Abstract
LIF (leukaemia inhibitory factor) is a key cytokine for maintaining self-renewal and pluripotency of mESCs (mouse embryonic stem cells). Upon binding to the LIF receptor, LIF activates three major intracellular signalling pathways: the JAK (Janus kinase)/STAT3 (signal transducer and activator of transcription 3), PI3K (phosphoinositide 3-kinase)/AKT and SHP2 [SH2 (Src homology 2) domain-containing tyrosine phosphatase 2]/MAPK (mitogen-activated protein kinase) pathways. These pathways converge to orchestrate the gene expression pattern specific to mESCs. Among the many signalling events downstream of the LIF receptor, activation and DNA binding of the transcription factor STAT3 plays a central role in transducing LIF’s functions. The fundamental role of LIF for pluripotency was highlighted further by the discovery that LIF accelerates the conversion of epiblast-derived stem cells into a more fully pluripotent state. In the present review, we provide an overview of the three major LIF signalling pathways, the molecules that interact with STAT3 and the current interpretations of the roles of LIF in pluripotency.
Keywords: cell differentiation, embryonic stem cell (ESC), kinase inhibitor, leukaemia inhibitory factor (LIF), protein phosphorylation, signal transducer and activator of transcription 3 (STAT3)
INTRODUCTION
ESCs (embryonic stem cells) are characterized by three distinguishing features: pluripotency (the capability of differentiating into tissues derived from all three germ layers), self-renewal (maintenance of an undifferentiated state) and limitless proliferation (see [1-5] for reviews). All three of these features are maintained in part in mESCs (mouse ESCs) by the cytokine LIF (leukaemia inhibitory factor). LIF is routinely added to the culture medium of mESCs and the removal of LIF results in a rapid differentiation of mESCs. Because of this critical requirement, the intracellular signalling of LIF has been a main focus of investigators studying the regulatory mechanisms of self-renewal and pluripotency in mESCs. In contrast with mESCs, FGF (fibroblast growth factor) 2, also called basic FGF, and activin A or its related cytokines are used to maintain self-renewal in hESCs (human ESCs) [4,6]. Recent studies have suggested that hESCs exist in a slightly differentiated ‘primed state’ of pluripotency rather than the ‘näive state’ characteristic of mESCs and that LIF facilitates the acquisition of the näive state of pluripotency [7-9]. These findings reinforce the notion that LIF is critical to the regulation of self-renewal and pluripotency in ESCs and that defining its role in ESC biology is important for a full understanding of the ‘stemness’ of ESCs. In the present review, we will discuss the biochemistry of LIF signalling pathways and their roles in maintenance and acquisition of pluripotency.
HISTORICAL BACKGROUND
As several independent lines of research converged, it became clear that LIF was an essential cytokine for self-renewal and pluripotency of mESCs [10]. ESCs are derived from blastocysts cultured on top of feeder cells commonly prepared from mouse embryonic fibroblasts [11,12]. It was later discovered that mESCs retained self-renewal and pluripotency in the absence of feeder cells when conditioned medium prepared from BRL (Buffalo rat liver) cells was added to the culture dish [13]. The essential component of the conditioned medium was biochemically purified and named DIA (differentiation inhibiting activity) [14]. Bacterially expressed recombinant DIA could also be used in lieu of feeder cells to maintain mESCs. Around the same time a novel cytokine that inhibited proliferation and induced differentiation of mouse myeloid leukaemia cells was purified from the conditioned medium of Krebs II ascites cells and was named LIF [15-17]. Another research group isolated a cDNA clone encoding a novel human cytokine that promoted proliferation of the mouse leukaemia cell line DA-1a and termed it HILDA (human interleukin for DA cells) [18,19]. Eventually, investigators realized that all three proteins DIA, LIF and HILDA were identical [14,20,21]. LIF turned out to be a member of the well-characterized IL (interleukin)-6 cytokine family, and knowledge of IL-6 signalling pathways greatly facilitated investigations into the intracellular signalling pathways of LIF.
