Abstract
Telomeres, the natural ends of eukaryotic chromosomes, prevent the loss of chromosomal sequences and preclude their recognition as broken DNA. Telomere length is kept under strict boundaries by the action of various proteins, some with negative and others with positive effects on telomere length. Recently, data have been accumulating to support a role for DNA replication in the control of telomere length, although through a currently poorly understood mechanism. Elg1p, a replication factor C (RFC)-like protein of yeast, contributes to genome stability through a putative replication-associated function. Here, we show that Elg1p participates in negative control of telomere length and in telomeric silencing through a replication-mediated pathway. We show that the telomeric function of Elg1 is independent of recombination and completely dependent on an active telomerase. Additionally, this function depends on yKu and DNA polymerase. We discuss alternative models to explain how Elg1p affects telomere length.
Keywords: DNA replication, replication factor C, Saccharomyces cerevisiae
Replication of a linear DNA molecule is carried out by DNA polymerases and additional proteins that duplicate the DNA by moving coordinately on both the lagging and leading strands (1). DNA attrition at the ends of chromosomes due to the “end replication problem” is avoided by the action of the specialized reverse transcriptase, called telomerase (2, 3). Telomerase adds G-rich DNA repeats termed telomeres onto the ends of chromosomes. In addition to their role in preventing loss of chromosomal sequences, telomeres serve as a boarding pad for a multiprotein complex that is implicated in several important cellular functions. Among these functions is capping telomeres, thus avoiding the recognition of the natural ends of chromosomes as broken DNA. This capping in turn prevents cell cycle arrest and chromosomal fusions, allowing proper chromosomal segregation (4).
Many proteins, most of which are part of the telomeric complex, regulate telomere length in both positive and negative fashions (5). The best-studied pathway of telomerase-negative regulation is the Rap1p pathway in yeast. Rap1p is bound throughout the telomeric sequence and interacts with two proteins, Rif1p and Rif2p (6, 7). The resulting complex is thought to prevent telomerase from accessing the telomeric DNA substrate by a yet unknown mechanism. Additional factors were shown to be involved in telomere length regulation. Tel1p and the yKu70p-yKu80p heterodimer act as positive regulators of telomerase (8–10). Tel1p is proposed to act by recruiting the MRX (Mer11/Rad50/Xrs2) complex (11). The yKu70p/yKu80p heterodimer influences telomere length by protecting telomere ends from degradation. It also has additional functions in telomeric silencing (through recruiting Sir proteins to the telomeres), telomeric nuclear localization, and telomere replication timing (12–15). Cdc13p binds the telomeric single strand, recruits telomerase to the telomere, and prevents degradation (16, 17). By these functions, Cdc13p acts as a positive regulator of telomere length. In addition, Cdc13p also regulates telomerase negatively by binding Stn1p, which prevents telomerase recruitment to the telomeres (18). In addition Cdc13p interact physically and genetically with polymerase (Pol) α, thus providing a link between telomere length regulation and the DNA replication machinery (19). This interaction might underline the significance of Cdc13p in recruiting both telomerase and DNA polymerase to the telomere. One of the models raised (19) suggested that proteins involved in DNA replication might compete with telomerase for the telomeric substrate.
When the replication machinery reaches the end of a linear DNA molecule, on its lagging strand, it might interact with the telomerase enzymatic complex. In agreement with this finding, some of the genes involved in DNA replication also influence telomere length regulation. Polα, Polδ, and DNA primase are required for telomere addition. It was suggested that in their absence telomerase is inactive (20). Mutations in the large subunit of replication factor C (RFC) (Rfc1p/Cdc44p) and of DNA polymerase &ααχντε (Polα/Cdc17p) cause telomere elongation (21, 22). The RFC complex is required for DNA replication and repair. It recognizes primer-template structures and acts as a “clamp-loader” to recruit proliferating cell nuclear antigen (PCNA), the DNA replication processivity factor (23). Mutations in Polα and in PCNA are also associated with loss of telomeric silencing (24, 25).
Elg1p exhibits sequence similarity to Rfc1p and interacts with Rfc2 to -5 proteins (but not with Rfc1p itself). These facts may point to the existence of an alternative RFC complex, in which Rfc1p is replaced by Elg1p (26, 27). At least two other similar RFC complexes exist in yeast; in these complexes, Rfc1p is replaced by either Ctf18p or Rad24p (28, 29). Deletion of CTF18 causes telomere shortening (30) whereas a RAD24 deletion does not affect telomere length. ELG1 was identified recently as a gene whose deletion causes increased levels of recombination. Mutations in ELG1 exhibit impaired growth or lethality in combination with mutations in genes involved in homologous recombination. These findings raised the possibility that ELG1 may act to suppress replication-related DNA damage, thereby functioning as a keeper of genome stability (26, 27).
