DNA Pol ε synthesizes the leading strands, following the CMG (Cdc45/Mcm2-7/GINS) helicase, although the N-terminal polymerase domain of the catalytic subunit, Cdc20 in fission yeast, is dispensable for viability. We show that the C-terminal domain of Cdc20 plays the noncatalytic essential roles in both the assembly and progression of CMG helicase.
Abstract
DNA polymerase epsilon (Pol ε) synthesizes the leading strands, following the CMG (Cdc45, Mcm2-7, and GINS [Go-Ichi-Nii-San]) helicase that translocates on the leading-strand template at eukaryotic replication forks. Although Pol ε is essential for the viability of fission and budding yeasts, the N-terminal polymerase domain of the catalytic subunit, Cdc20/Pol2, is dispensable for viability, leaving the following question: what is the essential role(s) of Pol ε? In this study, we investigated the essential roles of Pol ε using a temperature-sensitive mutant and a recently developed protein-depletion (off-aid) system in fission yeast. In cdc20-ct1 cells carrying mutations in the C-terminal domain of Cdc20, the CMG components, RPA, Pol α, and Pol δ were loaded onto replication origins, but Cdc45 did not translocate from the origins, suggesting that Pol ε is required for CMG helicase progression. In contrast, depletion of Cdc20 abolished the loading of GINS and Cdc45 onto origins, indicating that Pol ε is essential for assembly of the CMG complex. These results demonstrate that Pol ε plays essential roles in both the assembly and progression of CMG helicase.
INTRODUCTION
All the components of the replisome, including DNA helicase and DNA polymerases, are loaded onto chromosomal replication origins during the process of initiation (Bell and Dutta, 2002). In eukaryotes, the replicative helicase is composed of Cdc45, Mcm2-7, and GINS (Go-Ichi-Nii-San), referred to as the CMG complex (Gambus et al., 2006; Moyer et al., 2006; Pacek et al., 2006). Following the helicase, Pol ε and Pol δ are considered to synthesize the leading and lagging strands, respectively, after priming by Pol α (Pursell et al., 2007; Nick McElhinny et al., 2008; Miyabe et al., 2011).
Replisome assembly is tightly regulated by progression of the cell cycle. In G1-phase, Mcm2-7 complex is loaded onto replication origins by the origin recognition complex, Cdc6/Cdc18 and Cdt1, resulting in prereplicative complex (pre-RC) formation (Diffley et al., 1994; Bell and Dutta, 2002). On activation of cyclin-dependent kinase (CDK) and Dbf4-dependent kinase (DDK) at the onset of S-phase, DDK phosphorylates components of Mcm2-7 and stimulates origin loading of Sld3 (Yabuuchi et al., 2006; Heller et al., 2011; Tanaka et al., 2011). CDK phosphorylates Sld3 and Sld2/Drc1, and promotes formation of a complex of Sld3-Dpb11/Cut5-Sld2/Drc1 at replication origins (Masumoto et al., 2002; Tanaka et al., 2007; Zegerman and Diffley, 2007; Fukuura et al., 2011). Finally, these factors collectively recruit GINS and Cdc45 onto Mcm2-7, resulting in assembly of the CMG complex (Remus and Diffley, 2009; Araki, 2010; Labib, 2010). Origin DNA is unwound by the activated CMG helicase, and then RPA, a single-stranded DNA-binding protein complex, and DNA polymerases are recruited to the origin, resulting in assembly of the complete replisome.
Genetic analyses of mutator polymerases support a model wherein Pol ε is primarily responsible for synthesis of the leading strands at replication forks (Pursell et al., 2007; Nick McElhinny et al., 2008; Miyabe et al., 2011). The Pol ε holo-complex is composed of four subunits, the catalytic Pol2/Cdc20/p261 and accessories Dpb2/p59, Dpb3/p12, and Dpb4/p17 (Asturias et al., 2006; Pursell and Kunkel, 2008). Cdc20/Pol2 and Dpb2 are essential for cell viability (Morrison et al., 1990; Araki et al., 1991a; D'Urso and Nurse, 1997; Feng et al., 2003). Dpb3 is essential in fission yeast (Spiga and D'Urso, 2004), but not in budding yeast (Araki et al., 1991b). Dpb4 is dispensable for viability in both types of yeast (Ohya et al., 2000; Spiga and D'Urso, 2004). The Xenopus p261-p59 complex supports DNA replication in Xenopus egg extracts, whereas the p261 subunit alone, or in a complex with the p12 and p17 subunits, does not (Shikata et al., 2006). Recent experiments strongly suggest that the CMG complex unwinds the DNA duplex by translocating along the strand in a 3′–5′ direction, forming the template strand for Pol ε at replication forks (Moyer et al., 2006; Ilves et al., 2010; Yardimci et al., 2010; Fu et al., 2011). It has been reported that GINS associates with the Pol ε complex and stimulates the polymerase activity of Pol ε in vitro (Bermudez et al., 2011). These findings suggest functional interplay between CMG helicase and Pol ε.
In budding yeast and fission yeast, the N-terminal polymerase domain of the catalytic subunit of Pol ε is dispensable for cell viability, whereas the C-terminal domain (CTD) is essential (Dua et al., 1999; Kesti et al., 1999; Feng and D'Urso, 2001). This suggests that Pol ε has essential role(s) in reactions other than synthesis of the leading strands. Similarly, the CTD but not the N-terminal polymerase domain of the catalytic subunit of Pol ε is essential for DNA replication in the Drosophila eye imaginal disk cells (Suyari et al., 2012), suggesting that the noncatalytic function(s) of Pol ε is conserved from yeast to higher eukaryotes. In budding yeast, a fragile complex containing Pol ε, GINS, Dpb11, and Sld2, called as the preloading complex (pre-LC), is prerequisite for loading of these factors onto replication origins (Muramatsu et al., 2010). In contrast, DNA replication is initiated in Pol ε–depleted Xenopus egg extracts, although DNA synthesis is distorted (Waga et al., 2001; Fukui et al., 2004). In human cells, small interfering RNA depletion of Pol ε causes delay in S-phase and progression of replication forks (Bermudez et al., 2011). Therefore it remains unclear whether Pol ε is required for replisome assembly in organisms other than budding yeast.
To elucidate the essential function of Pol ε in fission yeast, we first isolated a temperature-sensitive mutant harboring mutations in Cdc20 CTD. At the restrictive temperature of cdc20-ct1, the CMG components, Pol α, and Pol δ were efficiently localized at origins, although Pol ε subunits did not form a stable complex or associate with the origin. Interestingly, Cdc45, which was loaded onto the origins, did not migrate from the origin, indicating that Pol ε is required for the progression of CMG helicase. This is the specific function of Pol ε, because Pol α depletion by a protein degradation system, the Auxin-Inducible-Degron (AID), in combination with transcriptional repression (the off-aid system) (Nishimura et al., 2009; Kanke et al., 2011), which abolished origin loading of Pol δ and DNA synthesis, allowed extensive migration of Cdc45 and Pol ε. These findings suggest that Pol ε has a role in the progression of CMG helicase in a DNA synthesis–independent manner. Finally, we investigated whether Pol ε is required for replisome assembly by depleting Cdc20 using the off-aid system. On depletion, neither GINS, Cdc45, Cut5, nor Drc1 was loaded onto replication origins, whereas Mcm6 and Sld3 were localized, indicating that Pol ε is required for assembly of the CMG complex at replication origins. From these results, we argue that Pol ε plays essential roles in both the assembly and progression of CMG helicase.
RESULTS
A temperature-sensitive cdc20-ct1 mutant exhibits a defect in an early step of DNA replication
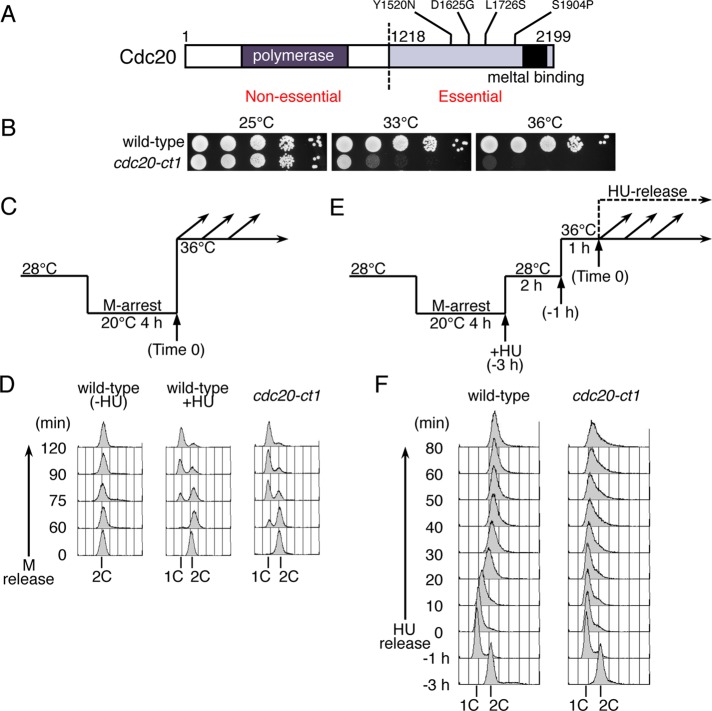
To investigate the essential function of Pol ε in fission yeast, we first created temperature-sensitive mutants carrying mutations in the CTD of Cdc20. One of these mutants, cdc20-ct1, carrying four amino acid substitutions, Y1520N, D1625G, L1726S, and S1904P (Figure 1, A and B), was used for detailed analysis because it exhibited clear cell-cycle arrest. To examine the possible defect in DNA replication, wild-type and cdc20-ct1 cells arrested at M-phase by the cold-sensitive nda3-KM311 mutation (Hiraoka et al., 1984) were released synchronously at 36°C (Figure 1C), and the DNA contents were analyzed by flow cytometry (Figure 1D). Cells with 1C DNA were accumulated in cdc20-ct1, similarly with wild-type cells released in the presence of hydroxyurea (HU), which retards progression of the replication fork due to depletion of deoxyribonucleotide pools (Figure 1D). These results indicate that Cdc20 plays an essential role in the early stages of DNA replication. To examine whether cdc20-ct1 has a defect in the elongation process after the initiation of replication, we analyzed the DNA contents of cells released from HU arrest (Figure 1E). Wild-type and cdc20-ct1 cells were arrested at M-phase by incubation at 20°C for 4 h (Time –3 h) and released in the presence of HU at 28°C, which is the permissive temperature for cdc20-ct1. This allowed initiation of DNA replication from early replication origins but prevented progression of replication forks. After inactivation of Cdc20-ct1 at 36°C for 1 h, HU-treated cells were released into fresh medium without HU (Time 0), and the DNA contents of cells collected at indicated time points were analyzed using flow cytometry. In wild-type cells, DNA contents increased from 1C to 2C in 40 min after HU release, indicating resumption of DNA synthesis (Figure 1F, wild type). In contrast, in cdc20-ct1 cells, DNA content did not increase extensively (Figure 1F, cdc20-ct1). These results suggest that the cdc20-ct1 mutant has a defect in the elongation step of DNA replication.