BIOCHEMISTRY OF LIF SIGNALLING PATHWAYS
The IL-6 cytokine family comprises eight cytokines: IL-6, IL-11, IL-27, oncostatin M, LIF, ciliary neurotrophic factor, cardiotrophin-1 and NNT/BSF-3/CLC (neurotrophin-1/B-cell-stimulating factor-3/cardiotrophin-like cytokine), all of which use gp130 (glycoprotein 130) as a signal transducing protein (see [22-30] for reviews of the IL-6 family; [25,26] thoroughly cover the signalling pathways of the IL-6 family, and [30] provides a detailed discussion of the ESC-specific roles of LIF). When LIF binds to the LIF receptor, it triggers three signalling pathways: (i) the JAK (Janus kinase)/STAT3 (signal transducer and activator of transcription 3) pathway; (ii) the PI3K (phosphoinositide 3-kinase)/AKT pathway; and (iii) the SHP2 [SH2 (Src homology 2) domain-containing tyrosine phosphatase 2]/MAPK (mitogen-activated protein kinase) pathway. Analyses of these signalling pathways have greatly benefited from the use of chemical inhibitors specific to different kinases involved in them. Commonly used inhibitors in the study of LIF are listed in Table 1.
Table 1. Protein kinase inhibitors.
See [154,155] for comprehensive review articles describing the specificity of each of these protein kinase inhibitors. CDK1, cyclin-dependent kinase 1.
| Kinase | Inhibitor | Concentration | Reference(s) |
|---|---|---|---|
| JAK | JAK inhibitor 1 | 0.6–10 μM | [66,150] |
| PI3K | LY294002 | 5 μM (40 μM in [60]) | [59,60,63,66,72] |
| PI-103 | 10 μM | [60] | |
| AKT | AKTV | 5 μM | [60] |
| GSK3 | BIO | 2 μM | [60,84,151] |
| GSK inhibitor XV | 10 nM | [60] | |
| CHIR99021/CT99021 | 3 μM | [60,73,147,150,152] | |
| Kenpaullone (also inhibits CDK1/cyclin B) | 5 μM | [150] | |
| MEK | PD98059 | 1–50 μM | [59,66,99,121] |
| PD184352 | 0.8–25 μM | [73,80,150,153] | |
| PD0325901 | 0.4–1 μM | [73,80,147,152] | |
| U0126 | 10 μM | [59,66] | |
| FGF receptor tyrosine kinase | SU5402 | 2–5 μM | [73,80,153] |
| PD173074 | 100–190 nM | [73,80,153] | |
| p38 | SB203580 | 1–2 μM | [121] |
LIF/JAK/STAT3 signalling pathway
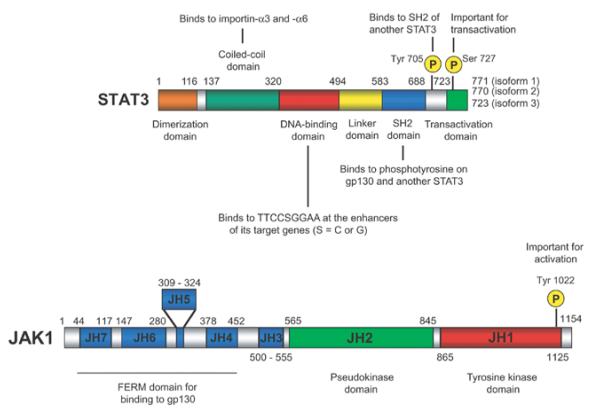
The LIF/JAK/STAT3 pathway has been extensively studied [22,24-26,28,29,31], and its central role in self-renewal has been well established by the fact that activation of STAT3 is sufficient to maintain mESC self-renewal [22,24-26,28,29,31]. STAT3 is a ubiquitously expressed protein containing six domains: a dimerization domain, a coiled-coil domain, a DNA-binding domain, a linker domain, an SH2 domain and a transactivation domain (Figure 1) [24,28,29]. It is activated by JAKs. JAKs contain seven domains called JH (JAK homology) domains 1–7, which are conserved among family members (Figure 1) [24,28,29]. JH1, located at the C-terminus, is the catalytic domain for tyrosine kinase activity. JH2 is a kinase-like (pseudokinase) domain without enzymatic activity, but appears to be required for the kinase activity of JH1 [29]. These two adjacent domains were thought to resemble the image of Janus, the Roman god with two heads, hence the name Janus kinases. JH4–JH7 are collectively called the FERM (band 4.1, ezrin, radixin and moesin) domain, which is involved in binding to gp130.
