Abstract
The genome of Neurospora crassa encodes two different cellobiose dehydrogenases (CDHs) with a sequence identity of only 53%. So far, only CDH IIA, which is induced during growth on cellulose and features a C-terminal carbohydrate binding module (CBM), was detected in the secretome of N. crassa and preliminarily characterized. CDH IIB is not significantly upregulated during growth on cellulosic material and lacks a CBM. Since CDH IIB could not be identified in the secretome, both CDHs were recombinantly produced in Pichia pastoris. With the cytochrome domain-dependent one-electron acceptor cytochrome c, CDH IIA has a narrower and more acidic pH optimum than CDH IIB. Interestingly, the catalytic efficiencies of both CDHs for carbohydrates are rather similar, but CDH IIA exhibits 4- to 5-times-higher apparent catalytic constants (kcat and Km values) than CDH IIB for most tested carbohydrates. A third major difference is the 65-mV-lower redox potential of the heme b cofactor in the cytochrome domain of CDH IIA than CDH IIB. To study the interaction with a member of the glycoside hydrolase 61 family, the copper-dependent polysaccharide monooxygenase GH61-3 (NCU02916) from N. crassa was expressed in P. pastoris. A pH-dependent electron transfer from both CDHs via their cytochrome domains to GH61-3 was observed. The different properties of CDH IIA and CDH IIB and their effect on interactions with GH61-3 are discussed in regard to the proposed in vivo function of the CDH/GH61 enzyme system in oxidative cellulose hydrolysis.
INTRODUCTION
The extracellular fungal flavocytochrome cellobiose dehydrogenase (CDH) (EC 1.1.99.18) constitutes a considerable fraction of the lignocellulolytic enzymes secreted by many cultures of wood-degrading basidiomycetes, e.g., 0.5% in Phanerochaete chrysosporium (11), up to 1.2% in Trametes spp. (20), 2.4% in Ceriporiopsis subvermispora (9), and 2.2% in Sclerotium rolfsii (19), and ascomycetes, e.g., 12% in Corynascus thermophilus (8), 2.3% in Myriococcum thermophilum (7), and 2.4% in Neurospora crassa (23). The widespread appearance of CDH implies an important function of this enzyme in wood degradation. Since its discovery in 1972, the exact catalytic role of CDH and its interaction with other fungal lignocellulolytic enzymes remained unclear. Several in vivo functions have been proposed (2, 11, 42). The most widely supported mechanism in the last 2 decades is related to the ability of CDH to produce hydrogen peroxide and concomitantly reduce the level of Fe3+, which potentially generates hydroxyl radicals by a Fenton-type reaction. However, the catalytic efficiency of oxygen reduction is very low and about 100 times slower than the reduction of other electron acceptors such as quinones. Similarly, the reduction of weakly complexed Fe3+ species (e.g., by carboxylic acids) occurring in plant material is much slower than ferricyanide turnover and additionally limited to pH values below 4.5. While white rot fungi generate such acidic pH values during growth, the majority of ascomycete CDH producers degrade lignocellulose at higher pH values (34).
Evidence is growing that the main purpose of CDH in vivo is to transfer electrons not to low-molecular-weight electron acceptors like oxygen and Fe3+ but to members of the glycoside hydrolase 61 (GH61) protein family. Recently, a polysaccharide monooxygenase (PMO) activity for three members of the GH61 protein family in N. crassa was described (17, 22), and copper was specified as an active-site constituent of these enzymes. Therefore, the name PMO was suggested previously by Phillips and coworkers for the investigated GH61 proteins (22). The GH61 family is widely distributed in fungi. BLAST searches of 7 CDH-producing basidiomycetous and ascomycetous fungi revealed that cdh and gh61 genes occur together in the genomes of the CDH-producing fungi Trametes versicolor, P. chrysosporium, Chaetomium globosum, Thielavia terrestris, Podospora anserina, Glomerella graminicola, and Aspergillus fumigatus. On the other hand, numerous fungi with gh61 genes in their genomes do not have cdh genes (e.g., Hypocrea jecorina, anamorph [ana.] Trichoderma reesei). It was also shown that the addition of certain T. terrestris or Thermoascus aurantiacus GH61 proteins has a high stimulatory effect on the lignocellulose hydrolysis performance of T. reesei cellulases without involving CDH (10). Additionally, many of the tested members of the GH61 family have been shown not to cleave cellulose under the tested conditions. Therefore, it remains to be elucidated if the polysaccharide monooxygenase activity is a feature of all GH61 proteins.
The CDH enzyme family is a heterogeneous group of proteins with sequence identities as low as 35%. Phylogenetic analyses of these sequences (8, 41, 42) showed several well-supported branches, which correlate partially with the species classification. Basidiomycete CDH sequences (from the Atheliales, Corticiales, and Polyporales) form the well-supported branch of class I CDHs. Class II consists only of sequences of ascomycete origin (the Sordariales, Xylariales, and Hypocreales). This class of CDHs partitions into two subclasses: class IIA CDHs contain a carbohydrate binding module (CBM), whereas class IIB CDHs do not. N. crassa is a member of the Sordariales, and two cdh genes are found in its genome: one with a C-terminal CBM (CDH IIA) and one without (CDH IIB) (8). For members of the Eurotiales, Helotiales, and Pleosporales, CDH-encoding sequences of a separate phylogenetic branch were found by genome sequencing projects. The secretion of these class III CDHs has not yet been confirmed. Class II CDHs prefer cellobiose and cellooligosaccharides as substrates but can also oxidize other mono- and disaccharides although with lower catalytic efficiencies. This difference from class I CDHs might be an adaption to different habitats and substrates.
In the genome of N. crassa, 14 gh61 genes are found (27), which are induced to various extents during growth on cellulose and xylan (28). Marletta and coworkers previously showed by high-performance liquid chromatography (HPLC) and liquid chromatography-mass spectrometry (LC-MS) measurements that the class IIB CDH from Myceliophthora thermophila interacts with N. crassa GH61s (NCU01050, NCU07898, and NCU08760) (22) and investigated the reaction pathway for the oxidative cleavage of cellulose by these GH61s (1). CDH is supposed to act as a reductase (via its heme b) for these PMOs, which catalyze the insertion of oxygen into C-H bonds adjacent to the glycosidic linkage. The oxygen atoms destabilize the glycosidic bond, which leads to its elimination and the formation of a sugar lactone or ketoaldose (22). Previous structural studies of copper-containing GH61 proteins revealed that their putative active sites showed structural homologies to the active site of chitin binding protein 21 (CBP21), which is capable of cleaving chitin. The proposed reaction mechanism for CBP21 suggests a chitin oxygenase activity, which consumes both oxygen and a reducing agent as cosubstrates (35–37). The first GH61 structure was obtained from Cel61A, a protein with endoglucanase activity found in H. jecorina. The structure does not show a copper atom in the active site but shows nickel, which was added to obtain better crystals and as a source for anomalous scattering to obtain near-atomic resolution (16). However, the authors of that study stated that nickel-containing enzymes are quite unusual and suggested that other transition metals could bind to Cel61A in vivo. The first structure showing a copper in the active site was reported by Quinlan et al. (24) and Westereng et al. (39), which was followed by biochemical support for the function of the copper (22).
It was demonstrated previously that the addition of M. thermophila CDH IIA (designated CDH-1 by Phillips et al. [22]) restores the cellulolytic activity of a Δcdh-1 N. crassa strain, whereas a 10-fold-higher concentration of CDH IIB (CDH-2) is necessary to replace CDH IIA (22). Earlier studies by Langston et al. showed that purified GH61 proteins have no demonstrable direct hydrolase activity, but Thermoascus aurantiacus GH61A in combination with Humicola insolens CDH cleaves cellulose into soluble, oxidized oligosaccharides (17).
We selected the model organism N. crassa for the first characterization of both CDH subclasses from one organism and to investigate their different physical and catalytic properties. To study the interaction of CDH IIA and CDH IIB with their proposed natural electron acceptor, GH61-3 from the same organism was expressed and purified. N. crassa GH61-3 is found in the same phylogenetic branch of GH61 proteins that showed PMO activity, and its encoding gene, NCU02916, is strongly upregulated during N. crassa cultivation on Miscanthus (32). Pichia pastoris was chosen to express all three enzymes without possibly interfering purification tags. After elucidating the physical and catalytic properties, the CDH/GH61-3 interaction was studied. Previously reported data for N. crassa transcriptome and secretome analyses are used to explore the induction and regulation of CDH IIA and CDH IIB as well as of members of the GH61 protein family.
MATERIALS AND METHODS
Organism, vectors, and culture conditions.
The genes coding for CDH IIA and CDH IIB from N. crassa were isolated previously (8) and were cloned into the CloneJET vector (Fermentas). The N. crassa gene NCU02916, encoding the GH61-3 protein, was codon optimized for expression in Pichia pastoris (see Fig S1 in the supplemental material) and commercially synthesized by Invitrogen. P. pastoris strain X-33 and the vectors pPICZαA and pPICZB are components of the Pichia Easy Select expression system from Invitrogen. P. pastoris transformants were grown on YPD plates (10 g liter−1 yeast extract, 20 g liter−1 peptone, 10 g liter−1 glucose, and 15 g liter−1 agar) containing 100 mg liter−1 zeocin.