FIGURE 1:
Defect at an early step of DNA replication in the cdc20-ct1 temperature-sensitive mutant. (A) Schematic representation of the Cdc20, the catalytic subunit of Pol ε in S. pombe. Locations of the conserved B-family DNA polymerase domain (polymerase) and cysteine-rich metal binding motifs (metal binding) are shown. The numbering refers to the amino acid residues in the Cdc20 polypeptide. The locations of amino acid alterations in the cdc20-ct1 are shown. (B) Temperature-sensitive growth of the cdc20-ct1 mutant. Tenfold serial dilutions of wild-type and cdc20-ct1 (HM1317) mutant cells were spotted onto EMM plates, and the plates were then incubated at 25°C, 33°C, or 36°C. (C) For synchronous release from M-phase at the restrictive temperature of cdc20-ct1, HM2970 nda3-KM311 (wild-type) and HM1320 cdc20-ct1 nda3-KM311 (cdc20-ct1) cells were arrested at metaphase by incubation at 20°C for 4 h, and then released at 36°C (Time 0). Wild-type cells were released in the absence (wild-type -HU) or presence of HU (15 mM) (wild-type +HU). (D) Aliquots of cells were taken at the indicated time points and analyzed by flow cytometry. Positions of 1C and 2C DNA contents are shown. (E) For synchronous release from HU arrest, wild-type (HM2970) and cdc20-ct1 (HM1320) cells were arrested at metaphase by incubation at 20°C for 4 h and released at 28°C in the presence of HU (13 mM, –3 h). After cultured with HU for 2 h (–1 h), the cells were incubated at 36°C for 1 h and then released into fresh medium without HU (Time 0). (F) Aliquots taken at indicated time points were analyzed by flow cytometry. Positions of 1C and 2C DNA contents are indicated.
Cdc20 CTD is required for efficient progression of the CMG helicase
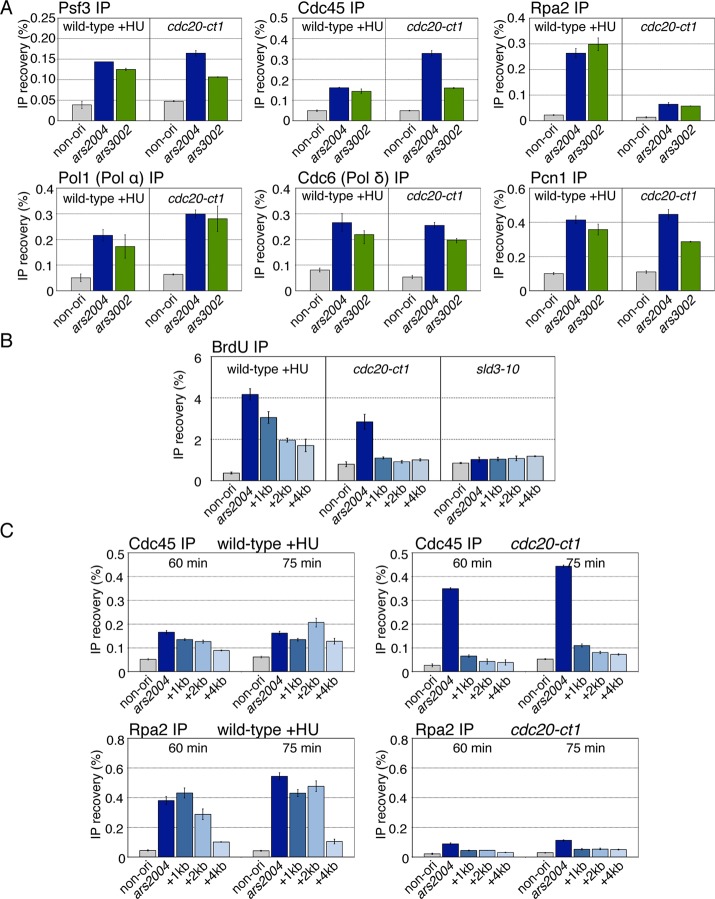
To clarify the reaction that requires the function of Cdc20 CTD, we examined whether replisome components were assembled at replication origins in cdc20-ct1 using chromatin immunoprecipitation (ChIP) assays. We first carried out ChIP assays for a GINS subunit, Psf3, and Cdc45 to examine whether components of the CMG complex are recruited to origins. Wild-type and cdc20-ct1 cells were released synchronously from M-phase (see Figure 1C). Wild-type cells were released in the presence of HU (15 mM), allowing the detection of transient localization of replication factors at the origin at this high temperature. DNA immunoprecipitated with Psf3 or Cdc45 at 60 min after M release was analyzed by quantitative PCR (qPCR) for two early origins, ars2004 and ars3002, and a nonorigin region located 30 kb from ars2004. In HU-treated wild-type cells, the two origin DNAs were preferentially enriched in comparison to the nonorigin region, indicating that Psf3 and Cdc45 were assembled onto origins (Figure 2A, wild-type +HU). In cdc20-ct1, Psf3 and Cdc45 were localized to the origins, as in the HU-treated wild-type cells (Figure 2A, cdc20-ct1). These results show that the cdc20-ct1 mutant does not have a defect in loading of the CMG components onto origins. We next performed ChIP assays for Rpa2 (RPA), Pol1, the catalytic subunit of Pol α, Cdc6, the catalytic subunit of Pol δ, and Pcn1 (PCNA) to examine whether origin DNA was unwound and other DNA polymerases associated with the origin in cdc20-ct1. Pol1 (Pol α), Cdc6 (Pol δ), and Pcn1 were localized at the origins in cdc20-ct1 at levels similar to those in HU-treated wild-type cells (Figure 2A), indicating that the cdc20-ct1 mutant does not have defective origin association of Pol α, Pol δ, and PCNA. In contrast, localization of Rpa2 at the origins was decreased in the mutant relative to HU-treated wild-type cells, although it was significantly higher than in the nonorigin region (Figure 2A, Rpa2 immunoprecipitation [IP]). These results suggest that origin DNA is unwound but less extensively in cdc20-ct1. Because origin association of Pol α depends on DNA unwinding (Walter and Newport, 2000) and RPA (Tanaka and Nasmyth, 1998; Mimura et al., 2000), it is likely that a small amount of RPA bound to the single-stranded DNA around the replication origin is sufficient for recruitment of Pol α.
FIGURE 2:
Pol ε is required for efficient progression of CMG helicase. (A) Derivatives of nda3-KM311 (wild-type) and nda3-KM311 cdc20-ct1 (cdc20-ct1) carrying flag-cdc45, pol1-flag, or cdc6-flag were analyzed by ChIP assay at 60 min after M-phase release (see Figure 1C), with addition of HU (15 mM) in the case of wild type. ChIP recoveries of two early origins, ars2004 and ars3002, and the nonorigin fragments located 30 kb from ars2004, were measured by qPCR. Mean ± SD obtained from multiple measurements in qPCR is presented. The reproducible results obtained in biologically independent experiments are shown in C, Figure 4C, and Supplemental Figure S1A. (B) HM1427 Pnmt1-TK Padh1-hENT nda3-KM311 (wild type), HM1431 cdc20-ct1 Pnmt1-TK Padh1-hENT nda3-KM311 (cdc20-ct1), and HM1603 sld3-10 Pnmt1-TK Padh1-hENT nda3-KM311 (sld3-10) cells were synchronously released from M-phase at 36°C in the presence of BrdU (200 mM), with addition of HU (15 mM) in the case of wild type. Samples taken at 60 min after M release were analyzed using the BrdU-IP assay. BrdU-immunoprecipitated DNAs were analyzed by qPCR using primers for ars2004, the regions located 1, 2, and 4 kb from ars2004, denoted as “+1kb,” “+2kb,” or “+4kb,” and the nonorigin region. Mean ± SD obtained from multiple measurements in qPCR is presented. The reproducible results obtained in biologically independent experiments are shown in Supplemental Figure S1B. (C) Aliquots of flag-cdc45 nda3-KM31 (wild-type, HM631) and cdc20-ct1 flag-cdc45 nda3-KM311 (cdc20-ct1, HM1322), obtained at indicated time points, were analyzed by ChIP assays with anti-FLAG or anti-Rpa2 as described in (A) and were quantified using primers for ars2004, the regions located 1, 2, and 4 kb from ars2004 and the nonorigin region. The reproducible results obtained in biologically independent experiments are shown in Figure 4C.
Because, in the cdc20-ct1 mutant, both Pol1 (Pol α) and Cdc6 (Pol δ) were efficiently recruited onto the origins (Figure 2A), there was a possibility that DNA synthesis might have been initiated. Therefore we carried out the 5-bromo-2′-deoxyuridine (BrdU)-labeling assay to examine whether, in fact, DNA synthesis is initiated in the cdc20-ct1 mutant. Wild-type, cdc20-ct1, and sld3-10, which has a defect in the initiation process (Nakajima and Masukata, 2002), were labeled with BrdU for 60 min after M release, and BrdU-DNA recovered by immunoprecipitation with anti-BrdU antibody was analyzed by qPCR using primers that amplify the nonorigin, the ars2004 origin, and the regions 1, 2, and 4 kb from the origin. In HU-treated wild-type cells, BrdU was robustly incorporated in the ars2004 and the surrounding regions, and the amount incorporated gradually decreased as the distance from the origin increased (Figure 2B, wild-type +HU). These results indicated that replication initiated at the origin and then propagated from it. In the sld3-10 mutant, BrdU was not incorporated significantly in any regions (Figure 2B, sld3-10), due to defective initiation. In contrast, in cdc20-ct1, BrdU was specifically incorporated at the ars2004 origin but not in the 1–4 kb region (Figure 2B, cdc20-ct1), indicating that DNA synthesis was initiated at the origin but did not propagate. Thus Cdc20 CTD is essential for the progression of replication forks.
To determine whether CMG complex that assembled at the origin was subsequently translocated, we examined the localization of Cdc45 and Rpa2 in the regions 1, 2, and 4 kb distant from ars2004. In HU-treated wild-type cells at an early time point (60 min), ars2004 DNA was recovered with Cdc45-IP more efficiently than at the nonorigin region, and then the DNA in the 2-kb region became more enriched than that at ars2004 (75 min), suggesting that Cdc45 first associates with the origin and then translocates (Figure 2C, wild-type +HU, Cdc45 IP). At a later time point, Rpa2 also became localized efficiently in the 2-kb region, in addition to the ars2004 region, indicating progression of DNA unwinding (Figure 2C, wild-type +HU, Rpa2 IP). In contrast, in cdc20-ct1, Cdc45 was highly enriched at ars2004, but greatly reduced in the 1–4 kb region (Figure 2C, cdc20-ct1, Cdc45 IP). These results suggest that Cdc45 does not migrate efficiently from the origin. Consistent with Cdc45 localization, Rpa2 was localized only in the ars2004 region (Figure 2C, cdc20-ct1, Rpa2 IP). The decreased localization of Rpa2 may be due to the limited amount of single-stranded DNA even around the origin. From these results, we conclude that Cdc20 CTD is required for the efficient progression of CMG helicase from the origin.