Figure 1. Domain structures of STAT3 and JAK1.
Amino acid numbers are shown above each rectangle.
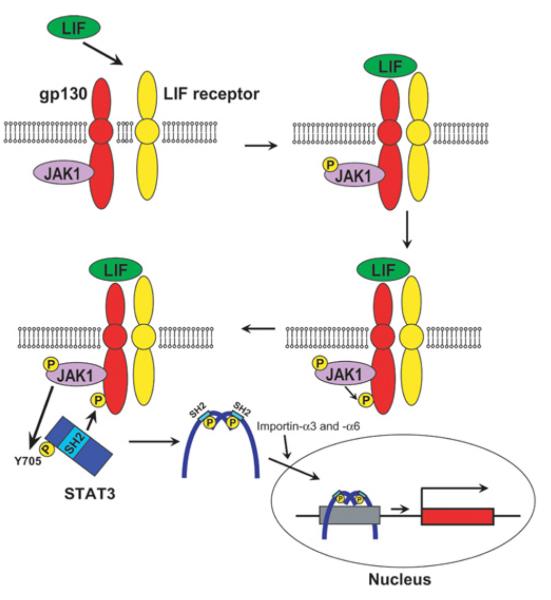
When LIF binds to the LIF receptor, the receptor recruits the membrane protein gp130 to form a heterodimer (Figure 2). This heterodimer then activates the gp130-bound JAK through transphosphorylation within a single JAK or between two JAKs [32]. Phosphorylation of the tyrosine at position 1022 is critical for activation of JAK1. Among the four JAKs, JAK1 and JAK2 are the ones primarily involved in the LIF signalling pathway [29]. In particular, JAK1 is important for the self-renewal of mESCs as shown by the higher concentration of LIF required in culture medium to maintain Jak1−/− mESCs in comparison with its wild-type counterpart [33]. Activated JAKs then phosphorylate four tyrosine residues on gp130, creating a docking site for the SH2 domain of STAT3 [34-36]. JAKs also phosphorylate the recruited STAT3 on Tyr705, resulting in the homodimerization of STAT3. This homodimerization is mediated by two identical interactions between phosphorylated Tyr705 of one STAT3 and the SH2 domain of the other. The N-terminus dimerization domain is also important for the interaction between two STAT3s, although the details of this interaction need to be investigated. Bromberg et al. [37] replaced both the alanine residue at position 661 and the asparagine at position 663 with cysteine residues to induce STAT3 dimerization independent of Tyr705. This mutant STAT3 called STAT3C spontaneously dimerizes, binds to DNA and activates its target genes, thus demonstrating that spontaneous dimerization can create a constitutively active STAT3. Dimerized STAT3 is then imported into the nucleus via the binding of nuclear import proteins, importin-α3 and importin-α6, to the coiled-coil domain of STAT3 [38,39]. STAT3 is constantly shuttled between the cytoplasm and nucleus independently of Tyr705 phosphorylation. Dimerized STAT3 finally binds to the consensus sequence TTCCSGGAA (S C or G) at the enhancer of its target genes to regulate their expression = [40,41]. LIF also induces phosphorylation of Ser727 on STAT3, but its significance for self-renewal is unknown [42]. Ser727 is phosphorylated by several kinases including p38, ERK (extracellular-signal-regulated kinase) and JNK (c-Jun N-terminal kinase) [27,43] in other settings, and this phosphorylation is important for the transactivation potency of STAT3, although the details of this mechanism remain to be studied [44,45].
Figure 2. LIF/JAK/STAT3 signalling pathway.