Construction of CDH IIA, CDH IIB, and NCU02916 expression vectors.
Previously reported plasmids pNCIIA and pNCIIB (8) were used as templates for the amplification of cdhIIA and cdhIIB with primers 5NCa-BstBI (5′-TATTTCGAAACGATGAGGACCACCTCGGCC-3′) and 3NCa-XbaI (5′-TATCACGTGCTACACACACTGCCAATACC-3′) and primers 5NCb-EcoRI (5′-TATGAATTCATGAAGGTCTTCACCCGC-3′) and 3NCb-NotI (5′-TATGCGGCCGCTCATCTTCTCCATTTTCCC-3′), respectively. PCR was performed with Phusion high-fidelity DNA polymerase from New England BioLabs, a deoxynucleoside triphosphate (dNTP) mix from Fermentas, oligonucleotide primers from VBC Biotech (Vienna, Austria), and a C-1000 thermocycler from Bio-Rad Laboratories. The resulting cdhIIA cDNA and the NCU02916 gene in the cloning vector were digested with BstBI and XbaI and cloned into the equally treated vector pPICZα A. cDNA from cdhIIB and vector pPICZ B were digested with EcoRI and XbaI and ligated by using the Rapid DNA ligation kit from Fermentas. The procedures resulted in genes encoding proteins with their native signal sequences cloned under the control of the methanol-inducible AOX1 promoter. C-terminal tags for purification or antibody detection were omitted. The correct insertion of the genes and the absence of mutations were confirmed by DNA sequencing. Linearized, verified plasmids were used for transformation into electrocompetent P. pastoris cells.
Microscale screening for high-producing CDH transformants.
The cultivation and expression of both CDHs were done with 96-deep-well plates according to methods described previously by Weis et al. (38), with small modifications. Cells were grown in 250 μl BMD1 (13.4 g liter−1 yeast nitrogen base, 0.4 mg liter−1 biotin, 10 g liter−1 glucose, 200 mM potassium phosphate [pH 6.0]) at 25°C, 385 rpm, and 60% humidity for approximately 60 h to reach the stationary growth phase. Induction was started by the addition of 250 μl of BMM2 medium (13.4 g liter−1 yeast nitrogen base, 0.4 mg liter−1 biotin, 1% methanol, 200 mM potassium phosphate [pH 6.0]) to reach a final concentration of 0.5% methanol. After 70, 82, and 108 h of incubation, 50 μl BMM10 (BMM2 with 5% methanol) was added to maintain inducing conditions. The cultivation was stopped after 130 h by the centrifugation of the deep-well plates at 2,600 × g at 4°C for 10 min. The supernatant of each well was analyzed for enzymatic activity with a 2,6-dichloroindophenol (DCIP) assay.
Enzyme production and purification.
CDH IIA and CDH IIB were produced in a 7-liter bioreactor filled with 4 liters of basal salts medium. After sterilization, the pH of the medium was adjusted to pH 5.0 with 28% ammonium hydroxide and maintained at this level throughout the whole fermentation process. The cultivation was started by the addition of a 0.4-liter (9% [vol/vol]) preculture grown on YPD medium in 1-liter baffled shaking flasks at 125 rpm at 30°C overnight. The cultivation was performed according to the Pichia Fermentation Process guidelines of Invitrogen, and the expression of the recombinant protein was induced with methanol. The Invitrogen protocol was altered at the transition phase from glycerol to methanol according to methods described previously by Sygmund et al. (30). The cultivation temperature was 30°C, the airflow rate was 6 liter min−1, and the stirrer speed was 800 rpm. Samples were taken regularly.
GH61-3 was produced in a Multifors fermentor (Infors HT, Bottmingen, Switzerland) with a total volume of 500 ml. The fed-batch fermentation was done according the Pichia Fermentation Process guidelines of Invitrogen, with slight modifications. The basal salts medium was supplemented with 0.1 mM CuSO4. The batch fermentation (300-ml starting volume) was inoculated with 25 ml of preculture at 30°C. The airflow was kept constant at 2 liters min−1, and the stirrer speed was set to 1,000 rpm. The pH was maintained at 5.0 with ammonium hydroxide. After the depletion of glycerol in the batch medium, the fed-batch phase was started with a constant feed of 2.4 ml h−1 of 50% glycerol containing 12 ml liter−1 Pichia trace metal (PTM1) salts for 8 h. For induction, the feed was switched to 100% methanol containing 12 ml liter−1 PTM1 salts at a low flow rate of 0.6 ml h−1 overnight for the adaptation of the culture to methanol, and the temperature was lowered to 25°C. Afterwards, the feed rate was adjusted to keep the dissolved oxygen concentration at around 4%. Antifoam was injected manually as required throughout the fermentation. Samples were taken for measurements of wet biomass and total soluble protein content.
Protein purification was started by a centrifugation (6,000 × g for 30 min at 4°C) of the fermentation broth. A saturated ammonium sulfate solution was slowly added to the clear culture supernatants containing CDH IIA (4.7 liters), CDH IIB (4.5 liters), and GH61-3 (0.35 liters), to give a 20%-saturated (CDH IIA) or 30%-saturated (CDH IIB and GH61-3) solution. The precipitate was removed by centrifugation (6,000 × g for 20 min at 4°C), and the clear supernatant was loaded onto a 600-ml PHE-Sepharose Fast Flow column (chromatographic equipment and materials from GE Healthcare Biosciences) equilibrated with 50 mM phosphate buffer (pH 5.5) containing 20% (CDH IIA) or 30% (CDH IIB and GH61-3) ammonium sulfate. Proteins were eluted within a linear gradient from 20 to 0% ammonium sulfate within 5 column volumes or from 30 to 0% within 7.5 column volumes, and fractions were collected in aliquots. Fractions containing the enzymes of interest were pooled and diafiltrated with a hollow-fiber cross-flow module (Microza UF module SLP-1053, 10-kDa cutoff; Pall Corporation). The diafiltrated CDH pools (∼3 mS cm−1) were applied onto a 20-ml column packed with Q15-Source preequilibrated with 50 mM (CDH IIA) or 20 mM (CDH IIB) sodium acetate buffer (pH 5.5). Proteins were eluted within a linear salt gradient from 0 to 1 M NaCl within 10 column volumes. To obtain a homogenous preparation of CDH IIB, gel filtration with Superdex 200 was added as a final polishing and buffer exchange step with 50 mM sodium acetate buffer (pH 5.5). Fractions from hydrophobic interaction chromatography (HIC) containing GH61-3 (judged by A280 readings and SDS-PAGE) were concentrated and loaded onto a 470-ml gel filtration column (Sephacryl S-300) equilibrated with 100 mM sodium acetate buffer (pH 5.0). The purest CDH and GH61-3 fractions were pooled, concentrated, sterile filtered (0.2 μm), aliquoted, and stored at −80°C.
Enzyme assays and protein determination.
The activity of intact CDH was specifically determined by monitoring the reduction of 20 μM cytochrome c (cyt c) (ϵ550 = 19.6 mM−1 cm−1). This electron acceptor is exclusively reduced at the cytochrome domain. The activities of both the intact holoenzyme and its catalytically active proteolytic cleavage product, the dehydrogenase domain, were spectrophotometrically assayed by using 0.3 mM DCIP (ϵ520 = 6.8 mM−1 cm−1) as an electron acceptor. The reaction was monitored for 180 s at 30°C in a Lambda 35 UV-visible (UV-Vis) spectrophotometer featuring a temperature-controlled 8-cell changer (Perkin-Elmer). All assay mixtures were measured by using McIlvaine buffer (21), at the indicated pH, containing 30 mM lactose as the saturating substrate. One unit of CDH activity was defined as the amount of enzyme that oxidizes 1 μmol of the electron acceptor per minute under the assay conditions. The interaction of GH61-3 with CDH IIA or CDH IIB was measured by its interference with the cyt c assay. Molar ratios of cyt c to GH61-3 from 1:0.25 to 1:4 were measured, and the highest interference was found at a ratio of 1:4, which was used to obtain pH profiles of this interaction (buffer with sodium acetate [pH 3 to 6] and sodium phosphate [pH 6 to 8]). The interaction of GH61-3 is given as the reduction of the cyt c activity (percent inhibition) and was measured in triplicates against a reference reaction without GH61-3. Protein concentrations were determined by the Bradford method using a prefabricated assay from Bio-Rad Laboratories and bovine serum albumin (BSA) as the calibration standard. Spectra of homogeneously purified proteins in both the oxidized and reduced states were recorded at room temperature by using a Hitachi U-3000 spectrophotometer. The proteins were diluted in McIlvaine buffer (pH 6.0) to an absorbance at 280 nm of ∼1, and the spectrum was recorded before and immediately after the addition of the reductant (cellobiose for CDH and ascorbate for GH61-3) to the cuvette.
Electrophoresis.
