Abstract
Every month, numerous publications appear that include neuroanatomic volumetric observations. The current and past literature that includes volumetric measurements is vast, but variable with respect to specific species, structures, and subject characteristics (such as gender, age, pathology, etc.). In this report we introduce the Internet Brain Volume Database (IBVD), www.nitrc.org/projects/ibvd, a site devoted to facilitating access to and utilization of neuroanatomic volumetric observations as published in the literature. We review the design and functionality of the site. The IBVD is the first database dedicated to integrating, exposing and sharing brain volumetric observations across species and disease. It offers valuable functionality for quality assurance assessment of results as well as support for meta-analysis across large segments of the published literature that are obscured from traditional text-based search engines.
Keywords: Brain, Volume, Quantitative neuroanatomy, Morphometry, Database, Website
Introduction
In this report we introduce the Internet Brain Volume Database (IBVD). Instantiated as a web-accessible database (www.cma.mgh.harvard.edu/ibvd and www.nitrc.org/projects/ibvd), this site is devoted to facilitating discovery and access to neuroanatomic volumetric observations as published in the literature. The IBVD is the first database dedicated to integrating, exposing, and sharing brain volumetric observations across species and disease. The current objective is to demonstrate the utility of a database of neuroanatomic volumetric observations from the literature. This database is initiated through a manual, retrospective review and entry of the pertinent literature in order to evaluate the system design for this endeavor. Once successful, future extensions will be necessary to enable a long term, sustainable, prospective data capture solution that will likely involve the active participation of the authors and publishers.
Every month, on the order of 10 publications appear that include neuroanatomic volumetric observations. The current and past literature that includes volumetric measurements is vast, but variable with respect to specific species, structures, and subject characteristics (such as gender, age, pathology, etc.). In general, much more has been published regarding quantitative neuroanatomy in the human as compared to other species. The advent of magnetic resonance imaging (MRI) opened a impressive opportunity to make precise, noninvasive, in-vivo measurements in many subject populations (Caviness et al. 1999).
Quantitative morphometric studies have been performed in virtually every neurologic and psychiatric disorder (for example (Seidman et al. 2002; Courchesne and Pierce 2005; Rosas et al. 2005; Seidman et al. 2005; Leow et al. 2009; Schuff et al. 2009)). Of course, tantamount to understanding the neuroanatomic changes that are related to pathological conditions is the need to have a precise understanding of normative neuroanatomic development (Caviness et al. 1996; Thompson et al. 2000; Saitoh et al. 2001; Salat et al. 2004; Casey et al. 2005; Giedd et al. 2006; Lenroot et al. 2007). While substantial imaging archives of raw data are being assembled (Mazziotta et al. 2001; Marcus et al. 2007; Waber et al. 2007; Jack et al. 2008), access to the quantitative derived data, specifically as it becomes utilized in the published literature, is still greatly underserved.
Since thousands of individual measurements of brain structures have been made and reported, it is impossible for individual researchers to maintain a working knowledge of the effective ‘view’ supported by the literature of exactly how large a specific neuroanatomic structure should be (given a species, age, gender, imaging technique, analysis technique, etc.). Since there are many factors that effect the measurements and results, there is a substantial need to be able to evaluate the relative contributions of these factors to a true, unbiased estimate of brain structural volume. Such an assessment is served through the application of meta-analysis to the results of individual studies in order to derive the prevalence of true underlying biological effects in relation to methodological (subject, sampling, acquisition, analysis, etc.) sources of variance (Bzdok et al. 2011; Jardri et al. 2011). Sites like the IBVD are designed to specifically support such broad, literature-based assessment.
In this report we review the database design and web-based user interface for the IBVD. We describe the current status of data entry into the system, and provide examples of the types of meta-analyses that are subsequently supported by the system. The prospects and challenges for moving this type of analysis scheme forward are then discussed.
Methods
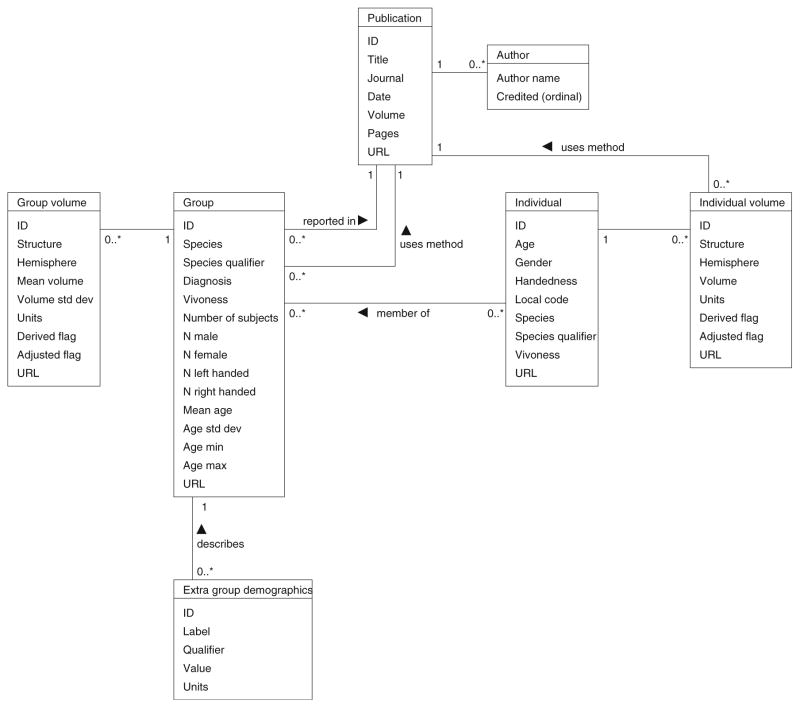
IBVD is a web application implemented in PHP on top of a PostgreSQL database. The database schema, shown in Fig. 1, is designed to capture the complete set of information necessary to describe a volumetric observation in the literature. The conceptual starting point for this description is the ‘publication’ itself. Publications typically describe the demographic characteristics of ‘Groups’ of subjects, and then report ‘Group Volumes’ for some set of anatomic structures. In addition, individual subjects that comprise the Groups may be described in terms of their ‘Individual Demographic’ and ‘Individual Volume’ observations. While bulk uploading of standardized text representations of data is supported, Fig. 2 shows the user interface for each of these web-based entry forms.