Although LIF is the only member of the IL-6 family routinely used to culture mESCs, other family members such as oncostatin M, ciliary neurotrophic factor and cardiotrophin 1 can also maintain self-renewal of mESCs because of their shared signalling mechanisms that converge on STAT3 [46-48]. STAT3 activation is necessary and sufficient for self-renewal and pluripotency of mESCs cultured in the presence of FBS (fetal bovine serum) without feeder cells. Niwa et al. [49] showed the essential requirement of STAT3 using a dominant-negative mutant of STAT3 in which Tyr705 was replaced with a phenylalanine residue. Sufficiency was proven by introducing a chimaeric protein called STAT3ER between full-length STAT3 and the ligand-binding domain of the oestrogen receptor [31]. Addition of the synthetic ligand 4-hydroxytamoxifen to mESC culture induces phosphorylation of Tyr705 in STAT3ER, leading to self-renewal of mESCs in the presence of FBS without either LIF or feeder cells. Although mESCs secrete LIF in an autocrine manner at a low level [50], this was insufficient for self-renewal of mESCs in the study reported by Matsuda et al. [31] because mESCs without STAT3ER differentiated in the absence of exogenous LIF.
The JAK/STAT pathway is subject to negative regulation by at least three protein families [25,32]. First, several phosphatases dephosphorylate tyrosine residues in JAKs and STAT3 (Figure 3a). The phosphatases include SHPs, PTP1B (protein tyrosine phosphatase 1b) and PTP-BL (protein tyrosine phosphatase basophil-like). Secondly, a group of proteins called PIAS (protein inhibitor of activated STAT) directly binds to STAT and inhibits its activity through several mechanisms, including SUMOylation and inhibition of DNA binding [51] (Figure 3b). It is not known whether these phosphatases and PIAS are involved in the LIF/STAT3 pathway in mESCs. A third inhibitory protein family is SOCS (suppressors of cytokine signalling), which are up-regulated upon cytokine stimulation unlike the two other inhibitors of the JAK/STAT pathway [52-54] (Figure 3c). Among the eight members of this family, SOCS1 and SOCS3 inhibit JAKs by inserting their kinase inhibitory region into the activation domain of JAKs [32,55]. Loss of SOCS1 or SOCS3 does not affect the development of blastocysts from which mESCs are derived, but is lethal in later stages of development. ESCs established from Socs3−/− mice display prolonged retention of phosphorylated STAT3 upon re-addition of LIF after its temporary depletion [56], which is consistent with the role of SOCS3 as an inhibitor of JAK activity. Phenotypically, Socs3−/− ESCs retain pluripotency as shown by teratoma (benign tumour containing tissues derived from all three germ layers) formation upon injection into immunocompromized mice, but some cells tend to spontaneously differentiate into endodermal cells even in the presence of LIF in culture. This paradoxical result, differentiation via loss of a JAK inhibitor, is explained by the concomitant activation of the SHP2/MAPK pathway, which facilitates mESC differentiation, as described below, by the loss of SOCS3 [56]. Thus depletion of SOCS3 disrupts the balance between the two opposing signalling pathways (self-renewal by JAK/STAT3 and differentiation by SHP2/MAPK), resulting in differentiation of mESCs. Overexpressed SOCS3 promotes the differentiation of mESCs as expected, in particular to haematopoietic lineages, potentially through the inhibition of the LIF/STAT3 pathway [57].
Figure 3. Three inhibitory mechanisms of the LIF/JAK/STAT3 pathway.
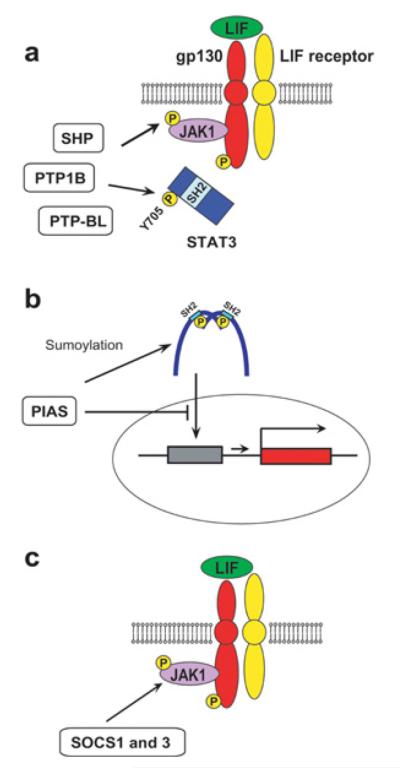
(A) Inhibition of JAK1 and STAT3 by SHP, PTP1B and PTP-BL. (B) Inhibition of STAT3 by PIAS. (C) Inhibition of JAK1 by SOCS1 and SOCS3.