Here, we show that deletion of ELG1 causes telomere elongation, unraveling a role for Elg1p in limiting telomere length. We further characterize the telomeric pathway in which Elg1p participates and suggest a model to explain the relation between the roles of Elg1p in DNA replication and in determination of telomere length.
Materials and Methods
Plasmid Construction. pELG1 was constructed by cloning a 2.3-kb BamHI-PstlI PCR-amplified fragment of ELG1 in the BamHI-PstlI site of pCM189 (31).
Strain Construction. All deletion strains were of BY4741 background. Strains containing the temperature-sensitive alleles cdc17-1 (22) and cdc44-1 (32) were transferred to the BY4741 genetic background by backcrosses. The strain containing the ADE2 telomeric gene was created by three backcrosses of the UCC41 strain provided by D. Gottschling (33). All deletion strains used contain replacement of the gene ORF by a KanMX cassette, with the exception of CTF18, which is replaced by HygB. Diploids heterozygous for ELG1 and a second gene were dissected, and tetra-type tetrads were selected. The genotypes of the spores were confirmed by PCR. A minimum of two tetrads from each cross was analyzed. All strains were grown at 30°C and on yeast extract/peptone/dextrose (YPD) media, unless stated differently. Strains transformed with pELG1 were grown on synthetic dextrose medium lacking uracil (SD-Ura) media to avoid plasmid loss.
Silencing Tests. Cells were grown on YPD liquid media supplemented with adenine (2 mg/ml) for 1 day. Cultures were plated on YPD plates and incubated until development of red color.
Southern Blots. Genomic DNA was prepared from saturated cultures by using a modified version of the zymolase method and hybridized as described (34). Saccharomyces cerevisiae-specific telomeric probe was prepared by phosphorylation with T4 polynucleotide kinase of a telomeric G-strand oligonucleotide. Hybridizations (1 h) and washes (total 20 min) were performed at 45°C. The G-strand oligonucleotide was 5′-TGGGTGTGGGTGTGGGTGTGGGTG-3′; the C strand 12 oligonucleotide was 5′-CACCCACACCCACACCCACACCCA-3′.
Results
We have performed a systematic screen of the entire collection of viable S. cerevisiae haploid gene deletion mutants for genes whose deletion causes telomeric shortening or elongation, with the goal of obtaining a full genomic view of genes involved in telomere metabolism (A.K. and M. McEachern, unpublished results). Among the deletion strains that exhibited a deviation from the wild-type telomeric pattern, we identified Δelg1 as one of the strains with elongated telomeres.
To confirm that the telomere elongation phenotype was indeed the result of deletion of the ELG1 gene, we performed several genetic tests. The first was a tetrad analysis test, which showed that the telomeric phenotype indeed cosegregated with the kanamycin resistance phenotype conferred by the deletion marker. The phenotype appeared by the first passage after sporulation but reached its full potential by the fifth passage in all spore clones tested (data not shown). As a second test, we expressed ELG1 from a plasmid in a Δelg1 strain. This expression resulted in telomeres shortening back to the wild-type length (data not shown). These two independent tests confirmed that disruption of ELG1 was responsible for the observed telomere length elongation in the Δelg1 strain.
Our aim was to characterize the cellular pathway in which ELG1 acts in preventing telomere elongation. As mentioned, ELG1 was identified as a gene responsible for suppression of various types of chromosomal recombination (26, 27).
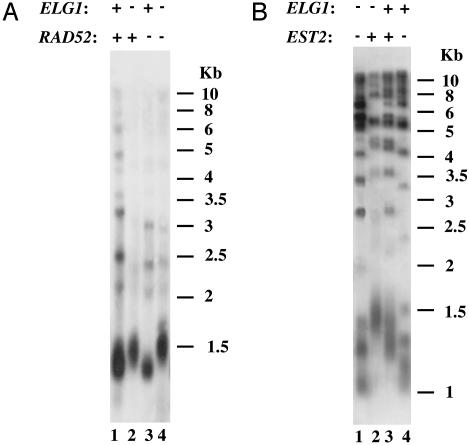
Therefore, one possibility was that deletion of ELG1 promoted recombination between telomeric repeats, thereby resulting in elongated telomeres. We thus tested the involvement of RAD52, a key component of the homologous recombination pathway, in telomere elongation of the Δelg1 strain. As shown, Δelg1 Δrad52 (Fig. 1A, lane 4) had the same telomere elongation phenotype as the Δelg1 strain (Fig. 1 A, lane 2), demonstrating that telomere elongation in the Δelg1 strain was not dependent on recombination.
