Abstract
Multifrequency electron-spin echo envelope modulation (ESEEM) spectroscopy is used to ascertain the nature of the bonding interactions of various active site amino acids with the Mn ions that compose the oxygen-evolving cluster (OEC) in photosystem II (PSII) from the cyanobacterium Synechocystis sp. PCC 6803 poised in the S2 state. Spectra of natural isotopic abundance PSII (14N-PSII), uniformly 15N-labeled PSII (15N-PSII), as well as 15N-PSII containing 14N-histidine (14N-His/15N-PSII) are compared. These complementary data sets allow for a precise determination of the spin Hamiltonian parameters of the postulated histidine nitrogen interaction with the Mn ions of the OEC. These results are compared to those from a similar study on PSII isolated from spinach. Upon mutation of His332 of the D1 polypeptide to a glutamate residue, all isotopically sensitive spectral features vanish. Additional Ka- and Q-band ESEEM experiments on the D1-D170H site-directed mutant give no indication of new 14N-based interactions.
A pentanuclear metal cluster serves at the catalytic site for water oxidation in photosystem II (PSII)1. This oxygen-evolving complex (OEC) is composed of four Mn ions and one Ca ion bridged to one another via solvent-derived oxo (or hydroxo) bridges. This cluster is presumably bound to the PSII protein by several active-site amino acid residues. Spectroscopic and kinetic studies of PSII prepared with site-specific isotopically labeled amino acids and site-directed mutations have suggested a number of possible candidates for such protein-derived ligands.(2–4) These include D1-Asp170, D1-Glu189, D1-His332, D1-Glu333, D1-His337, D1-Asp342, CP43-Glu354, and the carboxy terminus of the D1 polypeptide at D1-Ala344. The recent spate of X-ray crystal structures of PSII from the thermophilic cyanobacteria Thermosynechococcus elongatus and Thermosynechococcus vulcanus indicate that all the above residues are indeed near enough to the OEC to be considered within the first coordination sphere.(5–13) However, each of these structures also shows subtle and in some cases significant differences in relative positions of these residues to the Mn cluster. No doubt, these discrepancies are due in large part to the photoreduction of the Mn ions that is known to occur during acquisition of the diffraction data.(14,15) A much higher resolution (1.9 Å) crystal structure was recently reported2 and was obtained with a lower flux of X-rays combined with crystal rotation to minimize radiation-induced damage (the structure has since been deposited in the Protein Data Bank (accession code 3ARC) and discussed in a corresponding paper (16)). Their model of the OEC and its ligands is consistent with much of the previous crystallographic results. At the time of this writing, however, it is unknown if sufficient resolution exists to make Mn oxidation state assignments. Furthermore, the evolution of this geometry as a function of S-state is currently not available from the diffraction data. Thus there remains a strong need for quantitative characterization of the native protein—OEC bonding interactions.
Electron paramagnetic resonance (EPR) spectroscopy has been a particularly useful tool for characterizing the electronic and geometric structure features of the OEC and does so without deleteriously affecting the protein.(17–26) Of the five S-states of the Kok cycle (S0…S4); the best characterized by EPR spectroscopy is the S2 state. Two distinct EPR signals, both attributed to the OEC in the S2 state, are detected at X-band (≈9 GHz) excitation frequencies. The first signal consists of 18–22 55Mn (I = 5/2) hyperfine-induced features and is centered at g = 1.98. This so-called multiline signal (MLS) arises from an S = 1/2 ground spin state of the OEC achieved by a combination of ferromagnetic and antiferromagnetic exchange interactions between the three Mn(IV) ions and one Mn(III) ion.(27–30) The second signal, a derivative-shaped feature centered at g = 4.1, has been assigned to an S = 5/2 ground spin state that arises from an alternative exchange coupling scheme between the four Mn ions.(26,31) Researchers have studied how chemical treatment of PSII-containing preparations (e.g., with small alcohols, fluoride, or ammonia) affects the equilibrium between these two S2 forms.(32,33) Additional studies have focused on the effect of site-directed mutations on the generation and appearance of the MLS.(34–37) These results are crucial to determining the involvement of certain amino acids in securing the OEC to the peptide and tuning its redox properties for proper S-state advancement.
In particular, mutation of D1-His332 to glutamate (D1-H332E) in PSII from Synechocystis sp. PCC 6803 abolishes O2 evolving activity, but short (30 s) illumination at 273 K of dark-adapted (S1 state) particles leads to formation of a MLS with 24 narrowly spaced peaks (the usual, 8–10 min 195 K illumination procedure surprisingly yields no MLS).(34) These results indicate that the Mn-cluster is assembled in these mutants but that the magnetic properties of the OEC are altered. A more detailed insight into the nature of protein—OEC interactions is gained from the application of advanced EPR methods such as electron spin echo envelope modulation (ESEEM) spectroscopy. ESEEM spectroscopy detects transition frequencies between nuclear spin levels that are hyperfine-coupled to the unpaired electron spin, which, in this case, is localized on the OEC. Early X-band ESEEM studies established that a feature appearing at 4.8 MHz was due to magnetic coupling of a 14N nucleus to the OEC of PSII from Synechococcus sp. in its S2 state.(38) Later studies on Synechocystis sp. PCC 6803 that contained histidine residues labeled with 15N at both imidazole nitrogen positions showed a dramatically changed ESEEM spectrum.(39) This nitrogen isotope sensitivity confirms that features observed using X-band ESEEM arise from histidine (though, no new features assigned to coupling from a 15N nucleus were identified). Then an ESEEM study comparing wild-type*. Wild-type* refers to PSII isolated from a control strain of Synechocystis sp. PCC 6803 constructed in identical fashion as the D1-H332E mutant, but containing the wild-type psbA-2 gene. and D1-H332E PSII from Synechocystis sp. PCC 6803 found that the 4.8 MHz peak decreased or disappeared upon this mutation (see Figure S1).(40) A similar feature has now been observed in X-band ESEEM spectra of PSII isolated from higher plants (spinach(40–42) and pea seedlings(43)) as well as mesophilic(38,40) and thermophilic cyanobacteria.(36) Unfortunately, rather little information as to the strength of the magnetic interaction between this histidine nitrogen and the OEC can be derived from just one peak.
Ideally, the ESEEM spectrum of an I = 1 nucleus coupled to an S = 1/2 electron spin possesses six clearly resolved peaks (Figure 1). Three of these arise from transitions between nuclear spin levels in the mS = +1/2 (α) electron spin manifold while the other three features are due to nuclear spin-flip transitions in the mS = −1/2 (β) manifold. In practice, however, rarely are all these transitions evident. The magnitudes of the isotropic hyperfine and nuclear Zeeman interactions are not equal, which typically leads to the appearance of just two peaks in the frequency spectrum. These are assigned to the double quantum (ΔmI = 2) transitions in each electron spin manifold, νdqα and νdqβ. Accompanying these peaks could also be features at approximately half the frequencies of the νdq transitions corresponding to single quantum transitions νsq, however, these peaks are more susceptible to broadening by hyperfine anisotropy and nuclear quadrupole interaction and thus often go undetected. As the nuclear Zeeman and hyperfine interactions (HFI) become equal in magnitude they cancel each other out in one electron spin manifold (which manifold depends on the relative signs of the interactions).(44) This leads to a coalescence of nuclear spin levels in that manifold that are split by only the nuclear quadrupole interaction (NQI). Transitions between these nominally pure quadrupole states are labeled ν0, ν−, and ν+. Experimentally, this cancellation condition can be met for arbitrary hyperfine coupling by simply changing the magnitude of the applied magnetic field and performing the ESEEM experiment at the corresponding excitation frequency (maintaining a constant g-value). The cancellation condition cannot be met if the magnitude of the hyperfine interaction is larger than the excitation bandwidth of the microwave pulses.
Figure 1.
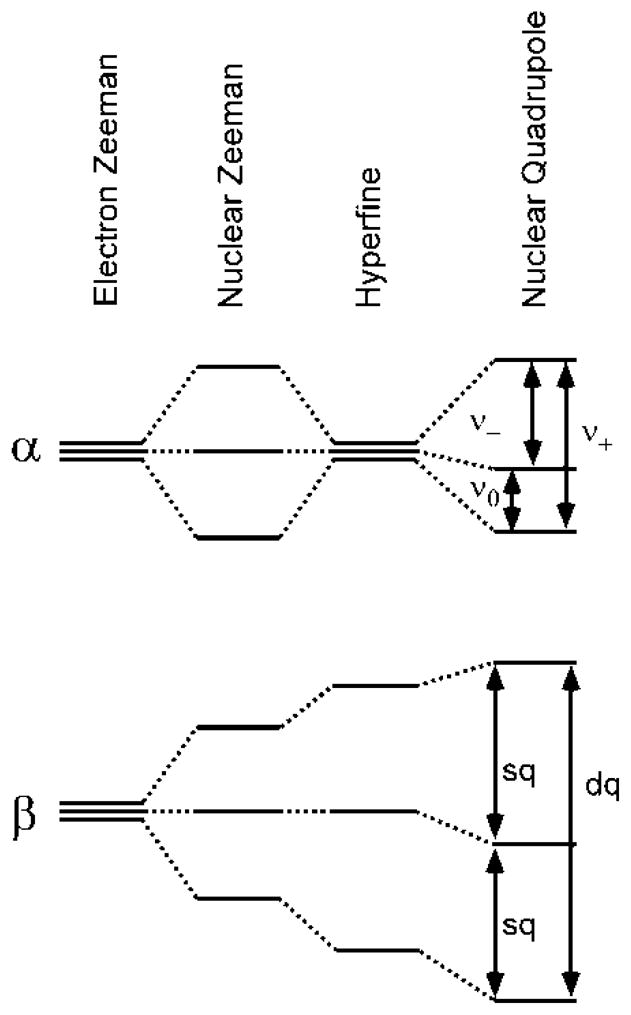
Energy level diagram of electron and nuclear spin levels of an S = 1/2, I = 1 spin system. As the magnitude of the applied magnetic field B0 is increased from zero to a value where nuclear Zeeman interaction ≈ Aeff/2, the spin levels in one spin manifold (α) collapse and are only split by the nuclear quadrupole interaction. This case leads to maximal modulation depth in the ESEEM spectrum.
Recently, our laboratory performed such a multi-frequency (from X- to Ka-band frequencies, 9.75 to 30.76 GHz) ESEEM study of PSII-containing thylakoids isolated from spinach.(41) The Ka-band spectra possessed prominent peaks at 0.88, 1.98, 6.90, and 14.00 MHz that were well-simulated with the following magnetic parameters: Aiso = 7.3 MHz, Aaniso = [+0.70 +0.70 −1.40] MHz, e2Qq/h= 1.98 MHz, η = 0.84. The results from this study afforded, for the first time, quantitative insight into the strength of the HFI of this 14N nucleus to the OEC in the S2 state. Further, it allowed for the characterization of the electric quadrupole tensor, a key parameter in assessing the electronic structure of the nitrogen atom and its chemical environment. As all observed features were fully accounted for in the simulations of the data and no combination peaks were present in the three-pulse ESEEM spectrum, we concluded that only a single 14N nucleus was strongly interacting with the Mn cluster.
To further complement this new, more quantitative understanding of the proteinaceous nitrogen coordination of the OEC we have extended these studies of spinach PSII to Q-band (34.5 GHz) and compare these findings to analogous spectra of native PSII from Synechocystis sp. PCC 6803 (14N-PSII). We also present results from ESEEM studies of fully 15N-labeled Synechocystis (15N-PSII), which allow for a more precise determination of the nitrogen HFI as energy shifts of the nuclear spin levels due to NQI are absent. Notably, the ESEEM spectra of 15N-PSII collected at Ka- and Q-band, in contrast to the results obtained at X-band, are the first data to show obvious features from hyperfine-coupled 15N nuclei.(38,39) To verify again that the observed isotope sensitive features arise from histidine nitrogen(s) coupling to the OEC, we have prepared 15N-labeled PSII containing natural-abundance histidine (14N-His/15N-PSII). All these data are then compared to those of two site-directed mutants, D1-H332E and D1-D170H. Differences between these data sets are discussed in the context of recent ESEEM spectroscopic results on a D1-H332S mutant from Thermosynechococcus elongatus(36) and an amino acid ligation scheme for the OEC in the S2 state is proposed.
Materials and Methods
Construction of Mutant Strains and Propagation of Cultures
The D1-D170H and D1-H332E mutant strains of the cyanobacterium Synechocystis sp. PCC 6803 have been described previously.(40,45) Each mutation was constructed in the psbA-2 gene and the mutation-bearing plasmid was transformed into a host strain of Synechocystis that lacks all three psbA genes and contains a hexahistidine-tag (His-tag) fused to the C-terminus of CP47. Cells were maintained on solid BG-11 media(46) containing 5 mM TES-NaOH (pH 8.0), 0.3% sodium thiosulfate, 5 mM glucose, 10 μM 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), 5 μg/mL kanamycin monosulfate, and 20 μg/mL gentamycin sulfate. The DCMU, thiosulfate, and antibiotics were omitted from liquid cultures. Large-scale liquid cultures (each consisting of two or three 7 L cultures held in glass carboys) were propagated as described previously.(47) For the purification of PSII core complexes uniformly labeled with 15N, liquid cultures were propagated in the presence of 10 mM Na15NO3 as the sole nitrogen source (98% 15N enrichment, Cambridge Isotope Laboratories, Andover, MA).(37,48) For the purification of PSII core complexes containing natural abundance 14N-histidine in a uniformly 15N-labeled background, liquid cultures were propagated in the presence of both 240 μM L-histidine(39) and 10 mM Na15NO3.
