Abstract
Estrogen receptor (ER) β counteracts the activity of ERα in many systems. In agreement with this, we show in this study that induced expression of ERβ in the breast cancer cell line T47D reduces 17β-estradiol-stimulated proliferation when expression of ERβ mRNA equals that of ERα. Induction of ERβ reduces growth of exponentially proliferating cells with a concomitant decrease in components of the cell cycle associated with proliferation, namely cyclin E, Cdc25A (a key regulator of Cdk2), p45Skp2 (a key regulator of p27Kip1 proteolysis), and an increase in the Cdk inhibitor p27Kip1. We also observed a reduced Cdk2 activity. These findings suggest a possible role for ERβ in breast cancer and imply that ERβ-specific ligands may reduce proliferation of ER-positive breast cancer cells through actions on the G1 phase cell-cycle machinery.
Keywords: cell cycle, p27, cyclin E, Cdc25A, Cdk2
The proliferative actions of 17β-estradiol (E2) mediated via estrogen receptor (ER) α can be opposed by ERβ. The mechanism behind this antagonism seems to be related to the different actions of E2–ERα and E2–ERβ complexes at AP-1 sites, respectively (1). This antiproliferative effect of ERβ might explain protection by estrogen against development of colon cancer, which has been seen in women on hormone replacement therapy (2) and may be of significance in the progression of colon cancer, where it has been shown that the malignant cells in the colon lose expression of ERβ (1). The ventral prostate and the uterus are other tissues where ERβ exerts antiproliferative actions. In the absence of ERβ (in ERβ-/- mice), there is hyperplasia of the prostate epithelium (2) and hypersensitivity of the uterus to the growth effects of E2 (3).
Breast cancer cell lines are extensively used as model systems to study aberrant E2-dependent and E2-independent growth. One commonly used cell line whose growth is stimulated by E2 is the ERα-positive cell line T47D (3, 4). E2 regulates expression of key cell-cycle genes such as c-Myc, cyclin D1, cyclin E, A, Cdc25A, p45Skip2, and p27Kip1 (5–10). The cyclin D promoter is one site where ERβ opposes ERα-mediated activation and the ERβ–antiestrogen complex can stimulate transcription (1). Increased transcription of the cyclin D genes occurs 3–4 h after E2 exposure of cells (10). Its transcription is reduced by antiestrogens, and this reduction in levels of cyclin D1 contributes to reduced cell proliferation (11). Another indication of the importance of cyclin D1 in E2 signaling is that its overexpression in breast cancer cells leads to resistance to antiestrogens (12).
Cyclins E and A are important later in the G1 phase of the cell cycle when they participate in activation of Cdk2, a crucial step in moving the cell into the S phase of the cell cycle (13). Another factor that is essential for activation of Cdk2 activity is the phosphatase Cdc25A, which is itself induced by E2 (10). Inhibitors of Cdk2 also play an important role in cell-cycle regulation. p27Kip1 is one such inhibitor. In its turn, level of p27Kip1 is regulated by ubiquitin-induced degradation, which is mediated by p45Skip2 (14); when p45Skip2 is overexpressed, levels of the cell-cycle inhibitor p27Kip1 are reduced and the cell becomes resistant to antiestrogens.
In a recent report, Omoto et al. (15) expressed ERβ stably in MCF-7 cells under the control of a cytomegalovirus promoter and found that the receptor had a negative effect on proliferation of these cells and reduced the number of colonies in an anchorage–independence assay.
In the present study, we have investigated how ERβ affects cellular proliferation in response to E2 in T47D cells stably transfected with tetracycline-regulated ERβ expression plasmid. We have investigated the specific effects of ERβ expression on the components of the cell cycle machinery Cdk2, cyclin D1, Cdc25A, cyclin E, and p27Kip1 in these cells.
Materials and Methods
Cell Culture. T47D cells were cultured in DMEM/Ham's F-12 (1:1) supplemented with 5% FBS, penicillin, and streptomycin. For experiments using E2, DMEM without phenol red and FBS treated with Dextran-coated charcoal (DCCFBS) were used.
