Abstract
Rival exposure causes Drosophila melanogaster males to prolong mating. Longer-Mating-Duration (LMD) may enhance reproductive success, but its underlying mechanism is currently unknown. Here we report that LMD is context-dependent and can be induced solely via visual stimuli. We further show that LMD involves neural circuits important for visual memory, including central neurons in the ellipsoid body but not the mushroom bodies or the fan-shaped bodies, and may rely on the rival exposure memory lasting several hours. LMD is affected by a subset of learning and memory mutants. LMD depends on the circadian clock genes timeless and period but not Clock or cycle, and persists in many arrhythmic conditions. Moreover, LMD critically depends on a subset of pigment dispersing factor (PDF) neurons rather than the entire circadian neural circuit. Our study thus delineates parts of the molecular and cellular basis for LMD – a plastic social behavior elicited by visual cues.
Competition between males contributes to ‘sexual selection’, which Charles Darwin defined as the “struggle between the individuals of one sex, generally the males, for the possession of the other sex”1. In Drosophila, males vigorously compete with one another and this aggressive behavior may help with the acquisition or defense of food resources as well as gaining access to mates2.
In addition to aggression, male–male competition may take the form of ‘sperm competition’3, a process for the males’ sperm to compete in fertilizing the ova4. One of the tactics for this process is prolonged copulation. In Drosophila, whereas there appears to be little correlation between the mating duration and fertility based on counting progeny numbers3, lengthening a male’s mating duration increases its paternity share among the progeny, indicative of greater reproductive success4.
Mating duration can be influenced by many factors, including environment5; age6, body size7, infection status of the male8, and whether the female is a virgin9. The mating duration is plastic and depends on the male’s social experience; Drosophila melanogaster males respond to the presence of other males by prolonging the mating duration10. Since little is known about the molecular and cellular basis of rival-induced longer mating duration (Longer-Mating-Duration: LMD), we took advantage of the fact that mating duration traits are under male control11 and hence more amenable to genetic analysis than other social behavior12, and characterized the underlying circuitry.
LMD in Drosophila melanogaster is a plastic behavior dependent on the environmental context
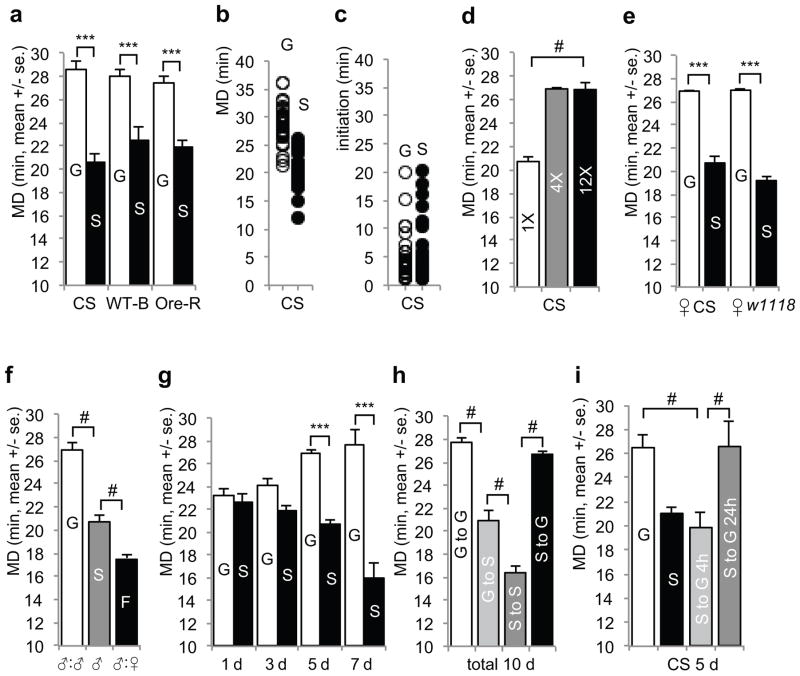
The mating duration for the Dahomey strain is longer for males housed together with rivals for 5 days prior to mating10. By using the same experimental protocol, we found the mating duration of group-reared wild type (WT) Canton-S (CS) males also lasted longer (by more than 5 min) than that of singly reared males (Fig. 1a–b), though they had comparable timing for mating initiation (Fig. 1c) and mating success ratio over the course of 60 min (~85%). Similar LMD was observed in WT Berlin and Oregon R strains (Fig. 1a) as well as another species of fruit flies, Drosophila simulans (Supplementary Fig. 1a). LMD did not seem to depend on the number of rivals present during group rearing (Fig. 1d) as reported for the Dahomey strain13, nor did LMD vary with the number of males present during the mating duration assay (Supplementary Fig. 1g–h), in contrast to what has been reported for the Dahomey strain10. The white mutant (w1118) males showed no LMD (Supplementary Fig. 1b–f).
Figure 1.
General characteristics of the ‘Longer-Mating-Duration (LMD)’ behavior. Males were kept for 5 days after eclosion with single housing or in groups of 4 males and then placed in the mating chamber with one 5 day-old virgin female; for detailed experimental procedure, see Methods. a. Mating duration assays of Canton-S (CS), wild type Berlin (WT-B) and Oregon R (Ore-R) males. The mating partners’ genotype was the same as that of the males. G represents group-reared and S represents single-reared males. From left to right, number of tested animals (n): 30, 27, 29, 37, 26, and 35. t values (t): 9.134, 8.490, and 7.891. b. Distribution of the same data of CS shown in a. Each circle (white: group-reared; n=30, black: singly reared; n=27) represents the mating duration of a single male. c. Distribution of copulation initiation time of CS in group- (n=30) and singly (n=27) reared males shown in b. d. Mating duration assays of CS males, singly reared (white bar), group-reared with 3 other males (grey), or group-reared with 11 other males (black). n=33, 30, and 45 (left to right). F value (F): 69.44. e. Mating duration assays of group- or singly reared CS males with 5 day-old CS or w1118 female partners. The genotypes of the female partner are indicated below the bars. From left to right, n=128, 110, 42, and 43. t=12 and 8.446. f. Mating duration assays of group- (G: 4 CS males reared together), or singly reared (S: 1 CS male singly reared), and males reared with 3 virgin females (F: 1 CS male reared with 3 CS females, which were replaced with virgins daily). From left to right, n=30, 27, and 41. F=113.5. g. Mating duration assays of CS males after 1, 3, 5, or 7 days of group- or singly rearing. From left to right, n=35, 39, 31, 35, 26, 31, 29, and 31. t=0.9817, 4.799, 5.064, and 8.985. h. Mating duration assays of CS males. After 5 days, group- or singly rearing conditions were either maintained (white and dark gey bars) or changed (light grey and black bars) for the subsequent 5 days. From left to right, n=30, 17, 18, and 17. F=49.62. i. Mating duration assays of CS males. Group-rearing condition was shifted to singly rearing for 4 h (light grey bar) or 24 h (dark grey bar) before the mating duration assay. From left to right, n=22, 13, 16, and 30. F=31.70. Bars represent mean of the mating duration (MD) with error bars representing the s.e.m. Asterisks represent significant differences revealed by Student’s t test (* p<0.05, ** p<0.01, *** p<0.001). Number signs represent significant differences revealed by Dunn’s Multiple Comparison Test (# p<0.05). The same symbols for statistical significance are used in all other figures.
LMD was evident for CS males with either CS or w1118 female partners (Fig. 1e), consistent with the findings that the mating duration is largely under male control in Drosophila melanogaster and Drosophila simulans11. When we introduced 3 CS virgin females to be housed together with a single CS male and replaced the females with virgin females daily over the course of 5 days, we found the mating duration for these CS males with prior exposure to females was shorter than that of group- or singly reared males (Fig. 1f), probably reflecting the fact that the males’ resource for mating is not unlimited13. By quantifying the mating duration every other day, we found the group-reared males began to show LMD by the 5th day of cohabitation (Fig. 1g), while the mating duration of singly reared males became progressively shorter after 5 days of isolation (Fig. 1g).
