Abstract
The rol (designated for resorcinol) gene cluster rolRHMD is involved in resorcinol catabolism in Corynebacterium glutamicum, and RolR is the TetR-type regulator. In this study, we investigated how RolR regulated the transcription of the rol genes in C. glutamicum. The transcription start sites and promoters of rolR and rolHMD were identified. Quantitative reverse transcription-PCR and promoter activity analysis indicated that RolR negatively regulated the transcription of rolHMD and of its own gene. Further, a 29-bp operator rolO was located at the intergenic region of rolR and rolHMD and was identified as the sole binding site for RolR. It contained two overlapping inverted repeats and they were essential for RolR-binding. The binding of RolR to rolO was affected by resorcinol and hydroxyquinol, which are the starting compounds of resorcinol catabolic pathway. These two compounds were able to dissociate RolR-rolO complex, thus releasing RolR from the complex and derepressing the transcription of rol genes in C. glutamicum. It is proposed that the binding of RolR to its operator rolO blocks the transcription of rolHMD and of its own gene, thus negatively regulated resorcinol degradation in C. glutamicum.
INTRODUCTION
Based on the exploration of DNA and protein databases, TetR family regulators are particular abundant in microbes exposed to environmental changes, e.g., soil bacteria such as Nocardia, Streptomyces, Pseudomonas, and Bacillus species (26). Consequently, members of the TetR family are believed to be involved in the adaptation to complex and changing environments. It has been reported that TetR family regulators function as negative regulators for physiological processes such as efflux pumping and biosynthesis of antibiotics (9, 10, 15, 18, 29), osmotic stress (27), and solvent resistance (6). Involvement of a TetR-type regulator in cymene and biphenyl catabolism was reported in Pseudomonas putida (7, 8, 24, 32). Recently, the gene rolR (tagged as ncgl1110 in GenBank), encoding a TetR-type regulator and involved in resorcinol degradation, was identified in Corynebacterium glutamicum (16). However, the regulatory mechanism of this TetR-type regulator for aromatic catabolism has not been revealed.
Resorcinol and its derivatives occur in soil and other environments due to their extensive applications in adhesive production, tire and rubber processing, and antiseptic and disinfectant preparations (23). Besides industrial production via chemical synthesis, resorcinol and its derivatives are produced in nature as secondary plant products (3, 5). Exposure of microbes to resorcinolic compounds in environment leads to the evolution of metabolic pathways and regulatory machines for resorcinol catabolism. Investigations indicated that resorcinol was degraded via different pathways in bacteria. In Azotobacter vinelandii, resorcinol was converted into pyrogallol, and subsequently the aromatic ring was cleaved by a pyrogallol 1,2-dioxygenase (13). P. putida, Rhizobium species, and C. glutamicum converted resorcinol into hydroxyquinol, and hydroxyquinol was subsequently degraded by either 2,3,5-trihydroxytoluene 1,2-dioxygenase (meta-cleavage) or by hydroxyquinol 1,2-dioxygenase (ortho-cleavage) (4, 16, 33). Our previous study showed that the rolR gene, encoding a TetR-type transcriptional regulator, together with rolH, rolM, and rolD that encode resorcinol hydroxylase, maleylacetate reductase, and hydroxyquinol 1,2-dioxygenase, respectively, compose a gene cluster involved in resorcinol catabolism in C. glutamicum (16). The apo- and effector-bound (with resorcinol) crystal structures of RolR were reported, showing that conformation variation of DNA-binding domain occurs due to resorcinol binding (20). In the present study we sought to reveal the regulatory mechanism of RolR on the transcription of rol genes in C. glutamicum.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in the present study are listed in Table 1. Escherichia coli was grown aerobically on a rotary shaker (180 rpm) at 37°C in Luria-Bertani (LB) broth or on an LB plate with 1.5% (wt/v) agar. C. glutamicum was routinely grown on a rotary shaker (150 rpm) at 30°C in LB medium or in mineral salts medium (19), which was adjusted to pH 8.4 and supplemented with 0.05 g of yeast extract liter−1. Resorcinol was added at final concentrations of 2 mM (sterilized by filtration through 0.2-μm-pore-size filters) when they served as carbon and energy sources. Cellular growth was monitored by measuring the turbidity at 600 nm. Antibiotics were used at the following concentrations: kanamycin at 50 μg ml−1 for E. coli and 25 μg ml−1 for C. glutamicum and chloramphenicol at 20 μg ml−1 for E. coli and 10 μg ml−1 for C. glutamicum.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| JM109 | recAl supE44 endAI hsdR17 gyrA96 relAI thi Δ(lac-proAB) F′(traD36 proAB lacIq lacZΔM15) | Stratagene (catalog no. 200235) |
| BL21(DE3) | hsdS gal (λcIts857 ind-l Sam7 nin-5 lacUV5-T7 gene 1) | Novagen (catalog no. 69387–3) |
| C. glutamicum | ||
| RES167 | Restriction-deficient mutant of ATCC 13032, Δ(cglIM-cglIR-cglIIR) | University of Bielefeld |
| RES167ΔrolR | A fragment of DNA encoding for amino acids 105 to 215 of rolR was deleted | 16 |
| RES167ΔrolR/pXMJ19-ProlR-lacZ | For ProlR activity determination in the absence of RolR | This study |
| RES167ΔrolR/pTRCmob-rolR/pXMJ19-ProlR-lacZ | For ProlR activity determination in the presence of RolR | This study |
| Plasmids | ||
| pXMJ19 | E. coli-C. glutamicum shuttle expression vector (Camr Ptac lacIq pBL1 oriVC.g. pK18 oriVE.c.) | 17 |
| pXMJ19-lacZ | pXMJ19 carrying lacZ | 34 |
| pXMJ19-ProlR-lacZ | pXMJ19 carrying ProlR and lacZ, and lacIq and Ptac were deleted | This study |
| pET28a | Expression vector with N-terminal hexahistidine affinity tag | Novagen |
| pET28a-rolR | pET28a derivative for expression of N-His6-tagged RolR in E. coli | This study |
| pTRCmob | E. coli-C. glutamicum shuttle expression vector (derived from pEC-XK99E, harboring mob region) | 21 |
| pTRCmob-rolR | pTRCmob derivative for expression of rolR in C. glutamicum | This study |
Analysis of sequence data.