SDS-PAGE was carried out by using Mini-Protean TGX precast gels (Bio-Rad Laboratories) with a gradient of 4 to 15%. Protein bands were visualized by staining with Bio-Safe Coomassie. An unstained Precision Plus protein standard was used for mass determinations. All procedures were done according to the manufacturer's recommendations (Bio-Rad Laboratories). To estimate the degree of glycosylation, homogenous CDH samples were treated with PNGase F (New England BioLabs) under denaturing conditions, according to the manufacturer's instructions. GH61-3 was deglycosylated with 1,000 U mg−1 endoglycosidase Hf (New England BioLabs) and 0.02 mg mg−1 α-mannosidase (Sigma) in 50 mM sodium acetate buffer (pH 5.0) containing 10 mM ZnCl2. Chromatofocusing was used to determine the isoelectric points of both CDHs. Dialyzed sample solutions containing CDH and glucose oxidase (Sigma) with a known pI of 4.25 as an internal standard were loaded onto a 10-ml Mono P column (GE Healthcare) equilibrated with 0.025 M imidazole-HCl (pH 7.4). The protein concentrations of the applied samples were approximately 0.1 mg ml−1. Proteins were eluted within 10 column volumes with a linear gradient from 0 to 100% Polybuffer 74 (pH 3.6). Absorbances at 280 nm, 420 nm (heme b), and 450 nm (flavin adenine dinucleotide [FAD]) were measured online along with the pH values.
Voltammetric measurements.
The preparation of thiol-modified gold electrodes (diameter of 1.6 mm and area of 0.02 cm2; BASi, West Lafayette, IN) for cyclic voltammetry and square-wave voltammetry started with the dipping of the electrodes into a piranha solution (H2SO4-H2O2 ratio of 3:1 [note that piranha solution is highly corrosive and a strong oxidizer, the mixing of the solutions is exothermic, and explosions might occur if the peroxide concentration exceeds 50%]) for 10 min, followed by electrochemical cleaning by cycling in 0.1 M NaOH with a scan rate of 100 mV s−1 between 0 and −1,000 mV versus the standard hydrogen electrode (SHE) (10 cycles). Afterwards, the electrodes were cleaned mechanically by polishing on Microcloth (Buehler, Lake Bluff, IL) in a Masterprep polishing suspension (0.05 μm; Buehler). The electrodes were rinsed with water and sonicated for 10 min in HQ water, followed by cycling in 0.5 M H2SO4 for 20 cycles with a scan rate of 200 mV s−1 between 0 and +1,950 mV versus SHE, and finally rinsed with HQ water. Thiol self-assembled monolayer (SAM) formation at the electrode surfaces was done by immersing the electrodes in a 10 mM thioglycerol solution at room temperature overnight. Before exposure to CDH, the electrodes were carefully rinsed with water. The electrodes were then covered with a Teflon cap, which formed a cell volume of about 30 μl on the thiol-modified gold electrodes. Modification with CDH was done by filling the cavity with enzyme solution (20 mg ml−1). A dialysis membrane (molecular mass cutoff of 14,000 Da; Carl Roth) was used to trap the enzyme in the cells (6). The dialysis membrane (presoaked in buffer) was pressed onto the electrode and fixed tightly with a rubber O ring. All measurements were performed at room temperature. Cyclic voltammetry (scan rate of 10 mV s−1) and square-wave voltammetry were performed by using a Gamry Reference 600 potentiostat (Gamry Instruments, Warminster, PA) scanning between −50 and 350 mV versus SHE. The square-wave voltammograms were recorded at a frequency of 1 Hz, a step potential of 2 mV, and an amplitude of 20 mV. A standard three-electrode configuration was used with an Ag|AgCl reference electrode (saturated KCl; Gamry Instruments) and a platinum wire as a counterelectrode. The buffers (McIlvaine buffer, optionally containing 10 mM lactose for catalytic experiments) used as electrolytes were carefully degassed under a vacuum and purged with argon prior to all experiments. To maintain the inert atmosphere, argon was blown over the solution during the measurements.
Kinetic measurements.
Initial rates for the determination of pH profiles of various electron acceptors were determined at 30°C with McIlvaine buffer ranging from pH 2.5 to 9. Additional electron acceptors not mentioned above for the enzyme assays were 1,4-benzoquinone (ϵ290 = 2.24 mM−1 cm−1) and ferrocenium hexafluorophosphate (FcPF6) (ϵ300 = 4.3 mM−1 cm−1). Due to the instability of FcPF6 at alkaline pH values, the buffer, lactose, and enzyme solutions were prewarmed, and the reaction was started by the addition of FcPF6 to the mixture. The relative activities of both CDHs for carbohydrates (lactose, cellobiose, maltose, maltotriose, mannose, glucose, galactose, and xylose) were measured with a 100 mM substrate concentration using DCIP (300 μM), cyt c (20 μM), 1,4-benzoquinone (1,000 μM), or FcPF6 (100 μM) as the electron acceptor. Catalytic constants for various carbohydrates were determined with 1,4-benzoquinone at pH 6.0. Catalytic constants were calculated by using nonlinear least-squares regression by fitting the observed data to the Michaelis-Menten equation (Sigma Plot 11; Systat Software).
RESULTS
Production and purification of recombinant CDH IIA, CDH IIB, and GH61-3.
The fermentations of CDH IIA (Fig. 1A) and CDH IIB (Fig. 1B) were performed as uniformly as possible. By the end of the glycerol feed, both cultures showed similar cell densities (170 g liter−1 with CDH IIA and 180 g liter−1 with CDH IIB). No CDH activity was detected at this time. After inducing enzyme expression by feeding with methanol, the specific growth rate (μ) was reduced to 25 to 30%, and the final cell densities were 270 g liter−1 (after 84 h) and 310 g liter−1 (after 96 h), respectively. The secretion of extracellular protein correlated with biomass production. The secreted protein concentration was higher for CDH IIB (0.65 g liter−1 after 98 h) than for CDH IIA (0.28 g liter−1 after 96 h). CDH IIA was expressed with a higher volumetric activity (1,700 U liter−1 by a DCIP assay at pH 5.0 and 360 U liter−1 by a cyt c assay at pH 6.0) than CDH IIB (410 U liter−1 by a DCIP assay at pH 5.0 and 130 U liter−1 by a cyt c assay at pH 6.0).
Fig 1.
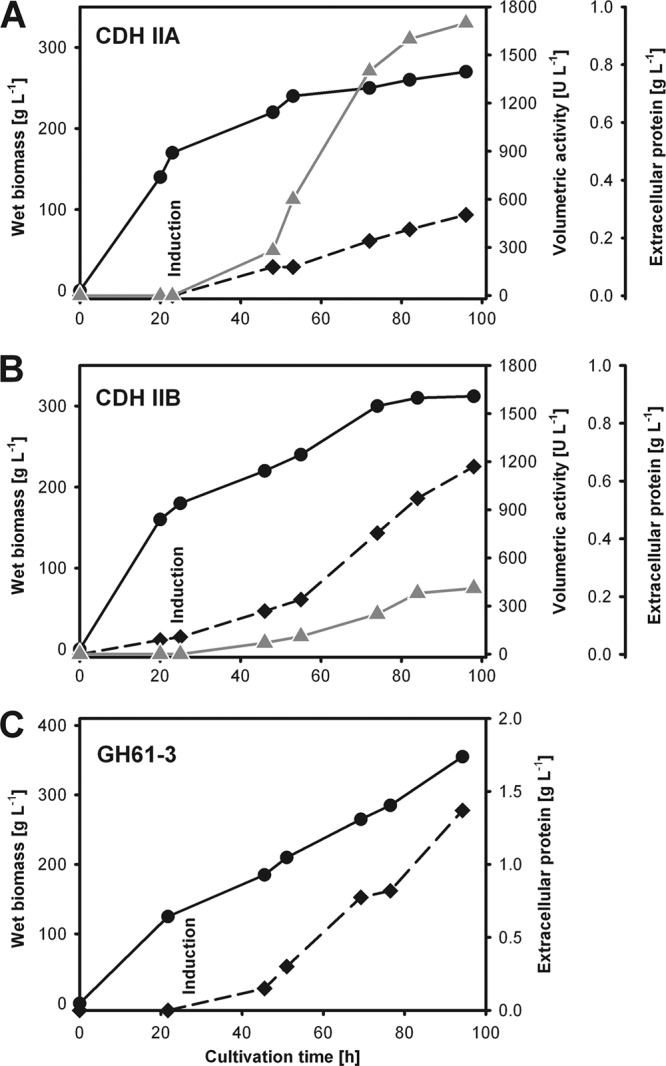
Production of recombinant CDH IIA (A), CDH IIB (B), and GH61-3 (C) in P. pastoris. Black circles, wet biomass; gray triangles, volumetric activity (DCIP assay at pH 5.0); black diamonds, extracellular protein concentration (Bradford assay). The measurements were done in duplicates; the difference between the values was <5%.
With no activity assay for GH61-3 at hand, NCU02916 transformants were checked for the successful integration of the gene into the P. pastoris genome by PCR with standard primers 5′AOX and 3′AOX. A positive clone was chosen for small-scale fermentation. A steady increase of the wet biomass could be observed throughout the fermentation, reaching a wet biomass concentration of 355 g liter−1 at the time of harvest (Fig. 1C). The specific growth rate was reduced by two-thirds after induction. The final extracellular protein concentration was 1.37 g liter−1. Following the expression of GH61-3 by SDS-PAGE (see Fig. S2 in the supplemental material), it was found that it represents the major protein (band at ∼50 kDa) in the culture supernatant. Its amount increased steadily during induction, and the final concentration of GH61-3 in the culture supernatant was ∼450 mg liter−1.