To test for the reversibility of LMD, we shifted singly reared males to group rearing, and also moved group-reared males to subsequent housing in isolation. The mating duration was shorter for males which were group-reared for the first 5 days then shifted to singly rearing for 5 days as compared with males group-reared for 10 days (Fig. 1h). In contrast, the mating duration was longer for males singly reared for the first 5 days then group-reared for 5 days as compared to males singly reared for 10 days (Fig. 1h). Exposure of males that had been singly reared for 5 days to rivals for 24 h was sufficient to induce LMD (Fig. 1i). Thus LMD is plastic; it can change over time in a way that depends on the male’s experience with his rivals.
Visual stimuli are required for inducing LMD
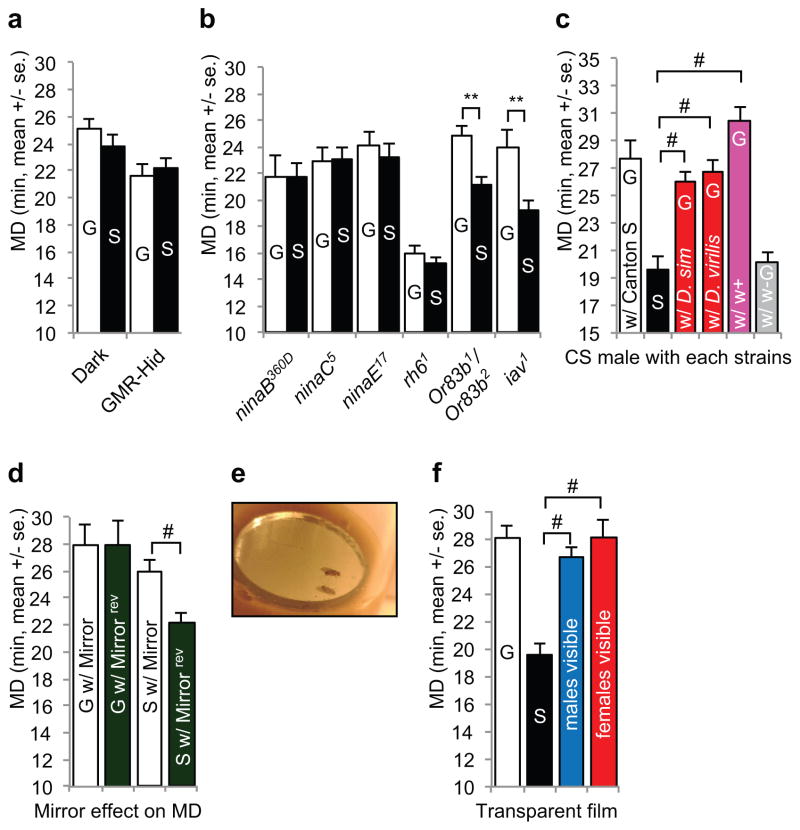
LMD appears to require visual inputs, because CS males that were group- or singly reared for 5 days in the dark did not show LMD (Fig. 2a). LMD was absent in blind males with photorecepters ablated via GMR-hid (Fig. 2a and Supplementary Fig. 2e), or mutants with impaired vision such as ninaB360D mutants (which do not synthesize the rhodopsin chromophore14), ninaC5 mutants (with degeneration of all rhabdomeres), ninaE17 mutants (which lack opsin in R1-6 photoreceptors), and rh61 mutants (which lack opsin in R8 photoreceptors15) (Fig. 2b). To explore the possible involvement of other sensory stimuli, we tested the Or83b1/Or83b2 mutants that show no behavioral or electrophysiological responses to many odorants, and found group-reared males displayed LMD as compared with singly reared males (Fig. 2b) or males that were housed with 4 CS females for 1 day (Supplementary Fig. 2d). In the w1118 genetic background with impaired vision, however, neither Or83b1/Or83b2 mutants nor the heterozygous controls displayed LMD (Supplementary Fig. 2g), indicating that Or83b-dependend olfactory inputs are dispensable whereas the visual inputs are essential for LMD. Inactivation of subsets of neurons for sensing olfactory and gustatory stimuli did not abolish LMD either (Fig. 5b). Moreover, presentation of male odors to singly reared males could not induce LMD (Supplementary Fig. 2f). We also tested the iav1 auditory mutants16 and found that they showed LMD (Fig. 2b). Thus, visual but not other sensory stimuli are required for LMD, although we cannot rule out the possible involvement of non-visual sensory stimuli.
Figure 2.
LMD is induced by visual stimuli. a. Mating duration assays of group- or singly reared CS males in constant darkness for 5 days and of GMR-Hid, blind animal. From left to right, n=29, 29, 15, and 19. t=1.247 and 0.4406. b. Mating duration assays of various vision, olfactory, and auditory mutants. The genotypes are indicated below the bars. From left to right, n=17, 20, 29, 29, 23, 27, 19, 26, 44, 25, 20, and 23. t=0.2103, 0.1664, 0.7524, 1.672, 3.436, and 4.724. c. Mating duration assays of group- (white bars), or singly reared (black) males, or males reared with 3 Drosophila simulans or Drosophila virilis (red), w+ strains (pink), or w1118 males (grey). The genotype of the w+ strain was w*;tub-GAL80ts. Eye color of w*;tub-GAL80ts strain is bright red. From left to right, n=22, 22, 27, 27, 30, and 24. F=20.83. d. Mating duration assays of group-reared males with a mirror or reversed mirror (G w/Mirror and G w/Mirrorrev), or singly reared males with a mirror or reversed mirror (S w/Mirror and S w/Mirrorrev) in place for 5 days. Reversed mirror is a mirror placed upside-down at the bottom of the vial, so that the male cannot see his reflection in the mirror. The color of the backside of the mirror is dark green. From left to right, n=15, 25, 24, and 23. F=4.891. e. The presentation of a mirror to a male fly. A small round mirror was placed at the bottom of the food vial to generate the male’s reflection. f. Mating duration assays of group- (white bars), or singly reared (black) males, and males reared with 3 other males (blue) or with 3 females (red) separated by transparent film. From left to right, n=28, 22, 18, and 20. F=19.19.
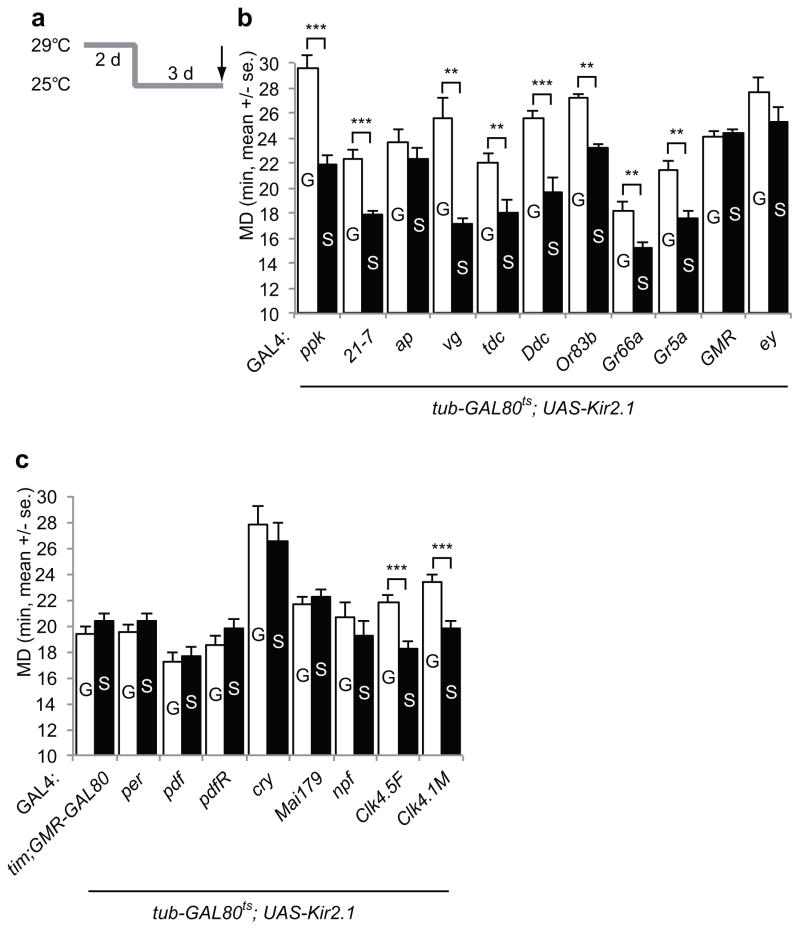
Figure 5.