Transfection and Plasmids. T47D cells stably transfected with tetracycline-regulated ERβ expression plasmid were generated in two steps. The cells were first transfected with pTet-tTAk (GIBCO/BRL) modified to contain puromycin resistance by using Lipofectin according to the manufacturer's instructions (GIBCO/BRL). Selection was performed with 0.5 μg/ml puromycin in the presence of 1 μg/ml tetracycline. A clone showing high levels of induction upon tetracycline withdrawal and low basal activity was selected by using the pUHC13-3 control plasmid (GIBCO/BRL). The short form of ERβ encoding 485 aa was fused to the flag tag (ERβ 485) and cloned into PBI-EGFP (Clontech). This construct was then transfected into the previously described inducible clone together with a neomycin resistance plasmid, and selection was performed with 500 μg/ml G418 (Calbiochem). For transient transfections of promoter constructs, normal T47D cells were used. Cells were plated in six-well plates at 50% confluency and synchronized as described under real-time PCR and primers (see below). The plasmids were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Real-Time PCR and Primers. Cells were added to six-well plates at a confluency of 40%; after 1 day, the normal medium was replaced by phenol red-free medium supplemented with 5% DCCFBS. After 24 h, 10 nM ICI 182,780 was added to the cultures, and incubation proceeded for an additional 48 h. For expression of ERβ, tetracycline was removed 12 h before initiation of treatment with E2. At time 0 h, the medium was changed to 0.5% DCCFBS, and E2 was added to a final concentration of 10 nM. RNA was prepared by adding 1 ml of TRIzol (Invitrogen) to each 35-mm plate at different time points after the start of treatment, and RNA was prepared according to the manufacturer's instructions. cDNA (100 ng) was amplified in a real-time PCR using TaqMan Universal Master Mix (PE Applied Biosystems) or, for cyclin E, QPCR Master Mix for Cybergreen (Medprobe). The real-time PCRs were performed in an ABI PRISM model 7700 sequence detector (Perkin–Elmer Applied Biosystems) under the following conditions: 50°C for 2 min, 95°C for 1 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 1 min. The optimum concentration of primers and probes was determined in preliminary experiments. All probes were labeled with 6-carboxyfluorescein as the 5′ reporter. The sequences of primers and probes are as follows. H cyclin D1 (16): F, 5′-CCGTCCATGCGGAAGATC-3′; R, 5′-ATGGCCAGCGGGAAGAC-3′; probe, 5′-CTTCTGTTCCTCGCAGACCTCCAGCAT; 200 nM primers, 200 nM probe. H cdc25A: F, 5′-TTGTTGTGTTTCACTGCGAGTTTT-3′;R,5′-AGGGTAGTGGAGTTTGGGGTATTC-3′; probe, 5′-CGATCTCTCTCTCTCACATACCGGCACAT; 200 nM primers, 200 nM probe. H ERβ (wt): F, 5′-TCCATGCGCCTGGCTAAC-3′; R, 5′-CAGATGTTCCATGCCCTTGTTA-3′; probe, 5′-TCCTGATGCTCCTGTCCCACGTCA; 100 nM primers, 100 nM probe. H ERa: F, 5′-ATCCTGATGATTGGTCTCGTCT-3′; R, 5′-GGATATGGTCCTTCTCTTCCAGA-3′; probe, 5′-GGCTACATCATCTCGGTTCCGCAT; 300 nM primers, 300 nM probe. H cyclin E: F, 5′-GGAAGAGGAAGGCAAACGTGA; R, 5′-TCGATTTTGGCCATTTCTTCAT; 300 nM primers. Real-time PCR was done in triplicate. The 18S rRNA (PDAR, Perkin–Elmer Applied Biosystems) was used as a reference gene.
Proliferation Assay. Cells were plated at a density of 10,000 cells per cm2. After 24 h, the medium was replaced with stripped medium (phenol red-free medium supplemented with 5% DCCFBS, 1% penicillin-streptomycin, 0.1% kanamycin) and 10 nM ICI 182,780. After an additional 48 h, the cells were washed with PBS, and medium containing 0.5% DCCFBS was added in combination with different treatments (with/without E2, 4OH-tamoxifen, raloxifene, and ICI 182,780). Tetracycline was maintained in the medium or removed as indicated in the figure legends. After 6 days of growth, proliferation assay was performed as described.
Western Blot, Antibodies, and Kinase Assays. Cells were grown to 50% confluency on a 100-mm tissue culture plate (Corning). Cultures were mitogen-starved as described above. After treatment with E2 or 4OH-tamoxifen for the indicated times, the cells were washed with PBS, and 200 μl of Nonidet P-40 lysis buffer (0.2% Nonidet P-40/225 mM NaCl/25 mM Tris, pH 7.4) was added. The cells were harvested with a rubber policeman, and the cell slurry was sonicated briefly before centrifugation at 13,000 × g for 5 min at 4°C. The supernatant was collected, and aliquots of 25 μg of protein were separated on SDS/PAGE. Antibodies used were as follows: mouse mAbs directed against Cdc25A (Ab-3) and E2F-1 (Ab-1) (NeoMarkers, Fremont, CA), cyclin D1 rabbit polyclonal antibody (sc-753), mouse monoclonal anticyclin E (HE12) and cyclin A (BF683), goat polyclonal anti-p27Kip1 (C19), and rabbit polyclonal anti-Skp2 (H435) (Santa Cruz Biotechnology). Histone kinase assays were performed as described (9, 17), using rabbit polyclonal antibodies to Cdk2 (M2) (Santa Cruz Biotechnology).
Flow Cytometric Analysis. For flow cytometric analysis, T47D cells were harvested in saline-EDTA, fixed in cold 70% ethanol, and stored at -20°C. Fixed cells were subsequently washed, treated with 100 μg/ml RNase A, and stained with 50 μg/ml propidium iodide. Analysis of DNA content was performed in a Becton Dickinson FACScan with a minimum of 15,000 events collected for analysis with Becton Dickinson cellquest software. The proliferative fraction in the culture was determined as cells in the S and G2/M phases of the cell cycle based on DNA content.
Results
ERβ Expression Inhibits Proliferation of T47D Cells. T47D cells proliferate in response to treatment with E2 (5). In breast cancer cell lines, ERα has been shown to be the receptor responsible for the proliferative effect of E2, whereas, based on studies from knockout mice, ERβ has been proposed to reduce ERα-induced proliferation (18). With real-time PCR using oligos specific for ERα and ERβ, respectively, we found that the predominant ER mRNA in T47D cells is ERα, the ratio ERα/ERβ mRNA being 9:1. We could not detect any endogenous expression of ERβ protein in these cells (data not shown).