LIF/PI3K/AKT signalling pathway
In a second LIF signalling pathway, JAKs activate PI3Ks, potentially through phosphorylation of the regulatory subunit p85 [58], which then activates the AKT serine/threonine kinases [59] (Figure 4). AKTs inhibit their major target protein GSK3β (glycogen synthase kinases 3β) by two independent mechanisms, resulting in an increase of the levels of Nanog and c-Myc, both of which are important for self-renewal of mESCs [60]. First, AKTs directly inactivate GSK3β by phosphorylation of Ser9 [61]. Secondly, AKTs facilitate nuclear export of GSK3β independently of phosphorylation, preventing access to target proteins in the nucleus, most prominently c-Myc [60]. GSK3β promotes ubiquitin-dependent degradation of c-Myc by phosphorylation of Thr58 [60,62]. GSK3β also inhibits Nanog expression, although the exact mechanism remains unknown [63]. Although GSK3α and GSK3β are structurally and functionally distinct [61,64], they are functionally redundant for self-renewal and differentiation of mESCs [65]. Therefore, although previous studies have focused on GSK3β, GSK3α might function in a similar way in the LIF/PI3K/AKT signalling pathway. The importance of GSK3α/β inhibition for self-renewal of mESCs is most strongly demonstrated using a chemical inhibitor in the ‘2i’ protocol in the absence of LIF (see below). Another pluripotency gene Tbx3 (T-box3) is also up-regulated by AKTs, although the involvement of GSK3α/β remains unknown [66].
Figure 4. LIF/PI3K/AKT signalling pathway.
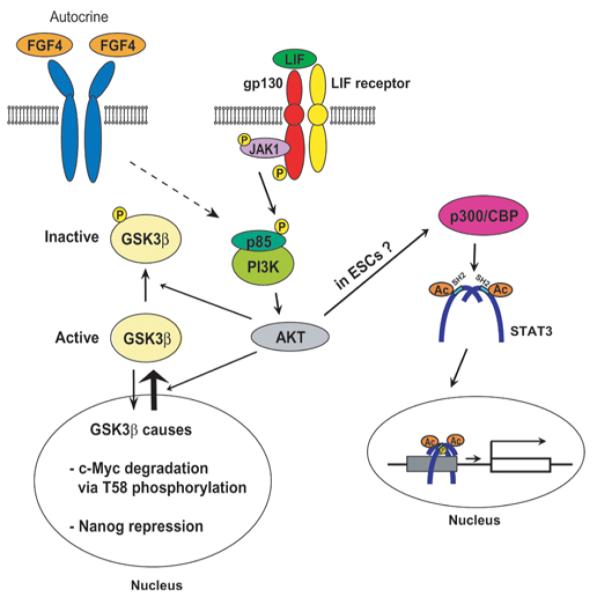
Ac, acetylation.
The LIF/PI3K/AKT pathway also induces acetylation of lysine residues on STAT3 as demonstrated using the PI3K inhibitor LY294002 [67,68], although this has not been shown in ESCs. One of these acetylations occurs on Lys685 and is mediated by the acetyltrasferase p300/CBP [CREB (cAMP-response-element-binding protein)-binding protein] independently of STAT3 phosphorylation [69]. Acetylated STAT3 forms more stable dimers and more actively transcribes target genes without phosphorylation of Tyr705 [70]. Thus LIF activates STAT3 through two mechanisms: phosphorylation of Tyr705 and acetylation of Lys685. The lysine residues at positions 49 and 87 are also acetylated by p300/CBP upon stimulation by IL-6 and oncostatin M [71], resulting in binding between the N-terminal domain of STAT3 and the bromodomain of p300/CBP. Although this leads to increased recruitment of p300/CBP to the enhancers of STAT3 target genes, the role of these two acetylations in the LIF signalling system in ESCs remains unclear. Watanabe et al. [72] have reported that AKT activation is sufficient for self-renewal of feeder-free mESCs in the absence of LIF by transfecting a constitutively active form of the AKT gene. Given the fact that STAT3 activation is essential for self-renewal [49], it is possible that STAT3 acetylation induced by Akt is playing a key role in this case. Down-regulation of STAT3 by siRNA (short interfering RNA) or shRNA (small-hairpin RNA) should be able to verify this possibility.
