Particular cis-Golgi proteins accumulate in novel punctate structures close to ERES by BFA treatment in tobacco BY-2 cells. These structures reassemble first to form cis-Golgi after BFA removal, and the Golgi stacks regenerate in the cis-to-trans order. This indicates that the punctate structures act as the scaffold for Golgi regeneration.
Abstract
The Golgi apparatus forms stacks of cisternae in many eukaryotic cells. However, little is known about how such a stacked structure is formed and maintained. To address this question, plant cells provide a system suitable for live-imaging approaches because individual Golgi stacks are well separated in the cytoplasm. We established tobacco BY-2 cell lines expressing multiple Golgi markers tagged by different fluorescent proteins and observed their responses to brefeldin A (BFA) treatment and BFA removal. BFA treatment disrupted cis, medial, and trans cisternae but caused distinct relocalization patterns depending on the proteins examined. Medial- and trans-Golgi proteins, as well as one cis-Golgi protein, were absorbed into the endoplasmic reticulum (ER), but two other cis-Golgi proteins formed small punctate structures. After BFA removal, these puncta coalesced first, and then the Golgi stacks regenerated from them in the cis-to-trans order. We suggest that these structures have a property similar to the ER-Golgi intermediate compartment and function as the scaffold of Golgi regeneration.
INTRODUCTION
The Golgi apparatus plays essential roles in modification, sorting, and transport of proteins destined for secretory organelles and the extracellular space. In animal and plant cells, the Golgi apparatus consists of several stacked, flattened membrane sacs called cisternae. In each Golgi stack, the cisternae are polarized between the cis side, receiving cargo from the endoplasmic reticulum (ER), and the trans side, sending cargo forward to post-Golgi organelles.
There was a long debate between two models about how cargo proteins are transported through the Golgi stack: the vesicular transport (stable compartments) model and the cisternal maturation model (Marsh and Howell, 2002; Malhotra and Mayor, 2006; Glick and Nakano, 2009). In the vesicular transport model, the Golgi stack was viewed as a series of distinct suborganelles, each of which had a characteristic set of resident proteins. Cargo proteins were supposed to travel from one cisterna to the next in anterograde vesicles, whereas Golgi-resident enzymes were to be excluded from the vesicles and retained in the cisternae. In the cisternal maturation model, on the other hand, Golgi cisternae were considered to be transient structures. A new cisterna forms at the cis-most face of the Golgi stack, traverses through the stack in the cis-to-trans direction as it matures, and dissipates into the trans-Golgi network at the trans-most face. Cargo proteins can be transported without exiting the cisternae, and Golgi-resident enzymes are postulated to move back to the earlier cisternae. Recent data from the yeast Saccharomyces cerevisiae (Losev et al., 2006; Matsuura-Tokita et al., 2006), algae (Becker et al., 1995), and mammals (Bonfanti et al., 1998) provide strong support for the cisternal maturation model.
Golgi biogenesis has been studied for a variety of cells. New Golgi stacks are shown to form de novo from the ER in yeast Pichia pastoris (Bevis et al., 2002), whereas the Golgi stack in the parasitic flagellate Toxoplasma gondii multiplies by fission (Pelletier et al., 2002). In mammalian cells, the fate of the Golgi apparatus during mitosis has been disputed, whether fragmented or absorbed into the ER (Zaal et al., 1999; Axelsson and Warren, 2004; Persico et al., 2009). On the other hand, the Golgi stacks in plant cells are maintained in the cytoplasm throughout mitosis to transport materials to new cell plates (Nebenführ et al., 2000). The number of Golgi stacks duplicates before cytokinesis in higher plants (Garcia-Herdugo et al., 1988; Seguí-Simarro and Staehelin, 2006). However, the mechanisms that govern the formation of the stacked structure of the Golgi apparatus remain mostly unknown.
Brefeldin A (BFA), an antibiotic compound produced by fungi, is known as a potent inhibitor of guanine nucleotide exchange factors (GEFs) for ARF GTPases (McCloud et al., 1995; Chardin and McCormick, 1999). ARF1 regulates formation of COPI-coated vesicles from the Golgi, which mediate retrograde transport from the Golgi to the ER (Donaldson et al., 2005). Studies in mammalian cells have shown that ARF1 and COPI coat proteins are released from the Golgi membranes by BFA treatment (Donaldson et al., 1990; Robinson and Kreis, 1992). After the release of COPI coats, the Golgi apparatus disassembles, and Golgi-resident enzymes are absorbed into the ER (Scheel et al., 1997; Barzilay et al., 2005). These effects of BFA are reversible. Golgi stacks regenerate after removal of the compound (Alcalde et al., 1992). Similar effects of BFA have also been reported for tobacco and Arabidopsis cultured cells (Takeuchi et al., 2000, 2002; Ritzenthaler et al., 2002).
In vertebrate cells, the Golgi stacks are centralized to the perinuclear region in a microtubule-dependent manner and form a complex ribbon-like structure, which makes it difficult to observe the stacked structure in living cells by light microscopy (Lowe, 2011). In contrast, many Golgi stacks with distinct structure are dispersed in the cytoplasm in plant cells, moving along actin filaments, which provides a great advantage for detailed investigation of the process of cisternal stacking by live imaging (Nakano and Luini, 2010). In this study, we examined the dynamic behavior of the Golgi stacks during BFA treatment in living tobacco BY-2 cells, whose Golgi cisternae are visualized with different fluorescent proteins at high resolution. We also observed regeneration of the Golgi stacks after BFA removal. Our results indicate that particular membrane proteins located at the cis-Golgi cisternae accumulate in small punctate structures upon BFA treatment, which are in close vicinity to the ER exit sites. We propose that these structures act as the scaffold for Golgi regeneration after BFA removal.
RESULTS
Golgi disassembly upon BFA treatment occurs after formation of aggregates from Golgi stacks
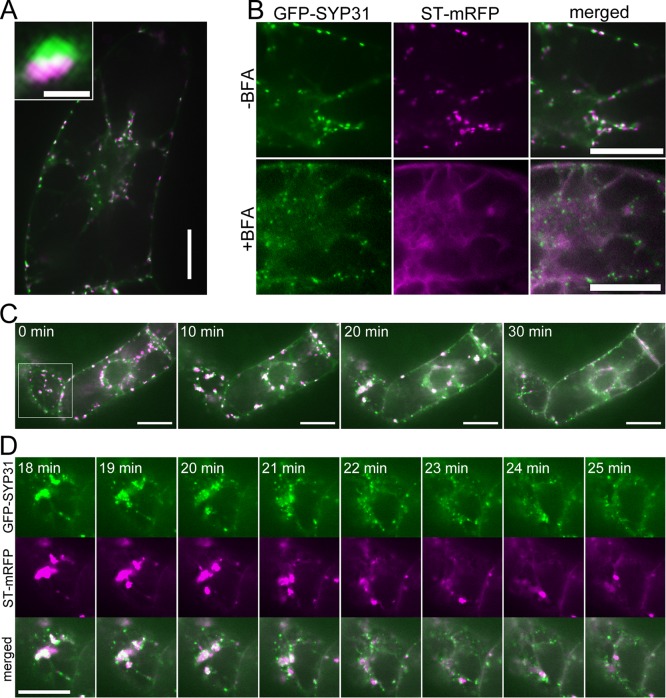
To observe the dynamics of each cisterna of the Golgi apparatus in living cells, we established a tobacco BY-2 cell line expressing the cis-Golgi marker SYP31 (Qa-SNARE; Uemura et al., 2004) tagged with green fluorescent protein (GFP-SYP31) and the trans-Golgi marker ST (the cytoplasmic, transmembrane, and stem regions of rat sialyltransferase; Boevink et al., 1998) tagged with monomeric red fluorescent protein (ST-mRFP). Observation by confocal laser scanning microscopy indicated that many dual-colored punctate structures were moving actively in the cytoplasm. Figure 1A shows the fluorescence of GFP and mRFP from cis- and trans-cisternae, respectively, which were located side by side with partial overlap, as previously observed in BY-2 cells coexpressing ManI-GFP and ST-mRFP (Saint-Jore-Dupas et al., 2006). These dual-colored punctate structures are typical images of the Golgi stacks. The effects of overexpression of these fluorescent markers were carefully assessed by looking at localization patterns. For example, overexpression of GFP-SYP31, which was reported to perturb the endomembrane system (Bubeck et al., 2008), did not affect the normal localizations of ST-mRFP and mRFP-VAM3 (Supplemental Figure S1) in our experimental system.
FIGURE 1:
Effects of BFA on the cis-Golgi marker GFP-SYP31 and the trans-Golgi marker ST-mRFP. Confocal images of BY-2 cells expressing GFP-SYP31 (cis, green) and ST-mRFP (trans, magenta). (A) Double-colored, dot-like structures represent the Golgi stacks. The inset image shows a higher magnification of a single stack. (B) Effects of BFA treatment. Before BFA treatment (top) and after 2 h of 50 μM BFA treatment (bottom). (C) Time-lapse observation during BFA treatment. BY-2 cells were treated with 50 μM BFA, and the fluorescent images were captured at 1-min intervals from just after the addition of BFA. The indicated times mean the elapsed time after the observation was started. (D) Magnified images of the boxed region in C. Scale bars, 20 μm; inset of A, 1 μm.