Neural circuitry mapping for LMD. a. The paradigm for identification of relevant neural circuitry. group- or singly reared animals expressing UAS-Kir2.1 with tub-GAL80ts using the various GAL4 drivers were kept at 29°C for 2 days (strong induction) then moved to 25°C for 3 days (mild expression). b. Mating duration assays for GAL4 screening using tub-GAL80ts; UAS-Kir2.1. Names of the GAL4 drivers are indicated below the bars. GAL4 control experiments are shown in Supplementary Fig. 4e. From left to right, n=12, 11, 14, 12, 15, 19, 15, 15, 12, 17, 11, 13, 32, 30, 13, 11, 21, 24, 28, 14, 10, and 12. t=5.9679, 5.2268, 1.0224, 5.0351, 2.7528, 4.0677, 9.4194, 3.2807, 3.5202, 0.4352, and 1.3235. c. Mating duration assays of males bearing GAL4 drivers that label various circadian clock neurons using tub-GAL80ts; UAS-Kir2.1. Names of the GAL4 drivers are indicated below the bars. GAL4 control experiments are shown in Supplementary Fig. 4f. From left to right, n=10, 11, 9, 11, 15, 18, 13, 12, 19, 16, 30, 28, 12, 12, 16, 17, 17, and 18. t=0.9737, 0.8136, 0.5334, 1.4835, 1.2317, 0.7242, 1.0560, 4.0560, and 4.3961.
We next investigate the nature of the visual inputs that elicit LMD. Drosophila can detect color and motion17,18. We presented different color stimuli to singly reared males and could not induce LMD (Supplementary Fig. 2a–b). Interestingly, rearing a Drosophila melanogaster CS male together with 3 Drosophila simulans or Drosophila virilis males induced LMD of the CS male (Fig. 2c). In contrast to the male’s indifference to w1118 male rivals, LMD was induced by w1118 male rivals bearing the mini-white transgene that bestowed the red eye color (Fig. 2c), as well as male rivals with orange eyes (Supplementary Fig. 2c). As stationary color dots could not induce LMD but moving males with red compound eyes could, we hypothesized that both color and motion are important to induce LMD. As a test, we placed a small mirror at the bottom of the vial that housed a single male (Fig. 2e). The mating duration is longer for singly reared males with a mirror that reflects the male’s image, as compared to control singly reared males with the mirror turned upside-down so that the male could not see his reflection (Fig. 2d). Whereas the singly reared male visited the area occupied by the mirror or the upside-down mirror with similar frequency (8.1 visits per hour with a mirror, 7.5 visits per hour with a reversed mirror), it lingered for an average of 60 sec over the mirror yielding its reflection but only about 10 sec over the reversed, upside-down mirror (Supplementary Fig. 2h). Next, we tested the effect of viewing moving females with red eyes, and found LMD was displayed by singly reared males that were separated by a transparent film from 3 other male or female flies (Fig. 2f). Thus, the visual stimulus that induces LMD in nature likely corresponds to flies with red compound eyes in motion.
The circadian clock genes timeless and period but not Clock or cycle are involved in LMD
Transcriptional feedback loops are critical for circadian clocks. In Drosophila, the Clock (Clk) and cycle (cyc) gene products activate the period (per), timeless (tim), vrille (vri), PAR domain protein 1 (Pdp1) and clockwork orange (cwo) genes, which in turn inhibit CLK-activated transcription or regulate Clk transcription19. Clock genes also regulate non-circadian phenomena, such as the male courtship song frequency, developmental time, sleep length, delayed cocaine sensitization, and the time course of giant fiber habituation20. Moreover, tim and per regulate the mating duration of Drosophila melanogaster21.
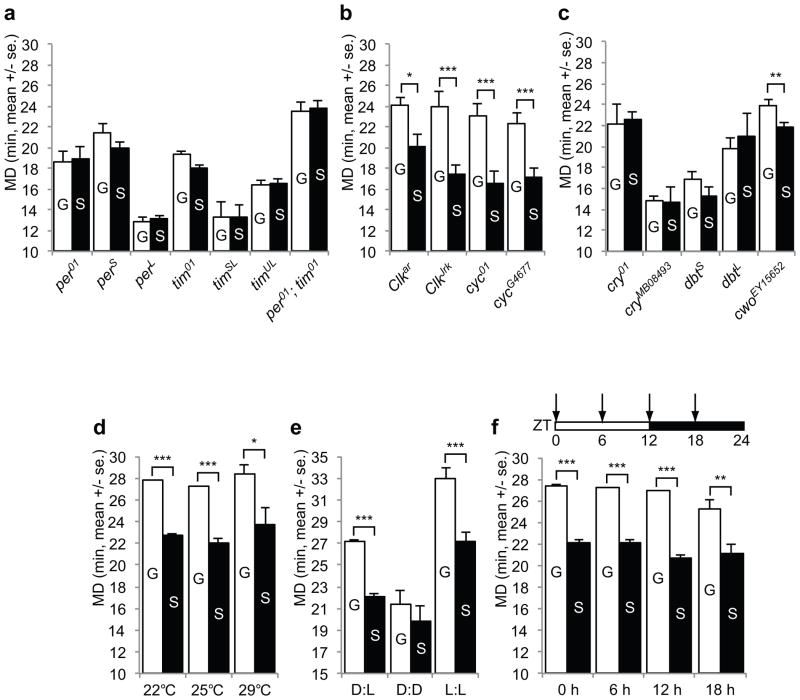
LMD was exhibited by Clk and cyc mutant males, but not tim and per mutant males (Fig. 3a–b) or males with mutation of cryptochrome (cry), which regulates the function of TIM, or mutation of doubletime (dbt), which encodes the kinase that phosphorylates PER (Fig. 3c). Moreover, LMD persisted in males with mutation of cwo, which encodes a putative bHLH transcription factor that acts preferentially at night to help terminate CLK-CYC-mediated transcription of target genes (Fig. 3c). Thus, PER/TIM but not CLK/CYC are required for LMD, even though these core clock genes act together to regulate circadian rhythm.
Figure 3.
LMD is affected in tim and per but not in Clk or cyc mutants. a. Mating duration assays of various tim and per mutants. The genotypes are indicated below the bars. From left to right, n=42, 32, 18, 15, 21, 18, 21, 24, 21, 27, 21, 24, 28, and 28. t=0.3659, 0.9971, 1.869, 1.534, 0.02206, 0.7068, and 0.3400. b. Mating duration assays of various Clk and cyc mutants. The genotypes are indicated below the bars. From left to right, n=16, 20, 28, 24, 24, 26, 27, and 26. t=5.475, 3.742, 4.758, and 5.754. c. Mating duration assays of various cry, dbt and cwo mutant animals. The genotypes are indicated below the bars. From left to right, n=21, 20, 21, 20, 22, 16, 20, 24, 28, and 27. t=0.3774, 0.09724, 1.989, 0.9722, and 3.081. d. Mating duration assays of group- or singly reared CS males at 22°C, 25°C, and 29°C. From left to right, n=83, 65, 46, 46, 22, and 20. t=8.108, 9.143, and 3.060. e. Mating duration assays of group- or singly reared CS males in standard 12 h dark 12 h light condition (D:L), in constant dark condition (D:D), and in constant light condition (L:L) at 25°C. From left to right, n=78, 62, 30, 30, 23, and 18. t=7.706, 1.247, and 4.554. f. Mating duration assays of group- or singly reared CS males at various time points of the 24 h circadian cycle (ZT=0 at the onset of light for 12 h dark 12 h light condition). Mating duration assays were performed at ZT0, ZT6, ZT12, or ZT18. From left to right, n=24, 24, 16, 21, 18, 16, 30, and 17. t=6.350, 4.951, 6.599, and 4.140.