We examined whether the function of Cdc20 CTD in progression of CMG helicase is independent of the DNA polymerase activity of Pol ε by constructing cdc20ΔN cells expressing Cdc20 CTD lacking the N-terminal polymerase domain (Supplemental Figure S2). In HU-treated cdc20ΔN cells, Cdc45 and Rpa2 were localized at the ars2004 origin and in the region 2–4 kb from the origin, similar to HU-treated wild-type cells (Supplemental Figure S2B), indicating progression of CMG helicase from the origin in the absence of Pol ε DNA polymerase activity. Moreover, localizations of Cdc20 CTD in the ars2004 and 2–4 kb regions were significantly higher than that in the nonorigin region (Supplemental Figure S2B), indicating progression of Cdc20 CTD with the replication forks. Reduced localization of Cdc20 CTD at the origin and the surrounding regions in comparison with those in wild-type cells suggests that DNA polymerization by Pol ε may contribute to stable association of Cdc20 with replication forks. We were unable to examine whether the cdc20-ct1 mutation impairs the progression of CMG helicase in cdc20ΔN, because cdc20-ct1ΔN was lethal. However, cdc20-S1904PΔN cells harboring one of four substitutions of cdc20-ct1 exhibited temperature sensitivity and defective DNA replication at the high temperature (36°C; Supplemental Figure S2, C and D). These results suggested that the function of Cdc20 CTD independent of the DNA polymerization activity is impaired by one of the mutations in cdc20-ct1.
Progression of CMG helicase is not dependent on Pol α but on Cdc20 CTD
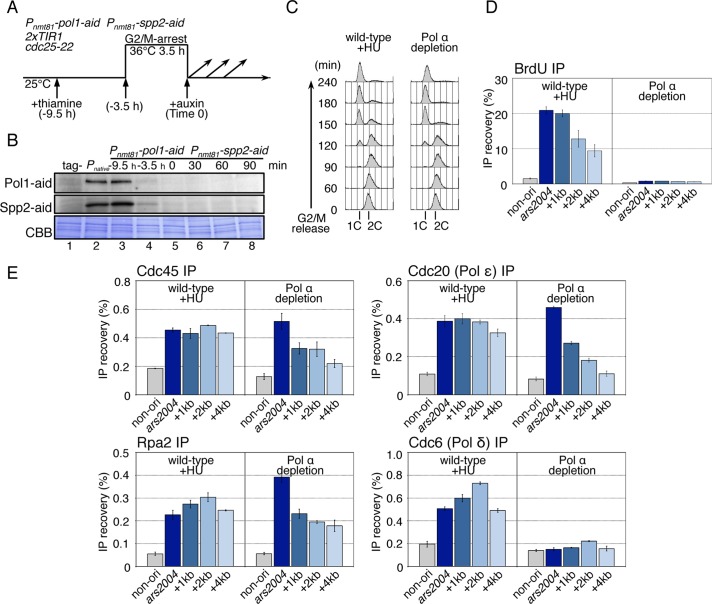
We next investigated whether Pol ε, among the replicative DNA polymerases, is specifically required for the progression of CMG helicase. To this end, Pol α-primase was conditionally depleted using an off-aid system, in which transcriptional repression was combined with a protein degradation system, the AID (Nishimura et al., 2009; Kanke et al., 2011). The pol1+ and spp2+ genes, which encode the polymerase-catalytic subunit and the large subunit of primase, respectively, were replaced with the aid-tagged genes under control of the thiamine-repressible nmt81 promoter (Pnmt81). Pnmt81-pol1-aid Pnmt81-spp2-aid (Pol α depletion) and pol1+ spp2+ (wild-type) cells cultured with thiamine to shut off the transcription were arrested at the G2/M boundary using the temperature-sensitive cdc25-22 mutation (Russell and Nurse, 1986), and auxin was added upon release into the synchronous cell cycle (Figure 3A). Under the conditions of off-aid, the Pol1-aid and Spp2-aid proteins were efficiently decreased to a level <5% of the wild-type amount (Figure 3B and unpublished observations), and cells containing 1C DNA accumulated, as observed in HU-treated wild-type cells, indicating a defect in the early stage of DNA replication (Figure 3C). For more detailed analysis of DNA synthesis, BrdU-labeled DNA was immunoprecipitated from Pol α–depleted cells and HU-treated wild-type cells, and analyzed by qPCR. In HU-arrested wild-type cells, BrdU was efficiently incorporated at both the ars2004 origin and the 1–4 kb regions, but not in the nonorigin region (Figure 3D, wild-type +HU). In Pol α–depleted cells, BrdU incorporation at the origin was greatly decreased (Figure 3D, Pol α depletion), showing that DNA was barely synthesized even at the early origin.
FIGURE 3:
CMG helicase and Pol ε can migrate from the origin in the absence of Pol α and DNA synthesis. (A) The experimental scheme for Pol α depletion is shown. HM3021 2xTIR1 cdc25-22 (wild-type) or the HM4011 Pnmt81-pol1-aid Pnmt81-spp2-aid 2xTIR cdc25-22 (Pol α depletion) strain grown at 25°C were cultured in the presence of thiamine (10 μg/ml) for 6 h and arrested at G2/M-phase by incubation at 36°C for 3.5 h, and then released at 25°C (Time 0). Auxin (0.5 mM) was added just before release. (B) The Pol1-aid and Spp2-aid proteins in WCE were analyzed by immunoblotting using anti-IAA17 antibody. Tag- (lane 1) and Pnative (lane 2) indicate the samples of the untagged strain (HM3021) and the strain for Pol1-aid and Spp2-aid, expressed from the native promoter (HM4031, pol1-aid spp2-aid 2xTIR1 cdc25-22) grown at 25°C, respectively. The samples of Pol α depletion (HM4011) were prepared at –9.5 h (lane 3), –3.5 h (lane 4), 0 (lane 5), 30 (lane 6), 60 (lane 7), and 90 min (lane 8) after release from G2/M. CBB represents total proteins bound on the membrane stained with Coomassie brilliant blue. (C) Wild-type (HM3696 Padh1-TK Padh1-hENT 2xTIR1 cdc25-22) and Pol α depletion (HM4111 Pnmt81-pol1-aid Pnmt81-spp2-aid Padh1-TK Padh1-hENT 2xTIR1 cdc25-22) cells were synchronously released from the G2/M boundary in the presence of BrdU (200 mM), with the addition of HU (10 mM) in the case of wild type. Aliquots taken at the indicated time points were analyzed by flow cytometry. Positions of 1C and 2C DNA contents are shown. (D) Samples taken at 90 min after G2/M release in (C) were analyzed by BrdU-IP assay as described in the legend to Figure 2B. The reproducible results obtained in biologically independent experiments are shown in Supplemental Figure S3A. (E) Wild-type and Pol α-depletion derivatives carrying flag-cdc45, cdc20-flag, or cdc6-flag were analyzed by ChIP assay at 80 min after G2/M release. In case of the wild type, HU was added at 10 mM upon G2/M release. ChIP DNAs with anti-FLAG or anti-Rpa2 were analyzed as described in the legend to Figure 2C. The reproducible results obtained in biologically independent experiments are shown in Figure 4C and Supplemental Figure S3B.
To examine whether the components of the replisome migrate from the origin in the absence of Pol α, we performed ChIP assays for Cdc45, Rpa2, Cdc20 (Pol ε), and Cdc6 (Pol δ). In HU-treated wild-type cells, all of Cdc45, Rpa2, Cdc20, and Cdc6 were localized at the ars2004 origin and in the region 1–4 kb from the origin at 80 min after G2/M release (Figure 3E, wild-type +HU). In the Pol α–depleted cells, Cdc45, Rpa2, and Cdc20 (Pol ε) were enriched at the origin and also localized in the 1–4 kb region with a gradual decrease in distal regions (Figure 3E, Pol α depletion). These results indicated that Pol α is not necessary for migration of the CMG helicase and Pol ε several kb from the origin. Gradual reduction in the localization of CMG components in distal regions suggests that the absence of DNA synthesis may reduce the rate of progression of the CMG helicase. Cdc6 (Pol δ) was not localized significantly in any region in Pol α–depleted cells (Figure 3E, Cdc6 (Pol δ) IP), suggesting that recruitment of Pol δ to replication forks requires primer synthesis by Pol α. Taken together with the results obtained for the cdc20-ct1 mutant, it is likely that progression of the CMG complex requires Pol ε but not other DNA polymerases or DNA synthesis, and that there might be some link between DNA synthesis and the rate of helicase unwinding.
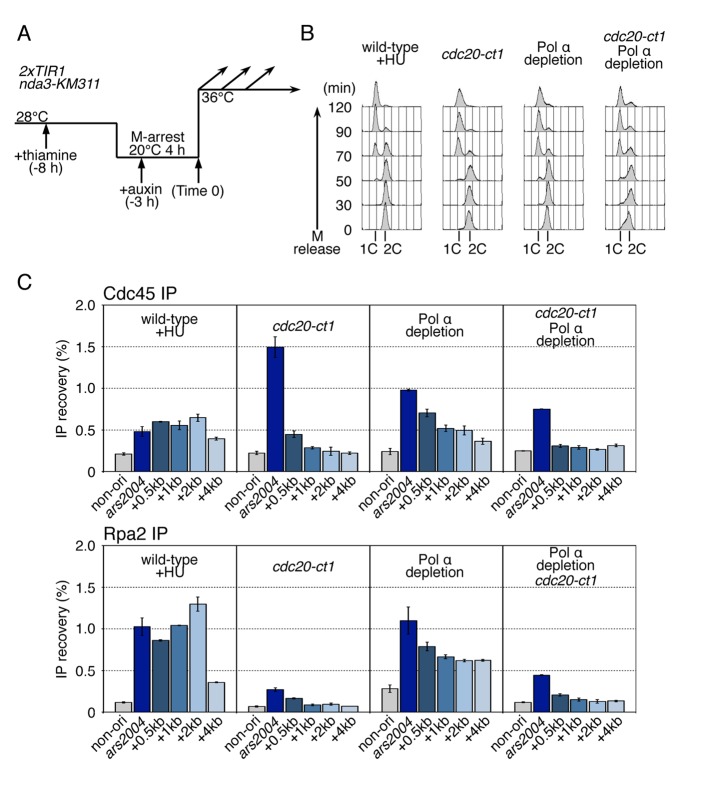
Next we investigated whether migration of CMG in the absence of Pol α depends on the function of Cdc20 CTD by introducing the cdc20-ct1 mutation under conditions of Pol α depletion (Figure 4). In cdc20-ct1 Pol α–depleted cells, Cdc45 and Rpa2 were preferentially localized at the origin, and localization in the 1–4 kb region was greatly reduced (Figure 4C). These results indicate that progression of CMG helicase requires the function of Cdc20 CTD.