Fig. 1.
IBVD database schema. Demonstrating the structure and relationship of the database tables for Publication, Group, Group volume, Individual, and Individual volume. The Appendix provides a brief definition for each database element. The “0…” notation represents that multiple records (indexed 0, 1, 2, etc.) are permissible (i.e. a Group can have multiple group volume records associated with it). Conversely, the ‘1’ represents an exclusive relationship (i.e. a Group Volume can be associated with one and only one group)
Fig. 2.
The user interface for each of these web-based entry forms. Includes forms for Publication, Group Demographic and Group Volume entry
Data fields are a mixture of forced choice selection, automated entry, and free text of various formats. For example, given the PubMed identifier (PMID) of a publication, the IBVD gathers information about the publication using the NCBI Entrez Utilities Web Services and automatically populates the IBVD Publication data fields. In addition, species, anatomic region, units or measurement, and numerous of the clinical and behavioral characteristics are filled from specific selection lists. The use of a structured vocabulary is necessary in order to maximize the interoperability of this site with other data resources. Structures, from the whole brain to major subdivisions (e.g. cerebrum) to components of these major regions (e.g. cerebral cortex, white matter) to subcomponent within these regions (e.g. precentral gyrus), are mapped into this ontology. All concepts, such as diagnosis, anatomic region, etc. also can have a concept identification code associated with them (i.e. from Unified Medical Language System, UMLS; or NeuroLEX) to facilitate database mediation and integration (Lindberg et al. 1993; Bowden and Dubach 2003; Bug et al. 2008; Gardner et al. 2008; Larson and Martone 2009). Figure 3 shows a completed example of the linked data pages that span from the publication to the individual volumetric measurements. A data dictionary with the field definitions is provided in the Appendix.
Fig. 3.
Linked example pages from publication record to individual data. Follows the sequential relationships between a specific publication entry, and drills down to the group volume record for one of the reported groups, and subsequently into the individual subject volume record for an individual within that group
Website Functions
From the website homepage, the user can access all major site functions. These include: registered user login (required only for data entry), data entry, data display and database search functions. Results of each search can be visualized by a plot of volume by age with various display options, or exported for additional off-line processing and analysis. Searching is facilitated by a specification form, where the user can specify specific group or individual criteria, including: diagnosis; anatomic region; age range; handedness; gender; species; and hemisphere. Visualization plots feature volume as a function of age, where x and y error bars represent age and volume standard deviations, respectively. Plot symbols and colors can be selected to represent the various characteristics of the search result, such as diagnosis, gender, structure, etc.
Results
In this section we will demonstrate the current status and some of the features of the database.
Data Entry
Data entry statistics (as of 06/2011) include: 647 publications, 1,598 subject groups, 9,699 group volume entries, over 80 clinical diagnoses, 1,939 individuals, 10,215 individual structural volume entries, and over 90 discrete brain structures. Note that at this time, most data is reported in the literature for groups of subjects, and inclusion of the volumetric data for each of the individuals within a group is relatively uncommon (although this should be encouraged). Tables 1 and 2 list the most common diagnoses and structures in order of prevalence in the database.
Table 1.
IBVD content by diagnosis. Includes the number of groups with the diagnosis and the UMLS and DSM-IV correspondence
| IBVD diagnosis | # Groups | UMLS CUI | DSM-IV code |
|---|---|---|---|
| Normal | 767 | NA | NA |
| Schizophrenia | 97 | C0392322 | 295.90 |
| Autistic Disorder (Autism) | 57 | C0004352 | 299.00 |
| Bipolar Disorder | 45 | C0005586 | 296.80 |
| Major Depressive Disorder (Unipolar) | 41 | C0154409 | 296.30 |
| Alzheimer’s Disease | 37 | C0002395 | 290.0 |
| ADHD | 18 | C0004269 | 314.9 |
| Alcohol Dependence | 17 | C0001973 | 303.9 |
| Dementia | 12 | C0497327 | 294.8 |
| Traumatic Brain Injury | 9 | ||
| Obsessive Compulsive Disorder (OCD) | 7 | C0028768 | 300.3 |
| Borderline Personality Disorder | 7 | C0006012 | 301.83 |
| Asperger’s Syndrome | 7 | C0236792 | 299.80 |
Table 2.