Circadian clocks may be synchronized by light:dark cycles or by temperature fluctuations22. We tested CS males reared at different temperature for 5 days, and found they all exhibited LMD (Fig. 3d). LMD was also intact for males exposed to light constantly (Fig. 3e), a condition known to induce arrhythmic activities23, and at different time of the day (Fig. 3f). Although there may be a tendency for the mating duration to be shorter in the evening, the variability of this measurement – in contrast to the consistent display of LMD – precluded any quantitative comparisons of mating durations under different conditions. Moreover, expression of PER or TIM in per and tim mutants with the pan-neuronal GAL4 driver (elav-GAL4) could rescue LMD (Fig. 4a–b), even though the transgene mRNA expression is not rhythmic24, indicating that LMD depends on TIM and PER function rather than their cyclic expression during circadian rhythm.
Figure 4.
PER but not CLK protein in PDF neurons is required to generate LMD. a. Mating duration assays of tim rescue experiments. The genotypes are indicated with the corresponding color boxes above the graph. tim 2-1, tim2-5, and tim 2-7 indicates different UAS insertion lines. From left to right, n=29, 32, 31, 32, 22, 23, 12, 10, 15, 16, 25, 26, 21, and 22. t=0.7206, 1.1305, 4.3467, 0.0148, 6.6467, 1.6624, and 3.2969. b. Mating duration assays of per rescue experiments. The genotypes are indicated with corresponding numbers below the graph. From left to right, n=35, 24, 18, 24, 18, 10, 18, 18, 16, 14, 32, 30, 32, and 34. t=1.3398, 0.7028, 1.6866, 0.0012, 1.4871, 0.1249, and 7.6178. c. Mating duration assays of per rescue experiment. GAL4 drivers that rescue LMD in per mutant are marked in red. GAL4 control experiments are shown in Supplementary Fig. 5a. From left to right, n=17, 18, 40, 39, 16, 16, 19, 17, 15, 13, 27, 23, 42, 42, 41, 38, 18, 18, 15, 16, 12, and 13. t=3.7814, 5.2929, 0.5668, 0.0311, 0.6360, 3.8086, 0.4341, 5.4656, 5.1253, 0.0595, and 1.4451. d. Mating duration assays of UAS-ClkDN transgene expressed animals. From left to right, n=18, 12, 31, and 35. t=4.9471 and 3.9306. e. Distribution of circadian clock neurons in the fly brain. Locations of clock neurons are from a review43.
To identify the neurons that require the PER activity for LMD, we used different GAL4 drivers to express PER in various subsets of circadian pacemaker neurons in per mutants. Circadian pacemaker neurons are divided into 6 groups: 3 dorsal clusters (DN1-3) and 3 lateral clusters (LNd, l-LNv, and s-LNv)25 (Fig. 4e). LMD was restored in per01 mutants expressing the UAS-per transgene via GAL4 drivers in broad circadian cells, PDF-expressing neurons, and lateral clock neurons including PDF-expressing cells (Fig. 4c and Supplementary Fig. 3f), and it was sufficient to have adult specific expression of UAS-per via tub-GAL80ts (Fig. 4b). However, LMD was not rescued by expressing the UAS-per transgene in all neuronal progenitors except for the PDF-expressing neurons via a pan-neuronal GAL4 driver combined with pdf-GAL80 (Fig. 4b). Moreover, LMD was abolished in pdf01 mutant flies lacking the neuropeptide PDF (Supplementary Fig. 3e). Thus, the PER and PDF function in PDF neurons of the adult male is crucial to generate LMD.
To address the question whether LMD requires the function of per and tim but not Clk and cyc in PDF neurons, we used a dominant-negative Clk transgene (UAS-ClkDN) that includes the protein interaction domains but not the basic DNA-binding domain26. Disrupting the Clk function in PDF neurons in an adult-specific manner using tub-GAL80ts had no effect on LMD (Fig. 4d), suggesting that the Clk function in PDF neurons is not required to generate LMD, even though disrupting Clk function in the PDF neurons abolishes circadian rhythm26. Taken together, these findings indicate that circadian rhythm is not crucial for LMD.
LMD involves only a subset of the neurons of the circadian circuits
To further characterize the neural circuits that are important for LMD, we electrically silenced defined groups of cells in a temperature dependent manner by expressing Kir2.1 potassium channels via various GAL4 drivers together with the tub-GAL80ts, so that Kir2.1 expression could be elevated by temperature shifts in order to silence the GAL4 expressing cells. Flies were reared at 29°C for the first 2 days to strongly induce Kir2.1 expression and then shifted to 25°C for the next 3 days for mild induction of Kir2.1 before testing (Fig. 5a).
LMD was not altered by expressing Kir2.1 in some of the peripheral sensory neurons (ppk-GAL4 and 21-7-GAL4), the wing boundary tissues that might be involved in male courtship songs (vg-GAL4), the octopamine/tyramine neurons (tdc-GAL4) or the dopaminergic neurons (Ddc-GAL4) involved in aggression (Fig. 5b). In contrast, expression of Kir2.1 in the visual system (GMR-GAL4 and ey-GAL4) eliminated LMD (Fig. 5b and Supplementary Fig. 3a). Thus, neurons involved with visual information processing are important for the generation of LMD, consistent with our visual mutant studies (Fig. 2).
Since LMD depends on a subset of the clock gene products including the PER function in PDF neurons, we next investigated the role of electrical activity in circadian pacemaker neurons. LMD was abolished upon inactivation of all clock neurons by using tim-GAL4 or per-GAL4 drivers to drive Kir2.1 expression (Fig. 5c). Inactivation of PDF-expressing cells, which include l-LNv and s-LNv, abolished LMD (Fig. 5c). Inactivation of CRY-positive cells, which include most of the lateral neurons and a small subset of dorsal neurons, abolished LMD as well (Fig. 5c). Inactivation of lateral neurons via the Mai179-GAL4 driver27 abolished LMD (Fig. 5c). Inactivation of a subset of LNd and a subset of LNv neurons that express neuropeptide F (NPF), a homolog of mammalian neuropeptide Y (NPY), via npf-GAL4 also abolished LMD (Fig. 5c). These findings implicate the electrical activity of lateral clock neurons for the generation of LMD.
It is well known that s-LNvs, which project dorsally to DNs, are the main circadian pacemaker cells. Moreover, the DN1 and DN3 cells send projections toward the s-LNv cell bodies28. To test whether dorsal neurons are involved with LMD, we used Clk-GAL4 drivers that specifically target a subset of DNs29. Interestingly, inactivation of a subset of dorsal neurons using Clk4.1M-GAL4 or Clk4.5F-GAL4 drivers had no effect on LMD (Fig. 5c). Thus, whereas the electrical activity required for LMD generation involves lateral neurons, it does not require this subset of dorsal neurons important for circadian rhythm generation. For all the experiments involving UAS-Kir2.1, normal LMD was displayed by control flies carrying GAL4 or UAS-Kir2.1 reared at the non-permissive temperature, and by flies carrying GAL4, tub-GAL80ts and UAS-Kir2.1 that were reared at the permissive temperature (Supplementary Fig. 4a–h).