We first observed dynamics of the Golgi stacks upon BFA treatment (50 μM BFA for 2 h). As previously reported (Saint-Jore et al., 2002), ST-mRFP was relocated to the ER membrane after the BFA treatment (Figure 1B). This relocation of ST was confirmed by observation of BY-2 cells expressing the ER luminal marker SP-GFP-HDEL (Takeuchi et al., 2000) with ST-mRFP (Supplemental Figure S2). On the other hand, GFP-SYP31 was localized to punctate structures, which were smaller in size and larger in number as compared with the cis-Golgi cisternae in untreated cells (Figure 1B). This punctate signal of GFP-SYP31 was also different in its shape from the untreated cis-Golgi cisternae. Whereas GFP-SYP31 resided on the flat, disk-like cisternae in untreated cells, it marked small dot-like structures in BFA-treated cells. A similar result was obtained when BFA treatment was performed with cycloheximide, a general inhibitor of protein synthesis (Supplemental Figure S3A).
Other than the major localization to the punctate structures, GFP-SYP31 signal seemed to increase slightly in the cytoplasm by BFA treatment (Figure 1B). Because SYP31 is a membrane protein, the cytosolic GFP signal may arise from tiny membrane vesicles. To test this possibility, we performed total internal reflection fluorescence microscopy (TIRFM), which enables selective visualization of sample surface regions with high resolution and high signal-to-noise ratio (Fujimoto et al., 2007; Konopka and Bednarek, 2008). TIRFM could detect many small punctate GFP signals in BFA-treated cells (Supplemental Figure S4B) but only a few in control cells (Supplemental Figure S4A). This result indicates that a significant population of SYP31 protein is also present on small vesicles after BFA treatment.
For detailed investigation of the response of Golgi cisternae to BFA, we performed time-lapse observation. At 10 min after BFA addition, the double-colored Golgi stacks began to congregate and coalesce into several aggregates in the cytoplasm (Figure 1C). Subsequently, the aggregates gradually disassembled. GFP-SYP31 was relocated to the small punctate structures, whereas ST-mRFP was absorbed into the ER membrane (Figure 1D). GFP-SYP31 and ST-mRFP in each aggregate seemed to disperse almost simultaneously from 20 to 25 min after BFA addition (Figure 1D), and most aggregates disappeared at 30 min after BFA addition (Figure 1C; see also Supplemental Movie S1). These results indicate that the disassembly of the Golgi by BFA treatment follows formation of Golgi aggregates and that cis- and trans-Golgi components show different behaviors subsequently.
Distinct behaviors of cis-Golgi markers upon BFA treatment
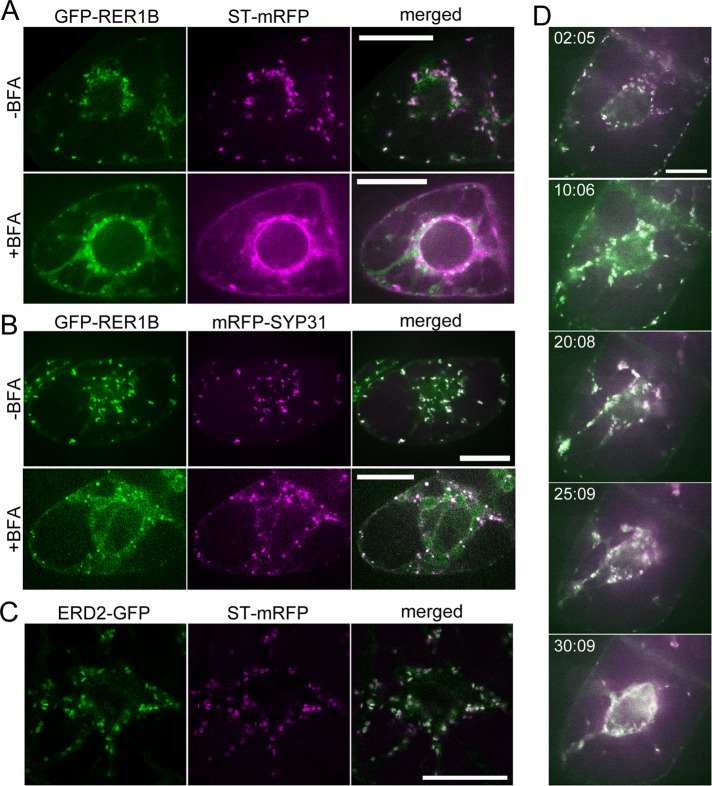
Next we investigated other cis-Golgi proteins. RER1B is a cis-Golgi protein, and a homologue of yeast Rer1p, which is responsible for retrieving a subset of ER membrane proteins from the Golgi to the ER (Sato et al., 1995, 1999). We established BY-2 cells expressing GFP-RER1B and ST-mRFP and observed them by confocal microscopy. The fluorescence patterns of GFP-RER1B and ST-mRFP were similar to those of GFP-SYP31 and ST-mRFP in untreated (control) and BFA-treated cells (Figure 2A). GFP-RER1B was also distributed to small punctate structures by BFA treatment. To examine whether the punctate structures with GFP-RER1B were identical to those of GFP-SYP31 observed after BFA treatment, we observed a BY-2 cell line expressing GFP-RER1B and mRFP-SYP31. These two cis-Golgi markers were colocalized almost completely on small punctate structures after BFA treatment (Figure 2B). Thus SYP31 and RER1B appear to share the common sensitivity to BFA. In contrast, another cis-Golgi protein, ERD2 tagged with GFP, responded to BFA in a distinct manner from the former two cis-Golgi proteins. ERD2 protein is a Golgi receptor of ER proteins, first identified in yeast, and acts in retrieval of KDEL-harboring proteins from the Golgi apparatus to the ER (Lewis et al., 1990; Semenza et al., 1990; Lee et al., 1993). Homologues of ERD2 are widely conserved among eukaryotes, and Arabidopsis thaliana ERD2 protein was also shown localized to the Golgi apparatus (Boevink et al., 1998; Takeuchi et al., 2000, 2002; Saint-Jore et al., 2002). When coexpressed in BY2 cells, ERD2-GFP and ST-mRFP exhibited fluorescence patterns similar to the cases for GFP-SYP31/ST-mRFP and GFP-RER1B/ST-mRFP (Figure 2C). On BFA treatment, however, ERD2-GFP redistributed to the ER, colocalizing with ST-mRFP after formation of aggregates (Figure 2D). This behavior was clearly distinct from the other cis-Golgi markers described earlier (compare Figure 2D with Figures 1B and 2A). Thus there appear to be at least two types of membrane proteins in the cis-Golgi cisternae: one relocates to the ER membrane, and the other localizes to small punctate structures by BFA treatment.
FIGURE 2:
Effects of BFA on the cis-Golgi markers GFP-RER1B and ERD2-GFP. (A) BY-2 cell expressing GFP-RER1B (cis, green) and ST-mRFP (trans, magenta). Before BFA treatment (top) and after 1.5 h of 50 μM BFA treatment (bottom). (B) BY-2 cells expressing GFP-RER1B (cis, green) and mRFP-SYP31 (cis, magenta). Before BFA treatment (top) and after 1.5 h of 50 μM BFA treatment (bottom). (C) BY-2 cell expressing ERD2-GFP (cis, green) and ST-mRFP (trans, magenta). (D) Time-lapse observation of ERD2-GFP/ST-mRFP–expressing cells during BFA treatment. The cells were treated with 50 μM BFA, and the fluorescent images were captured at 30-s intervals after BFA addition. The indicated times mean the elapsed time after BFA addition. Scale bars, 20 μm.
The disassembly of Golgi stacks by BFA is independent of actin filaments
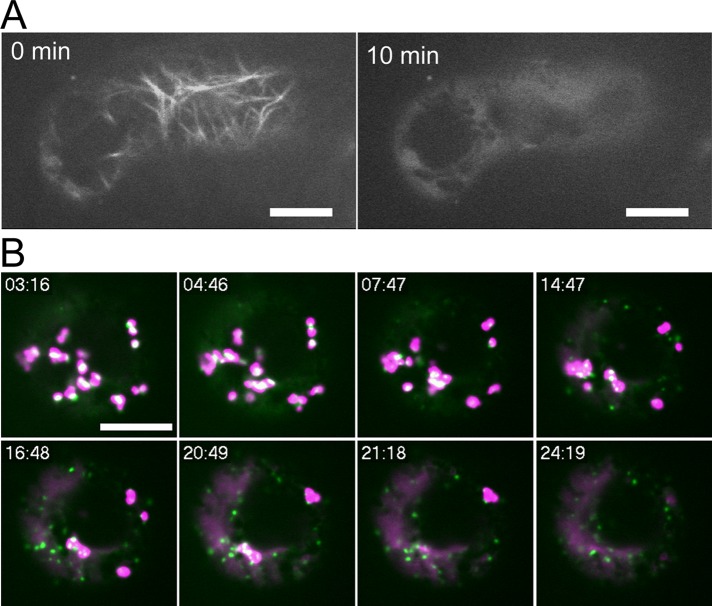
In plant cells, the dynamic movement of organelles including the Golgi apparatus is supposed to depend on actin filaments and myosin motors (Boevink et al., 1998; Nebenführ et al., 1999). To examine whether actin filaments play roles in disassembly of the Golgi upon BFA treatment, we examined the effect of BFA under an actin-depolymerizing condition. When treated with the actin-depolymerizing reagent latrunculin B (LatB), actin filaments visualized with Lifeact-Venus disappeared almost completely by 10 min in BY-2 cells (Figure 3A). After incubating the BY-2 cells expressing GFP-SYP31 and ST-mRFP with LatB for 30 min, we added BFA and performed time-lapse observation of the Golgi apparatus. We found no substantial differences in the response of the Golgi markers to BFA between LatB-treated and control cells (Figure 3B, Supplemental Movie S2, and Supplemental Figure S3B). Thus the response of the Golgi stacks to BFA was independent of actin filaments.