CDH IIA was purified to a specific activity of 21.2 U mg−1 (by a DCIP assay at pH 5.0) or 8.3 U mg−1 (by a cyt c assay at pH 6.0), with a high yield (73%) by a two-step chromatographic purification procedure (see Table S1 in the supplemental material). The purification of CDH IIB was more difficult because of its very low affinity for HIC resin as well as anion exchange resin, which resulted in weak binding and losses. Whereas CDH IIA bound already at 20% ammonium sulfate saturation (125 mS cm−1) to the PHE-Sepharose resin and was eluted at 11% (70 mS cm−1), CDH IIB needed 30% saturation (180 mS cm−1) and eluted at 20% (125 mS cm−1). On Q-Source resin, CDH IIA eluted at a higher NaCl concentration, ∼150 mM (15 mS cm−1), than did CDH IIB, ∼110 mM (11 mS cm−1). An additional gel filtration step had to be introduced to obtain homogenous CDH IIB, which resulted in a poor yield of only 11%. Purified CDH IIB had a specific activity of 5.1 U mg−1 (by a DCIP assay at pH 5.0) or 3.0 U mg−1 (by a cyt c assay at pH 6.0). GH61-3 bound to the HIC resin at a 30% ammonium sulfate saturation and eluted at 8% (50 mS cm−1). The fractions of the peak at 280 nm were pooled and concentrated, giving 5.7 ml of a green solution having a protein content of 9 mg ml−1. After a final gel filtration step, the purest fractions were pooled and concentrated, giving 2 ml of a light-blue solution with a protein content of 22.5 mg ml−1.
Physical properties.
Molecular masses were determined by SDS-PAGE. Both CDHs showed diffuse bands between 110 and 130 kDa, which can be caused by heterogeneous glycosylation and/or the commonly observed smearing of glycoproteins on SDS-PAGE gels (see Fig. S3 in the supplemental material). After deglycosylation under denaturing conditions with PNGase F, single, sharp bands with molecular masses of 85 kDa for CDH IIA and 88 kDa for CDH IIB were observed. The additional bands in the deglycosylated samples at 35 kDa originated from PNGase F. GH61-3 formed a diffuse band at ∼55 kDa. After 48 h of deglycosylation with endoglycosidase Hf and α-mannosidase, the molecular mass was reduced to ∼45 kDa. Chromatofocusing was used to determine the isoelectric points for CDH IIA and CDH IIB, which were at pH 5.14 and pH 4.93, respectively. The UV-Vis spectra of the purified CDHs (Fig. 2A and B) are characteristic. Upon reduction with lactose, a typical Soret band shift was observed, while the α- and β-peaks appeared at 563 nm and 533 nm, respectively. Concomitantly, the absorbance in the region between 450 to 500 nm decreased due to a reduction of FAD. The molar absorption coefficients at 280 nm for all proteins were calculated by using the mature amino acid sequence and the ProtParam program (http://web.expasy.org/protparam/). The molar absorption coefficients for CDH IIA (ϵ280 = 174 mM−1 cm−1, ϵ420(ox) = 103 mM−1 cm−1, ϵ430(red) = 142 mM−1 cm−1, ϵ532(red) = 15.3 mM−1 cm−1, and ϵ563(red) = 24.7 mM−1 cm−1) and for CDH IIB (ϵ280 = 157 mM−1 cm−1, ϵ420(ox) = 87 mM−1 cm−1, ϵ430(red) = 119 mM−1 cm−1, ϵ532(red) = 12.3 mM−1 cm−1, and ϵ563(red) = 19.7 mM−1 cm−1) were calculated from the spectra. The molar absorption coefficients for GH61-3 were an ϵ280 of 47 mM−1 cm−1 and an ϵ630(ox) of 0.04 mM−1 cm−1. The absorbance ratio (A630/A280) of GH61-3 was 0.00079 (Fig. 2C).
Fig 2.
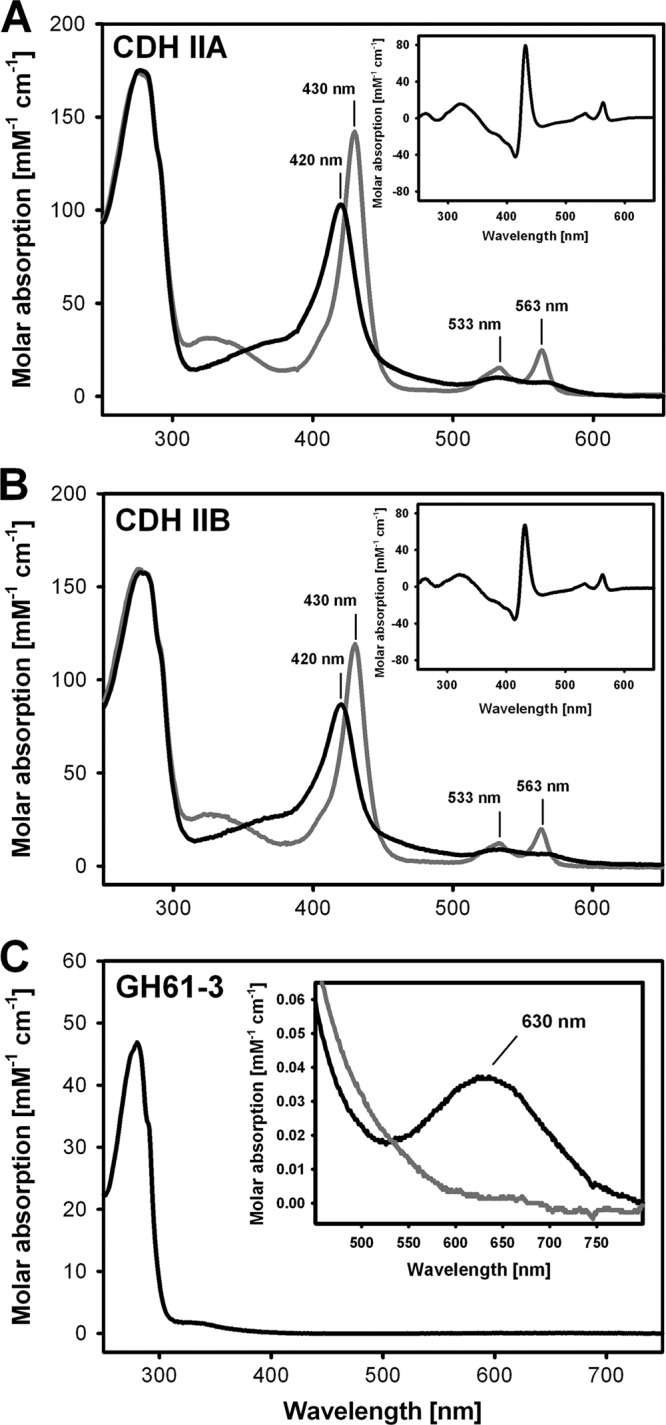
(A and B) Spectral characterization of CDH IIA (A) and CDH IIB (B) showing the oxidized (black) and reduced (gray) spectra. Lactose was used for reduction. The difference spectra (oxidized-reduced) are given in the insets. (C) Spectrum of oxidized GH61-3, with the inset showing the spectrum of the type 2 copper atom in its oxidized (black) and reduced (gray) states. Ascorbate was used to reduce the enzyme.
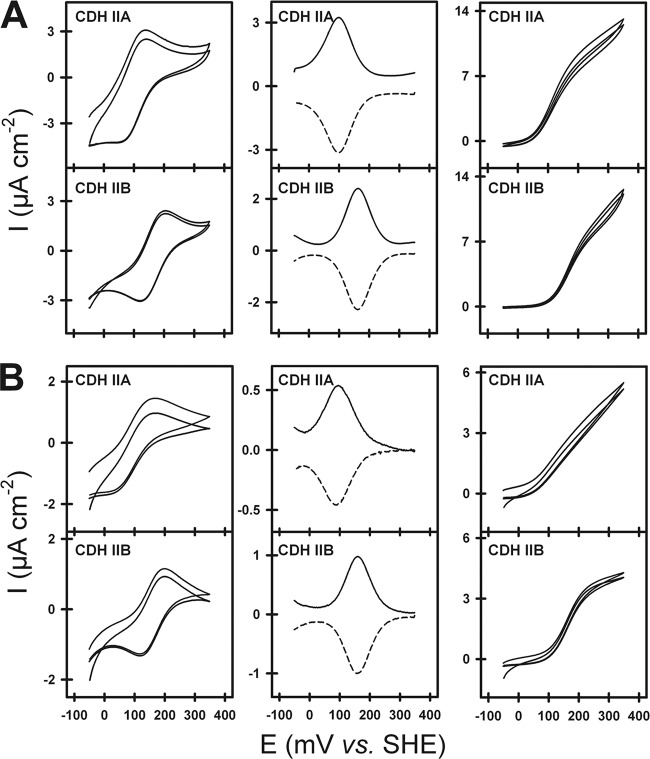
Cyclic voltammetry and square-wave voltammetry were employed to determine the redox properties of both CDH cytochrome domains at pH 6.0 and 7.5 (Fig. 3A and B). At pH 6.0, the specific catalytic currents were higher than those at pH 7.5, and also, the oxidative and reductive waves in the cyclic voltammograms were better defined than those at pH 7.5. The determined midpoint potentials of the cytochrome domain's heme b cofactor were 99 mV versus SHE at pH 6.0 and 93 mV versus SHE at pH 7.5 for CDH IIA and 163 mV versus SHE at pH 6.0 and 158 mV versus SHE at pH 7.5 for CDH IIB. The catalytic currents at 350 mV versus SHE in the presence of cellobiose were similar for both CDHs at pH 6.0 (12.5 μA cm−2). At pH 7.5, the current was lower, and slight differences were observed (5.3 μA cm−2 for CDH IIA and 4.5 μA cm−2 for CDH IIB). The onset of the catalytic currents on the gold electrodes started at around 60 mV versus SHE for CDH IIA and at around 100 mV versus SHE for CDH IIB. The onset was defined as the potential where 5% of the maximum current was measured. With GH61-3, no direct electron transfer (DET) could be observed on a thioglycerol-modified gold electrode.