Purification of PSII core complexes
PSII core complexes were purified under dim green light at 4 °C with Ni-NTA superflow affinity resin (Qiagen, Valentia, CA) as described previously.(49) The purification buffer consisted of 1.2 M betaine, 10% (v/v) glycerol, 50 mM MES-NaOH (pH 6.0), 20 mM CaCl2, 5 mM MgCl2, 50 mM histidine, 1 mM EDTA, and 0.03% (w/v) n-dodecyl β-D-maltoside. The purified PSII core complexes were concentrated to ≈9 mg of Chl/mL by ultrafiltration, frozen in liquid N2, and stored at −80 °C. DCMU was added to 1 mM to samples of D1-H332E mutant. No electron acceptor was added to any PSII samples. Subsequently, the concentrated PSII core complexes were loaded into 2.5 mm diameter and 1.5 mm diameter quartz tubes for study by Ka- and Q-band EPR spectroscopy, respectively. To verify the integrity of the mutant PSII core complexes, an aliquot of each large-scale culture was set aside and the sequence of the relevant domains of the D1 polypeptide was obtained after PCR amplification of genomic DNA.(50) No trace of the wild-type codon was detected in any of the mutant cultures.
PSII from spinach (BBY) was purified according to the method of Berthold, Babcock, and Yocum(51) modified to remove adventitiously bound Mn(II).(52,53)
Hydrolysis, Derivatization, and Mass Analysis of Histidine
The mass distribution of histidine residues in the 14N-His/15N-PSII preparation was assessed by mass spectrometry. The effluent from the Ni-NTA affinity column (consisting of thylakoid proteins other than PSII) was adsorbed onto a Pharmacia Q-Sepharose Fast Flow ion-exchange column; washed with 50 mM MES-NaOH (pH 6.0) 0.03% DM to remove excess n-dodecyl β-D-maltoside and other contaminants; then eluted with 50 mM MES-NaOH (pH 6.0), 0.03% n-dodecyl β-D-maltoside, 0.5 M NaCl. The effluent was then passed through a Pharmacia HR26/10 desalting column to remove NaCl, and concentrated to about 10 mg of Chl/mL with Centricon-100 concentrators. For hydrolysis, an aliquot of concentrated 14N-His/15N-protein solution was precipitated with 0.700 mL of 95% acetone (HPLC grade) followed by 30 s of vortexing. The sample was incubated for 90 min in a −20 °C freezer prior to centrifugation at 14 000×g for 10 min at 4 °C. The supernatant was removed, and the remaining pellet was dried under gaseous N2 for 3 hr. The dried pellet was then transferred to a glass vial, flushed with N2 gas, and 0.30 mL of 6 M HCl was added. Following acidification, the sample was then sealed and placed in an oil bath at 120 °C for 24 hr. Once cooled to room temperature, the residual solvent was evaporated with flowing N2 gas. The hydrolyzed sample was then derivatized with tert-butyldimethylsilylate as previously described.(54) Gas chromatography to mass spectrometry (GC/MS) spectroscopic analysis was performed at the UC Davis Proteomics/Genomics Center.
EPR Spectroscopy
The Ka-band (30.5–31.5 GHz) experiments were performed using a laboratory-built pulse EPR spectrometer and ESEEM probe that have been described previously.(55) Low-temperature operation was achieved using a Janis Research Company (Wilmington, MA) SuperVariTemp cryostat in conjunction with a Lakeshore Cryotronics (Westerville, OH) temperature controller. Q-band (34.5 GHz) experiments were performed with a Bruker Elexsys E580 pulse EPR spectrometer (Bruker BioSpin, Billerica, MA) fitted with a SuperQ-FT multi-frequency extension and using an EN 5107D2 resonator. Cryogenic temperatures were achieved and maintained with an Oxford CF-935 cryostat and Oxford ITC temperature controller.
All presented EPR spectra were generated by subtracting the spectrum of dark-adapted PSII particles from that obtained following 8 minutes of continuous illumination (using a Sylvania ELH 300 W halogen-tungsten lamp, color temperature = 3350 K) at 200 K. D1-H332E samples were illuminated at 273 K (ice/water) for 1 min as described previously.(34,40) These light-minus-dark spectra ideally contain only features associated with the OEC trapped in the S2 state. However, the redox-active tyrosine YD found in the D2 polypeptide of PSII, the Fe/QA site on the acceptor side of PSII, and cytochromes bound in nearby proteins all have EPR signals that can change upon illumination.(21) Therefore, two-pulse (π/2-τ- π-τ-echo) and three-pulse (π/2-τ-π/2-T-π/2-τ-echo) ESEEM experiments were performed at five magnetic field positions across the EPR envelope of the MLS. The signal from YD• is centered at g = 2 and only 4 mT wide. The signals from oxidized cytochrome and Fe(II)/QA− are broader than the MLS (the photoinduced Fe(II)/QA− signal appears at g = 1.90 and 1.64 or g = 1.82 and 1.67 depending on the presence or absence of CO2/HCO3−)(21), though anisotropic and have intensity maxima on the low-field and high-field sides, respectively, of the g = 2 region of the spectrum. Thus, only peaks from the OEC in the S2 state should appear in all five spectra. Furthermore, the Fe(II)/QA− spin system is known to relax very quickly and does not contribute to ESE EPR spectra collected at 4.5 K.(56) Indeed, the ESEEM features analyzed below are present in all five resonant field positions and appear at frequencies that are different due solely to the change in the nuclear Zeeman interaction as B0 changes. Three-pulse ESEEM spectra were acquired using multiple τ-values to ensure that no spectral features were suppressed. Other relevant instrument settings are given in the corresponding figure captions. Frequencies contributing to observed modulation patterns were visualized using cosine-Fourier transforms back-filled with only the two highest frequency components of the spectrum.(57) Spectral simulations were performed with the Matlab 7.5.0.342 (R2007b) software package (The Mathworks Inc., Natick, MA) using the EasySpin 3.1.5 toolbox.(58,59)
Results
Two-pulse Electron-Spin-Echo-Detected (ESE) EPR
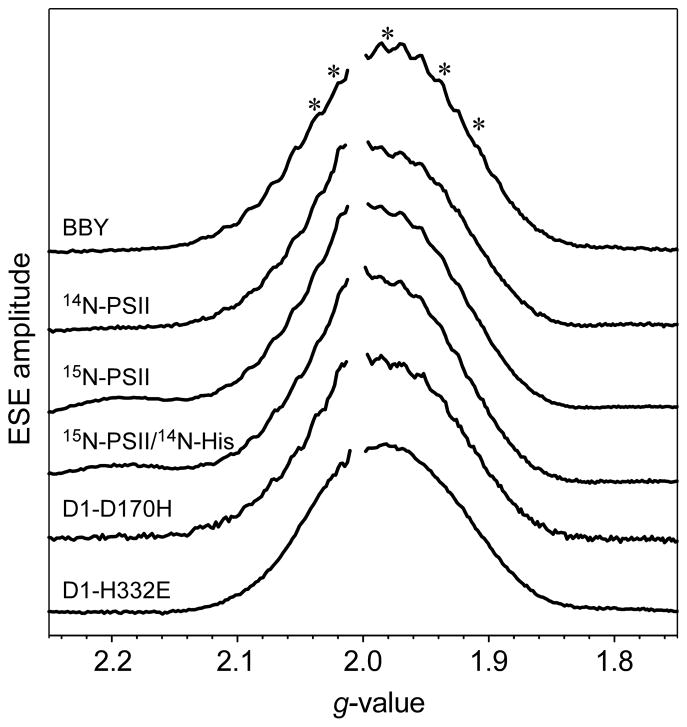
The Ka-band two-pulse ESE field-swept EPR spectrum for each PSII sample described in the Materials and Methods section is presented in Figure 2. The spectra are, for comparison, plotted versus the effective g-values instead of B0, to account for the slightly different microwave frequencies at which they were acquired. For clarity, the signal from YD• was removed from each trace. Excepting the data of the D1-H332E mutant, all the EPR spectra possess partially resolved 55Mn hyperfine couplings.
Figure 2.
Ka-band two-pulse ESE field-swept light-minus-dark spectra of different preparations of PSII. (A) Ethanol-treated PSII from spinach (BBY); and from Synechocystis (B) 14N-PSII, (C) 15N-PSII, (D) 15N-PSII/14N-His, (E) D1-D170H, and (F) D1-H332E. The asterisks (*) indicate the positions (g-factors kept constant) at which two- and three-pulse ESEEM experiments were performed. All Ka-band data presented in Figures 3–6 were acquired at g = 1.98. Microwave excitation frequency: (A–C) νMW = 30.757 GHz, (D) νMW = 30.783 GHz, (E) νMW = 30.847, and (F) νMW = 30.562 GHz. Additional instrument settings: π/2 = 30 ns, τ = 220 ns, ΔB0 = 1 mT, repetition time = 5 ms, T = 4.5 K.
The spectrum of ethanol-treated PSII from spinach (BBY) has a symmetric, Gaussian line-shape centered at g = 1.98, while the Synechocystis samples have spectra that have a slight asymmetry with greater intensity evident on the low field side of the YD• signal. This difference could be due to the contribution from of a small amount of photooxidized cytochrome (g-values = [2.98, 2.25, 1.53]).(60) All high(er)-field EPR studies (Q-band and W-band (94 GHz)) of plant(61) and cyanobacterial(62,63) PSII frozen solutions and single crystals have shown that principal g-values of the OEC in the S2 state are very nearly axial (e.g., g = [1.988, 1.981, 1.965] for PSII from Thermosynechococcus vulcanus(63)). Therefore, the observed asymmetry in the EPR spectrum of some of the Synechocystis samples cannot be attributed to changes in g anisotropy of the S2 signal between plant and cyanobacterial PSII.
The ESE-EPR spectra of wild-type PSII in Figure 2 were scaled according to the nominal concentration of OEC (inferred on the basis of spectrophotometrically determined chlorophyll concentration). The actual integrated areas of the signals of PSII core particles from Synechocystis are each within 10–15% of the ethanol-treated samples from spinach suggesting that the respective peptides are fully loaded with OEC and able to advance to at least the S2 state.
The star symbols (*) in Figure 2 denote the g-values at which two- and three-pulse ESEEM experiments were performed. All spectra shown in subsequent figures were acquired in resonance with the magnetic field position corresponding to g = 1.98. As described in the Materials and Methods section, all features observed in the presented frequency spectra were seen at all field positions examined.
Two-Pulse and Three-Pulse ESEEM
Natural Abundance PSII (BBY and 14N-PSII)
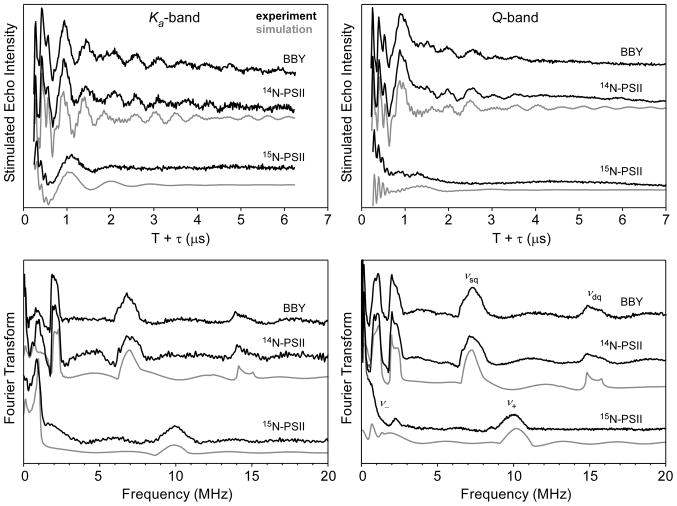
The light-minus-dark Ka-band two-pulse ESEEM spectra of BBY, 14N-PSII and 15N-PSII are shown in Figure S2. The corresponding frequency spectra achieved by cosine-backfilled Fourier transform are presented at the bottom of Figure S2. For each species, time-domain spectra acquired at Ka- and Q-band (see Supporting Information for Q-band data) excitation frequencies are quite similar, with both data sets showing deep modulation. The Ka-band spectrum of BBY (B0 = 1107.9 mT) is the same as that published previously and is shown again here for comparison to that of Synechocystis PSII.(41) The BBY two-pulse ESEEM spectrum has peaks at 2.09, 6.9, and 14.2 MHz that are nearly identical to features found in the 14N-PSII spectrum (B0 = 1107.9 mT) at 2.11, 6.9, and 14.3 MHz. Also present in the 14N-PSII spectrum are two features at 4.6 and 9.4 MHz with negative intensity that flank the positive peak at 6.9 MHz. Such negative intensity features are common in two-pulse ESEEM data and indicate that the corresponding peaks arise from combination frequencies (sum and difference) of the 2.11 MHz and 6.9 MHz transitions. The lifetime of the spin-echo in a two-pulse ESEEM experiment is limited by the phase memory time, which, for the OEC at 4.5 K, is relatively short (Tm ≈ 0.5 μs). As a result, low-frequency modulations (< 2 MHz) are poorly resolved.