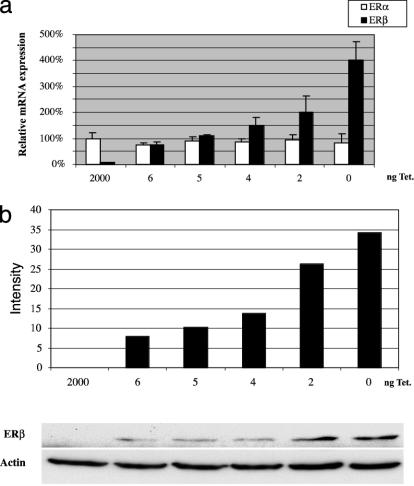
There is a lack of suitable breast cancer cell lines for studies on the function of endogenous ERβ. Our approach has been to stably transfect the breast cancer cell line T47D with a tetracycline-regulated vector for expression of ERβ. In these cells, we can regulate the expression level of the receptor by using different amounts of tetracycline (Fig. 1). We found that E2 inhibited proliferation of T47D cells completely when the mRNA levels of both receptors are equal (Figs. 1a and 2) in contrast to parental T47D cells where ERα predominates and E2 acts as a mitogen. Levels of ERβ protein detected with FLAG antibodies correlated well with ERβ mRNA expression (Fig. 1b).
Fig. 1.
Estimation of relative levels of ERα and ERβ using real-time PCR on cDNA prepared from T47D ERβ cells. (a) T47D ERβ cells cultured on six-well plates were treated with different tetracycline concentrations. After 12 h, RNA was prepared, cDNA was synthesized, and real-time PCR was performed by using ERα- and ERβ-specific primers. Each point represents three different treatments. (b) Whole-cell extracts were prepared from 100-mm plates with T47D ERβ cells treated with different concentrations of tetracycline for 12 h. Western blotting was done with 25 μg of protein FLAG antibody to detect tagged ERβ, and β-actin antibody was used as loading control.
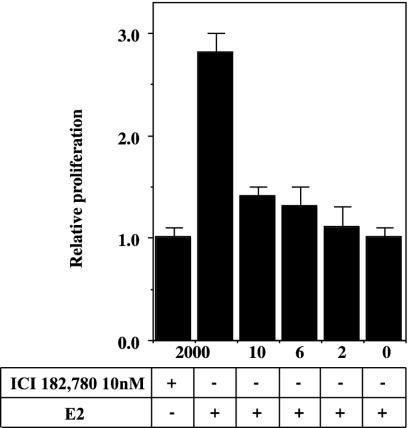
Fig. 2.
Expression of ERβ inhibits E2-stimulated proliferation. T47D ERβ cells were spread on 24-well plates and synchronized as described. The wells were washed with PBS and medium containing 10 nM E2, 0.5% DCCFBS, and 2,000, 10, 6, 2, or 0 ng of tetracycline/ml. The plates were then placed in an incubator for 5 days, whereupon proliferation assay was performed as described in Materials and Methods.
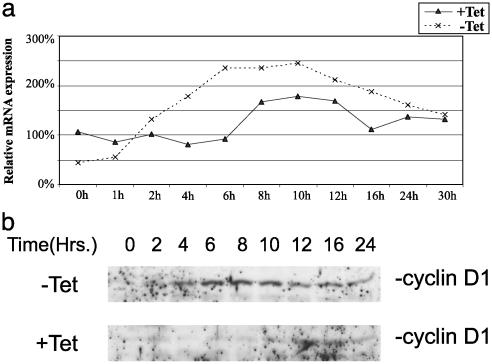
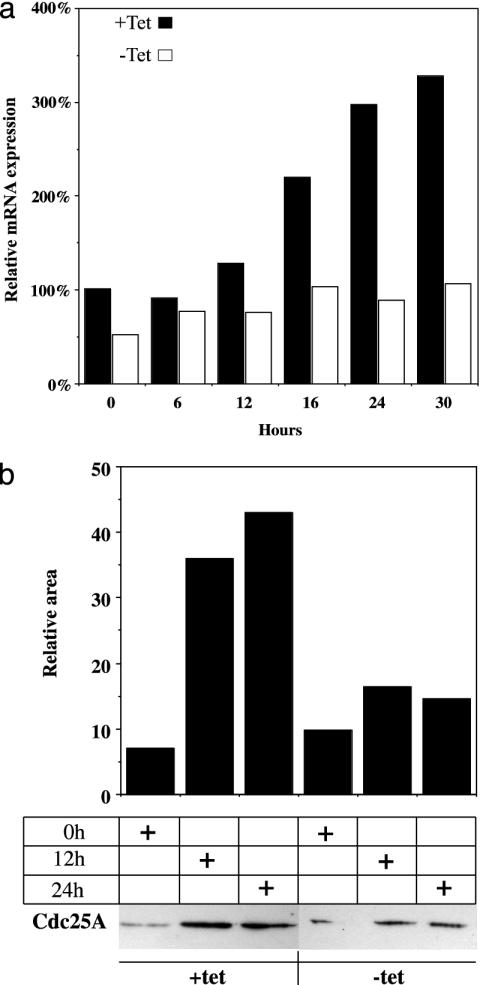
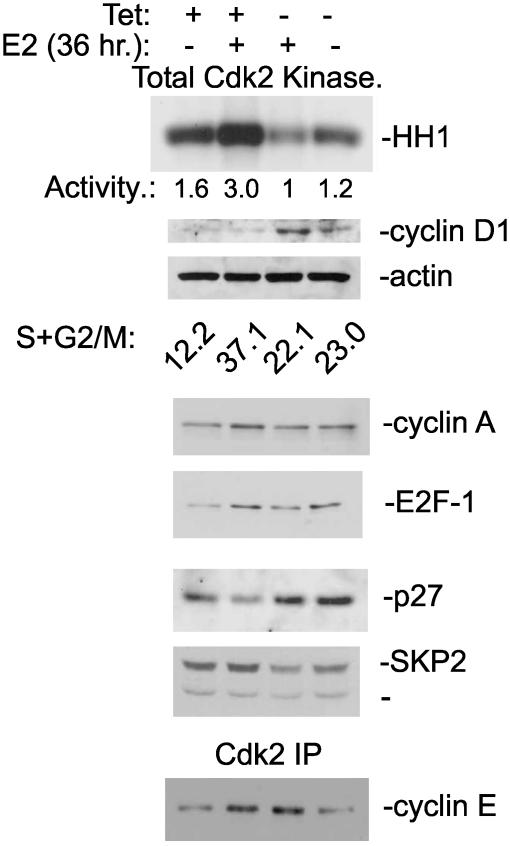
Cyclin D1 and Cdc25A mRNA Levels Are Affected by ERβ Expression. Real-time PCR and Western blot analysis of cyclin D1 and Cdc25A expression were performed on extracts of mitogen-deprived T47D cells treated with E2 for different lengths of time. We could show that cyclin D1 mRNA and protein are induced in response to E2 treatment, when the ratio of ERβ to ERα is 4:1 (Fig. 3). Furthermore, in the presence of tetracycline (i.e., with ERβ expression suppressed), Cdc25A mRNA and protein are induced by E2 within 12 h of treatment of ICI 182,780 pretreated cells. When tetracycline was absent (ERβ expressed), induction of Cdc25A mRNA and protein was weak (Fig. 4). To investigate whether regulation occurred at the transcriptional level, we transiently transfected a luciferase reporter regulated by a minimal Cdc25A promoter into T47D cells. The cells