The genome sequence of C. glutamicum ATCC 13032 (accession no. NC_003450 and NC_006958) was retrieved from the GenBank database (http://www.ncbi.nlm.nih.gov/). Sequence comparisons and database searches were carried out using BLAST programs at the BLAST server of the National Center for Biotechnology Information (NCBI) website. For identification of rolO-like sequences, motif-based sequence analysis tool MEME (http://meme.sdsc.edu/meme/cgi-bin/meme.cgi) was used according to the default parameters (1). The continuous 600-bp fragments encompassing 300 bp upstream and downstream of the start codon of the rolR-like genes were used.
Nucleic acid extraction and manipulation.
Genomic DNA of C. glutamicum was isolated using DNA extraction kit (SBS, Beijing, China). DNA manipulation, plasmid isolation, and agarose gel electrophoresis were routinely carried out according to standard methods. Total RNA was extracted from cells grown at late log phase by use of TRIzol reagent RNA extraction kit (Invitrogen, Carlsbad, CA). Cells were disrupted with liquid nitrogen grinding. The total RNA was quantified by NanoDrop ND-100 UV/Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE). Restriction endonucleases, ligase, and DNA polymerase were used according to the manufacturer's instructions. Vectors were electroporated into cells of E. coli and C. glutamicum according to previously described methods (31). The restricted DNA fragments amplified by PCR were separated by agarose gel electrophoresis and purified using an agarose gel DNA fragment recovery kit (Tiangen, Beijing, China). Target DNA fragments were PCR amplified using Pyrobest DNA polymerase (TaKaRa, Dalian, China) or Taq DNA polymerase (Promega, Madison, WI). The PCR products were ligated into T-vector using a pEASY-T1 cloning kit (Transgen, Beijing, China).
Primer extension.
Prior to reverse transcription, RNA was treated with DNase I (Promega) for 30 min at 37°C, followed by heat inactivation of DNase I. For primer extension of the rolHMD and rolR genes, primer PE11 and PE10 (see Table SA1 in the supplemental material) were labeled with [γ-32P]ATP using T4 polynucleotide kinase (Promega) according to the manufacturer's protocol. Reverse transcription was carried out with 10 pmol of 5′-32P-labeled primers and 30 μg of total RNA extracted from C. glutamicum cells cultivated with resorcinol. After annealing at 65°C for 10 min, deoxynucleoside triphosphates and Moloney murine leukemia virus transcriptase (Promega) were added, followed by incubation at 45°C for 60 min and then at 65°C for 10 min for denaturing the transcriptase. The reaction mixture was treated with phenol, precipitated with ethanol, and resuspended with loading buffer. The dissolved sample was separated on a 6% polyacrylamide gel containing 8 M urea. The DNA-sequencing reaction mixtures were set up using the same labeled primers and a silver stain sequencing kit (Promega).
Promoter activity assay.
The putative promoter ProlR was PCR amplified from genomic DNA and ligated into plasmid pXMJ19-lacZ (34) to generate pXMJ19-ProlR-lacZ. In order to overexpress rolR, plasmid pTRCmob-rolR was constructed by ligating PCR-amplified rolR into pTRCmob vector (21) and was electroporated into C. glutamicum. Primers for PCR amplification are listed in Table SA1 in the supplemental material. The C. glutamicum strains RES167ΔrolR/pXMJ19-ProlR-lacZ (rolR mutant carrying plasmid pXMJ19-ProlR-lacZ) and RES167ΔrolR/pTRCmob-rolR/pXMJ19-ProlR-lacZ (rolR mutant carrying plasmid pTRCmob-rolR and pXMJ19-ProlR-lacZ) were grown in LB broth, and the cells were harvested at late log phase to determine the β-galactosidase activity. The β-galactosidase activity was determined as previously described (10).
RT-PCR.
One microgram of total RNA was incubated with gDNA Eraser (TaKaRa) at 42°C for 2 min to remove genomic DNA and then reverse transcribed using PrimeScript RT Enzyme (TaKaRa) in a volume of 20 μl with random primer to generate first cDNA. For quantitative reverse transcription-PCR (RT-qPCR), the cDNA was diluted 20 times as a template, and PCR was performed by use of SYBR Premix Ex Taq II kit (TaKaRa) according to the instructions in an ABI Prism 7000 Fast Real-Time PCR System. Housekeeping gene rpoB of C. glutamicum was used as an internal control. The relative expression levels of target genes of wild-type and mutant under defined culture conditions to their expression levels of wild-type RES167 grown in the presence of glucose were evaluated by using the 2−ΔΔCT method (22). To determine whether genes rolH, rolM, and rolD are cotranscribed, the first cDNA of strain RES167 grown with resorcinol as the sole carbon source was used as a template to amplify the fragments spanning the intergenic region between rolH and rolM or between rolM and rolD. Primers used in RT-PCR are listed in Table SA1 in the supplemental material.
Purification of His6-RolR.
The coding sequence of rolR was amplified by PCR from genomic DNA of C. glutamicum. After endonuclease treatment, the rolR fragment was ligated into pET28a, generating plasmid pET28a-rolR. Expression of protein RolR with a His6 tag at its N terminus was induced in E. coli BL21(DE3) by 0.6 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 30°C for 3 h. His6-RolR was purified by Ni2+-NTA chromatography, followed by fast protein liquid chromatography with a gel filtration column and then quantified by using the Bradford method (2). The native molecular mass of purified His6-RolR was determined by gel filtration on a Superdex 75 10/300 GL column using a buffer (50 mM Tris-HCl, 150 mM NaCl [pH 7.5]) with a gel filtration calibration kit (low molecular weight; GE, United Kingdom). The calibration curve was plotted by use of the Kav versus the logarithm of the molecular weight.