Light-minus-dark three-pulse ESEEM spectra of BBY, 14N-PSII and 15N-PSII are shown in Figure 3. In contrast to the two-pulse ESEEM data, the intensity of the three-pulse ESEEM stimulated echo is not limited by phase memory but rather by spin-lattice relaxation (T1), which is much longer in this case. Thus modulations can persist for much longer making low frequency modulations easier to detect and the higher frequency features narrow compared to corresponding features in the two-pulse ESEEM spectrum. Additionally, combination peaks do not appear in three-pulse ESEEM spectra unless nearly equivalent hyperfine-coupled nuclei are present. Similar sets of peaks appear in both the BBY and 14N-PSII Ka-band three-pulse ESEEM spectra at 0.93, 2.01, 6.9 and 14.0 MHz; and 0.97, 2.00, 6.9, and 14.0 MHz, respectively (Figure 3, left). No additional spectral features are present in respective Q-band three-pulse ESEEM spectra (Figure 3, right). However, the two highest frequency peaks in the Ka-band spectra have shifted higher in Q-band spectra—Δ = 0.34 MHz and 0.70 MHz—due to an increase in the 14N nuclear Zeeman field. These shifts are consistent with the features being assigned to the νsq and νdq transitions, respectively. That the other, lower frequency features at approximately 1 and 2 MHz do not significantly change frequency with increasing B0 is diagnostic of their arising from nuclear transitions between nearly pure quadrupole states.
Figure 3.
Time-domain (top) and corresponding cosine-backfilled Fourier transformed (bottom) Ka- (left) and Q-Band (right) three-pulse ESEEM spectra of BBY, and 14N-PSII and 15N-PSII from Synechocystis. Ka-band instrument settings νMW = 30.757 GHz, B0 = 1.1079 T, π/2 = 15 ns, τ= 170 ns, ΔT = 15 ns, repetition time = 5 ms, T = 4.5 K. Q-band instrument settings for Synechocystis spectra: νMW = 33.927 GHz, B0 = 1.2229 T, π/2 = 16 ns, τ = 172 ns, ΔT = 16 ns, repetition time = 5 ms, T = 4.5 K. For spinach PSII spectra: νMW = 33.888 GHz, B0 = 1.2225 T, π/2 = 16 ns, τ = 172 ns, ΔT = 16 ns, repetition time = 5 ms, T = 4.5 K Spectral simulations were performed using the parameters given in Table 1.
Uniformly 15N-labeled PSII (15N-PSII)
The Ka- and Q-band ESEEM spectra of PSII from Synechocystis grown with 15NO3− as the sole nitrogen source are compared to those of BBY and 14N-PSII in Figures 3 and S2. The Ka-band two-pulse ESEEM spectrum of 15N-PSII lacks all features found in the natural abundance isotopologue spectra (Figure S2). Instead, a strong peak appears at 9.7 MHz. Possible combination peaks (negative features) at 8.6 and 10.8 MHz are present as well as a narrow feature at 0.9 MHz. The two-pulse ESEEM spectrum for 15N-PSII was very sensitive to background correction (cf. traces in Figure S3). For the Q-band data, a strong feature is observed at 10.30 MHz with a positive shoulder centered at 9.4 MHz. Two negative features appear at 7.75 and 11.27 MHz, and a broad, less intense, positive feature is centered at ≈18 MHz.
The corresponding three-pulse ESEEM spectra are more illuminating. A low-frequency modulation is now evident in the spectra at both excitation frequencies (Figure 3). The Ka-band spectrum has a narrow, intense feature at 0.85 and a broader peak at 9.9 MHz. These features are assigned to the νa and νb 15N nuclear spin-flip transitions in the two electron spin manifolds. That the latter peak appears at roughly twice the 15N Larmor frequency at B0 = 1108 mT (νL(15N) = 4.78 MHz) indicates that the corresponding 15N nucleus is experiencing near-cancellation conditions. Thus the isotropic (Aiso) and dipolar (Adip) contributions to the hyperfine matrix can be estimated directly from the frequencies of the two peaks seen Figure 3 (0.85 and 9.9 MHz) using the following relations: νa ≈ 3/4*Adip and νb ≈ Aiso + Adip/2.(64) We estimate Aiso(15N) = 9–10 MHz and Adip(15N) = 0.85–1.2 MHz. Peaks in the Q-band spectrum are more structured yet broader than those in the Ka-band spectrum signaling a departure from cancellation conditions (i.e., νL(15N) is no longer approximately equal to Aeff/2).
Additional, though small, spectral features at 2.25 and 8.45 MHz are apparent in Q-band three-pulse ESEEM light-minus-dark spectra. These features are centered at the 15N Larmor frequency of 5.28 MHz (at 1222.9 mT) and split by approximately 6 MHz. No corresponding peaks are obvious in any of the Ka-band ESEEM spectra of 15N-PSII. While these features may arise from an additional 15N nucleus that is hyperfine coupled to the OEC, it is more likely that they are from the four heme nitrogens of cytochrome b559 (Cyt b559). Indeed, these peaks are also seen in the spectrum of dark-adapted PSII, and the simulated ESEEM spectrum generated using reported 15N HFI parameters for Cyt b559 perfectly reproduces the frequencies observed here (see Supporting Information).(60) The intensity of the Cyt b559 EPR spectrum in cyanobacterial PSII preparations is known to be sensitive to light.(21) This could lead to incomplete subtraction of the Cyt b559 signal and allow nuclei coupled to the low-spin heme to be a minor contribution to the light-minus-dark ESEEM spectra.
ESEEM Simulations
In both the spectra of 14N-PSII and 15N-PSII from Synechocystis, all features are assumed to arise from the coupling of a single nitrogen nucleus to the S = 1/2 electron spin of the OEC poised in the S2 state. The precise magnitude of this magnetic coupling reports on not only the electronic structure of the nitrogen atom but the nature of the coordinating Mn ion—namely its oxidation state—as well. Beginning with the 15N hyperfine values estimated from the Ka-band ESEEM spectra above, we have simultaneously fit the 15N- and 14N-PSII ESEEM time traces obtained at both Ka- and Q-band excitation frequencies using a least-squares optimization routine implemented in EasySpin.(58,59) Spectra acquired using other values of τ were also used to constrain the simulation.
These best-fit three-pulse simulations were achieved with the following spin Hamiltonian parameters for the coupled 14N center (the parameters for the 15N-PSII ESEEM simulations are given simply by scaling the hyperfine values determined for 14N by the ratio of the respective nuclear gyromagnetic ratios; γ(15N)/γ(14N) = −1.4028): Aiso = 6.95 MHz, and Aaniso = [0.20 1.30 −1.50] MHz, e2Qq/h = −1.98 MHz, and η = 0.815. The signs of the contributing terms to the hyperfine interaction are only relative. The sign of e2Qq/h is assumed to be negative based on electron nuclear double resonance (ENDOR) data of other metal-coordinated imidazoles.(65)
The time-domain ESEEM data and corresponding Fourier transforms for BBY and 14N-PSII are essentially identical as is shown in Figure 3. As a result, the magnetic parameters determined here for Synechocystis PSII are very similar to those determined for the nitrogen coupled to the OEC in spinach PSII.(41) The modest differences in the nitrogen hyperfine tensor—a 0.35 MHz reduction in Aiso and an increase in hyperfine anisotropy—were required to properly simulate the ESEEM spectrum of 15N-PSII, an isotopologue that was not available for our earlier study on BBY. We therefore argue that the strength of the 14N hyperfine interaction with the unpaired electrons on the OEC in the S2 state is virtually identical between spinach and Synechocystis PSII.
Natural Abundance Histidine in 15N-PSII (14N-His/15N-PSII)
To assign the nitrogen-isotope sensitive features found in the above ESEEM data, we have also examined samples of uniformly 15N-labeled PSII that contain natural abundance 14N-histidine (14N-His/15N-PSII). These data should allow for facile distinction between spectroscopic features from nitrogens in histidine and those from other sources such as the polypeptide amide backbone or other nitrogen-containing amino acid side chains (e.g. lysine, glutamine, arginine, etc.).
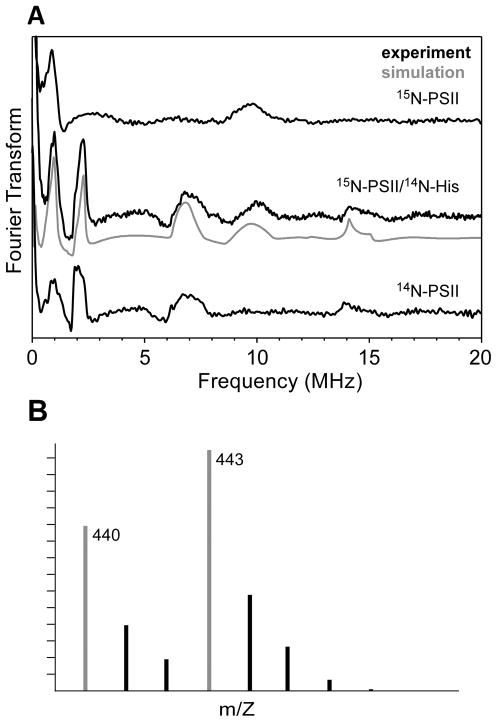
The Ka-band ESEEM spectrum of 14N-His/15N-PSII is presented in Figure 4A and has features at 1.0, 2.0, 7.0, 9.9, and 14.0 MHz. Upon comparison of this spectrum to analogous data of 14N-PSII and 15N-PSII, it is clear that 14N-His/15N-PSII has spectral features common to both isotopically pure samples. Mass spectrometric analysis of the TBS-derivatized hydrolyzed 14N-His/15N-PSII shows that ≈60% of histidine residues in the sample remain 15N-labeled as indicated by the peak at 443 m/z in Figure 4B and the remaining 40% are 14N-labeled (peak at 440 m/z). Therefore, simulation of the three-pulse ESEEM spectrum of 14N-His/15N-PSII (Figure 4A) requires a combination of the 14N and 15N computed signals from above (Figure 3). The two contributing time-domain signals were each generated using the magnetic parameters given in Table 1. The resulting 14N and 15N spectra were scaled by the factors 0.4 and 0.6, respectively, then added together. The resultant simulated spectrum models the experimental data extremely well. Though this degree of isotope contamination is quite high given that previous similar experiments showed the histidine isotopologue heterogeneity to be ≈15%,(39,66,67) the agreement of the GS/MS results with the observed ratio in the 14N-His/15N-PSII ESEEM spectrum confirms that all features arise from histidine nitrogen that is strongly interacting with the OEC.
Figure 4.
(A) Cosine-backfilled Fourier transformed Ka-band three-pulse ESEEM spectrum of 15N-PSII/14N-His (center) compared to those for 14N-PSII (top) and 15N-PSII (bottom). The 14N-PSII and 15N-PSII spectra are the same as those presented in Figure 3, bottom left. Acquisition of the 15N-PSII/14N-His spectrum employed the following instrument settings: νMW = 30.837 GHz, B0 = 1.1118 T, π/2 = 10 ns, ΔT = 15 ns, repetition time = 5 ms, T = 4.5 K and is the sum of spectra obtained using three different values for τ = 170, 200, and 210 ns. (B) GC/MS mass spectrum of the TBS-derivatized hydrolyzed 15N-PSII/14N-His.
Table 1.
Magnetic Coupling Parameters for Nitrogens bound to Biological Mn-clusters
| Species | Aiso (MHz) | Aaniso (MHz) | e2Qq/h (MHz) | η | Ref. |
|---|---|---|---|---|---|
| 14N-PSII (Synech.)a | 6.95 | 0.2 1.3 −1.5 | 1.98 | 0.82 | this work |
| 15N-PSII (Synech.) | −9.75 | −0.3 −1.8 2.1 | n.a. | n.a. | this work |
| PSII (BBY) | 7.3 | +0.5 +0.5 −1.0 | 1.98 | 0.84 | (41) |
| superoxidized MnCat (L. plantarum) | +2.67 | +0.41 +0.16 −0.57 | 2.25 | 0.58 | (75) |
| −5.75 | −0.2 −0.6 +0.8 | 2.01 | 0.79 | ||
| superoxidized MnCat (T. thermophilus) | +2.28 | +0.42 +0.28 −0.70 | 2.29 | 0.50 | (75) |
| −5.2 | −0.3 −0.7 +1.0 | 2.25 | 0.65 |
Euler rotation angles between the hyperfine and nuclear quadrupole tenors [α, β, γ] = [−30, 0, 40]°.
Site-Directed Mutants
To identify amino acids that could be coordinating to the OEC, we have examined the EPR spectra of two site-directed PSII mutants, D1-H332E and D1-D170H. On the basis of mutagenesis and crystallographic studies, both D1-His332 and D1-Asp170 have been suggested as ligands to the Mn cluster.(4) The ESE EPR spectra of the D1-H332E and D1-D170H mutants (Figure 2) are reduced in intensity by 65% and 45%, respectively, compared to that of 14N-PSII. Previously, EPR spectra of D1-H332E PSII have been shown to have only 60% of the MLS found for wild-type*.(34) Notably, the Ka-band D1-H332E spectrum lacks 55Mn hyperfine features that are present in all other data. The S2 state of this mutant is known to have a narrower X-band EPR spectrum with 55Mn hyperfine lines that are split by much less than is observed for the wild-type* enzyme. A similarly altered MLS has also been found for ammonia-treated(68) and strontium-substituted(69) PSII preparations.