had been treated with ICI 182,780 in low serum for 48 h. The basal activity of the Cdc25A promoter was unchanged by cotransfection with ERβ, but upon treatment with 10 nM E2, there was a substantial decrease in promoter activity (Fig. 5). In agreement with a study by Hodges et al. (11), 4OH-tamoxifen induced the promoter to the same extent as E2 in cells maintained in tetracycline, whereas expression of ERβ eliminated this induction (Fig. 5). Activity of Cdk2 complexes precipitated from cells expressing ERβ was markedly decreased relative to those from cells treated with E2 in the absence of ERβ, causing a decrease of cells in the proliferative fraction S+G2/M (Fig. 6). These complexes could not be activated by adding purified GSTCdc25A (data not shown). A similar inability of Cdc25A to activate Cdk2 was observed by Musgrove et al. (19) with progestin inactivated Cdk2. Levels of the Cdk inhibitor p27kip1 increased in ERβ-expressing cells and did not decline in response to E2 (Fig. 6) as we have demonstrated in previous studies with MCF-7 cells (9, 17). Expression of p45Skp2, a key component in ubiquitin/proteosome-mediated p27Kip1 proteolysis in breast cancer cells (8), was decreased as were cyclin A and E2F-1 (Fig. 6).
Fig. 3.
ERβ regulates cyclin D1 expression in response to E2 treatment. (a) T47D ERβ cells were spread on six-well plates at a low confluency (40%) and grown as described in Materials and Methods. Tetracycline was removed 12 h before start of treatment with 10 nM E2. Cells was harvested in TRIzol at different time points, and cDNA for real-time PCR was prepared; each point represents an average of two different cDNA preparations. (b) Whole-cell extracts were prepared from synchronized T47D ERβ cells grown on 100-mm plates as described in Materials and Methods. Proteins (50 μg) were separated on SDS/PAGE and electrotransferred to nitrocellulose membrane, and cyclin D1 protein was detected by using antibody directed against cyclin D1 (rabbit polyclonal antibody sc753, Santa Cruz Biotechnology).
Fig. 4.
ERβ regulates Cdc25A expression. (a) T47D ERβ cells were spread on six-well plates at a low confluency of 40% and synchronized as described in Materials and Methods. Tetracycline was removed 12 h before start of treatment with 10 nM E2. Cells were harvested in TRIzol at different time points, and cDNA for real-time PCR was prepared. (b) Whole-cell extracts were prepared from synchronized T47D ERβ cells grown on 100-mm plates as described in Materials and Methods. Proteins (50 μg) were separated on SDS/PAGE, electrotransferred to nitrocellulose membrane, and detected with antibody directed against Cdc25A (mouse monoclonal Ab-3, NeoMarkers Fremont, CA). Evaluation of signal strength was done with Bio-Rad chemidoc 1.0 (gel-imaging system).
Fig. 5.