FIGURE 3:
Effects of LatB during BFA treatment. (A) Time-lapse observation of a BY-2 cell expressing Lifeact-Venus during LatB treatment. The indicated times represent the elapsed time after the addition of LatB at 2 μM. (B) Time-lapse observation of a BY-2 cell expressing GFP-SYP31 (cis, green) and ST-mRFP (trans, magenta) during BFA treatment. The cell was treated with 2 μM LatB for 30 min before the addition of BFA at 50 μM. The fluorescent images were captured at 30-s intervals after BFA addition. The indicated times mean the elapsed time after BFA addition. Scale bars, 10 μm.
Yellow fluorescent protein–ATCASP, a medial-Golgi marker, is mostly relocated to the ER by BFA treatment
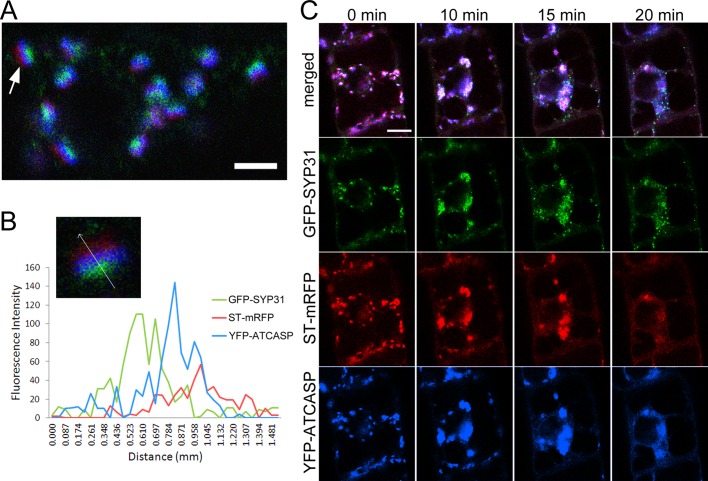
To gain further insight into the behavior of the Golgi stacks, we searched for a Golgi-resident protein with a property different from the aforementioned Golgi markers. ATCASP is a Golgi protein, which was reported as a putative Golgi matrix protein (Renna et al., 2005; Latijnhouwers et al., 2007). When coexpressed in BY-2 cells, yellow fluorescent protein (YFP)–ATCASP was localized at intermediate cisternae between GFP-SYP31 and ST-mRFP with partial overlap (Figure 4A). As shown in Figure 4B, the peak of the fluorescence signal from YFP-ATCASP was located between those of cis-Golgi GFP-SYP31 and trans-Golgi ST-mRFP. This localization pattern indicates that ATCASP is enriched in the medial-Golgi cisternae. BFA treatment resulted in relocalization of most of the YFP-ATCASP fluorescence to the ER, and a very weak signal was observed on the punctate structures of GFP-SYP31 (Figure 4C). In time-lapse observation, YFP-ATCASP and ST-mRFP behaved in a similar time course. At 20 min after BFA treatment, YFP-ATCASP and ST-mRFP were absorbed into the ER membrane, whereas GFP-SYP31 was relocated to the small punctate structures.
FIGURE 4;
Localization of YFP-ATCASP and its BFA treatment. Confocal images of BY-2 cells expressing GFP-SYP31 (cis, green), ST-mRFP (trans, red), and YFP-ATCASP (blue). (A) Triple-colored Golgi stacks. Arrow indicates the Golgi stack used for the analysis in B. (B) The fluorescence profile along the arrow across a Golgi stack. (C) Time-lapse observation during BFA treatment. Cells were treated with 50 μM BFA, and the fluorescent images were captured at 30-s intervals after the addition of BFA. The indicated times mean the elapsed time after BFA addition. Scale bars, 2 μm (A), 10 μm (C).
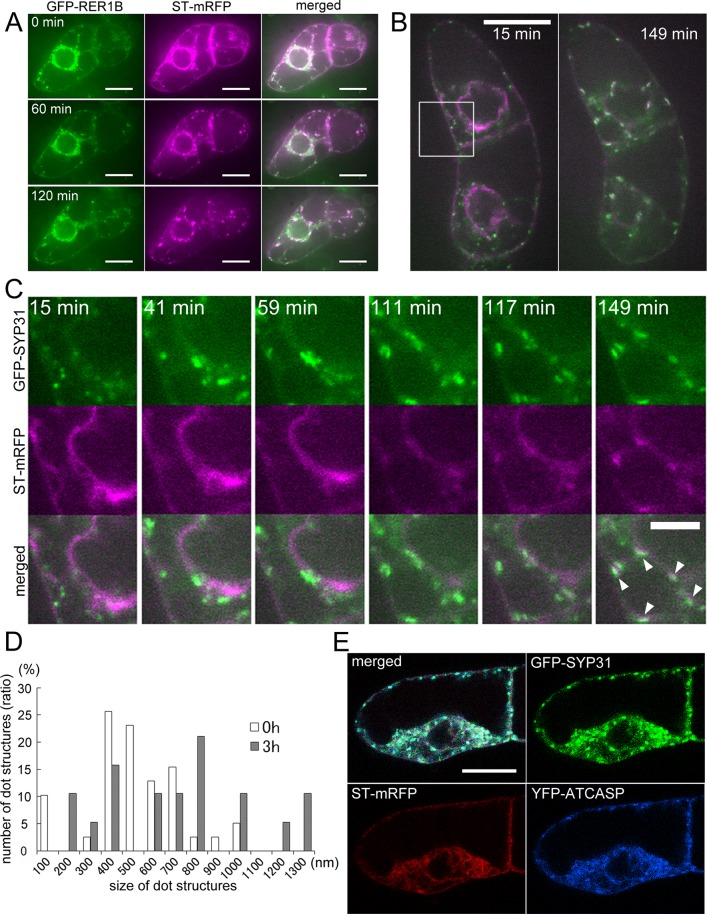
Golgi stacks regenerate in a cis-to-trans direction from the punctate structures of GFP-SYP31/GFP-RER1B
To observe the process of Golgi regeneration, we washed out BFA after 2 h of treatment and observed behaviors of the Golgi markers. In BY-2 cells coexpressing GFP-RER1B and ST-mRFP, ST-mRFP that had been localized to the ER gradually formed punctate structures probably as cisternae, and GFP-RER1B accompanied these regenerated cisternae (Figure 5A). At 2 h after BFA removal, most of the mRFP signal in the ER was relocalized to the Golgi cisternae (Figure 5A). Next we treated cells expressing GFP-SYP31 and ST-mRFP with LatB to stop the movement of organelles during Golgi regeneration, so that detailed processes of the regeneration of the Golgi apparatus could be observed. LatB was added to the cell suspension 30 min before BFA removal, and the regeneration of the Golgi stacks was observed in the presence of LatB and cycloheximide to avoid the involvement of newly synthesized Golgi marker proteins. The time-lapse observation demonstrated that the punctate structures of GFP-SYP31 gathered first, and subsequently ST-mRFP became concentrated at the structures (Figure 5, B and C, and Supplemental Movie S3). The numbers of dotty structures of GFP-SYP31 were compared between short (0 h) and long (3 h) times after BFA removal. It decreased by about half in 3 h (39 dots at 0 h and 18 dots at 3 h). On the other hand, the average of the longest diameter of each dot increased by 200 nm (750 nm at 0 h and 960 nm at 3 h; Figure 5D). We also found that YFP-ATCASP accumulated on the punctate structures of GFP-SYP31 at 10 min after BFA removal, whereas ST-mRFP was still dispersed throughout the ER at this time point (Figure 5E). These results presumably indicate that small punctate structures bearing GFP-SYP31 reassembled after BFA removal to generate the cis-Golgi cisternae, and medial-Golgi cisternae (YFP-ATCASP) are then regenerated before trans-Golgi cisternae (ST-mRFP) regeneration.
FIGURE 5:
Golgi regeneration after BFA removal. (A) Time-lapse observation of BY-2 cells expressing GFP-RER1B (cis, green) and ST-mRFP (trans, magenta) after BFA removal. The cells were treated with 50 μM BFA for 2 h, and then BFA was washed out. The fluorescent images were captured at 2-min intervals after BFA removal. The indicated times mean the elapsed time after the observation was started. (B) Time-lapse observation of BY-2 cells expressing GFP-SYP31 (cis, green) and ST-mRFP (trans, magenta) after BFA removal. The cells were treated with 50 μM BFA for 2 h, and then BFA was washed out. LatB (2 μM) was added 30 min before BFA removal, and cycloheximide (100 μM) was added at the point of BFA removal. The fluorescent images were captured at 2-min intervals after BFA removal. At 15 min after BFA removal (left) and 149 min after BFA removal (right). (C) Magnified images of the boxed region in B. The indicated times mean the elapsed time after BFA removal. (D) Comparison of the longest diameter of each dot of GFP-SYP31 between 15 min after BFA removal (0 h, white bars) and 193 min after BFA removal (3 h, gray bars). (E) BFA removal experiment of GFP-SYP31 (cis, green)/ST-mRFP (trans, red)/YFP-ATCASP (medial, blue)–expressing cells. The cells were treated with 50 μM BFA for 2 h, and then BFA was washed out. LatB was added 30 min before BFA removal. The fluorescent image was captured 10 min after BFA removal. Scale bars, 20 μm (A, B, E), 10 μm (C).