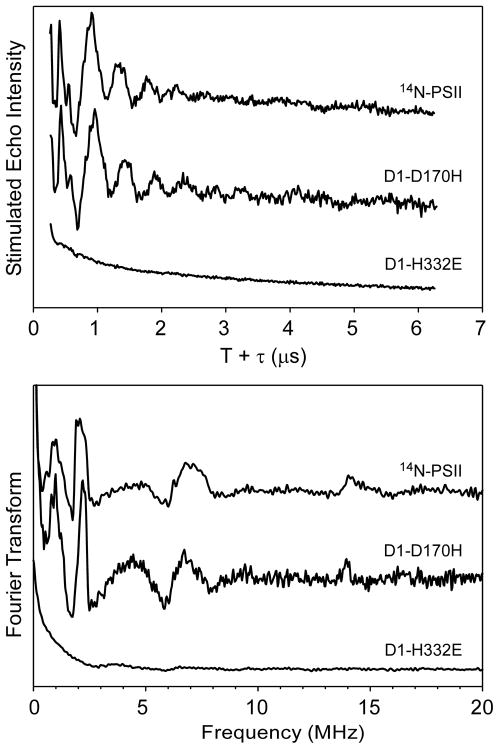
The ESE EPR spectrum of D1-D170H, on the other hand, is very similar to that of 14N-PSII. Its reduced intensity is correlated with a decrease in oxygen evolution activity (60% that of wild-type*).(70) One cause of these effects could be a decrease in the success of OEC photoassembly during which D1-Asp170 has been directly implicated as a participant.(70–74) The Ka- and Q-band ESEEM data of D1-D170H PSII show deep modulation patterns that are identical to those observed for 14N-PSII. Additional three-pulse ESEEM data sets for D1-D170H acquired using other τ-values, and at other magnetic field positions, found no differences compared to analogous data of 14N-PSII.
Earlier ESEEM studies performed at X-band showed that a nitrogen isotope sensitive feature at ≈5 MHz is greatly affected by the D1-H332E mutation (see Figure S1).(38–40) The Ka-band three-pulse ESEEM data of D1-H332E PSII presented here are devoid of any modulations (Figure 5, top) and thus give rise to no peaks in the Fourier transform (Figure 5, bottom; see also Figure S8).
Figure 5.
Time-domain (top) and corresponding cosine-backfilled Fourier transformed (bottom) Q-Band three-pulse ESEEM spectra of D1-H332E and D1-D170H PSII compared to those of 14N-PSII. The 14N-PSII spectrum is the same as that presented in Figure 3, bottom right. Instrument settings for D1-D170H data collection: νMW = 30.783 GHz, B0 = 1.1098 T, π/2 = 10 ns, τ = 200 ns, ΔT = 15 ns, repetition time = 5 ms, T = 4.5 K. For D1-H332E data collection: νMW = 30.562 GHz, B0 = 1.1063 T, ν/2 = 10 ns, τ = 210 ns, ΔT = 15 ns, repetition time = 5 ms, T = 5.0 K.
Discussion
Quantitative Analysis of Nitrogen Coupling to OEC
Simultaneous fitting of the Ka- and Q-band ESEEM spectra of 14N-PSII and 15N-PSII from Synechocystis sp. PCC 6803 acquired at multiple resonant field positions yielded precise magnetic parameters for a single nitrogen nucleus coupled to the Mn-cluster in the S2 state. These parameters—Aiso = 6.95 MHz, and Aaniso = [0.20 1.30 −1.50] MHz, e2Qq/h = 1.98 MHz, and η = 0.815—are reminiscent of those found for the imidazole nitrogen from histidine that is coordinated to the Mn(III) ion of the dimanganese-containing catalase (MnCat) poised in the Mn(III)Mn(IV) state.(75) These features must arise from a histidine imidazole group because the ESEEM spectrum of 15N-PSII containing 14N-His as the sole source of 14N possesses features found in the spectrum of the isotopically pure 14N-PSII (see Figure 4).
The effective hyperfine interaction measured here is dominated by a large isotropic contribution Aiso. The value of Aiso depends on (i) the degree of overlap between the nitrogen nucleus and the Mn-centered orbitals containing unpaired electrons (Fermi contact, fs); (ii) the hybridization of the nitrogen 3s-2pn orbitals and (iii) the projection factor c for the coordinating Mn ion (vide infra).
Aiso(14N) for PSII is larger than all other known Mn—N effective hyperfine couplings except for those resulting from a Mn—N bonding vector that is coincident with a lobe of a singly occupied metal-centered 3d-orbital.(75–77) For example, broken-symmetry density functional theory (BS DFT) computations on mixed-valence Mn(III)Mn(IV) dimers have shown that when the nitrogen nucleus lies along the local z-axis of the Mn(III) ion, there is a strong hyperfine interaction with the half-occupied 3dz2-orbital.(78) ENDOR studies of such compounds have detected a 14N HFI on the order of 9–13 MHz that is ascribed to such a coupling mode.(76,79) Also contributing to this large effective HFI is the projection factor of c = +2 for the coordinating Mn(III) ion in the dimer.(80) These projection factors are computed by determining the expectation value of the local electron spin projected onto the total electron spin of the molecule.(81) The Fermi contact can then be calculated using
where a° is the HFI of one electron spin in a 14N 2s orbital (+1811 MHz)(82) and S is the local electron spin of the coordinating ion, 2 in the case of Mn(III). For Mn(III)Mn(IV) MnCat from Lactobacillus plantarum, we found that the 14N atom bound to the Mn(III) ion had an unpaired spin in the 2s-orbital (f2s) equal to 0.64%.(75)
That the OEC is composed of exchange-coupled paramagnetic ions leads to a ladder of possible spin states—the lowest of which has a total spin of S = 1/2, for the S2 state. Results from pulse EPR inversion recovery studies established a range of values for the energy gap between the ground and first-excited spin state manifolds (24–36.5 cm−1, depending on the origin of the and method of preparation of the PSII).(29,30,83) Using this value combined with data from 55Mn ENDOR spectroscopic studies,(27,29,30) we and others have proposed a scheme for the exchange coupling of the four Mn centers and assigned formal oxidation states. We believe that in the S2 state the lone Mn(III) ion is part of the [Mn3Ca] cuboidal/pyramidal cluster that is weakly exchange coupled to MnD (Figure 6). The isotropic projection factors computed by Peloquin et al.(27) for this arrangement are cMn(III) = 1.77 and cMn(IV) ranges from 1.00 to 1.27. We note that the projection factors determined in references (29,30) are not substantially different from these values. If histidine is coordinated to the Mn(III) ion, the resulting f2s(14N) is 0.87%. If instead the imidazole is bound to one of the remaining Mn(IV) ions, this would correspond to a range of f2s(14N) is 0.91–1.15%.
Figure 6.
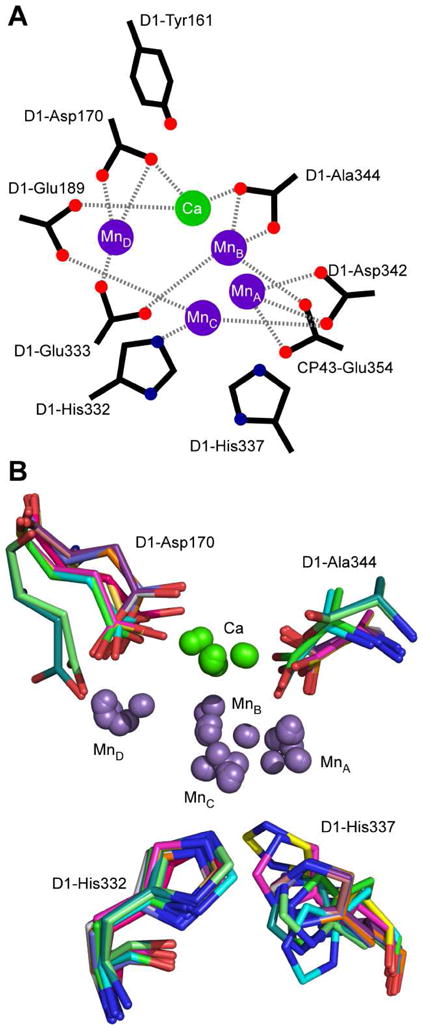
(A) Schematic representation of OEC including several possible liganding amino acids residues. (B) Superposition of [Mn4Ca] clusters and D1-Asp170, D1-His332, and D1-His337 from all available PDB-deposited X-ray crystallographic structures.
That the 14N Fermi contact computed above is so much larger than those measured for the equatorially liganded histidines in MnCat suggests that histidine is bound along an axis that contains a lobe of a half-occupied metal-centered orbital. Such a binding geometry seems unlikely for a high-spin d3 Mn(IV) ion as disruption of a pseudo-octahedral ligand field would lead to a significant loss of ligand field stabilization energy. Therefore, we prefer the description of the bonding partner to histidine as being a Mn(III) ion. In this case, the larger projection factor for a Mn(III) bound nitrogen rationalizes the observed large 14N HFI.
The nuclear quadrupole coupling parameters e2Qq/h and η reported above are at the extreme of the range of those found previously for d10 M(II)-coordinated imidazoles,(84) though almost identical to that found for the histidine bound to the Mn(III) ion in superoxidized MnCat, e2Qq/h = 2.01 MHz (Table 1).(75) The largest part of the electric field gradient at the nitrogen nucleus (q) is expected to coincide with the direction of the nitrogen lone pair—presumably lying along the Mn—N bonding axis. Though, in the absence of ESEEM data on oriented single crystals, we cannot be certain of this assignment. The magnitude of e2Qq/h measured for 14N-PSII indicates significant charge donation from the nitrogen lone-pair to the Mn-center reducing its population from 2.00 to approximately 1.65 electrons. This depopulation is driven by the Lewis acidity and high formal charge of the coordinating metal as well as the σ-donor strength of the histidine ligand.
Earlier studies have shown that the S2 MLS for BBY preparations has nearly identical spectral and spin-relaxation properties as those found for PSII derived from cyanobacteria. (29,30,62,63,85) As their magnetic properties are so similar, one can presume that structures of the respective OECs are also similar. Thus it not surprising that the magnetic parameters for the histidine nitrogen determined in this study of Synechocystis PSII are comparable to those found in our earlier Ka-band ESEEM study of PSII from spinach (cf. Table 1 and Figures 3 and 4).(41) As such, we expect that the origin of the nitrogen coupling is the same for each protein.
Identification of Proteinaceous Ligands to the OEC
Despite the advent of numerous X-ray structures, the identity of the proteinaceous ligands to the OEC in each of the five S-states remains somewhat controversial. Analysis of these PSII crystal structure data in conjunction with results from spectroscopic and biochemical studies of site-directed mutants suggest a number of possible ligands (see Figure 6A). However, the X-ray beam-induced reduction of the Mn sites to the +2 oxidation state leads to changes in the metal coordination environment.(86,87) One possible manifestation of this effect is illustrated in Figure 6B which compares the positions of D1-Asp170, D1-His332, D1-His337, D1-Ala344, and the five metal ions of the OEC determined in each of the crystal structures of PSII from T. elongatus and T. vulcanus (PDB entries 1S5L, 1W5C, 2AXT, 3A0B, 3A0H, 3BZ1, 3BZ2, and 3KZI).(5–13) While the position of D1-His332 is conserved with its imidazole in close proximity to MnC, D1-Asp170 and D1-His337 seem to sample a wide conformational space. Thus, we must rely on results from non-destructive spectroscopic techniques such as Fourier transform infrared (FTIR) difference and EPR to assess the native coordination environment of the OEC.
A recent Q-band (35 GHz) ENDOR study of Synechocystis PSII for which all alanine residues were 13C-labeled at the carbonyl carbon position showed conclusively that D1-Ala344 is coordinated to Mn with Aiso ≈ 1.0–1.2 MHz.(35) These findings corroborated earlier results from FTIR difference spectra which indicated that D1-Ala344 is bound to a Mn ion that undergoes a change in oxidation state upon advancement from S1 to S2.(49,66,88) Analogous ENDOR data on uniformly 13C-labeled PSII particles indicated that several other carbon-containing moieties are magnetically coupled to the OEC in the S2 state. However, the assigning of these additional spectral features to specific amino acid ligands is ongoing.
One oft-posited carboxylate ligand candidate is the aspartate residue at position 170 in the D1-polypeptide.(45,50,70–73,89–92) During photoassembly, D1-Asp170 has been shown to be of great importance in the binding of the first equivalent of Mn(II) ion to the Mn-depleted PSII protein.(72–74,91) This ion is then photo-oxidized to Mn(III) by the same electron transfer machinery responsible for water oxidation as the first step in generating the active OEC. Several of the X-ray structures show D1-Asp170 as being closest to MnD (Figure 6B).