EMSA.
The primers listed in Table SA1 in the supplemental material were labeled with [γ-32P]ATP using T4 polynucleotide kinase (Promega) according to the manufacturer's protocol. The 32P-labeled DNA fragments used in an electrophoretic mobility shift assay (EMSA) were obtained by PCR amplification with labeled primers and C. glutamicum genomic DNA as a template. Approximate 0.5 pmol of 32P-labeled DNA fragment and His6-RolR protein with different amounts were mixed in a total volume of 20 μl, followed by incubation at room temperature (about 25°C) for 30 min. The binding buffer contained 10 mM Tris-HCl (pH 7.5), 50 mM KCl, and 1 mM dithiothreitol (DTT). For the addition of effectors, all chemicals were added at 10 pmol and incubated together with 32P-labeled DNA and His6-RolR protein. The binding mixture was loaded onto a native gel (5% polyacrylamide). Electrophoresis was performed at room temperature with Tris-borate-EDTA buffer (0.5×) as a running buffer. After electrophoresis, the gels were dried and subjected to autoradiography.
DNase I footprinting.
The DNA fragment containing the operator and promoters of rolR and rolHMD was amplified with 5′-32P-labeled FP11F and FP11R (see Table SA1 in the supplemental material) from genomic DNA. The obtained 32P-labeled DNA fragment (1 pmol) was mixed with His6-RolR (10 pmol) in 50 μl of binding buffer (10 mM Tris-HCl, 50 mM KCl, 1 mM DTT [pH 7.5]). The mixture was incubated for 30 min at 25°C. Then, 10 mU of DNase I (Promega) was added for 3 min at 25°C. Then, 50 μl of DNase I stop buffer was added to the reaction mixture. After the treatment with phenol, the mixture was precipitated with ethanol at −70°C overnight. The sediments were resuspended with loading buffer. The dissolved sample was separated on a 6% polyacrylamide gel containing 8 M urea. The DNA-sequencing reaction mixtures were set up using the same 32P-labeled FP11F and a silver stain sequencing kit (Promega).
SPR.
All DNA fragments used in surface plasmon resonance (SPR) experiments are double stranded and are listed in Table SA1 in the supplemental material. The oligonucleotides were synthesized and annealed with equal amounts of sense and antisense DNA strands to produce double-stranded fragments. SPR experiments were performed on a Biacore 3000 apparatus (GE) with a running buffer composed of 25 mM HEPES, 100 mM NaCl, and 0.005% Tween 20 (pH 7.4). To determine the binding of RolR with different double-stranded DNA (dsDNA) fragments, His6-RolR was immobilized on a CM5 sensor chip (GE, Sweden), and the individual DNA fragment (50 μM, 20 μl) was injected at a flow rate of 20 μl/min. For the detailed analysis of the association and dissociation of operator rolO and RolR with the addition of effectors, a 5′-end biotinylated rolO dsDNA fragment was immobilized on a streptavidin-coated SA sensor chip (GE) and His6-RolR (38.8 nM, 60 μl) with resorcinol (1 mM, 60 μl) or hydroxyquinol (1 mM, 60 μl) or running buffer were injected with a “COINJECT” pattern at a flow rate of 30 μl/min. At the end of each cycle, the sensor chip was regenerated by injecting 5 μl of running buffer plus 0.01% sodium dodecyl sulfate. The dissociation rate (kd) were determined using BIAevaluation 4.1 software.
RESULTS
The rolHMD gene cluster is transcribed as one whole transcript.
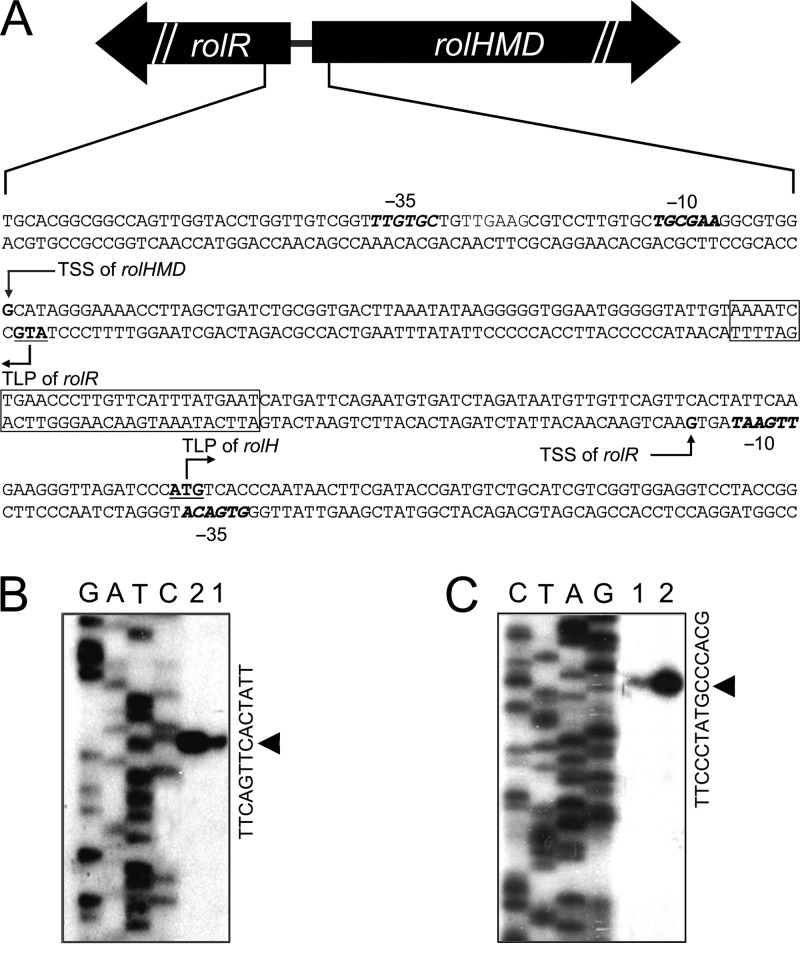