Fig 3.
Electrochemical measurements of both CDHs on thiol-modified gold electrodes at pH 6.0 (A) and pH 7.5 (B). Midpoint potentials of both CDHs were calculated from cyclic voltammograms (left) and square-wave voltammograms (middle) in the absence of a substrate. The cyclic voltammetry (right) in the presence of the substrate was used to measure the current of direct electron transfer to a gold electrode.
Catalytic properties.
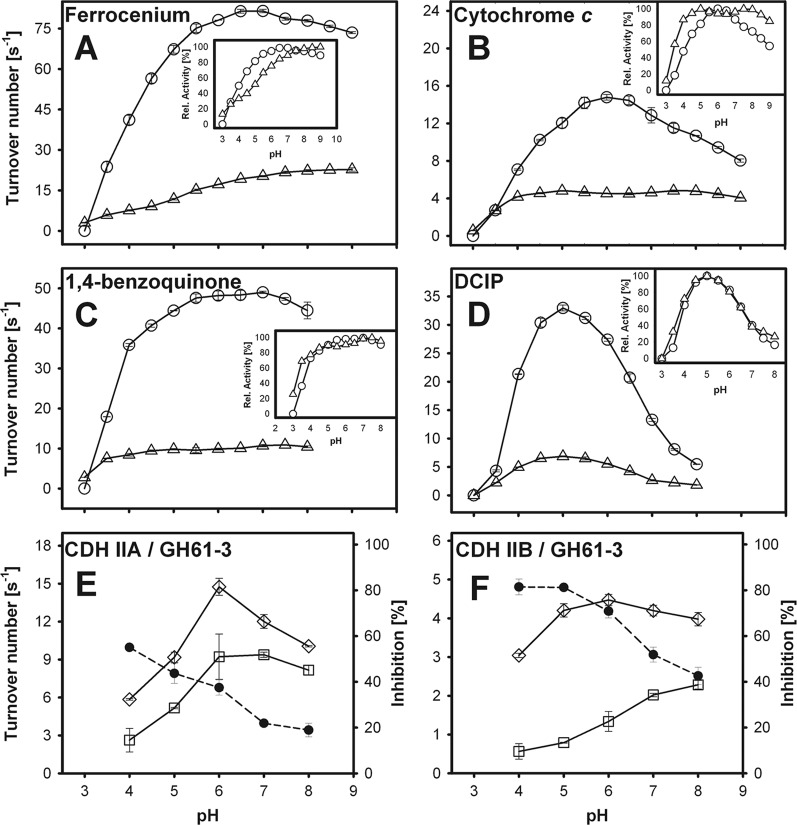
The pH dependencies of the two-electron-acceptor reduction by CDH IIA and CDH IIB (Fig. 4A to D) were similar for DCIP (optima at pH 5.0; >50% relative activity from pH 3.8 to 6.5) and 1,4-benzoquinone (optima at pH 7.0; >50% relative activity from pH 3.7 to >8.0). Due to quinhydrone formation in the alkaline milieu, pH values above 8.0 were not measured with 1,4-benzoquinone.
Fig 4.
(A to D) pH dependency of CDH IIA and CDH IIB activities for the artificial electron acceptors ferrocenium hexafluorophosphate (A), cytochrome c (B), 1,4-benzoquinone (C), and DCIP (D), using lactose as the electron donor. Circles, CDH IIA; triangles, CDH IIB. Activities from pH 2.5 to 9 were measured with McIlvaine buffer. (E and F) pH-dependent interaction of CDH IIA (E) and CDH IIB (F) with the competitive substrates cyt c and GH61-3. Diamonds, turnover rates of cyt c in the absence of GH61-3; squares, turnover rates of cyt c in the presence of GH61-3; filled circles, ratio of the turnover numbers, which is used as a measure of the inhibition of cyt c reduction by GH61-3.
The catalytic constants for the reduction of electron acceptors were determined at pH 5.0 for DCIP and at pH 6.0 for the other electron acceptors (Table 1). In a preliminary screening, we found that the relative activities of each CDH for various carbohydrates were independent of the use of the electron acceptor DCIP, cyt c, or 1,4-benzoquinone (see Table S2 in the supplemental material). In this experiment, the most obvious difference between CDH IIA and CDH IIB was a 5-times-higher glucose turnover rate than that of cellobiose by CDH IIB. By measuring the catalytic constants for cellobiose and lactose with DCIP as the cosubstrate, we observed substrate inhibition for both CDHs (see Table S3 and Fig. S4 and S5 in the supplemental material). This behavior was found only with DCIP as the electron acceptor. As a consequence, the apparent catalytic constants of CDHs for carbohydrates were determined with the electron acceptor 1,4-benzoquinone (Table 2).
Table 1.
Apparent kinetic constants of CDH IIA and CDH IIB for electron donors determined with lactose (30 mM) in McIlvaine buffer at the indicated pH valuesa
| Electron acceptor | Stoichiometry | pH | CDH IIA |
CDH IIB |
||||
|---|---|---|---|---|---|---|---|---|
| Mean Km (μM) ± SD | Mean kcat (s−1) ± SD | kcat/Km (mM−1 s−1) | Mean Km (μM) ± SD | Mean kcat (s−1) ± SD | kcat/Km (mM−1 s−1) | |||
| Cytochrome c | 2 | 6 | 67.9 ± 5.8 | 64.8 ± 1.7 | 954 | 6.7 ± 1.0 | 9.4 ± 0.3 | 1,403 |
| FcPF6 | 2 | 6 | 5.2 ± 0.2 | 75.4 ± 2.1 | 14,500 | 31.5 ± 7.5 | 19 ± 0.7 | 603 |
| 1,4-Benzoquinone | 1 | 6 | 26.2 ± 3.0 | 49.2 ± 0.7 | 1,878 | 24.3 ± 0.1 | 9.9 ± 0.4 | 407 |
| DCIP | 1 | 5 | 33.6 ± 4.8 | 38.1 ± 0.8 | 1,134 | 25.3 ± 5.8 | 7.4 ± 0.6 | 292 |
The sample standard deviation was calculated from data from 3 experiments.
Table 2.
Apparent kinetic constants of CDH IIA and CDH IIB for electron donors determined with 1,4-benzoquinone (1 mM) in McIlvaine buffer at pH 6.0c
| Carbohydrate | CDH IIA |
CDH IIB |
||||
|---|---|---|---|---|---|---|
| Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) | |
| Cellobiose | 0.090 | 45.8 | 508 | 0.022 | 11.4 | 527 |
| Cellotriose | 0.230 | 49.7 | 216 | 0.045 | 12.4 | 278 |
| Cellotetraose | 0.204 | 46.0 | 226 | 0.061 | 11.6 | 189 |
| Cellopentaose | 0.196 | 46.6 | 238 | 0.043 | 10.2 | 238 |
| Lactose | 0.290 | 49.3 | 170 | 0.089 | 11.8 | 133 |
| Maltose | 17.2 | 0.4 | 0.023 | 3.44 | 3.5 | 1.02 |
| Maltotriose | 34.3 | 1.8 | 0.053 | 1.2 | 1.0 | 0.8 |
| Maltotetraose | 35.7 | 2.2 | 0.061 | 4.5 | 1.4 | 2.74 |
| Glucose | 3,700a | 55a | 0.015 | 550 | 8.1 | 0.014 |
| Galactose | 1.5b | 3,600a | 3.6a | 0.001 | ||
| Mannopentaose | 39.2 | 19.8 | 0.506 | 8.4 | 7.4 | 0.88 |
| Mannose | 10,000a | 4.7 | 0.0004 | 13,300a | 4.3 | 0.0003 |
| Xylobiose | 3.6 | 45.4 | 12.8 | 1.31 | 5.8 | 4.45 |
| Xylotriose | 3 | 6 | 2 | 0.81 | 2.3 | 2.8 |
| Xylose | 2,770a | 1.5 | 0.0005 | 1,545a | 1.9 | 0.0012 |
| Arabinose | 0.1b | 3,200a | 0.6 | 0.0002 | ||
Extrapolated Km and kcat values.
Turnover number measured with 1,000 mM galactose.
The relative standard error (n = 3) for all reported Km and kcat values was <10%.
Interaction of CDH IIA and CDH IIB with GH61-3.