As it was not possible to site-specifically introduce magnetic nuclei (e.g., 13C (I = 1/2) or 17O (I = 5/2)) into the carboxylate side-chain of D1-Asp170, we instead specifically mutated into this position a histidine to use its two 14N atoms (I =1) of the imidazole side-chain to probe possible magnetic couplings with the OEC. The effects of the D1-D170H mutation were first evaluated by FTIR difference studies.(45,50,93) These spectra showed no changes compared to those of the wild-type* enzyme in the 1000–1800 cm−1 region implying that the carboxylate of aspartate is not bound to a Mn center that is oxidized from S0 to S3. It is noteworthy, however, that computational studies of OEC models have suggested carboxylate vibrational modes would be insensitive to metal ion oxidation if the corresponding change in charge is delocalized over the entire OEC.(94)
Earlier X-band(70) and the present Ka- and Q-band ESEEM spectra of the D1-D170H mutant show no new features compared to analogous spectra of the wild-type* enzyme (see Figure 5). Based on analogy to our EPR studies of MnCat,(75) if D1-His170 were coordinated to a Mn(IV) ion we would have expected to see evidence of this coupling in the X-band ESEEM spectrum. If it were instead bound equatorially to a Mn(III) ion, we should have seen features in the Ka- and Q-band ESEEM spectra. That we see no features at all these excitation frequencies may suggest that the D1-His170 imidazole is not a ligand to Mn. However, given the presence of other magnetic nuclei that are coupled to the OEC and are strongly modulating (e.g. 14N from either D1-His332 or D1-His337, see below), modulations from a weakly-coupled D1-His170 could be lost due to cross-suppression effects.(95) Alternatively, the hyperfine coupling could be sufficiently anisotropic as to broaden the corresponding ESEEM features beyond detection. Another possibility is that D1-His170 samples a wide conformational space when the OEC is poised in the S2 state as implied by the varied positions D1-Asp170 adopts in X-ray crystallographic results (Figure 6). This would lead to inhomogeneous broadening of any corresponding ESEEM features. Finally, the magnitude of hyperfine coupling could be so large as to make the energy of nuclear spin-flip transitions—necessary to observe the ESEEM effect—beyond the excitation bandwidth of the microwave pulses. This last possibility, though, is unlikely. X-band CW EPR spectra of WT* and D1-D170H samples poised in both the S1 and S2 states have essentially identical linewidths.(70) An earlier comparison of PSII from spinach grown hydroponically on 14N-nitrate and 15N-nitrate showed no discernible broadening of the EPR resonances.(96) Using simulations, the author argued that this lack of broadening sets a maximum on the isotropic hyperfine of a coupled 14N nucleus at ≈8–9 MHz, a value within our experimentally detectable range.(97)
Current proposals for native nitrogenous ligands to the OEC are limited to a Mn-coordinating imidazole nitrogen from either D1-His332 or D1-His337. It has been suggested that one or both of these residues may participate in the photoassembly of the OEC by coordinating with high-affinity, a Mn(II) ion.(92,98) In fully assembled PSII, D1-His337 has been proposed to act as the initial holding site for proton translocation, shuttling protons liberated from the OEC-substrate complex to the thylakoid lumen.(99) All mutations of D1-His332 in Synechocystis either greatly retard or abolish O2-evolution activity.(89) The D1-H332E mutant is able to assemble the cluster but cannot advance past the S2 state.(34,100) The corresponding S2 MLS observed previously at X-band(40) and here at Ka- and Q-band is slightly narrower than that found for native or wild-type* PSII preparations (see Figure 2) due to a reduction in the effective 55Mn hyperfine splitting.
This spectral change could be due to a change in the electronic ground state of the Mn(III) ion from 5B1g (corresponding to an empty 3dx2−y2 metal-centered orbital) to 5A1g (corresponding to an empty 3dz2-based orbital).(27,73,101) In the mutant, it is possible that the binding of the D1-Glu332 to MnC changes the relative energies of the eg orbitals leading to depopulation of the 3dz2-based molecular orbital. Alternatively, replacing a neutral imidazole ligand with the anionic sidechain of glutamate may lower the reduction potential of MnC relative to the other Mn-centers leading to a redistribution of oxidizing equivalents within the OEC. This intracluster electron transfer could place the formal Mn(III) ion in a sufficiently different bonding environment to yield a 5A1g ground state and give rise to the altered MLS. Interestingly, Cox et al. showed that the altered MLS generated by substituting Sr2+ for Ca2+ in PSII from T. elongatus does not result from a change in electronic ground state description of the Mn(III) ion.(30) Rather, a slight diminution of the Mn(III) ion zero-field splitting constant leads to reduction in both g-tensor and 55Mn hyperfine anisotropy for the OEC. Thus, there appear to be several mechanisms that could give rise to the altered MLS.
The X-, Ka-, and Q-band ESEEM spectra of D1-H332E mutant PSII from Synechocystis show none of the nitrogen isotope sensitive features observed in the spectra of wild-type* PSII (Figures 5 and S1). Most simply this indicates that the source of the nitrogen coupling is from D1-His332 and upon mutation to glutamate, there is no longer a nitrogen ligand available.
Comparison of Thermophilic and Mesophilic Cyanobacterial PSII
In contrast to these findings, Boussac and coworkers recently found that mutation of D1-His332 (to either Ser or Gln) in PSII from T. elongatus only slightly attenuates O2-evolution kinetics (activity is 80% of wild-type* enzyme) and does not alter the S2 MLS.(36) Furthermore, the corresponding X-band ESEEM spectrum of T. elongatus D1-H332S PSII possesses what appears to be the same feature at ≈5 MHz that has been previously attributed to the OEC-coordinating histidine.(38,39) The authors thus concluded that it is D1-His337 that provides the nitrogen that is hyperfine coupled to the OEC in the S2 state. The origin of this discrepancy between the Synechocystis and T. elongatus ESEEM results remains unclear. We do note, however, that using the 14N magnetic parameters given in Table 1 to simulate an ESEEM spectrum acquired at 352 mT we expect to observe a prominent feature 4.8 MHz with a shoulder at 2.5 MHz (see Figure S4). This strongly suggests that the features previously observed at X-band excitation frequencies and those obtained by excitation at Ka- and Q-band arise from the same hyperfine-coupled nucleus.
It is unlikely that the two native enzymes have different ligand sets for the OEC. However, there is no X-ray diffraction data available for PSII isolated from Synechocystis. Instead, all the available crystal structures of PSII are for enzyme isolated from thermophilic cyanobacteria including T. elongatus. In each of the PDB files, the D1-His332 ε-N is in close proximity to MnC (rMn—Nε = 1.99–2.49 Å) and the δ-N is H-bonded to the backbone carbonyl of D1-Glu329, locking the imidazole ring into place. Recently, the carboxylate sidechain of D1-Glu329 has been postulated to take part in an extended H-bonding network involved in proton translocation.(102) Mutating D1-His332 to glutamate may perturb this network and contribute to the minor loss of O2-evolution activity observed in T. elongatus.
The D1-H332E and D1-H332D mutants in Synechocystis are inactive, yet Synechocystis D1-H332S has 10–15 % activity of wild-type*. The corresponding change in fluorescence and thermo-luminescence properties suggests that mutating D1-His332 to a residue with a carboxylic side chain affects the communication between the electron donor side of PSII (the OEC) and the acceptor side 45 Å away, where plastoquinone is bound.(100) The serine mutant does not suffer from these effects again pointing to the role of this side chain as a H-bond donor to the backbone of D1-Glu329 that preserves the structural relationship between the donor and acceptor sides of the charge separation pathway.
It is interesting to note that of the 112 residues within 12 Å of the [Mn4Ca] cluster3 the only significant difference between the corresponding sequences of amino acids for the two organisms is immediately adjacent to D1-Glu329 (Table 2). In T. elongatus as well as T. vulcanus and spinach, D1-328 is a methionine whereas in Synechocystis this residue is a phenylalanine. Nonetheless, it is difficult to imagine how such a difference could give rise to a change in OEC ligand set.
Table 2.
Non-Identical Amino Acid Residues within 12 Å of OEC.
| polypeptide→ | D1 | CP43 | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| species/residue | 85 | 87 | 290 | 326 | 328 | 307 | 312 | 411 |
| Synechocystis | S | N | V | I | F | P | S | I |
| T. elongatus | S | N | I | L | M | M | A | I |
| spinach | T | A | I | L | M | P | A | A |
At this time, we cannot rule out that the nitrogen responsible for the observed ESEEM is on the imidazole ring of D1-His337. Results from recent Mn-edge XAS studies that compared wild-type Synechocystis PSII and the D1-H332E mutants suggest that there could be significant structural perturbations—a 0.05 Å elongation of Mn···Mn internuclear distances—of the OEC upon this mutation.(103) The authors offer that such geometric changes and the corresponding changes in electronic structure (described by the altered MLS and different behaviors for electron transfer)(34,100,104) could eliminate the hyperfine coupling from D1-His337. Somehow, the PSII mutants D1-H332S and D1-H332Q from T. elongatus do not similarly affect the electronic structure and the X-band nitrogen ESEEM spectrum is apparently unchanged.(36) Higher frequency studies on these T. elongatus mutants as well as on complementary mutants in Synechocystis are clearly warranted, as the results will afford a quantitative evaluation of any changes in nitrogen magnetic parameters.
Other T. elongatus PSII D1 mutants seem to preserve characteristics of the wild-type enzyme whereas the corresponding mutants in Synechocystis exhibit dramatically different properties. One such example is D1-His198, which serves as the axial ligand to Mg in chlorophyll D1 that makes up part of P680, the primary electron donor in PSII. In Synechocystis, the mutants D1-H198A and D1-H198Q change the absorption spectrum of P680 and affect its reduction potential.(105) In T. elongatus, these mutations have little effect.(106) The sidechain of D1-Glu130 is a H-bonding partner to pheophytin bound in D1 (PheoD1). The D1-Q130E substitution in T. elongatus has no effect on the kinetics and thermodynamics of electron transfer involving PheoD1(106) whereas, in Synechocystis, this mutant changes the pheophytin reduction potential by ≈38 meV.(107,108) In these cases, it seems that mutation of ligands to redox-active cofactors leads to significant changes in the mesophilic cyanobacterium Synechocystis, but through some unknown mechanism, the thermophilic T. elongatus is largely immune.
Why is Histidine a Ligand to the OEC?
Excepting PSII, all known Mn-based oxidoreductases (e.g. Mn-superoxide dismutase,(109) Mn-ribonucleotide reductase (RNR),(110) and the dimanganese catalase(111)) employ at least one histidine residue as a ligand to each Mn ion that makes up the active site. One possible reason for this is that the neutral charge and intermediate Lewis softness of the imidazole ring (assuming the distal nitrogen retains its proton) prevents the coordinating Mn ion from being oxidized beyond the 3+ oxidation state. Anionic ligands such as carboxylates and cystienates, on the other hand, bind strongly to and help stabilize the charge of higher oxidation states of coordinating metal ions. For example, in 2-Cys/2-His liganded Rieske-type(112,113) and even 3-Cys/1-His liganded [Fe2S2] clusters,(97) the Fe center coordinated by histidine(s) is not the site of oxidation as the more electronegative imidazole stabilizes the ferrous ion. The relatively large amount of unpaired spin in the 14N 2s-orbital (f2s(14N) = 0.87%) is a sign of the strength of interaction between the OEC and its active-site histidine. Perhaps then, PSII exploits this relationship to keep one Mn reduced (in the 3+ oxidation state), at least until the other Mn-centers of the OEC are oxidized. Support for this idea is gained by the hypothesis that alternative distribution schemes of the oxidizing equivalents over the four Mn ions abort the water-oxidation mechanism at the S2 state. For example, the altered MLS generated by D1-H332E site-directed mutagenesis of Synechocystis PSII (Figure 2) can be rationalized in terms of a change in the location of the Mn(III) ion within the cluster.(27) This would of course change the location of the Jahn-Teller axis and possibly lead to the perturbation of Mn333Mn distances seen in the EXAFS of the D1-H332E mutant.(103)
Protic ligands such as histidine can also activate metal-bound substrates. As one example, kinetic studies of eukaryotic arginase revealed that mutations of amino acids hydrogen-bonded to one of the metal-coordinating histidines could dramatically affect the Lewis acidity of the metal ion and enhance the rate of arginine hydrolysis.(114) Deprotonation of imidazole-like ligands on a Ru(II) complex was found to destabilize occupied metal-centered π-donor orbitals allowing for stronger interaction with the formally unoccupied C—H anti-bonding orbital of the substrate.(115) If MnC also coordinated one of the substrate waters, one can imagine that the above mechanisms would (i) enhance formation of a metal-bound hydroxo species and (ii) activate the O—H bond for insertion of an oxygen atom derived from the other OEC-bound substrate moiety. This would generate a Mn-peroxo intermediate of the type that has been proposed based on results from studies that attempted to drive S0 back to an S4-like state using high O2 pressure.(116,117)
Conclusions
The nitrogen isotope sensitive features found in the Ka- and Q-band ESEEM spectra of PSII from Synechocystis PCC 6803 are conclusively assigned to an active site histidine residue. In particular, our analysis of the effective hyperfine coupling points to the lone Mn(III) ion present in the S2 state as being the bonding partner with histidine. Whether the observed coupling arises from an imidazole nitrogen of D1-His332 or D1-His337 cannot be determined definitively from our data. What is known is that mutation of D1-His332 in PSII from Synechocystis leads to significant reduction in oxygen-evolving activity and dramatic changes in electronic structure—loss of all nitrogen isotope sensitive features and possibly a redistribution of oxidizing equivalents around the OEC. For currently unknown reasons, mutations of the same residue in T. elongatus have only modest effect on these fronts.