In mESCs, the PI3K/AKT pathway can also be activated by FGF4 [73], insulin, IGF1 (insulin-like growth factor 1) [74,75], retinol [76] and the ESC-specific Ras-like protein Eras [77,78], and inhibited by the tumour suppressor protein PTEN (phosphatase and tensin homologue deleted on chromosome 10) [79]. FGF4 functions as an autocrine stimulator for differentiation of mESCs by inducing several signalling pathways, including the PI3K/AKT pathway. However, the primary pathway involved in differentiation seems to be the Ras/MEK (MAPK/ERK kinase)/ERK pathway rather than the PI3K/AKT pathway because inhibition of MEK plays a decisive role for the maintenance of self-renewal of mESCs as shown in the 2i and 3i protocols discussed below [73,80]. The significance of the PI3K/AKT pathway induced by FGF4 for the loss of self-renewal of mESCs has not been characterized. Retinol-mediated PI3K signalling does not appear to be relevant to STAT3 phosphorylation [76]. Beyond this observation, the relationship between LIF and other PI3K stimulations has not been elucidated.
It should be noted that GSK3β is also involved in the self-renewal of mESCs as one of the key components of the canonical Wnt pathway (see [81-83] for reviews). Several groups have reported that activation of Wnt signalling pathways prevents differentiation and maintains pluripotency of ESCs [84-89] and promotes acquisition of pluripotency by mouse embryonic fibroblasts during iPSC (induced pluripotent stem cell) generation [90], although controversy still exists in the interpretation of some of the results as discussed in detail in [91]. In the absence of Wnt, β-catenin is phosphorylated by CKI (casein kinase I) and GSK3β with the support of the scaffold proteins Axin and APC (adenomatous polyposis coli) [81,92]. Phosphorylation of β-catenin triggers ubiquitination and subsequent degradation of the protein by the proteosome. This mechanism keeps the cytoplasmic level of β-catenin low in the absence of Wnt. When Wnt binds to the cell surface receptor complex Frizzled/LRP (LDL-receptor-related protein), the receptor complex induces three main signalling pathways, including the most extensively studied canonical Wnt pathway. When this pathway is activated, GSK3β is removed from Axin, resulting in stabilized and increased β-catenin. β-Catenin then enters into the nucleus and forms a complex with TCF/LEF (T-cell factor/lymphoid-enhancer-binding factor) transcription factors, activating transcription of Wnt target genes. In the absence of Wnt signalling, TCF/LEF generally functions as a transcriptional repressor by forming a complex with Groucho/Grg (Groucho-related gene)/TLE (transducin-like enhancer of split) proteins. Among the four family members of the TCF/LEF family, TCF3 is the most abundant member in mESCs [91]. This protein is colocalized with Oct4 (octamer-binding protein 4) and Nanog to more than 1000 promoters in mESCs, including those of the three master transcription factor genes for pluripotency: Oct4, Sox2 (SRY-box-containing gene 2) and Nanog, and contributes to reduced expression of these three pluripotency genes when mESCs are cultured under standard conditions where the endogenous Wnt activity is low [85]. Another study also demonstrated a suppressive role of TCF3 in the expression of Nanog [93]. Cole et al. [85] proposed that a TCF3-containing repressive complex is converted into an activating complex by Wnt, facilitating self-renewal, but this hypothesis has not been tested.
Although the LIF/PI3K/AKT pathway and the canonical Wnt pathway use the common protein GSK3β, these two pathways operate independently of each other. Acute treatment of mESCs with LIF does not alter phosphorylation of β-catenin at GSK3β-dependent sites or the level of β-catenin [59]. Additionally, enhanced activation of AKT does not increase nuclear β-catenin [72]. Moreover, although activation of β-catenin in mESCs increases the Stat3 mRNA level 2-fold, there is no consensus binding site for TCF/LEF in the promoter region of Stat3 and thus direct involvement of TCF/LEF in the increased Stat3 mRNA level remains unclear [89]. As a related issue, whether Wnt is sufficient for self-renewal in the absence of LIF is still controversial. Ogawa et al. [86] reported that Wnt is insufficient for self-renewal, but facilitates the effect of LIF; however, Takao et al. [87] showed that an activated β-catenin mutant can maintain self-renewal of mESCs in the absence of LIF or in the presence of a neutralizing antibody against LIF. Takao et al. [87] observed that this LIF-independency is specific to a certain mESC line and the expression level of the mutant β-catenin might also explain the discrepancy.