The interaction between CDH and GH61-3 was studied by the inhibition of cyt c activity in the presence of various concentrations of GH61-3. The rationale of the experiments was to treat cyt c and GH61-3 as competing substrates for the intermolecular electron transfer from the cytochrome domain of CDH. In preliminary experiments with CDH IIA, a constant concentration of 10 μM cyt c with various GH61-3 concentrations was tested. Even with a low molar ratio of cyt c to GH61-3 of 1:0.25, a significant reduction of the competing cyt c activity (10%) was found for both CDHs, which increased up to 60% at a ratio of 1:4. Experiments using BSA and copper sulfate (in a 4-fold excess over cyt c) as unspecific substitutes for GH61-3 showed little influence on the reduction of cyt c activity by CDH (see Fig. S6 in the supplemental material).
Initial rates were measured to guarantee steady-state conditions. A linear change in the absorbance indicates that, indeed, neither cyt c nor GH61-3 was depleted during the reactions. The measured pH dependency of the CDH IIA/GH61-3 (Fig. 4E) and CDH IIB/GH61-3 (Fig. 4F) interactions is expressed as the percent inhibition of cyt c activity. The highest level of inhibition of cyt c by the CDH/GH61-3 interaction was observed at an acidic pH. The pH optimum of the CDH IIB/GH61-3 interaction was more acidic for CDH IIA than for CDH IIB, but a much higher level of cyt c inhibition was found for the CDH IIB/GH61-3 interaction (81%) than for the CDH IIA/GH61-3 interaction (55%).
DISCUSSION
N. crassa is a model organism in the fields of genetics, biochemistry, and fungal biology and has been studied for many decades. It is also recognized as a potent producer of lignocellulolytic and hemicellulolytic enzymes (5, 25, 28). When grown under cellulolytic conditions, a major part of the secretome consists of CDH IIA (2.4%) and members of the GH61 (14.6%) family, altogether 17% (23) of the totally secreted protein mass, which indicates their importance in biomass degradation by N. crassa. In a previous transcriptome analysis of N. crassa by Tian et al. (32), a tremendous increase of the cdhIIA gene transcription level during cultivation on Miscanthus was found (160-fold after 16 h), whereas the level of transcription of cdhIIB increased only 2 to 3 times compared to that with growth on minimal medium. Some of the 14 gh61 genes found in N. crassa are also strongly upregulated during growth on Miscanthus, whereas others are not (Fig. 5). The transcriptions of 8 of the 14 putative gh61 genes found in the N. crassa genome were upregulated more than 5-fold, while 4 genes showed a weak or no change (for 2 genes, no transcriptional data are available). In comparison, a similar number of GH61s with or without a C-terminal CBM was upregulated, whereas only the CBM carrying CDH IIA was induced. The induction of CDH IIA during growth under cellulolytic conditions was shown previously to be even more pronounced in a strain carrying a deletion of cre-1. The authors of that study identified cdhIIA as a part of the CRE-1 regulon in N. crassa (27). cdhIIB seems to be unaffected by CRE-1 repression during growth on preferred carbon sources such as glucose. So far, the expression of CDH IIB in N. crassa cultures could not be verified (8, 23). A comparison of CDH IIA and CDH IIB amino acid sequences (for an alignment, see reference 8) showed a low level of identity (53.2%). Interestingly, the flavodehydrogenase domains share more identical amino acids (59.2%) than the cytochrome domains (39%). In other ascomycete species, CDH IIB is, however, an active enzyme and expressed solely (e.g., Corynascus thermophilus) or at much higher levels than CDH IIA (e.g., Hypoxylon haematostroma) (8). The reason for being missed in the purification of N. crassa culture supernatants reported previously by Harreither et al. (8) is most likely its small amount and low affinity for chromatography resins, where the small amount of CDH IIB was presumably not bound in the capture step and was lost with the flowthrough.
Fig 5.
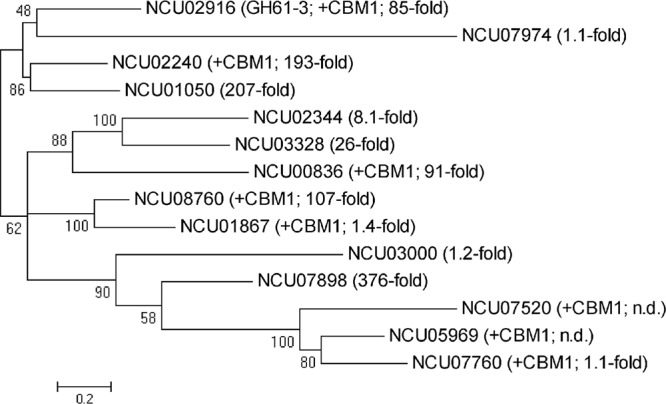
Phylogenetic tree of the N. crassa GH61 protein family. Sequence alignment was performed with Clustal using the default parameters, and phylogenetic analysis was done with MEGA 5 using the maximum likelihood method and 1,000 bootstrap replicates. Relative expression levels of N. crassa gh61 genes during growth on Miscanthus (16 h) are shown. Data are taken from data reported previously by Tian et al. (32). n.d., not determined.
To investigate if N. crassa CDH IIB is an active enzyme and in which aspects it differs from CDH IIA, P. pastoris was chosen for the heterologous overexpression of both CDHs. Since a previous attempt to express N. crassa CDH IIB in P. pastoris had failed (43), we chose a different approach for the construction of the expression cassette. Instead of using the Saccharomyces cerevisiae α-factor pre-pro leader peptide, the native leader sequence was used as the secretion signal. This approach was previously successfully applied for other FAD-dependent glucose-methanol-cholin (GMC) oxidoreductases (29, 30). To obtain both CDHs as similar as possible to the wild-type enzymes, any tags for purification or antibody detection were omitted. These changes in the expression strategy enabled the production of both CDHs in reasonable amounts. The higher volumetric activity in the culture supernatant of CDH IIA is due to the higher specific activity, because the protein concentrations of the CDHs were equal (80 mg liter−1, including its proteolytic degradation product, the flavodehydrogenase domain). In comparison, the percentage of the total extracellular protein representing the recombinant enzyme was highest for GH61-3 (33%), slightly lower for CDH IIA (17% plus 11% flavodehydrogenase domain), and lowest for CDH IIB (7% plus 5% flavodehydrogenase domain).
The first difference in the physical properties of both CDHs was observed during purification. CDH IIA showed a strong interaction with the phenyl groups of the hydrophobic-interaction resin, whereas CDH IIB was bound more weakly and needed a higher ammonium sulfate concentration. A similar behavior was found with the anion-exchange resin, where CDH IIB was also bound more weakly despite its lower pI. Since the number of putative glycosylation sites is identical in both CDHs (eight surface-exposed sequons each) and the amino acid compositions are also similar, the different binding/desorption behaviors are likely to originate from a different composition of surface regions or the presence of the CBM. The low affinity of CDH IIB for chromatography matrices not only reduced the yield by excessive tailing but also necessitated a further purification step by size-exclusion chromatography. GH61-3, like CDH IIB, needs 30% ammonium sulfate saturation to bind on the HIC matrix. It also bound poorly on an anion-exchange matrix (Q-Sepharose) and was therefore purified by size-exclusion chromatography. All enzymes were stable during purification and storage. The absorbance ratios (A420/A280) for CDH IIA (0.58) and CDH IIB (0.55) are high and confirm the purity of the obtained CDH preparations. Values for other ascomycetous CDHs are, e.g., 0.63 for Humicola insolens CDH IIB (13) and 0.60 for C. thermophilus CDH IIB (8). For GH61-3, the very low absorption of the copper ion at 630 nm indicates a type 2 copper complex.
The measured molecular masses of CDH IIA and CDH IIB were higher than that of CDH IIA purified from the N. crassa proteome (90 kDa), which seemed to be less glycosylated than the recombinant CDHs. The mass difference between the glycosylated and deglycosylated enzymes originates from N-glycans, which can be cleaved off by PNGase F. The bands of the deglycosylated CDHs are in perfect agreement with the masses calculated from the mature amino acid sequences, which are 86.2 kDa for CDH IIA and 86.6 kDa for CDH IIB. Recombinant GH61-3 is also glycosylated. However, since no reduction of the molecular mass was observed after PNGase treatment, we conclude that the contribution of N-glycans to the molecular masses of the glycoproteins is small. The two N-glycosylation consensus sequences (NXS/T) present in the GH61-3 sequence could increase the molecular mass by ∼5 kDa, if one assumes the typical high-mannose-type N-glycan commonly reported for secreted proteins from P. pastoris. Prolonged deglycosylation with a mixture of endoglucanase F and α-mannosidase showed a decrease in the mass by 10 kDa to ∼45 kDa, which is still higher than the calculated mass of the mature protein sequence (34.3 kDa). We conclude that GH61-3 is mostly O-glycosylated by mannose oligosaccharides (18, 33), possibly at the Ser/Thr-rich linker before the C-terminal CBM. The isoelectric points of 5.14 for CDH IIA and 4.93 for CDH IIB, as determined by chromatofocusing, are notably higher than those of most other CDHs (pI of ∼4.1), with the exception of Chaetomium sp. strain INBI(2-26−) CDH (pI of 5.0) (15). In comparison to the theoretically calculated pI values of 6.7 for CDH IIA and 7.9 for CDH IIB, this indicates a clear imbalance of surface-exposed anionic and buried cationic amino acids. The previously reported pI of 6.8 for recombinantly expressed CDH IIA (43) is much higher and reflects most likely the addition of a His6 tag.