The nitrogen magnetic coupling parameters reported here (see Table 1) are valuable in evaluating DFT models of the OEC in the S2 state.(118–120) Recently, such calculations have been assessed based on agreement of computed 55Mn hyperfine coupling constants with those derived from multi-frequency ENDOR studies.(121,122) Ligand HFI, however, is a more discriminatory means by which to judge the computed structures. By selectively introducing magnetic nuclei in positions about the OEC(35) (or taking advantage of naturally occurring ones such as the nitrogens in histidine), we can characterize magnetic interactions that will be extremely sensitive to the spin topology of the cluster. Interestingly, an energetically competent computational model of the OEC developed by Siegbahn shows that MnC can be a substrate binding site, is the location of the Mn(III) ion in the S2 state, and is coordinated D1-His332.(119) This model is consistent with many spectroscopic results including those from EXAFS and 55Mn ENDOR studies.(29,30) Importantly, the Siegbahn model also comes closest to predicting the strength of the 14N hyperfine and nuclear quadrupole interactions reported here.(120)
Note added during review
While this manuscript was under review, the 1.9 Å resolution crystal structure of PSII from T. vulcanus was deposited in the Protein Data Bank (accession code 3ARC) and discussed in a corresponding paper.(16) These data were collected under conditions in which reduction of the cluster due to the incident X-ray beam is greatly diminished compared to earlier crystallography studies. This technique afforded unparalleled resolution of the electron density of the OEC including the identification of five oxo bridges. However, the reported dosing rate still suggests that some Mn-centered reduction occurs (personal communication with Junko Yano and Johannes Messinger).(14) As with all preceding crystal structures, the imidazole sidechain of D1-His332 is shown to be bound directly to one of the Mn centers of the cluster (MnC, Figure 6). The ε-nitrogen of D1-His337, on the other hand, appears to be H-bonded to the μ3-oxo group that bridges the three Mn ions found within the cuboidal part of the OEC. Perpendicular to the MnC—NHis332 bonding vector is a very long MnC—oxo bond of 2.60 Å, leading to an almost square-pyramidal ligand field for MnC. As described in the above Discussion Section, the strength of the 14N hyperfine interaction reported here and previously(41,42) is consistent with the histidine being an equatorial ligand to either a square pyramidal or tetragonally elongated pseudo-octahedral Mn(III) ion (i.e., with a 5B1g ground state).
Supplementary Material
Acknowledgments
We are grateful to Anh P. Nguyen for preparing the thylakoid membranes from Synechocystis used by R.J.S. and to Robert M. McCarrick for preparing the thylakoid membranes from spinach used by G.J.Y.
Footnotes
Some of these results were presented earlier in a conference proceedings report that was not peer-reviewed.(1)
Abbreviations: EPR, electron paramagnetic resonance; ENDOR, electron nuclear double resonance; ESEEM, electron-spin echo envelope modulation; EXAFS, extended X-ray absorption fine structure; FTIR, Fourier transform infrared spectroscopy; HFI, hyperfine interaction, MLS, multiline signal; NQI, nuclear quadrupole interaction; PSII, photosystem II; OEC, oxygen-evolving complex; XAS, X-ray absorption spectroscopy.
The initial report was presented at the 15th International Congress of Photosynthesis in Beijing, China, by J.-R. Shen, Y. Umena, K. Kawakami, and N. Kamiya (poster PS6.5).
Specifically, this sphere includes D1-58–67, 83–90,112, 157–173, 178–191, 290–297, 325–344; D2-312–321, 349–352; and CP43-306–313, 352–358, 399–401, 410–412 (using 3KZI.pdb). Overall, there are 21 differences in the D1 sequence between Synechocystis sp. PCC 6803 and T. elongatus.
This work was funded by the National Institutes of Health grant GM-048242 to R.D.B. and grant GM-076232 to R.J.D and the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences, U.S. Department of Energy, grant DE-FG02-10ER16191 to R.J.D. The EPR spectrometers used in this study are part of the CalEPR center and were funded by NIH grants GM-061211 and S10-RR021075 and the University of California at Davis.
Supporting Information. Additional Ka-band ESEEM spectra collected at other field positions. Q-band ESE EPR and ESEEM spectra along with corresponding simulations. Simulations of cyt b559 contributions to ESEEM data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Yeagle GJ, Gilchrist ML, Jr, Walker LM, Debus RJ, Britt RD. Multifrequency electron spin-echo envelope modulation studies of nitrogen ligation to the manganese cluster of photosystem II. Philos Trans R Soc, B. 2008;363:1157–1166. doi: 10.1098/rstb.2007.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diner BA. Amino acid residues involved in the coordination and assembly of the manganese cluster of photosystem II. Proton-coupled electron transport of the redox-active tyrosines and its relationship to water oxidation. Biochim Biophys Acta Bioenerget. 2001;1503:147–163. doi: 10.1016/s0005-2728(00)00220-6. [DOI] [PubMed] [Google Scholar]
- 3.Debus RJ. Amino acid residues that modulate the properties of tyrosine YZ and the manganese cluster in the water oxidizing complex of photosystem II. Biochim Biophys Acta Bioenerget. 2001;1503:164–186. doi: 10.1016/s0005-2728(00)00221-8. [DOI] [PubMed] [Google Scholar]
- 4.Debus RJ. Protein ligation of the photosynthetic oxygen-evolving center. Coord Chem Rev. 2008;252:244–258. doi: 10.1016/j.ccr.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zouni A, Witt H-T, Kern J, Fromme P, Krauss N, Saenger W, Orth P. Crystal structure of photosystem II from Synechococcus elongatus at 3.8 Å resolution. Nature. 2001;409:739–743. doi: 10.1038/35055589. [DOI] [PubMed] [Google Scholar]
- 6.Kamiya N, Shen JR. Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7 Å resolution. Proc Natl Acad Sci U S A. 2003;100:98–103. doi: 10.1073/pnas.0135651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004;303:1831–1838. doi: 10.1126/science.1093087. [DOI] [PubMed] [Google Scholar]
- 8.Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature. 2005;438:1040–1044. doi: 10.1038/nature04224. [DOI] [PubMed] [Google Scholar]
- 9.Kern J, Loll B, Zouni A, Saenger W, Irrgang KD, Biesiadka J. Cyanobacterial photosystem II at 3.2 Å resolution - the plastoquinone binding pockets. Photosynth Res. 2005;84:153–159. doi: 10.1007/s11120-004-7077-x. [DOI] [PubMed] [Google Scholar]
- 10.Kawakami K, Umena Y, Kamiya N, Shen JR. Location of chloride and its possible functions in oxygen-evolving photosystem II revealed by X-ray crystallography. Proc Natl Acad Sci U S A. 2009;106:8567–8572. doi: 10.1073/pnas.0812797106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guskov A, Kern J, Gabdulkhakov A, Broser M, Zouni A, Saenger W. Cyanobacterial photosystem II at 2.9 Å resolution and the role of quinones, lipids, channels and chloride. Nat Struct Mol Biol. 2009;16:334–342. doi: 10.1038/nsmb.1559. [DOI] [PubMed] [Google Scholar]
- 12.Murray JW, Maghlaoui K, Kargul J, Ishida N, Lai TL, Rutherford AW, Sugiura M, Boussac A, Barber J. X-ray crystallography identifies two chloride binding sites in the oxygen-evolving centre of photosystem II. Energ Environ Sci. 2008;1:161–166. [Google Scholar]
- 13.Guskov A, Broser M, Gabdulkhakov A, Kern J, Zouni A, Loll B, Biesiadka J, Saenger W. X-ray crystallographic analysis of PSII from T. elongatus at 3.0 Åresolution. Photosynth Res. 2007;91:PS422. [Google Scholar]
- 14.Yano J, Kern J, Irrgang KD, Latimer MJ, Bergmann U, Glatzel P, Pushkar Y, Biesiadka J, Loll B, Sauer K, Messinger J, Zouni A, Yachandra VK. X-ray damage to the Mn4Ca complex in single crystals of photosystem II: A case study for metalloprotein crystallography. Proc Natl Acad Sci U S A. 2005;102:12047–12052. doi: 10.1073/pnas.0505207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grabolle M, Haumann M, Müller C, Liebisch P, Dau H. Rapid loss of structural motifs in the manganese complex of oxygenic photosynthesis by X-ray irradiation at 10–300 K. J Biol Chem. 2006;281:4580–4588. doi: 10.1074/jbc.M509724200. [DOI] [PubMed] [Google Scholar]
- 16.Umena Y, Kawakami K, Shen J-R, Kamiya N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature. 2011;473:55–60. doi: 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- 17.Dismukes GC, Siderer Y. EPR spectroscopic observations of a manganese center associated with water oxidation in spinach chloroplasts. FEBS Lett. 1980;121:78–80. [Google Scholar]
- 18.Dismukes GC, Siderer Y. Intermediates of a polynuclear manganese center involved in photosynthetic oxidation of water. Proc Natl Acad Sci U S A. 1981;78:274–278. doi: 10.1073/pnas.78.1.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dismukes GC, Ferris K, Watnick P. EPR spectroscopic evidence for a tetranuclear manganese cluster as the site for photosynthetic oxygen evolution. Photobiochem Photobiophys. 1982;3:243–256. [Google Scholar]
- 20.Rutherford AW, Boussac A, Zimmermann JL. EPR studies of the oxygen evolving enzyme. New J Chem. 1991;15:491–500. [Google Scholar]
- 21.Miller AF, Brudvig GW. A guide to electron paramagnetic resonance spectroscopy of photosystem II membranes. Biochim Biophys Acta Bioenerget. 1991;1056:1–18. doi: 10.1016/s0005-2728(05)80067-2. [DOI] [PubMed] [Google Scholar]
- 22.Britt RD, Peloquin JM, Campbell KA. Pulsed and parallel-polarization EPR characterization of the photosystem II oxygen-evolving complex. Annu Rev Biophys Biomol Struct. 2000;29:463–495. doi: 10.1146/annurev.biophys.29.1.463. [DOI] [PubMed] [Google Scholar]
- 23.Peloquin JM, Britt RD. EPR/ENDOR characterization of the physical and electronic structure of the OEC Mn cluster. Biochim Biophys Acta. 2001;1503:96–111. doi: 10.1016/s0005-2728(00)00219-x. [DOI] [PubMed] [Google Scholar]
- 24.Kawamori A. Electron transfer and structure of plant photosystem II. Prog Theor Chem Phys. 2003;10:529–563. [Google Scholar]
- 25.Britt RD, Campbell KA, Peloquin JM, Gilchrist ML, Aznar CP, Dicus MM, Robblee J, Messinger J. Recent pulsed EPR studies of the Photosystem II oxygen-evolving complex: implications as to water oxidation mechanisms. Biochim Biophys Acta. 2004;1655:158–171. doi: 10.1016/j.bbabio.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 26.Haddy A. EPR spectroscopy of the manganese cluster of photosystem II. Photosynth Res. 2007;92:357–368. doi: 10.1007/s11120-007-9194-9. [DOI] [PubMed] [Google Scholar]
- 27.Peloquin JM, Campbell KA, Randall DW, Evanchik MA, Pecoraro VL, Armstrong WH, Britt RD. 55Mn ENDOR of the S2-state multiline EPR signal of photosystem II: Implications on the structure of the tetranuclear Mn cluster. J Am Chem Soc. 2000;122:10926–10942. [Google Scholar]
- 28.Kulik LV, Epel B, Lubitz W, Messinger J. 55Mn pulse ENDOR at 34 GHz of the S0 and S2 states of the oxygen-evolving complex in photosystem II. J Am Chem Soc. 2005;127:2392–2393. doi: 10.1021/ja043012j. [DOI] [PubMed] [Google Scholar]
- 29.Su JH, Cox N, Ames W, Pantazis DA, Rapatskiy L, Lohmiller T, Kulik LV, Dorlet P, Rutherford AW, Neese F, Boussac A, Lubitz W, Messinger J. The electronic structures of the S2 states of the oxygen-evolving complexes of photosystem II in plants and cyanobacteria in the presence and absence of methanol. Biochim Biophys Acta Bioenerget. 2011;107:829–840. doi: 10.1016/j.bbabio.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 30.Cox N, Rapatskiy L, Su JH, Pantazis DA, Sugiura M, Kulik L, Dorlet P, Rutherford AW, Neese F, Boussac A, Lubitz W, Messinger J. Effect of Ca2+/Sr2+ substitution on the electronic structure of the oxygen-evolving complex of photosystem II: A combined multifrequency EPR, 55Mn-ENDOR, and DFT Study of the S2 State. J Am Chem Soc. 2011;133:3635–3648. doi: 10.1021/ja110145v. [DOI] [PubMed] [Google Scholar]
- 31.Haddy A, Lakshmi KV, Brudvig GW, Frank HA. Q-band EPR of the S2 state of photosystem II confirms an S = 5/2 origin of the X-band g = 4.1 signal. Biophys J. 2004;87:2885–2896. doi: 10.1529/biophysj.104.040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beck WF, Brudvig GW. Binding of amines to the oxygen-evolving center of photosystem II. Biochemistry. 1986;25:6479–6486. doi: 10.1021/bi00369a021. [DOI] [PubMed] [Google Scholar]