FIGURE 4:
Cdc20 CTD is required for efficient progression of CMG helicase under Pol α depletion. (A) The experimental scheme for Pol α depletion used for the experiments in this figure is shown. HM3914 flag-cdc45 2xTIR1 nda3-KM311 (wild-type), HM3910 cdc20-ct1 flag-cdc45 2xTIR1 nda3-KM311 (cdc20-ct1), HM4017 Pnmt81-pol1-aid Pnmt81-spp2-aid flag-cdc45 2xTIR1 nda3-KM311 (Pol α depletion), and HM4095 cdc20-ct1 Pnmt81-pol1-aid Pnmt81-spp2-aid flag-cdc45 2xTIR nda3-KM311 (cdc20-ct1 Pol α depletion) strains grown at 28°C were cultured in the presence of thiamine (10 μg/ml) for 4 h and arrested at M-phase by incubation at 20°C for 4 h, and then released at 36°C (Time 0). Auxin (0.5 mM) was added 3 h before release. HU (15 mM) was added upon release in the case of the wild type. (B) Aliquots taken at the indicated time points were analyzed by flow cytometry. Positions of 1C and 2C DNA contents are shown. (C) Samples taken at 50 min after M release were analyzed by ChIP assays. Chromatin-immunoprecipitated DNAs with anti-FLAG or anti-Rpa2 were analyzed by qPCR using primers for ars2004, the regions located 0.5 kb, 1 kb, 2 kb, and 4 kb from ars2004, denoted as “+0.5kb,” “+1kb,” “+2kb,” or “+4kb,” and the nonorigin region. Mean ± SD obtained from multiple measurements in qPCR is presented.
Complex formation and origin association of Cdc20 and Dpb2 are impaired by the cdc20-ct1 mutation
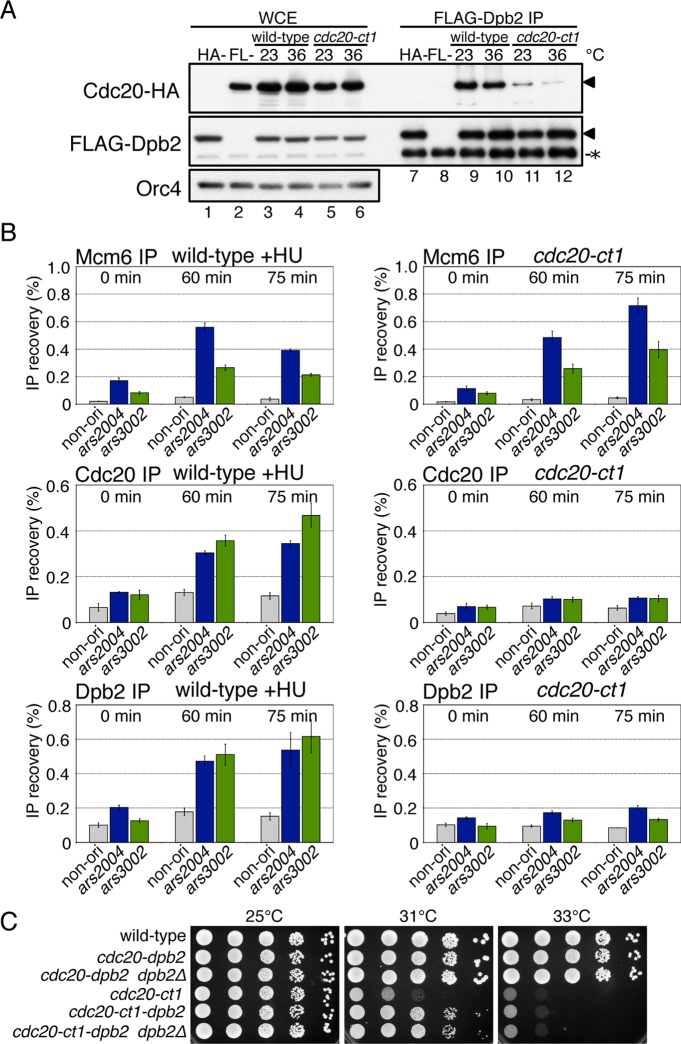
Pol2/Cdc20/p261 and Dpb2/p59, the second largest subunit of Pol ε, are both essential for DNA replication in budding yeast, fission yeast, and Xenopus egg extracts (Morrison et al., 1990; Araki et al., 1991a; D'Urso and Nurse, 1997; Feng et al., 2003; Shikata et al., 2006). It has been reported that Pol2/p261 interacts directly with Dpb2/p59 through the conserved zinc finger motifs (metal binding motifs, Figure 1A) at the C-terminal end (Dua et al., 1999; Shikata et al., 2006; Bermudez et al., 2011). A recent study has indicated that one of the postulated zinc finger motifs required for formation of Pol ε complex is an iron-sulfur cluster (Netz et al., 2012). To examine whether the cdc20-ct1 mutation affects the interaction of Cdc20 with Dpb2, we examined the complex formation by Cdc20 and Dpb2, using hemagglutinin (HA)-tagged cdc20-ct1 and FLAG-tagged dpb2. The results of immunoblotting of the total cellular proteins showed that the mutation did not significantly affect the amounts of Cdc20 and Dpb2 at both 23°C or 36°C (Figure 5A, lanes 3–6). By immunoprecipitation of FLAG-Dpb2 in the wild type, Cdc20 was coimmunoprecipitated at both temperatures, indicating that Cdc20 and Dpb2 form a complex (Figure 5A, lanes 9 and 10). In contrast, in cdc20-ct1, the amount of Cdc20 protein coimmunoprecipitated with Dpb2 at 23°C was smaller than that in the wild type, and was further decreased at 36°C, showing that the interaction with Dpb2 is impaired by the cdc20-ct1 mutation (Figure 5A, lanes 11 and 12).
FIGURE 5:
Complex formation and origin association of Cdc20 and Dpb2 are impaired by the cdc20-ct1 mutation. (A) HM1770 cdc20-HA dpb2-flag (wild-type, lanes 3, 4, 9, and 10) and HM4217 cdc20-ct1-HA dpb2-flag (cdc20-ct1, lanes 5, 6, 11, and 12) were grown at 23°C, and half of each of the cell types were shifted to 36°C for 2 h. Proteins in WCE (lanes 1–6) and double the corresponding amounts of immunoprecipitates obtained with anti-FLAG (FLAG-Dpb2 IP, lanes 7–12) were analyzed by immunoblotting with anti-HA, anti-FLAG, or anti-Orc4 antibodies. HA- (lanes 1 and 7) and FL- (lanes 2 and 8) indicate the samples of the Cdc20-untagged strain (HM3445 flag-dpb2) and the Dpb2-untagged strain (HM1675 cdc20-HA), respectively, grown at 23°C. Positions of Cdc20-HA and FLAG-Dpb2 are indicated by triangles (◂), and nonspecific protein bands are indicated by an asterisk (*). (B) Aliquots of nda3-KM311 (wild-type) and cdc20-ct1 nda3-KM311 (cdc20-ct1), derivatives carrying cdc20-flag or flag-dpb2, were synchronously released from M-phase and subjected to ChIP analysis at the indicated time points as described in Figure 2A. The reproducible results obtained in biologically independent experiments are shown in Supplemental Figure S4. (C) Wild-type, cdc20-dpb2 (HM4417), cdc20-dpb2 dpb2Δ (HM4439), cdc20-ct1 (HM1317), cdc20-ct1-dpb2 (HM4421), and cdc20-ct1-dpb2 dpb2Δ (HM4441) cells in log phase were serially diluted tenfold, spotted onto YE plates, and incubated at the indicated temperatures.
To examine whether stable complex formation is required for the origin association of Cdc20 and Dpb2, respectively, we performed ChIP assays in wild-type and cdc20-ct1 cells carrying cdc20-flag or flag-dpb2. In HU-treated wild-type cells, Mcm6, Cdc20, and Dpb2 were localized at ars2004 and ars3002 at 60–75 min (Figure 5B, left). In contrast, the localization of Cdc20-ct1 protein and Dpb2 was greatly decreased in cdc20-ct1, whereas Mcm6 was associated with origins (Figure 5B, right), suggesting that a complex of Cdc20 and Dpb2 is required for stable association with the origins.
If the essential function of the Cdc20 CTD, which is impaired by the cdc20-ct1 mutation, is to interact with Dpb2, tethering of the Cdc20-ct1 with Dpb2 would suppress the defect in cdc20-ct1. To test this possibility, we constructed cells expressing Cdc20-Dpb2 or Cdc20-ct1-Dpb2 fusion proteins at the native cdc20+ locus. In the presence of cdc20-dpb2, it was possible to delete the dpb2+ gene without causing any defect in its growth, indicating that the fused protein is functional as Cdc20 and Dpb2 (Figure 5C). When Cdc20-ct1-Dpb2 was expressed, the temperature sensitivity was partly suppressed (Figure 5C). These results suggest that the interaction of Cdc20 CTD with Dpb2 is required for the essential function of Cdc20 CTD. Because the growth recovery was partial, it is possible that the cdc20-ct1 mutation impaired some other essential functions of the CTD.
Pol ε is required for loading of CMG components onto replication origins
We showed that Pol ε is required for the progression of CMG helicase after the initiation of DNA replication. However, it is possible that the cdc20 temperature-sensitive mutants might impair some specific function of Pol ε and that Pol ε has another essential role in the initiation process, as has been shown in budding yeast (Masumoto et al., 2000; Muramatsu et al., 2010). To examine this possibility, we depleted Cdc20 using the off-aid system as described earlier in text. Under the off-aid conditions, Cdc20-aid protein was decreased to a level <10% of the wild-type amount at 30 min after addition of auxin (Figure 6A and unpublished observations). In flow cytometry analysis of wild-type cells without depletion, a 4C DNA peak was generated within 80–120 min after G2/M release (Figure 6B, left), indicative of DNA replication in S-phase, because the cytokinesis of fission yeast occurs in late S-phase under standard laboratory conditions (Knutsen et al., 2011). In contrast, upon Cdc20 depletion, cells with 1C DNA accumulated, indicating a defect in the early stage of DNA replication (Figure 6B, right). Interestingly, BrdU was not incorporated even at the origins under Cdc20 depletion (Figure 6C). These results are different from those obtained using cdc20-ct1 (Figure 2B), and suggest that Cdc20 depletion causes a defect in an initiation process of DNA replication.
FIGURE 6:
Pol ε is required for the loading of CMG components onto replication origins. Cdc20 proteins were depleted, similarly to the procedures used for Pol α depletion described in Figure 3A, and thiamine was added 12 h before G2/M arrest (–15.5 h). (A) Cdc20-aid proteins in WCE prepared at the indicated time points were analyzed by immunoblotting with anti-IAA17 antibody. Tag- (lane 1) and Pnative (lane 2) indicate the samples of the Cdc20-untagged strain (HM3021) and the strain for Cdc20-aid expressed from the native promoter (HM3503, cdc20-aid 2xTIR1 cdc25-22) grown at 25°C, respectively. Position of Cdc20-aid is shown by a triangle (◂), and nonspecific protein bands are indicated by an asterisk (*). CBB represents total proteins bound on the membrane stained with Coomassie brilliant blue. (B) DNA contents of the wild-type cells without HU (left) and Cdc20-depleted cells (right) were analyzed by flow cytometry. Positions of 1C, 2C, and 4C DNA contents are shown. (C) Wild-type (HM3696) and Cdc20-depletion (HM4129 Pnmt81-cdc20-aid Padh1-TK Padh1-hENT 2xTIR1 cdc25-22) cells were synchronously released from the G2/M boundary in the presence of BrdU (200 mM), with the addition of HU (10 mM) in the case of wild type. Aliquots taken at 90 min after G2/M release were analyzed by the BrdU-IP assay as described in Figure 3D and quantified using primers for ars2004, ars3002, and the nonorigin region. The similar results obtained from an independent culture are presented in Supplemental Figure S3A. (D) Aliquots of HM3452 psf2-flag cdc45-myc 2xTIR1 cdc25-22 (wild-type) and HM3450 Pnmt81-cdc20-aid psf2-flag cdc45-myc 2xTIR cdc25-22 (Cdc20 depletion) cultures taken at the indicated time points were analyzed using ChIP assays with anti-Mcm6, anti-FLAG, or anti-myc. In case of wild type, HU was added at 10 mM upon G2/M release. Mean ± SD obtained from multiple measurements in qPCR is presented. The reproducible results obtained in biologically independent experiments are shown in Supplemental Figure S5A. (E) Wild-type and Cdc20-depletion derivatives carrying sld3-flag cut5-myc or drc1-flag were analyzed by ChIP assay, as described in (D), except HU-free wild-type cells were used as a control. The reproducible results are shown in Supplemental Figure S5B.