Previous study found that the gene cluster rolRHMD (ncgl1110 to ncgl1113) is involved in resorcinol catabolism (16). According to DNA sequence analysis, the rolR gene is located upstream of rolH gene and has a transcriptional direction opposite to that of rolHMD. There is an intergenic DNA fragment of 153 bp between the coding regions of rolR and rolHMD genes (Fig. 1A). To identify whether rolH, rolM, and rolD are cotranscribed, RT-PCR analysis was performed using the total RNA of RES167 cells grown in the presence of resorcinol. With the addition of reverse transcriptase, the fragments spanning the intergenic region between rolH and rolM or between rolM and rolD were amplified, whereas no product was found without reverse transcriptase (see Fig. SA1 in the supplemental material), indicating that rolH, rolM, and rolD are transcribed as one continuous transcript, so we named the transcript rolHMD. In order to determine the transcription start sites of rolR and rolHMD, primer extension was carried out (Fig. 1B and C). The results indicated that the transcription of rolHMD starts at a G (corresponding to a C of the complementary chain in Fig. 1C), which is located 157 nucleotides (nt) upstream of the rolH translational start point (ATG). The transcription of rolR in the opposite direction also starts with a G (corresponding to a C of the complementary chain in Fig. 1B), which is located 129 nt upstream of its own translational start point. Therefore, the rolR and rolHMD genes are transcribed in opposite directions, and their transcripts overlap for 132 nt. With the identified transcription start sites, the promoters (−10 and −35 regions) of rolR and rolHMD were deduced (Fig. 1A). It is noteworthy that ProlHMD (the promoter of rolHMD) is located in the coding region of rolR and that ProlR (the promoter of rolR) overlaps with the start codon of rolHMD.
Fig 1.
Promoters and transcription start site (TSS) analysis of rolR and rolHMD. (A) Depiction of the regulatory region of rolR and rolHMD genes. The TSS, the −10 and −35 regions, and the translational start points (TLP) of rolR and rolH genes are shown. The −10 and −35 regions of rolR and rolHMD genes are indicated in boldface italics. The rolO sequence is boxed. The TSSs were determined using primer extension of rolR (B) and rolHMD (C). Lanes 1 and 2 of panels B and C show reverse transcription products loaded with different concentrations.
RolR negatively regulates the transcription of rolHMD and of its own gene.
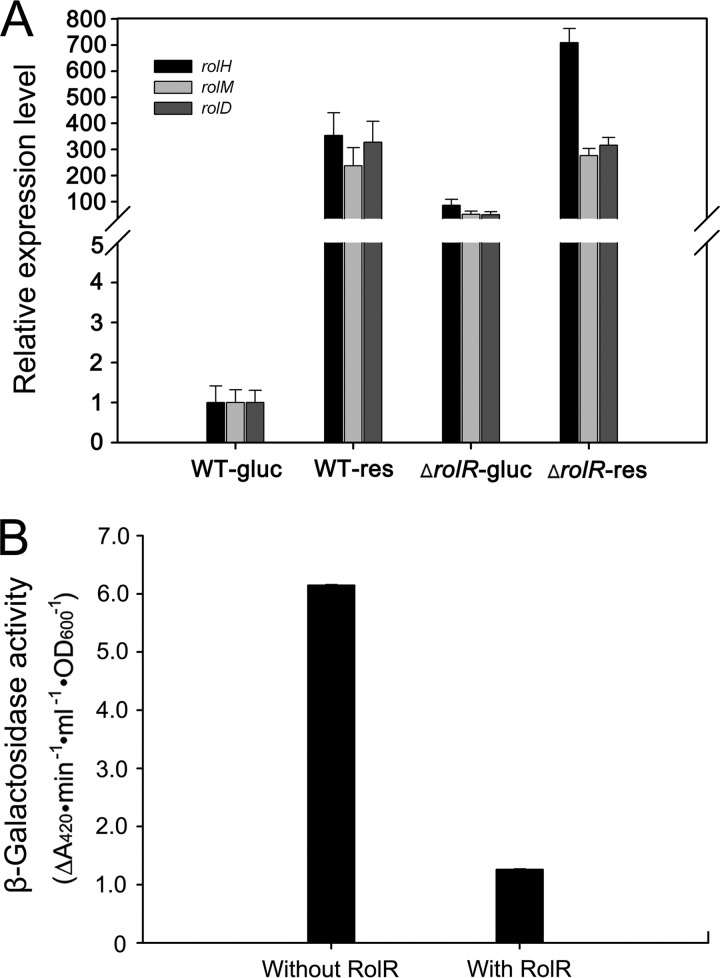
Our previous study suggested that RolR might negatively regulate rol genes (16). In order to obtain direct evidence for the regulation of RolR on rolHMD, quantitative RT-PCR was performed with wild-type strain RES167 and the ES167ΔrolR mutant strain grown with glucose or resorcinol at concentration of 2 mM as the sole carbon source. When RES167 was grown with resorcinol, the expressions of rolH, rolM, and rolD increased ∼300-fold compared to growth with glucose (Fig. 2A). Regardless of glucose or resorcinol, the expressions of rolH, rolM, and rolD in the mutant RES167ΔrolR were much higher than in RES167 grown with glucose (Fig. 2A), indicating that RolR indeed negatively regulated the expression of rolHMD gene. Further, the deduced promoter ProlR was PCR amplified and ligated into pXMJ19-lacZ to generate pXMJ19-ProlR-lacZ. The effect of RolR on promoter (ProlR) was evaluated by determination of the β-galactosidase activity in the cells of RES167ΔrolR/pTRCmob-rolR/pXMJ19-ProlR-lacZ (with RolR) and RES167ΔrolR/pXMJ19-ProlR-lacZ (without RolR). The ProlR promoter showed lower transcriptional activities in the presence of RolR than in the absence of RolR (Fig. 2B), indicating that RolR negatively regulated the transcription of its own gene.