Fig. 1.
Elongation of telomeres in Δelg1 depends on telomerase, but not on recombination. Shown is Southern blot analysis with an S. cerevisiae-specific telomeric probe. (A) Genomic DNA was prepared from spore derivatives dissected from ELG1/Δelg1 RAD52/Δrad52 tetrads. Lane 1, ELG1 RAD52; lane 2, Δelg1 RAD52; lane 3, ELG1 Δrad52 single mutants; lane 4, Δelg1 Δrad52 double mutant. (B) Genomic DNA was prepared from spore derivatives dissected from ELG1/Δelg1 EST2/Δest2 tetrads. Lane 1, Δelg1 Δest2 double mutant; lane 2, Δelg1 EST2; lane 3, ELG1 EST2; lane 4, ELG1 Δest2.
To further understand the mechanism underlying the telomere elongation phenotype caused by the deletion of ELG1, we tested the involvement of several genes mapped to different telomeric length regulation pathways: EST2, RIF2, RIF1, YKU70, and TEL1.
To test whether telomere elongation in the Δelg1 strain involved the activity of telomerase, we examined the telomeric phenotype of the double mutant Δelg1 Δest2. EST2 encodes the catalytic subunit of telomerase (35). Therefore, if telomerase were involved in telomere elongation of Δelg1, we would expect the deletion of EST2 to suppress this phenotype. Indeed, as shown in Fig. 1B, lane 1, deletion of EST2 prevented telomere elongation of Δelg1. These results show that telomere elongation in the Δelg1 mutant is telomerase-dependent.
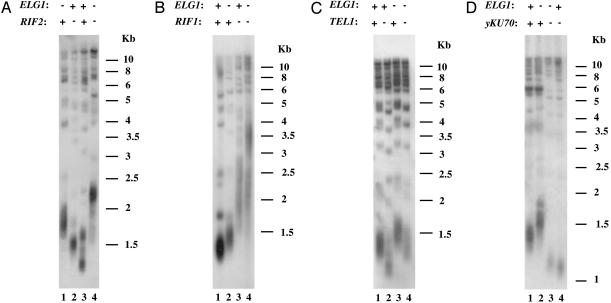
As mentioned above, Rif2p and Rif1p were found to regulate telomerase negatively by binding to the C-terminal domain of Rap1p. Deletion of either gene results in telomere elongation (7). Therefore, it was possible that ELG1 acts in the same genetic pathway as RAP1 and the associated factors RIF1 and RIF2. As shown in Fig. 2A, lane 4, deletion of both RIF2 and ELG1 resulted in telomere lengthening that was more severe than that observed in each of the single mutant (Fig. 2 A, lanes 1 and 2). Double deletion of ELG1 and RIF1 also seemed to have a similar effect on telomere length although, because the effects of deletion of RIF1 were so severe, the additive effect of the double deletion on telomere length was less clear-cut (Fig. 2B, etc.). Therefore, we conclude that the negative regulation of telomere length by ELG1 and RIF2, and probably RIF1, is carried out through distinct pathways.
Fig. 2.
Elongation of telomeres in Δelg1 depends on KU70, but not on RIF2, RIF1,or TEL1. Southern blot analysis with an S. cerevisiae-specific telomeric probe was done. (A) Genomic DNA was prepared from spore derivatives dissected from ELG1/Δelg1 RIF2/Δrif2 tetrads. Lane 1, Δelg1RIF2; lane 2, ELG1 Δrif2; lane 3, ELG1 RIF2; lane 4, Δelg1 Δrif2 double mutant. (B) Genomic DNA was prepared from spore derivatives dissected from ELG1/Δelg1 RIF1/Δrif1 tetrads. Lane 1, ELG1 RIF1; lane 2, Δelg1RIF1; lane 3, ELG1 Δrif1 double mutant; lane 4, Δelg1 Δrif1.(C) Genomic DNA was prepared from spore derivatives dissected from ELG1/Δelg1 TEL1/Δtel1 tetrads. Lane 1, ELG1 TEL1; lane 2, ELG1 Δtel1; lane 3, Δelg1TEL1; lane 4, Δelg1 Δte1l double mutant. (D) Genomic DNA was prepared from spore derivatives dissected from ELG1/Δelg1 YKU70/Δyku70 tetrads. Lane 1, ELG1 YKU70; lane 2, Δelg1 YKU70; lane 3, Δelg1 Δyku70 double mutant; lane 4, ELG1 Δyku70.