IBVD content by neuroanatomic structure. For each structure, the number of groups and individual volume records are indicated. Of these records, the numbers that are from ‘normal’ or ‘pathological’ diagnoses is indicated for group and individual are indicated
| Structure | Groups | (Normal/Pathological) | Individuals | (Normal/Pathological) |
|---|---|---|---|---|
| Hippocampus | 1263 | (619/644) | 513 | (171/342) |
| Amygdala | 790 | (386/404) | 429 | (102/327) |
| Brain | 624 | (410/214) | 1654 | (1605/49) |
| Caudate | 461 | (237/224) | 579 | (252/327) |
| Putamen | 304 | (159/145) | 429 | (102/327) |
| Thalamus | 301 | (144/157) | 82 | (52/30) |
| Lateral Ventricle | 299 | (183/116) | 576 | (249/327) |
| Cerebrum | 292 | (160/132) | 174 | (161/13) |
| Gray Matter | 260 | (161/99) | 2 | (0/2) |
| White Matter | 244 | (160/84) | 22 | (0/22) |
| Intracranial | 215 | (102/113) | 0 | (0/0) |
| Temporal Lobe | 208 | (98/110) | 0 | (0/0) |
| Cerebellum | 193 | (113/80) | 183 | (183/0) |
| CSF | 161 | (96/65) | 2 | (0/2) |
| Pallidum | 158 | (90/68) | 429 | (102/327) |
| Cerebral White Matter | 130 | (79/51) | 521 | (194/327) |
| Ventricular System | 130 | (94/36) | 234 | (234/0) |
| Third Ventricle | 119 | (73/46) | 143 | (34/109) |
| Hippo-Amygdala Complex | 100 | (55/45) | 0 | (0/0) |
| Nucleus Accumbens | 88 | (46/42) | 0 | (0/0) |
| Corpus Callosum | 85 | (52/33) | 92 | (92/0) |
| Temporal Lobe (gm) | 85 | (49/36) | 92 | (92/0) |
| Cerebral Cortex | 83 | (54/29) | 429 | (102/327) |
| Cerebral Gray Matter | 78 | (52/26) | 92 | (92/0) |
| Frontal Lobe (gm) | 69 | (41/28) | 92 | (92/0) |
| Parietal Lobe (gm) | 69 | (41/28) | 92 | (92/0) |
| Fourth Ventricle | 64 | (48/16) | 143 | (34/109) |
| Occipital Lobe (gm) | 63 | (36/27) | 92 | (92/0) |
| Frontal Lobe | 60 | (34/26) | 0 | (0/0) |
| Inferior Lateral Ventricle | 56 | (37/19) | 429 | (102/327) |
| Brainstem | 51 | (37/14) | 9 | (9/0) |
| Entorhinal Cortex | 50 | (27/23) | 0 | (0/0) |
Example Queries
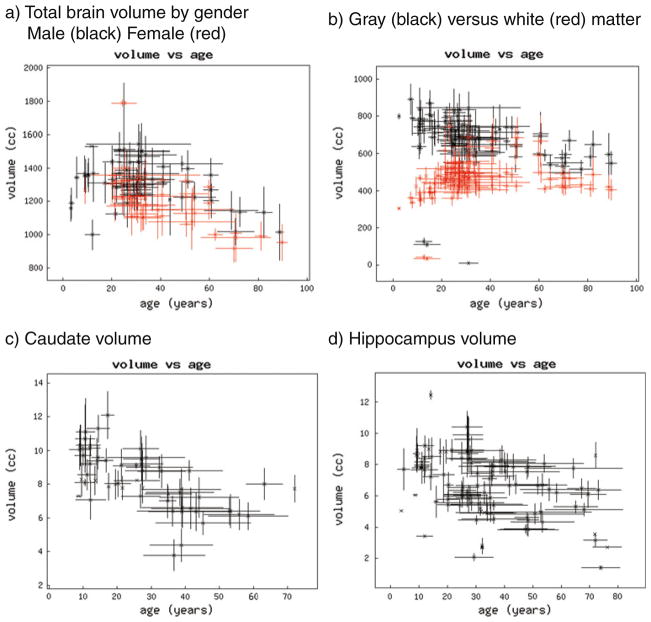
The most efficient way to see the effectiveness of the database is to view the graphical results of typical queries. Figure 4 demonstrates the results of the volumetric results returned for subject groups indicated as ‘normal’ for total brain, total cerebral gray and white matter, right and left caudate nucleus, and right and left hippocampus. Note that Fig. 4b–d did not select for the gender makeup or specific methodology employed for the reported volumes, although each of these factors can be included in the search criteria.
Fig. 4.
Graphical plots of sample query results. For these plots of group volume, each group is represented by the mean age (on the x axis) and mean volume (on the y axis). The x-axis bars represent the standard deviation of the reported group age; the y-axis bar represents the reported standard deviation of the volume. a Results of the volumetric results returned for subject groups indicated as ‘normal’ for total brain, plotted by gender. b Co-plot of total cerebral gray and white matter. c Plot of total caudate nucleus volume by age. d Plot of total hippocampus volume by age
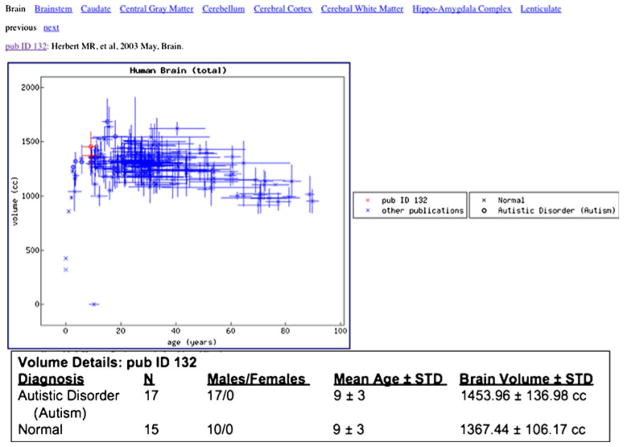
Additional insight into the content of the database comes from viewing results of studies of specific pathologies, in isolation or taken across the literature. An example of this is shown in Fig. 5, where the ‘Paper View’ functionality is used to show the results of Herbert et al. 2003 (Herbert et al. 2003) relative to the rest of the literature. In this example, for the anatomic structure ‘total brain volume’, in one plot we can see the consistency for the normal group from this paper with the rest of the literature.