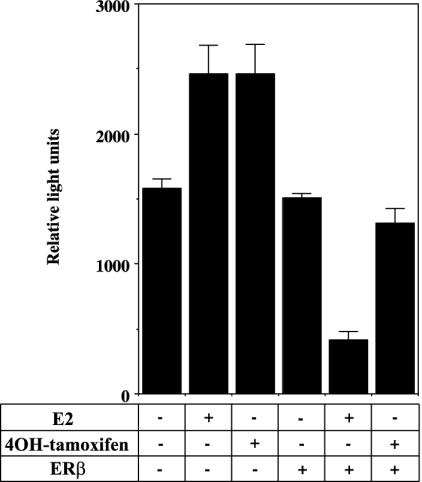
Transient transfection of cdc25A promoter into T47D cells. Cdc25A promoter -450 to +126 in pGL3 basic (1 μg) was transfected into normal T47D cells with or without cotransfection with 100 ng of pcDNA3 or Flag 485 ERβ. Transfected cells were treated with 10 nM E2 or 100 nM 4OH-tamoxifen for 24 h before harvest and luciferase assay. Each bar represents an average of measurements from three wells.
Fig. 6.
ERβ inhibits Cdk2 activity and prevents reduction of p27Kip1 protein level by reducing expression of p45Skip2. T47D cells with inducible ERβ expression were maintained as an asynchronous proliferating culture. Tetracycline was washed out, and whole-cell extracts were made after 36 h. The proliferative fraction in the culture is reported as S+G2/M.
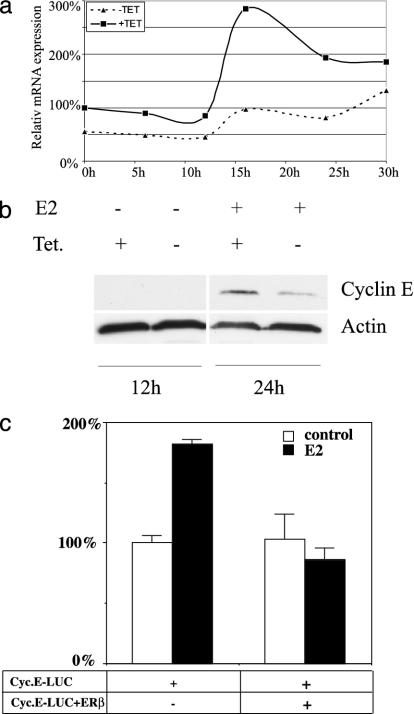
ERβ Inhibits E2 Induction of Cyclin E. Cyclin E is necessary for activation of Cdk2. Because we found that purified GST Cdc25A could not rescue the inhibited Cdk2 complex precipitated from ERβ-expressing cells, we investigated whether the level of cyclin E was reduced by the activation of ERβ. We found that, as expected, cyclin E was up-regulated by E2 at both mRNA and protein levels in control T47D cells. However, expression of ERβ inhibited the E2 induction of cyclin E mRNA/protein (Figs. 6 and 7 a and b). Cyclin A mRNA was regulated in the same way as cyclin E by ERβ (data not shown). In transient transfections, the cyclin E promoter was induced by E2 in control cells, but in ERβ-expressing cells it was repressed (Fig. 7c).
Fig. 7.
ERβ regulates cyclin E expression at mRNA and protein levels. (a) Real-time PCR on cyclin E1 mRNA in T47D cells treated with 10 nM E2 for different times. (b) Western blot using antibody to cyclin E on extracts from T47D cells treated with E2 for 24 h compared to nontreated control. (c) Transient transfection of T47D cells with cyclin E promoter luciferase construct ± transfected pcDNA3 FLAG ERβ.
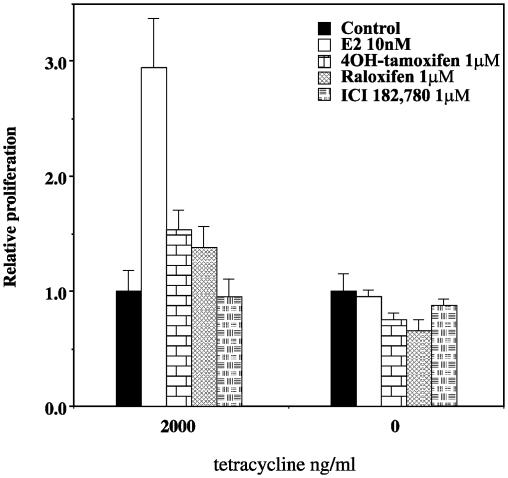
Antiestrogens Do Not Induce Proliferation in the Presence of ERβ in T47D Cells. Because ERβ can activate AP-1 in the presence of antiestrogens (20), it could be argued that the receptor would stimulate proliferation in the presence of antiestrogens. In the presence of 2 μg/ml tetracycline (no ERβ expression), E2 induced proliferation >2-fold, whereas 1 μM 4OH-tamoxifen and raloxifene stimulated proliferation only weakly, and ICI 182,780 did not. The weak stimulation by 4OH-tamoxifen is likely caused by the weak agonistic of this agent on ERα (21). Expression of ERβ completely prevented proliferation in response to E2, 4OH-tamoxifen, raloxifene, and ICI 182,780 (Fig. 8).
Fig. 8.
ERβ inhibits proliferation using 10 nM E2 or 1 μM solutions of the antiestrogens 4OH-tamoxifen, raloxifene, and ICI182,780. T47D ERβ cells were spread onto 24-well plates and synchronized as described in Materials and Methods. Treatment was started as indicated, the cells were harvested after 5 days, and proliferation assay was performed as described. Each bar represents an average of measurements from six wells.
