. Suppressor analysis of mutations in the loop 2-3 motif of lactose permease: evidence that glycine-64 is an important residue for conformational changes. J. Bacteriol. 179: 2616–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones DC, Cooper RA. 1990. Catabolism of 3-hydroxybenzoate by the gentisate pathway in Klebsiella pneumoniae M5a1. Arch. Microbiol. 154: 489–495 [DOI] [PubMed] [Google Scholar]
- 21. Kashket ER. 1985. The proton motive force in bacteria: a critical assessment of methods. Annu. Rev. Microbiol. 39: 219–242 [DOI] [PubMed] [Google Scholar]
- 22. Keen NT, Tamaki S, Kobayashi D, Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70: 191–197 [DOI] [PubMed] [Google Scholar]
- 23. Law CJ, Maloney PC, Wang DN. 2008. Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 62: 289–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leveau JH, Zehnder AJ, van der Meer JR. 1998. The tfdK gene product facilitates uptake of 2,4-dichlorophenoxyacetate by Ralstonia eutropha JMP134(pJP4). J. Bacteriol. 180: 2237–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin LX, Liu H, Zhou NY. 2010. MhbR, a LysR-type regulator involved in 3-hydroxybenzoate catabolism via gentisate in Klebsiella pneumoniae M5a1. Microbiol. Res. 165: 66–74 [DOI] [PubMed] [Google Scholar]
- 26. Liu DQ, Liu H, Gao XL, Leak DJ, Zhou NY. 2005. Arg169 is essential for catalytic activity of 3-hydroxybenzoate 6-hydroxylase from Klebsiella pneumoniae M5a1. Microbiol. Res. 160: 53–59 [DOI] [PubMed] [Google Scholar]
- 27. Liu H, Wang SJ, Zhou NY. 2005. A new isolate of Pseudomonas stutzerithat degrades 2-chloronitrobenzene. Biotechnol. Lett. 27: 275–278 [DOI] [PubMed] [Google Scholar]
- 28. Liu TT, et al. 2011. Functional characterization of a gene cluster involved in gentisate catabolism in Rhodococcus sp. strain NCIMB 12038. Appl. Microbiol. Biotechnol. 90: 671–678 [DOI] [PubMed] [Google Scholar]
- 29. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- 30. Luo S, Liu DQ, Liu H, Zhou NY. 2006. Site-directed mutagenesis of gentisate 1,2-dioxygenases from Klebsiella pneumoniae M5a1 and Ralstonia sp. strain U2. Microbiol. Res. 161: 138–144 [DOI] [PubMed] [Google Scholar]
- 31. Miguez CB, Greer CW, Ingram JM, Macleod RA. 1995. Uptake of benzoic acid and chloro-substituted benzoic acids by Alcaligenes denitrificans BRI 3010 and BRI 6011. Appl. Environ. Microbiol. 61: 4152–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mooney A, O'Leary ND, Dobson AD. 2006. Cloning and functional characterization of the styE gene, involved in styrene transport in Pseudomonas putida CA-3. Appl. Environ. Microbiol. 72: 1302–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nichols NN, Harwood CS. 1997. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J. Bacteriol. 179: 5056–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pao SS, Paulsen IT, Saier MH., Jr 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62: 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Park M, et al. 2007. Molecular and biochemical characterization of 3-hydroxybenzoate 6-hydroxylase from Polaromonas naphthalenivorans CJ2. Appl. Environ. Microbiol. 73: 5146–5152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pogulis RJ, Vallejo AN, Pease LR. 1996. In vitro recombination and mutagenesis by overlap extension PCR. Methods Mol. Biol. 57: 167–176 [DOI] [PubMed] [Google Scholar]
- 37. Rapp M, et al. 2004. Experimentally based topology models for E. coli inner membrane proteins. Protein Sci. 13: 937–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robson ND, Parrott S, Cooper RA. 1996. In vitro formation of a catabolic plasmid carrying Klebsiella pneumoniae DNA that allows growth of Escherichia coli K-12 on 3-hydroxybenzoate. Microbiology 142: 2115–2120 [DOI] [PubMed] [Google Scholar]
- 39. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 40. Shen XH, Jiang CY, Huang Y, Liu ZP, Liu SJ. 2005. Functional identification of novel genes involved in the glutathione-independent gentisate pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 71: 3442–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walker AW, Keasling JD. 2002. Metabolic engineering of Pseudomonas putida for the utilization of parathion as a carbon and energy source. Biotechnol. Bioeng. 78: 715–721 [DOI] [PubMed] [Google Scholar]
- 42. Wang LH, Hamzah RY, Yu YM, Tu SC. 1987. Pseudomonas cepacia 3-hydroxybenzoate 6-hydroxylase: induction, purification, and characterization. Biochemistry 26: 1099–1104 [DOI] [PubMed] [Google Scholar]
- 43. Williams PA, Murray K. 1974. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J. Bacteriol. 120: 416–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xiao Y, Zhang JJ, Liu H, Zhou NY. 2007. Molecular characterization of a novel ortho-nitrophenol catabolic gene cluster in Alcaligenes sp. strain NyZ215. J. Bacteriol. 189: 6587–6593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu Y, Yan DZ, Zhou NY. 2006. Heterologous expression and localization of gentisate transporter Ncg12922 from Corynebacterium glutamicum ATCC 13032. Biochem. Biophys. Res. Commun. 346: 555–561 [DOI] [PubMed] [Google Scholar]
- 46. Yang YF, Zhang JJ, Wang SH, Zhou NY. 2010. Purification and characterization of the ncgl2923-encoded 3-hydroxybenzoate 6-hydroxylase from Corynebacterium glutamicum. J. Basic Microbiol. 50: 599–604 [DOI] [PubMed] [Google Scholar]
- 47. Yin Y, et al. 2010. Characterization of catabolic meta-nitrophenol nitroreductase from Cupriavidus necator JMP134. Appl. Microbiol. Biotechnol. 87: 2077–2085 [DOI] [PubMed] [Google Scholar]
- 48. Youn JW, Jolkver E, Kramer R, Marin K, Wendisch VF. 2008. Identification and characterization of the dicarboxylate uptake system DccT in Corynebacterium glutamicum. J. Bacteriol. 190: 6458–6466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zheng P, Sun J, van den Heuvel J, Zeng AP. 2006. Discovery and investigation of a new, second triose phosphate isomerase in Klebsiella pneumoniae. J. Biotechnol. 125: 462–473 [DOI] [PubMed] [Google Scholar]
- 50. Zhou NY, Fuenmayor SL, Williams PA. 2001. nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J. Bacteriol. 183: 700–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.