Since either electrical silencing or excessive excitation of clock neurons can abolish the rhythmic behavior30,31, we sought to activate these same neurons by overexpression of the bacterial voltage-gated sodium channel NachBac using the pdf-GAL4 driver. Surprisingly, we found no effect on LMD (Supplementary Fig. 4j), even though UAS-NaChBac expression in LNV pacemaker neurons abolishes the cyclic accumulation of PDF and induces complex behavioral rhythms with multiple superimposed periods31. However, overexpression of the temperature-sensitive dominant-negative dynamin mutant shibirets (UAS-shits) in PDF neurons eliminated LMD (Supplementary Fig. 4i). Thus electrical silencing or inhibition of dynamin-dependent endocytosis of PDF neurons may affect LMD, but activating those neurons has no effect on LMD. As summarized in Supplementary Fig. 3f for the expression patterns of the GAL4 lines used with the distribution of the clock neurons indicated in Fig. 4e, our study implicates a subset of the lateral neurons in LMD generation, which likely involves processes distinct from the regulation of circadian rhythm.
LMD requires neurons in the ellipsoid body that is important for visual memory
In Drosophila, there are several phases of memory: short-term memory lasting for less than an hour, mid-term memory lasting from one to three hours, and two forms of long-term memory that are distinguishable by training procedures, namely the anesthesia-resistant memory and long-term memory32.
To test how long the rival exposure memory for LMD lasts, we shifted group-reared flies to single housing at various time points before the mating duration assay. LMD persisted at the same level for up to 6 h of single housing but began to fade between 6 and 12 h of housing in isolation (Fig. 6a). It thus appears the rival exposure memory for LMD lasts longer than short-term or mid-term memory. Furthermore, because every mating duration assay was performed after mild CO2 anesthesia, LMD likely relies on a form of rival exposure memory that is resistant to anesthesia.
Figure 6.
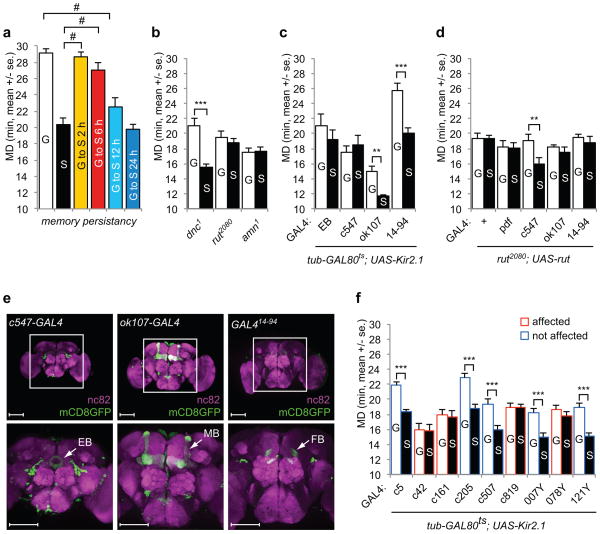
LMD requires visual memory. a. Flies were group- (white) or singly reared (black) for five days. Group-reared flies were shifted to single rearing condition for 2 h (yellow), 6 h (red), 12 h (sky blue), or 24 h (blue) before mating duration assay. From left to right, n=46, 23, 31, 27, 22, and 24. F=30.54. b. Mating duration assays of memory mutants. The genotypes are indicated below the bars. From left to right, n=21, 22, 14, 15, 12, and 14. t=5.2061, 0.5560, and 0.2420. c. Mating duration assays of GAL4 drivers which label the ellipsoid body (EB-GAL4 and c547), mushroom bodies (ok107-GAL4), or fan-shape bodies (GAL414–94) using tub-GAL80ts; UAS-Kir2.1. The experimental paradigm was the same as described in Fig. 5a. GAL4 control experiments are shown in Supplementary Fig. 4g. From left to right, n=21, 18, 21, 20, 30, 28, 32, and 28. t=1.2925, 0.9622, 4.8442, and 4.3464. d. Mating duration assays of rut2080 mutant with UAS-rut transgene expression driven by GAL4 drivers as indicated below the bars. GAL4 control experiments are shown in Supplementary Fig. 3d. From left to right, n=24, 24, 32, 31, 32, 35, 17, 17, 17, and 16. t=0.0664, 0.1289, 2.8196, 0.7981, and 0.8027. e. GAL4 expression profiles in the brain. Flies expressing each of the GAL4 drivers were crossed with UAS-mCD8GFP flies then immunostained with anti-GFP (green) and nc82 (magenta) antibodies (c547-GAL4, ok107-GAL4, and GAL414–94). Top panels show 200X images and bottom panels show 400X images of the boxed central brain region from top panels. White arrows indicate the brain regions labeled by GAL4 drivers (EB: ellipsoid body, MB: mushroom bodies, FB: fan-shape bodies). Scale bars represent 100 μM. f. Mating duration assays for GAL4 drivers that label the central complex using tub-GAL80ts; UAS-Kir2.1. GAL4 drivers that affect LMD are marked in red, and those with no effect on LMD are in blue. Names of the GAL4 drivers are indicated below the bars. GAL4 control experiments are shown in Supplementary Fig. 5b. From left to right, n=50, 33, 17, 15, 16, 14, 31, 31, 15, 13, 13, 16, 16, 16, 27, 32, 32, and 29. t=6.8137, 0.1023, 0.2649, 4.7739, 3.5733, 0.0471, 4.4566, 1.0110, and 4.6859.
Next, we tested the dnc, rut, and amn mutants, which suffer from defective learning and memory. LMD was impaired in rut and amn mutants, but not in dnc mutants (Fig. 6b). To confirm that the LMD defects of amn1 mutants is indeed due to amn mutation, we rescued the LMD defect of males bearing the amn mutation on the X chromosome by introducing a third chromosome that carries a short duplication of the X chromosome including the amn gene33 (Supplementary Fig. 3c). Genetic intervention has provided strong evidence that the mushroom bodies act as the seat of memory for odours32. In contrast, the visual pattern memory of Drosophila melanogaster involves the central complex, which includes the ellipsoid body and fan-shape bodies34,35. To identify brain regions likely involved in memory processing in LMD, we drove expression of UAS-Kir2.1 in the ellipsoid body, fan-shaped bodies, or mushroom bodies using several GAL4 lines (Fig. 6e). LMD was not altered by UAS-Kir2.1 expression in the mushroom bodies via ok107-GAL4 (Fig 6c), but was abolished by Kir2.1 expression in the ellipsoid body via EB-GAL4 and c547-GAL4 (Fig. 6c). We also tested GAL4 drivers for expression of Kir2.1 in fan-shaped bodies via GAL414–94, and found no effect on LMD (Fig. 6c). Given that the mushroom bodies inputs in Drosophila are predominantly olfactory36, the dependence of LMD on the ellipsoid body rather than mushroom bodies is consistent with our observation that LMD requires visual stimuli.
Having found that LMD depends on the rut gene and central neurons in the ellipsoid body, we used different GAL4 drivers to express the rut gene product in various subsets of neurons in rut mutants. Expression of the UAS-rut transgene with GAL4 drivers for expression in lateral clock neurons, mushroom bodies or fan-shaped bodies did not restore LMD to rut2080 mutants (Fig. 6d). However, UAS-rut expression in the ellipsoid body driven by c547-GAL4 could rescue the LMD defect of rut2080 mutants (Fig. 6d) and rutMB2769 mutants (Supplementary Fig. 3b). Taken together with the previous report that UAS-rut expression in the ellipsoid body driven by c547-GAL4 could successfully restore the visual memory defect of rut2080 mutants35, our observations suggest that the expression of rut in the ellipsoid body is likely important for processing the visual memory required for LMD.
The central complex is composed of four interconnected neuropils: the protocerebral bridge, the fan-shaped bodies, the ellipsoid body, and the noduli. By using several GAL4 lines with known expression patterns in the central complex37 to drive the expression of Kir2.1, we found the LMD was eliminated by Kir2.1 expression via the c42-GAL4 line that labels R2 and R4m neurons in the ellipsoid body, the c161-GAL4 and 078Y-GAL4 lines that label a small field in the ellipsoid body, and the c819-GAL4 line that labels R2 and R4m neurons in a pattern similar to that of c547-GAL4 (Fig. 6f). Since the c507-GAL4 line that labels R3 and R4d neurons had no effect on LMD, LMD likely requires the activity of R2 neurons in the ellipsoid body. The GAL4 lines that label other central complex areas had no effect on LMD (Fig. 6f). As summarized in Supplementary Fig. 5c for the expression patterns of the GAL4 lines used, our study implicates a subset of neurons within the ellipsoid body in the generation of LMD.