Two additional genes known to affect telomere length are YKU70 and TEL1. Deletion of either gene results in dramatic telomere shortening, but the two genes were mapped to two distinct pathways of telomerase positive regulation (10). To test whether ELG1 acts at one of these genetic pathways, the telomeric phenotypes of the corresponding double mutants were tested. As shown in Fig. 2C, lane 4, telomere length of the Δtel1 Δelg1 mutant exhibited an intermediate phenotype, resembling that of the wild-type strain, demonstrating that TEL1 and ELG1 act in distinct pathways affecting telomere length. In contrast, telomere length of the double mutant Δyku70 Δelg1 (Fig. 2D, lane 3) resembled that of the single mutant Δyku70 (Fig. 2D, lane 4). This finding shows that ELG1 and YKU70 act in the same pathway of telomere length regulation and that YKU70 is required for the elongated telomere phenotype of the Δelg1 mutant.
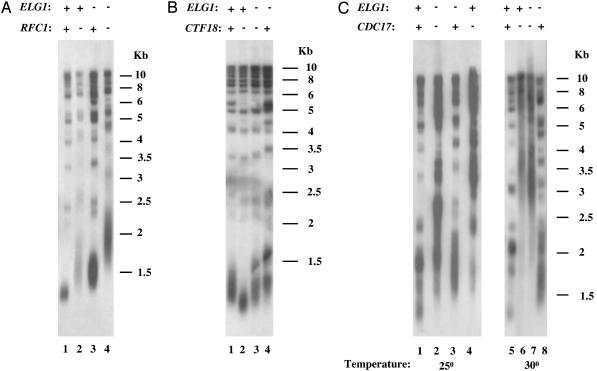
If ELG1 participated in an alternative RFC complex, distinct from those described for RFC1, RAD24, and CTF18, it is expected that its effect on telomere length might also be carried out through a distinct genetic pathway. As seen in Fig. 3A, lane 4, the phenotype of the double mutant cdc44-1 Δelg1 is much more severe than the phenotypes of either single mutant (lanes 2 and 3). The telomeric length of the double mutant Δelg1 Δctf18 (Fig. 3B, lane 3) is shorter than that of the single Δelg1 mutant (Fig. 3B, lane 4) and longer than that of the single Δctf18 mutant (Fig. 3B, lane 2).
Fig. 3.
ELG1 operates in telomere elongation in the same genetic pathway as POLα, but in a distinct genetic pathway from CTF18 and RFC1. DNA was analyzed by Southern blot hybridized with an S. cerevisiae-specific telomeric probe. (A) Genomic DNA was prepared from spore derivatives dissected from ELG1/Δelg1 CDC44/cdc44-1 tetrads. Lane 1, ELG1 CDC44; lane 2, ELG1 cdc44-1; lane 3, Δelg1 CDC44; lane 4, Δelg1 cdc44-1 double mutant. (B) Genomic DNA was prepared from spore derivatives dissected from ELG1/Δelg1 CTF18/Δctf18 tetrads. Lane 1, ELG1 CTF18; lane 2, ELG1/Δctf18; lane 3, Δelg1/Δctf18 double mutant; lane 4, Δelg1 CTF18. (C) Genomic DNA was prepared from spore derivatives dissected from ELG1/Δelg1 CDC17/cdc17-1 tetrads. Spores 1–4 were grown at 25°C, and spores 5–8 were grown at 30°C. Lane 1, ELG1 CDC17; lane 2, Δelg1 cdc17-1 double mutant; lane 3, Δelg1 CDC17; lane 4, ELG1 cdc17-1; lane 5, ELG1 CDC17; lane 6, ELG1 cdc17-1; lane 7, Δelg1 cdc17-1 double mutant; lane 8, Δelg1 CDC17.