Fig 2.
RolR negatively regulates the expression of rolHMD and of its own gene. (A) RT-qPCR analysis of gene relative expression level in wild-type RES167 grown with glucose (WT-gluc) or resorcinol (WT-res) or in mutant RES167ΔrolR grown with glucose (ΔrolR-gluc) or resorcinol (ΔrolR-res) as the sole carbon source. The housekeeping gene rpoB of C. glutamicum was used as an internal control. The relative expression levels of target genes of the wild-type and mutant under defined culture conditions to their expression levels of wild-type RES167 grown in the presence of glucose are shown. (B) rolR promoter activity analysis at different backgrounds using β-galactosidase reporter gene (lacZ). C. glutamicum strains RES167ΔrolR/pXMJ19-ProlR-lacZ (without RolR) and RES167ΔrolR/pTRCmob-rolR/pXMJ19-ProlR-lacZ (with RolR) were grown in LB broth, and the cells were harvested at late log phase to determine the β-galactosidase activity. In panel A, error bars show the standard deviations of three technical replicates. In panel B, standard deviations of three technical replicates are <0.01.
RolR specifically binds to a 29-bp sequence (rolO) located at the intergenic region of rolR and rolHMD.
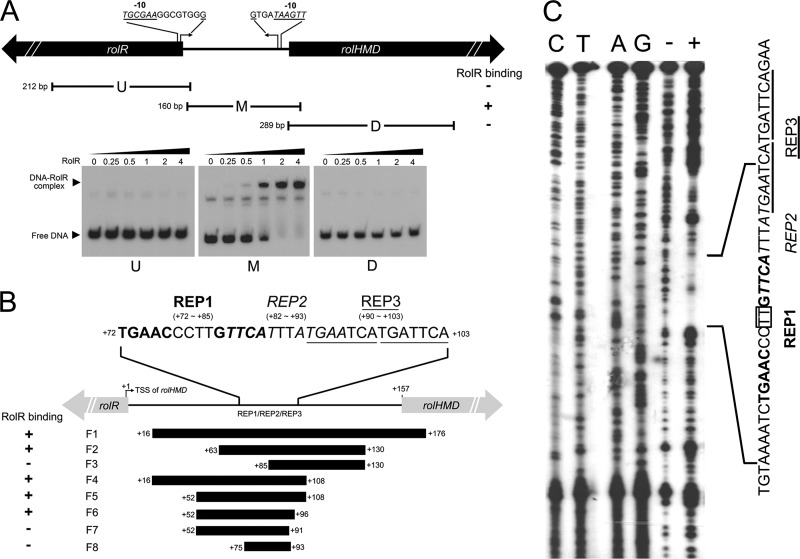
To further analyze the function of RolR, the rolR gene was cloned and expressed in E. coli BL21(DE3) cells. N-terminal His6-tagged RolR (His6-RolR) was purified by Ni2+-NTA chromatography and gel filtration (see Fig. SA2A in the supplemental material). The purified, native His6-RolR showed a molecular mass of 64.8 kDa (see Fig. SA2B in the supplemental material), suggesting a homodimer of native RolR. To identify the binding site of RolR regulator in promoter region, EMSA was performed with His6-RolR protein and three DNA fragments (Fig. 3A). The EMSA results showed that RolR specifically bound to the DNA fragment M, but not fragments U and D, suggesting that RolR binding site is located in the intergenic region of rolR and rolHMD genes.
Fig 3.
Pinpointing the binding site of RolR by use of EMSA and DNase I footprinting. (A) Delineation of chromosomal DNA regions interacting with RolR. Distances are approximately drawn to scale. Further below are shown the EMSA results. Approximate 0.5 pmol of 32P-labeled DNA fragment was used, and the loading of His6-RolR protein to each lane was 0, 0.25, 0.5, 1, 2, and 4 pmol, respectively. (B) Involvement of three inverted repeats in the binding of RolR to DNA. REP1, REP2, and REP3 are shown in boldface, italics, and underlined, respectively. (C) DNase I footprinting assay of RolR on chromosomal DNA region. Lanes C, T, A, and G are sequencing ladders. Lane “+” and “−” are DNase I digestion patterns of the intergenic fragment between rolR and rolHMD genes with or without RolR, respectively. One pmol of 32P-labeled DNA fragment and 10 pmol of His6-RolR protein were used. The sequences of three REPs and the protected region of RolR are shown on the right. The two bases (TT) that are not protected by RolR are shown in the box.
The binding sites (operators) of TetR-type regulators are featured by palindrome sequences (26). Careful examination of fragment M revealed three inverted repeats—REP1 (TGAACCCTTGTTCA), REP2 (TTCATTTATGAA), and REP3 (TGAATCA TGATTCA)—which might be related to the RolR operator. Different combinations of the inverted repeats were synthesized and used for EMSA analysis. The EMSA results showed that DNA fragments containing the inverted repeats REP1 and REP2 (F1∼F2 and F4∼F6) could bind His6-RolR protein (Fig. 3B). When REP1 and/or REP2 were truncated, as represented by F3, F7, and F8 fragments, binding between His6-RolR and DNA fragments was not observed (Fig. 3B). However, disruption of the inverted repeat REP3 (as in the case of F6) did not affect the binding of His6-RolR, indicating that REP3 was not essential for RolR binding (Fig. 3B). This result demonstrates that REP1 and REP2 are essential for the binding of RolR and may be part of the operator.
Furthermore, DNase I footprinting results revealed that a 29-bp DNA region was protected by His6-RolR protein from digestion (Fig. 3C). The 29-bp DNA fragment was termed as operator rolO. The rolO is located between the transcription start sites and translational start points of both rolR and rolHMD genes and covered the entire REP1 and REP2 and partial REP3 sequences (Fig. 3C). Further SPR assay indicated that His6-RolR could only bind to the rolO fragment rather than any single inverted repeat (REP1, REP2, or REP3) (see Fig. SA3 in the supplemental material). These results demonstrated that the binding of RolR with DNA needed the whole rolO region rather than a single inverted repeat.