Another difference between both CDHs was found in the electrochemically measured redox potentials of the heme b cofactor in the cytochrome domain. CDH IIA showed, with 95 mV versus SHE, a 65-mV-lower midpoint potential than the CDH IIB counterpart. The two values are in good agreement with other previously reported midpoint potentials but represent the upper and the lower values reported so far. The lower value for CDH IIA is in good agreement with data reported previously for C. thermophilus CDH IIB (31). For other class II CDHs, higher values of around 130 mV under neutral-pH conditions were reported. The midpoint potential changed by only 5 mV between pH 6.0 and 7.5, which was also found previously for CDH IIB from H. insolens and CDH IIA from M. thermophilum (3, 13).
Steady-state kinetic measurements for various electron acceptors revealed identical pH optima of both CDHs for the two-electron acceptors DCIP (pH 5.0) and 1,4-benzoquinone (pH 7.0), which are reduced at the flavodehydrogenase domain. However, the pH profiles for the one-electron acceptors cyt c and FcPF6 differed, which can be seen from the insets showing relative activities. Cyt c is strictly dependent on the presence of the cytochrome domain of CDH for intermolecular electron transfer. CDH IIA has a bell-shaped optimum for cyt c turnover (pH 6.0), while CDH IIB shows a broad plateau from pH 4.0 to 9.0 with two optima at pH 5.0 and 7.5. Thus, CDH IIB has an extended activity in the acidic and alkaline pH ranges. The pH optimum of CDH IIB for FcPF6 is much higher than that of CDH IIA. The pH optima of various N. crassa cellulolytic enzymes range from pH 5.0 to 7.0, with significant residual activities from pH 3.0 to 8.5 (4, 40). Judged from the pH profiles of the one-electron acceptors, CDH IIB works over a broader pH range and at alkaline pH values.
For all tested electron acceptors, the Km values were in the low-micromolar range. The catalytic efficiencies for the two-electron acceptors DCIP and 1,4-benzoquinone were 3.9 and 4.6 times higher for CDH IIA, respectively, and the catalytic efficiency for FcPF6 was even 24 times higher for CDH IIA. Cyt c is the exception: it was 1.5 times more efficiently reduced by CDH IIB. For the determination of the catalytic constants for carbohydrates, 1,4-benzoquinone was used to avoid the limitation of the oxidative FAD cycle by the contribution of intramolecular electron transfer (IET) on the measurements when using cyt c or to limit the reaction by the inhibition found with DCIP for N. crassa CDH turnover. Substrate inhibition in the presence of DCIP was observed for both CDHs but was stronger for CDH IIB. The Ki values for cellobiose and lactose remained relatively constant between pH 4.0 and 7.0.
The highest catalytic efficiencies for electron donors were found for the β-1,4-linked disaccharide cellobiose and cellooligosaccharides. Although the catalytic efficiencies are very similar, the CDHs differ again in their apparent kcat and Km values. CDH IIA has approximately 4- to 5-times-higher kcat values, whereas CDH IIB has approximately 4- to 5-times-lower Km values. Nevertheless, the observed catalytic constants for a number of sugar substrates revealed a broad substrate spectrum. Beside the cellobiose-mimicking substrate lactose, both CDHs can convert the β-1,4-linked hemicelluloses xylobiose and xylotriose with Km values in the low-millimolar range and catalytic efficiencies only 50 to 100 times lower than those for cellobiose. This demonstrates that CDH IIA and CDH IIB can be efficiently reduced by xylobiose and xylotriose in vivo and might reflect the life-style of N. crassa as a degrader of hemicellulose-rich plant cell walls. The catalytic efficiencies for the monomers glucose and xylose are 25,000 to 35,000 times lower than those for the di- or polysaccharides built of them. The discrimination of glucose with respect to cellobiose is nearly as high as that in basidiomycetous class I CDHs (e.g., an 87,000-fold-higher catalytic efficiency for P. chrysosporium CDH [12]). Starch-derived maltose and maltooligosaccharides featuring α-1,4 linkages are moderate substrates for CDH IIB but poor substrates for CDH IIA. 2α-Mannobiose with an α-1,2 linkage was not converted, which stands in contrast to previously reported data (43). To verify measurements, 2α-mannobiose was obtained from two different suppliers (Sigma and Santa Cruz Biotechnology Inc.). Mannopentaose was, however, a modest substrate. Comparisons of further catalytic constants for N. crassa CDH IIA reported here with data reported previously by Zhang et al. are limited because of the different electron acceptors and pH values used (DCIP at pH 4.5 [43]). Other hemicellulose constituents, such as mannose, arabinose, and galactose, are not in vivo substrates.
Cyt c, which is exclusively oxidized at the cytochrome domain of CDH, provides a measure of the IET rate between the flavodehydrogenase and the cytochrome domains when it is assumed that the cytochrome domain/cyt c interaction is very fast and not rate limiting. High bimolecular rate constants have indeed been reported for P. chrysosporium CDH (1.75 × 107 M−1 s−1 [26] and 6.6 × 106 M−1 s−1 [14]). From the catalytic efficiencies measured for cyt c with CDH IIA (9.5 × 105 M−1 s−1) and CDH IIB (1.4 × 106 M−1 s−1), a similarly fast electron transfer reaction can be assumed. Interestingly, the kcat value was 6.9 times higher for CDH IIA despite the 65-mV-lower midpoint potential of its cytochrome domain. Both CDHs showed IET at up to pH 9.0. Fast-kinetic studies will be necessary to elucidate the IET and the cytochrome domain/cyt c interaction in more detail.
The second investigated reaction, in which IET is involved in the transfer of reduction equivalents to a macroscopic, terminal electron acceptor, was the interaction of CDH with SAM gold electrodes. It was found that at pH 6.0, direct electron transfer (DET) to the macroscopic electron acceptor limits the whole reaction. This is assumed from the similar catalytic currents obtained from both CDHs at pH 6.0 and 7.5. If this last electron transfer step would not be rate limiting, the different IET rates indicated by the above-discussed cyt c turnover should result in a higher current for CDH IIA. The lower midpoint potential of the CDH IIA cytochrome domain is also reflected by the lower onset potential of CDH IIA's catalytic current. This different midpoint potential might be of importance for the interaction with GH61 polysaccharide monooxygenases.
Experimental data from a growing body of literature emphasize the concept that CDH enhances the depolymerization of crystalline cellulose in a synergistic mechanism together with GH61 polysaccharide monooxygenases. Not much is yet known about the interaction of CDH and GH61 enzymes, but it was shown previously that electron transfer proceeds via the cytochrome domain, since the flavodehydrogenase domain alone showed no effect (22). However, those studies used CDHs and GH61 enzymes from different organisms. N. crassa GH61-3 (NCU02916), used in this study, is a close relative of NCU02240 and NCU01050, which were both identified previously as polysaccharide monooxygenases (22). The interaction with both N. crassa CDHs was measured by using a competing-substrate approach. Cyt c was chosen as the competing substrate for GH61-3, since in both reactions, IET is involved, as postulated. From the strong inhibition of the reaction with cyt c, which is an electron acceptor with a high catalytic efficiency/bimolecular rate constant, it is obvious that the interaction of CDH with GH61-3 is of a similarly high efficiency. This effect was found to be more pronounced for CDH IIB. The cyt c reaction was inhibited by 81% in the presence of GH61-3 at pH 4.0 and 5.0, whereas for CDH IIA, an inhibition of only 55% at pH 4.0 was found. Similar to the pH profiles of the one-electron acceptors of cyt c and FcPF6, CDH IIB also showed a more efficient interaction with GH61-3 at neutral/alkaline pHs. At pH 8.0, GH61-3 inhibited the cyt c reduction of CDH IIB to 43% but that of CDH IIA to only 19%. Whether CDH IIB generally interacts more efficiently with N. crassa polysaccharide monooxygenases from the GH61 family or just in the case of GH61-3 remains to be elucidated. The higher pH optimum of the CDH IIB/GH61-3 interaction is probably a result of the higher redox potential of CDH IIB's cytochrome domain.
This work characterized in detail the properties of N. crassa CDH IIA, CDH IIB, and GH61-3 and their interactions for electron transfer. The two CDHs differ strongly in their transcription levels, the presence of a CBM, the redox potentials of the cytochrome domain, and the pH optima with positively charged one-electron acceptors and GH61-3. We suggest that CDH IIA and CDH IIB fulfill different functions at different fungal growth phases, under different pH conditions, or with different GH61 enzymes. Their ability to obtain electrons from cellobiose or cellooligosaccharides and xylobiose or xylooligosaccharides suggests that, depending on the reduced GH61 polysaccharide monooxygenase, CDHs are involved in cellulose and hemicellulose degradation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Viktoria Hell for superb technical assistance.
This work has been supported by project L395-B11 and BioTop (Biomolecular Technology of Proteins) (grant FWF-W1224) of the Austrian Science Fund (FWF).