Fig. 5.
Demonstration of the ‘Paper View’ display for Herbert et al. 2003. Total brain volume is plotted; with the observations of this specific paper shown in red. Comparison volumes from the remainder of the database are shown in blue, and represent data for normal subjects (x) as well as groups with the diagnosis of Autistic Disorder (o)
Discussion
The IBVD is a work in progress. The principal objective is to provide a database of neuroanatomic volumetric data as reported in the literature. In addition, we provide a mechanism to facilitate its review, discovery and extension as part of an overall neuroinformatics infrastructure designed to expose deeper, non-textual aspects of the published neuroscience data. The current instantiation provides a proof of concept for the utility of the site and generates substantial feedback and guidance for design of a long term, sustainable solution to this type of specialized data repository.
In its current state, while covering only a small percentage of the total set of published volumetric observations, the database can be shown to be useful for performing confirmative and integrative assessments of new and existing data. Despite the myriad methods for generating volumetric observations (different imaging, analysis, subject characterization, etc.), the IBVD demonstrates that specific neuroanatomic structures are relatively insensitive to the specific assessment methodological details. In contrast, other structures are demonstrated to be quite sensitive to the analysis methodological details. Qualitatitively, for each structure we can assess the between group volume standard deviation relative to the within group standard deviation, regardless of the analysis method employed. Examples of each of these classes of structure are seen in Fig. 4 with the cerebrum and hippocampus volume plots, respectively. The potential reasons for substantial methodological variability in the hippocampus are numerous. First, it is a relatively small structure (approximately 8 cc in young adults), which magnifies sensitivity to all acquisition and analyses sources of variance (Filipek et al. 1994). Second, numerous of its borders are open to methodological interpretation (Konrad et al. 2009; Rodionov et al. 2009). These are also often modulated by available imaging resolution. Specifically, the nature of the handling of the hippocampus—amygdala border can be variable, as can the extent to which the hippocampus definition includes entorhinal cortex, subiculum, perirhinal cortex, etc. In providing the ability to present a literature-wide normative ‘growth curve’ of specific neuroanatomic structure volume, the observer has quick access to an assessment of that structures volumetric measurement ‘stability’. In this case, stability refers to the relative insensitivity of the volumetric measures with respect to the methodological variables. Extra caution is warranted in the interpretation of the relative literature values for structures that demonstrate clear methodological dependence.
It is important to note that the proof of concept of this class of data extraction and mining from the historical literature is feasible and informative, but it is clear that new published reports would benefit greatly from a prospective approach to data entry. In addition, continued harvesting of information from historical publications, as well as new publications, could take advantage of automated text recognition software, designed to identify neuroanatomic structures in association with volumetric observations in the literature (Muller et al. 2008). The most feasible approach to systematic and automated incorporation of volumetric data from future publications is to promote the analytic capture in a standardized format of the detailed metadata of the volumetric observations in machine-readable form at time of publication. It is at the time of the accepted, but not yet published, manuscript where the authors are most familiar with, and amenable to, content markup. Establishment of a standardized markup language for volumetric observations (VolML), and creation of simple, easy-to-use tools for expressing data in this form, using volume tables or output of volumetric analysis software directly as input, will be critical to moving this endeavor forward.
Interoperability
While the free-standing IBVD website provides valuable functionality to the user, its utility is also demonstrated through its interoperations with other websites. The IBVD is used to provide a "Size Differences by Sex, Age and Diagnosis” link out of the Braininfo website1 (Bowden and Dubach 2003). In addition, since all IBVD publications have an associated PubMed ID, the IBVD publication page can be associated with the PubMed page for each article in the ‘LinkOut’ resource listing.2 This capability is mediated by the Neuroscience Information Framework (NIF) DISCO LinkOut Broker framework (Marenco et al. 2008). Finally, IBVD is part of the NIF ‘data federation’, enabling direct query of the database in the context of the integrated NIF search portal3 (Gardner et al. 2008).
The IBVD is the first, to our knowledge, database specifically designed to host neuroanatomic volumetric observations from the published literature. As such, it captures a specific class of derived result that is reported in the literature; such results are not specifically searchable by the existing text-based indexing services. Other databases have been developed to capture different classes of derived data in the literature, of particular note are the databases designed hold to the Talairach coordinates of foci identified in functional neuroimaging activation studies (i.e. BrainMap (Laird et al. 2005), SumsDB (Van Essen 2009)).