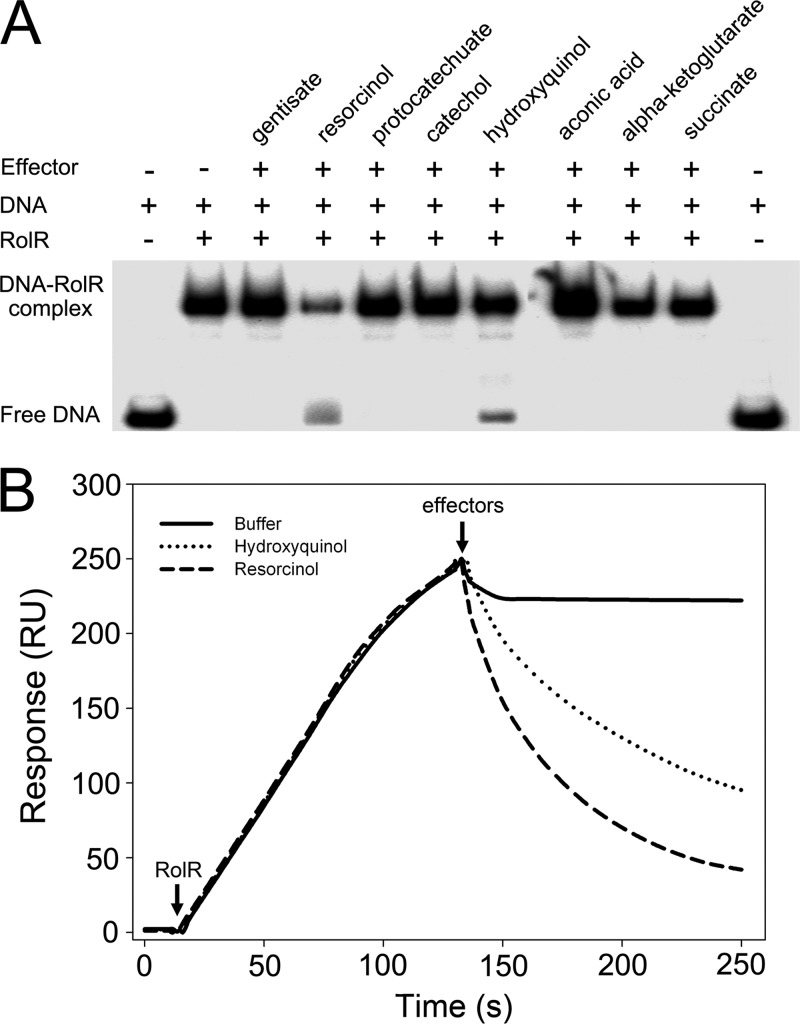
Resorcinol and hydroxyquinol affect the binding of RolR to rolO.
Previous study has reported that resorcinol is an effector of RolR (20). To identify other effectors that affect the binding of RolR with rolO, a range of chemicals, including metabolic intermediates (hydroxyquinol, succinate, aconitic acid, and α-ketoglutarate) and chemical analogs (protocatechuate, catechol, and gentisate), were tested by EMSA. The results showed that resorcinol and hydroxyquinol clearly reduced the binding of His6-RolR with rolO, and the other chemicals did not show any observable effect (Fig. 4A). The effect of resorcinol and hydroxyquinol on dissociation of His6-RolR-rolO complex was further examined by SPR assay. As shown in Fig. 4B, both resorcinol and hydroxyquinol initiated the dissociation of His6-RolR-rolO complex. The kd values for His6-RolR-rolO complex on resorcinol and hydroxyquinol were determined to be 1.41 × 10−2 s−1 and 7.34 × 10−3 s−1, respectively, indicating that resorcinol had a stronger effect on dissociation of His6-RolR-rolO complex than did hydroxyquinol. These results indicate that resorcinol and hydroxyquinol are able to dissociate the binding of RolR with its operator rolO and that they are the effectors for RolR regulation of resorcinol metabolism in C. glutamicum.
Fig 4.
Resorcinol and hydroxyquinol affect the binding of RolR to rolO. (A) Effects of different chemicals on the binding affinity of RolR to the intergenic fragment between rolR and rolHMD, as determined by EMSA. The DNA used in this EMSA was fragment M in Fig. 3A. (B) Resorcinol or hydroxyquinol can dissociate RolR-rolO complex. The 5′-end-biotinylated rolO dsDNA fragment was immobilized on a streptavidin-coated SA sensor chip (GE). His6-RolR (38.8 nM, 60 μl) in running buffer was injected at a flow rate of 30 μl/min. The effector resorcinol (1 mM, 60 μl) or hydroxyquinol (1 mM, 60 μl) was injected immediately after His6-RolR by using the “COINJECT” pattern. Running buffer alone or combined with His6-RolR was injected as a blank controls at the same flow rate (30 μl/min). The arrows show the start points of injection of His6-RolR or effectors. The response is measured in resonance unit (RU) and is proportional to the mass and numbers of molecules binding to the sensor chip.
DISCUSSION
In this study, we investigated how RolR regulated the transcription of the rol genes in C. glutamicum. The results indicated that RolR repressed the transcription of rolHMD and of its own gene by binding to a 29-bp operator rolO. The rolO was the sole binding site for RolR, and it was located at the intergenic region of rolR and rolHMD. The binding of RolR to rolO was affected by resorcinol and hydroxyquinol, which are the metabolic substrates of resorcinol catabolic pathway. These two compounds were able to dissociate RolR-rolO complex, thus releasing RolR from the complex and derepressing the transcription of rol genes in C. glutamicum.
