Abstract
In animal mitochondria, several codons are non-universal and their meanings differ depending on the species. In addition, the tRNA structures that decipher codons are sometimes unusually truncated. These features seem to be related to the shortening of mitochondrial (mt) genomes, which occurred during the evolution of mitochondria. These organelles probably originated from the endosymbiosis of an aerobic eubacterium into an ancestral eukaryote. It is plausible that these events brought about the various characteristic features of animal mt translation systems, such as genetic code variations, unusually truncated tRNA and rRNA structures, unilateral tRNA recognition mechanisms by aminoacyl-tRNA synthetases, elongation factors and ribosomes, and compensation for RNA deficits by enlarged proteins. In this article, we discuss molecular mechanisms for these phenomena. Finally, we describe human mt diseases that are caused by modification defects in mt tRNAs.
Keywords: genetic code, tRNA, animal mitochondria, translation system, modified nucleotides, mitochondrial diseases
Introduction
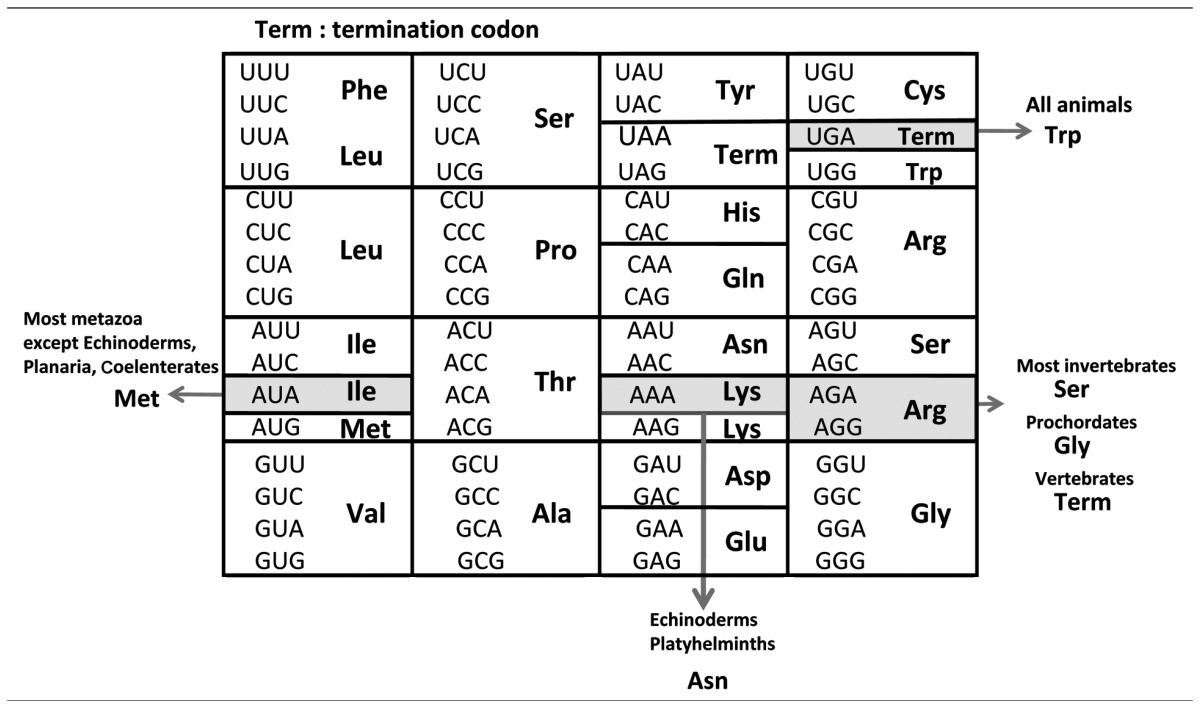
It was long believed that the genetic code is universal for all organisms (Table 1). However, in 1979, Barrel et al. first found the non-universal genetic code by comparing the HeLa cell mt DNA sequence of the cytochrome oxydase subunit II gene and the amino acid sequence of the corresponding beef heart protein, which showed that UGA is used as a tryptophan (Trp) codon instead of as a termination codon, and that AUA may be a methionine (Met) instead of isoleucine (Ile) codon.1) Anderson et al. determined the complete genome sequence of HeLa cell mitochondria and defined characteristic features of the mt genome2): it is small, circular and AT-rich, consisting of a total of 16,569 bp, and gene organization is very compact, consisting of 13 proteins and two rRNA and 22 tRNA genes with scarce spacer regions. An unusually truncated tRNASer of 63 nucleotides lacking the D loop and stem was found in the tRNA gene sequence3) and also at the RNA level.4)
Table 1.
Standard genetic code (inside the box) and variations in animal mitochondrial genetic code (outside)
These findings are intriguing, and an exploration of the translation systems of animal mitochondria is warranted, because they could provide a key to understanding the mechanisms of evolution of life, especially the processes involved in the transition between the “RNA world” to the “RNP (ribonucleoprotein) world”.5) We intended to pursue the variations of the genetic code in various animals, to carry out structural and functional studies of truncated mt tRNAs, and to construct an in vitro mt translation system to examine how truncated tRNAs function and how a non-universal genetic code is decoded by tRNAs.
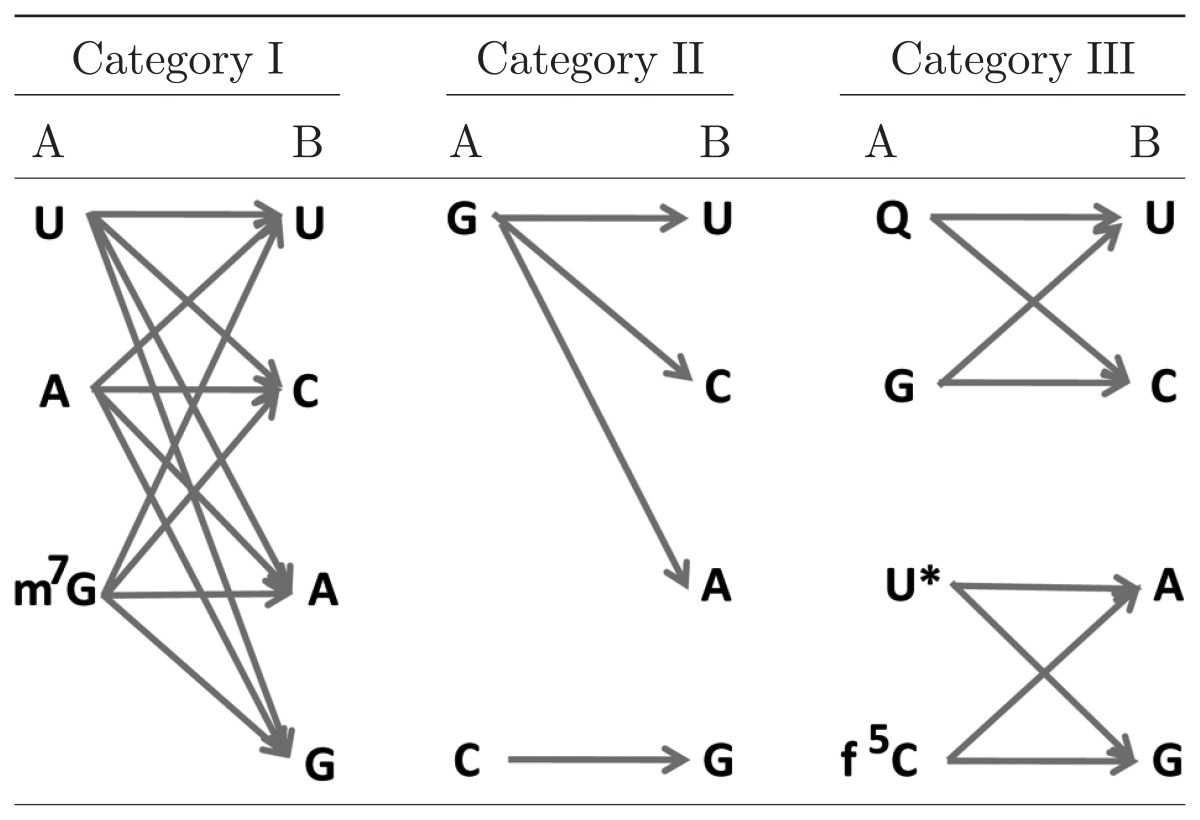
Since 1983, we have discovered non-universal genetic codes in several animals and examined how such variations of the genetic code have emerged during the course of animal evolution (Table 2). To pursue the decoding mechanism of non-universal genetic codes by mt tRNAs in extant animal mitochondria, we have analyzed mt tRNAs at the RNA level and discovered novel modified nucleosides located in the anticodon wobble position of various tRNAs. Thus, we have further expanded the “Wobble rule”6),7) to the latest version (Table 3), in which other modified nucleosides in addition to unmodified U, A and G at the anticodon wobble position are taken into consideration for codon recognition. We have succeeded in constructing an in vitro translation system of bovine mitochondria and have confirmed that these modified nucleosides are indeed involved in the recognition of non-universal codons. We have also proved that even truncated tRNAs can form cloverleaf-like tertiary structures and function in the in vitro bovine mt translation system. We have observed that mt enzymes and ribosomes are able to recognize both mt and E. coli tRNAs; however, E. coli enzymes and ribosomes are unable to recognize mt tRNAs in either in vitro translation system. We termed these phenomena “a unilateral recognition mechanism”. We also observed how truncated RNA segments in tRNAs and rRNAs are compensated for structurally and functionally by enlarged proteins in mt translation systems. Finally, we have found novel modified nucleosides in human mt tRNAs and have revealed that they are highly relevant to human mt diseases.
Table 2.
Relationship between genetic code of animal mitochondria and animal phyla
| Codon (Universal codon) | UGA (Term) | AUA (Ile) | AAA (Lys) | AGA (Arg) | AGG (Arg) |
|---|---|---|---|---|---|
| Vertebrates (human, bovine, rat, mouse, chicken, frog) | Trp | Met | Lys | Term | Term |
| Prochordates (ascidian, asymmetron) | Trp | Met | Lys | Gly | Gly |
| Echinoderms (sea urchin, starfish) | Trp | Ile | Asn | Ser | Ser |
| Arthropods | |||||
| Most (shrimp, dalhina) | Trp | Met | Lys | Ser | Ser |
| Insect (Drosophila) | Trp | Met | Lys | Ser | – |
| Molluscs (squid, octopus, Liolophura, Mesogastropoda) | Trp | Met | Lys | Ser | Ser |
| Nematodes (nematodes, ascaris) | Trp | Met | Lys | Ser | Ser |
| Platyhelminths | |||||
| Most (Echinostomida, Trematoda) | Trp | Met | Asn | Ser | Ser |
| Rhabditiophora (Planaria) | Trp | Ile | Asn | Ser | Ser |
| Coelenterates (jellyfish, coral, sea anemone, hydrozoa) | Trp | Ile | Lys | Arg | Arg |
| Protozoans (trypanosome, Paramecium) | Trp | Ile | Lys | Arg | Arg |
Bold letter: non-universal codon; Term: termination codon.
Table 3.
An expanded wobble rule: possible pairings between the wobble nucleoside of tRNA (A) and the codon third nucleoside of mRNA (B) found in animal mitochondria
References U: 7, 38, 58, 59; A: 58, 60; m7G: 43, 46; G: 17, 23, 47; Q: 61; U*: cmnm5(s2)U, 50, 51, 54, mnm5U, 51, τm5U, τm5s2U, 57; f5C: 47, 48, 55, 56.
All these features observed for mt translation systems will be helpful to elucidate how animal mitochondria have emerged over the course of evolution. In addition, the discoveries of novel modified nucleosides in human mitochondria and their roles in human mt diseases will provide a key to medical treatment of these diseases.
I. Genetic code variations and the anticodon structure of the corresponding tRNA
1. Genetic code variations in animal mitochondria
Since 1981, when Anderson et al. first determined a complete nucleotide sequence of HeLa cell mitochondria,2) the complete nucleotide sequences of metazoan mt genomes have been reported by several groups including our own; these include the mt genomes of human,2) bovine,8) rat,9) mouse,10) chicken,11) frog,12) ascidian,13) sea urchin,14),15) starfish,16) fruit fly,17) nematode,18) and others. Several other metazoan mt genomes have been partially sequenced.19) Complete or partial sequences of some protozoan mt genomes have also been determined; they are not single circles but are segmented, with their genome sizes being much larger than those of metazoans.20)–22)
To track the evolutionary variation of genetic codes, we have sequenced mt genomes from various animal phyla, such as starfish,16),23) coelacanth,24) lung-fish,24) ascidian,25) mollusc,26) and squid,27) and found some non-universal genetic codes. Together with data obtained by other groups,1)–3),8)–12),14),15),17)–22),28) the genetic code variations that have been identified in animal mitochondria are summarized in Table 2.29) Five codons are changeable in their meanings depending on the animal phyla.30) UGA is a termination codon in the standard genetic code, but it specifies Trp in all animal mitochondria.1) The UAA termination codon was once assumed to be used as a Tyr codon in planarian mitochondria,31) but there are no data for other platyhelminth mitochondria in the literature to confirm this,32) and there is no structural information on the planarian mt tRNATyr that decodes the UAA codon. In the present eubacterial release factors RF1 and RF2, RF1 recognizes UAG and UAA codons, while RF2 recognizes UGA and UAA codons,33) so that it is difficult to assume that UAA is changeable than UAG and UGA. Thus, I omitted contradictory result on the UUA codon in Table 2. AUA codes for Ile in the standard genetic code and in mitochondria of Protozoa,20)–22),28) Cnidaria,34) Rhabditophora in Platyhelminthes,31) and Echinodermata,23) but it encodes Met in the mitochondria of Platyhelminthes except for Rhabditophora,34) Nematoda,18) Mollusca, Arthropoda,35),36) Prochordata,13),25) and Vertebrata.1) AGA/AGG codons are unique in that they change their meanings in four ways: Arg, Ser, Gly and termination, depending on the animal taxon (Arg in protozoans,21),22),28) Ser in most metazoans,18),23),30) Gly in prochordates,25) and a termination codon in vertebrates2)). The AAA codon is changed to an Asn codon only in echinoderms23) and platyhelminths.32)
2. Codon-anticodon relationships in the mitochondrial genetic code
The codon variations thus obtained in various animal mitochondria are only inferred from comparison of DNA sequences with the corresponding protein sequences.1) The simplest way to ascertain that these changed codons are actually used in mt translation systems is to compare the codon with the anticodon of the corresponding tRNA.30) We have analyzed mt tRNAs isolated from various sources such as bovine,37)–41) rat, chicken, frog, starfish,42),43) ascidian,44),45) squid,46) arthropod,47) and nematode,48)–51) to confirm that the assignment of codon variations is reasonable.
For isolating individual tRNAs in an amount sufficient for sequencing and functional analysis, if possible, we used conventional column chromatography techniques (BD-cellulose, DEAE-cellulose, etc.) followed by 15% polyacrylamide-gel electrophoresis. The tRNA fraction was detected by aminoacylation with crude bovine mt aminoacyl-tRNA synthetase (S-100 fraction). For example, 5~20 kg of bovine heart or liver was needed to isolate about 5 A260 units (250 μg) of mt tRNASerGCU, and complete isolation required a few months.37) Later, we adopted a hybridization assay instead of an aminoacylation assay to detect tRNA, which enhanced detection sensitivity by 100 fold and enabled us to distinguish even isoacceptor tRNAs such as tRNASerGCU and tRNASerUGA.38)
From the sequence analysis of mt tRNAs involved in decoding non-universal genetic codes, it has been determined that the tRNA anticodon, especially the first nucleoside (wobble nucleoside), is deeply involved in decoding.29) The anticodon of tRNATrp that deciphers the UGA termination codon as Trp has been found to be U*CA in bovine52) and rat53) mitochondria. U* seems to be 5-carboxymethylaminomethyl-(2-thio)U (cmnm5(s2)U),50),51),54) which is known to pair with G as well as A at the codon third position. We found a novel modified nucleoside, 5-formylcytidine (f5C), at the wobble position of tRNAMet from bovine55) and nematode mitochondria, and also from squid, frog, chicken, rat and fruit fly47) mitochondria. Recently, using an in vitro mt translation system, we confirmed that tRNAMet with the anti-codon f5CAU can decode both AUG and AUA codons.56) Thus, it turned out that the change in the AUA codon from Ile to Met is brought about by formylation of C at the wobble position of tRNAMet. In echinoderm and platyhelminth mitochondria,23),32) not only the usual Asn codons AAU and AAC, but also the usual Lys codon AAA, are translated as Asn by a single mt tRNAAsn with the anticodon GUU. The nucleotide sequencing of starfish mt tRNAAsn revealed that the anticodon is GΨU and that U35 at the anticodon second position is modified to pseudouridine (Ψ).42) In contrast, mt tRNALys, corresponding to another Lys codon, AAG, has the anti-codon CUU. Mt tRNAs possessing anticodons closely related to that of tRNAAsn, but responsible for decoding only two codons each (tRNAHis, tRNAAsp and tRNATyr) were found to possess unmodified U35 in all cases, suggesting the importance of Ψ35 in tRNAAsn for decoding the AAU, AAC and AAA codons. Experiments with an in vitro translation system confirmed that tRNAAsnGΨU has a ~two-fold higher translational efficiency than tRNAAsnGUU.42) It is exceptional that modification at the anticodon second nucleoside is involved in the codon change. AGA and AGG codons are read as Ser in most metazoan mitochondria and as Gly in prochordate mitochondria. In vertebrate mitochondria these codons are changed to termination codons. We found that 7-methylguano-sine (m7G) is present at the wobble position of tRNASerGCU in most invertebrate mitochondria.43),46) Therefore, the anticodon m7GCU of tRNASerGCU is most likely responsible for reading the four AGN codons as Ser.46) On the other hand, the AGG codon is absent from only insect mitochondria (Table 2), and the anticodon wobble position of tRNASerGCU is un-modified G. In this case, the unmodified GCU anticodon of tRNASerGCU seems to read the three codons AGU, AGC and AGA as Ser.47) These interpretations need to be confirmed in an in vitro translation system. The anticodon of tRNAGlyUCU is U*CU in ascidian mitochondria. U* is probably 5-taurino-methyl U (τm5U)57) (Kondow, A., Suzuki, T. and Watanabe, K., in preparation). It is apparent that the U*CU anticodon of tRNAGlyUCU reads the AGR (AGA and AGG) codons as Gly, similar to the case of tRNATrp in which the anticodon U*CA reads both UGR codons as Trp and similar to the case for tRNALeu(UUR), as discussed in Chapter III.