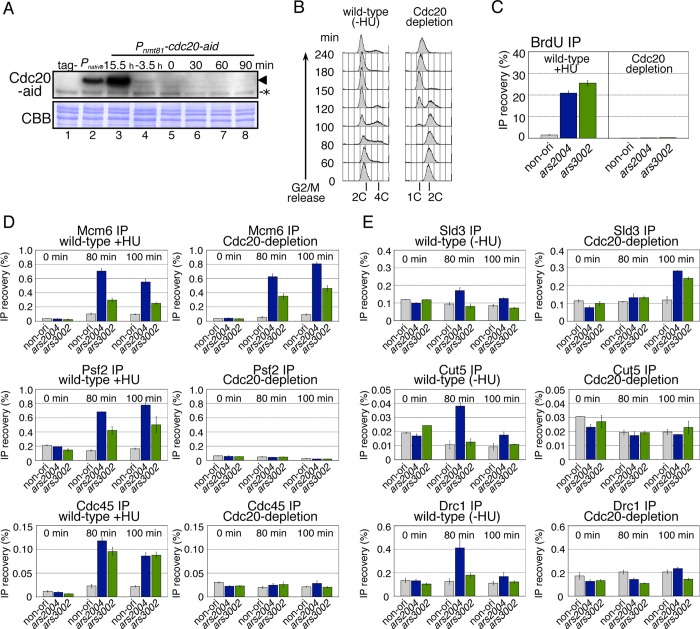
To clarify the reaction in the initiation process that requires Cdc20, we examined whether replisome components assembled at replication origins in Cdc20-depleted cells using the ChIP assay. We first carried out ChIP assays for Mcm6, Psf2 (GINS), and Cdc45 to examine assembly of the CMG complex. In HU-treated wild-type cells, Mcm6, Psf2, and Cdc45 were preferentially enriched at the origins relative to the nonorigin region at 80–100 min after release from the G2/M block, indicating the localization of CMG components at the origins (Figure 6D, left). In contrast, in Cdc20-depleted cells, origin association of Psf2 or Cdc45 was abolished, although Mcm6 was accumulated at the origins (Figure 6D, right). These results indicate that Cdc20 is required for loading of GINS and Cdc45 onto the origins.
We next investigated whether Sld3, Cut5, and Drc1, which are required for loading of GINS and Cdc45 (Takayama et al., 2003; Yabuuchi et al., 2006; Fukuura et al., 2011; Heller et al., 2011), bound to the origins in the absence of Cdc20. In wild-type cells without HU treatment to avoid binding of Cut5 to stalled replication forks, Sld3, Cut5, and Drc1 were transiently localized at ars2004 at 80 min (Figure 6E, left). We were unable to detect significant localization of these factors at ars3002, where replication initiates less efficiently than ars2004. In Cdc20-depleted cells, Sld3 was accumulated at ars2004 and ars3002 at 100 min, the timing being slightly later than that in wild-type cells (Figure 6E, right). However, specific binding of Cut5 and Drc1 was not observed at these origins (Figure 6E, right), indicating that Cdc20 is required for loading of Cut5 and Drc1 onto the origins. These results are consistent with previous observations that Sld3 is loaded onto origins at a distinct step before loading of GINS and Cut5 (Yabuuchi et al., 2006).
We examined whether Dpb2, which forms a complex with Cdc20, is also required for replisome assembly in fission yeast (Supplemental Figure S6). Under the off-aid conditions for Dpb2, localizations of Cut5, Drc1, Psf2, and Cdc45 at the origins were greatly decreased, whereas Mcm6 and Sld3 were localized there (Supplemental Figure S6C). Although the amount of Dpb2 or Cdc20 did not significantly decrease in Cdc20-depleted or Dpb2-depleted cells, respectively (Supplemental Figure S7, A and D), their localizations at the origins were abolished (Supplemental Figure S7, C and F), indicating that Cdc20 and Dpb2 depend on each other in origin binding. These results indicate that Dpb2 as well as Cdc20 play an essential role in the assembly of replisome components during the initiation process of DNA replication.
Pol ε is required for formation of complexes containing GINS and other replication factors
The results of protein depletion experiments showed that Cdc20 (Pol ε) is required for loading of GINS, Cut5, Drc1, and Cdc45 onto replication origins (Figure 6, D and E). GINS and Dpb11/Cut5 associate with origins in a mutually dependent manner (Takayama et al., 2003; Yabuuchi et al., 2006). To examine the role of Pol ε (Cdc20) in the formation of complexes containing GINS and other replication factors, we precipitated FLAG-tagged Psf2, a subunit of GINS, from cells with or without Cdc20 depletion. Cells were treated with formaldehyde to induce cross-linking, allowing detection of any fragile complex using antibodies against various replication proteins (Figure 7A). The strain carrying untagged Psf2 (tag-) was used as a control. The results of immunoblotting of whole-cell extracts (WCE) showed that the amounts of proteins tested did not change significantly in Cdc20-depleted cells (Figure 7A, lanes 1–6). We were unable to detect Cdc20-aid protein with anti-IAA17 antibody after formaldehyde treatment (unpublished observation). On immunoprecipitation of Psf2-FLAG, the other components of the GINS complex, Sld5, Psf1, and Psf3, were coimmunoprecipitated under all the conditions used, except in the untagged strain (Figure 7A, lanes 7–12), indicating that the GINS complex forms independently of Cdc20. In cells without depletion, Cut5 was coimmunoprecipitated at both the G2/M boundary (0 min) and S-phase (100 min), showing that GINS forms a complex with Cut5 even at the G2/M boundary when GINS does not associate with origins (Figure 7A, lanes 9 and 10). In contrast, Cdc45 and Mcm6 were coimmunoprecipitated specifically in S-phase (Figure 7A, lanes 9 and 10), indicating that the CMG complex is assembled in S-phase. In Cdc20-depleted cells, the amount of Cut5 coimmunoprecipitated with Psf2-FLAG was significantly decreased at both G2/M and S-phase (Figure 7A, lanes 11 and 12), showing that Cdc20 is required for GINS to form a complex with Cut5. Moreover, Cdc45 and Mcm6, which coimmunoprecipitated with Psf2-FLAG in S-phase, were greatly decreased under Cdc20 depletion (Figure 7A, lanes 11 and 12). These results indicate that Cdc20 (Pol ε) is required for GINS to form complexes with Cdc45 and Mcm2-7. We next investigated whether Pol ε interacts with GINS, Cut5, and Cdc45 by immunoprecipitating FLAG-tagged Dpb2 (Figure 7B). In cells without depletion, Psf1 (GINS) and Cut5 were detected in FLAG-Dpb2 immunoprecipitates at the G2/M boundary and were increased at S-phase (Figure 7B, lanes 9 and 10). Cdc45 was coimmunoprecipitated with FLAG-Dpb2 preferentially in S-phase (Figure 7B, lanes 9 and 10). These results show that Pol ε forms complexes with GINS and Cut5 at the G2/M boundary and then stably associates with the CMG complexes in S-phase. In contrast, in Cdc20-depleted cells, none of Psf1, Cut5, or Cdc45 was coimmunoprecipitated with FLAG-Dpb2 at the G2/M boundary or S-phase (Figure 7B, lanes 11 and 12). These results indicate that Cdc20 is required for formation of complexes containing GINS, Cut5, and Dpb2 at the G2/M boundary and those additionally containing Mcm2-7 and Cdc45 in S-phase.
FIGURE 7:
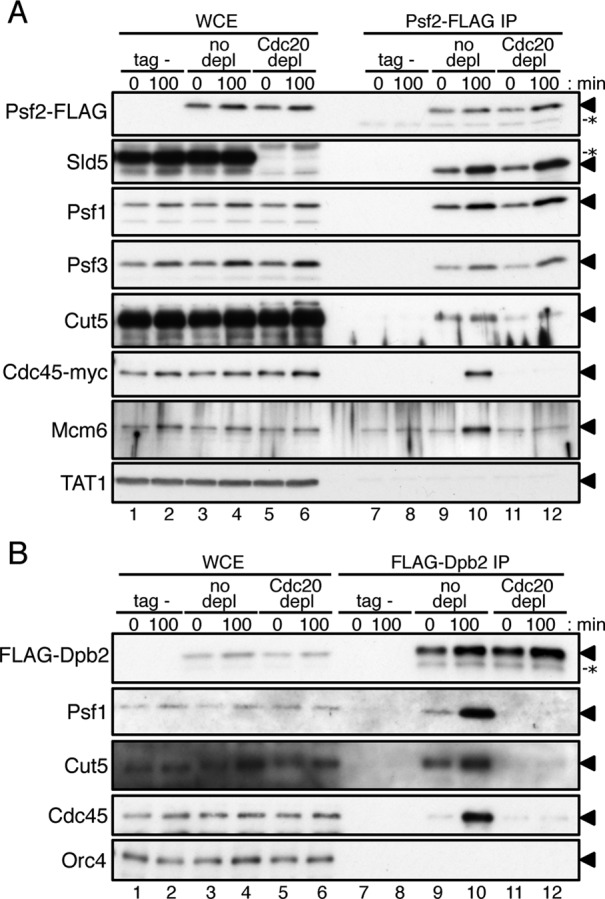
Pol ε is required for GINS to form complexes with Cut5, Cdc45, and Mcm6. Cdc20 protein was depleted as described in Figure 6. (A) Aliquots of HM3454 Pnmt81-cdc20-aid cdc45-myc 2xTIR1 cdc25-22 (tag-) and HM3450 Pnmt81-cdc20-aid psf2-flag cdc45-myc 2xTIR cdc25-22 without or with depletion (no depletion or Cdc20 depletion) were collected at 0 min and 100 min without addition of HU after G2/M release (see Figure 6) and treated with 1% formaldehyde. Proteins in WCE (lanes 1–6) and immunoprecipitates obtained with anti-FLAG (Psf2-FLAG IP, lanes 7–12) were analyzed by immunoblotting with anti-FLAG, anti-Sld5, anti-Psf1, anti-Psf3, anti-Cut5, anti-myc (Cdc45-myc), anti-Mcm6, or anti-tubulin antibodies. Triangles (◂) on the right of the panels indicate positions of the relevant proteins. Nonspecific protein bands are indicated by asterisks (*). It is noteworthy that thiamine-repressible proteins in WCE (lanes 1–4 in Sld5 panel) cross-reacted with the anti-Sld5 antibody (unpublished observation). The samples used for WCE (lanes 1–6) corresponded to 20% (Psf2-FLAG, Sld5, Psf1, Psf3, and tubulin), 1% (Cut5, Cdc45-myc), and 0.1% (Mcm6) of the proteins used for Psf2-FLAG IP. (B) Aliquots of HM4661 Pnmt81-cdc20-aid drc1-HA 2xTIR1 cdc25-22 (tag-) and HM4664 Pnmt81-cdc20-aid flag-dpb2 drc1-HA 2xTIR cdc25-22 without or with depletion (no depletion or Cdc20 depletion) at 0 min and 100 min after G2/M release were analyzed as in (A). Proteins in WCE (lanes 1–6) and immunoprecipitates obtained with anti-FLAG (FLAG-Dpb2 IP, lanes 7–12) were analyzed by immunoblotting with anti-FLAG, anti-Psf1, anti-Cut5, anti-Cdc45, or anti-Orc4 antibodies. Positions of the relevant proteins and a nonspecific protein are indicated by triangles (◂) and an asterisk (*), respectively. The samples used for WCE (lanes 1–6) corresponded to 20% (FLAG-Dpb2, Orc4) and 1% (Psf1, Cut5, and Cdc45) of the proteins used for FLAG-Dpb2 IP.