RolR was reported to be a member of the TetR family regulators (16). The typical TetR in Gram-negative bacteria represses the transcriptions of tetA and of its own gene (tetR) by binding to two operators that locate at respective promoter region of tetA and of tetR and overlap RNA polymerase binding site, so that RNA polymerase cannot bind to promoters to start transcription (25, 28). A different regulatory mechanism was found for QacR in the Gram-positive Staphylococcus aureus, which negatively regulated the qacA that confers resistance to monovalent and bivalent cationic lipophilic antiseptics/disinfectants such as quaternary ammonium compounds (qac), but not qacR itself (11). The repression happens through QacR binding to the operator IR1 (overlapping the region from −6 to +21) and thus not affecting the binding of RNA polymerase to qacA promoter but preventing the transition of RNA polymerase-promoter complex into a productively transcribing state (11, 12). Based on our results, the regulatory mechanism of RolR in C. glutamicum is different from that of the previously known TetR-type repressors. First, RolR binds to only one operator rolO and simultaneously represses the transcription of both rolR and rolHMD. Second, the operator rolO is located in the intragenic region of rolR and rolHMD and is not at the promoter region or overlapping with the transcription start site. Due to the position of rolO distant to the transcription start sites of both genes (the overlapping +66 to +94 region for rolHMD and the +40 to +68 region for rolR), it is likely that RolR represses the transcription of rolHMD and of its own gene by a roadblock mechanism as PurR in E. coli (14).
The TetR-type transcriptional repressors usually form homodimer and then bind to target promoters (26). It is likely that there is a direct relationship between the number of multimerization of TetR-type repressor and the length of operator. For example, the typical TetR binds to a 15-bp operator tetO as one homodimer (25). Unlike the TetR, QacR recognizes the 28-bp operator IR1, which is nearly twice the length of tetO, and binds to it as a pair of homodimers (30). The EthR of Mycobacterium tuberculosis binds to a 55-bp operator IG-55 cooperatively as a homo-octamer (9). Our results showed that RolR recognized the 29-bp operator rolO, which is nearly the same length to the operator IR1 (28 bp) of S. aureus (30). Since, in addition, we found that RolR occurred as a homodimer in its native form (20), we propose that RolR binds to rolO as a pair of homodimers. Given the DNase I footprinting result that the bases “TT” in the middle of rolO was not protected by RolR, it is very likely that each RolR homodimer docks on one arm of rolO.
Recently, the crystal structure of RolR was determined in its effector-bound (with resorcinol) and apo forms. The structure revealed that resorcinol-binding initiated a 4.5-Å decrease in center-to-center separation of the two recognition helices (h3-h3′) with the result that RolR was not suitable for DNA binding (20). Based on previous data and data presented here, a model explaining the regulation of resorcinol catabolism by RolR in C. glutamicum is proposed and illustrated in Fig. 5. When there is no resorcinol in environment (state 1), a pair of RolR dimers bind to the operator rolO and block the advancement of RNA polymerase, thus repressing the transcription of both rolR and rolHMD. When resorcinol occurs in the environment (state 2), it binds to the RolR dimer and results in the res-RolR dimer released from the operator rolO. This derepresses the transcriptions of rolR and of rolHMD. Due to the discovery that hydroxyquinol could disassociate the RolR-rolO complex and functions as an effector alternative to resorcinol, we deduce that hydroxyquinol plays the same role that resorcinol does.
Fig 5.
Proposed working model of the regulation of resorcinol catabolism in C. glutamicum by RolR.
Previously, sequence similarity analysis revealed 29 RolR homologs (e-value < 10−4) by using the C-domain (73 to 229 amino acids) of RolR in the nonredundant protein sequence database from the genomes of Burkholderia, Mycobacterium, Rhodococcus, and Streptomyces species (20). Motif searches on continuous 600-bp fragments around the start codon of rolR-like genes identified 29-bp sequences (Fig. 6A) that shared a similar structure and were named as a rolO-like motif (Fig. 6B). The rolO-like motif is composed of a long inverted repeat with two 13-bp arms separated by two bases in the middle, and there is one imperfect short inverted repeat within each 13-bp arm (Fig. 6B). This structure of rolO-like motif might be an important configuration for RolR-like regulator recognition and binding. When the genetic organization around the rolO-like motif was examined, it was found that 24 of the 27 genomes were similar to the rol gene cluster in C. glutamicum. The rolR-like regulatory genes are oriented to the opposite direction of its potential target genes, and the rolO-like motifs are located at the intergenic regions of the rolR-like and structural genes (see Fig. SA4 in the supplemental material). The identification of rolO-like motifs and similar genetic organizations in those bacteria suggests that they might share a similar regulatory mechanism.
Fig 6.
Identification of rolO-like motifs. (A) Identified rolO-like motifs from Corynebacterium, Burkholderia, Mycobacterium, Rhodococcus, and Streptomyces. The positions of each rolO-like motif with respect to the translational start point of each rolR-like gene are given. The first nucleotide A of coding region is designated as +1. “−” indicates the position upstream of the first nucleotide A of coding region. (B) rolO-like motif identified by the motif-based sequence analysis tool MEME (1). The long inverted repeat with two 13-bp arms separated by two bases is indicated by bold arrows. The short imperfect inverted repeats in each 13-bp arm are boxed in white or gray, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants (30730002 and 30725001) from the National Natural Science Foundation of China and by a grant from MOST of China (973 project 2011CBA00805).