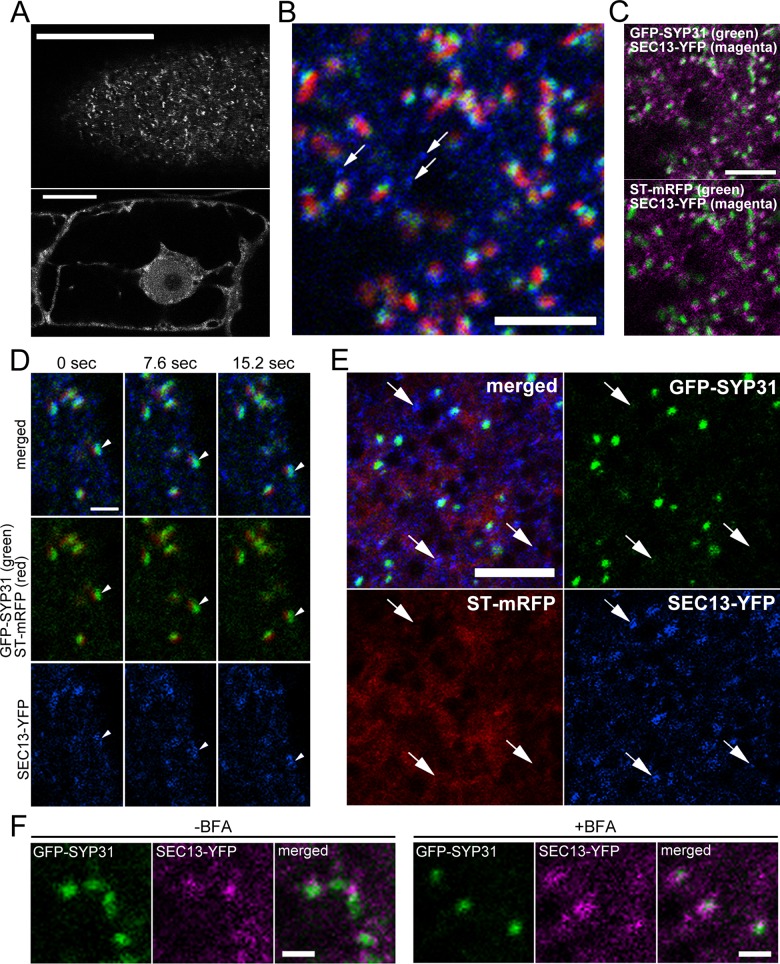
Punctate structures of GFP-SYP31/GFP-RER1B lie in the close vicinity of ER exit sites
We considered it likely that the small puncta with GFP-SYP31 and GFP-RER1B observed after BFA treatment were the ER exit sites (ERES), the domains on the ER membrane where COPII vesicles destined for the Golgi apparatus are produced. To test this possibility, we visualized ERES by SEC13-YFP, the similar construct used successfully to visualize yeast ERES (Rossanese et al., 1999; Sato and Nakano, 2007; Okamoto et al., 2012). In BY-2 cells, SEC13-YFP was observed in the cytosol and on the nuclear envelope in medial optical sections and on many dot-like structures in confocal sections near the plasma membrane (Figure 6A). The distribution on the nuclear envelope is consistent with previous reports because SEC13 is also a member of the nuclear pore complex (Siniossoglou et al., 1996; Alber et al., 2007). Thus focusing at the cell periphery is appropriate to observe only ERES. We established a cell line coexpressing SEC13-YFP, GFP-SYP31, and ST-mRFP and examined spatial relationship between these markers. The Golgi stacks were closely associated with the SEC13 signals (Figure 6B). The number of the SEC13 spots was larger than that of the Golgi stacks. There were SEC13 spots that were free from the Golgi stacks (Figure 6B, arrows). Similar distribution patterns of ERES were reported by immunodetection (Yang et al., 2005). GFP-SYP31 and SEC13-YFP puncta were localized closer than in the case between ST-mRFP and SEC13-YFP (Figure 6C). This perhaps reflects the role of the cis-Golgi cisternae receiving COPII vesicles from the ER. Short-interval time-lapse imaging demonstrated that one ERES signal moved together with a Golgi stack (Figure 6D, arrowheads), which is consistent with the previous observation that the ERES and the Golgi behave as a unit (daSilva et al., 2004; Hanton et al., 2009). We saw no dissociation of the Golgi from its partner ERES. In the cells expressing YFP-SEC13 and mRFP-SYP31, the rapid motion of the punctate signals of SEC13-YFP that were not associated with Golgi stacks was unaffected by LatB treatment. In contrast, the puncta of SEC13-YFP accompanied by Golgi stacks stopped together with their partner Golgi (Supplemental Figure S5A). This suggests that the SEC13-YFP spots with associated Golgi stacks are the active ERES that are exporting cargo to the Golgi.
FIGURE 6:
Relationship between Golgi markers and ERES. (A) Confocal images of BY-2 cells expressing SEC13-YFP. Cell periphery (top) and the confocal plane passing through the nucleus (bottom). (B–E) Confocal images of BY-2 cells expressing GFP-SYP31 (cis), ST-mRFP (trans), and SEC13-YFP (ERES). (B) Image of cell periphery. GFP-SYP31 (green)/ST-mRFP (red)/SEC13-YFP (blue). Arrows indicate the ERES without partner Golgi stacks. (C) Images of two colors extracted from A. Top, GFP-SYP31 (green)/SEC13-YFP (magenta); bottom, ST-mRFP (green)/SEC13-YFP (magenta). (D) Time-lapse confocal images with short interval (7.6 s). GFP-SYP31 (green)/ST-mRFP (red)/SEC13-YFP (blue). Arrowheads indicate a unit of a Golgi stack and an ERES moving toward the lower right. (E) Effects of BFA treatment. GFP-SYP31 (green)/ST-mRFP (red)/SEC13-YFP (blue). The cells were treated with 50 μM BFA for 150 min. Arrows indicate the dots of SEC13-YFP without partner GFP-SYP31 signal. (F) Comparison of positional relationship between GFP-SYP31 (green) and SEC13-YFP (magenta) signals before and after BFA treatment. Scale bars, 20 μm (A), 5 μm (B, C, E), 2 μm (D), 1 μm (F).
Using this transgenic cell line, we examined the relationship between the ERES and the BFA-induced, SYP31-positive punctate domains. After 2.5 h of BFA treatment, ST-mRFP dispersed into the ER membrane, and GFP-SYP31 was localized to small dot-like structures as described earlier. On the other hand, the fluorescence pattern of SEC13-YFP did not substantially change by BFA treatment. Almost all punctate structures of GFP-SYP31 appeared to associate with the SEC13-YFP spots, but these signals did not completely overlap (Figure 6E). We also occasionally observed SEC13-YFP signals without accompanying GFP-SYP31 signals (Figure 6E, arrows). The LatB-sensitive motion of SEC13 spots was observed only for those associated with SYP31, as was the case of control cells (Supplemental Figure S5A). Thus the relationship between the SEC13 spots and the punctate structures of SYP31 remained similar before and after the BFA treatment (Figure 6F).
In the cell line expressing only SEC13-YFP, we observed similar LatB-sensitive and LatB-insensitive motions of SEC13 puncta with or without BFA treatment (Supplemental Figure S5B). These results suggest that it is unlikely that the association between SYP31 and ERES is caused by SYP31 overexpression.
The relationships between the SYP31 puncta and the ER or ERES were also examined by using superresolution confocal live imaging microscopy (SCLIM) that we developed, which enables two-color observation with extremely high speed and sensitivity (Matsuura-Tokita et al., 2006; Nakano and Luini, 2010; Okamoto et al., 2012). The GFP-SYP31/ST-mRFP–expressing cells were observed by SCLIM with optical slices 0.2 μm apart on the Z-axis, and the images were reconstructed into three dimensions with deconvolution to achieve high spatial resolution (Nakano and Luini, 2010). After 2 h of BFA treatment, the GFP-SYP31 puncta were located very close to the ER labeled by ST-mRFP, but the GFP signal did not entirely overlap with the mRFP signal (Figure 7A and Supplemental Movie S4). The punctate structures were frequently clinging to the tips of the protruding regions of the tubular ER (Figure 7A, arrowheads). Next the BY-2 cell line expressing SEC13-YFP and mRFP-SYP31 was observed in the same way. In control cells, mRFP-SYP31 signals were closely associated with those of SEC13-YFP, as observed by two-dimensional (2D) confocal images (Figure 7B). Moreover, by SCLIM, we found that the signal of SEC13-YFP surrounded the Golgi stacks at the cisternal rims, making ring-shaped fluorescence patterns (Figure 7B, insets). This may suggest that the Golgi stacks interact with ERES at their cisternal rims. By 3D time-lapse imaging, we observed the Golgi stacks moving together with the SEC13 ring (Supplemental Movie S5). After BFA treatment, the punctate structures of mRFP-SYP31 were still in the proximity of SEC13-YFP spots without full overlap (Figure 7B). The ring patterns of SEC13-YFP around the Golgi cisternae, which were often observed in control cells, rarely appeared in BFA-treated cells (Figure 7B, insets).