Discussion
We have shown here that expression of ERβ in the breast cancer cell line T47D inhibits proliferation in response to E2 treatment. By monitoring cell-cycle components, we found that, surprisingly, cyclin D1 is induced earlier and to a higher level if ERβ is expressed. This finding is in apparent contradiction with the current hypothesis (based on analysis with the -936 cyclin D1 promoter) that ERβ inhibits proliferation of breast cancer cells by reducing E2-mediated induction of cyclin D1 (1, 22). The reason for the discrepancy might be that additional regulatory elements situated further upstream or downstream of the cyclin D1 gene are targets of regulation by ERβ. Furthermore, the studies referred to (13, 22) used HeLa cells, which lack endogenous ER, and, in this system, the activity of ER may be different from that in T47D cells where ER is naturally expressed. Although it is well established that E2 activates the cyclin D1 promoter (22), E2 induces cyclin D1 mRNA and protein in MCF-7 cells within 3–4 h of stimulation of quiescent cells (10), indicating that induction of cyclin D1 is not an early event in entry of cells into the cell cycle. The importance of cyclin D1 expression is illustrated by studies indicating that the major antiproliferative effect of tamoxifen is related to its inhibition of cyclin D1 expression (11) and that overexpression of cyclin D1 or c-Myc is sufficient to increase the proliferation of MCF-7 cells (23). However, this is not the case in all situations: when ERα was expressed from an adenovirus vector in the breast cancer cell line MDA-MB-231, there was increased expression of cyclin D1 but a reduction in proliferation (24).
In MCF-7 cells, ERα is known to induce expression of c-Myc and E2F-1, which are, in turn, important for E2 induction of Cdc 25A mRNA and protein levels (9, 25). In contrast, we found that ERβ expression in T47D cells results in repression of Cdc 25A mRNA and protein expression. The inhibited Cdk2 complex precipitated from ERβ-expressing cells could not be activated with purified GST-Cdc25A (data not shown), indicating that, as shown in other studies (11, 19), Cdc25A is not solely responsible for the lack of Cdk2 activation. A further examination of the regulation by ERβ of other cell-cycle factors revealed that expression of both cyclin E and cyclin A mRNA was decreased by ERβ. Cyclins E and A are critical components of the active Cdk2 complex driving passage from G1 into S phase. Overexpression of cyclins E or A is associated with poor prognosis in breast cancers (26–29). We cannot at this stage discriminate between the alternative possibilities that the reduction in cyclin E and A levels is caused (i) by ERβ's inhibition of ERα's transcriptional activity or (ii) by ERβ directly repressing the activity of the cyclin E and A promoters. There is no evidence at this time that ERα or ERβ binds directly or indirectly to enhancer elements in the cyclin E or A promoters.
Inhibition of expression of cyclin E, cyclin A, Cdc25A, and E2F-1 does not necessarily indicate a direct regulation by ERβ. It may simply be an indication that the cells are arrested in G1 phase (9). Because expression of all of these cell cycle-associated factors is regulated by E2F-dependent mechanisms, the effect of ERβ activation on G1-phase regulators may be the consequence of cell-cycle inhibition and a lack of release from E2F/pocket protein transcriptional repression. Such cell-cycle inhibition may result from a failure to down-regulate Cdk inhibitors such as p27Kip1 (Fig. 6), whose regulation in MCF-7 breast cancer cells is cell cycle independent (8, 9). This failure in down-regulation of p27Kip1 may, in turn, reflect the observed decrease in p45Skp2 levels (Fig. 6). We have previously shown that E2 up-regulates p45Skp2 expression and elicits nuclear export of p27Kip1 in MCF-7 cells, leading to degradation of this Cdk inhibitor in both nucleus and cytoplasm (8).
It is clear that active cyclin E-Cdk2 is essential for the formation of the origins of replication complexes when cells reenter the cell cycle from a quiescent state, but several recent studies have raised questions about the primacy of cyclin E-Cdk2 in regulating G1/S transition in continuously proliferating mouse embryo fibroblasts and colon cancer cells (30–34). In breast cancer cells, Cdk2 activity does not necessarily correlate with cyclin E levels (35) and may instead reflect the relative amounts of cyclin E and p27Kip1 (36). Cdk2 activity is turned on in quiescent MCF-7 cells by E2 without significant changes in cyclin E expression and instead relates to changes in association of cyclin E-Cdk2 with CIP/KIP Cdk inhibitors (37).
Our observation that the cyclin E content of Cdk2 immunoprecipitates was unchanged by ERβ expression (Fig. 6) leaves it unclear as to whether cyclin E suppression is the primary cause of the low Cdk2 activity in ERβ-expressing cells. The basis of Cdk2 and cell-cycle inhibition in ERβ-expressing T47D cells thus remains to be fully characterized.
Under our experimental conditions, antiestrogens like 4OH-tamoxifen, raloxifene, and ICI 182,780 did not increase proliferation of cells expressing ERβ at high levels. This finding was surprising because the antiestrogen–ERβ complex is known to activate AP-1 sites. Clearly, the interaction of ERβ with antiestrogens is not sufficient to increase proliferation in T47D cells.