Thus, the codon-anticodon pairing relationships in animal mt systems can be classified into three categories (Table 3). In the first category, a single species of tRNA can read all four codons (four-way wobble). It is well known that tRNAs possessing an unmodified U at the wobble position can read all four codons in a codon box,7),38),58),59) and the same holds for tRNAs with an unmodified A of tRNAArg58),60) or m7G of tRNASerGCU43),46) at the wobble position. The second category of pairing relationships is one in which the codon box is divided 3:1. Within a box, a codon ending with G is read by a tRNA possessing an unmodified C at the wobble position, whereas the remaining three codons ending with U, C and A are read by a tRNA possessing an unmodified G at the wobble position (three-way wobble). Such cases are observed in the codon boxes of starfish (AAU, AAC and AAA)23),47) and Drosophila (AGN).17) In this case there is no tRNA that corresponds to AGG, so AGG is an unassigned codon. In the third category, the codon box is divided 2:2. In this case, two codons ending with U and C are read by a tRNA possessing an unmodified G or queosine (Q)61) at the wobble position, whereas the other two codons ending with A and G are read by a tRNA possessing either a modified uridine such as cmnm5(s2)U,50),51),54) 5-methylaminomethyl U (mnm5U),51) τm5U and 5-taurinomethyl-2-thio U (τm5s2U),57) or f5C47),55) at the wobble position (two-way wobble). This situation is observed in the UGN codon boxes of all animals, AUN of vertebrates, Drosophila and nematodes, AAN of vertebrates, Drosophila and nematodes, and AGN of vertebrates (in this case AGR codons are termination codons) and ascidians.
The molecular mechanisms by which such modifications function to alter codon recognition remain to be solved. In the case of four-way wobble, m7G as well as unmodified U58) and A58),62) at the wobble position of tRNA could decode all 4 codons, which may conform to “two out of three” reading method proposed by Lagerkvist.63),64) Existence of no competitor tRNA in the 4 codon boxes would facilitate such reading method. Why m7G but not unmodified G is contained in tRNASerGCU of most invertebrate mitochondria43),46) would be an intriguing problem in connection with the codon change and animal lineage. Ψ present at the middle position of anticodon of tRNAAsnGΨU in insect mitochondria47) would strengthen the codon-anticodon interaction, otherwise there are two A-U pairings between the first and second codons and the third and second anticodons. Q and U* are known to facilitate codon-anticodon base-parings with pyrimidine-ending61) and purine-ending codons,59) respectively. f5C is newly found modified nucleoside at the anticodon wobble position of tRNAMet from most metazoan mitochondria,47),55) whose function to decode AUA as well as AUG56) would also be intriguing in connection with the codon change and animal lineage.
Through these studies, we expanded the “Wobble rule”6),7) to the latest version (Table 3), including new modified nucleosides at the anticodon wobble position of tRNAs, as has been discussed.
3. Genetic code variations and animal evolution.29)
In considering the evolution of the genetic code in animal mitochondria, the codon-capture hypothesis based on AT pressure, proposed by Osawa and Jukes,7),65) has been most helpful. Codon capture means that any codon can be read by the corresponding tRNA, but if a competitor tRNA or a release factor arises that has stronger affinity toward the codon than does the original tRNA, the codon will then be read by the competitor. Thus, the codon is reassigned or captured. As for the UGA codon, there has been so far no report of a release factor recognizing this codon in animal mitochondria.66),67) Such a release factor corresponding to eubacterial RF2 was probably lost in the animal mt system, so that the UGA codon became unassigned. When a residue of the anticodon for tRNATrp changed from C to modified U (U*), and a part of the Trp UGG codon in reading frames was changed to UGA by AT pressure, the UGA codon was captured by tRNATrpU*CA and read as Trp. In fact, in the bacterium Mycoplasma caplicolum, where both the UGG and UGA codons are used as Trp codons, there are two tRNA genes with similar sequences that are arranged in tandem, one with the anticodon TCA and the other with CCA.68) The transcripts of these genes (tRNATrpU*CA and tRNATrpCCA) have also been detected.54) These observations suggest that the tRNA with the CCA anticodon was duplicated and that one copy mutated to a tRNATrp with the anticodon UCA. This situation is suggestive of an intermediate state in the evolution from the ancestral bacterium to mitochondria.
To explain the evolutionary change of other non-universal codons in metazoan mitochondria, the genome economization effect (especially, that tRNA genes are restricted to 22~23 species, and that tRNAs are not imported from the cytoplasm in almost all metazoan mitochondria52)) should be taken into consideration, in addition to AT pressure. The AUA codon is read as Ile in protozoans, coelenterates, some platyhelminths, and echinoderms, but it is read as Met in most metazoans. f5C at the wobble position of tRNAMet in most metazoans47),48),55) may be a key to understanding the codon reassignment from AUA-Ile to AUA-Met. Since the interaction between tRNAIleGAU and AUA on the ribosome might be more unstable than that between the tRNAIleGAU and AUY codons, when tRNAMet acquired the capacity to decode AUA by 5-formylation of C at the wobble position, tRNAMetf5CAU may have prevailed over tRNAIleGAU in the interaction with the AUA codon. Thus, the reassignment of Ile to Met could have easily occurred. In echinoderm mitochondria, f5C was lost in tRNAMet, so that AUA is read as Ile by tNAIleGAU. The AAA codon is read as Lys in protozoans and most metazoans, but only in platyhelminths and echinoderms is it read as Asn. In this case, pseudouridylation at the second position of the anticodon of tRNAAsn42) was critical for the codon reassignment. The interaction between tRNAAsnGΨU and AAA may have prevailed over that between tRNALysUUU or tRNALysCUU (in echinoderms and Drosophila) and AAA. In the case of AGR codons, in the evolutionary process from protozoa to invertebrates, tRNAArgU*CU disappeared from the mt genome because of the reduction in genome size (in protozoa some tRNA genes are absent from the mt genome and most tRNAs are imported from the cytoplasm, but in almost all metazoan mitochondria, importation of tRNA has not been reported52)). This has created a situation in which AGR codons cannot be translated. Since the AGR-Arg sites in the mitochondrial genomes of protozoa such as Trypanosoma are mostly replaced by CGN-Arg codons and partially by a few other codons throughout metazoan mitochondria,69) AGR codons were converted mainly to CGN upon deletion of tRNAArgU*CU, so that AGR became unassigned (first step). Once the anticodon first letter (G) of tRNASerGCU was modified to m7G, all four AGN codons were read as Ser.43),46) AGR codons pairing with this tRNASerG*CU then appeared in reading frames because of the mutation of AGY-Ser codons or other codons, and they were captured by Ser (second step). In ancestors of prochordates and vertebrates, demethylation of m7G of tRNASerG*CU may have occurred, so the resulting tRNASerGCU no longer reads the AGG codon. Strong selective constraints resulting from the lost translation of AGG caused the AGG codon to change mainly to AGY or other codons, so that the AGG codon became unassigned (third step). After prochordates were separated from vertebrate ancestors, a fourth event may have occurred: the tRNAGlyUCC gene was duplicated in mitochondrial DNA (mtDNA). In fact, the ascidian mt genome has two tRNAGly genes,13) and the anticodon of one species of tRNAGlyUCC was converted from TCC to TCT because of AT pressure, resulting in tRNAGlyUCU, which might have occurred in prochordate genomes. Then, the anticodon wobble position of tRNAGlyUCU must have been modified to U* (tRNAGlyU*CU) so as to decode AGR codons. At the same time, AT pressure caused GGN codons to change to AGR codons. Since the interaction of tRNAGlyU*CU with AGR is stronger than that of tRNASerGCU with AGR, AGR codons are captured by tRNAGlyU*CU, resulting in the translation of AGR codons as Gly. The low GC content of the CO I region in ascidian mitochondria in comparison to that of vertebrates is consistent with this speculation. In the ancestors of vertebrates, AGR codons may have appeared in the reading frames by deletion of U from the UAG termination codon, concomitantly with a functional change in the vertebrate release factor so as to recognize AGR codons. A possible candidate for an mt release factor capable of recognizing AGR codons was reported.67) The release factor prevails over tRNASerGCU in decoding the AGA codon and thus changes AGR codons to termination codons.25)
II. Structure and function of the translation apparatus
To clarify the molecular basis of genetic code variations in animal mitochondria, it is necessary to dissect their translation apparatus at the molecular level. The following are results of our experiments carried out for this purpose.
1. Higher-order structures of mt tRNAs
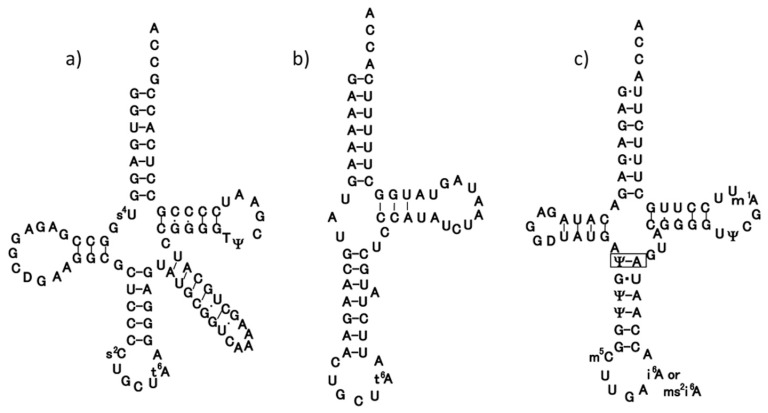
Many animal mt tRNAs are thought to have unusual secondary structures as inferred from their gene sequences; they seem to lack the interaction between the D and T arms that usually occurs in canonical tRNAs.70),71) Serine tRNA specific for codon AGY (tRNASerGCU) has the most unusual secondary structure in that it lacks the entire D arm,3),4) but it still exhibits serine-accepting activity in vitro.37) The codon AGY has been found to be translated as Ser in almost all protein-encoding genes so far identified in mammalian mitochondria.2) Thus, tRNASerGCU possessing a truncated secondary structure is presumed to function in the mt translation system in the same way as other mt tRNAs possessing both the D and T arms. If this presumption is correct, all mt tRNAs should share common structural features allowing them to function in mt ribosomes.72),73) An early work proposing a tertiary structural model for mt tRNASerGCU on the basis of chemical probing suggested that this tRNA also possesses an L-shaped structure74) that is slightly smaller than those of other canonical tRNAs.75) Steinberg et al.72) carried out computer modeling of the tRNA under the condition that the distance and orientation between the anticodon and the CCA-3′ end were constant in order that the tRNA can participate in the mt translation system,73) which resulted in a model with a “boomerang” shape rather than an “L” shape. However, no concrete experimental analysis of the tertiary structure of mt tRNAs has been reported so far. To determine the higher-order structure of tRNASerGCU by means of NMR spectroscopy, we synthesized a bovine mt tRNASerGCU transcript using T7 RNA polymerase and measured its 1H-NMR spectrum in the imino proton region.41) Although the imino proton signals heavily overlapped, we succeeded in assigning all seven proton signals of the G-C base pairs by a combination of base replacement and 15N-labeling of the G residues of a whole tRNA molecule or of the 3′ half fragment (Fig. 1b). The results indicate that the tRNA possesses the secondary structure that was proposed on the basis of biochemical studies to discriminate between single strand and double strand regions using nuclease digestion and/or chemical probing.37),74),76) Analysis of the effect of the magnesium concentration on the G-C pairs suggests that the acceptor stem and T stem do not form a co-axial helix, and that the core region of the tRNA does not interact with magnesium ions. These features are significantly different from those of canonical tRNAs. Despite this, it is very likely that the tRNA as a whole adopts a nearly L-shaped tertiary structure, rather than the “boomerang” model of Steinberg et al.72)
Fig. 1.
Clover-leaf structures of tRNASerGCU of E. coli (a) and bovine mitochondria (b), and tRNASerUGA of bovine mitochondria (c), in which an additional base-pair in the anticodon stem is boxed.
Bovine mt tRNASerUGA was thought to have two U-U mismatches at the top of the acceptor stem, as inferred from its gene sequence.8) We isolated the tRNA by a hybridization method and determined its complete sequence including the modified nucleotides.39) Analysis of the 5′-terminal nucleotide and enzymatic determination of the whole sequence of tRNASerUGA revealed that the tRNA started from the third nucleoside G of the putative tRNASerUGA gene, which had been formally proposed.39) Enzymatic probing of tRNASerUGA suggested that the tRNA possesses an unusual cloverleaf structure with the following characteristics (Fig. 1c). 1) There is only one nucleotide between the acceptor stem and the D stem. 2) The anticodon stem seems to consist of six base pairs. Since the same type of cloverleaf structure as above could be constructed only for mt tRNAsSerUGA of mammals such as human, rat and mouse, but not for those of non-mammals such as chicken and frog, this unusual secondary structure seems to be conserved only in mammalian mitochondria.39)
1H-NMR analysis of the tRNASerUGA transcript77) showed that it adopts a nearly L-shaped tertiary structure with tertiary base pairings similar to those found in yeast tRNAPhe, which is representative of canonical tRNAs. However, magnesium ion titration revealed that Mg2+ affects the chemical shifts of the tRNASerUGA transcript differently than those of canonical tRNAs so far studied; the former is less sensitive toward Mg2+, especially in the D arm region. This observation was confirmed by NMR analysis with paramagnetic manganese ion titration. Hill plots derived from circular dichroism spectral changes caused by titration with Mg2+ suggested that the tRNASerUGA transcript has fewer Mg 2+ binding sites than does yeast tRNAPhe as well as its transcript, a finding that was consistent with the NMR data. We thus assume that the thermal instability of both the transcript and tRNASerUGA itself originates from a reduction in the number of divalent ion binding sites within the tRNA molecule. These results suggest a new type of thermal instability for mt tRNA. Recently, the three-dimensional structure of the pyrro-lysyl-tRNA synthetase-tRNAPyl complex was determined by X-ray analysis at 3.1 Å resolution, which showed that the tertiary structure of tRNAPyl has an L-shaped structure similar to that of canonical tRNAs.78) The structural characteristics of tRNAPyl are quite similar to those of mt tRNASerUGA, except for the absence of the D loop-T loop interaction, in that there is an elongated anticodon stem of six base pairs, only one nucleotide at the junction connecting the acceptor and D stems, a short variable region of only three bases, and a small D loop of only five bases. This is also consistent with the result of the 1H-NMR analysis of mt tRNASerUGA described here.77)
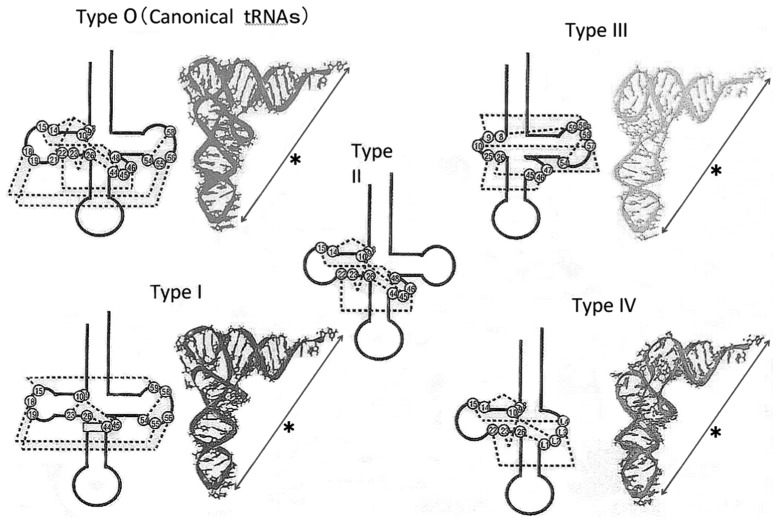
We have analyzed other mt tRNAs from bovine (tRNAPhe40)) and nematode (tRNASerUCU,48) tRNAMet49),79) and tRNAPhe48)) sources and came to the following conclusions. 1) The mt tRNAs so far known can be folded into a similar L-shaped tertiary structure, in which the distance and mutual orientation between anticodon and CCA terminus are preserved. This structure is prerequisite for mt tRNAs to function on the ribosome in the translation process as described below. 2) mt tRNAs can be classified into five groups on the basis of their structural characteristics72),73),77) (Fig. 2): mt tRNAs in the first group (type I), which includes tRNASerUGA, possess an almost normal cloverleaf secondary structure, in which the conserved GG and UΨC sequences are present in the D and T loops, respectively, but an additional base pair is in the anticodon stem.39) A unique point is that they have the lowest melting temperature among the mt tRNAs so far examined,39),73) probably because they lack two Mg2+-binding sites. tRNAs in the second group (Type II) apparently have the usual cloverleaf structure, but lack the conserved GG and UUC sequences in the D and T loops, respectively, that are necessary for the D loop-T loop interaction.40) Those in the third group (Type III), such as tRNASerGCU,74) are without the entire D arm. The fourth group (Type IV) entirely lacks the T loop, and this absence is characteristic of most nematode mt tRNAs.48),49) The fifth group (Type 0) includes a few mt tRNAs with canonical secondary/tertiary structures possessing the conserved GG and UUC sequences that form the D loop-T loop interaction (for example, tRNASerUGA in non-mammalian mitochondria39)), which are not discussed here.