FIGURE 7:
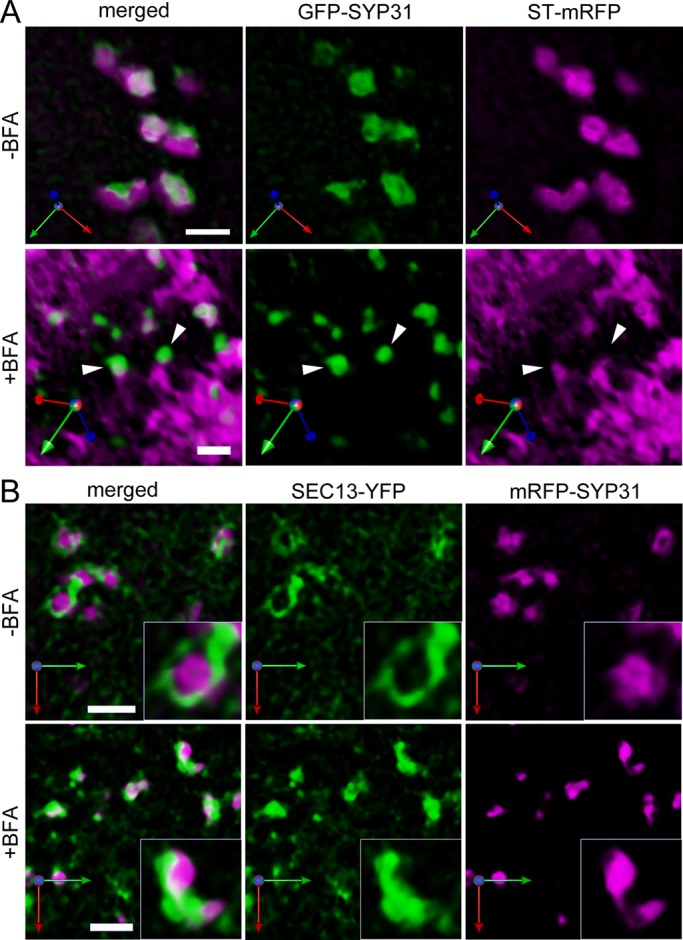
Three-dimensional deconvolution observations of the Golgi markers and ERES. BY-2 cells were observed by SCLIM with optical slices 0.2 μm apart on the Z-axis. 3D images were reconstructed and deconvolved by parameters optimized for the Yokogawa spinning-disk confocal scanner. (A) GFP-SYP31 (cis, green) and ST-mRFP (trans, magenta). Before BFA treatment (top) and after 2 h of 50 μM BFA treatment (bottom). Arrowheads indicate the punctate compartments of GFP-SYP31 on the tips of the tubular ER. (B) SEC13-YFP (ERES, green) and mRFP-SYP31 (cis, magenta). Before BFA treatment (top) and after 1.5 h of 50 μM BFA treatment (bottom). Scale bars, 2 μm.
These results indicate that the punctate structures depicted by GFP-SYP31 after BFA treatment are not the ERES themselves but unique structures existing in the close vicinity to the ERES, which have not been characterized before. Considering all the results we obtained in this study, we propose that these novel punctate structures act as the scaffold in Golgi regeneration after removal of BFA.
DISCUSSION
Dynamic behaviors of the Golgi apparatus in plant cells
Biogenesis of the Golgi apparatus has been an issue of keen interest in cell biology and has evoked many disputes (Glick and Nakano, 2009; Nakano and Luini, 2010). The significance of the stacked structure and molecular mechanisms of its formation have drawn particular attention, but no clear answers have been provided. Comparative studies should give valuable hints because the organization of the Golgi differs widely from species to species. We propose that plant cells serve as a very nice system because individual Golgi stacks are separated from each other in the cytoplasm, which presents an ideal opportunity to apply high-resolution, live-imaging techniques.
Because the cisternal maturation of the Golgi apparatus has been accepted as a favorable basic scheme, one remaining puzzle is how a new cis-Golgi compartment is created. COPII vesicles that are formed from the ER must give rise to such a new compartment, and thus the relationship between the domain of the ER specialized for COPII budding (ERES) and the Golgi apparatus is a focus in the efforts to understand Golgi biogenesis.
BFA response
BFA provides a nice way to study Golgi organization because in mammalian cells its treatment causes relocation of Golgi proteins into the ER, and its washout leads to reformation of the Golgi. Similar effects have been observed with plant cells as well (Takeuchi et al., 2000, 2002; Ritzenthaler et al., 2002; Saint-Jore et al., 2002; daSilva et al., 2004; Yang et al., 2005; Hanton et al., 2009; Schoberer et al., 2010). Reversibility of the effects after BFA washout would give a good opportunity to observe Golgi stack reorganization by live imaging. In previous studies on the effects of BFA in plant cells, Ritzenthaler et al. (2002) reported that Golgi stacks in BY-2 cells lost their cisternae in a cis-to-trans direction by ultrastructural level. Schoberer et al. (2010) looked at tobacco leaf epidermal cells and showed that Golgi markers redistributed to the ER in the trans-to-cis order by BFA treatment. trans-Golgi proteins were shifted to the ER quickly, whereas a much longer time was required for the complete relocation of the cis-Golgi marker to the ER. They also observed the behavior of ATCASP, which did not fully distribute to the ER by BFA treatment. Madison and Nebenführ (2011) also examined the effect of BFA on Golgi proteins in BY-2 cells. Both cis- and trans-cisternae fused with the ER upon BFA treatment, but no significant difference was found for different cisternae.
In the present study, we performed detailed time-lapse observation by high-resolution confocal laser scanning microscopy with short time intervals (30–60 s) and demonstrated that the Golgi stacks formed aggregates first, and then breakdown of GFP-SYP31 labeled cis-cisternae into small compartments and the dispersion of the trans-Golgi marker ST-mRFP to the ER occurred almost at the same time. The reason for the difference from previous studies is unknown, but the resolution in time and space of microscopic observation could be a critical factor. Our data clearly indicate that the collapse of cis- and trans-cisternae by BFA occurs almost at the same time after the aggregation of Golgi apparatus in tobacco BY-2 cells.
Difference among cis-Golgi proteins
In the present study, we noticed a difference in the behaviors of the cis-Golgi proteins upon BFA treatment. That is, one membrane protein (KDEL receptor ERD2) is almost completely absorbed into the ER, whereas others (Qa-SNARE SYP31/SED5 and retrieval receptor RER1) shift their localization to novel structures in the vicinity to the ERES. Although they are closely associated with the ERES, their localizations do not completely overlap with ERES. What do these structures represent?
We obtained relevant interesting results in our previous work. Takeuchi et al. (2002) examined the effects of the transient expression of dominant mutants of ARF1 on localization of Golgi proteins (ERD2, RER1B, and SYP31/SED5) in tobacco BY-2 and Arabidopsis cultured cells. In summary, the ARF1 constitutive active (GTP fixed) mutant caused ER relocation of ERD2 and some alteration of Golgi-like punctate structure for RER1B and SYP31. The ARF1 dominant negative (GDP fixed) mutant also caused ER relocation of ERD2 but had almost no effect for RER1B and SYP31. Furthermore, BFA treatment phenocopied the expression of the ARF1 dominant-negative mutant. We interpreted these results as follows. The steady-state localization of Golgi proteins was shifted to the ER by the blockade of anterograde ER-to-Golgi traffic. ARF1 mutants have primary defects in the retrograde Golgi-to-ER traffic, but they are also impaired in the anterograde ER-to-Golgi traffic indirectly (Gaynor and Emr, 1997) and thus relocate ERD2 to the ER. We explained the reason that RER1B and SYP31 were not significantly affected by the ARF1 mutants as due to the difference of timing between the folding of GFP-tagged proteins and the invocation of the secondary anterograde block, because we were using a transient expression system in this work. We believed that ERD2 might be more slowly folded than RER1B and SYP31, so that when the exogenously expressed GFP-ERD2 became fluorescent the secondary ER-to-Golgi blockade was already in effect, whereas earlier-folding RER1B and SYP31 could escape this block. The difference of BFA effects between ERD2 and RER1B/SYP31 was explained similarly.
In revisiting these results, we were surprised to see the reproducibility of the difference between ERD2 and RER1B/SYP31. We judged in the former work that the Golgi structures of RER1B and SYP31 were slightly or not significantly affected by the ARF1 mutants or by BFA, but the data now seem to indicate that RER1B and SYP31 may be relocated to smaller dotty structures like those we see in the present study. The difference was most probably not due to the folding problem. Considering the possible role of SYP31/RER1B in rebuilding the Golgi stacks (see later discussion), these two classes of cis-Golgi proteins might have an intrinsic difference in their properties or their precise localizations within the region that looks like cis-Golgi. It is also possible that two or more routes exist in ER-Golgi transport, which are regulated by different ARF-GEFs with different sensitivity to BFA.
The Golgi apparatus regenerates in a cis-to-trans order
Golgi regeneration after BFA removal has attracted attention in mammalian research, especially to reveal the mechanisms of Golgi biogenesis and reassembly after mitosis (Seemann et al., 2000; Puri and Linstedt, 2003; Glick and Nakano, 2009). With plants, attempts also have been made by electron microscopy and live-cell imaging to observe the Golgi after BFA removal (Ritzenthaler et al., 2002; Langhans et al., 2007; Schoberer et al., 2010), but time-dependent regeneration processes of the Golgi stacks have not been sufficiently described.
In the present study, we performed multicolor time-lapse live-cell imaging with short intervals, which enabled us to analyze detailed behaviors of multiple Golgi membrane proteins in the same cells with the passage of time. From time-lapse confocal images, we found that the punctate structures of the cis-Golgi marker GFP-SYP31 gather first, and the trans-Golgi marker ST-mRFP concentrates later during Golgi stack regeneration after BFA removal. Furthermore, YFP-ATCASP, which is found by tricolor fluorescence imaging to localize mainly at the medial-Golgi, begins to concentrate before ST-mRFP does. These results suggest that the Golgi stacks regenerate in the cis-to-trans order, as reported before in mammalian cells and tobacco leaf epidermal cells (Alcalde et al., 1992; Puri and Linstedt, 2003; Schoberer et al., 2010). This is consistent with the cisternal maturation model, in which a new cisterna is formed at the cis-side of the stack by ER-derived carriers and progresses from the cis- to the trans-Golgi pole as it matures (Nakano and Luini, 2010).