Palmieri et al. (38) have studied the expression of both ERα and ERβ in ductal cancers of various grades and shown that although ERβ is the dominant receptor in normal breasts, it is not expressed in grade 1 ductal cancer. Grade 1 ductal cancer shows high expression of ERα, and these are the cancers that respond well to tamoxifen. In grade 2 ductal cancer, both ERα and ERβ are highly expressed, whereas in grade 3 ductal cancer, which has the poorest prognosis, there is commonly neither ERα nor ERβ (38). Fuqua et al. (39), analyzing 242 breast tumors with a monoclonal antibody against the N terminus of ERβ, showed that this receptor isoform is present in 62% of the cases expressing ERα. No correlation of ERβ expression with tumor grade or S-phase fraction was observed; the study, however, did not take the relative levels of the receptors into consideration. Another study by Iwao et al. (40) using 112 breast tumors showed that, whereas ERα mRNA is up-regulated during the progression of ER-positive breast cancers, ERβ mRNA is down-regulated. It is now clear that it is insufficient to measure only one form of ERβ in breast cancer. Another ERβ splice variant, ERβ cx (41), is well expressed in breast cancer (42), which is important because if ERβ cx is expressed in the same cells as ERα, it quenches ERα action (41). Any meaningful study on the relationship between ER expression and prognosis with antiestrogen treatment must measure ERα, ERβ, and ERβ cx and, in addition, must examine the cellular localization of these receptors.
Our studies were done in cells coexpressing ERα and ERβ and show that when these two receptors are together in a cell ERβ can inhibit the proliferative response of ERα to E2.
Acknowledgments
This work was supported by the Swedish Cancer Fund, KaroBio, and the National Institutes of Health.
Abbreviations: ER, estrogen receptor; E2, 17β-estradiol; DCCFBS, Dextran-coated charcoal-treated FBS.
References
- 1.Liu, M. M., Albanese, C., Anderson, C. M., Hilty, K., Webb, P., Uht, R. M., Price, R. H., Jr., Pestell, R. G. & Kushner, P. J. (2002) J. Biol. Chem. 277, 24353-24360. [DOI] [PubMed] [Google Scholar]
- 2.Nelson, H. D., Humphrey, L. L., Nygren, P., Teutsch, S. M. & Allan, J. D. (2002) J. Am. Med. Assoc. 288, 872-881. [DOI] [PubMed] [Google Scholar]
- 3.Lin, C. Q., Singh, J., Murata, K., Itahana, Y., Parrinello, S., Liang, S. H., Gillett, C. E., Campisi, J. & Desprez, P. Y. (2000) Cancer Res. 60, 1332-1340. [PubMed] [Google Scholar]
- 4.Strom, A., Arai, N., Leers, J. & Gustafsson, J.-Å. (2000) Oncogene 19, 5951-5953. [DOI] [PubMed] [Google Scholar]
- 5.Dubik, D., Dembinski, T. C. & Shiu, R. P. (1987) Cancer Res. 47, 65117-65121. [PubMed] [Google Scholar]
- 6.Carroll, J. S., Swarbrick, A., Musgrove, E. A. & Sutherland, R. L. (2002) Cancer Res. 62, 3126-3131. [PubMed] [Google Scholar]
- 7.Watts, C. K., Sweeney, K. J., Warlters, A., Musgrove, E. A. & Sutherland, R. L. (1994) Breast Cancer Res. Treat. 31, 95-105. [DOI] [PubMed] [Google Scholar]
- 8.Foster, J. S., Fernando, R. I., Ishida, N., Nakayama, K. I. & Wimalasena, J. (2003) J. Biol. Chem. 278, 41355-41366. [DOI] [PubMed] [Google Scholar]
- 9.Foster, J. S., Henley, D. C., Bukovsky, A., Seth, P. & Wimalasena, J. (2001) Mol. Cell. Biol. 21, 794-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster, J. S., Henley, D. C., Ahamed, S. & Wimalasena, J. (2001) Trends Endocrinol. Metab. 12, 320-327. [DOI] [PubMed] [Google Scholar]
- 11.Hodges, L. C., Cook, J. D., Lobenhofer, E. K., Li, L., Bennett, L., Bushel, P. R., Aldaz, C. M., Afshari, C. A. & Walker, C. L. (2003) Mol. Cancer Res. 1, 300-311. [PubMed] [Google Scholar]
- 12.Hui, R., Finney, G. L., Carroll, J. S., Lee, C. S., Musgrove, E. A. & Sutherland, R. L. (2002) Cancer Res. 62, 6916-6923. [PubMed] [Google Scholar]
- 13.Doisneau-Sixou, S. F., Sergio, C. M., Carroll, J. S., Hui, R., Musgrove, E. A. & Sutherland, R. L. (2003) Endocr. Relat. Cancer 10, 179-186. [DOI] [PubMed] [Google Scholar]
- 14.Signoretti, S., Di Marcotullio, L., Richardson, A., Ramaswamy, S., Isaac, B., Rue, M., Monti, F., Loda, M. & Pagano, M. (2002) J. Clin. Invest. 110, 633-641. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Omoto, Y., Eguchi, H., Yamamoto-Yamaguchi, Y. & Hayashi, S. (2003) Oncogene 22, 5011-5020. [DOI] [PubMed] [Google Scholar]