The IBVD is not a replacement or alternative to data storage efforts designed to host raw imaging data. Indeed, the IBVD can be a useful conduit to help connect volumetric observations in the literature to the raw data sources, when available. While reanalysis of raw data is important for many reasons, the archiving of raw data is a different informatics problem than the one addressed here. Each group and individual volume data record includes a ‘url’ field that permits association between the volume observation and the raw data that leads to the observation. It is important to note, however, that it is the conclusions and observations that are made in a paper that persist and become the points for comparison in future publications, even decades later. This database supports the specific retention of the resultant volumetric data that supports these discussion and conclusion points for easier future reference.
Only limited graphics and no statistical analysis are offered via the IBVD website itself. This is intentional, as providing full-featured capabilities in either of these areas would entail a substantial investment in infrastructure development. It was felt that, since graphical and statistical treatment are much better handled by numerous separate applications, the focus of the IBVD should be to provide interoperable export of data for use in these other packages. The complete results of any search can be exported, as a tab (or other custom separator) separated value file, for incorporation into any statistics or visualization software. As an example of this, Fig. 6 demonstrates the results of an analysis in the ‘R’ statistical package4 for the generation of a continuous ‘growth curve’ for the total cerebellum volume in humans. To accomplish this, any dataset found as the result of a search in IBVD can be downloaded as a PHP table, which can be imported into R. In this example, the contributed package ‘ggplot2’ (Wickham 2009) is used to model the collection of volume data by a third order polynomial of age to estimate growth across the lifespan. Confidence bands and a weighting factor of study size are also incorporated into the graphic. In summary, while there is no fully featured web service, IBVD provides a complete text-based access to the publication, group demographic, and group volume tables. The relative simplicity of the database schema makes this a viable solution. Development of a more complex API is awaiting future release.
Fig. 6.
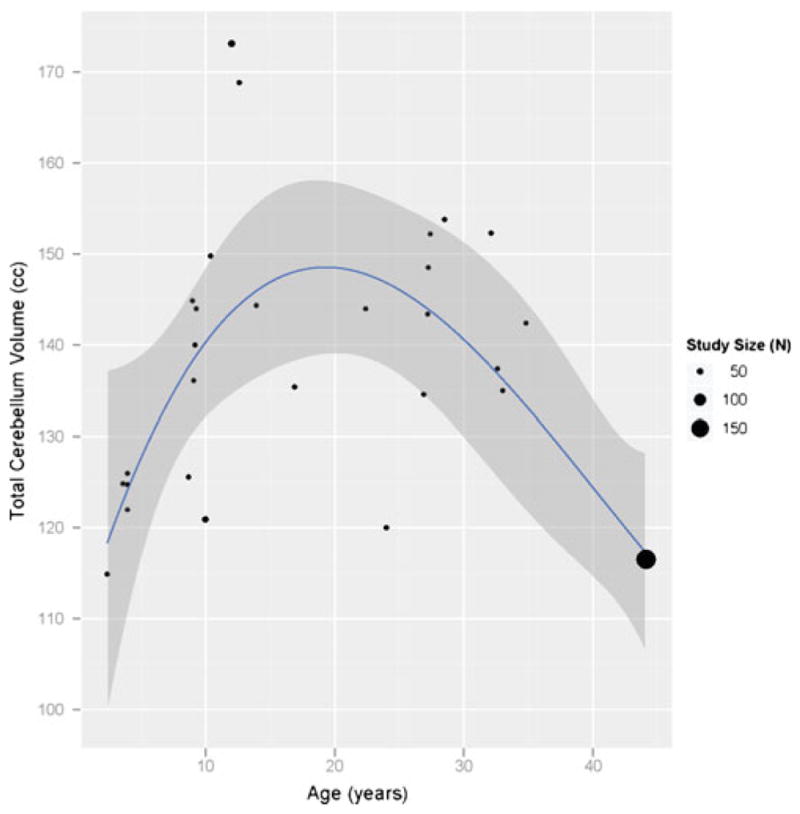
Growth curve for total cerebellum volume. Example statistical analysis for 3rd order polynomial fit in R for volume as a function of age from data exported from the IBVD
Limitations
While this report focuses on the underlying design and implementation of the database, support of broad and numerous studies of meta-analysis are envisioned for future publications. However, support of meta-analysis is predicated on coverage of the appropriate literature for the specific topic. To date, the data entry approach has been to enter example literature for the most prevalent volumetric structures and disorders. Efforts are ongoing to increase the content of the database, and targeted data entry will be necessary to facilitate specific meta-analytic investigation of the historical literature on the volumetric observations of the developing brain. The manual data entry described here is moderately time consuming. A publication takes approximately 10 s to enter (only the PubMed ID needs to be typed). A typical ‘group’ record takes about 2 min, depending on the number of demographic values reported. Most papers have between 2 and 10 groups, with an average of 3.2 groups per publication. Each volumetric record takes approximately 30 s to enter. Groups have from 1 to as many as 52 volumetric observations (with a mean of 6.1 volumes per group). With the addition of site navigation time, a typical paper takes 20–30 min of data entry. Data entry is preceded, however, by careful study and identification of all relevant data items in the paper. Volume and demographic data must be identified in both tabular and text contexts. This identification phase can take 30 min for typical volumetric papers.
Numerous extensions to the basic database can readily be envisioned. Inclusion of a new table to monitor the statistical comparisons between group volumetric reports can be envisioned. This would characterize the nature of the statistical test, the notation of which volumetric observations were tested and what covariates were included, and capture the significance of the reported findings. In addition, since growth curves and comparison of curves between diagnostic classes are a very common end use for the data, a better integration of the data export into a dedicated statistical interface to support growth curve generation would greatly facilitate database usage.