Fig. 2.
Secondary (black) and tertiary structures (colored) of canonical tRNAs (Type 0) and animal mitochondrial tRNAs (Type I~Type IV). All tRNAs share the constraint that the distance (shown by*) and the mutual orientation between the anticodon and the CCA terminus are constant. Dotted lines in the secondary structure show internal base interactions.
Why almost all animal mt tRNAs have such unorthodox structures and exhibit thermal instability, and why such characteristics are necessary for the functioning of the mt translation system, remains unclear. However, all the mt components used for mt translation, such as mt tRNAs, aminoacyl-tRNA synthetases, elongation factors, ribosomes and rRNAs, appear to have been simplified during the evolution of mitochondria, probably in parallel with genome economization,80) which would have brought about the structural characteristics of mt tRNAs that exist today.
2. Do aberrant mt tRNAs function in the translation system?
Clarification that mt tRNAs are indeed functional requires an in vitro mt translation system. For this purpose, we asked L. Spremulli, University of North Carolina, US, to collaborate with us, since she is a specialist in mitochondrial translation enzymes and ribosomes. Under her instruction we constructed an in vitro translation system of bovine mitochondria. The first version was a homologous in vitro poly(U)-directed poly(Phe) synthesis system using bovine mt ribosomes, elongation factors and phenylalanyl-tRNAPhe81) (Type II in Fig. 2). The efficiency of incorporation of Phe into poly(Phe) in this mt system was several times lower than that for the homologous E. coli system. However, we found later that the addition of 1 mM spermine to the translation system enhanced the efficiency to a level similar to that observed in the homologous E. coli system.56)
Next, we tried to determine if mt tRNAMet (Type II) possessing f5C at the anticodon wobble position decodes not only the AUG codon, but also the AUA codon using the above-mentioned mt translation system.56),82) The native mt tRNAMet could translate both AUA and AUG codons as Met, but the corresponding synthetic tRNAMet lacking f5C (anticodon CAU) translated only the AUG codon in codon-dependent ribosomal binding and in vitro translation assays. Furthermore, the E. coli elongator tRNAMetm with the anticodon ac4CAU (ac4C: 4-acetylcytidine) and the bovine cytoplasmic initiator tRNAMet (anti-codon CAU) translated only the AUG codon for Met on the mt ribosome.56) These results demonstrate that the f5C modification in mt tRNAMet plays a crucial role in decoding the non-universal AUA codon as Met. Since this translation reaction also works with the E. coli ribosome, we concluded that genetic code variation is compensated for by a change in the tRNA anticodon, not by a change in the ribosome.
Finally, the translation activities of tRNASerGCU (Type III) and tRNASerUCN (Type I) were examined.83) At first, both tRNAs were altered so as to be charged with Ala by E. coli Ala-tRNA synthetase and to decode poly(U) as a messenger, by replacing the third base-pair of the acceptor stem with G ·U84) and the anticodon with GAA. Such altered tRNAs should be able to synthesize poly(Ala) depending on poly(U). The results were that both tRNAs could translate poly(U) to synthesize poly(Ala) on the ribosome, but the efficiency with the tRNASerGCU analogue was several times lower than that with the tRNASerUGA analogue. Although the tRNASerUGA analogue produced oligopeptides longer than 10mers, the product of the tRNASerGCU analogue was mostly short oligopeptides, up to a tetramer.83) Both tRNASer analogues had almost the same ability to form ternary complexes with the mt elongation factor Tu (EF-Tu) and GTP, but the ability to bind the ribosomal A site was several times lower for the tRNASerGCU analogue than for the tRNASerUGA analogue. These results show that the tRNASerGCU analogue has a molecular disadvantage on the ribosome, which is probably due to the lack of a D arm. Second, using a synthetic mRNA consisting of a Shine-Dalgarno sequence followed by an AUG codon, a ten-fold repeat of the UUC codon, a serine codon (UCA or AGC), a CAU codon and a stop codon (UAA), we compared Ser-tRNASerUGA and Ser-tRNASerGCU with respect to the synthetic mRNA-dependent incorporation of [3H]Ser in the presence of Met-tRNAMet, Phe-tRNAPhe and His-tRNAHis. We observed that the incorporation efficiency of tRNASerGCU was less than half that of tRNASerUGA. Third, we used a dihydrofolate reductase (DHFR) mRNA in which the sixth GCG codon was replaced by the UAG amber codon, and tested whether tRNASerUGA and tRNASerGCU analogues in which the anticodon was replaced by CUA so as to correspond to the amber codon could suppress the amber codon and produce the full-length DHFR protein. Both tRNASerUGA and tRNASerGCU analogues could form the DHFR band on an agarosegel, although the band formed by the tRNASerGCU analogue was much thinner than that formed by the tRNASerUGA analogue (Hanada, T., Suzuki, T. and Watanabe, K., in preparation). All these experimental results indicate that both mt tRNASerGCU and tRNASerUGA with unusual secondary structures are capable of translation on the ribosome, although the translation efficiency is much lower for tRNASerGCU than for tRNASerUGA, which is probably due to the absence of a D arm for tRNASerGCU.
Thus, it was clear that the Type II mt tRNAs, tRNAPhe and tRNAMet, tRNASerGCU (a Type III tRNA) and tRNASerUGA (a Type I tRNA) (see Fig. 2), can all function in the mt in vitro translation system, irrespective of the presence or absence of the D loop-T loop interaction or of a D arm. Therefore, these results provide experimental evidence for the presumption that if the distance and orientation between the anticodon and CCA terminus are preserved in all mt tRNAs with unusual structures (Fig. 2), these could function in the mt translation system.
3. Unilateral recognition mechanism of enzymes and ribosomes in mitochondrial and E. coli translation systems
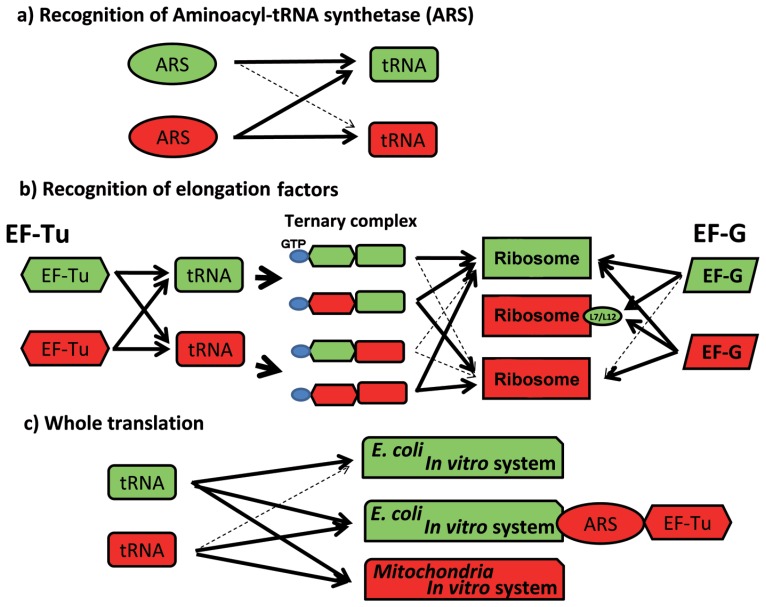
To elucidate the molecular basis by which aberrant mt tRNAs function in the mt translation system, we compared interactions of tRNAs with enzymes (aminoacyl-tRNA synthetase [ARS] and EF-Tu) and ribosomes in the mt system and the E. coli system, focusing on the exchangeability of components in these two systems.
In the recognition of tRNA by aminoacyl-tRNA synthetase, a unilateral aminoacylation specificity was observed for bovine mitochondria and eubacteria:85) mt phenylalanyl-, threonyl-, arginyl- and lysyl-tRNA synthetases (PheRS, ThrRS, ArgRS and LysRS) were shown to charge and stringently distinguish cognate E. coli tRNA species from noncognate ones, as did the corresponding E. coli synthetases. In contrast, mt seryl-tRNA synthetase (SerRS) not only charged cognate E. coli tRNASer species, but also extensively misacylated noncognate E. coli tRNA species. These results suggest a certain conservation of tRNA recognition mechanisms between the mt and E. coli ARS’s in the former four synthetases which are most likely to recognize anticodon sequences, but such a conservation of tRNA recognition mechanism does not exist in SerRS which recognize no anticodon sequence. On the other hand, these eubacterial ARS’s could not charge cognate mt tRNAs85) (Fig. 3a). This unilateral bias in aminoacylation implies that the tRNA recognition mechanism of mt ARSs may have evolved to be, to some extent, simpler than their eubacterial counterparts in response to reductions in the number of animal mt tRNA species, and to simplifications of their structures. Any combination of mt and E. coli or Thermus thermophilus86) components is functional in ternary complex formation among EF-Tu, GTP and aminoacyl-tRNA (aa-tRNA);81) however, in the ribosomal A site binding of aa-tRNA from the ternary complex, mt EF-Tu functions on either ribosome, but E. coli EF-Tu is inactive on the mt ribosome81) (Fig. 3b). This also shows the unilateral bias of the ribosomal A site binding of aa-tRNA, suggesting a dual function of mt EF-Tu.
Fig. 3.
Unilateral recognition relationships of tRNA with ARS (a), EF-Tu and EF-G (b), and ribosomes in whole translation reactions (c) in E. coli (green) and bovine mt (red) translation systems. Bold arrows and dotted arrows show functional and nonfunctional reactions, respectively.
It has been reported that while mt EF-G can function on both the mt ribosome (55S) and the E. coli ribosome (70S), E. coli EF-G cannot function on the mt ribosome.87),88) Again, this demonstrates unilaterality in the translocase activity of EF-G. We prepared hybrid mt and E. coli ribosomes to investigate their functional equivalency. A hybrid mt ribosome containing E. coli L7/L12 instead of mt L7/L12 clearly activated the GTPase of E. coli EF-G and efficiently promoted its translocase activity in an in vitro translation system89) (Fig. 3b). This result demonstrates that the functional compatibility between EF-G and the L7/L12 protein in the ribosome governs its translational specificity. Thus, the mt ribosome is functionally equivalent to the E. coli ribosome despite their distinct compositions.
To summarize these results, E. coli tRNA can function in both E. coli and mt systems, but mt tRNA cannot function in an E. coli translation system. However, the E. coli system into which mt ARSs and EF-Tu are added can accept mt tRNAs as substrates (Fig. 3c). Ribosomes are functionally compatible between mitochondria and E. coli, but components such as ARS and EF-Tu, which interact directly with mt tRNAs, need specific assistance to compensate structurally and functionally for unusually truncated mt tRNAs, a few example of which are described below.
4. Structural and functional compensation of truncated RNA segments by proteins in mitochondrial translation systems
In this study, the interactions of ARS, EF-Tu and ribosomal proteins with mt tRNAs and ribosomal RNA (rRNA) are viewed as examples of compensation.
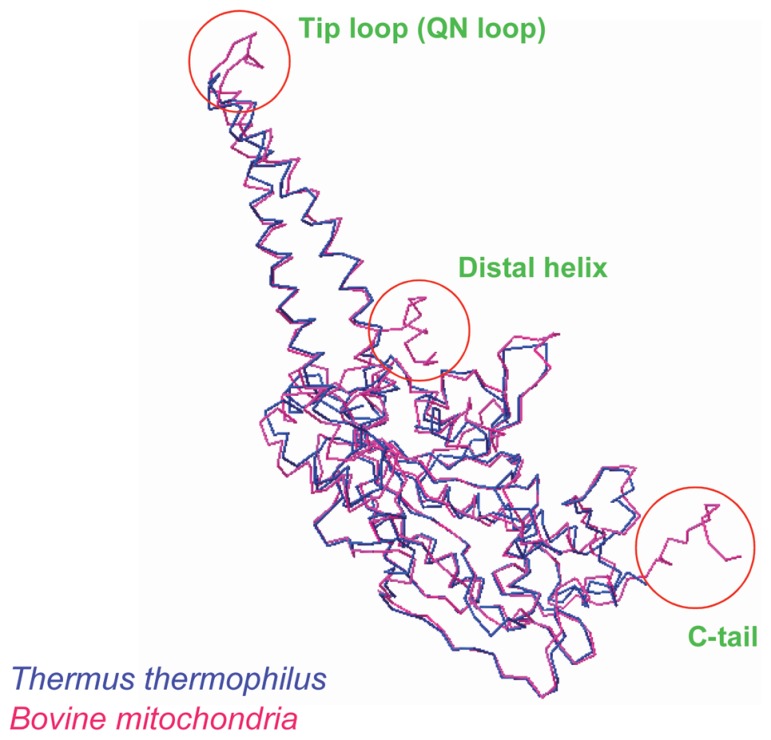
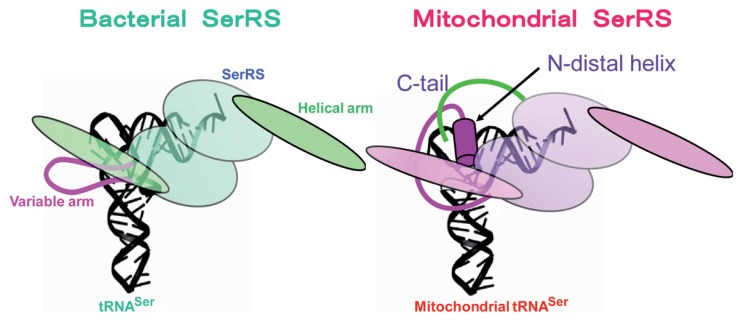
As described above, mt ARS’s can charge both mt and E. coli tRNAs, but E. coli ARS’s cannot charge mt tRNAs (Fig. 3a). We focused on the structures of both mt and bacterial SerRS, because bacterial SerRS is known to recognize the large variable arm of tRNASer as an identity determinant,90),91) but none of the mt serine isoacceptor tRNAs possesses the elongated variable arm, and their secondary structures are quite different from each other (Fig. 1). As the crystal structure of bacterial (T. thermophilus) SerRS has been determined,92) we analyzed bovine mt SerRS complexed with seryl adenylate at an atomic resolution of 1.65 Å,93) in collaboration with the laboratory of J. Nyborg at Aarhus University, Denmark. Figure 4 shows a superposition of the Cα chain of mt SerRS (red line) onto the known structure of T. thermophilus SerRS (blue line), showing that the tertiary structures of the two SerRS’s are almost identical, except for three domains in mt SerRS that are absent from bacterial SerRS: the Distal helix in the N-terminal region (39 amino acids), the Tip loop (QN loop) at the top of the two long α helical arms (10 amino acids) and the C tail in the C-terminal region (24 amino acids). Although the two long α helical arms are responsible for recognition by the long variable arm of bacterial tRNASer,92) replacement of several Arg residues with Ala in these regions minimally influenced the aminoacylation activity of mt SerRS. However, the aminoacylation activities of mt SerRS towards mt tRNASerGCU and tRNASerUGA were severely influenced by deletion mutants of the Distal helix and the C tail in a different manner: the Ser acceptor activity of tRNASerGCU was lowered to one-fifth of control levels by deletion of the Distal helix. On the other hand, the Ser acceptor activity of tRNASerUGA was lowered to one-fifth and one-half of control levels by deletion of the Distal helix and the C tail, respectively.93) Namely, the Distal helix is indispensable for both tRNAsSer, but the C tail is necessary for only tRNASerUGA, which may mean a dual-mode recognition of SerRS toward two distinct aberrant mt tRNAsSer by an alternative combination of interaction sites. Thus, it seems that mt SerRS compensates for the truncated parts of mt tRNASer as compared with the canonical tRNAs, with extensions of both the N and C termini (Fig. 5).
Fig. 4.
Superposition of the Cα chain of bovine mt SerRS (red) onto the known structure of T. thermophilus SerRS (blue). The N-terminal helical region (Distal helix), the top region of the two long α helical arms (Tip loop) and the C-terminal region (C-tail) are additional regions in bovine mitochondria (circled) not present in T. thermophilus SerRS.
Fig. 5.
Model of tRNA recognition by SerRS in bacterial (left) and mitochondrial systems (right). The truncated parts of mt tRNA are compensated for by the Distal helix (red rod) for both tRNASerGCU and tRNASerUGA and the C tail (green line) for tRNASerUGA of mt SerRS.