The SYP31/RER1B compartment acts as the scaffold for Golgi regeneration and has an ER-Golgi intermediate compartment–like property
As described here, there are two types of cis-Golgi proteins; upon BFA treatment, one (SYP31 and RER1B) localizes to the punctate structures and the other (ERD2) to the ER membrane. By super-high-resolution 3D analysis with the high-speed and high-sensitivity confocal microscope that we developed (Matsuura-Tokita et al., 2006; Nakano and Luini, 2010), we conclude that the punctate structures of SYP31 are in the very close vicinity of the ERES (represented by SEC13-YFP) but not the ERES themselves. Because these structures reassemble first to regenerate cis-Golgi cisternae after BFA removal, we propose that they function as “seeds” of the Golgi stacks by receiving the materials for cis-cisternae.
In mammalian cells, Golgi enzymes are absorbed to the ER by BFA treatment, but GM130 and GRASP65 (Golgi matrix proteins) and some Golgi membrane proteins (including mammalian homologues of SYP31, RER1, and ERD2) remain at punctate structures that localize adjacent to ERES (Tang et al., 1993; Nakamura et al., 1995; Füllekrug et al., 1997a, b; Seemann et al., 2000; Ward et al., 2001). These structures are often called Golgi remnants because they appear to be left behind after the Golgi absorption to the ER. The punctate structures of SYP31 and RER1B we observe in BY-2 are similar to these Golgi remnants in terms of behavior after BFA treatment. However, it is not evident whether these structures are indeed left behind or are re-formed after fusion with the ER.
The role of the SYP31 compartment as a scaffold in rebuilding the Golgi stacks also calls to mind its relationship to the so-called ER-Golgi intermediate compartment (ERGIC) in animal cells. In mammalian cells, ERGIC is considered a kind of specialized compartment that stands between the ERES in the cell periphery and the Golgi ribbon near the centrosome, which are separated by a long distance but are connected by microtubules (Nakano and Luini, 2010). Such a microtubule-dependent organization of the Golgi is developed only in vertebrate cells. In invertebrates, plants, and fungi, Golgi stacks are scattered in the cytoplasm and are frequently found in the vicinity of the ERES. However, a recent report on Caenorhabditis elegans Golgi suggests that an ERGIC-like compartment also exists in invertebrate cells, in the region sandwiched between the ERES and the Golgi stack (Witte et al., 2011). If a similar situation could be envisioned in plant cells, such an ERGIC compartment might be exaggerated when the assembly of new cis-cisternae is blocked. This compartment would mature into cis-cisternae when the blockage is removed. The SYP31 compartment produced by BFA treatment appears to satisfy these conditions and may be deemed an ERGIC-like compartment in plant cells. ERGIC is believed to form from ER-derived COPII vesicles, and the vesicles with SYP31 observed by TIRFM in BFA-treated cells might well be such vesicles before assembling into ERGIC.
Mammalian Golgi matrix proteins such as GM130 and GRASPs are believed to play important roles in maintenance of the Golgi structure and perhaps function as the template for Golgi reassembly after mitosis (Glick, 2002; Seemann et al., 2002). However, their homologues have not been found in plant genomes (Latijnhouwers et al., 2007; Lowe, 2011). Plant cells must have used different molecules to establish Golgi scaffolds. The novel punctate compartment of SYP31/RER1B identified in this study would be a nice clue to understanding molecular mechanisms of Golgi biogenesis.
MATERIALS AND METHODS
Construction of plasmids
The DNA fragment coding ST-mRFP and mRFP-SYP31 was cloned in pDD302 and pDD301, respectively, and recombined into pGWB3501 (a kind gift from T. Nakagawa, Shimane University) by LR clonase (Invitrogen, Carlsbad, CA). The DNA fragment coding P35S-GFP-RER1B-TNOS was digested from RER1B/pSKP1 (Takeuchi et al., 2000) by SalI and NotI and entered into pENTR1A. P35S-GFP-RER1B-TNOS in pENTER1A and mRFP-SYP31 in pDD301 or ST-mRFP in pDD302 were recombined into pGWB3501 by LR clonase, and the resulting plasmids were designated GFP-RER1B/ST-mRFP/pGWB3501 and GFP-RER1B/mRFP-SYP31/pGWB3501, respectively. SP-GFP-HDEL and Lifeact-Venus were described in Takeuchi et al. (2000) and Era et al. (2009), respectively.
To generate ERD2-GFP in pGWB1, the DNA fragment coding P35S-ERD2-GFP-TNOS was amplified by PCR using a primer set (5′-CACC-GATTAGCCTTTTCAATTTCAGAAAG-3′ and 5′-TTGCGGGACTCTAATCATAA-3′). The fragment was cloned into pENTR/D-TOPO (Invitrogen) and recombined into pGWB1 by LR clonase. The fragment coding mRFP-VAM3 was similarly cloned into pENTER/D-TOPO and recombined into pGWB1. The DNA fragment coding ST was amplified by PCR using a primer set (5′-CACC-ATGATTCATACCAACTTGAAGAAAAAGTTC-3′ and 5′-CATGGCCACTTTCTCCTGG-3′). The fragment was cloned into pENTR/D-TOPO and recombined into pH7RWG2 (Karimi et al., 2005) by LR clonase. The fragments coding GFP-SYP31 and mRFP-SYP31 similarly were cloned into pENTR/D-TOPO and recombined into pHGW (Karimi et al., 2002).
For the construction of SEC13-YFP and YFP-ATCASP, each cDNA was amplified by PCR and cloned into pENTR/D-TOPO. The primer sets (5′-CACCATGCCTCCTCAGAAGATTGA-3′ and 5′-TGGCTCAACAACAGTCACTT-3′ for SEC13) and (5′-CACCATGGAGGTTTCGCAAGATGG-3′ and 5′-TTAAAGACCGTGAGGAAGGTTT-3′ for ATCASP) were used. The entry clones of SEC13 and ATCASP were then recombined with pK7YWG2 and pK7WGY2, respectively (Karimi et al., 2005).
The establishment of transgenic BY-2 cell lines
WT bright yellow-2 (BY-2) tobacco (Nicotiana tabacum) cell suspension cultures were grown in modified Murashige and Skoog (MS) medium enriched with 0.2 mg/l 2-4-D and were maintained as described in a previous study (Nagata et al., 1992). Three- and four-day-old BY-2 cells were used in the microscopic observations.
Ti plasmids were individually transformed into Agrobacterium tumefaciens strain EHA105. Three 5-ml aliquots of 3-d-old BY-2 cells were incubated with 40, 100, and 400 μl of the overnight culture of the transformed A. tumefaciens by a method described previously (An, 1985). After 48 h of incubation at 28°C, cells were washed three times in 5 ml of modified MS medium and resuspended in modified MS medium containing 200 mg/l Claforan with suitable antibiotics for selection: 50 mg/l hygromycin for ST-mRFP, mRFP-SYP31, GFP-SYP31, GFP-SYP31/ST-mRFP, GFP-RER1B/ST-mRFP, and GFP-RER1B/mRFP-SYP31 and 50 mg/l kanamycin for Lifeact-Venus and SEC13-YFP. Then the suspended cells were plated onto solid modified MS medium containing the same antibiotics as the medium used for suspension (excluding Claforan). After ∼3 wk, selected calli were transferred onto new plates and cultured for additional ∼2 wk. As each callus reached a size of 1–2 cm in diameter, suitable lines expressing marker proteins at the appropriate level for microscopic observation were selected. Each selected line was transferred to 30 ml of new modified MS medium and cultured with continuous shaking similar to WT cells. Transgenic BY-2 cells expressing SEC13-YFP/GFP-SYP31/ST-mRFP were established by introducing the SEC13-YFP into the cell line expressing GFP-SYP31/ST-mRFP by the method described above with the A. tumefaciens EHA105 with Ti plasmids harboring SEC13-YFP. Transformants were suspended in modified MS medium containing 200 mg/l Claforan with 50 mg/l kanamycin and selected on plates of solid modified MS medium containing 50 mg/l kanamycin. The selected calli were transferred onto plates of modified MS medium containing 25 mg/l hygromycin and 25 mg/l kanamycin. Transgenic BY-2 cells expressing YFP-ATCASP/GFP-SYP31/ST-mRFP were established similarly by using Ti plasmids harboring YFP-ATCASP. Transgenic BY-2 cell lines expressing ERD2-GFP/ST-mRFP and SP-GFP-HDEL/ST-mRFP were also established similarly by introducing ERD2-GFP or SP-GFP-HDEL into the cell line expressing ST-mRFP with the A. tumefaciens EHA105 with Ti plasmids harboring ERD2-GFP or SP-GFP-HDEL, respectively. BY-2 cells expressing GFP-SYP31/mRFP-VAM3 were also established similarly by introducing mRFP-VAM3 into the cell line expressing GFP-SYP31. BY-2 cells expressing SEC13-YFP/mRFP-SYP31 were also established by introducing SEC13-YFP into the cell line expressing mRFP-SYP31.
BFA treatment and removal
BFA, 50 mM (Sigma-Aldrich, St. Louis, MO), diluted in dimethyl sulfoxide (DMSO), was prepared as stock solution. Aliquots of stock solution were added to suspension cultures at 50 μM in the final concentration. For Golgi-regeneration experiments, BY-2 cells treated with BFA were washed twice by fresh modified MS medium (BFA free) and resuspended in the modified MS medium containing drugs indicated in the figure legends.
Drug treatments
To depolymerize actin filaments, LatB (Sigma-Aldrich) stock solution was prepared at 2 mM in DMSO and added to suspension cultures at 2 μM in the final concentration. To inhibit the protein synthesis, cycloheximide (Sigma-Aldrich) stock solution was prepared at 100 mM in DMSO and used at 100 μM in the final concentration. The timings when these drugs were added are indicated in the figure legends.