- 33.Cole J, Yachandra VK, Guiles RD, McDermott AE, Britt RD, Dexheimer SL, Sauer K, Klein MP. Assignment of the g = 4.1 EPR signal to manganese in the S2 state of the photosynthetic oxygen-evolving complex: An x-ray absorption edge spectroscopy study. Biochim Biophys Acta Bioenerget. 1987;890:395–398. doi: 10.1016/0005-2728(87)90169-1. [DOI] [PubMed] [Google Scholar]
- 34.Debus RJ, Campbell KA, Peloquin JM, Pham DP, Britt RD. Histidine 332 of the D1 polypeptide modulates the magnetic and redox properties of the manganese cluster and tyrosine YZ in photosystem II. Biochemistry. 2000;39:470–478. doi: 10.1021/bi9917737. [DOI] [PubMed] [Google Scholar]
- 35.Stull JA, Stich TA, Service RJ, Debus RJ, Mandal SK, Armstrong WH, Britt RD. 13C ENDOR reveals that the D1 polypeptide C-terminus is directly bound to Mn in the photosystem II oxygen evolving complex. J Am Chem Soc. 2010;132:446–447. doi: 10.1021/ja908688t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugiura M, Rappaport F, Hillier W, Dorlet P, Ohno Y, Hayashi H, Boussac A. Evidence that D1-His332 in photosystem II from Thermosynechococcus elongatus interacts with the S3 state and not with the S2 state. Biochemistry. 2009 doi: 10.1021/bi901067b. [DOI] [PubMed] [Google Scholar]
- 37.Service RJ, Yano J, McConnell I, Hwang HJ, Niks D, Hille R, Wydrzynski T, Burnap RL, Hillier W, Debus RJ. Participation of glutamate-354 of the CP43 polypeptide in the ligation of manganese and the binding of substrate water in photosystem II. Biochemistry. 2011;50:63–81. doi: 10.1021/bi1015937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeRose VJ, Yachandra VK, McDermott AE, Britt RD, Sauer K, Klein MP. Nitrogen ligation to manganese in the photosynthetic oxygen-evolving complex: Continuous-wave and pulsed EPR studies of photosystem II particles containing nitrogen-14 or nitrogen-15. Biochemistry. 1991;30:1335–1341. doi: 10.1021/bi00219a025. [DOI] [PubMed] [Google Scholar]
- 39.Tang XS, Diner BA, Larsen BS, Gilchrist ML, Jr, Lorigan GA, Britt RD. Identification of histidine at the catalytic site of the photosynthetic oxygen-evolving complex. Proc Natl Acad Sci U S A. 1994;91:704–708. doi: 10.1073/pnas.91.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Debus RJ, Campbell KA, Gregor W, Li ZL, Burnap RL, Britt RD. Does histidine 332 of the D1 polypeptide ligate the manganese cluster in photosystem II? An electron spin echo envelope modulation study. Biochemistry. 2001;40:3690–3699. doi: 10.1021/bi002394c. [DOI] [PubMed] [Google Scholar]
- 41.Yeagle GJ, Gilchrist ML, McCarrick RM, Britt RD. Multifrequency pulsed electron paramagnetic resonance study of the S2 state of the photosystem II manganese cluster. Inorg Chem. 2008;47:1803–1814. doi: 10.1021/ic701680c. [DOI] [PubMed] [Google Scholar]
- 42.Milikisiyants S, Chatterjee R, Weyers A, Meenaghan A, Coates C, Lakshmi KV. Ligand environment of the S2 state of photosystem II: A study of the hyperfine interactions of the tetranuclear manganese cluster by 2D 14N HYSCORE spectroscopy. J Phys Chem B. 2010;114:10905–10911. doi: 10.1021/jp1061623. [DOI] [PubMed] [Google Scholar]
- 43.Åhrling KA, Evans MCW, Nugent JHA, Ball RJ, Pace RJ. ESEEM studies of substrate water and small alcohol binding to the oxygen-evolving complex of photosystem II during functional turnover. Biochemistry. 2006;45:7069–7082. doi: 10.1021/bi052146m. [DOI] [PubMed] [Google Scholar]
- 44.Flanagan HL, Gerfen GJ, Singel DJ. Multifrequency electron spin-echo envelope modulation: The determination of nitro group nitrogen-14 hyperfine and quadrupole interactions of DPPH in frozen solution. J Chem Phys. 1988;88:20–24. [Google Scholar]
- 45.Chu H-A, Debus RJ, Babcock GT. D1-Asp170 is structurally coupled to the oxygen evolving complex in photosystem II as revealed by light-induced Fourier transform infrared difference spectroscopy. Biochemistry. 2001;40:2312–2316. doi: 10.1021/bi0022994. [DOI] [PubMed] [Google Scholar]
- 46.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. Generic assignments, strain histories, and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 47.Strickler MA, Walker LM, Hillier W, Britt RD, Debus RJ. No evidence from FTIR difference spectroscopy that aspartate-342 of the D1 polypeptide ligates a Mn ion that undergoes oxidation during the S0 to S1, S1 to S2, or S2 to S3 transitions in photosystem II. Biochemistry. 2007;46:3151–3160. doi: 10.1021/bi062195e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamanari T, Kimura Y, Mizusawa N, Ishii A, Ono T. Mid- to low-frequency Fourier transform infrared spectra of S-state cycle for photosynthetic water oxidation in Synechocystis sp. PCC 6803. Biochemistry. 2004;43:7479–7490. doi: 10.1021/bi0362323. [DOI] [PubMed] [Google Scholar]
- 49.Strickler MA, Walker LM, Hillier W, Debus RJ. Evidence from biosynthetically incorporated strontium and FTIR difference spectroscopy that the C-terminus of the D1 polypeptide of photosystem II does not ligate calcium. Biochemistry. 2005;44:8571–8577. doi: 10.1021/bi050653y. [DOI] [PubMed] [Google Scholar]
- 50.Chu HA, Nguyen AP, Debus RJ. Site-directed photosystem II mutants with perturbed oxygen-evolving properties. 1 Instability or inefficient assembly of the manganese cluster in vivo. Biochemistry. 1994;33:6137–6149. doi: 10.1021/bi00186a013. [DOI] [PubMed] [Google Scholar]
- 51.Berthold DA, Babcock GT, Yocum CF. A highly resolved, oxygen-evolving photosystem II preparation from spinach thylakoid membranes. EPR and electron-transport properties. FEBS Lett. 1981;134:231–234. [Google Scholar]
- 52.Ford RC, Evans MCW. Isolation of a photosystem 2 preparation from higher plants with highly enriched oxygen evolution activity. FEBS Lett. 1983;160:159–164. [Google Scholar]
- 53.Campbell KA, Gregor W, Pham DP, Peloquin JM, Debus RJ, Britt RD. The 23 and 17 kDa Extrinsic Proteins of Photosystem II Modulate the Magnetic Properties of the S1-State Manganese Cluster. Biochemistry. 1998;37:5039–5045. doi: 10.1021/bi9800552. [DOI] [PubMed] [Google Scholar]
- 54.Dasneves HJC, Vasconcelos AMP. Capillary gas chromotography of amino acids, including asparagine and glutamine - Sensitive gas chromatographic mass spectrometric and selected ion monitoring gas chromatographic mass spectrometric detection of the N, O(S)-tert(S)-tert-butyldimethylsilyl derivatives. J Chromatogr. 1987;392:249–258. doi: 10.1016/s0021-9673(01)94270-0. [DOI] [PubMed] [Google Scholar]
- 55.Stich TA, Lahiri S, Yeagle G, Dicus M, Brynda M, Gunn A, Aznar C, DeRose VJ, Britt RD. Multifrequency pulsed EPR studies of biologically relevant manganese(II) complexes. Appl Magn Reson. 2007;31:321–341. doi: 10.1007/BF03166263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peloquin JM, Tang XS, Diner BA, Britt RD. An Electron Spin-Echo Envelope Modulation (ESEEM) Study of the QA Binding Pocket of PS II Reaction Centers from Spinach and Synechocystis. Biochemistry. 1999;38:2057–2067. doi: 10.1021/bi982033l. [DOI] [PubMed] [Google Scholar]
- 57.Mims WB. Elimination of the dead-time artifact in electron spin-echo envelope spectra. J Magn Reson. 1984;59:291–306. [Google Scholar]
- 58.Stoll S, Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J Magn Reson. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 59.Stoll S, Britt RD. General and efficient simulation of pulse EPR spectra. Phys Chem Chem Phys. 2009;11:6614–6625. doi: 10.1039/b907277b. [DOI] [PubMed] [Google Scholar]
- 60.García-Rubio I, Martínez JI, Picorel R, Yruela IL, Alonso PJ. HYSCORE spectroscopy in the cytochrome b559 of the photosystem II reaction center. J Am Chem Soc. 2003;125:15846–15854. doi: 10.1021/ja035364g. [DOI] [PubMed] [Google Scholar]
- 61.Smith PJ, Åhrling KA, Pace RJ. Nature of the S2 state electron paramagnetic resonance signals from the oxygen-evolving complex of photosystem II: Q-band and oriented X-band studies. J Chem Soc, Faraday Trans. 1993;89:2863–2868. [Google Scholar]
- 62.Teutloff C, Kessen S, Kern J, Zouni A, Bittl R. High-field (94-GHz) EPR spectroscopy on the S2 multiline signal of photosystem II. FEBS Lett. 2006;580:3605–3609. doi: 10.1016/j.febslet.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 63.Matsuoka H, Furukawa K, Kato T, Mino H, Shen JR, Kawamori A. g-Anisotropy of the S2-state manganese cluster in single crystals of cyanobacterial photosystem II studied by W-band electron paramagnetic resonance spectroscopy. J Phys Chem B. 2006;110:13242–13247. doi: 10.1021/jp055008f. [DOI] [PubMed] [Google Scholar]
- 64.Lai A, Flanagan HL, Singel DJ. Multifrequency electron spin echo envelope modulation is S = 1/2, I = 1/2 systems - Analysis of the spectral amplitudes, line-shapes, and linewidths. J Chem Phys. 1988;89:7161–7166. [Google Scholar]
- 65.Scholes CP, Lapidot A, Mascarenhas R, Inubushi T, Isaacson RA, Feher G. Electron nulcear double resonance (ENDOR) from heme and histidine nitrogens in single crystals of aquometmyoglobin. J Am Chem Soc. 1982;104:2724–2735. [Google Scholar]
- 66.Chu H-A, Hillier W, Debus RJ. Evidence that the C-terminus of the D1 polypeptide of photosystem II is ligated to the manganese ion that undergoes oxidation during the S1 to S2 transition: An isotope-edited FTIR study. Biochemistry. 2004;43:3152–3166. doi: 10.1021/bi035915f. [DOI] [PubMed] [Google Scholar]
- 67.Campbell KA, Peloquin JM, Diner BA, Tang XS, Chisholm DA, Britt RD. The τ-nitrogen of D2 histidine 189 is the hydrogen bond donor to the tyrosine radical YD• of photosystem II. J Am Chem Soc. 1997;119:4787–4788. [Google Scholar]
- 68.Beck WF, De Paula JC, Brudvig GW. Ammonia binds to the manganese site of the oxygen-evolving complex of photosystem II in the S2 state. J Am Chem Soc. 1986;108:4018–4022. [Google Scholar]
- 69.Boussac A, Rutherford AW. Nature of the inhibition of the oxygen-evolving enzyme of photosystem II induced by sodium chloride washing and reversed by the addition of calcium(2+) or strontium(2+) Biochemistry. 1988;27:3476–3483. [Google Scholar]
- 70.Debus RJ, Aznar C, Campbell KA, Gregor W, Diner BA, Britt RD. Does aspartate 170 of the D1 polypeptide ligate the manganese cluster in photosystem II? An EPR and ESEEM study. Biochemistry. 2003;42:10600–10608. doi: 10.1021/bi034859f. [DOI] [PubMed] [Google Scholar]
- 71.Boerner RJ, Nguyen AP, Barry BA, Debus RJ. Evidence from directed mutagenesis that aspartate 170 of the D1 polypeptide influences the assembly and/or stability of the manganese cluster in the photosynthetic water-splitting complex. Biochemistry. 1992;31:6660–6672. doi: 10.1021/bi00144a005. [DOI] [PubMed] [Google Scholar]
- 72.Nixon PJ, Diner BA. Aspartate 170 of the photosystem II reaction center polypeptide D1 is involved in the assembly of the oxygen-evolving manganese cluster. Biochemistry. 1992;31:942–948. doi: 10.1021/bi00118a041. [DOI] [PubMed] [Google Scholar]
- 73.Campbell KA, Force DA, Nixon PJ, Dole F, Diner BA, Britt RD. Dual-mode EPR detects the initial intermediate in photoassembly of the photosystem II Mn cluster: The influence of amino acid residue 170 of the D1 polypeptide on Mn coordination. J Am Chem Soc. 2000;122:3754–3761. [Google Scholar]
- 74.Hwang HJ, McLain A, Debus RJ, Bumap RL. Photoassembly of the manganese cluster in mutants perturbed in the high affinity Mn-binding site of the H2O-oxidation complex of photosystem II. Biochemistry. 2007;46:13648–13657. doi: 10.1021/bi700761v. [DOI] [PubMed] [Google Scholar]
- 75.Stich TA, Whittaker JW, Britt RD. Multifrequency EPR studies of manganese catalases provide a complete description of proteinaceous nitrogen coordination. J Phys Chem B. 2010;114:14178–14188. doi: 10.1021/jp908064y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tan XL, Gultneh Y, Sarneski JE, Scholes CP. EPR-ENDOR of the electronic structure from 2 nitrogenously ligated bis(μ-oxo)MnIIIMnIV model complexes spectroscopically relevant to the multi-manganese center of photosytem II. J Am Chem Soc. 1991;113:7853–7858. [Google Scholar]
- 77.Sinnecker S, Neese F, Lubitz W. Dimanganese catalase-spectroscopic parameters from broken-symmetry density functional theory of the superoxidized MnIII/MnIV state. J Biol Inorg Chem. 2005;10:231–238. doi: 10.1007/s00775-005-0633-9. [DOI] [PubMed] [Google Scholar]