Precise identification of the volumetric analysis method employed is one of the most complex tasks associated with the characterization of a volumetric publication. In the absence of a formal ontology or description language for a set of anatomic definitions, we have chosen to represent the method of analysis by reference to the publication that best characterizes the anatomic method employed in the paper. ‘Method ID’ is available as a plotting factor for the results of searches in the database. The database does include a free text notes field for each publication. Methodological details can be included in this field. Standard terms and descriptions that should be should be considered for method description include imaging descriptors (i.e. field strength, imaging resolution, pulse sequence, software, etc. for MR data) as well as software description and the anatomic conventions employed, etc. Additional work is ongoing to create a more formal methodological description framework.
Despite the prevalence of volumetric observations in the literature, the presence of ‘voxel-based’ morphometric observations are rapidly eclipsing the volumetric observations in terms of sheer observations due in part to the ease of application of VBM-style analyses. While VBM between-group statistical inferences do not result in volumetric results for anatomic structures per se, the presence or absence of VBM findings localized to specific structures is a very valuable adjunct to the observations of structural differences between the same contrast groups. Fortunately, there are existing databases for capture of voxel-based group differences that have been developed for the capture of fMRI foci (i.e. BrainMap (Laird et al. 2005), SumsDB (Van Essen 2009)), and these have on-going extensions to capture voxel-based morphometric results as well. Future efforts to create blended reports of volumetric and voxel-based morphometric observations are proceeding.
Conclusion
In summary, the IBVD is an operational proof of concept of a domain-specific metadata capture system designed to operate on the published literature of neuroanatomic structural volumetric observations. Even in its prototypic stage, it supports quality assurance interrogation of results as well as meta-analysis across large segments of the published literature that are obscured from traditional text-based search engines.
Acknowledgments
This work was supported in part by grant NS34189 (PI Kennedy) as part of the Human Brain Project, MH083320 (Co PIs Kennedy and Frazier) and NS064354 (PI Gao). Thanks for tireless data entry efforts are owed to Julianne Steele, Aliya Dincer and Thomas Buckley. Marilyn Albert and Bruce Fischl provided access to individual data from their published work. Robert Williams provided group and individual mouse brain volumes from the Mouse Brain Library (MBL—http://www.mbl.org/).
Appendix
In this appendix we review the details of the various database tables.
Publication
Data sources
In most cases, publications are journal articles, but they may be other sources of data, such as on-line databases.
| ID | Integer | IBVD unique identifier for this publication. |
| Title | Text | Name of data source. Typically the title of the journal article, but this may be the name of a special data source (e.g. “Mouse Brain Library”). |
| Journal | Text | Journal name. |
| Date | Text | Date of publication. |
| Volume | Text | Journal volume/number. |
| Pages | Text | Page range for journal article. |
| URL | Text | URL for this publication or data source. |
Author
Journal article authors.
| Author name | Text | The name of an author |
| Credited | Integer | An ordinal giving the position of the author in the list of authors. The first author will have credited = 1, second author credited = 2, etc. |
Group
Subject groups
Groups are reported on in publications. The groups can be associated with a reference for the volume measurement methods employed (manual techniques for measurement, algorithms, software packages, etc) by reference to the most representative publication.
| ID | Integer | IBVD unique identifier for this subject group. |
| Method ID | Integer | IBVD Publication ID for publication(s) indicating methodology employed for this group. |
| Species | Text | Species of subjects in this group. |
| Species qualifier | Text | Subspecies of subjects in this group, for instance mouse strain. |
| Diagnosis | Text | Diagnosis of subjects in this group. |
| Vivoness | Text | Whether the volume measurement was done in vivo, in vitro, or ex vivo. |
| Number of subjects | Integer | Number of subjects in this group. |
| N male | Integer | Number of male subjects in this group. |
| N female | Integer | Number of female subjects in this group. |
| N left handed | Integer | Number of left handed subjects in this group. |
| N right handed | Integer | Number of right handed subjects in this group. |
| Mean age | Float | Mean age, in years, of subjects in this group. |
| Age std dev | Float | Standard deviation of ages, in years, of subjects in this group. |
| Age min | Float | Minimum age, in years, of subjects in this group. |
| Age max | Float | Maximum age, in years, of subjects in this group. |
| URL | Text | URL for this group (for on-line data sources or publications sharing data on-line). |
Group volume
Reported volumes for subject groups.
| ID | Integer | IBVD unique identifier for this reported volume. |
| Structure | Text | Structure being measured. |
| Hemisphere | Text | Hemisphere of the structure being measured (left, right, or total). |
| Mean volume | Float | Mean of volumes in this subject group. |
| Volume std dev | Float | Standard deviation of volumes in this subject group. |
| Units | Text | Units (ml, cc, etc) of the volume measurement. |
| Derived flag | Boolean | Whether this volume was derived from reported individual volumes. False for volumes directly reported in publications. |
| Adjusted flag | Boolean | Whether the reported measurement is stated to be ‘adjusted’ (for total brain volume, for example). |
| URL | Text | URL for this volume (for on-line data sources or publications sharing data on-line). |
Extra group demographics
Reported demographics for subject groups. Basic demographic information is stored in the group table; this table provides extensibility for storing demographics.