To further narrow down the relevant subset of neurons, we used various specific promoters to drive the expression of GAL80, which blocks the transcription of GAL4, in combination with the GAL4 driver, so that only those neurons expressing GAL4 but not GAL80 would express Kir2.1. We used GMR-GAL80 to suppress expression of UAS-Kir2.1 in the compound eye, pdf-GAL80 to suppress expression in lateral clock neurons, or cry-GAL80 to suppress expression in lateral clock neurons and a subset of the dorsal clusters of clock neurons. LMD was not affected by combining UAS-Kir2.1 together with GMR-GAL80 and ey-GAL4 or ap-GAL4 (Fig. 7a), indicating that the compound eye is crucial for LMD. In contrast, LMD was eliminated by the expression of UAS-Kir2.1 driven by pdf-GAL4 or cry-GAL4, even in the presence of GMR-GAL80 (Fig. 7a), indicating that the clock neurons labeled by these GAL4 lines are important for LMD. Moreover, LMD was eliminated by Kir2.1 expression via GAL4 drivers that label lateral clock neurons, such as cry-GAL4, npf-GAL4 and Mai179-GAL4, but not when these GAL4 drivers were combined with pdf-GAL80 (Fig. 7b), indicating that PDF neurons play important roles in generating LMD. Similar results were obtained when we used cry-GAL80, which labels a broader subset of clock neurons including PDF neurons (Fig. 7c). When combined with GMR-GAL80, pdf-GAL80, or cry-GAL80, only c547 GAL4 could still drive Kir2.1 expression to eliminate LMD (Fig. 7a–d), indicating that neuronal activity of the ellipsoid body is critical to generate LMD. Taken together, these studies implicate the compound eye, PDF neurons, and neurons in the ellipsoid body in the generation of LMD. Whereas the compound eye processes visual inputs, the ellipsoid body is likely involved in visual memory. Given that PDF neurons may be connected with the ellipsoid body through l-LNv38 (Supplementary Fig. 5d), it would be interesting to test in future studies how PDF neurons might be involved in conveying the information regarding rival exposure to other central neurons.
Figure 7.
Neural circuitry important for LMD. a. Mating duration assays for GAL4 using GMR-GAL80, UAS-Kir2.1. Names of the GAL4 drivers are indicated below the bars. From left to right, n=30, 29, 44, 36, 35, 33, 34, 32, 30, 28, 18, and 16. t=6.5826, 3.9368, 5.2981, 0.7026, 0.1782, and 0.0700. b. Mating duration assays for GAL4 using pdf-GAL80; UAS-Kir2.1. Names of the GAL4 drivers are indicated below the bars. From left to right, n=17, 15, 22, 20, 23, 26, 32, 32, 30, and 27. t=3.0698, 5.1976, 4.5501, 5.4580, and 1.2663. c. Mating duration assays for GAL4 using cry-GAL80; UAS-Kir2.1. Names of the GAL4 drivers are indicated below the bars. From left to right, n=18, 12, 23, 29, 34, 33, 15, and 13. t=4.9471, 5.6204, 5.4312, and 0.6286. d. Mating duration assays for GAL4 using cry-GAL80; GMR-GAL80, UAS-Kir2.1. Names of the GAL4 drivers are indicated below the bars. From left to right, n=30, 28, 50, 43, 54, 58, 16, and 17. t=4.7684, 5.6077, 8.0481, and 0.9262.
Our study provides evidence that males retain the memory of rival exposure, based primarily on visual stimuli, for several hours and lengthen mating duration accordingly. Indeed, LMD could be induced by allowing a male to view flies of either sex through a transparent partition, or flies of different species, or images of themselves in a mirror (Fig. 2), indicating that LMD could be generated by visual stimuli without chemical communication. We further show that the circadian clock genes tim and per but not Clk or cyc are important for LMD (Fig. 3). Not only can the LMD defects of per and tim mutants be rescued by the expression of PER or TIM via the non-rhythmic GAL4 driver (Fig. 4a–b), expression of PER in PDF neurons is sufficient to restore LMD to per mutants (Fig. 4c). Moreover, LMD requires electrical activity in lateral neurons but not some of the dorsal neurons important for circadian rhythm (Fig. 5). It thus seems unlikely that circadian rhythm regulation is crucial for LMD. We further show that LMD involves the memory of rival exposure that lasts for several hours and is resistant to anesthesia (Fig. 6a) and it requires the rut function in the ellipsoid body (Fig. 6c–d). Finally, we have shown that LMD generation depends on the activity of the compound eye, the PDF neurons, and a subset of neurons in the ellipsoid body (Fig. 7).
Recent studies of social experience-mediated and context-dependent sexual behaviors of the fruit fly39,40 implicate chemical communication of males via pheromones. In this study we show that vision in a social setting is important to generate the LMD. Whereas a recent report suggests that males use multiple redundant cues to detect mating rivals16, we found LMD can be elicited by visual cues because (1) rearing flies in constant darkness eliminated LMD (Fig. 2a), (2) blind mutants and males with defective vision showed no LMD (Fig. 2a–b), (3) LMD can be generated simply by placing a mirror to allow a singly reared male fruit fly to see his reflection for 5 days (Fig. 2d). The visual stimulus for LMD likely derives from the red compound eye in motion because LMD can be induced by males of different species or females visible through a transparent film (Fig. 2c, f), but not by mutant males without red pigment in their compound eyes (Fig. 2c).
LMD provides a new paradigm for the visual memory. Learning and memory studies in flies thus far focus primarily on the memory circuits in mushroom bodies32, however, LMD requires a subset of neurons in the ellipsoid body rather than mushroom bodies (Fig. 6c–f). The ellipsoid body is the central brain region required for visual learning and memory34,35,41, whereas mushroom bodies are not required for memory formation in visual learning in a flight simulator42. Since the mating duration assay is simpler than the flight simulator for the investigation of visual memory, it can be useful for large-scale genetic screens to identify mutants with altered visual memory.
METHODS
Fly Rearing and Strains
D. melanogaster were raised on cornmeal-yeast medium at similar densities to yield adults with similar body sizes7. Flies were kept in 12 h light: 12 h dark cycles (LD) at 25°C (ZT 0 is the beginning of the light phase, ZT12 beginning of the dark phase) except for some experimental manipulation (constant dark, constant light, and experiments with the flies carrying tub-GAL80ts). Wild-type flies were Canton-S. To reduce the variation from genetic background, all flies were backcrossed for at least 3 generations to CS strain. All mutants and transgenic lines used here have been described previously.
The following lines were obtained from Dr. Alex C. Keene and Dr. Justin Blau (New York University, USA)44: perS, perL, cry01, cyc01, ClkJrk, Clkar, tim-GAL4;GMR-GAL80, pdf-GAL80;pdf-GAL80, cry-GAL80;cry-GAL80, UAS-ClkDN.
The following lines were obtained from Dr. Martin Heisenberg (Universität Würzburg, Germany)18: WT Berlin, rh61, ninaE17.
The following line was obtained from Dr. Charlotte Helfrich-Förster (Universität Würzburg, Germany)27: Mai179-GAL4.
The following line was obtained from Dr. Leslie C. Griffith (Brandeis University, USA)45: pdfR-GAL4.
The following lines were obtained from Dr. Ravi Allada (Northwestern University, USA)29: Clk4.5F-GAL4, Clk4.1M-GAL4.
The following line was obtained from Dr. Markus Noll (University of Zürich, Switzerland)46: ninaB360D
The following lines were obtained from Dr. Jeffrey L. Price (University of Missouri, USA)47: dbtL, dbtS.