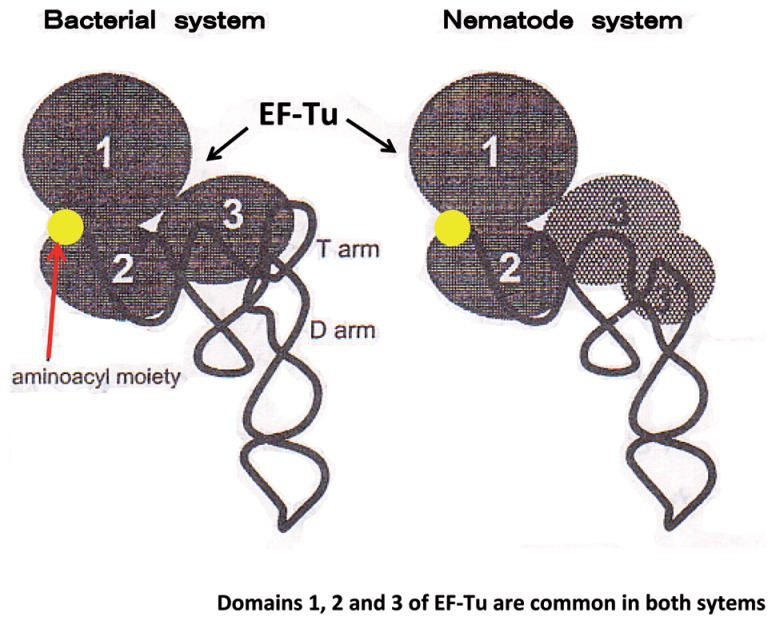
It is well known that conventional EF-Tu from prokaryotes and eukaryotes bind to tRNAs by recognizing amino acid-acceptor stems and T stems of aminoacyl tRNAs.94),95) However, in nematodes, 20 mt tRNA species of a total of 22 species lack the T arm, and the remaining two species (tRNASerUCU and tRNASerUGA) lack the D arm.18) T. Ohtsuki et al. in my laboratory found two different sequences that encode mt EF-Tu homologs in the nematode (C. elegans) mt genomic and cDNA partial sequence databases. The corresponding proteins designated EF-Tu196) and EF-Tu297) were over-expressed in E. coli. Both proteins have domains 1–3 in common with bacterial EF-Tu, but possess additional amino acid extensions (57 and 17 residues) at their C termini. An extremely long C-terminal extension in EF-Tu1 was named Domain 3′. In examining the interaction of the EF-Tu’s with tRNAs, we found that EF-Tu1 binds to T arm-lacking tRNAs, but not to D arm-lacking tRNAs or canonical tRNAs having a normal cloverleaf structure.96) An experiment in which domains between the mt and T. thermophilus systems were exchanged confirmed that Domain 3′ is prerequisite for the binding of EF-Tu1 to T arm-lacking tRNA.96) The results of these experiments indicated that Domain 3′ of EF-Tu1 compensates for the T arm-lacking space in the T arm-lacking tRNA, so that the EF-Tu1-T arm-lacking tRNA complex forms a structure similar to that of the usual EF-Tu-tRNA complex, which enables the complex to bind to the ribosome in a manner similar to that seen in conventional systems (Fig. 6). On the other hand, EF-Tu2 binds to D arm-lacking tRNAs, but not to T arm-lacking tRNAs.97) Judging from the amino acid sequence of EF-Tu2 and the biochemical data, it is very likely that EF-Tu2 recognizes not only the tRNA molecule itself, but also the aminoacyl moiety attached to the tRNA. It turned out that nematode EF-Tu2 has a strong specificity for the aminoacyl moiety of seryl-tRNA.97) Thus, it became clear that nematode mitochondria use two distinct forms of EF-Tu’s for recognizing the two different classes of tRNAs. Each of the nematode mt EF-Tu species appears to have acquired a specific mechanism to compensate for the structural deficiencies of nematode mt tRNAs. In particular, the case of the binding of EF-Tu1 with T arm-lacking tRNA is a typical second example of structural and functional compensation of truncated RNA segments by proteins.
Fig. 6.
Schematic representation of tRNA complexed with EF-Tu in bacterial (left) and nematode mt systems (right). An extended C-terminal region (57 amino acids) of nematode mt EF-Tu, Domain 3′, compensates for the absence of the T arm in nematode mt tRNA.
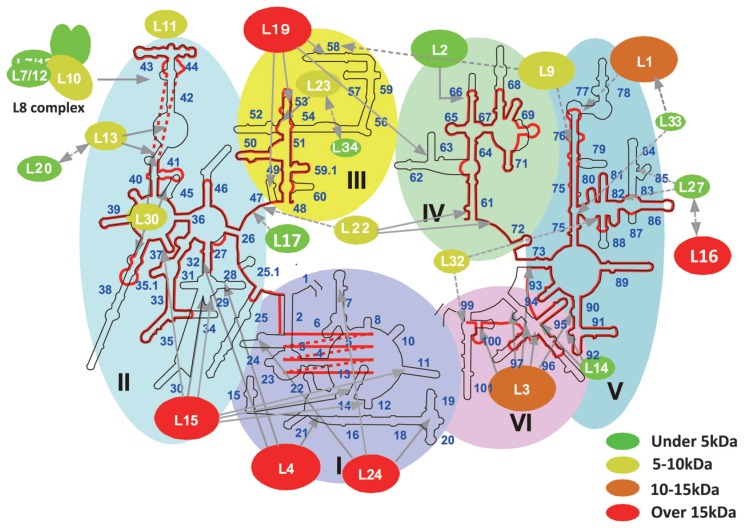
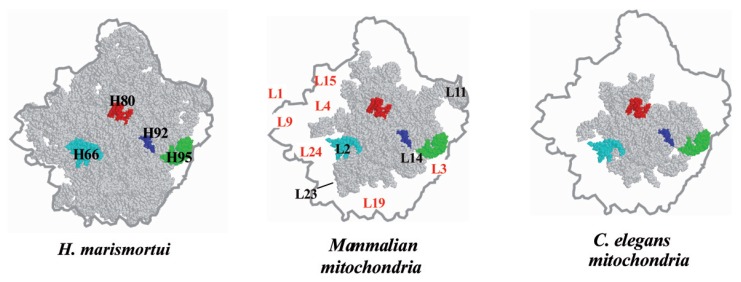
The third case concerns the mt ribosome. The mammalian mt ribosome is a smaller 55S particle compared with the E. coli 70S particle, but the molecular mass of the mt ribosome is 3.57 MDa, larger than that of E. coli (2.49 MDa), as estimated in a physiological study.98) The mt ribosome consists of a large 39S and a small 28S subunit, each of which contains 16S and 12S rRNAs, respectively,99) but there is no small rRNA equivalent to the 5S RNA in the E. coli ribosome. The protein to RNA ratio is completely reversed between these two ribosomes, and the protein composition of the mt ribosome is estimated to be about 75%. T. Suzuki and his coworkers in our laboratory identified 55 mt ribosomal proteins by using two-dimensional electrophoresis and LC/MS/MS analysis, and further searching of the EST database.100)–109) They succeeded in specifying the proteins in the mt genome. Since the crystal structure of the ribosomal 50S subunit of an archaeon, Haloarcula marismortui has been determined,110) it is possible to identify where each ribosomal protein binds in the rRNA. Figure 7 shows the secondary structure of a large mt ribosomal RNA (16S rRNA) (red line) superimposed on that of a corresponding bacterial large subunit rRNA (black line),111) on which mt ribosomal protein subunits are mapped.108),110) An intriguing finding is that mt ribosomal proteins that bind to the well-conserved binding sites of mt 16S RNA have molecular masses similar to those of their bacterial counterparts. Among the supporting evidence for conservation are the following observations: helices 66, 61, 53, 43/44 and 95 of rRNA in the bacterial large ribosomal subunit,110) which are well conserved in mt rRNA, serve as the main binding sites for proteins L2, L22, L23, L11 and L14, respectively, whose mt ribosomal homologues are of similar sizes. In contrast, mt ribosomal proteins, having reduced or no distinct binding sites on mt rRNA, have larger masses, which are caused by extensions at their N or C termini.109) L24, L4 and L15 contact many sites of Domains I and II of bacterial large subunit rRNA,110) whereas the binding sites for mt homologues are absent from mt rRNA (Fig. 7). Thus, the molecular masses of the mt L24, L4 and L15 homologues are more than 15 kDa greater than those of bacterial L24, L4 and L15. The mt L19 homologue, of which binding sites on mt rRNA have been almost completely lost, also has a much greater mass than that of its bacterial counterpart. The absence of helices 77 and 78 in mt rRNA seems to be compensated for by the extended termini of the mt L1 and L9 homologues. The binding sites of L3 on helices 94 and 100 of domain VI in bacterial 23S rRNA are also missing from mt 16S rRNA, and thus the mt L3 homologue possesses long N-and C-terminal extensions. These observations strongly suggest that enlarged mt ribosomal proteins can compensate for the deficit in mt ribosomal RNAs.108),109) To confirm these observations, three-dimensional models of human and nematode (C. elegans) mt rRNAs were constructed based on crystal structure data of the bacterial 50S ribosome110) (Fig. 8). We have measured the sedimentation coefficient (55S), buoyant density (1.40 g/cm3) and diffusion coefficient ((1.48 ±0.04) × 10−7cm2s−1) of another nematode, Ascaris suum, and determined that the nematode mt ribosome does not differ significantly in size from the mammalian mt ribosome, although the nematode mt ribosome has a much higher protein composition than do other mt ribosomes.112) The rRNA that almost overwhelms the inner surface of the bacterial 50S particle (gray part in Fig. 8) is greatly reduced in mammalian and C. elegans mt ribosome particles, while the region containing the functional center with peptidyl transferase, which is surrounded by the principal helices H66 (light blue), H80 (red), H92 (blue) and H95 (green) is conserved. Several rRNA portions missing from the mt ribosome, which are localized discontinuously in the secondary structure (Fig. 7), form large missing domains as shown by the surrounding white portions in the figure, and these are located at the central protuberance, at the bottom, the back and the left side. Several ribosomal proteins can be seen to occupy the missing rRNA portions in the mt ribosomes. The rRNA portions binding to the ribosomal protein sites seem to be shortened and replaced by other enlarged ribosomal proteins. Thus, the extended N or C termini of mt ribosomal proteins fill up the empty spaces in mt rRNAs that are occupied by rRNA in bacteria. According to sequence comparisons of mt rRNAs from various organisms, the rRNA shortening can be found in mitochondria from protozoan, Drosophila, Xenopus and mammalian sources, but yeast and plant mitochondria have mt rRNAs that are similar in size to bacterial rRNA. Thus, it can be assumed that compensation for the RNA deficit in mt rRNAs by proteins occurred in the early stage of animal mt evolution.
Fig. 7.
Secondary structures for large ribosomal RNAs of bacteria and mammalian mitochondria, shown with interacting ribosomal proteins.108) The secondary structure of human mt 16S RNA (red line) was superimposed on the secondary structure of E. coli 23S rRNA (black line), which is shown according to the format of Ban et al.110) The 5′ region of mt 16S rRNA (about 160 bases) could not be aligned with domain I of bacterial 23S rRNA. Ovals that represent large ribosomal proteins are mapped onto the secondary structure of rRNA, with interactions indicated by gray arrows. Solid arrows show interaction maps that were identified by the crystal structure of the 50S subunit.110) Broken arrows indicate interaction maps obtained from biochemical studies. The oval size of each protein indicates its molecular weight relative to that of its E. coli counterpart. The difference in molecular weight between the mt ribosomal protein and its E. coli counterpart is indicated by color: red, more than 15 kDa bigger than the E. coli counterpart; orange, 10~15 kDa; yellow, 5~10 kDa; green, less than 5 kDa. The colored large shaded regions represent the six domains from which the ribosome was constructed.
Fig. 8.
Three-dimensional models for large mt ribosomal RNA (gray) from mammalian (middle) and C. elegans (right) mitochondria, based on the crystal structure of a bacterial 50S subunit108) (left). The outline shows an edge line of the crystal structure of the 50S subunit from the crown view. Some functional rRNA domains are colored: red, P loop; blue, A loop; green, S/R loop; light blue, L2 binding helix (H66). The topological orientation of the ribosomal protein is based on the model for the mammalian mt ribosome.
In summary, structural and functional compensations of truncated RNA segments by proteins are seen in ARS, EF-Tu and ribosomes in the mt translation system.
III. Novel modified nucleotides found in human mitochondrial tRNA are relevant to mitochondrial diseases
With the development of a new technique, “chaplet” column chromatography, to isolate individual mt tRNAs in a short time with sufficient recovery, we extended our analyses to human mt tRNAs, which has led us to elucidate the molecular mechanisms of human mt diseases caused by point mutations in mt tRNA genes.
1. Novel modified uridines containing a taurine side-chain are found in human mt tRNAs
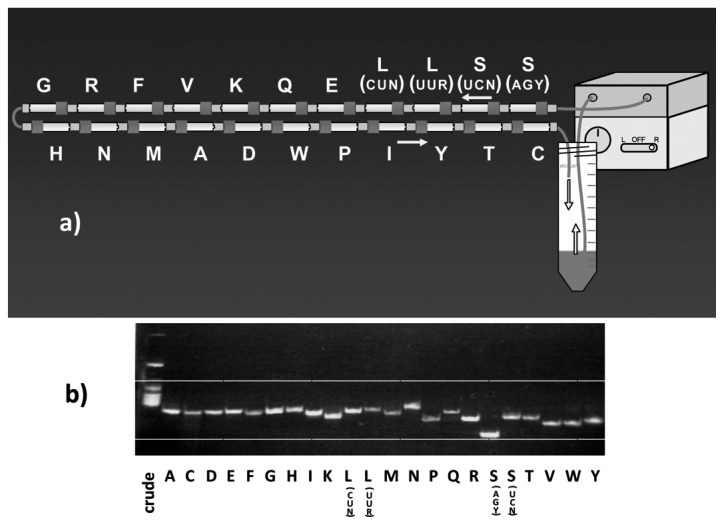
For isolation of mt tRNAs, we have adopted a hybridization assay as the detection method (Chapter I-2). However, the conventional column chromatography techniques followed by detection using a hybridization assay with a DNA probe were very troublesome and time-consuming for isolating mt tRNAs with a low cellular abundance. To overcome this difficulty, T. Suzuki and his coworkers in my laboratory developed a new tRNA isolation technique57),113),114) employing a solid-phase DNA probe method that takes an affinity chromatographic approach, the principle of which was originally devised by H. Tsurui et al.115) In this method, to entrap the desired tRNA, a biotinylated DNA probe complementary to the target mt tRNA is immobilized on streptavidin Sepharose, and a crude tRNA fraction that includes about 0.1% of each mt tRNA is circulated through the column by a pump at 65 °C. Continuous circulation was proved to be the most important factor in maximizing the individual tRNA yields. To isolate individual mt tRNAs, 22 columns, each specific for a different tRNA, were connected in tandem (Fig. 9a), and the crude tRNA sample was loaded onto the columns by circulation. After circulation, each column was isolated and each tRNA was eluted. Thus, it was possible to simultaneously isolate all 22 individual mt tRNA species from the same crude tRNA preparation using this approach (Fig. 9b). This system was named “chaplet” column chromatography.114) A crude tRNA fraction from bovine liver was then circulated through the chaplet columns, and 0.5~1 mg of individual mt tRNAs could be isolated from 1 kg of bovine liver as well as from human placenta by this single chromatographic step within one week. The purity of each tRNA was close to 100%.
Fig. 9.
a) Schematic of “chaplet” column chromatography. The DNA columns, in which a 3′-biotinylated DNA probe complementary to each mt tRNA is immobilized on streptavidin sepharose, are tandemly connected. Crude tRNA fractions from bovine liver or human placenta are circulated through this chaplet column to entrap each mt tRNA. b) Polyacrylamide gel electrophoretic pattern of purified mt tRNAs obtained by chaplet column chromatography. Each tRNA is isolated with almost 100% purity.
Thus, we obtained 22 species of mt tRNA corresponding to 60 sense codons, which forms the minimal decoding system in extant living organisms (Table 1). First, the tRNA anticodons were analyzed by conventional nuclease digestion followed by the two-dimensional thin-later chromatographic analysis, in order to relate them to the codons. Four tRNAs (tRNAPhe, tRNAIle, tRNACys and tRNASer(AGY)) responsible for two pyrimidine-ending codon sets have G at the wobble position (Category III in Table 3), while eight tRNAs (tRNALeu(CUN), tRNAVal, tRNASer(UCN), tRNAPro, tRNAThr, tRNAAla, tRNAArg and tRNAGly) corresponding to family boxes have U (Category I). The remaining ten tRNAs have four kinds of modified nucleosides at the wobble position, which were identified by LC/MS analysis. Four tRNAs (tRNATyr, tRNAHis, tRNAAsn and tRNAAsp) have Q,61) which corresponds to U or C in the two pyrimidine-ending codon sets (Category III). tRNAMet has f5C at its wobble position, as described in Chapter II-2, which is responsible for the codon change from AUA-Ile to AUA-Met56) (Category III). The remaining five tRNAs have two new modified nucleosides at the wobble position. tRNALeu and tRNATrp have a novel uridine derivative with a molecular mass of 381 Da, while tRNALys, tRNAGlu and tRNAGln have a 2-thiouridine derivative with 397 Da.57) These modified uridines can be included in Category III in Table 3.