DISCUSSION
It is widely accepted that DNA Pol ε is primarily responsible for synthesis of the leading strand at the replication fork in eukaryotes. Nonetheless, it remains paradoxical that the DNA polymerase domain of the catalytic subunit, Pol2 and Cdc20 in budding and fission yeasts, respectively, is dispensable for viability. In budding yeast, Pol ε has an essential role before initiation of replication, as it is required for the loading of the initiation factors, GINS, Dpb11, and Sld2 onto replication origins. However, it remains unknown whether this is the only essential role of Pol ε. In this study using the off-aid system, we demonstrated that the catalytic Cdc20 subunit and the second largest Dpb2 subunit are essential for assembly of the CMG complex at the origin in fission yeast. Moreover, using a temperature-sensitive allele of cdc20 bearing mutations in the CTD, we showed that the CTD of Cdc20 is required for progression of the CMG helicase. This function seems to be specific to Pol ε and distinct from catalytic DNA polymerization activity. These findings have shed light on a functional link between polymerase and helicase in the replisome.
The role of Pol ε in replisome assembly
The results of protein depletion experiments show that Cdc20 and Dpb2 of the Pol ε complex are required for loading of GINS, Cut5, Drc1, and Cdc45 onto replication origins (Figure 6 and Supplemental Figures S5 and S6). Moreover, origin loading of Pol ε is dependent on GINS (Yabuuchi et al., 2006; Pai et al., 2009) and Cut5/Dpb11 (Masumoto et al., 2000), which are recruited to the origin, together with Drc1/Sld2, in a mutually dependent manner (Masumoto et al., 2002; Takayama et al., 2003; Yabuuchi et al., 2006; Fukuura et al., 2011; Heller et al., 2011). Consequently, Pol ε, GINS, Cut5, and Drc1 are mutually dependent regarding association with origins. The results of coimmunoprecipitation after cross-linking showed that Dpb2 formed complexes with GINS and Cut5 even at the G2/M boundary (Figure 7B), when none of them associated with replication origins (Figure 6 and Yabuuchi et al., 2006). Interestingly, complex formation of GINS with Cut5 is dependent on Cdc20 (Figure 7A). These results suggest that a ternary complex containing Pol ε, GINS, and Cut5 is formed, although it might be fragile, at the G2/M boundary. Consistently, a recent study has demonstrated that Pol ε, GINS, Dpb11, and Sld2 form a fragile complex (the pre-LC) in a CDK-dependent but pre-RC–independent manner in budding yeast (Muramatsu et al., 2010). The results of the two-hybrid analysis suggest that Cdc20 interacts with Sld5 and Psf3 of GINS and that Dpb2 interacts with Cut5, Drc1, and all subunits of GINS (Supplemental Figure S8). These interactions might be involved in the formation of the fragile complex and in its recruitment to replication origins. It is plausible to consider that such a complex is required specifically for initiation of replication, because neither Cut5/Dpb11 nor Drc1/Sld2 travels with replication forks after DNA synthesis initiates (Gambus et al., 2006; Kanemaki and Labib, 2006; Taylor et al., 2011). Recently an interesting model of a role of initiation specific factors has been proposed: Dpb11, Sld2, and Sld3 are less abundant in comparison with the number of replication origins, and they are involved in a temporal program of origin firing in budding yeast (Mantiero et al., 2011; Tanaka et al., 2011). Cut5/Dpb11, Sld3, and Drc1/Sld2 seem to dissociate from the origin after initiation and recycle for late replicating origins.
A noncatalytic role of Pol ε in the progression of CMG helicase after initiation
In contrast to the results obtained under Cdc20-depleted conditions, the cdc20-ct1 mutation did not result in a defect in the loading of CMG components onto replication origins. Moreover, RPA, Pol α, and Pol δ were localized at origins (Figure 2A) and initiated DNA synthesis. Interestingly, however, the replisome did not propagate extensively from the origin in the mutant, because Cdc45 did not translocate to the region 1 kb from the origin (Figure 2C). These results imply that Cdc20 CTD is required for progression of the CMG helicase from the origin, although we were unable to exclude the possibility that the CMG complex assembled in cdc20-ct1 would not be fully functional. The requirement of Cdc20 for elongation is supported by the fact that depletion of Cdc20 from HU-arrested cells caused severe inhibition of DNA replication after HU removal (Supplemental Figure S9). Because the Cdc20 CTD lacking the N-terminal domain is sufficient for progression of Cdc45 and Rpa2 from the origin (Supplemental Figure S2B), the function of Cdc20 CTD in CMG progression is independent of the DNA polymerase activity. Moreover, the progression of CMG helicase from the origin is independent of Pol α or DNA synthesis, as shown in Pol α–depleted cells (Figures 3 and 4). A recent in vitro study has shown that human Pol ε and the CMG complex act in a coordinated manner for synthesis of long DNA (Kang et al., 2012). This result supports our argument that Pol ε may stimulate the progression of the CMG helicase.
The defect of cdc20-ct1 seems to be at least partly due to its decreased interaction with Dpb2, because the temperature sensitivity of cdc20-ct1 was suppressed by tethering Cdc20 with Dpb2 (Figure 5C). Because Dpb2 was shown to interact with many components of CMG in two-hybrid analyses (Supplemental Figure S8C), some of these interactions may be required for stimulation of the helicase activity of the CMG complex. Furthermore, it is possible that cdc20-ct1 is defective in interactions with factors other than Dpb2, because tethering Dpb2 with Cdc20-ct1 did not completely suppress temperature-sensitive growth (Figure 5C). If this is the case, then the interaction between Cdc20 and Mcm7, which is decreased by the cdc20-ct1 mutation (Supplemental Figure S8A), may contribute to CMG progression.
The argument for a functional link between helicase and polymerase in the replisome can be extended further to organisms other than fission yeast. Structural and biochemical studies using various viral and prokaryotic replication systems have provided a detailed understanding of the mechanisms of replisome function (Langston et al., 2009; Patel et al., 2011). In the T4 phage replisome, the polymerase (gp43) together with the helicase (gp41) can carry out DNA synthesis at physiological rates, suggesting that a direct interaction between the polymerase and the helicase may be central to the progression of replisome (Dong et al., 1996). In the T7 phage replisome, dynamic interactions of the polymerase (gp5) with the helicase (gp4) ensure the highly progressive DNA unwinding and synthesis (Hamdan et al., 2007). In the Escherichia coli replisome, the τ subunit of the Pol III holoenzyme forms a bridge between Pol III polymerase subunits and the DnaB helicase, and stimulates the helicase activity of DnaB (Kim et al., 1996). The framework of this mechanism (i.e., how DNA polymerases ensure the highly progressive action of the helicase) may be conserved through viruses, bacteria, and eukaryotes.
Requirement of Pol ε from initiation through elongation of DNA replication
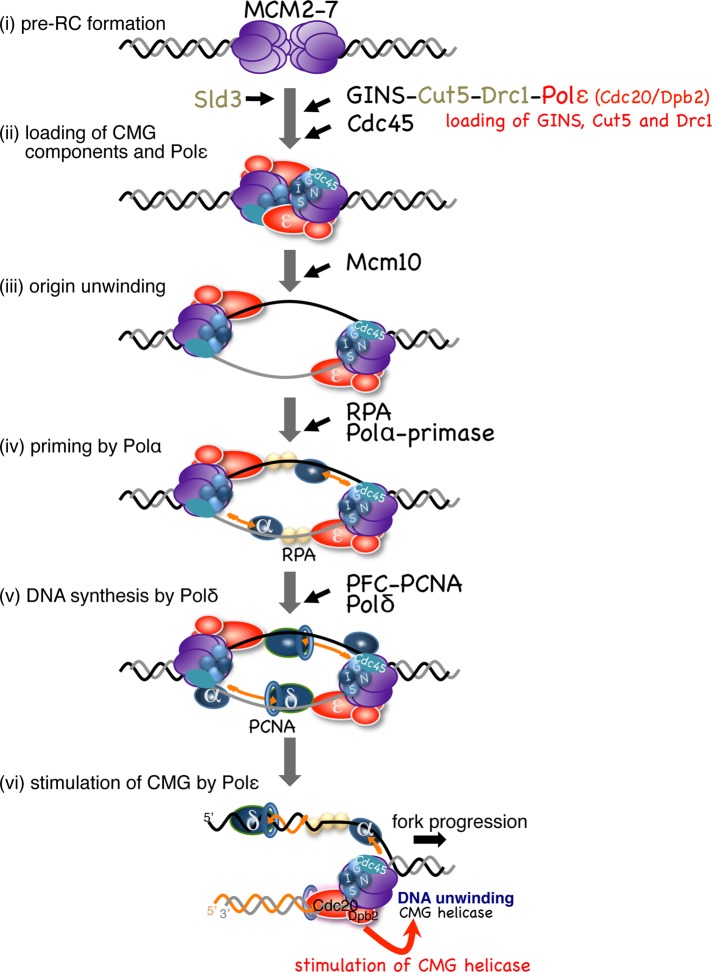
In the present study, we showed that Pol ε is required for both assembly and progression of the CMG helicase complex. It is suitable that the leading-strand polymerase Pol ε associates with the CMG helicase components during the initiation process, because both complexes translocate on the same leading-strand template at the replication fork. On the basis of observations described in this study, we propose a model of assembly and progression of the replisome, in which Pol ε plays two distinct roles (Figure 8). At the onset of S-phase, CMG components and Pol ε are loaded onto the origin in a mutually dependent manner (Figure 8, ii). Mcm10 plays an essential role in origin DNA unwinding after assembly of the CMG components (Kanke et al., 2012; van Deursen et al., 2012; Watase et al., 2012; Figure 8, iii). On activation of the helicase, Pol ε continues to associate with the CMG helicase that translocates on the leading-strand template. RPA and then Pol α are recruited to the origin and initiate DNA synthesis (Figure 8, iv). Pol δ binds to the primed DNA and extends it toward the translocating CMG and Pol ε (Figure 8, v). Pol ε that associates with the CMG helicase takes over to extend the leading strand (Figure 8, vi). In the absence of the N-terminal polymerase domain of the catalytic subunit of Pol ε, the CTD and Dpb2 are sufficient for the essential roles in the assembly and stimulation of the CMG helicase, and Pol δ could potentially substitute for synthesis of the leading strand. The tight link between Pol ε and the CMG is maintained throughout the process from initiation through elongation.