Footnotes
Published ahead of print 22 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Beeson WT, Phillips CM, Cate JHD, Marletta MA. 2012. Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J. Am. Chem. Soc. 134: 890–892 [DOI] [PubMed] [Google Scholar]
- 2. Cameron MD, Aust SD. 2001. Cellobiose dehydrogenase—an extracellular fungal flavocytochrome. Enzyme Microb. Technol. 28: 129–138 [DOI] [PubMed] [Google Scholar]
- 3. Coman V, Harreither W, Ludwig R, Haltrich D, Gorton L. 2007. Investigation of electron transfer between cellobiose dehydrogenase from Myriococcum thermophilum and gold electrodes. Chem. Analityczna 52: 945–960 [Google Scholar]
- 4. Eberhart BM, Beck RS, Goolsby KM. 1977. Cellulase of Neurospora crassa. J. Bacteriol. 130: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fan Z, et al. 2012. A novel biochemical route for fuels and chemicals production from cellulosic biomass. PLoS One 7: e31693 doi: 10.1371/journal.pone.0031693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haladjian J, Bianco P, Nunzi F, Bruschi M. 1994. A permselective-membrane electrode for the electrochemical study of redox proteins. Application to cytochrome C552 from Thiobacillus ferrooxidans. Anal. Chim. Acta 289: 15–20 [Google Scholar]
- 7. Harreither W, Coman V, Ludwig R, Haltrich D, Gorton L. 2007. Investigation of graphite electrodes modified with cellobiose dehydrogenase from the ascomycete Myriococcum thermophilum. Electroanalysis 19: 172–180 [Google Scholar]
- 8. Harreither W, et al. 2011. Catalytic properties and classification of cellobiose dehydrogenases from ascomycetes. Appl. Environ. Microbiol. 77: 1804–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harreither W, et al. 2009. Cellobiose dehydrogenase from the ligninolytic basidiomycete Ceriporiopsis subvermispora. Appl. Environ. Microbiol. 75: 2750–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harris PV, et al. 2010. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry 49: 3305–3316 [DOI] [PubMed] [Google Scholar]
- 11. Henriksson G, Johansson G, Pettersson G. 2000. A critical review of cellobiose dehydrogenases. J. Biotechnol. 78: 93–113 [DOI] [PubMed] [Google Scholar]
- 12. Henriksson G, Sild V, Szabo IJ, Pettersson G, Johansson G. 1998. Substrate specificity of cellobiose dehydrogenase from Phanerochaete chrysosporium. Biochim. Biophys. Acta 1383: 48–54 [DOI] [PubMed] [Google Scholar]
- 13. Igarashi K, et al. 1999. Cellobiose dehydrogenase from the fungi Phanerochaete chrysosporium and Humicola insolens. A flavohemoprotein from Humicola insolens contains 6-hydroxy-fad as the dominant active cofactor. J. Biol. Chem. 274: 3338–3344 [DOI] [PubMed] [Google Scholar]
- 14. Igarashi K, et al. 2005. Electron transfer chain reaction of the extracellular flavocytochrome cellobiose dehydrogenase from the basidiomycete Phanerochaete chrysosporium. FEBS J. 272: 2869–2877 [DOI] [PubMed] [Google Scholar]
- 15. Karapetyan KN, et al. 2006. Properties of neutral cellobiose dehydrogenase from the ascomycete Chaetomium sp. INBI 2-26(−) and comparison with basidiomycetous cellobiose dehydrogenases. J. Biotechnol. 121: 34–48 [DOI] [PubMed] [Google Scholar]
- 16. Karkehabadi S, et al. 2008. The first structure of a glycoside hydrolase family 61 member, Cel61B from Hypocrea jecorina, at 1.6 Å resolution. J. Mol. Biol. 383: 144–154 [DOI] [PubMed] [Google Scholar]
- 17. Langston JA, et al. 2011. Oxidoreductive cellulose depolymerization by the enzymes cellobiose dehydrogenase and glycoside hydrolase 61. Appl. Environ. Microbiol. 77: 7007–7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Letourneur O, et al. 2001. Characterization of Toxoplasma gondii surface antigen I (SAGI) secreted from Pichia pastoris: evidence of hyper O-glycosylation. Biotechnol. Appl. Biochem. 33: 35–45 [DOI] [PubMed] [Google Scholar]
- 19. Ludwig R, Haltrich D. 2002. Cellobiose dehydrogenase production by Sclerotium species pathogenic to plants. Lett. Appl. Microbiol. 35: 261–266 [DOI] [PubMed] [Google Scholar]
- 20. Ludwig R, et al. 2004. Characterisation of cellobiose dehydrogenases from the white-rot fungi Trametes pubescens and Trametes villosa. Appl. Microbiol. Biotechnol. 64: 213–222 [DOI] [PubMed] [Google Scholar]
- 21. McIlvaine TC. 1921. A buffer solution for colorimetric comparison. J. Biol. Chem. 49: 183–186 [Google Scholar]
- 22. Phillips CM, Beeson WT, Cate JH, Marletta MA. 2011. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem. Biol. 6: 1399–1406 [DOI] [PubMed] [Google Scholar]
- 23. Phillips CM, Iavarone AT, Marletta MA. 2011. Quantitative proteomic approach for cellulose degradation by Neurospora crassa. J. Proteome Res. 10: 4177–4185 [DOI] [PubMed] [Google Scholar]
- 24. Quinlan RJ, et al. 2011. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. U. S. A. 108: 15079–15084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rao M, Mishra C, Keskar S, Srinivasan MC. 1985. Production of ethanol from wood and agricultural residues by Neurospora crassa. Enzyme Microb. Technol. 7: 625–628 [Google Scholar]
- 26. Rogers MS, Jones GD, Antonini G, Wilson MT, Brunori M. 1994. Electron transfer from Phanerochaete chrysosporium cellobiose oxidase to equine cytochrome c and Pseudomonas aeruginosa cytochrome c-551. Biochem. J. 298: 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun J, Glass NL. 2011. Identification of the CRE-1 cellulolytic regulon in Neurospora crassa. PLoS One 6: e25654 doi: 10.1371/journal.pone.0025654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun J, Tian C, Diamond S, Glass NL. 2012. Deciphering transcriptional regulatory mechanisms associated with hemicellulose degradation in Neurospora crassa. Eukaryot. Cell 11: 482–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sygmund C, et al. 2012. Simple and efficient expression of Agaricus meleagris pyranose dehydrogenase in Pichia pastoris. Appl. Microbiol. Biotechnol. 94: 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sygmund C, et al. 2011. Heterologous overexpression of Glomerella cingulata FAD-dependent glucose dehydrogenase in Escherichia coli and Pichia pastoris. Microb. Cell Fact. 10: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tasca F, et al. 2011. A third generation glucose biosensor based on cellobiose dehydrogenase from Corynascus thermophilus and single-walled carbon nanotubes. Analyst 136: 2033–2036 [DOI] [PubMed] [Google Scholar]
- 32. Tian C, et al. 2009. Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc. Natl. Acad. Sci. U. S. A. 106: 22157–22162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trimble RB, et al. 2004. Characterization of N- and O-linked glycosylation of recombinant human bile salt-stimulated lipase secreted by Pichia pastoris. Glycobiology 14: 265–274 [DOI] [PubMed] [Google Scholar]
- 34. Tuomela M, Vikman M, Hatakka A, Itävaara M. 2000. Biodegradation of lignin in a compost environment: a review. Bioresour. Technol. 72: 169–183 [Google Scholar]
- 35. Vaaje-Kolstad G, Horn SJ, Van Aalten DMF, Synstad B, Eijsink VGH. 2005. The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J. Biol. Chem. 280: 28492–28497 [DOI] [PubMed] [Google Scholar]
- 36. Vaaje-Kolstad G, Houston DR, Riemen AHK, Eijsink VGH, Van Aalten DMF. 2005. Crystal structure and binding properties of the Serratia marcescens chitin-binding protein CBP21. J. Biol. Chem. 280: 11313–11319 [DOI] [PubMed] [Google Scholar]
- 37. Vaaje-Kolstad G, et al. 2010. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330: 219–222 [DOI] [PubMed] [Google Scholar]
- 38. Weis R, et al. 2004. Reliable high-throughput screening with Pichia pastoris by limiting yeast cell death phenomena. FEMS Yeast Res. 5: 179–189 [DOI] [PubMed] [Google Scholar]
- 39. Westereng B, et al. 2011. The putative endoglucanase PcGH61D from Phanerochaete chrysosporium is a metal-dependent oxidative enzyme that cleaves cellulose. PLoS One 6: e27807 doi: 10.1371/journal.pone.0027807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yazdi MT, Woodward JR, Radford A. 1990. The cellulase complex of Neurospora crassa: activity, stability and release. J. Gen. Microbiol. 136: 1313–1319 [DOI] [PubMed] [Google Scholar]
- 41. Zámocký M, Hallberg M, Ludwig R, Divne C, Haltrich D. 2004. Ancestral gene fusion in cellobiose dehydrogenases reflects a specific evolution of GMC oxidoreductases in fungi. Gene 338: 1–14 [DOI] [PubMed] [Google Scholar]
- 42. Zámocký M, et al. 2006. Cellobiose dehydrogenase—a flavocytochrome from wood-degrading, phytopathogenic and saprotropic fungi. Curr. Protein Pept. Sci. 7: 255–280 [DOI] [PubMed] [Google Scholar]
- 43. Zhang R, Fan Z, Kasuga T. 2011. Expression of cellobiose dehydrogenase from Neurospora crassa in Pichia pastoris and its purification and characterization. Protein Expr. Purif. 75: 63–69 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.