The following lines were obtained from Dr. Amita Sehgal (University of Pennsylvania Medical School, USA)24: per01, tim01, per01;tim01, as well as the lines used for rescuing per and tim mutants phenotype in Fig. 4a- and b.
The following lines were obtained from Dr. Michael W. Young (The Rockefeller University, USA): timSL, timUL.
The following lines were obtained from Dr. J. Douglas Armstrong (University of Edinburgh): c547-GAL4, c507-GAL4, 121Y-GAL4.
The following lines were obtained from Dr. Makoto Sato (Kanazawa University)48: bsh-GAL4, drf-GAL4, hth-GAL4.
The following lines were obtained from Bloomington Stock Center: GMR-Hid, Or83b1, Or83b2, ninaC5, cycG4677, dnc1, rut2080, amn1, cwoEY15652, cryMB08493, UAS-tubGAL80ts, UAS-Kir2.1, UAS-shits, UAS-NachBac, wa, iav1, rut2080;UAS-rut, rutMB2769;UAS-rut, c5-GAL4, c42-GAL4, c161-GAL4, c205-GAL4, c819-GAL4, 007Y-GAL4, 078Y-GAL4, w1118; Dp(1;3)DC368, PBac[DC368]VK00033.
The following lines were obtained from Kyoto Stock Center (DGRC): D. simulans, D. virilis.
Small mirrors were obtained from Factory Direct Craft (3/4″ Round Glass Mosaic Tile Mirrors).
Mating Duration Assays
Males of the appropriate strain were collected individually and placed into vials with food. In the case of group-reared condition, 4 males from the same strain were placed into vials except for some experimental manipulation (for example, Fig. 2d). The day of eclosion was designated as day 1 of adult life. CS females were collected from bottles and placed into vials for 5 days. Each vial contained 5 females. These females were used as mating partners for mating duration assays.
Mating duration assay procedure is performed as previously described49. In short, at the fifth day after eclosion, males of the appropriate strain and CS females were mildly anaesthetized by CO2. Then a single female was placed to the mating chamber. After placing the females, we inserted a transparent film then placed a single male to the other side of the film in each chamber. The mating chamber was then covered and placed in a 25°C incubator for recovery. After 1 h of recovery, the transparent film was removed then mating activities are recorded and counted. Only the males that succeeded to mate within 1 h were included for analyses. Initiation and completion of copulation were recorded with an accuracy of 10 sec, and total mating duration was calculated for each couple.
Immunostaining
To examine the expression pattern of GAL4 in adult brains, GAL4 transgenic lines were crossed with UAS-mCD8-GFP flies. Brains of adult flies were dissected 5 days after eclosion. Dissected brains were subjected to immunostaining as described before50. Briefly, dissected adult brains were fixed in 4% formaldehyde for 30 min at room temperature. After fixation, brains were washed with 1% PBT three times (30 min each) and blocked in 5% normal donkey serum for 30 min. The brains were then incubated with primary antibodies in 1% PBT at 4°C overnight followed with fluorophore-conjugated secondary antibodies for 2 hours at room temperature. Brains were mounted with anti-fade mounting solution (Invitrogen, catalog #S2828) on slides for imaging.
Statistical Analysis
We performed 3 independent tests for all the experiments. More than 12 males (group-or singly reared) were used for each independent experiment. Besides assessment of significance from statistical test (Student’s t test), we included the data set, which showed statistical significance for at least 2 of these three independent tests. Our experience suggests that relative mating duration value differences between group- and singly reared are always consistent; however, both absolute value and magnitude of the difference in each strain can vary. So we always include internal controls for each treatment and believe that each test should be considered independently as suggested by previous studies16. Therefore, statistical comparisons were made between groups that were exposed or not exposed to rivals within each experiment. As mating duration of males showed normal distribution (Kolmogorov-Smirnov tests, p > 0.05), we used two-sided Student’s t tests. Each figure shows the mean +/− standard error (s.e.m) (*** = p < 0.001, ** = p < 0.01, * = p < 0.05).
When we compare the difference of mating duration in experiments without internal control built in, (for example, Fig. 2c), we always performed control experiments of wild type for each independent experiment for internal comparison. And in this case, we analyzed data using ANOVA for statistically significant differences (at a 95.0% confidence interval) between the means of mating duration for all conditions. If a significant difference between the means was found by Kruskal-Wallis test, then the Dunn’s Multiple Comparison Test was used to compare the mean mating duration of each condition to determine which conditions were significantly different from condition of interest. (# = p < 0.05)
Supplementary Material
Acknowledgments
We thank Dr. Sijun Zhu for the unpublished fly line GAL414–94. We also thank Drs. A. Keene, J. Blau, M. Heisenberg, C. Helfrich-Förster, L. Griffith, R. Allada, M. Noll, Jeffrey L. Price, A. Sehgal, M. Young, J. D. Armstrong, and M. Sato for kindly providing valuable flies. We are grateful to Dr. A. Keene for valuable discussion on this project and Dr. Jim Berg for writing with the manuscript. The work was supported by NIH grant 2R37NS040929 to YNJ. LYJ and YNJ are investigators of Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
W.J.K. designed and performed the experiments; W.J.K., Y.N.J. and L.Y.J. wrote the manuscript; Y.N.J. and L.Y.J. supervised the project.
References
- 1.Darwin C. The Descent of Man, and Selection in Relation to Sex. John Murray; 1871. [Google Scholar]
- 2.Kim Y-K. Handbook of Behavior Genetics. Springer; New York: 2009. pp. 317–330. [Google Scholar]
- 3.Ridley M. Sperm competition and sexual selection. Nature. 1999;397:576–577. [Google Scholar]
- 4.Parker GA. Sperm Competition and Its Evolutionary Consequences in Insects. Biological Reviews of the Cambridge Philosophical Society. 1970;45:525. [Google Scholar]
- 5.Markow TA. Evolutionary biology. 1996;29:73–106. [Google Scholar]
- 6.Koref-Santibanez S. Effects of age and experience on mating activity in the sibling species Drosophila pavani and Drosophila gaucha. Behav Genet. 2001;31:287–297. doi: 10.1023/a:1012279325621. [DOI] [PubMed] [Google Scholar]
- 7.Lefranc A, Bundgaard J. The influence of male and female body size on copulation duration and fecundity in Drosophila melanogaster. Hereditas. 2000;132:243–247. doi: 10.1111/j.1601-5223.2000.00243.x. [DOI] [PubMed] [Google Scholar]
- 8.De Crespigny FEC, Pitt TD, Wedell N. Increased male mating rate in Drosophila is associated with Wolbachia infection. Journal of Evolutionary Biology. 2006;19:1964–1972. doi: 10.1111/j.1420-9101.2006.01143.x. [DOI] [PubMed] [Google Scholar]
- 9.Luck N, Joly D. Sexual selection and mating advantages in the giant sperm species, Drosophila bifurca. J Insect Sci. 2005;5:10. doi: 10.1093/jis/5.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bretman A, Fricke C, Chapman T. Plastic responses of male Drosophila melanogaster to the level of sperm competition increase male reproductive fitness. Proc Biol Sci. 2009;276:1705–1711. doi: 10.1098/rspb.2008.1878. Epub 2009 Feb 1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazzi D, Kesaniemi J, Hoikkala A, Klappert K. Sexual conflict over the duration of copulation in Drosophila montana: why is longer better? BMC Evol Biol. 2009;9:132. doi: 10.1186/1471-2148-9-132. 1471-2148-9-132 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macbean IT, Parsons PA. Directional Selection for Duration of Copulation in Drosophila Melanogaster. Genetics. 1967;56:233. doi: 10.1093/genetics/56.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bretman A, Fricke C, Hetherington P, Stone R, Chapman T. Exposure to rivals and plastic responses to sperm competition in Drosophila melanogaster. Behavioral Ecology. 21:317–321. doi: 10.1093/beheco/arp189. [DOI] [Google Scholar]