Confocal microscopy
For 2D observation of BY-2 cells expressing fluorescent proteins, two microscopic settings were used: 1) an Olympus IX81-ZDC fluorescence microscope (Olympus, Tokyo, Japan) equipped with a confocal scanner unit (model CSU10; Yokogawa Electric, Tokyo, Japan) and a cooled digital charge-coupled device (CCD) camera (model ORCA-R2; Hamamatsu Photonics, Hamamatsu, Japan) and 2) a confocal laser scanning microscope (model LSM710; Zeiss, Jena, Germany). For long time-lapse observations, cells were placed on 35-mm glass-bottom dishes with poly-l-lysine coating (Matsunami, Osaka, Japan). Images were processed and analyzed with MetaMorph (Molecular Devices, Sunnyvale, CA), ImageJ 1.43 (National Institutes of Health, Bethesda, MD), and Photoshop CS5 (Adobe Systems, San Jose, CA).
SCLIM was used for high-resolution 3D imaging. In the SCLIM system that we developed (Matsuura-Tokita et al., 2006; Nakano and Luini, 2010), an Olympus IX-70 microscope was equipped with a custom-made, super-low-noise, high-speed confocal scanner (Yokogawa Electric), custom-made, cooled image intensifiers (Hamamatsu Photonics), and high-speed, high-sensitivity electron-multiplying CCD cameras (Hamamatsu Photonics). The objective lens was oscillated vertically to the coverslip by a custom-made, high-speed, low-vibration piezo actuator system (Yokogawa Electric). Data were oversampled and subjected to a deconvolution analysis with Volocity (Improvision, PerkinElmer, Waltham, MA) by using the point-spread function optimized for the Yokogawa spinning-disk confocal system.
Supplementary Material
Acknowledgments
We thank members of the Nakano laboratory and Kiminori Toyooka of the RIKEN Plant Science Center for valuable discussions. This work was supported by a Grant-in-Aid for Specially Promoted Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by funds from the Extreme Photonics and the Cellular Systems Biology Projects of RIKEN. Y.I. is a recipient of the Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science.
Abbreviations used:
- BFA
brefeldin A
- ER
endoplasmic reticulum
- ERES
ER exit site
- ERGIC
ER-Golgi intermediate compartment
- GFP
green fluorescent protein
- LatB
latrunculin B
- mRFP
monomeric red fluorescent protein
- SCLIM
superresolution confocal live imaging microscopy
- TIRFM
total internal reflection fluorescence microscopy
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-01-0034) on June 27, 2012.
REFERENCES
- Alber F, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Alcalde J, Bonay P, Roa A, Vilaro S, Sandoval IV. Assembly and disassembly of the Golgi complex: two processes arranged in a cis-trans direction. J Cell Biol. 1992;116:69–83. doi: 10.1083/jcb.116.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G. High efficiency transformation of cultured tobacco cells. Plant Physiol. 1985;79:568–570. doi: 10.1104/pp.79.2.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson MAB, Warren G. Rapid, endoplasmic reticulum-independent diffusion of the mitotic Golgi haze. Mol Biol Cell. 2004;15:1843–1852. doi: 10.1091/mbc.E03-07-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzilay E, Ben-Califa N, Hirschberg K, Neumann D. Uncoupling of brefeldin A-mediated coatomer protein complex-I dissociation from Golgi redistribution. Traffic. 2005;6:794–802. doi: 10.1111/j.1600-0854.2005.00317.x. [DOI] [PubMed] [Google Scholar]
- Becker B, Bölinger B, Melkonian M. Anterograde transport of algal scales through the Golgi complex is not mediated by vesicles. Trends Cell Biol. 1995;5:305–307. doi: 10.1016/s0962-8924(00)89047-9. [DOI] [PubMed] [Google Scholar]
- Bevis BJ, Hammond AT, Reinke CA, Glick BS. De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat Cell Biol. 2002;4:750–756. doi: 10.1038/ncb852. [DOI] [PubMed] [Google Scholar]
- Boevink P, Oparka K, Cruz SS, Martin B, Betteridge A, Hawes C. Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 1998;15:441–447. doi: 10.1046/j.1365-313x.1998.00208.x. [DOI] [PubMed] [Google Scholar]
- Bonfanti L, Mironov AA Jr, Martínez-Menárguez JA, Martella O, Fusella A, Baldassarre M, Buccione R, Geuze HJ, Mironov AA, Luini A. Procollagen traverses the Golgi stack without leaving the lumen of cisternae: evidence for cisternal maturation. Cell. 1998;95:993–1003. doi: 10.1016/s0092-8674(00)81723-7. [DOI] [PubMed] [Google Scholar]
- Bubeck J, Scheuring D, Hummel E, Langhans M, Viotti C, Foresti O, Denecke J, Banfield DK, Robinson DG. The syntaxins SYP31 and SYP81 control ER–Golgi trafficking in the plant secretory pathway. Traffic. 2008;9:1629–1652. doi: 10.1111/j.1600-0854.2008.00803.x. [DOI] [PubMed] [Google Scholar]
- Chardin P, McCormick F. Brefeldin A: the advantage of being uncompetitive. Cell. 1999;97:153–155. doi: 10.1016/s0092-8674(00)80724-2. [DOI] [PubMed] [Google Scholar]
- daSilva LL, Snapp EL, Denecke J, Lippincott-Schwartz J, Hawes C, Brandizzi F. Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. Plant Cell. 2004;16:1753–1771. doi: 10.1105/tpc.022673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Honda A, Weigert R. Multiple activities for Arf1 at the Golgi complex. Biochim Biophys Acta. 2005;1744:364–373. doi: 10.1016/j.bbamcr.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Lippincott-Schwartz J, Bloom GS, Kreis TE, Klausner RD. Dissociation of a 110-kD peripheral membrane protein from the Golgi apparatus is an early event in brefeldin A action. J Cell Biol. 1990;111:2295–2306. doi: 10.1083/jcb.111.6.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Era A, Tominaga M, Ebine K, Awai C, Saito C, Ishizaki K, Yamato KT, Kohchi T, Nakano A, Ueda T. Application of Lifeact reveals F-actin dynamics in Arabidopsis thaliana and the liverwort, Marchantia polymorpha. Plant Cell Physiol. 2009;50:1041–1048. doi: 10.1093/pcp/pcp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Arimura S, Nakazono M, Tsutsumi N. Imaging of plant dynamin-related proteins and clathrin around the plasma membrane by variable incidence angle fluorescence microscopy. Plant Biotechnol. 2007;24:449–455. [Google Scholar]
- Füllekrug J, Boehm J, Röttger S, Nilsson T, Mieskes G, Schmitt HD. Human Rer1 is localized to the Golgi apparatus and complements the deletion of the homologous Rer1 protein of Saccharomyces cerevisiae. Eur J Cell Biol. 1997a;74:31–40. [PubMed] [Google Scholar]
- Füllekrug J, Sönnichsen B, Schäfer U, Nguyen Van P, Söling HD, Mieskes G. Characterization of brefeldin A induced vesicular structures containing cycling proteins of the intermediate compartment/cis-Golgi network. FEBS Lett. 1997b;404:75–81. doi: 10.1016/s0014-5793(97)00097-5. [DOI] [PubMed] [Google Scholar]
- Garcia-Herdugo G, González-Reyes JA, Gracia-Navarro F, Navas P. Growth kinetics of the Golgi apparatus during the cell cycle in onion root meristems. Planta. 1988;175:305–312. doi: 10.1007/BF00396335. [DOI] [PubMed] [Google Scholar]
- Gaynor EC, Emr SD. COPI-independent anterograde transport: cargo-selective ER to Golgi protein transport in yeast COPI mutants. J Cell Biol. 1997;136:789–802. doi: 10.1083/jcb.136.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BS. Can the Golgi form de novo? Nat Rev Mol Cell Biol. 2002;3:615–619. doi: 10.1038/nrm877. [DOI] [PubMed] [Google Scholar]
- Glick BS, Nakano A. Membrane traffic within the Golgi apparatus. Annu Rev Cell Dev Biol. 2009;25:113–132. doi: 10.1146/annurev.cellbio.24.110707.175421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanton SL, Matheson LA, Chatre L, Brandizzi F. Dynamic organization of COPII coat proteins at endoplasmic reticulum export sites in plant cells. Plant J. 2009;57:963–974. doi: 10.1111/j.1365-313X.2008.03740.x. [DOI] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P. Modular cloning in plant cells. Trends Plant Sci. 2005;10:103–105. doi: 10.1016/j.tplants.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inz D, Depicker A. GATEWAY (TM) vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Konopka CA, Bednarek SY. Variable-angle epifluorescence microscopy: a new way to look at protein dynamics in the plant cell cortex. Plant J. 2008;53:186–196. doi: 10.1111/j.1365-313X.2007.03306.x. [DOI] [PubMed] [Google Scholar]
- Langhans M, Hawes C, Hillmer S, Hummel E, Robinson DG. Golgi regeneration after brefeldin A treatment in BY-2 cells entails stack enlargement and cisternal growth followed by division. Plant Physiol. 2007;145:527–538. doi: 10.1104/pp.107.104919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latijnhouwers M, Gillespie T, Boevink P, Kriechbaumer V, Hawes C, Carvalho CM. Localization and domain characterization of Arabidopsis golgin candidates. J Exp Bot. 2007;58:4373–4386. doi: 10.1093/jxb/erm304. [DOI] [PubMed] [Google Scholar]
- Lee HI, Gal S, Newman TC, Raikhel NV. The Arabidopsis endoplasmic reticulum retention receptor functions in yeast. Proc Natl Acad Sci USA. 1993;90:11433–11437. doi: 10.1073/pnas.90.23.11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, Sweet DJ, Pelham HRB. The ERD2 gene determines the specificity of the luminal ER protein retention system. Cell. 1990;61:1359–1363. doi: 10.1016/0092-8674(90)90699-f. [DOI] [PubMed] [Google Scholar]