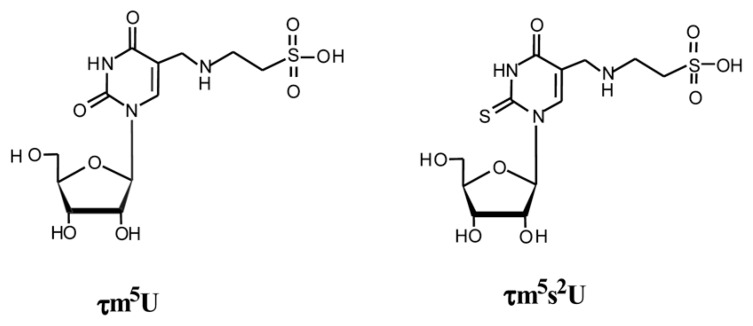
NMR analysis of the purified nucleosides showed the absence of an H6 proton cross peak in the 1H-COSY spectrum, which indicates the presence of a substituent at position 5 in the uracil ring. By Fourier transform-ion cyclotron resonance mass spectrometry, the molecular weight of the 2-thiouridine derivative was determined and its atomic composition was ascertained with excellent accuracy (0.03 p.p.m.) to be C12H19N3O8S2. These findings indicate that the main modification occurs at position 5 in the uracil base, and the most plausible structure is a taurinomethyl moiety possessing a sulfonic acid group derived from taurine. The 2-thiouridine derivative possesses an additional sulfur at position 2. The two nucleosides were named 5-taurinomethyluridine (τm5U) and 5-taurinomethyl-2-thiouridine (τm5s2U) (Fig. 10). Then, we asked K. Saigo and T. Wada of the University of Tokyo, to chemically synthesize the former compound, and we finally identified the novel uridine derivative as τm5U by comparing the synthetic products using LC/MS and NMR.57) Thus, it was clear that τm5U is contained in tRNALeu and tRNATrp, whereas τm5s2U is contained in tRNALys, tRNAGlu and tRNAGln at their anticodon wobble positions.
Fig. 10.
Chemical structures of τm5U (left) and τm5s2U (right).
Our observations show that while there are only four species of wobble modifications, Q, f5C, τm5U, and τm5s2U, that are required for deciphering the minimal mt decoding system, they play essential roles in mt translation. This suggests that if the wobble base is not modified, mt translation will not function correctly, and the resulting proteins may be altered. This may lead to mt diseases. Supporting this notion, we have found human mitochondrial diseases that are caused by wobble modification defects of τm5U and τm5s2U in tRNALeu (UUR) and tRNALys, respectively (see section 3).
2. Biosynthetic mechanism of taurinome-thyluridines
In τm5U and τm5s2U, taurine is attached at the C5 position of the uracil ring through a methylene group. These taurine-containing uridines were found at the wobble position of mt tRNAs from human, bovine, cat, fish, and ascidian sources (our unpublished observations), whereas mt tRNAs from C. elegans had 5-carboxymethylaminomethyluridine (cmnm5U) at the corresponding wobble position, so that taurine-containing uridines seem to be common in vertebrate and prochordate mitochondria.
Taurine is one of the most abundant free amino acids in human body fluids including plasma, as well as intracellular contents,116) and it is thought to play physiological roles in bile salt synthesis, modulation of calcium fluxes, cardiac contractility, maintenance of photoreceptor cells, modulation of neuronal excitability, osmoregulation, and cell proliferation and viability in primates including humans and cats. However, the exact roles of taurine in these functions have not been clarified and there is no evidence that taurine is incorporated into macromolecules such as protein and RNA. To confirm that taurine is indeed a direct component of the modified uridines in the above-mentioned five species of mt tRNAs, we cultured HeLa cells with [18O]-labeled taurine for two days, isolated mt tRNA from the cells and subjected it to LC/MS analysis. Mass chromatogram analysis of total nucleosides in mt tRNALeu(UUR) and tRNALys revealed that τm5U and τm5s2U had increased mass due to the incorporation of [18O] taurine.57) This was the first observation that intracellular taurine is incorporated into mt tRNA, which indicated that the inner membrane of mitochondria has an active taurine transporter. This enables the transport of taurine from the cytoplasm to mitochondria, while plasma taurine is known to be transported into the cytoplasm through the high affinity taurine transporter on the cytoplasmic membrane. Thus, the taurine imported into mitochondria seems to be used as a constituent for τm5U synthesis in mt tRNAs.
3. Relevance to human mitochondrial disease – a modification defect of taurine in mt tRNAs is found in mitochondrial encephalomyopathies
It has been known that mtDNA mutations are responsible for a wide spectrum of human diseases that are caused by mt dysfunction. In particular, point mutations in mt tRNA genes are frequently found in mt diseases.117),118) In 1990, S. Ohta and his coworkers, at the Nippon Medical School, Japan, revealed that mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS), one of the major clinical subgroups of the mitochondrial encephalomyopathies, is caused by point mutation in the tRNALeu gene that is responsible for the translation of the UUR leucine codons (tRNALeu(UUR)).119),120) The majority (80%) of MELAS patients possess an A to G transition at nucleotide position (np) 3243,119),121) whereas in about 10% of the patients, a T to C transition is observed at position 3271 in maternally inherited diabetes with deafness,122) and in progressive external ophthalmoplegia.123),124) On the other hand, J. M. Shoffner et al. found an A to G transition at np 8344 in the tRNALys gene, in most patients with myoclonus epilepsy associated with ragged-red fibers (MERRF),125) another major clinical subgroup of the mitochondrial encephalomyopathies. Thus, it is clear that the clinical features of the mt diseases depend on the tRNA species and/or positions of the mutations (Fig. 11). However, the exact relationship between the location of the mutations and their clinical phenotypic consequences are not fully understood. To demonstrate that the three mutations in mt tRNA genes (A3243G, T3271C, and A8344G) are directly involved in the mt dysfunction associated with these mutations, cybrid cell lines have been studied, which were made by transferring mutant mtDNA derived from patients into human cells lacking mtDNA (ρ0 cells).126),127) In cybrid cells containing a high ratio of mutated MELAS mtDNA, both respiratory enzyme activity and protein synthesis were decreased.128),129) It was proposed in several papers that the MELAS mutations directly impair the proper function of tRNALeu(UUR) molecules, resulting in a decrease of mt respiratory activity in these patients.130),131) However, there has been no conclusive evidence showing that the point mutations are directly responsible for the mt dysfunction.
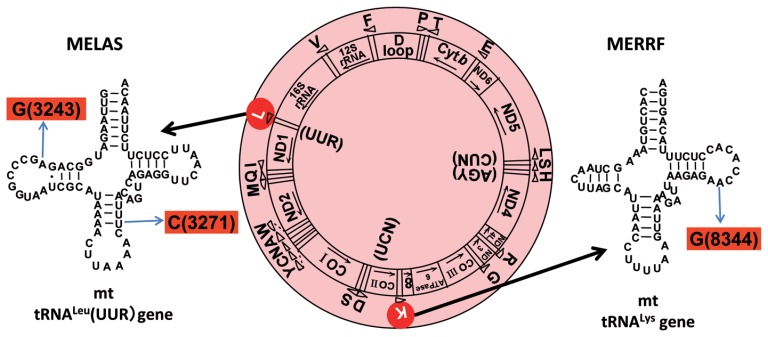
Fig. 11.
Gene organization of the human mt genome (center), in which MELAS (A3243G and U3271C) and MERRF (A8344G) point mutations are indicated in the tRNALeu(UUR) (left) and tRNALys genes (right), respectively.
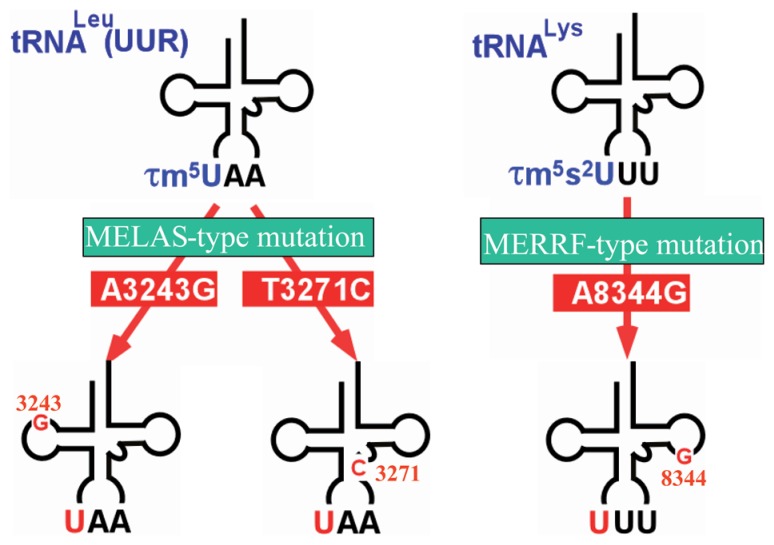
Therefore, by using cybrid cells possessing homo-plasmic pathogenic mutations, we, in collaboration with S. Ohta and his colleagues, isolated tRNALeu(UUR) and tRNALys by utilizing chaplet column chromatography. Through nucleotide analysis of these tRNAs, we found that mt tRNALeu(UUR) bearing the A3243G or T3271C mutation lacks the 5-taurinomethyl modification of τm5U, leaving an unmodified uridine at the anticodon wobble position (Fig. 12).132) All the other nucleotides, except for that of the mutation point, were identical between wild-type and mutant tRNAs. This result clearly explains why these different mutations (A3243G and T3271C) are associated with the same clinical phenotype. Using cybrid cells derived from MERRF patients, we also found that the mutant mt tRNALys bearing the A8344G mutation lacks both the 5-taurinomethyl and 2-thio modifications of τm5s2U at the wobble position (Fig. 12),57),133) the other residues being unchanged. We also detected taurine modification defects in tRNAs, not only from cybrid cells, but also from tissues of MELAS and MERRF patients.
Fig. 12.
Wobble modification defect in mutant tRNAs, tRNALeu(UUR) (left) and tRNALys (right) from the mt diseases MELAS3243 and 3271, and MERRF8344. These point mutations act as negative determinants for taurine-containing wobble uridines in the tRNAs.
Thus, it turned out that the two mt diseases MELAS and MERRF both lack the taurine modification of their respective mutant tRNAs. It is most likely that the point mutations in the mt tRNALeu(UUR) and tRNALys genes inhibit the biosynthetic pathway of taurine modification at the wobble position of the relevant tRNAs. As uridine modifications at the wobble position are known to be important for precise and efficient codon recognition59),134) (Table 3), a wobble modification deficiency may cause a serious defect in decoding.
4. Abnormal translation caused by modification deficiency of mutated tRNAs
Since MELAS cybrid cells bearing the A3243G or U3271C point mutations exhibit translational activity to different extents, the effect of point mutations in mt tRNAs on translation needs to be taken into consideration129),135) apart from the effect of the wobble modification deficiency. To separate these two effects, we first prepared four kinds of mt tRNAs: a wild-type tRNALeu(UUR) possessing τm5U at the wobble position, an “operated” tRNALeu(UUR) with a nucleotide sequence identical to that of wild type tRNALeu(UUR) except that it lacks only τm5U, an A3243G mutant tRNALeu(UUR) that lacks τm5U and possesses an A to G point mutation at position 3243, and a U3271C mutant tRNALeu(UUR) that lacks τm5U and possesses a U to C point mutation at position 3271. The latter two tRNAs are MELAS mutant tRNAsLeu(UUR) purified from the relevant mutant cybrid cells. The operated tRNALeu(UUR) was constructed by a molecular surgery technique136) as follows: the mt tRNALeu(UUR) (160 μg), isolated from human placenta (27 kg) by chaplet column chromatography, was cut in half at the wobble position with a hammerhead ribozyme, and τm5U in the 5′ half of the fragment was removed by periodate oxidation and replaced with an unmodified uridine by enzymatic ligation. The altered 5′ half was then religated with the 3′ half.137)
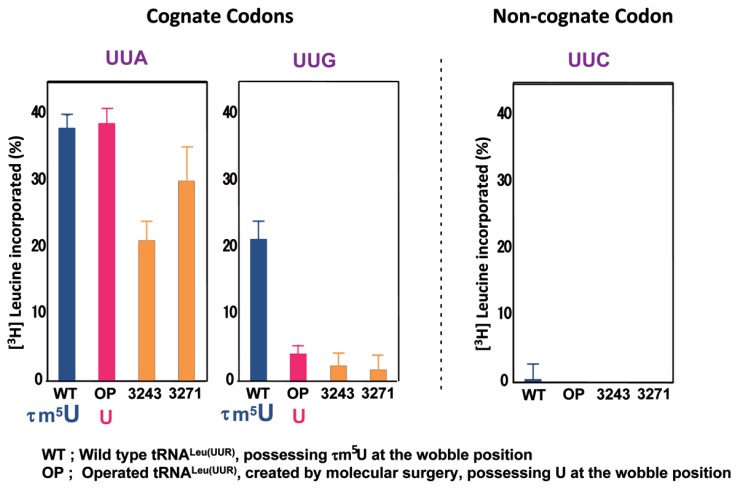
We then examined the translation activities of these four species of tRNALeu(UUR) using a mt in vitro translation system83) with synthetic mRNAs possessing 30 triplet repeats for the Leu codons UUA and UUG, as well as UUC as a negative control.137) As shown in Fig. 13, the wild-type tRNALeu(UUR) efficiently decoded both the UUA and UUG codons, although the maximum values were considerably different (the value for UUG decoding was ~60% of that for UUA decoding). The operated tRNALeu(UUR) showed the same activity as the wild-type tRNALeu(UUR) in UUA decoding, but a severe reduction (~82%) in UUG decoding. The A3243G mutant tRNALeu(UUR) showed a considerable reduction (~44%) in UUA decoding, as well as a severe reduction in UUG decoding (~89%). The U3271C mutant tRNALeu(UUR) also showed a moderate reduction (~21%) in UUA decoding, as well as a severe reduction (~92%) in UUG decoding.137) All four species of tRNAsLeu(UUR) had no activity with the noncognate UUC codon, implying that the experiments were appropriately controlled.
Fig. 13.
Translational activities of wild-type tRNALeu(UUR) (WT), operated tRNALeu(UUR) (OP), A3243G mutant tRNALeu(UUR) (3243), and U3271C mutant tRNALeu(UUR) (3271), in an in vitro mt translation assay using test mRNAs containing the UUA (left), UUG (center) or UUC (right) (negative control) codons.137) The radioactivity of the [3H]Leu-tRNA input in the reaction mixture was defined arbitrarily as 100%. The averages of three independent experiments with SD values are shown.
From these results, the following conclusions could be drawn. First, the severe reduction in UUG decoding by the MELAS mutant tRNAs can be mainly attributed to the lack of wobble modification. Second, the MELAS point mutations themselves impose a certain negative effect on translation, because considerable or moderate reductions in UUA decoding were observed for both the A3243G and U3271C mutant tRNAsLeu(UUR), but not for the operated tRNALeu(UUR), all of which lack τm5U and possess identical nucleotide sequences except for the mutations. The negative effect might arise from the fragile relaxed structure caused by the mutations, because the A3243G mutation could disrupt the potential tertiary interaction U8-A14-A21 observed in canonical tRNA,138) and the T3271C mutation destabilizes the anticodon stem.139) The MELAS tRNALeu(UUR) with the A3243G mutation showed a more severe reduction in UUA decoding than did the tRNALeu(UUR) with the T3271C mutation. This result is consistent with the translational activities of MELAS cybrid cells with these point mutations.129),135) To confirm that the wobble modification is responsible for UUG decoding, we carried out a ribosomal A site binding experiment. Native mt tRNALeu(UUR) bound efficiently to both the UUA and UUG codons, while the operated tRNA showed the same binding affinity to the UUA codon as compared with the native tRNALeu(UUR), but weak binding affinity (~18%) for the UUG codon.137) This finding suggests that the UUG codon-specific translational defect of the mt tRNALeu(UUR) lacking the wobble modification is caused by incomplete codon-anticodon base pairing on the ribosomal A site. The modified wobble uridine thus plays a functional role in decoding the UUG codon by stabilizing U:G wobble base-pairing on the ribosomal A site. These results therefore suggest that deficient decoding of the UUG codon arising from the lack of wobble modification is one of the primary causes of MELAS.
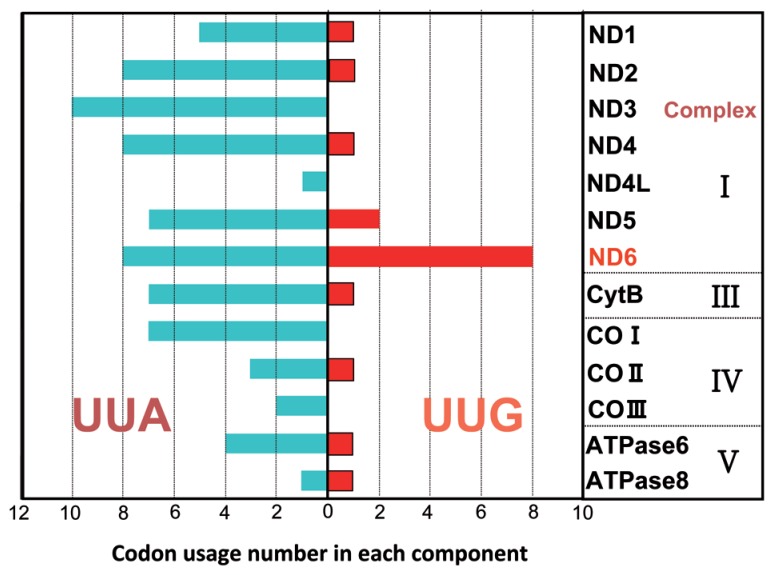
We have noticed a specific bias of Leu codon usage in the 13 proteins encoded by human mtDNA genes137) (Fig. 14). For example, despite the minor usage (0~2) of the UUG codon in most proteins, the ND6 gene, which is a component of respiratory chain complex I (NADH-coenzyme Q reductase), contains eight UUG codons that constitute 42.1% of the total Leu codons and 4.6% of the total codons in ND6. It has been reported that when the A3243G or T3271C mtDNA levels in cybrid cells are increased, the translational activity of ND6 is specifically and markedly reduced without a decrease in total mitochondrial protein synthesis.129),130) Furthermore, a point mutation (G14453A) in the structural gene for ND6 was found to be associated with a severe MELAS syndrome.140) Considering the UUG codon-specific translational defect described in this study, these facts support the idea that MELAS patients experience a translational depression of ND6. This idea nicely explains why a specific reduction of Complex I activity is characteristic of MELAS patients.141),142) These results indicate that the UUG codon-specific translational defect caused by defective wobble taurine modification is primarily responsible for the molecular pathogenesis of MELAS. In addition, our study suggests that the point mutation itself, in particular the A3243G mutation, contributes to the tRNALeu(UUR) translational defect to a considerable extent. Thus, the extent of the decoding disorder for each MELAS mutant tRNA varies with the effect of each pathogenic point mutation.