Footnotes
Published ahead of print 15 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers, p 28–36 In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, vol 2 ICISMB, San Diego, CA: [PubMed] [Google Scholar]
- 2. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- 3. Cazorla FM, et al. 2006. Biocontrol of avocado dematophora root rot by antagonistic Pseudomonas fluorescens PCL1606 correlates with the production of 2-hexyl 5-propyl resorcinol. Mol. Plant-Microbe Interact. 19: 418–428 [DOI] [PubMed] [Google Scholar]
- 4. Chapman PJ, Ribbons DW. 1976. Metabolism of resorcinylic compounds by bacteria: alternative pathways for resorcinol catabolism in Pseudomonas putida. J. Bacteriol. 125: 985–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dayan FE, Watson SB, Nanayakkara NPD. 2007. Biosynthesis of lipid resorcinols and benzoquinones in isolated secretory plant root hairs. J. Exp. Bot. 58: 3263–3272 [DOI] [PubMed] [Google Scholar]
- 6. Duque E, Segura A, Mosqueda G, Ramos JL. 2001. Global and cognate regulators control the expression of the organic solvent efflux pumps TtgABC and TtgDEF of Pseudomonas putida. Mol. Microbiol. 39: 1100–1106 [DOI] [PubMed] [Google Scholar]
- 7. Eaton RW. 1996. p-Cumate catabolic pathway in Pseudomonas putida Fl: cloning and characterization of DNA carrying the cmt operon. J. Bacteriol. 178: 1351–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eaton RW. 1997. p-Cymene catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. J. Bacteriol. 179: 3171–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Engohang-Ndong J, et al. 2004. EthR, a repressor of the TetR/CamR family implicated in ethionamide resistance in mycobacteria, octamerizes cooperatively on its operator. Mol. Microbiol. 51: 175–188 [DOI] [PubMed] [Google Scholar]
- 10. Gristwood T, Fineran PC, Everson L, Salmond GPC. 2008. PigZ, a TetR/AcrR family repressor, modulates secondary metabolism via the expression of a putative four-component resistance-nodulation-cell-division efflux pump, ZrpADBC, in Serratia sp. ATCC 39006. Mol. Microbiol. 69: 418–435 [DOI] [PubMed] [Google Scholar]
- 11. Grkovic S, Brown MH, Roberts NJ, Paulsen IT, Skurray RA. 1998. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem. 273: 18665–18673 [DOI] [PubMed] [Google Scholar]
- 12. Grkovic S, Brown MH, Schumacher MA, Brennan RG, Skurray RA. 2001. The staphylococcal QacR multidrug regulator binds a correctly spaced operator as a pair of dimers. J. Bacteriol. 183: 7102–7109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Groseclose EE, Ribbons DW. 1981. Metabolism of resorcinylic compounds by bacteria: new pathway for resorcinol catabolism in Azotobacter vinelandii. J. Bacteriol. 146: 460–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He B, Zalkin H. 1992. Repression of Escherichia coli purB is by a transcriptional roadblock mechanism. J. Bacteriol. 174: 7121–7127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hillen W, Berens C. 1994. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 48: 345–369 [DOI] [PubMed] [Google Scholar]
- 16. Huang Y, et al. 2006. Genetic characterization of the resorcinol catabolic pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 72: 7238–7245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jakoby M, Nolden L, Meier-Wagner J, Krämer R, Burkovski A. 2000. AmtR, a global repressor in the nitrogen regulation system of Corynebacterium glutamicum. Mol. Microbiol. 37: 964–977 [DOI] [PubMed] [Google Scholar]
- 18. Kitani S, Yamada Y, Nihira T. 2001. Gene replacement analysis of the butyrolactone autoregulator receptor (FarA) reveals that FarA acts as a novel regulator in secondary metabolism of Streptomyces lavendulae FRI-5. J. Bacteriol. 183: 4357–4363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Konopka A. 1993. Isolation and characterization of a subsurface bacterium that degrades aniline and methylanilines. FEMS Microbiol. Lett. 111: 93–99 [Google Scholar]
- 20. Li D-F, et al. 2011. Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator. PLoS One 6: e19529 doi:10.1371/journal.pone.0019529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu Q, Ouyang S-P, Kim J, Chen G-Q. 2007. The impact of PHB accumulation on l-glutamate production by recombinant Corynebacterium glutamicum. J. Biotechnol. 132: 273–279 [DOI] [PubMed] [Google Scholar]
- 22. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- 23. Lynch BS, Delzell ES, Bechtel DH. 2002. Toxicology review and risk assessment of resorcinol: thyroid effects. Regul. Toxicol. Pharmacol. 36: 198–210 [DOI] [PubMed] [Google Scholar]
- 24. Ohta Y, Maeda M, Kudo T. 2001. Pseudomonas putida CE2010 can degrade biphenyl by a mosaic pathway encoded by the tod operon and cmtE, which are identical to those of P. putida F1 except for a single base difference in the operator-promoter region of the cmt operon. Microbiology 147: 31–41 [DOI] [PubMed] [Google Scholar]
- 25. Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W. 2000. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat. Struct. Biol. 7: 215–219 [DOI] [PubMed] [Google Scholar]
- 26. Ramos JL, et al. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69: 326–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rokenes TP, Lamark T, Strom AR. 1996. DNA-binding properties of the BetI repressor protein of Escherichia coli: the inducer choline stimulates BetI-DNA complex formation. J. Bacteriol. 178: 1663–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saenger W, Orth P, Kisker C, Hillen W, Hinrichs W. 2000. The tetracycline repressor: a paradigm for a biological switch. Angew. Chem. Int. Ed. 39: 2042–2052 [DOI] [PubMed] [Google Scholar]
- 29. Sanchez P, Alonso A, Martinez JL. 2002. Cloning and characterization of SmeT, a repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. Antimicrob. Agents Chemother. 46: 3386–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schumacher MA, et al. 2002. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J. 21: 1210–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tauch A, et al. 2002. Efficient electrotransformation of Corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr. Microbiol. 45: 362–367 [DOI] [PubMed] [Google Scholar]
- 32. Tropel D, van der Meer JR. 2004. Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol. Mol. Biol. Rev. 68: 474–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yoshida M, et al. 2007. Biochemical and genetic analysis of the γ-resorcylate (2,6-dihydroxybenzoate), catabolic pathway in Rhizobium sp. strain MTP-10005: identification and functional analysis of its gene cluster. J. Bacteriol. 189: 1573–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao K-X, Huang Y, Chen X, Wang N-X, Liu S-J. 2010. PcaO positively regulates pcaHG of the β-ketoadipate pathway in Corynebacterium glutamicum. J. Bacteriol. 192: 1565–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.