- 14.von Lintig J, Dreher A, Kiefer C, Wernet MF, Vogt K. Analysis of the blind Drosophila mutant ninaB identifies the gene encoding the key enzyme for vitamin A formation invivo. Proc Natl Acad Sci U S A. 2001;98:1130–1135. doi: 10.1073/pnas.031576398031576398. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook T, Pichaud F, Sonneville R, Papatsenko D, Desplan C. Distinction between color photoreceptor cell fates is controlled by Prospero in Drosophila. Dev Cell. 2003;4:853–864. doi: 10.1016/s1534-5807(03)00156-4. S1534580703001564 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Bretman A, Westmancoat JD, Gage MJ, Chapman T. Males use multiple, redundant cues to detect mating rivals. Curr Biol. 21:617–622. doi: 10.1016/j.cub.2011.03.008. S0960-9822(11)00280-6 [pii] [DOI] [PubMed] [Google Scholar]
- 17.Menne D, Spatz HC. Colour vision in Drosophila melanogaster. Journal of Comparative Physiology A: Neuroethology, Sensory, Neural, and Behavioral Physiology. 1977;114:301–312. doi: 10.1007/bf00657325. [DOI] [Google Scholar]
- 18.Yamaguchi S, Wolf R, Desplan C, Heisenberg M. Motion vision is independent of color in Drosophila. Proc Natl Acad Sci U S A. 2008;105:4910–4915. doi: 10.1073/pnas.0711484105. 0711484105 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng X, Sehgal A. Probing the relative importance of molecular oscillations in the circadian clock. Genetics. 2008;178:1147–1155. doi: 10.1534/genetics.107.088658. 178/3/1147 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall JC. Systems approaches to biological rhythms in Drosophila. Methods Enzymol. 2005;393:61–185. doi: 10.1016/S0076-6879(05)93004-8. S0076687905930048 [pii] [DOI] [PubMed] [Google Scholar]
- 21.Beaver LM, Giebultowicz JM. Regulation of copulation duration by period and timeless in Drosophila melanogaster. Curr Biol. 2004;14:1492–1497. doi: 10.1016/j.cub.2004.08.022S0960982204006050. [pii] [DOI] [PubMed] [Google Scholar]
- 22.Glaser FT, Stanewsky R. Temperature synchronization of the Drosophila circadian clock. Current biology : CB. 2005;15:1352–1363. doi: 10.1016/j.cub.2005.06.056. [DOI] [PubMed] [Google Scholar]
- 23.Emery P, Stanewsky R, Hall JC, Rosbash M. Drosophila cryptochromes: A unique circadian-rhythm photoreceptor. Nature. 2000;404:456–457. doi: 10.1038/35006558. [DOI] [PubMed] [Google Scholar]
- 24.Yang Z, Sehgal A. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. S0896-6273(01)00218-5 [pii] [DOI] [PubMed] [Google Scholar]
- 25.Peschel N, Helfrich-Forster C. Setting the clock - by nature: Circadian rhythm in the fruitfly Drosophila melanogaster. FEBS Lett. doi: 10.1016/j.febslet.2011.02.028. S0014-5793(11)00132-3 [pii] [DOI] [PubMed] [Google Scholar]
- 26.Tanoue S, Krishnan P, Krishnan B, Dryer SE, Hardin PE. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Current biology : CB. 2004;14:638–649. doi: 10.1016/j.cub.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Rieger D, Wulbeck C, Rouyer F, Helfrich-Forster C. Period gene expression in four neurons is sufficient for rhythmic activity of Drosophila melanogaster under dim light conditions. J Biol Rhythms. 2009;24:271–282. doi: 10.1177/0748730409338508. 24/4/271 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Shafer OT, Helfrich-Forster C, Renn SC, Taghert PH. Reevaluation of Drosophila melanogaster’s neuronal circadian pacemakers reveals new neuronal classes. J Comp Neurol. 2006;498:180–193. doi: 10.1002/cne.21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, et al. DN1(p) circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Curr Biol. 20:591–599. doi: 10.1016/j.cub.2010.02.056. S0960-9822(10)00326-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nitabach MN, Blau J, Holmes TC. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. S0092867402007377 [pii] [DOI] [PubMed] [Google Scholar]
- 31.Nitabach MN, et al. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. 26/2/479 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heisenberg M. Mushroom body memoir: from maps to models. Nat Rev Neurosci. 2003;4:266–275. doi: 10.1038/nrn1074nrn1074. [pii] [DOI] [PubMed] [Google Scholar]
- 33.Popodi E, et al. 2010 [Google Scholar]
- 34.Liu G, et al. Distinct memory traces for two visual features in the Drosophila brain. Nature. 2006;439:551–556. doi: 10.1038/nature04381. nature04381 [pii] [DOI] [PubMed] [Google Scholar]
- 35.Pan Y, et al. Differential roles of the fan-shaped body and the ellipsoid body in Drosophila visual pattern memory. Learn Mem. 2009;16:289–295. doi: 10.1101/lm.1331809. 16/5/289 [pii] [DOI] [PubMed] [Google Scholar]
- 36.Stocker RF. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res. 1994;275:3–26. doi: 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- 37.Renn SC, et al. Genetic analysis of the Drosophila ellipsoid body neuropil: organization and development of the central complex. J Neurobiol. 1999;41:189–207. [PubMed] [Google Scholar]
- 38.Shang Y, Griffith LC, Rosbash M. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19587–19594. doi: 10.1073/pnas.0809577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kent C, Azanchi R, Smith B, Formosa A, Levine JD. Social context influences chemical communication in D. melanogaster males. Curr Biol. 2008;18:1384–1389. doi: 10.1016/j.cub.2008.07.088. S0960-9822(08)01033-6 [pii] [DOI] [PubMed] [Google Scholar]
- 40.Krupp JJ, et al. Social experience modifies pheromone expression and mating behavior in male Drosophila melanogaster. Curr Biol. 2008;18:1373–1383. doi: 10.1016/j.cub.2008.07.089. S0960-9822(08)01034-8 [pii] [DOI] [PubMed] [Google Scholar]
- 41.Gong Z, Xia S, Liu L, Feng C, Guo A. Operant visual learning and memory in Drosophila mutants dunce, amnesiac and radish. J Insect Physiol. 1998;44:1149–1158. doi: 10.1016/s0022-1910(98)00076-6. S0022191098000766 [pii] [DOI] [PubMed] [Google Scholar]
- 42.Guo A, Gotz KG. Association of visual objects and olfactory cues in Drosophila. Learn Mem. 1997;4:192–204. doi: 10.1101/lm.4.2.192. [DOI] [PubMed] [Google Scholar]
- 43.Choi C, Nitabach MN. Circadian biology: environmental regulation of a multi-oscillator network. Curr Biol. 20:R322–324. doi: 10.1016/j.cub.2010.02.036. S0960-9822(10)00224-1 [pii] [DOI] [PubMed] [Google Scholar]
- 44.Keene AC, et al. Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr Biol. 20:1209–1215. doi: 10.1016/j.cub.2010.05.029. S0960-9822(10)00596-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parisky KM, et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008;60:672–682. doi: 10.1016/j.neuron.2008.10.042. S0896-6273(08)00942-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krstic D, Boll W, Noll M. Sensory integration regulating male courtship behavior in Drosophila. PLoS One. 2009;4:e4457. doi: 10.1371/journal.pone.0004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Preuss F, et al. Drosophila doubletime mutations which either shorten or lengthen the period of circadian rhythms decrease the protein kinase activity of casein kinase I. Mol Cell Biol. 2004;24:886–898. doi: 10.1128/MCB.24.2.886-898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hasegawa E, et al. Concentric zones, cell migration and neuronal circuits in the Drosophila visual center. Development. 138:983–993. doi: 10.1242/dev.058370. 138/5/983 [pii] [DOI] [PubMed] [Google Scholar]
- 49.Yang CH, et al. Control of the postmating behavioral switch in Drosophila females by internal sensory neurons. Neuron. 2009;61:519–526. doi: 10.1016/j.neuron.2008.12.021. S0896-6273(08)01093-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.