- 16.Bijwaard, K. E., Aguilera, N. S., Monczak, Y., Trudel, M., Taubenberger, J. K., & Lichy, J. H. (2001) Clin. Chem. 47, 195-201. [PubMed] [Google Scholar]
- 17.Foster, J. S. & Wimalasena, J. (1996) Mol. Endocrinol. 10, 488-498. [DOI] [PubMed] [Google Scholar]
- 18.Weihua, Z., Saji, S., Makinen, S., Cheng, G., Jensen, E. V., Warner, M. & Gustafsson, J.-Å. (2000) Proc. Natl. Acad. Sci. USA 97, 5936-5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musgrove, E. A., Swarbrick, A., Lee, C. S., Cornish, A. L. & Sutherland, R. L. (1998) Mol. Cell. Biol. 18, 1812-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paech, K., Webb, P., Kuiper, G. G., Nilsson, S., Gustafsson, J.-Å., Kushner, P. J. & Scanlan, T. S. (1997) Science 277, 1508-1510. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, T. A., Richer, J. K., Bain, D. L., Takimoto, G. S., Tung, L. & Horwitz, K. B. (1997) Mol. Endocrinol. 11, 693-705. [DOI] [PubMed] [Google Scholar]
- 22.Altucci, L., Addeo, R., Cicatiello, L., Dauvois, S., Parker, M. G., Truss, M., Beato, M., Sica, V., Bresciani, F. & Weisz, A. (1996) Oncogene 12, 2315-2324. [PubMed] [Google Scholar]
- 23.Prall, O. W., Rogan, E. M., Musgrove, E. A., Watts, C. K. & Sutherland, R. L. (1998) Mol. Cell. Biol. 18, 4499-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Licznar, A., Caporali, S., Lucas, A., Weisz, A., Vignon, F. & Lazennec, G. (2003) FEBS Lett. 553, 445-450. [DOI] [PubMed] [Google Scholar]
- 25.Wang, W., Dong, L., Saville, B. & Safe, S. (1999) Mol. Endocrinol. 13, 1373-1387. [DOI] [PubMed] [Google Scholar]
- 26.Kim, H. K., Park, I. A., Heo, D. S., Noh, D. Y., Choe, K. J., Bang, Y. J. & Kim, N. K. (2001) Eur. J. Surg. Oncol. 27, 464-471. [DOI] [PubMed] [Google Scholar]
- 27.Bukholm, I. R., Bukholm, G. & Nesland, J. M. (2001) Int. J. Cancer 93, 283-287. [DOI] [PubMed] [Google Scholar]
- 28.Landberg, G., Nielsen, N. H., Nilsson, P., Emdin, S. O., Cajander, J. & Roos, G. (1997) Cancer Res. 57, 549-554. [PubMed] [Google Scholar]
- 29.Keyomarsi, K., Tucker, S. L. & Bedrosian, I. (2003) Nat. Med. 9, 152. [DOI] [PubMed] [Google Scholar]
- 30.Geng, Y., Yu, Q., Sicinska, E., Das, M., Schneider, J. E., Bhattacharya, S., Rideout, W. M., Bronson, R. T., Gardner, H. & Sicinski, P. (2003) Cell 114, 431-443. [DOI] [PubMed] [Google Scholar]
- 31.Ortega, S., Prieto, I., Odajima, J., Martin, A., Dubus, P., Sotillo, R., Barbero, J. L., Malumbres, M. & Barbacid, M. (2003) Nat. Genet. 35, 25-31. [DOI] [PubMed] [Google Scholar]
- 32.Gladden, A. B. & Diehl, J. A. (2003) Cancer Cell, 4, 160-162. [DOI] [PubMed] [Google Scholar]
- 33.Hinds, P. W. (2003) Cancer Cell 3, 305-307. [DOI] [PubMed] [Google Scholar]
- 34.Tetsu, O. & McCormick, F. (2003) Cancer Cell 3, 233-245. [DOI] [PubMed] [Google Scholar]
- 35.Sweeney, K. J., Swarbrick, A., Sutherland, R. L. & Musgrove, E. A. (1998) Oncogene 16, 2865-2878. [DOI] [PubMed] [Google Scholar]
- 36.Loden, M., Nielsen, N. H., Roos, G., Emdin, S. O. & Landberg, G. (1999) Oncogene 18, 2557-2566. [DOI] [PubMed] [Google Scholar]
- 37.Prall, O. W., Sarcevic, B., Musgrove, E. A., Watts, C. K. & Sutherland, R. L. (1997) J. Biol. Chem. 272, 10882-10894. [DOI] [PubMed] [Google Scholar]
- 38.Palmieri, C., Cheng, G. J., Saji, S., Zelada-Hedman, M., Warri, A., Weihua, Z., Van Noorden, S., Wahlstrom, T., Coombes, R. C., Warner, M. & Gustafsson, J.-Å. (2002) Endocr. Relat. Cancer 9, 1-13. [DOI] [PubMed] [Google Scholar]
- 39.Fuqua, S. A., Schiff, R., Parra, I., Moore, J. T., Mohsin, S. K., Osborne, C. K., Clark, G. M. & Allred, D. C. (2003) Cancer Res. 63, 2434-2439. [PMC free article] [PubMed] [Google Scholar]
- 40.Iwao, K., Miyoshi, Y., Egawa, C., Ikeda, N. & Noguchi, S. (2000) Int. J. Cancer 88, 733-736. [DOI] [PubMed] [Google Scholar]
- 41.Ogawa, S., Inoue, S., Watanabe, T., Orimo, A., Hosoi, T., Ouchi, Y. & Muramatsu, M. (1998) Nucleic Acids Res. 26, 3505-3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saji, S., Omoto, Y., Shimizu, C., Horiguchi, S., Watanabe, T., Funata, N., Hayash, S., Gustafsson, J.-Å. & Toi, M. (2002) Breast Cancer 9, 303-307. [DOI] [PubMed] [Google Scholar]