- 78.Sinnecker S, Neese F, Noodleman L, Lubitz W. Calculating the electron paramagnetic resonance parameters of exchange coupled transition metal complexes using broken symmetry density functional theory: Application to a MnIII/MnIV model compound. J Am Chem Soc. 2004;126:2613–2622. doi: 10.1021/ja0390202. [DOI] [PubMed] [Google Scholar]
- 79.Randall DW. Pulsed EPR Studies of Tyrosine Radicals and Manganese Complexes: Insights into Photosynthetic Oxygen Evolution. University of California-Davis; Davis, CA: 1998. [Google Scholar]
- 80.Bencini A, Gatteschi D. EPR of Exchange Coupled Systems. Verlag; 1990. [Google Scholar]
- 81.Orio M, Pantazis DA, Petrenko T, Neese F. Magnetic and spectroscopic properties of mixed valence manganese(III, IV) dimers: A systematic study using broken symmetry density functional theory. Inorg Chem. 2009;48:7251–7260. doi: 10.1021/ic9005899. [DOI] [PubMed] [Google Scholar]
- 82.Morton JR, Preston KF. Atomic paramaters for paramagnetic resonance data. J Magn Reson. 1978;30:577–582. [Google Scholar]
- 83.Lorigan GA, Britt RD. Temperature-dependent pulsed electron paramagnetic resonance studies of the S2 state multiline signal of the photosynthetic oxygen-evolving complex. Biochemistry. 1994;33:12072–12076. doi: 10.1021/bi00206a009. [DOI] [PubMed] [Google Scholar]
- 84.Ashby CIH, Cheng CP, Brown TL. Nitrogen-14 nuclear quadrupole resonance spectra of coordinated imidazole. J Am Chem Soc. 1978;100:6057–6063. [Google Scholar]
- 85.Aasa R, Andreasson LE, Lagenfelt G, Vanngard T. A comparison between the multiline EPR signals of spinach and Anacystis nidulans and their temperature dependence. FEBS Lett. 1987;221:245–248. [Google Scholar]
- 86.Yano J, Yachandra V. Oxidation state changes of the Mn4Ca cluster in photosystem II. Photosynth Res. 2007;92:289–303. doi: 10.1007/s11120-007-9153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pushkar YL, Yano J, Sauer K, Boussac A, Yachandra VK. Structural changes in the Mn4Ca cluster and the mechanism of photosynthetic water splitting. Proc Natl Acad Sci U S A. 2008;105:1879–1884. doi: 10.1073/pnas.0707092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mizusawa N, Yamanari T, Kimura Y, Ishii A, Nakazawa S, Ono T-A. Changes in the functional and structural properties of the Mn cluster induced by replacing the side group of the C-terminus of the D1 protein of photosystem II. Biochemistry. 2004;43:14644–14652. doi: 10.1021/bi0486076. [DOI] [PubMed] [Google Scholar]
- 89.Chu HA, Nguyen AP, Debus RJ. Site-directed photosystem II mutants with perturbed oxygen-evolving properties. 2 Increased binding or photooxidation of manganese in the absence of the extrinsic 33-kDa polypeptide in vivo. Biochemistry. 1994;33:6150–6157. doi: 10.1021/bi00186a014. [DOI] [PubMed] [Google Scholar]
- 90.Chu HA, Nguyen AP, Debus RJ. Amino acid residues that influence the binding of manganese or calcium to photosystem II. 1 The lumenal interhelical domains of the D1 polypeptide. Biochemistry. 1995;34:5839–5858. doi: 10.1021/bi00017a016. [DOI] [PubMed] [Google Scholar]
- 91.Diner BA, Nixon PJ. The rate of reduction of oxidized redox-active tyrosine, Z+, by exogenous manganese(2+) is slowed in a site-directed mutant, at aspartate 170 of polypeptide D1 of photosystem II, inactive for photosynthetic oxygen evolution. Biochim Biophys Acta Bioenerget. 1992;1101:134–138. [Google Scholar]
- 92.Ghirardi ML, Lutton TW, Seibert M. Effects of carboxyl amino acid modification on the properties of the high-affinity, manganese-binding site in photosystem II. Biochemistry. 1998;37:13559–13566. doi: 10.1021/bi980358w. [DOI] [PubMed] [Google Scholar]
- 93.Debus RJ, Strickler MA, Walker LM, Hillier W. No evidence from FTIR difference spectroscopy that aspartate-170 of the D1 polypeptide ligates a manganese ion that undergoes oxidation during the S0 to S1, S1 to S2, or S2 to S3 transitions in photosystem II. Biochemistry. 2005;44:1367–1374. doi: 10.1021/bi047558u. [DOI] [PubMed] [Google Scholar]
- 94.Sproviero EM, Gascon JA, McEvoy JP, Brudvig GW, Batista VS. Computational studies of the O2-evolving complex of photosystem II and biomimetic oxomanganese complexes. Coord Chem Rev. 2008;252:395–415. doi: 10.1016/j.ccr.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stoll S, Calle C, Mitrikas G, Schweiger A. Peak suppression in ESEEM spectra of multinuclear spin systems. J Magn Reson. 2005;177:93–101. doi: 10.1016/j.jmr.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 96.Andreasson LE. Is nitrogen liganded to manganese in the photosynthetic oxygen-evolving system? EPR studies after isotopic replacement with nitrogen-15. Biochim Biophys Acta Bioenerget. 1989;973:465–467. [Google Scholar]
- 97.Dicus MM, Conlan A, Nechushtai R, Jennings PA, Paddock ML, Britt RD, Stoll S. Binding of histidine in the Cys3His1-coordinated 2Fe-2S cluster of human mitoNEET. J Am Chem Soc. 2010;132:2037–2049. doi: 10.1021/ja909359g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cohen RO, Nixon PJ, Diner BA. Participation of the C-terminal region of the D1-polypeptide in the first steps in the assembly of the Mn4Ca cluster of photosystem II. J Biol Chem. 2007;282:7209–7218. doi: 10.1074/jbc.M606255200. [DOI] [PubMed] [Google Scholar]
- 99.Kusunoki M. Mono-manganese mechanism of the photosystem II water splitting reaction by a unique Mn4Ca cluster. Biochim Biophys Acta Bioenerget. 2007;1767:484–492. doi: 10.1016/j.bbabio.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 100.Allahverdiyeva Y, Deak Z, Szilard A, Diner BA, Nixon PJ, Vass I. The function of D1-H332 in photosystem II electron transport studied by thermoluminescence and chlorophyll fluorescence in site-directed mutants of Synechocystis 6803. Eur J Biochem. 2004;271:3523–3532. doi: 10.1111/j.0014-2956.2004.04287.x. [DOI] [PubMed] [Google Scholar]
- 101.Campbell KA, Lashley MR, Wyatt JK, Nantz MH, Britt RD. Dual-mode EPR study of Mn(III) salen and the Mn(III) salen-catalyzed epoxidation of cis-beta-methylstyrene. J Am Chem Soc. 2001;123:5710–5719. doi: 10.1021/ja0027463. [DOI] [PubMed] [Google Scholar]
- 102.Service RJ, Hillier W, Debus RJ. Evidence from FTIR difference spectroscopy of an extensive network of hydrogen bonds near the oxygen-evolving Mn4Ca cluster of photosystem II Involving D1-Glu65, D2-Glu312, and D1-Glu329. Biochemistry. 2010;49:6655–6669. doi: 10.1021/bi100730d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yano J, Walker LM, Strickler MA, Service RJ, Yachandra VK, Debus RJ. Altered structure of the Mn4Ca cluster in the oxygen-evolving complex of photosystem II by a histidine ligand mutation. J Biol Chem. 2011;286:9257–9267. doi: 10.1074/jbc.M110.205740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chu HA, Nguyen AP, Debus RJ. Amino acid residues that influence the binding of manganese or calcium to photosystem II. 2 The carboxy-terminal domain of the D1 polypeptide. Biochemistry. 1995;34:5859–5882. doi: 10.1021/bi00017a017. [DOI] [PubMed] [Google Scholar]
- 105.Diner BA, Schlodder E, Nixon PJ, Coleman WJ, Rappaport F, Lavergne J, Vermaas WF, Chisholm DA. Site-directed mutations at D1-His198 and D2-His197 of photosystem II in Synechocystis PCC 6803: sites of primary charge separation and cation and triplet stabilization. Biochemistry. 2001;40:9265–9281. doi: 10.1021/bi010121r. [DOI] [PubMed] [Google Scholar]
- 106.Sugiura M, Boussac A, Noguchi T, Rappaport F. Influence of histidine-198 of the D1 subunit on the properties of the primary electron donor, P680, of photosystem II in Thermosynechococcus elongatus. Biochim Biophys Acta Bioenerget. 2008;1777:331–342. doi: 10.1016/j.bbabio.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 107.Merry SAP, Nixon PJ, Barter LMC, Schilstra M, Porter G, Barber J, Durrant JR, Klug DR. Modulation of quantum yield of primary radical pair formation in photosystem II by site-directed mutagenesis affecting radical cations and anions. Biochemistry. 1998;37:17439–17447. doi: 10.1021/bi980502d. [DOI] [PubMed] [Google Scholar]
- 108.Cser K, Vass I. Radiative and non-radiative charge recombination pathways in photosystem II studied by thermoluminescence and chlorophyll fluorescence in the cyanobacterium Synechocystis 6803. Biochim Biophys Acta Bioenerget. 2007;1767:233–243. doi: 10.1016/j.bbabio.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 109.Edwards RA, Baker HM, Whittaker MM, Whittaker JW, Jameson GB, Baker EN. Crystal structure of Escherichia coli manganese superoxide dismutase at 2.1 Å resolution. J Biol Inorg Chem. 1998;3:161–171. [Google Scholar]
- 110.Boal AK, Cotruvo JA, Stubbe J, Rosenzweig AC. Structural basis for activation of class Ib ribonucleotide reductase. Science. 2010;329:1526–1530. doi: 10.1126/science.1190187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Antonyuk SV, Melik-Adamyan VR, Popov AN, Lamzin VS, Hempstead PD, Harrison RM, Artymyuk PD, Barynin VV. Three-dimensional structure of dimanganese catalase from Thermus thermophilus at 1 Å resolution. Kristallografiya. 2000;45:111–122. [Google Scholar]
- 112.Fee JA, Findling KL, Yoshida T, Hille R, Tarr GE, Hearshen DO, Dunham WR, Day EP, Kent TA, Munck E. Purification and characterization of the Rieske iron-sulfur protein from Thermus thermophilus - Evidence for a 2Fe-2S cluster having non-cysteine ligands. J Biol Chem. 1984;259:124–133. [PubMed] [Google Scholar]
- 113.Gurbiel RJ, Batie CJ, Sivaraja M, True AE, Fee JA, Hoffman BM, Ballou DP. Electron nuclear double resonance spectroscopy of 15N-enriched phthalate dioxygenase from Pseudomonas cepacia proves that 2 histidines are oordinated to the [2Fe-2S] Rieske-type clusters. Biochemistry. 1989;28:4861–4871. doi: 10.1021/bi00437a051. [DOI] [PubMed] [Google Scholar]
- 114.Stone EM, Chantranupong L, Georgiou G. The second-shell metal ligands of human arginase affect coordination of the nucleophile and substrate. Biochemistry. 2010;49:10582–10588. doi: 10.1021/bi101542t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hashiguchi BG, Young KJH, Yousufuddin M, Goddard WA, Periana RA. Acceleration of nucleophilic CH activation by strongly basic solvents. J Am Chem Soc. 2010;132:12542–12545. doi: 10.1021/ja102518m. [DOI] [PubMed] [Google Scholar]
- 116.Clausen J, Junge W. Detection of an intermediate of photosynthetic water oxidation. Nature. 2004;430:480–483. doi: 10.1038/nature02676. [DOI] [PubMed] [Google Scholar]
- 117.Clausen J, Junge W, Dau H, Haumann M. Photosynthetic water oxidation at high O2 backpressure monitored by delayed chlorophyll fluorescence. Biochemistry. 2005;44:12775–12779. doi: 10.1021/bi051183a. [DOI] [PubMed] [Google Scholar]
- 118.Schinzel S, Kaupp M. Validation of broken-symmetry density functional methods for the calculation of electron paramagnetic resonance parameters of dinuclear mixed-valence MnIIIMnIV complexes. Can J Chem. 2009;87:1521–1539. [Google Scholar]
- 119.Siegbahn PEM. Structures and energetics for O2 formation in photosystem II. Acc Chem Res. 2009;42:1871–1880. doi: 10.1021/ar900117k. [DOI] [PubMed] [Google Scholar]
- 120.Schinzel S, Schraut J, Arbuznikov AV, Siegbahn PEM, Kaupp M. Density functional calculations of 55Mn, 14N and 13C electron paramagnetic resonance parameters support an energetically feasible model system for the S2 state of the oxygen-evolving complex of photosystem II. Chem Eur J. 2010;16:10424–10438. doi: 10.1002/chem.201000584. [DOI] [PubMed] [Google Scholar]
- 121.Pantazis DA, Orio M, Petrenko T, Zein S, Lubitz W, Messinger J, Neese F. Structure of the oxygen-evolving complex of photosystem II: Information on the S2 state through quantum chemical calculation of its magnetic properties. Phys Chem Chem Phys. 2009;11:6788–6798. doi: 10.1039/b907038a. [DOI] [PubMed] [Google Scholar]
- 122.Pantazis DA, Orio M, Petrenko T, Zein S, Bill E, Lubitz W, Messinger J, Neese F. A new quantum chemical approach to the magnetic properties of oligonuclear transition-metal complexes: Application to a model for the tetranuclear manganese cluster of photosystem II. Chem Eur J. 2009;15:5108–5123. doi: 10.1002/chem.200802456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.