Although deletion of RAD24 does not affect telomere length, we tested whether it affects telomere length in combination with Δelg1. The telomeric length of the double mutant Δelg1 Δrad24 resembled that of the single mutant Δelg1 (data not shown). These experiments demonstrate that the function of Elg1p is distinct from that of the other alternative RFC genes in relation to telomere length. We also tested the genetic interaction between the cdc17-1 allele of Polα and a deletion of ELG1. As shown in Fig. 3C, telomere length of the double mutant cdc17-1 Δelg1 was similar to that of the single cdc17-1 strain at 25°C and at 30°C (Fig. 3C). Therefore, we conclude that ELG1 and CDC17 regulate telomere length negatively through the same genetic pathway.
Mutations in genes involved in replication of the lagging strand (Polα, RAD27) cause an increase in the amount of telomeric single-stranded DNA (ssDNA) (24, 36). We examined the telomeric ssDNA in rad24, ctf18, rfc1, and elg1 mutants and found no accumulation of ssDNA (data not shown). Therefore, there is no direct correlation between the telomere length phenotype of RFC-like proteins, including Elg1p, and accumulation of telomeric ssDNA.
Genes involved in telomere length regulation often influence the structure of the telomeric heterochromatin, and these changes are sometimes associated with an increase or decrease in the transcription of genes placed adjacent to the telomere (13, 33, 37).
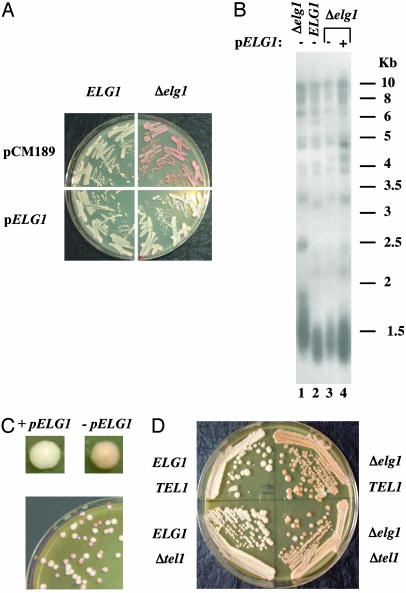
Telomere position effect can be easily monitored by placing the ADE2 gene near the telomere of chromosome VII (33). We generated a heterozygous strain (elg1/ELG1) containing a subtelomeric ADE2 gene. Spore derivatives of ELG1 genotype showed normal silencing at the ADE2 locus whereas spore derivatives of the Δelg1 genotype exhibited a substantial elevation of silencing, evident from the pink coloration of the colonies (Fig. 4A). This phenotype can be suppressed by a plasmid harboring ELG1 (Fig. 4A), demonstrating that the absence of ELG1 is the cause for the appearance of pink colonies.
Fig. 4.
Deletion of ELG1 elevates silencing at an ADE2 gene placed near the telomere in S. cerevisiae. (A) Complementation of the elevated telomeric silencing phenotype was tested in the indicated strains. Strains containing the Δelg::KanMX deletion allele or the ELG1 allele were transformed with control (pCM189) or pELG1 plasmids. The strains were grown on YPD media supplemented with adenine and plated on SD-Ura plates. (B) The uncoupling of silencing and length phenotype of Δelg1 was tested by picking separately Ura+ (lane 4) and Ura- (lane 3) colonies for extraction of genomic DNA. DNA was analyzed by Southern blot hybridized with an S. cerevisiae-specific telomeric probe. Telomeric length was compared with Δelg::KanMX strain (lane 1) and ELG1 (lane 2). (C) Silencing was tested in a strain containing the Δelg::KanMX deletion allele and pELG1 plasmid. This strain was transferred from selective media (SD-Ura) to nonselective media (YPD). Colonies were replica-plated on SD-Ura plates. (D) Genomic DNA was prepared from tetrad created by the dissection of an ELG1/Δelg1 TEL1/Δtel1 diploid with an ADE2 telomeric gene. Spores with all gene combinations (ELG1, Δelg1, Δtel1, and Δelg1 Δtel1) and a telomeric ADE2 gene were grown on YPD liquid media supplemented with adenine and plated on a YPD plate.
We further examined whether the elevation in telomere position effect arises directly from the elongation in telomere length. We tested the telomeric silencing phenotype of the Δelg1 strain immediately after loss of the pELG1 plasmid, while its telomeres were still only moderately elongated (Fig. 4B), and found that, even when telomere length is close to wild-type levels, elevation in telomere position effect is apparent (Fig. 4C). In addition, we observed that Δtel1 Δelg1 exhibited an elevated level of telomeric silencing that resembles that of the single mutant Δelg1 (Fig. 4D), despite the fact that its telomere length resembles that of the wild-type strain (Fig. 2C). The single mutant Δtel1 exhibited telomeric silencing levels that resembled that of the wild-type strain, as expected.