FIGURE 8:
Requirement of Pol ε from initiation through elongation of DNA replication. A model of assembly and progression of the replisome in fission yeast is presented. (i) In G1-phase, the double-hexameric Mcm2-7 is loaded onto the origin, forming pre-RC. (ii) At the onset of S-phase, Sld3 is loaded onto pre-RC, depending on DDK, and then the CMG components, GINS and Cdc45, are recruited with the aid of Sld3, Cut5, Drc1, and Pol ε. In this step, Pol ε binds to the origin in a manner mutually dependent on GINS, Cut5, and Drc1. (iii) After assembly of the CMG components and Pol ε, Mcm10 plays an essential role in origin DNA unwinding catalyzed by the CMG helicase (Kanke et al., 2012). On activation of the helicase, Pol ε continues to associate with the CMG helicase that translocates on the leading-strand template. (iv) RPA is loaded onto the single-stranded DNA, and then Pol α-primase initiates DNA synthesis. (v) Pol δ is recruited onto the primed-template DNA with the aid of RFC and PCNA (RFC is not shown for simplicity) to extend nascent DNA strands. (vi) When Pol δ collides with Pol ε, Pol ε takes over to extend the leading strand. Throughout steps (iii)–(vi), Cdc20 and Dpb2 may associate with the CMG helicase and stimulate its progression.
MATERIALS AND METHODS
Strains and media
Schizosaccharomyces pombe strains used in this study are listed in Supplemental Table S1. Details of strain construction are provided in the Supplemental Material. Strains were cultured in complete YE medium and minimal EMM medium (Moreno et al., 1991), all solid medium containing 2.0% agar.
Cell-cycle synchronization and flow cytometry
To synchronize the cell cycle, the temperature-sensitive cdc25-22 mutation was used for G2/M arrest (Russell and Nurse, 1986). Derivatives of cdc25-22 were incubated at 36°C for 3.5 h for the arrest, and then released at 25°C. To repress transcription, thiamine was added at a concentration of 10 μg/ml at the indicated time points before G2/M arrest. A synthetic auxin, NAA (Nacalai Tesque, Kyoto, Japan), was added at a concentration of 0.5 mM upon release from the G2/M boundary to induce protein degradation. For synchronization of the cell cycle of cdc20-ct1 derivatives, the cold-sensitive mutation nda3-KM311 was used for metaphase arrest (Hiraoka et al., 1984). nda3-KM311 derivatives grown at 28°C were incubated at 20°C for 4 h for the arrest, and then released at 36°C. For flow cytometry analysis, cells were fixed with 70% ethanol and incubated with propidium iodide at 0.5 μg/ml and RNase A at 50 μg/ml in 50 mM sodium citrate for 1 h at 37°C. Samples were then measured using a FACScan (Becton, Dickinson and Company, Franklin Lakes, NJ).
Preparation of cell extracts and immunoblotting
WCE were prepared as described previously (Kanke et al., 2011). Proteins in the extracts were separated by SDS–PAGE and transferred onto polyvinylidene difluoride membranes (Immobilon/Millipore, Billerica, MA). The membranes were incubated for 1 h at room temperature in PBS with Tween 20 (PBST) containing 5% skim milk and reacted in PBST containing 1% BSA overnight at 4°C with anti-IAA17 (1:1000, Nishimura et al., 2009), anti-Orc4 (1:1000, Takahashi and Masukata, 2001), anti-Sld5 (1:500, Yabuuchi et al., 2006), anti-Psf1 (1:1000, Yabuuchi et al., 2006), anti-Psf3 (1:500, Yabuuchi et al., 2006), anti-Cut5 (1:1000, Saka et al., 1994), anti-Mcm6 (1:2000, Ogawa et al., 1999) or anti-Cdc45 (1:1000, Nakajima and Masukata, 2002), anti-tubulin (1:2000; Woods et al., 1989), anti-FLAG (1:2000, M2 monoclonal; Sigma, St. Louis, MO), anti-HA (1:1000, 16B12), or anti-myc (1:1000, c-myc antibody-1 Clone 9E11) antibodies. Horseradish peroxidase–conjugated anti–mouse or anti–rabbit immunoglobulin G (IgG) was used as the secondary antibody (1:10,000; Jackson ImmunoResearch, West Grove, PA). Binding was visualized with West Pico or Femto Chemiluminescent Substrate (Pierce/Thermo Fisher Scientific, Rockford, IL) and an x-ray film development system (TCX-101; Konica Minolta, Tokyo, Japan).
ChIP assays
ChIP assays were performed as described previously (Hayashi et al., 2007) with some modifications. Immunoprecipitation was carried out with Dynal magnetic beads (Invitrogen, Carlsbad, CA) associated with mouse anti-FLAG (1:500, M2 monoclonal; Sigma), anti-myc (1:300, c-myc antibody-1 Clone 9E11), anti-Mcm6 (1:500, Ogawa et al., 1999) or anti-Rpa2/Ssb2 (1:500, Yabuuchi et al., 2006) antibodies, or with Protein A-Sepharose beads (GE Healthcare, Waukesha, WI) associated with anti-Pcn1 (1:200, a gift from T. Tsurimoto) or anti-Psf3 antibodies (1:200, Yabuuchi et al., 2006). The immunoprecipitated DNA and total DNA were quantitated by qPCR using SYBR green I in a 7300 Real-Time PCR System (Applied Biosystems, Carlsbad, CA). The primers used for qPCR are listed in Supplemental Table S2.
BrdU-labeling assay
BrdU incorporation assays were performed as described previously (Katou et al., 2003; Hayashi et al., 2007) with some modifications. In this study, cells expressing the herpes simplex virus thymidine kinase gene (TK+), expressed from Pnmt1 or Padh1, and the human equilibrative nucleoside transporter gene (hENT+) under control of Padh1 were used. S. pombe cells, incubated in the presence of 200 μM BrdU (Sigma), were fixed with 0.1% sodium azide, and the total cellular DNA was prepared as described previously (Hayashi et al., 2007). The cellular DNA was sonicated until the DNA had been sheared into fragments of ∼400 bp. DNA was denatured by boiling and used for immunoprecipitation with anti-BrdU antibody (500 ng; Becton Dickinson) associated with Dynal (Invitrogen) anti-mouse IgG magnetic beads. After rotation at 4°C for 4 h, the immunoprecipitated DNA was recovered and purified, similarly to the method used for ChIP except for reverse cross-linking.
Immunoprecipitation
Coimmunoprecipitation assays of Pol ε complex (Figure 5A) were carried out as described previously (Takahashi and Masukata, 2001) with some modifications. Cells (5 × 108) were suspended in 0.4 ml of HBNT buffer (HB buffer [ Moreno et al., 1991] supplemented with 50 mM NaCl and 0.1% Triton X-100) and disrupted with acid-washed glass beads using a Micro Smash (Tomy Seiko, Tokyo, Japan) seven times for 30 s each time. After the broken cells had been recovered by centrifugation at 5000 rpm for 1 min, 140 U of DNase I (TaKaRa, Shiba, Japan) was added, and incubation was conducted at 22°C for 15 min. The supernatant obtained by centrifugation at 15,000 rpm for 15 min was used for immunoprecipitation with agarose beads associated with mouse anti-FLAG M2 antibody (Sigma). After rotation at 4°C for 1 h, the immunoprecipitates were washed with HBNT buffer four times, and the proteins were eluted with SDS-sample buffer by incubation at 95°C for 5 min. A coimmunoprecipitation assay after cross-linking was carried out similarly to the method used for ChIP with some modification: cells (3 × 108) were suspended in 0.4 ml of ChIP lysis buffer (50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 0.1% Triton X-100, 0.1% sodium deoxycholate) supplemented with 2.5 mM phenylmethylsulfonyl fluoride and 0.5% proteinase inhibitor cocktail (Sigma) and disrupted with acid-washed glass beads using a Micro Smash (TOMY) five times for 30 s each time. After the broken cells had been recovered by centrifugation at 5000 rpm for 1 min, the samples were sonicated four times for 10 s each time and incubated with 140 U of DNase I (TaKaRa) for 30 min at 4°C. The supernatant obtained by centrifugation at 15,000 rpm for 15 min was used for immunoprecipitation with Dynal magnetic beads (Invitrogen) associated with mouse anti-FLAG (1:300, M2 monoclonal; Sigma). After rotation at 4°C for 2 h, the immunoprecipitates were washed with ChIP lysis buffer four times, and the proteins were eluted with SDS-sample buffer by incubation at 95°C for 10 min.
Supplementary Material
Acknowledgments
We thank Masayoshi Fukuura and Makoto Hayashi for critical reading of the manuscript. We also thank Mitsuhiro Yanagida, Toshiki Tsurimoto, and Gennaro D'Urso for providing anti-Cut5 antibody, anti-PCNA antibody, and flag-dpb2 strains, respectively. This study was supported by a Grant-in-Aid from the Ministry of Education, Science, Technology, Sports, and Culture, Japan (to H.M.) and by a Grant-in-Aid for JSPS Fellows (to T.H. and M.K.).
Abbreviations used:
- AID
Auxin-Inducible-Degron
- BrdU
5-bromo-2′-deoxyuridine
- CDK
cyclin-dependent kinase
- ChIP
chromatin immunoprecipitation
- CMG complex
Cdc45, Mcm2-7, and GINS (Go-Ichi-Nii-San) complex
- CTD
C-terminal domain
- DDK
Dbf4-dependent kinase
- HA
human influenza hemagglutinin
- HU
hydroxyurea
- IP
immunoprecipitation
- PBST
PBS with Tween 20
- Pol ε
polymerase epsilon
- pre-LC
preloading complex
- pre-RC
prereplicative complex
- qPCR
quantitative PCR
- WCE
whole-cell extracts
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-05-0339) on June 20, 2012.