- Losev E, Reinke CA, Jellen J, Strongin DE, Bevis BJ, Glick BS. Golgi maturation visualized in living yeast. Nature. 2006;441:1002–1006. doi: 10.1038/nature04717. [DOI] [PubMed] [Google Scholar]
- Lowe M. Structural organization of the Golgi apparatus. Curr Opin Cell Biol. 2011;23:85–93. doi: 10.1016/j.ceb.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Madison SL, Nebenführ A. Live-cell imaging of dual-labeled Golgi stacks in tobacco BY-2 cells reveals similar behaviors for different cisternae during movement and brefeldin A treatment. Mol Plant. 2011;4:896–908. doi: 10.1093/mp/ssr067. [DOI] [PubMed] [Google Scholar]
- Malhotra V, Mayor S. The Golgi grows up. Nature. 2006;441:939–940. doi: 10.1038/441939a. [DOI] [PubMed] [Google Scholar]
- Marsh BJ, Howell KE. The mammalian Golgi—complex debates. Nat Rev Mol Cell Biol. 2002;3:789–795. doi: 10.1038/nrm933. [DOI] [PubMed] [Google Scholar]
- Matsuura-Tokita K, Takeuchi M, Ichihara A, Mikuriya K, Nakano A. Live imaging of yeast Golgi cisternal maturation. Nature. 2006;441:1007–1010. doi: 10.1038/nature04737. [DOI] [PubMed] [Google Scholar]
- McCloud TG, Burns MP, Majadly FD, Muschik GM, Miller DA, Poole KK, Roach JM, Ross JT, Lebherz WB. Production of brefeldin-A. J. Ind Microbiol. 1995;15:5–9. doi: 10.1007/BF01570006. [DOI] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S. Tobacco BY-2 cell line as the “HeLa” cell in the cell biology of higher plants. Int Rev Cytol. 1992;132:1–30. [Google Scholar]
- Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano A, Luini A. Passage through the Golgi. Curr Opin Cell Biol. 2010;22:471–478. doi: 10.1016/j.ceb.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Nebenführ A, Frohlick JA, Staehelin LA. Redistribution of Golgi stacks and other organelles during mitosis and cytokinesis in plant cells. Plant Physiol. 2000;124:135–151. doi: 10.1104/pp.124.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A, Gallagher LA, Dunahay TG, Frohlick JA, Mazurkiewicz AM, Meehl JB, Staehelin LA. Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 1999;121:1127–1141. doi: 10.1104/pp.121.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Kurokawa K, Matsuura-Tokita K, Saito C, Hirata R, Nakano A (2012). High-curvature domains of the endoplasmic reticulum (ER) are important for the organization of ER exit sites in Saccharomyces cerevisiae. J Cell Sci. doi: 10.1242/jcs.100065. doi: 10.1242/jcs.100065. [DOI] [PubMed] [Google Scholar]
- Pelletier L, et al. Golgi biogenesis in Toxoplasma gondii. Nature. 2002;418:548–552. doi: 10.1038/nature00946. [DOI] [PubMed] [Google Scholar]
- Persico A, Cervigni RI, Barretta ML, Colanzi A. Mitotic inheritance of the Golgi complex. FEBS Lett. 2009;583:3857–3862. doi: 10.1016/j.febslet.2009.10.077. [DOI] [PubMed] [Google Scholar]
- Puri S, Linstedt AD. Capacity of the Golgi apparatus for biogenesis from the endoplasmic reticulum. Mol Biol Cell. 2003;14:5011–5018. doi: 10.1091/mbc.E03-06-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renna L, Hanton SL, Stefano G, Bortolotti L, Misra V, Brandizzi F. Identification and characterization of AtCASP, a plant transmembrane Golgi matrix protein. Plant Mol Biol. 2005;58:109–122. doi: 10.1007/s11103-005-4618-4. [DOI] [PubMed] [Google Scholar]
- Ritzenthaler C, Nebenführ A, Movafeghi A, Stussi-Garaud C, Behnia L, Pimpl P, Staehelin LA, Robinson DG. Reevaluation of the effects of brefeldin A on plant cells using tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. Plant Cell. 2002;14:237–261. doi: 10.1105/tpc.010237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS, Kreis TE. Recruitment of coat proteins onto Golgi membranes in intact and permeabilized cells: effects of brefeldin A and G protein activators. Cell. 1992;69:129–138. doi: 10.1016/0092-8674(92)90124-u. [DOI] [PubMed] [Google Scholar]
- Rossanese OW, Soderholm J, Bevis BJ, Sears IB, O'Connor J, Williamson EK, Glick BS. Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J Cell Biol. 1999;145:69–81. doi: 10.1083/jcb.145.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jore CM, Evins J, Batoko H, Brandizzi F, Moore I, Hawes C. Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 2002;29:661–678. doi: 10.1046/j.0960-7412.2002.01252.x. [DOI] [PubMed] [Google Scholar]
- Saint-Jore-Dupas C, Nebenführ A, Boulaflous A, Follet-Gueye ML, Plasson C, Hawes C, Driouich A, Faye L, Gomord V. Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell. 2006;18:3182–3200. doi: 10.1105/tpc.105.036400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Nakano A. Mechanisms of COPII vesicle formation and protein sorting. FEBS Lett. 2007;581:2076–2082. doi: 10.1016/j.febslet.2007.01.091. [DOI] [PubMed] [Google Scholar]
- Sato K, Nishikawa S, Nakano A. Membrane protein retrieval from the Golgi apparatus to the endoplasmic reticulum (ER): characterization of the RER1 gene product as a component involved in ER localization of Sec12p. Mol Biol Cell. 1995;6:1459–1477. doi: 10.1091/mbc.6.11.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Ueda T, Nakano A. The Arabidopsis thaliana RER1 gene family: its potential role in the endoplasmic reticulum localization of membrane proteins. Plant Mol Biol. 1999;41:815–824. doi: 10.1023/a:1006329828395. [DOI] [PubMed] [Google Scholar]
- Scheel J, Pepperkok R, Lowe M, Griffiths G, Kreis TE. Dissociation of coatomer from membranes is required for brefeldin A-induced transfer of Golgi enzymes to the endoplasmic reticulum. J Cell Biol. 1997;137:319–333. doi: 10.1083/jcb.137.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoberer J, Runions J, Steinkellner H, Strasser R, Hawes C, Osterrieder A. Sequential depletion and acquisition of proteins during Golgi stack disassembly and reformation. Traffic. 2010;11:1429–1444. doi: 10.1111/j.1600-0854.2010.01106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann J, Jokitalo E, Pypaert M, Warren G. Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature. 2000;407:1022–1026. doi: 10.1038/35039538. [DOI] [PubMed] [Google Scholar]
- Seemann J, Pypaert M, Taguchi T, Malsam J, Warren G. Partitioning of the matrix fraction of the Golgi apparatus during mitosis in animal cells. Science. 2002;295:848–851. doi: 10.1126/science.1068064. [DOI] [PubMed] [Google Scholar]
- Seguí-Simarro JM, Staehelin LA. Cell cycle-dependent changes in Golgi stacks, vacuoles, clathrin-coated vesicles and multivesicular bodies in meristematic cells of Arabidopsis thaliana: a quantitative and spatial analysis. Planta. 2006;223:223–236. doi: 10.1007/s00425-005-0082-2. [DOI] [PubMed] [Google Scholar]
- Semenza JC, Hardwick KG, Dean N, Pelham HR. ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell. 1990;61:1349–1357. doi: 10.1016/0092-8674(90)90698-e. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Ueda T, Sato K, Abe H, Nagata T, Nakano A. A dominant negative mutant of Sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 2000;23:517–525. doi: 10.1046/j.1365-313x.2000.00823.x. [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Ueda T, Yahara N, Nakano A. Arf1 GTPase plays roles in the protein traffic between the endoplasmic reticulum and the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 2002;31:499–515. doi: 10.1046/j.1365-313x.2002.01372.x. [DOI] [PubMed] [Google Scholar]
- Tang BL, Wong SH, Qi XL, Low SH, Hong W. Molecular cloning, characterization, subcellular localization and dynamics of p23, the mammalian KDEL receptor. J Cell Biol. 1993;120:325–338. doi: 10.1083/jcb.120.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH. Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct Funct. 2004;29:49–65. doi: 10.1247/csf.29.49. [DOI] [PubMed] [Google Scholar]
- Ward TH, Polishchuk RS, Caplan S, Hirschberg K, Lippincott-Schwartz J. Maintenance of Golgi structure and function depends on the integrity of ER export. J Cell Biol. 2001;155:557–570. doi: 10.1083/jcb.200107045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte K, Schuh AL, Hegermann J, Sarkeshik A, Mayers JR, Schwarze K, Yates JR, 3rd, Eimer S, Audhya A. TFG-1 function in protein secretion and oncogenesis. Nat Cell Biol. 2011;13:550–558. doi: 10.1038/ncb2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YD, Elamawi R, Bubeck J, Pepperkok R, Ritzenthaler C, Robinson DG. Dynamics of COPII vesicles and the Golgi apparatus in cultured Nicotiana tabacum BY-2 cells provides evidence for transient association of Golgi stacks with endoplasmic reticulum exit sites. Plant Cell. 2005;17:1513–1531. doi: 10.1105/tpc.104.026757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaal KJM, et al. Golgi membranes are absorbed into and reemerge from the ER during mitosis. Cell. 1999;99:589–601. doi: 10.1016/s0092-8674(00)81548-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.