Therefore, we conclude that the elevation of telomeric silencing in the Δelg1 strain was probably due to the lack of Elg1p in this strain and not to elongation of the telomere. Because telomeric silencing was shown to depend on the activity of silencing factors (38), we tested their involvement in elevation of silencing in the Δelg1 strain. As expected, telomeric silencing of the Δelg1 strain depended on SIR2, an essential component of the telomeric silencing complex (Fig. 5A). As mentioned, yKu70p is essential for telomeric silencing (12). Telomeric silencing of the Δelg1 strain was also dependent on YKU70 (Fig. 5B). In summary, elevation of silencing in the Δelg1 strain depended on the essential components of the telomeric silencing complex in the same manner as normal telomeric silencing.
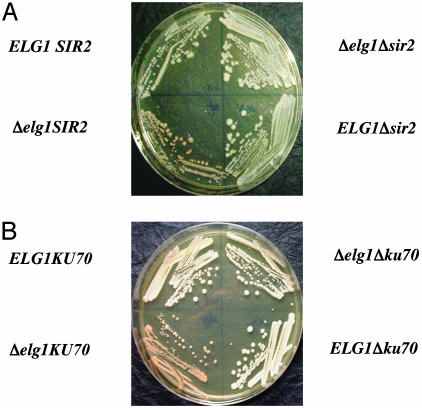
Fig. 5.
Increased silencing at an ADE2 gene placed near the telomere in aΔelg1 background depends on YKU70 and SIR2. (A) ELG1/Δelg1 SIR2/Δsir2 diploid with an ADE2 telomeric gene was dissected. Spores with all gene combinations (ELG1, Δelg1, Δsir2, and Δelg1 Δsir2) and a telomeric ADE2 gene were grown on YPD liquid media supplemented with adenine and plated on a YPD plate. (B) ELG1/Δelg1 YKU70/Δyku70 diploid with an ADE2 telomeric gene was dissected. Spores with all gene combinations (ELG1, Δelg1, Δyku70, and Δelg1 Δyku70) and a telomeric ADE2 gene were grown on YPD liquid media supplemented with adenine and plated on a YPD plate.
Discussion
The main finding of this article is that the alternative RFC component Elg1p participates in negative control of telomere length and telomeric silencing. We have characterized both phenotypes and discovered that the increase in telomere length observed in the absence of ELG1 requires an active telomerase and does not depend on homologous recombination although it was previously shown that deletion of ELG1 increases DNA recombination in other regions of the genome. Our study of the genetic pathway in which ELG1 acts showed that ELG1 is independent of TEL1, RIF1, or RIF2.
In contrast, we show that the effect of Elg1p on telomere length depends on yKu70p, a component of the NHEJ pathway and on Polα. We have also found that the increase in telomeric silencing seen in the absence of ELG1 depended on yKu70p, which is known to recruit the Sir complex to the telomeres (15, 39), and on Sir2p, a major component of the telomeric silencing complex (40). Elg1p probably affects silencing directly because we have shown that the telomere position effect enhancement in Δelg1 strains is not a consequence of telomere elongation.
ELG1 and DNA Replication. Mutations in genes that participate in various aspects of DNA replication result in increased telomere length (21, 22). It was previously shown that mutations in RFC1 and CTF18, an RFC1-like gene, affect telomere length (21, 30). Rad24p, another Rfc1-like protein, which was shown to participate in a telomere checkpoint in the absence of telomerase (40), does not affect telomere length. Therefore, not all RFC-like complexes are associated with telomere length control. Biochemical studies guided by sequence analysis have shown that Elg1p is part of an alternative RFC complex (26, 27, 41, 42).
Here, we show that deletion of an ELG1 results in an increase in telomere length and that the effect of ELG1 on telomeres is genetically unlinked from that of other RFC-like complexes. Our results point strongly to the involvement of DNA replication in the telomeric function of Elg1p. We show that, in the absence of ELG1, elevation of telomere length depends on CDC17/Polα and on YKU70. Mammalian Ku was found to interact with DNA polymerases α, δ, and ε, PCNA, and the RFC complex (43). These results may also indicate that yKu70p might be involved in DNA replication. ELG1 may operate as an alternative slidingclamp loader that loads an additional unknown PCNA-like complex according to particular cellular needs (26, 27, 41). It is possible that one of these functions associated with ELG1 also limits telomere length and telomeric silencing.