| ID | Integer | IBVD unique identifier for this extra demographic value. |
| Label | Text | The value being reported, for instance “Body Mass Index” or “Education Level.” |
| Qualifier | Text | Type of value being reported. This is unset for a direct measurement, but may indicate a mean, standard deviation, minimum, or maximum. In this way, an education level, say, of 12 +− 3 years may be stored. |
| Value | Text | The stored value. |
| Units | Text | Units of the stored value. |
Individual
Individual subject demographic data. Individual subjects can be reported in association with a group.
| ID | Integer | IBVD unique identifier for this subject. |
|---|---|---|
| Age | Float | Age, in years, of this subject. |
| Gender | Text | Gender of this subject. |
| Handedness | Text | Handedness of this subject. |
| Local code | Text | The source’s identifier for this subject. This is used for identifying a subject in an on-line database. |
| Species | Text | Species of the subject. |
| Species qualifier | Text | Subspecies of the subject, for instance mouse strain. |
| Vivoness | Text | Whether the volume measurements for this subject were done in vivo, in vitro, or ex vivo. |
| URL | Text | URL for this subject (for on-line data sources or publications sharing data on-line). |
Individual volume
Reported volumes for individual subjects. The individual volume record can report a volume measurement method (manual techniques for measurement, algorithms, software packages, etc) by reference to the most representative publication.
| ID | Integer | IBVD unique identifier for this reported volume. |
|---|---|---|
| Method ID | Integer | IBVD Publication ID for publication(s) indicating methodology employed for this individual. |
| Structure | Text | Structure being measured. |
| Hemisphere | Text | Hemisphere of the structure being measured (left, right, or total). |
| Volume | Float | Reported volume. |
| Units | Text | Units (ml, cc, etc) of the volume measurement. |
| Derived flag | Boolean | Whether this volume was derived from other volumes. False for volumes directly reported in publications. |
| Adjusted flag | Boolean | Whether the reported measurement is stated to be ‘adjusted’ (for total brain volume, for example). |
| URL | Text | URL for this volume (for on-line data sources or publications sharing data on-line). |
Footnotes
Information Sharing Statement
The website described in this report is currently accessible at www.cma.mgh.harvard.edu/ibvd and www.nitrc.org/projects/ibvd.
See http://www.ncbi.nlm.nih.gov/pubmed/18003631, for example.
Contributor Information
David N. Kennedy, Email: David.Kennedy@umassmed.edu, Division of Neuroinformatics and the Child and Adolescent NeuroDevelopment Initiative (CANDI), Department of Psychiatry, University of Massachusetts Medical School, 356 Plantation St, Biotech 1, Suite 100, Worcester, MA 01605, USA
Steven M. Hodge, Division of Neuroinformatics and the Child and Adolescent NeuroDevelopment Initiative (CANDI), Department of Psychiatry, University of Massachusetts Medical School, 356 Plantation St, Biotech 1, Suite 100, Worcester, MA 01605, USA
Yong Gao, Department of Neurology, Massachusetts General Hospital, Boston, MA, USA.
Jean A. Frazier, Division of Neuroinformatics and the Child and Adolescent NeuroDevelopment Initiative (CANDI), Department of Psychiatry, University of Massachusetts Medical School, 356 Plantation St, Biotech 1, Suite 100, Worcester, MA 01605, USA
Christian Haselgrove, Division of Neuroinformatics and the Child and Adolescent NeuroDevelopment Initiative (CANDI), Department of Psychiatry, University of Massachusetts Medical School, 356 Plantation St, Biotech 1, Suite 100, Worcester, MA 01605, USA.
References
- Bowden DM, Dubach MF. NeuroNames 2002. Neuroinformatics. 2003;1(1):43–60. doi: 10.1385/NI:1:1:043. [DOI] [PubMed] [Google Scholar]
- Bug WJ, Ascoli GA, et al. The NIFSTD and BIRNLex vocabularies: building comprehensive ontologies for neuroscience. Neuroinformatics. 2008;6(3):175–194. doi: 10.1007/s12021-008-9032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzdok D, Langner R, et al. ALE meta-analysis on facial judgments of trustworthiness and attractiveness. Brain Structure Function. 2011;215(3–4):209–223. doi: 10.1007/s00429-010-0287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey BJ, Tottenham N, et al. Imaging the developing brain: what have we learned about cognitive development? Trends in Cognitive Sciences. 2005;9(3):104–110. doi: 10.1016/j.tics.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Caviness VSJ, Kennedy DN, et al. The developing human brain: A morphometric profile. In: Thatcher RW, Lyon GR, Rumsey J, Krasnegor N, et al., editors. Developmental neuroimaging: Mapping the development of brain and behavior. New York: Academic; 1996. pp. 3–14. [Google Scholar]
- Caviness VS, Lange J, NT, et al. MRI-based brain volumetrics: emergence of a developmental brain science. Brain and Development. 1999;21(5):289–295. doi: 10.1016/s0387-7604(99)00022-4. [DOI] [PubMed] [Google Scholar]
- Courchesne E, Pierce K. Brain overgrowth in autism during a critical time in development: implications for frontal pyramidal neuron and interneuron development and connectivity. International Journal of Developmental Neuroscience. 2005;23(2–3):153–170. doi: 10.1016/j.ijdevneu.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Filipek PA, Richelme C, et al. The young adult human brain: an MRI-based morphometric analysis. Cerebral Cortex. 1994;4(4):344–360. doi: 10.1093/cercor/4.4.344. [DOI] [PubMed] [Google Scholar]