REFERENCES
- Araki H. Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr Opin Cell Biol. 2010;22:766–771. doi: 10.1016/j.ceb.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Araki H, Hamatake RK, Johnston LH, Sugino A. DPB2, the gene encoding DNA polymerase II subunit B, is required for chromosome replication in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991a;88:4601–4605. doi: 10.1073/pnas.88.11.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H, Hamatake RK, Morrison A, Johnston LH, Sugino A. Cloning DPB3, the gene encoding the third subunit of DNA polymerase II of Saccharomyces cerevisiae. Nucleic Acids Res. 1991b;19:4867–4872. doi: 10.1093/nar/19.18.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asturias FJ, Cheung IK, Sabouri N, Sedelnikova O, Wepplo D, Johansson E. Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat Struct Biol. 2006;13:35–43. doi: 10.1038/nsmb1040. [DOI] [PubMed] [Google Scholar]
- Bell SD, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Bermudez VP, Farina A, Raghavan V, Tappin I, Hurwitz J. Studies on human DNA polymerase ε and GINS and their role in DNA replication. J Biol Chem. 2011;286:28963–28977. doi: 10.1074/jbc.M111.256289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Urso G, Nurse P. Schizosaccharomyces pombe cdc20+ encodes DNA polymerase epsilon and is required for chromosomal replication but not for the S phase checkpoint. Proc Natl Acad Sci USA. 1997;94:12491–12496. doi: 10.1073/pnas.94.23.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Dong F, Weitzel SE, von Hippel PH. A coupled complex of T4 DNA replication helicase (gp41) and polymerase (gp43) can perform rapid and processive DNA strand-displacement synthesis. Proc Natl Acad Sci USA. 1996;93:14456–14461. doi: 10.1073/pnas.93.25.14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol ε and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 1999;274:22283–22288. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- Feng W, D'Urso G. Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase epsilon are viable but require the DNA damage checkpoint control. Mol Cell Biol. 2001;21:4495–4504. doi: 10.1128/MCB.21.14.4495-4504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Rodriguez-Menocal L, Tolun G, D'Urso G. Schizosaccharomyces pombe Dpb2 binds to origin DNA early in S phase and is required for chromosomal DNA replication. Mol Biol Cell. 2003;14:3427–3436. doi: 10.1091/mbc.E03-02-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Schärer OD, Walter JC. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui T, Yamauchi K, Muroya T, Akiyama M, Maki H, Sugino A, Waga S. Distinct roles of DNA polymerases delta and epsilon at the replication fork in Xenopus egg extracts. Genes Cells. 2004;9:179–191. doi: 10.1111/j.1356-9597.2004.00716.x. [DOI] [PubMed] [Google Scholar]
- Fukuura M, Nagao K, Obuse C, Takahashi TS, Nakagawa T, Masukata H. CDK promotes interactions of Sld3 and Drc1 with Cut5 for initiation of DNA replication in fission yeast. Mol Biol Cell. 2011;22:2620–2633. doi: 10.1091/mbc.E10-12-0995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, Van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- Hamdan SM, Johnson DE, Tanner NA, Lee J-B, Qimron U, Tabor S, van Oijen AM, Richardson CC. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol Cell. 2007;27:539–549. doi: 10.1016/j.molcel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- Hayashi MT, Katou Y-M, Itoh T, Tazumi A, Yamada Y, Takahashi TS, Nakagawa T, Shirahige K, Masukata H. Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast. EMBO J. 2007;26:1327–1339. doi: 10.1038/sj.emboj.7601585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146:80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes β-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaki M, Labib K. Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J. 2006;25:1753–1763. doi: 10.1038/sj.emboj.7601063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YH, Galal WC, Farina A, Tappin I, Hurwitz J. Properties of the human Cdc45/Mcm2-7/GINS helicase complex and its action with DNA polymerase ε in rolling circle DNA synthesis. Proc Natl Acad Sci USA. 2012;109:6042–6047. doi: 10.1073/pnas.1203734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanke M, Kodama Y, Takahashi TS, Nakagawa T, Masukata H. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J. 2012;31:2182–2194. doi: 10.1038/emboj.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanke M, Nishimura K, Kanemaki M, Kakimoto T, Takahashi TS, Nakagawa T, Masukata H. Auxin-inducible protein depletion system in fission yeast. BMC Cell Biol. 2011;12:8. doi: 10.1186/1471-2121-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou Y-M, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- Kesti T, Flick K, Keränen S, Syväoja JE, Wittenberg C. DNA polymerase ε catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 1999;3:679–685. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- Kim S, Dallmann HG, McHenry CS, Marians KJ. Coupling of a replicative polymerase and helicase: a τ-DnaB interaction mediates rapid replication fork movement. Cell. 1996;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- Knutsen JH, Rein ID, Rothe C, Stokke T, Grallert B, Boye E. Cell-cycle analysis of fission yeast cells by flow cytometry. PLoS One. 2011;6:e17175. doi: 10.1371/journal.pone.0017175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010;24:1208–1219. doi: 10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston LD, Indiani C, O'Donnell M. Whither the replisome: emerging perspectives on the dynamic nature of the DNA replication machinery. Cell Cycle. 2009;8:2686–2691. doi: 10.4161/cc.8.17.9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiero D, Mackenzie A, Donaldson A, Zegerman P. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J. 2011;30:4805–4814. doi: 10.1038/emboj.2011.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Muramatsu S, Kamimura Y, Araki H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature. 2002;415:651–655. doi: 10.1038/nature713. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Sugino A, Araki H. Dpb11 controls the association between DNA polymerases α and ε and the autonomously replicating sequence region of budding yeast. Mol Cell Biol. 2000;20:2809–2817. doi: 10.1128/mcb.20.8.2809-2817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura S, Masuda-Sasa T, Matsui T, Takisawa H. Central role for Cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells. 2000;5:439–452. doi: 10.1046/j.1365-2443.2000.00340.x. [DOI] [PubMed] [Google Scholar]
- Miyabe I, Kunkel TA, Carr AM. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genetics. 2011;7:e1002407. doi: 10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Morrison A, Araki H, Clark AB, Hamatake RK, Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1990;62:1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu S, Hirai K, Tak Y-S, Kamimura Y, Araki H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol ε, and GINS in budding yeast. Genes Dev. 2010;24:602–612. doi: 10.1101/gad.1883410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima R, Masukata H. SpSld3 is required for loading and maintenance of SpCdc45 on chromatin in DNA replication in fission yeast. Mol Biol Cell. 2002;13:1462–1472. doi: 10.1091/mbc.02-01-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, Stodola JL, Lill R, Burgers PM, Pierik AJ. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol. 2012;8:125–132. doi: 10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PMJ, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Takahashi TS, Masukata H. Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol Cell Biol. 1999;19:7228–7236. doi: 10.1128/mcb.19.10.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya T, Maki S, Kawasaki Y, Sugino A. Structure and function of the fourth subunit (Dpb4p) of DNA polymerase ε in Saccharomyces cerevisiae. Nucleic Acids Res. 2000;28:3846–3852. doi: 10.1093/nar/28.20.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- Pai CC, García I, Wang SW, Cotterill S, MacNeill SA, Kearsey SE. GINS inactivation phenotypes reveal two pathways for chromatin association of replicative α and ε DNA polymerases in fission yeast. Mol Biol Cell. 2009;20:1213–1222. doi: 10.1091/mbc.E08-04-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, Pandey M, Nandakumar D. Dynamic coupling between the motors of DNA replication: hexameric helicase, DNA polymerase, and primase. Curr Opin Chem Biol. 2011;15:595–605. doi: 10.1016/j.cbpa.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell ZF, Isoz I, Lundstrom E-B, Johansson E, Kunkel TA. Yeast DNA polymerase ε participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell ZF, Kunkel TA. DNA polymerase epsilon: a polymerase of unusual size (and complexity) Prog Nucleic Acid Res Mol Biol. 2008;82:101–145. doi: 10.1016/S0079-6603(08)00004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Diffley JF. Eukaryotic DNA replication control: lock and load, then fire. Curr Opin Cell Biol. 2009;21:771–777. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Russell P, Nurse P. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell. 1986;45:145–153. doi: 10.1016/0092-8674(86)90546-5. [DOI] [PubMed] [Google Scholar]
- Saka Y, Fantes P, Sutani T, McInerny C, Creanor J, Yanagida M. Fission yeast cut5 links nuclear chromatin and M phase regulator in the replication checkpoint control. EMBO J. 1994;13:5319–5329. doi: 10.1002/j.1460-2075.1994.tb06866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata K, Masuda-Sasa T, Okuno Y, Waga S, Sugino A. The DNA polymerase activity of Pol ε holoenzyme is required for rapid and efficient chromosomal DNA replication in Xenopus egg extracts. BMC Biochem. 2006;7:21. doi: 10.1186/1471-2091-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiga M-G, D'Urso G. Identification and cloning of two putative subunits of DNA polymerase epsilon in fission yeast. Nucleic Acids Res. 2004;32:4945–4953. doi: 10.1093/nar/gkh811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyari O, Kawai M, Ida H, Yoshida H, Sakaguchi K, Yamaguchi M. Differential requirement for the N-terminal catalytic domain of the DNA polymerase ε p255 subunit in the mitotic cell cycle and the endocycle. Gene. 2012;495:104–114. doi: 10.1016/j.gene.2011.12.056. [DOI] [PubMed] [Google Scholar]
- Takahashi TS, Masukata H. Interaction of fission yeast ORC with essential adenine/thymine stretches in replication origins. Genes Cells. 2001;6:837–849. doi: 10.1046/j.1365-2443.2001.00468.x. [DOI] [PubMed] [Google Scholar]
- Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 2003;17:1153–1165. doi: 10.1101/gad.1065903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr Biol. 2011;21:2055–2063. doi: 10.1016/j.cub.2011.11.038. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Nasmyth K. Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J. 1998;17:5182–5191. doi: 10.1093/emboj/17.17.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Moore K, Murray J, Aves SJ, Price C. Mcm10 interacts with Rad4/Cut5(TopBP1) and its association with origins of DNA replication is dependent on Rad4/Cut5(TopBP1) DNA Repair (Amst) 2011;10:1154–1163. doi: 10.1016/j.dnarep.2011.09.001. [DOI] [PubMed] [Google Scholar]
- van Deursen F, Sengupta S, De Piccoli G, Sanchez-Diaz A, Labib K. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J. 2012;31:2195–2206. doi: 10.1038/emboj.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga S, Masuda T, Takisawa H. DNA polymerase ε is required for coordinated and efficient chromosomal DNA replication in Xenopus egg extracts. Proc Natl Acad Sci USA. 2001;98:4978–4983. doi: 10.1073/pnas.081088798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter JC, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase α. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Watase G, Takisawa H, Kanemaki MT. Mcm10 plays a role in functioning of the eukaryotic replicative DNA helicase, Cdc45-Mcm-GINS. Curr Biol. 2012;22:343–349. doi: 10.1016/j.cub.2012.01.023. [DOI] [PubMed] [Google Scholar]
- Woods A, Sherwin T, Sasse R, MacRae TH, Baines AJ, Gull K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J Cell Sci. 1989;93(Pt 3):491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- Yabuuchi H, Yamada Y, Uchida T, Sunathvanichkul T, Nakagawa T, Masukata H. Ordered assembly of Sld3, GINS and Cdc45 is distinctly regulated by DDK and CDK for activation of replication origins. EMBO J. 2006;25:4663–4674. doi: 10.1038/sj.emboj.7601347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardimci H, Loveland AB, Habuchi S, van Oijen AM, Walter JC. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol Cell. 2010;40:834–840. doi: 10.1016/j.molcel.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.