The Role of Alternative Elg1p-RFC in Telomere Length Regulation and Telomeric Silencing. We propose two alternative models for the function of Elg1p at the telomere. The first model is based on the competition between telomerase and DNA polymerase. At the telomere, both enzymatic complexes compete for the same substrate, namely, telomeric DNA. According to this model, Elg1p functions to open telomeric chromatin structures that interfere with passage of the replication fork. In this manner, it functions as a processivity factor for replication of the telomeric region. In the absence of Elg1p, DNA polymerase is less able to compete with telomerase due to its reduced ability to pass through the telomeric chromatic region. As a result, telomerase gains better access to its substrate, leading to the observed increased telomere length. The telomeric chromatin structure remains at the telomere, resulting in increased levels of telomeric silencing.
The second model assumes that the telomeric phenotype observed in the absence of Elg1p is due to increased levels of DNA double-strand breaks. It has been proposed that the absence of Elg1p causes replication fork collapse or a defect in fork restart, resulting in the accumulation of genome-wide DNA lesions, including double-strand breaks (26, 27, 41). At the telomere, these breaks may recruit the nonhomologous end-joining protein complex, including yKu70p. Once the telomere associates with more yKu70p, telomerase is recruited more efficiently to the telomere, hence the observed increased telomere length in Δelg1 strains. The yKu complex has a well documented role in recruitment of the Sir proteins to the telomeres (39). It is therefore possible that, in the absence of Elg1p, excess of yKu complex at the telomere end leads to the increased telomeric silencing through more efficient recruitment of the Sir complex.
Further studies are required to determine whether ELG1 affects telomere length through a direct role in replication (e.g., by interacting with PCNA/PCNA-like complex). Alternatively, it may play a more indirect role, namely through effects on chromatin and thereby accessibility of the opposing enzymatic machineries working at the telomere.
Acknowledgments
This article is dedicated to the children of Anat Krauskopf, Yotam and Merav. We thank all members of the Krauskopf laboratory and Martin Kupiec for scientific ideas and for critically reading the manuscript. This work was supported by the Israeli Cancer Foundation and the Israeli Science Foundation.
Abbreviations: Pol, polymerase; RFC, replication factor C; PCNA, proliferating cell nuclear antigen; YPD, yeast extract/peptone/dextrose; SD-Ura, synthetic dextrose medium lacking uracil.
References
- 1.Waga, S. & Stillman, B. (1998) Annu. Rev. Biochem. 67, 721-751. [DOI] [PubMed] [Google Scholar]
- 2.Greider, C. W. & Blackburn, E. H. (1985) Cell 43, 405-413. [DOI] [PubMed] [Google Scholar]
- 3.Singer, M. S. & Gottschling, D. E. (1994) Science 266, 404-409. [DOI] [PubMed] [Google Scholar]
- 4.Cervantes, R. B. & Lundblad, V. (2002) Curr. Opin. Cell Biol. 14, 351-356. [DOI] [PubMed] [Google Scholar]
- 5.McEachern, M. J., Krauskopf, A. & Blackburn, E. H. (2000) Annu. Rev. Genet. 34, 331-358. [DOI] [PubMed] [Google Scholar]
- 6.Hardy, C. F., Sussel, L. & Shore, D. (1992) Genes Dev. 6, 801-814. [DOI] [PubMed] [Google Scholar]
- 7.Wotton, D. & Shore, D. (1997) Genes Dev. 11, 748-760. [DOI] [PubMed] [Google Scholar]
- 8.Lustig, A. J. & Petes, T. D. (1986) Proc. Natl. Acad. Sci. USA 83, 1398-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulton, S. J. & Jackson, S. P. (1996) Nucleic Acids Res. 24, 4639-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porter, S. E., Greenwell, P. W., Ritchie, K. B. & Petes, T. D. (1996) Nucleic Acids Res. 24, 582-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ritchie, K. B. & Petes, T. D. (2000) Genetics 155, 475-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laroche, T., Martin, S. G., Gotta, M., Gorham, H. C., Pryde, F. E., Louis, E. J. & Gasser, S. M. (1998) Curr. Biol. 8, 653-656. [DOI] [PubMed] [Google Scholar]