- Gardner D, Akil H, et al. The neuroscience information framework: a data and knowledge environment for neuroscience. Neuroinformatics. 2008;6(3):149–160. doi: 10.1007/s12021-008-9024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedd JN, Clasen LS, et al. Puberty-related influences on brain development. Molecular and Cellular Endocrinology. 2006;254–255:154–162. doi: 10.1016/j.mce.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Herbert MR, Ziegler DA, et al. Dissociations of cerebral cortex, subcortical and cerebral white matter volumes in autistic boys. Brain. 2003;126(Pt 5):1182–1192. doi: 10.1093/brain/awg110. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, Bernstein MA, et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. Journal of Magnetic Resonance Imaging. 2008;27(4):685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardri R, Pouchet A, et al. Cortical activations during auditory verbal hallucinations in schizophrenia: a coordinate-based meta-analysis. The American Journal of Psychiatry. 2011;168(1):73–81. doi: 10.1176/appi.ajp.2010.09101522. [DOI] [PubMed] [Google Scholar]
- Konrad C, Ukas T, et al. Defining the human hippocampus in cerebral magnetic resonance images–an overview of current segmentation protocols. NeuroImage. 2009;47(4):1185–1195. doi: 10.1016/j.neuroimage.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AR, Lancaster JL, et al. BrainMap: the social evolution of a human brain mapping database. Neuroinformatics. 2005;3(1):65–78. doi: 10.1385/ni:3:1:065. [DOI] [PubMed] [Google Scholar]
- Larson SD, Martone ME. Ontologies for neuroscience: what are they and what are they good for? Frontiers in Neuroscience. 2009;3(1):60–67. doi: 10.3389/neuro.01.007.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenroot RK, Gogtay N, et al. Sexual dimorphism of brain developmental trajectories during childhood and adolescence. NeuroImage. 2007;36(4):1065–1073. doi: 10.1016/j.neuroimage.2007.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leow AD, Yanovsky I, et al. Alzheimer’s disease neuroimaging initiative: a one-year follow up study using tensor-based morphometry correlating degenerative rates, bio-markers and cognition. NeuroImage. 2009;45(3):645–655. doi: 10.1016/j.neuroimage.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg DA, Humphreys BL, et al. The unified medical language system. Methods of Information in Medicine. 1993;32(4):281–291. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Wang TH, et al. Open Access Series of Imaging Studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. Journal of Cognitive Neuroscience. 2007;19(9):1498–1507. doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]
- Marenco L, Ascoli GA, et al. The NIF LinkOut broker: a web resource to facilitate federated data integration using NCBI identifiers. Neuroinformatics. 2008;6(3):219–227. doi: 10.1007/s12021-008-9025-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, et al. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 2001;356(1412):1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HM, Rangarajan A, et al. Textpresso for neuroscience: searching the full text of thousands of neuroscience research papers. Neuroinformatics. 2008;6(3):195–204. doi: 10.1007/s12021-008-9031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov R, Chupin M, et al. Evaluation of atlas-based segmentation of hippocampi in healthy humans. Magnetic Resonance Imaging. 2009;27(8):1104–1109. doi: 10.1016/j.mri.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Rosas HD, Hevelone ND, et al. Regional cortical thinning in preclinical Huntington disease and its relationship to cognition. Neurology. 2005;65(5):745–747. doi: 10.1212/01.wnl.0000174432.87383.87. [DOI] [PubMed] [Google Scholar]
- Saitoh O, Karns CM, et al. Development of the hippocampal formation from 2 to 42 years: MRI evidence of smaller area dentata in autism. Brain. 2001;124(Pt 7):1317–1324. doi: 10.1093/brain/124.7.1317. [DOI] [PubMed] [Google Scholar]
- Salat DH, Buckner RL, et al. Thinning of the cerebral cortex in aging. Cerebral Cortex. 2004;14(7):721–730. doi: 10.1093/cercor/bhh032. [DOI] [PubMed] [Google Scholar]
- Schuff N, Woerner N, et al. MRI of hippocampal volume loss in early Alzheimer’s disease in relation to ApoE genotype and biomarkers. Brain. 2009;132(Pt 4):1067–1077. doi: 10.1093/brain/awp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman LJ, Faraone SV, et al. Left hippocampal volume as a vulnerability indicator for schizophrenia: a magnetic resonance imaging morphometric study of nonpsychotic first-degree relatives. Archives of General Psychiatry. 2002;59(9):839–849. doi: 10.1001/archpsyc.59.9.839. [DOI] [PubMed] [Google Scholar]
- Seidman LJ, Valera EM, et al. Structural brain imaging of attention-deficit/hyperactivity disorder. Biological Psychiatry. 2005;57(11):1263–1272. doi: 10.1016/j.biopsych.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Giedd JN, et al. Growth patterns in the developing brain detected by using continuum mechanical tensor maps. Nature. 2000;404(6774):190–193. doi: 10.1038/35004593. [DOI] [PubMed] [Google Scholar]
- Van Essen DC. Lost in localization–but found with foci?! NeuroImage. 2009;48(1):14–17. doi: 10.1016/j.neuroimage.2009.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waber DP, De Moor C, et al. The NIH MRI study of normal brain development: performance of a population based sample of healthy children aged 6 to 18 years on a neuropsychological battery. Journal of the International Neuropsychological Society. 2007;13(5):729–746. doi: 10.1017/S1355617707070841. [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2: Elegant graphics for data analysis. New York: Springer; 2009. [Google Scholar]