Fig. 14.
Usage of UUR Leu codons in 13 proteins encoded in human mtDNA. The numbers of UUA/UUG codons are shown for each gene.
We also examined the translational ability of the mutant mt tRNALys from MERRF patients, which bears the A8344G mutation. This analysis showed that tRNALys lacking the τm5s2U modification could not translate either of the cognate codons AAA and AAG. This defect is due to a complete loss of codon-anticodon pairing on the ribosome,143) because 2-thio modification of the wobble base is known to be critical for decoding AAR codons144) (Category III in Table 3). This result explains why MERRF patients show a marked defect in whole mt translation.143),145),146) Thus, the different symptoms exhibited by MELAS and MERRF patients may be explained by the fact that the mutant tRNAs lacking the wobble modification in these patients show a distinct pattern of codon recognition.
In summary, the molecular mechanism causing the mt dysfunction in MELAS patients has been unraveled. A point mutation at nucleotide position 3243 or 3271 in mtDNA results in a taurine modification deficiency at the anticodon wobble position of the mutant tRNALeu(UUR), which subsequently causes a UUG codon-specific translational defect that may lead to a translational depression of ND6. This defective codon-specific translation indicates that the deficiency in taurine modification could be a key to the expression of clinical phenotypes in mt diseases. Molecular surgery that changes the identity of the taurine-modification enzyme might serve as a useful clinical treatment for patients with the MELAS and MERRF encephalomyopathies.
Summary and perspectives
It is thought that mitochondria arose from the endosymbiosis of an aerobic eubacterium into an ancestral eukaryote.147) Thus, animal mt translation systems would have their origin in the eubacterial system and since then evolved separately. In the course of mt evolution, the mt genome size was reduced through the loss of spacer regions and the transfer of mt genes to the host genome.148) Eventually, through this process, the size of the metazoan mt genome was reduced to about 12~16 kb, which now codes for a total of 37 genes, including all of the RNA genes necessary for translation (22 tRNAs and two rRNAs) and the 13 subunit proteins that make up the respiratory enzyme complex. The shortening of animal mt genomes would have some advantages for the maintenance and/or proliferation of mitochondria; the first advantage would be to enhance the rapidity of mt genome replication. The second advantage would be to ensure accurate replication. The mt genome is known to replicate in a strand-asymmetric manner and/or by a coupled synthesis of leading and lagging strands, but in growing cells where maintenance of the mt genome is essential, strand-asymmetric replication is predominant.149) Since the mt matrix is in a highly oxidative environment, single-stranded DNA resulting from strand-asymmetric replication150) would be easily oxidized, resulting in mutations. Thus, it would be advantageous to make the mt genome as small as possible to ensure accurate replication.
In the process of decreasing the mt genome, the number of mt tRNA genes was reduced to 22, together with the shortening of their lengths. Only 22 different species of tRNA are required for the translation of about 60 different codons in animal mitochondria, which presents a striking contrast to other canonical translation systems, such as prokaryotic and eukaryotic cytoplasmic systems, which utilize more than 50 species of tRNA.151) Twenty-two tRNA species are considered to be the minimal number capable of translating the genetic code. By determining the sequences of mt genomes and mt tRNAs from various animal species, we have constructed a table describing genetic code variations as a function of evolution (Table 2). In this process, we discovered several modified nucleosides in the wobble and middle positions of mt tRNA anticodons and determined that they are essential in decoding codons and also in causing genetic code variations. Specific modification systems may have developed to produce these modified nucleosides, making it possible for this minimal set of tRNAs to decode 60 sense codons. Thus, we have established a mitochondria-specific decoding table (an expanded wobble rule; Table 3). However, it remains to be determined why different modified nucleosides are used in mt tRNAs from different animal species; in other words, why different modification systems have emerged in individual animal species during evolution.
The average size of mt tRNAs decreased during evolution to less than 75 nucleotides. The conserved GG and TΨC sequences in the respective D and T loops were mostly lost, resulting in the absence of the D loop-T loop interaction. In particular, some species of tRNAs lost the D arm or T arm,151) which resulted in Type III and Type IV tRNAs (Fig. 2). Our summaries of the data so far lead us to the conclusion that all mt tRNAs seem to form similar L-shaped tertiary structures irrespective of divergence in their secondary structures, with the constraint that the distance and mutual orientation between the anticodon and the CCA terminus should remain constant. We have proved that even truncated and diversified tRNAs (at least, Type I, II, and III tRNAs in Fig. 2) can function in an mt in vitro translation system.
To elucidate the molecular basis by which aberrant mt tRNAs function in the mt translation system, we have compared interactions of tRNAs with enzymes (ARS and EF-Tu) and ribosomes in mt and E. coli systems, focusing on the exchangeability of components in these two systems. Unilateral recognition specificity was found, which demonstrates that mt ARS, EF-Tu and ribosomes can interact with both mt and E. coli tRNAs, while E. coli ARS and ribosomes cannot interact with mt tRNAs. Although E. coli EF-Tu forms a ternary complex with mt tRNA, this complex cannot deliver mt tRNA to the A site of either mt or E. coli ribosomes (Fig. 3). It is thought that mt tRNAs with truncated structures must be compensated for structurally and functionally by protein components. We found that the Distal helix and C tail of mt SerRS (Figs. 4, 5) and Domain 3′ of nematode mt EF-Tu1 (Fig. 6) compensate for such deficits in mt tRNAs.
As part of the process of the shortening of the mt genome, the number of rRNA genes was reduced to two, corresponding to 12S and 16S rRNAs, while the equivalent of the bacterial 5S RNA gene was discarded. Thus, mt rRNAs became half the size of their E. coli counterparts, but functional sites including the peptidyltransferase center were completely conserved. It was found that the empty spaces in mt rRNAs that correspond to spaces that are occupied by rRNA in bacteria are filled with enlarged mt ribosomal proteins with extended N or C termini (Figs. 7, 8). In summary, structural and functional compensations of truncated RNA segments by proteins are seen in ARS, EF-Tu and ribosomes in mt translation systems. To compensate for such losses in rRNAs and tRNAs, cytoplasmic proteins would have to be imported into mitochondria. The transport of proteins would have been much easier than that of RNAs, because proteins can be imported into mitochondria relatively easily by inserting specific signal sequences upstream of the relevant protein genes, whereas RNA transport requires complex systems involving several specific proteins.152) This might explain why RNAs were shortened and the resulting deficits in essential structures were compensated for by the importation of proteins from the cytoplasm. Thus, it is possible that the transfer of certain structural and functional roles from RNA to proteins occurred during evolution from ancestral to extant organisms. Further research into protein compensatory systems is expected to provide more in-depth knowledge about mt translation systems.
In the process of analyzing mt translation systems, we devised a new technique, chaplet column chromatography, for rapidly and efficiently isolating mt tRNAs. This technical advance allowed us to investigate human mt tRNAs and human mt diseases. In collaboration with S. Ohta’s group, we succeeded in determining the cause of the mt encephalomyopathies MELAS and MERRF to be the absence of the modified nucleosides τm5U and τm5s2U at the anticodon wobble positions of mt tRNALeu(UUR) and tRNALys, respectively. Particularly in MELAS patients, the A3243G or T3271C point mutation in the mt tRNALeu(UUR) gene causes a taurine modification deficiency at the anticodon wobble position of the mutant tRNALeu(UUR), which subsequently causes a UUG codon-specific translational defect that may lead to a translational depression of ND6. This conclusion is consistent with the experimental result that a point mutation (G14453A) in the structural gene for ND6 was found to be associated with a severe MELAS syndrome.140) Our observations of mt diseases provide a new understanding of the molecular causes of some human diseases. We have shown that wobble modification defects in mutant mt tRNAs are a primary cause for various mt diseases, although the point mutations in the tRNAs that perturb wobble modification are also pathogenic in their own right. These are the first reported instances of human diseases that have arisen from RNA modification disorders.
Finally, what are the goals and significance of the investigation of mt translation systems as described here? The first point of interest is to elucidate the factors that permitted changes in the genetic code in individual animal species, and to follow the evolution of animal mitochondria from protozoa to vertebrates with respect to genetic code changes in their mitochondria. The second point is to increase our knowledge about which RNA structures are required for proper mt functions and to define the minimal prerequisites for RNA structures in translation systems. The third point is to gain insights into how the transition from the “RNA world” to the “RNP world” occurred early in the evolution of life. The fourth point is to pursue mt diseases at the molecular level in more detail, for example, to determine the recognition mechanism of the taurine-modifying enzyme that acts on mt tRNALeu(UUR) and tRNALys, which could lead to the development of medical treatments for mt diseases. We strongly expect that further studies on mt translation systems will be pursued to provide more in-depth knowledge of the origin of life as well as to develop tools for medical applications.
Acknowledgements
I thank all the members in my laboratories of the Tokyo Institute of Technology (1988~1992) and the University of Tokyo (1980~1988, 1992~2004), whose endeavors have produced this work. Among them, I particularly thank Drs. Tsutomu Suzuki (now Professor of the University of Tokyo) and Takashi Ohtsuki (now Associate Professor of Okayama University). I also thank our collaborators, Professor Linda Spremulli at North Carolina University, USA, Professor Mathias Sprinzl at Universitat Bayreuth, Germany, Professor James A. McCloskey at University of Utah, USA, the late Professor Jens Nyborg, Aarhus University, Denmark, and Professor Shigeo Ohta at Nippon Medical School, whose close and vigorous collaboration made this research possible. I thank Drs. Syozo Osawa (Professor Emeritus of Na-goya University), the late Kin-ichiro Miura (Professor Emeritus of the University of Tokyo), Susumu Nishimura (Laboratory Animal Resource Center, University of Tsukuba), Yoshiro Shimura (Professor Emeritus of Kyoto University) and Tairo Oshima (Professor Emeritus of the Tokyo Institute of Technology) for their generous support and encouragements during the course of our research activities at the Tokyo Institute of Technology and the University of Tokyo. This work was supported by a grant-in-aid for scientific research from the Ministry of Education, Science, Sports, and Culture of Japan, a grant from the New Energy and Industrial Technology Development Organization (NEDO), and Human Frontier Science Program Grants, for which my sincere thanks are due.
Abbreviations
- tRNASer(UCN)
serine-specific tRNA corresponding to UCN codons
- tRNASerUGA
serine-specific tRNA with anti-codon UGA, the same as tRNASer(UCN); N stands for A, U, C or G; R stands for A or G; Y stands for U or C
- MELAS
mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes
- MERRF
myoclonus epilepsy associated with ragged-red fibers
- mt
mitochondrial
- mtDNA
mitochondrial DNA.
Profile
Kimitsuna Watanabe, Dr. Sci., and Professor Emeritus of the University of Tokyo, was born in Osaka in 1941. He graduated from the University of Tokyo, Faculty of Arts and Sciences, Department of Basic Science in 1967. He finished his PhD work in 1972 in the laboratory of Professor K. Imahori of the Graduate School of Arts and Sciences, the University of Tokyo. Then, he got a researcher position at Mistubishi-Kasei Institute of Life Sciences, in the laboratory of Dr. T. Oshima. From 1975, he spent 2 years at Max-Planck-Institut fur Experimentelle Medizin in West Germany as a post-doctoral fellow under the supervision of Prof. F. Cramer. In 1980 he became an associate professor in Faculty of Agriculture, the University of Tokyo, and in 1984 he moved to the Faculty of Engineering as an associate professor. In 1988, he was appointed to a professor in Tokyo Institute of Technology, Graduate School of Life Science and in 1991 he moved to the University of Tokyo, Graduate School of Engineering as a professor. In 2001 he moved to the Graduate School of Frontier Sciences at Kashiwa-Campus and in 2004 he retired from the University. Then, he became Director of Biological Information Center, National Institute of Advanced Industrial Science and Technology and in 2008 he became Research Advisor of Biomedicinal Information Research Center of the same institute. In addition to the studies depicted in this review, his research fields are 1) the mechanisms on the thermostability of tRNAs and translation system from an extreme thermophile, Thermus thermophilus and the biosynthetic mechanism of a modified nucleoside, 5-methyl-2-thiouridine, an element responsible for the thermostability of tRNA, 2) the molecular mechanism of amino acid reassignment of CUG codon in Candida yeast, and 3) construction of a robust cell-free protein-synthesizing system using a thermophilic bacterium. He was awarded Young Investigator Award of the Japan Biochemical Society in 1980, and the Chemical Society of Japan Award for 2003.