- 13.Mishra, K. & Shore, D. (1999) Curr. Biol. 9, 1123-1126. [DOI] [PubMed] [Google Scholar]
- 14.Cosgrove, A. J., Nieduszynski, C. A. & Donaldson, A. D. (2002) Genes Dev. 16, 2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boulton, S. J. & Jackson, S. P. (1998) EMBO J. 17, 1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, J. J. & Zakian, V. A. (1996) Proc. Natl. Acad. Sci. USA 93, 13760-13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans, S. K. & Lundblad, V. (1999) Science 286, 117-120. [DOI] [PubMed] [Google Scholar]
- 18.Chandra, A., Hughes, T. R., Nugent, C. I. & Lundblad, V. (2001) Genes Dev. 15, 404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi, H. & Zakian, V. A. (2000) Genes Dev. 14, 1777-1788. [PMC free article] [PubMed] [Google Scholar]
- 20.Diede, S. J. & Gottschling, D. E. (1999) Cell 99, 723-733. [DOI] [PubMed] [Google Scholar]
- 21.Adams, A. K. & Holm, C. (1996) Mol. Cell. Biol. 16, 4614-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carson, M. J. & Hartwell, L. (1985) Cell 42, 249-257. [DOI] [PubMed] [Google Scholar]
- 23.Davey, M. J., Jeruzalmi, D., Kuriyan, J. & O'Donnell, M. (2002) Nat. Rev. Mol. Cell. Biol. 3, 826-835. [DOI] [PubMed] [Google Scholar]
- 24.Adams Martin, A., Dionne, I., Wellinger, R. J. & Holm, C. (2000) Mol. Cell. Biol. 20, 786-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, Z., Shibahara, K. & Stillman, B. (2000) Nature 408, 221-225. [DOI] [PubMed] [Google Scholar]
- 26.Ben-Aroya, S., Koren, A., Liefshitz, B., Steinlauf, R. & Kupiec, M. (2003) Proc. Natl. Acad. Sci. USA. 100, 9906-9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellaoui, M., Chang, M., Ou, J., Xu, H., Boone, C. & Brown, G. W. (2003) EMBO J. 22, 4304-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayer, M. L., Gygi, S. P., Aebersold, R. & Hieter, P. (2001) Mol. Cell 7, 959-970. [DOI] [PubMed] [Google Scholar]
- 29.Green, C. M., Erdjument-Bromage, H., Tempst, P. & Lowndes, N. F. (2000) Curr. Biol. 10, 39-42. [DOI] [PubMed] [Google Scholar]
- 30.Hanna, J. S., Kroll, E. S., Lundblad, V. & Spencer, F. A. (2001) Mol. Cell. Biol. 21, 3144-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gari, E., Piedrafita, L., Aldea, M. & Herrero, E. (1997) Yeast 13, 837-848. [DOI] [PubMed] [Google Scholar]
- 32.Moir, D., Stewart, S. E., Osmond, B. C. & Botstein, D. (1982) Genetics 100, 547-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottschling, D. E., Aparicio, O. M., Billington, B. L. & Zakian, V. A. (1990) Cell 63, 751-762. [DOI] [PubMed] [Google Scholar]
- 34.Maddar, H., Ratzkovsky, N. & Krauskopf, A. (2001) Mol. Biol. Cell 12, 3191-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lingner, J., Cech, T. R., Hughes, T. R. & Lundblad, V. (1997) Proc. Natl. Acad. Sci. USA 94, 11190-11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parenteau, J. & Wellinger, R. J. (1999) Mol. Cell. Biol. 19, 4143-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, C., Mao, X. & Lustig, A. J. (1994) Genetics 138, 1025-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aparicio, O. M., Billington, B. L. & Gottschling, D. E. (1991) Cell 66, 1279-1287. [DOI] [PubMed] [Google Scholar]
- 39.Tsukamoto, Y., Kato, J. & Ikeda, H. (1997) Nature 388, 900-903. [DOI] [PubMed] [Google Scholar]
- 40.Enomoto, S., Glowczewski, L. & Berman, J. (2002) Mol. Biol. Cell 13, 2626-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanellis, P., Agyei, R. & Durocher, D. (2003) Curr. Biol. 13, 1583-1595. [DOI] [PubMed] [Google Scholar]
- 42.Neuwald, A. F., Aravind, L., Spouge, J. L. & Koonin, E. V. (1999) Genome Res. 9, 27-43. [PubMed] [Google Scholar]
- 43.Matheos, D., Ruiz, M. T., Price, G. B. & Zannis-Hadjopoulos, M. (2002) Biochim. Biophys. Acta 1578, 59-72. [DOI] [PubMed] [Google Scholar]