References
- 1).Barrell B. G., Bankier A. T., Drouin J. (1979) A different genetic code in human mitochondria. Nature 282, 189–194 [DOI] [PubMed] [Google Scholar]
- 2).Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H. L., Coulson A. R., Drouin J., et al. (1981) Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 [DOI] [PubMed] [Google Scholar]
- 3).de Bruijn M. H. L., Schreier P. H., Eepron I. C., Barrrell B. G., Chen E. Y., Armstrong P. W., et al. (1980) A mammalian mitochondrial serine transfer RNA lacking the “dihydrouridine” loop and stem. Nucleic Acids Res. 8, 5213–5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Arcari P., Brownlee G. G. (1980) The nucleotide sequence of a small (3S) seryl-tRNA (anticodon GCU) from beef heart mitochondria. Nucleic Acids Res. 8, 5207–5212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Gesteland R. F., Ceck T. R., Atkins J. F. (eds.) (1999) The RNA World Second Edition – The nature of modern RNA suggests a prebiotic RNA world. Cold Spring Harbor Laboratory Press., New York, pp. 1–709 [Google Scholar]
- 6).Crick F. H. C. (1966) Codon-anticodon pairings: The wobble hypothesis. J. Mol. Biol. 19, 548–555 [DOI] [PubMed] [Google Scholar]
- 7).Osawa S., Jukes T. H. (1989) Codon reassignment (codon capture) in evolution. J. Mol. Evol. 28, 271–278 [DOI] [PubMed] [Google Scholar]
- 8).Anderson S., de Bruijn M. H. L., Coulson A. R., Eperon I. C., Sanger F., Young I. G. (1982) Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J. Mol. Biol. 156, 683–717 [DOI] [PubMed] [Google Scholar]
- 9).Gadaleta G., Pepe G., de Candia G., Quagliariello C., Sbisà E., Saccone C. (1989) The complete 3 nucleotide sequence of the Rattus norvegicus mitochondrial genome. Cryptic signals revealed by comparative analysis between vertebrates. J. Mol. Biol. 28, 497–516 [DOI] [PubMed] [Google Scholar]
- 10).Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. (1981) Sequence and gene organization of mouse mitochondrial DNA. Cell 26, 167–180 [DOI] [PubMed] [Google Scholar]
- 11).Desjardins P., Morais R. (1990) Sequence and gene organization of the chicken mitochondrial genonme. A novel gene order in higher vertebrates. J. Mol. Biol. 212, 599–634 [DOI] [PubMed] [Google Scholar]
- 12).Roe B. A., Ma D-P., Wilson R. K., Wong J. F. H. (1985) The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J. Biol. Chem. 260, 9759–9774 [PubMed] [Google Scholar]
- 13).Yokobori S., Ueda T., Feldmaier-Fucks G., Paabo S., Ueshima R., Kondow A., et al. (1999) Complete DNA sequence of the mitochondrial genome of the ascidian Halocynthia roretzi (Chordata, Uro-chordata). Genetics 153, 1851–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Jacobs H. T., Elliot D. J., Math V. B., Farquharson A. (1988) Nucleotide sequence and gene organization of sea urchin mitochondrial DNA. J. Mol. Biol. 202, 185–217 [DOI] [PubMed] [Google Scholar]
- 15).Cantatore P., Roerti M., Rainaldi G., Gadaleta M. N., Saccone C. (1989) The complete nucleotide sequence, gene organization and the genetic code of the mitochondrial genome of Peracentrotus lividus. J. Biol. Chem. 264, 10965–10975 [PubMed] [Google Scholar]
- 16).Asakawa S., Himeno H., Miura K., Watanabe K. (1995) Nucleotide sequence and gene organization of the starfish Asterina pectinifera mitochondrial genome. Genetics 140, 1047–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Clary D. O., Wolstenholme D. R. (1985) The mitochondrial DNA molecule of Drosophila yakuba: nucleotide sequence, gene organization and the genetic code. Proc. Natl. Acad. Sci. USA 71, 2777–2781 [DOI] [PubMed] [Google Scholar]
- 18).Okimoto R., Macfarlane J. L., Clary D. O., Wolstenholme D. R. (1992) The mitochondrial genomes of two nematodes, Caenorhabditis elegans and Ascaris suum. Genetics 130, 471–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Attimonelli M., Altamura N., Benne R., Boyen C., Brennicke A., Carone A., et al. (1999) MitBASE: a comprehensive and integrated mitochondrial DNA database. Nucleic Acids Res. 27, 128–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Hensgens L. A. M., Brakenhoff J., De Vries B. F., Sloop P., Tromp M. C., Van Boon J. H., et al. (1984) The sequence of the gene for cytochrome c oxydase subunit I, a frameshift containing gene for cytochrome c oxidase subunit II and seven unassigned reading frames in Trypanosoma brucei mitochondrial maxi-circle DNA. Nucleic Acids Res. 12, 7327–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Pritchard A. E., Sable C. L., Venuti S. E., Cummings D. J. (1990) Analysis of NADH dehydrogenase proteins, ATPase subunit 9, cytochrome b and ribosomal protein 14 encoded in the mitochondrial DNA of Paramecium. Nucleic Acids Res. 18, 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Pritchard A. E., Seilhamer J. J., Mahalingam R., Sable C. L., Venuti S. E., Cummings D. J. (1990) Nucleotide sequence of the mitochondrial genome of Paramecium. Nucleic Acids Res. 18, 173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Himeno H., Masaki H., Kawai T., Ohta T., Kumagai I., Miura K., et al. (1987) Unusual genetic codes and a novel gene structure for tRNASerAGY in starfish mitochondrial DNA. Gene 56, 219–230 [DOI] [PubMed] [Google Scholar]
- 24).Yokobori S., Hasegawa M., Ueda T., Okada N., Nishikawa K., Watanabe K. (1994) Relationship among coelacanths, lungfishes and tetrapods: a phylogenetic analysis based on mitochondrial cytochrome oxidase gene sequences. J. Mol. Evol. 38, 602–609 [DOI] [PubMed] [Google Scholar]
- 25).Yokobori S., Ueda T., Watanabe K. (1993) Codons AGA and AGG are read as Glycine in ascidian mitochondria. J. Mol. Evol. 36, 1–8 [DOI] [PubMed] [Google Scholar]
- 26).Yamazaki N., Ueshima R., Terrett J. A., Yokobori S., Kaifu M., Segawa R., et al. (1997) Evolution of pulmonate gastropod mitochondrial genomes: comparisons of gene organization of euhadra, cepaea and albinaria and implications of unusual tRNA secondary structures. Genetics 145, 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Sasuga J., Yokobori S., Kaifu M., Ueda T., Nishikawa K., Watanabe K. (1999) Gene contents and organization of a mitochondrial DNA segment of the squid Loligo bleekeri. J. Mol. Evol. 48, 692– 702 [DOI] [PubMed] [Google Scholar]
- 28).Seilhamer J. J., Cummings D. J. (1982) Altered genetic code in Paramecium mitochondria. Possible evolutionary trends. Mol. Gen. Genet. 187, 236– 239 [DOI] [PubMed] [Google Scholar]
- 29).Watanabe K, Osawa S. (1995) tRNA sequences and variations in the genetic code. IntRNA: Structure, Biosynthesis and Function (eds. Söll D., RajBhandary U. L.). ASM Press, Washington, D.C. USA, pp. 215–250 [Google Scholar]
- 30).Yokobori S., Suzuki T., Watanabe K. (2001) Genetic code variations in mitochondria: tRNA as a major determinant of genetic code plasticity. J. Mol. Evol. 53, 314–326 [DOI] [PubMed] [Google Scholar]
- 31).Bessho Y., Ohama T., Osawa S. (1992) Planarian mitochondria II. The unique genetic code as deduced from cytochrome c oxidase subunit I gene sequences. J. Mol. Evol. 34, 331–335 [DOI] [PubMed] [Google Scholar]
- 32).Telford M. J., Herniou E. A., Russel R. B., Littlewood D. T. J. (2000) Changes in mitochondrial genetic codes as phylogenetic characters: Two examples from the flatworms. Proc. Natl. Acad. Sci. USA 97, 11359–11364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Nakamura Y., Ito K. (1998) How protein reads the stop codon and terminates translation. Genes Cells. 3, 263–278 [DOI] [PubMed] [Google Scholar]
- 34).Pont-Kingdom G. A., Beagley C. T., Okimoto R., Wolstenholme D. R. (1994) Mitochondrial DNA of the sea anemone, Metridium ssenile (Cnidaria): Prokaryote-like genes for tRNAf-Met and small-subunit ribosomal RNA and standard genetic code specificities for AGR and ATA codons. J. Mol. Evol. 39, 387–399 [DOI] [PubMed] [Google Scholar]
- 35).Batuecas B., Garesse R., Calleja M., Valverde J. R., Marco R. (1988) Genome organization of Artemia mitochondrial DNA. Nucleic Acids Res. 16, 6515–6529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).de Bruijn M. H. L. (1983) Drosophila melanogaster mitochondrial DNA, a novel organization and genetic code. Nature 304, 234–241 [DOI] [PubMed] [Google Scholar]
- 37).Ueda T., Ohta T., Watanabe K. (1985) Large scale isolation and some properties of AGY-specific serine tRNA from bovine heart mitochondria. J. Biochem. 98, 1275–1284 [DOI] [PubMed] [Google Scholar]
- 38).Yokogawa T., Kumazawa Y., Miura K., Watanabe K. (1989) Purification and characterization of two serine tRNAs from bovine mitochondria by using a hybridization assay method. Nucleic Acids Res. 17, 2623–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Yokogawa T., Watanabe Y., Kumazawa Y., Ueda T., Hirao I., Miura K., et al. (1991) A novel cloverleaf structure found in mammalian mitochondrial tRNASer(UCN). Nucleic Acids Res. 19, 6101–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Wakita K., Watanabe Y., Yokogawa T., Kumazawa Y., Nakamura S., Ueda T., et al. (1994) Higher-order structure of bovine mitochondrial tRNAPhe lacking the ‘conserved’ GG and TΨCG sequences as inferred by enzymatic and chemical probing. Nucleic Acids Res. 22, 347–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Hayashi I., Yokogawa T., Kawai G., Ueda T., Nishikawa K., Watanabe K. (1997) Assignment of imino proton signals of G-C base pairs and magnesium ion binding: An NMR study of bovine mitochondrial tRNASerGCU lacking the entire D arm. J. Biochem. 121, 1115–1122 [DOI] [PubMed] [Google Scholar]
- 42).Tomita K., Ueda T., Watanabe K. (1999) The presence of pseudouridine in the anticodon alters the genetic code; a possible mechanism for assignment of the AAA lysine codon as asparagines in echinoderm mitochondria. Nucleic Acids Res. 27, 1683–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43).Matuyama S., Ueda T., Crain P. F., McCloskey J. A., Watanabe K. (1998) A novel wobble rule found in starfish mitochondria. Presence of 7-methylguanosine at the anticodon wobble position expands decoding capability of tRNA. J. Biol. Chem. 273, 3363–3368 [DOI] [PubMed] [Google Scholar]
- 44).Kondow A., Yokobori S., Ueda T., Watanabe K. (1998) Ascidian mitochondrial tRNAMet possessing unique structural characteristics. Nucleosides Nucleotides 17, 531–539 [PubMed] [Google Scholar]
- 45).Kondow A., Suzuki T., Yokobori S., Ueda T., Watanabe K. (1999) An extra tRNAGly(U*CU) found in ascidian mitochondria responsible for decoding non-universal codons AGA/AGG as glycine. Nucleic Acids Res. 27, 2554–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46).Tomita K., Ueda T, Watanabe K. (1998) 7-Methylguanosine at the anticodon wobble position of squid mitochondrial tRNASerGCU. Molecular basis for assignment of AGA/AGG codons as serine in invertebrate mitochondria. Biochim. Biophys. Acta 1399, 78–82 [DOI] [PubMed] [Google Scholar]
- 47).Tomita K., Ueda T., Ishiwa S., Crain P. F., McCloskey J. A., Watanabe K. (1999) Codon reading patterns in Drosophila melenogaster mitochondria based on their tRNA sequences: a unique wobble rule in animal mitochondria. Nucleic Acids Res. 27, 4291–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Watanabe Y., Tsurui H., Ueda T., Furushima R., Takamiya S., Kita K., et al. (1994) Primary and higher order structures of nematode (Ascaris suum) mitochondrial tRNAs lacking either the T or D stem. J. Biol. Chem. 269, 22902–22906 [PubMed] [Google Scholar]
- 49).Ohtsuki T., Kawai G., Watanabe Y., Kita K., Nishikawa K., Watanabe K. (1996) Preparation of biologically active Ascaris suum mitochondrial tRNAMet with a TV-replacement loop by ligation of chemically synthesized RNA fragments. Nucleic Acids Res. 24, 662–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Sakurai M., Ohtsuki T., Suzuki T., Watanabe K. (2005) Unusual usage of wobble modification in mitochondrial tRNAs of the nematode Ascarisn suum. FEBS Lett. 579, 2767–2772 [DOI] [PubMed] [Google Scholar]
- 51).Sakurai M., Ohtsuki T., Watanabe K. (2005) Modification at position 9 with 1-methyladenosine is crucial for structure and function of nematode mitochondrial tRNAs lacking the entire T-arm. Nucleic Acids Res. 33, 1653–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52).Roe B. A., Wong J. F. H., Chen E. Y., Armstrong P. A. (1981) Sequence analysis of mammalian mitochondrial tRNAs. InRecombinant DNA: Proceedings of the Third Cleveland Symposium on Macromolecules. (ed. Walton A. G.). Elsevier Scientific Publishing Co., Amsterdam, pp. 167–176 [Google Scholar]
- 53).Randerath K., Agrawal H. P., Randerath E. (1983) tRNA alterations in cancer. Recent Results Cancer Res. 84, 103–120 [DOI] [PubMed] [Google Scholar]
- 54).Andachi Y., Yamao F., Muto A., Osawa F. (1989) Codon recognition patterns as deduced from sequences of the complete set of transfer RNA species in Mycoplasma capricolum. Resemblance to mitochondria. J. Mol. Biol. 209, 37–54 [DOI] [PubMed] [Google Scholar]
- 55).Moriya J., Yokogawa T., Wakita K., Ueda T., Nishikawa K., Crain P. F., et al. (1994) A novel modified nucleoside found at the first position of the anticodon of methionine tRNA from bovine liver mitochondria. Biochemistry 33, 2234–2239 [DOI] [PubMed] [Google Scholar]
- 56).Takemoto C., Spremulli L. L., Benkowski L. A., Ueda T., Yokogawa T., Watanabe K. (2009) Unconventional decoding of the AUA codon as methionine by mitochondrial tRNAMet with the anticodon f5CAU as revealed with a mitochondrial in vitro translation system. Nucleic Acids Res. 37, 1616–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).Suzuki T., Suzuki T., Wada T., Saigo K., Watanabe K. (2002) Taurine as a constituent of mitochondrial tRNA: new insights into the functions of taurine and human mitochondrial disease. EMBO J. 21, 6581–6589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58).Samuelsson T., Guindy Y. S., Lustig F., Borén T., Lagerkvist U. (1987) Apparent lack of discrimination in the reading of certain codons in Mycoplasma mycoides. Proc. Natl. Acad. Sci. USA, 84, 3166–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59).Yokoyama S, Nishimura S. (1995) Modified nucleosides and codon recognition. IntRNA: Structure, Biosynthesis and Function. (eds. Söll D., RajBhandary U. L.) ASM Press, Washington D.C., pp. 207–223 [Google Scholar]
- 60).Watanabe Y., Tsurui H., Ueda T., Furushima-Shimogawara, Takamiya S., Kita K., et al. (1997) Primary sequence of mitochondrial tRNAArg of a nematode Ascaris suum. Occurrence of unmodified adenosine at the first position of the anticodon. Biochim. Biophys. Acta 1350, 119–122 [DOI] [PubMed] [Google Scholar]
- 61).Nishimura S. (1983) Structure, biosynthesis and function of queosine in transfer RNA. Prog. Nucl. Acid Res. Mol. Biol. 28, 49–73 [DOI] [PubMed] [Google Scholar]
- 62).Borén T., Elias P., Samuelsson T., Claesson C., Barciszewska M., Gehrke C. W., et al. (1993) Undiscriminating codon reading with adenosine in the wobble position. J. Mol. Biol. 230, 739–749 [DOI] [PubMed] [Google Scholar]
- 63).Lagerkvist U. (1978) “Two out of three”: An alternative method for codon reading”. Proc. Natl. Acad. Sci. USA 75, 1759–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64).Mitra S. K., Lustig F., Åkesson B., Lagerkvist U., Strid L. (1977) Codon-anticodon recognition in the valine codon family. J. Biol. Chem. 252, 471–478 [PubMed] [Google Scholar]
- 65).Osawa S., Jukes T. H., Watanabe K., Muto A. (1992) Recent evidence for evolution of the genetic code. Microbiol. Rev. 59, 229–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66).Lee C. C., Timms K. M., Trotman C. N. A., Tate W. P. (1987) Isolation of a rat mitochondrial release factor. Accomodation of the changed genetic code for termination. J. Biol. Chem. 262, 3548– 3552 [PubMed] [Google Scholar]
- 67).Soleimanpour-Lichaei H. R., Kuhl I., Gaisne M., Passos J. F., Wydro M., Rorbach J., et al. (2007) mtRF1a is a human mitochondrial translation release factor decoding the major termination codons UAA and UAG. Mol. Cell 27, 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68).Yamao F., Iwagami S., Azumi Y., Muto A., Osawa S. (1988) Evolutionary dynamics of tryptophan tRNAs in Mycoplasma caplicolum. Mol. Gen. Genet. 212, 364–369 [DOI] [PubMed] [Google Scholar]
- 69).Osawa S., Ohama T., Jukes T. H., Watanabe K. (1989) Evolution of the mitochondrial genetic code. I. Origin of AGR serine and stop codons in metazoan mitochondria. J. Mol. Evol. 29, 202–207 [DOI] [PubMed] [Google Scholar]
- 70).Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J, et al. (1982) Comparison of the human and bovine mitochondrial genomes. InMitochondrial Genes. Cold Spring Harbor Laboratory, New York, USA, pp. 5–43 [Google Scholar]
- 71).Roe B. A., Wong J. F. H., Chen E. Y., Armstrong P. W., Stankierwicz A., Ma D. P., et al. (1982) Mammalian mitochondrial tRNAs: A modified nucleotide 3′ to the anticodon may modulate their codon response. InMitochondrial Genes. Cold Spring Harbor Laboratory, New York, USA, pp. 45–49 [Google Scholar]
- 72).Steinberg S., Gautheret D., Cedergren R. (1994) Fitting the structurally diverse animal mityochondrial tRNAsSer to common three-dimensional constraints. J. Mol. Biol. 236, 982–989 [DOI] [PubMed] [Google Scholar]
- 73).Watanabe Y., Kawai G., Yokogawa T., Hayashi N., Kumazawa Y., Ueda T., et al. (1994) Higher-order structure of bovine mitochondrial tRNASerUGA: chemical modification and computer modeling. Nucleic Acids Res. 22, 5378–5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74).de Bruijn M. H., Klug A. (1983) A model for the tertiary structure of mammalian mitochondrial transfer RNAs lacking the entire ‘dihydrouridine’ loop and stem. EMBO J. 2, 1309–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75).Wolstenholme D. R. (1992) Animal mitochondrial DNA: structure and evolution. Int. Rev. Cytol. 141, 173–216 [DOI] [PubMed] [Google Scholar]
- 76).Ueda T., Yotsumoto Y., Ikeda K., Watanabe K. (1992) The T-loop region of animal mitochondrial tRNASer(AGY) is a main recognition site for homologous seryl-tRNA synthetase. Nucleic Acids Res. 20, 2217–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77).Hayashi I., Kawai G., Watanabe K. (1998) Higher-order structure and thermal instability of bovine mitochondrial tRNASerUGA investigated by proton NMR spectroscopy. J. Mol. Biol. 284, 57–69 [DOI] [PubMed] [Google Scholar]
- 78).Nozawa K., O’Donoghue P., Gundllapalli S., Araiso Y., Ishitani R., Umehara T., et al. (2009) Pyrroly-syl-tRNA synthetase–tRNAPyl structure reveals the molecular basis of orthogonality. Nature 457, 1163–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79).Ohtsuki T., Kawai G., Watanabe K. (1998) Stable isotope-edited NMR analysis of Ascaris suum mitochondrial tRNAMet having a TV-replacement loop. J. Biochem. 124, 28–34 [DOI] [PubMed] [Google Scholar]
- 80).Gray M. W. (1992) The endosymbiont hypothesis revisited. Int. Rev. Cytol. 141, 233–357 [DOI] [PubMed] [Google Scholar]
- 81).Kumazawa Y., Schwartzbach C. J., Liao H-X., Mizumoto K., Kaziro Y., Miura K, et al. (1991) Interactions of bovine mitochondrial phenylalanyl-tRNA with ribosomes and elongation factors from mitochondria and bacteria. Biochim. Biophys. Acta 1090, 167–172 [DOI] [PubMed] [Google Scholar]
- 82).Takemoto C., Koike T., Yokogawa T., Benkowski L., Spremulli L. L., Ueda T., et al. (1995) The ability of bovine mitochondrial transfer RNAMet to decode AUG and AUA codons. Biochimie 77, 104–108 [DOI] [PubMed] [Google Scholar]
- 83).Hanada T., Suzuki T., Yokogawa T., Takemotohori C., Sprinzl M., Watanabe K. (2001) Translation ability of mitochondrial tRNAsSer with unusual secondary structures in an in vitro translation system of bovine mitochondria. Genes Cells 6, 1019–1030 [DOI] [PubMed] [Google Scholar]
- 84).Hou Y. M., Schimmel P. (1988) A simple structural feature is a major determinant of the identity of a transfer RNA. Nature 333, 140–145 [DOI] [PubMed] [Google Scholar]
- 85).Kumazawa Y., Himeno H., Miura K., Watanabe K. (1991) Unilateral aminoacylation specificity between bovine mitochondria and eubacteria. J. Biochem. 109, 421–427 [DOI] [PubMed] [Google Scholar]
- 86).Gebhardt-Singh E., Sprinzl M. (1986) Ser-tRNAs from bovine mitochondrion form ternary complexes with bacterial elongation factor Tu and GTP. Nucleic Acids Res. 14, 7175–7188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87).Denslow N. D., O’Brien T. W. (1979) Elongation factors EF-G from E. coli and mammalian mitochondria are not functionally interchangeable. Biochem. Biophys. Res. Commun. 90, 1257– 1265 [DOI] [PubMed] [Google Scholar]
- 88).Eberly S. L., Locklear V., Spremulli L. L. (1985) Bovine mitochondrial ribosomes. Elongation factor specificity. J. Biol. Chem. 260, 8721–8725 [PubMed] [Google Scholar]
- 89).Terasaki M., Suzuki T., Hanada T., Watanabe K. (2004) Functional compatibility of elongation factors between mammalian mitochondrial and bacterial ribosomes: characterization of GTPase activity and translation elongation by hybrid ribosomes bearing heterologous L7/L12 proteins. J. Mol. Evol. 336, 331–342 [DOI] [PubMed] [Google Scholar]
- 90).Himeno H., Hasegawa T., Ueda T., Watanabe K., Shimizu M. (1990) Conversion of aminoacylation specificity from tRNATyr to tRNASer. Nucleic Acids Res. 18, 6815–6819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91).Sampson J. R., Sacks M. E. (1993) Contribution of discrete tRNASer domains to aminoacylation by E. coli seryl-tRNA synthetase. A kinetic analysis using model RNA substrates. Nucleic Acids Res. 21, 4467–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92).Fujinaga M., Berthet-Colomonas C., Yaremchuk A. D., Tukalo M. A., Cusack S. (1993) Refined crystal structure of the seryl-tRNA synthetase from Thermus thermophilus at 2.5 Å resolution. J. Mol. Biol. 234, 222–233 [DOI] [PubMed] [Google Scholar]
- 93).Chimnaronk S., Jeppesen G. M., Suzuki T., Nyborg J., Watanabe K. (2005) Dual-mode recognition of noncanonical tRNAsSer by seryl-tRNA synthetase in mammalian mitochondria. EMBO J. 24, 3369–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94).Nissen P., Kjeldgaard M., Thirup S., Polenhina G., Reshetnikova L., Clark B. F., et al. (1995) Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270, 1464–1472 [DOI] [PubMed] [Google Scholar]
- 95).Nissen P., Thirup S., Kjeldgaard M., Nyborg J. (1999) The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure 7, 143–156 [DOI] [PubMed] [Google Scholar]
- 96).Ohtsuki T., Watanabe Y., Takemoto C., Kawai G., Ueda T., Kita K., et al. (2001) An “elongated” translation elongation factor Tu for truncated tRNAs in nematode mitochondria. J. Biol. Chem. 276, 21571–21577 [DOI] [PubMed] [Google Scholar]
- 97).Ohtsuki T., Sato A., Watanabe Y., Watanabe K. (2002) A unique serine-specific elongation factor Tu found in nematode mitochondria. Nature Str. Biol. 9, 669–673 [DOI] [PubMed] [Google Scholar]
- 98).Patel V. B., Cunningham C. C., Hantgan R. R. (2001) Physiochemical properties of rat liver mitochondrial ribosomes. J. Biol. Chem. 276, 6739– 6746 [DOI] [PubMed] [Google Scholar]
- 99).O’Brien T. W. (1971) The general occurrence of 55 S ribosomes in mammalian liver mitochondria. J. Biol. Chem. 246, 3409–3417 [PubMed] [Google Scholar]
- 100).Matthews D. E., Hessler R. A., Denslow N. D., Edwards J. S., O’Brien T. W. (1982) Protein composition of the bovine mitochondrial ribosome. J. Biol. Chem. 257, 8788–8794 [PubMed] [Google Scholar]
- 101).Goldschmidt-Reisin S., Kitakawa M., Herfurth E., Wittmann-Liebold B., Grohmann L., Graack H. R. (1998) Mammalian mitochondrial ribosomal proteins. N-terminal amino acid sequencing, characterization, and identification of corresponding gene sequences. J. Biol. Chem. 273, 34828–34836 [DOI] [PubMed] [Google Scholar]
- 102).Graack H. R., Bryant M. L., O’Brien T. W. (1999) Identification of mammalian mitochondrial ribosomal proteins (MRPs) by N-terminal sequencing of purified bovine MRPs and comparison to Data Bank sequences: The large ribosomal particle. Biochemistry 38, 16569–16577 [DOI] [PubMed] [Google Scholar]
- 103).O’Brien T. W., Fiesler S. E., Denslow N. D., Thiede B., Wittmann-Liebold B., Mougey E. B., et al. (1999) Mammalian mitochondrial ribosomal proteins (2). Amino acid sequencing, characterization and identification of corresponding gene sequencing. J. Biol. Chem. 274, 36043–36051 [DOI] [PubMed] [Google Scholar]
- 104).O’Brien T. W., Liu J., Sylvester J. E., Mougey E. B., Fischel-Ghodsian N., Thiede B., et al. (2000) Mammalian mitochondrial ribosomal proteins (4). Amino acid sequencing, characterization and identification of corresponding gene sequencing. J. Biol. Chem. 275, 18153–18159 [DOI] [PubMed] [Google Scholar]
- 105).Cavdar Koc E., Blackburn K., Burkhart W., Spremulli L. L. (1999) Identification of a mammalian mitochondrial homolog of ribosomal protein S7. Biochem. Biophys. Res. Commun. 266, 141– 146 [DOI] [PubMed] [Google Scholar]
- 106).Cavdar Koc E., Burkhart W., Blackburn K., Moseley A., Koc H., Spremulli L. L. (2000) A proteomics approach to the identification of mammalian mitochondrial small subunit ribosomal proteins. J. Biol. Chem. 275, 32585–32591 [DOI] [PubMed] [Google Scholar]
- 107).Cavdar Koc E., Burkhart W., Blackburn K., Moseley A., Spremulli L. L. (2001) The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J. Biol. Chem. 276, 19363–19374 [DOI] [PubMed] [Google Scholar]
- 108).Suzuki T., Terasaki M., Takemoto-Hori C., Hanada T., Ueda T., Wada A., et al. (2001) Structural compensation for the deficit of rRNA with proteins in the mammalian mitochondrial ribosome. Systematic analysis of protein components of the large ribosomal subunit from mammalian mitochondria. J. Biol. Chem. 276, 21724–21736 [DOI] [PubMed] [Google Scholar]
- 109).Suzuki T., Terasaki M., Takemoto-Hori C., Hanada T., Ueda T., Wada A., et al. (2001) Proteomic analysis of the mammalian mitochondrial ribosome. Identification of protein components in the 28S small subunit. J. Biol. Chem. 276, 33181–33195 [DOI] [PubMed] [Google Scholar]
- 110).Ban N., Nissen P., Hansen J., Moore P. B., Steitz T. A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920 [DOI] [PubMed] [Google Scholar]
- 111).Gutell R. R., Schnare M. N., Gray M. W. (1990) The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. Nucleic Acids Res. 18, 2319–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112).Zhao F., Ohtsuki T., Yamada K., Yoshinari S., Kita K., Watanabe Y., et al. (2005) Isolation and physicochemical properties of protein-rich nematode mitochondrial ribosomes. Biochemistry 44, 9232–9237 [DOI] [PubMed] [Google Scholar]
- 113).Kaneko T., Suzuki T., Kapushoc S. T., Rubio M. A., Ghahvini J., Watanabe K., et al. (2003) Wobble modification differences and subcellular localization of tRNAs in Leishmania tarentolae. Implication for tRNA sorting mechanism. EMBO J. 22, 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114).Suzuki T. (2005) Biosynthesis and function of tRNA wobble modifications. InTopics in Current Genetics, Springer-Verlag, New York, Vol. 12, pp. 24–69 [Google Scholar]
- 115).Tsurui H., Kumazawa Y., Sanokawa R., Watanabe Y., Kuroda T., Wada A., et al. (1994) Batchwise purification of specific tRNAs by a solid-phase DNA probe. Anal. Biochem. 221, 166–172 [DOI] [PubMed] [Google Scholar]
- 116).Huxtable R. J. (1992) Physiological actions of taurine. Physiol. Rev. 72, 101–163 [DOI] [PubMed] [Google Scholar]
- 117).Schon E. A., Bonilla E., DiMauro S. (1997) Mitochondrial DNA mutations and pathogenesis. J. Bioenerg. Biomembr. 29, 131–149 [DOI] [PubMed] [Google Scholar]
- 118).Wallace D. C., Lott M. T. (2003) MITOMAP: A human mitochondrial genome Database. http://www.mitomap.org/ [DOI] [PMC free article] [PubMed]
- 119).Kobayashi Y., Momoi M. Y., Tominaga K., Momoi T., Nihei K., Yanagisawa M., et al. (1990) A point mutation in the mitochondrial tRNA-Leu(UUR) gene in MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis and strokelike episodes). Biochem. Biophys. Res. Commun. 173, 816–822 [DOI] [PubMed] [Google Scholar]
- 120).Kobayashi Y., Momoi M. Y., Tominaga K., Shi-moizumi H., Nihei K., Yanagisawa M., et al. (1991) Respiration-deficient cells are caused by a single point mutation in the mitochondrial tRNA-Leu(UUR) gene in mitochondrial myopathy, encephalopathy, lactic acidosis and strokelike episodes (MELAS). Am. J. Hum. Genet. 49, 590–599 [PMC free article] [PubMed] [Google Scholar]
- 121).Goto Y., Nonaka I., Horai S. (1990) A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 348, 651–653 [DOI] [PubMed] [Google Scholar]
- 122).van den Ouweland J. M., Lemkes H. H., Ruiten-beek W., Sandkuijl L. A., de Vijlder M. F., Struyvenberg P. A., et al. (1992) Mutation in mitochondrial tRNALeu(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat. Genet. 1, 368–371 [DOI] [PubMed] [Google Scholar]
- 123).Johns D. R., Hurko O. (1991) Mitochondrial leucine tRNA mutation in neurological diseases. Lancet 337, 927–928 [DOI] [PubMed] [Google Scholar]
- 124).Moraes C. T., Ciacci F., Silvestri G., Shanske S., Sciacco M., Hirano M, et al. (1993) Atypical clinical presentations associated with the MELAS mutation at position 3243 of human mitochondrial DNA. Neuromuscul. Disord. 3, 43–50 [DOI] [PubMed] [Google Scholar]
- 125).Shoffner J. M., Lott M. T., Lezza A. M., Seibel P., Ballinger S. W., Wallace D. C. (1990) Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell 61, 931–937 [DOI] [PubMed] [Google Scholar]
- 126).King M. P., Attardi G. (1989) Human cells lacking mtDNA. Repopulation with exogeneous mitochondria by complementation. Science 246, 500–503 [DOI] [PubMed] [Google Scholar]
- 127).Hayashi J., Ohta S., Kikuchi A., Takemitsu M., Goto Y., Nonaka I. (1991) Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 88, 10614–10618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128).Chomyn A., Meola G., Bresolin N., Lai S. T., Scarlato G., Attardi G. (1991) In vitro genetic transfer of protein synthesis and respiration defects to mitochondrial DNA-less cells with myopathy-patient mitochondria. Mol. Cell. Biol. 11, 2236–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129).Hayashi J., Ohta S., Takai D., Miyabayashi S., Sakuta R., Goto Y., et al. (1993) Accumulation of mtDNA with a mutation at position 3271 in tRNA(Leu)(UUR) gene introduced from a MELAS patient to HeLa cells lacking mtDNA results in progressive inhibition of mitochondrial respiratory function. Biochem. Biophys. Res. Commun. 197, 1049–1055 [DOI] [PubMed] [Google Scholar]
- 130).Dunbar D. R., Moonie P. A., Zeviani M., Holt I. J. (1996) Complex I deficiency is associated with 3243G:C mitochondrial DNA in osteosarcoma cell hybrid. Hum. Mol. Genet. 5, 123–129 [DOI] [PubMed] [Google Scholar]
- 131).Jacobs H. T. (2003) Disorders of mitochondrial protein synthesis. Hum. Mol. Genet. 12, 293–301 [DOI] [PubMed] [Google Scholar]
- 132).Yasukawa T., Suzuki T., Suzuki T., Ueda T., Ohta S., Watanabe K. (2000) Modification defect at anticodon wobble nucleotide of mitochondrial tRNAs(Leu)(UUR) with pathogenic mutation of mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes. J. Biol. Chem. 275, 4251–4257 [DOI] [PubMed] [Google Scholar]
- 133).Yasukawa T., Suzuki T., Ishii N., Ueda T., Ohta S., Watanabe K. (2000) Defect in modification at the anticodon wobble nucleotide of mitochondrial tRNALys with the MERRF encephalopathy pathogenic mutation. FEBS Lett. 467, 175–178 [DOI] [PubMed] [Google Scholar]
- 134).Björk G. R. (1995) Biosynthesis and function of modified nucleosides. IntRNA: Structure, Biosynthesis and functions. (eds. Söll D., RajBhandary U. L.). ASM Press, Washington DC, pp. 165– 205 [Google Scholar]
- 135).Chomyn A., Martinuzzi A., Yoneda M., Daga A., Hurko O., Johns D., et al. (1992) MELAS mutation in mtDNA binding site for transcription termination factor causes defects in protein synthesis and in respiration but no change in levels of upstream and downstream mature transcripts. Proc. Natl. Acad. Sci. USA, 89, 4221–4225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136).Suzuki T., Ueda T., Watanabe K. (1997) The ‘polysemous’ codon-a codon with multiple amino acid assignment caused by dual specificity of tRNA identity. EMBO J. 16, 1122–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137).Kirino Y., Yasukawa T., Ohta S., Akira S., Ishihara K., Watanabe K., et al. (2004) Codon-specific translational defect caused by a wobble modification deficiency in mutant tRNA from a human mitochondrial disease. Proc. Natl. Acad. Sci. USA 101, 15070–15075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138).Moras D., Comarmond M. B., Fischer J., Weiss R., Thierry J. C., Ebel J. P., et al. (1980) Crystal structure of yeast tRNAAsp. Nature 288, 669–674 [DOI] [PubMed] [Google Scholar]
- 139).Wittenhagen L. M., Roy M. D., Kelley S. O. (2003) The pathogenic U3271C human mitochon-drial tRNALeu(UUR) mutation disrupts a fragile anticodon stem. Nucleic Acids Res. 31, 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140).Ravn K., Wibrand F., Hansen F. J., Horn N., Rosenberg T., Schwartz M. (2001) An mtDNA mutation, 14453G → A, in the NADH dehydrogenase subunit 6 associated with severe MELAS syndrome. Eur. J. Hum. Genet. 9, 805–809 [DOI] [PubMed] [Google Scholar]
- 141).Koga Y., Nonaka I., Kobayashi M., Tojyo M., Nihei K. (1988) Finding in muscle in complex I (NADH coenzyme Q reductase) deficiency. Ann. Neurol. 24, 749–756 [DOI] [PubMed] [Google Scholar]
- 142).Goto Y., Horai S., Matsuoka T., Koga Y., Nihei K., Kobayashi M., et al. (1992) Mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS): A correlative study of the clinical features and mitochondrial DNA mutation. Neurology 42, 545–550 [DOI] [PubMed] [Google Scholar]
- 143).Yasukawa T., Suzuki T., Ishii N., Ohta S., Watanabe K. (2001) Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease. EMBO J. 20, 4794–4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144).Ashraf S. S., Sochacka E., Cain R., Guenther R., Malkiewicz A., Agris P. F. (1999) Single atom modification (O → S) of tRNA confers ribosoma binding. RNA 5, 188–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145).Yoneda M., Miyatake T., Attardi G. (1994) Complementaiton of mutant and wild-type human mitochondrial DNAs coexisting since the mutation event and lack of complementation of DNAs introduced separately into a cell within distinct organelles. Mol. Cell. Biol. 14, 2699–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146).Enriquez J. A., Chomyn A., Attardi G. (1995) MtDNA mutation in MERRF syndrome causes defective aminoacylation of tRNA(Lys) and premature translation termination. Nat. Genet. 10, 47–55 [DOI] [PubMed] [Google Scholar]
- 147).Margulis L. (1981) Symbiosis in cell evolution. Life and its environment on the early earth. Freeman and Company, W. H., USA [Google Scholar]
- 148).Lang B. F., Burger G., O’Kelly C. J., Cedergren R., Golding G. B., Lemieux C., et al. (1977) An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 387, 493–497 [DOI] [PubMed] [Google Scholar]
- 149).Holt I. J., Lorimer H. E., Jacobs H. T. (2000) Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell 100, 515–524 [DOI] [PubMed] [Google Scholar]
- 150).Clayton D. A. (1982) Replication of animal mitochondrial DNA. Cell 28, 693–705 [DOI] [PubMed] [Google Scholar]
- 151).Sprinzl M., Horn C., Brown M., Ioudovitch A., Steinberg S. (1988) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26, 148–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152).Mattaj I. W., Englmeier L. (1998) Nucleoplasmic transport: The soluble phase. Ann. Rev. Biochem. 67, 265–306 [DOI] [PubMed] [Google Scholar]