LIF/SHP2/MAPK pathway and other LIF signalling pathways
The third LIF signalling pathway is less well characterized than the preceding two. When combined with the available evidence for other IL-6 family members, the third pathway is thought to signal through SHP2/MAPK (Figure 5) [94-97]. Binding of LIF to the LIF receptor induces JAK-mediated tyrosine phosphorylation on gp130, which leads to the recruitment and subsequent phosphorylation of SHP2 by JAKs. SHP2 then interacts with the Grb2 (growth-factor-receptor-bound protein 2)–SOS (son of sevenless) complex to activate the MAPK pathway involving the Ras/RAF/MEK/ERK cascade. Unlike the two pathways mentioned above, this system induces differentiation of mESCs by down-regulating Tbx3 and Nanog [66,98]. The Ras/RAF/MEK/ERK cascade is also activated by autocrine FGF4, inducing differentiation of mESCs as described above. It is known that the MEK inhibitor PD098059 facilitates self-renewal of mESCs [99] and this can be interpreted as inhibition of the converged differentiation-inducing pathway downstream of LIF and FGF4.
Figure 5. LIF/SHP2/MAPK signalling pathway.
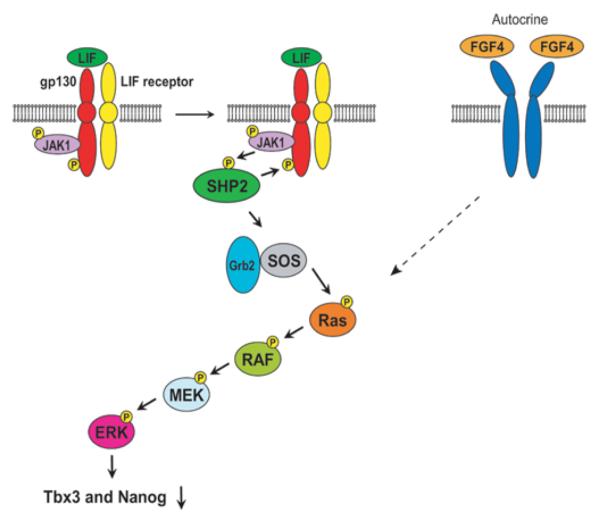
Additionally, LIF activates several other kinases, such as the Src family tyrosine kinases cYes and Hck (haemopoietic cell kinase). Addition of the Src kinase inhibitor SU6656 and siRNA against cYes induces differentiation of mESCs, indicating that it is essential for mESC self-renewal [99,100]. Importantly, cYes inhibition by SU6656 does not affect LIF-induced phosphorylation of STAT3 or ERK, suggesting that cYes is either located downstream of STAT3 or ERK, or functions independently of these two kinases. Hck is associated with gp130, but its relationship with other signalling proteins is not currently known [101].
FUNCTIONAL ANALYSES OF STAT3 IN ESCs
Many groups have directed research efforts to understand how STAT3 regulates self-renewal and pluripotency of mESCs. Two representative directions include identification of target genes of STAT3 and analysis of proteins that interact with STAT3. These two directions are discussed below.
STAT3 target genes
To understand which genes are regulated by STAT3, several research groups have employed genome-wide approaches (Table 2). Chen et al. [40] applied ChIP (chromatin immunoprecipitation) sequencing technology to map binding sites for STAT3 in mESCs. They found 2546 genomic sites where STAT3 was bound and approximately one third (718) of these loci were co-occupied by Oct4, Sox2 and Nanog. Among the STAT3-binding sites were many pluripotency genes including Oct4 and Nanog. These findings are consistent with the notion that STAT3 is tightly integrated into the gene regulatory mechanism of pluripotency. Kidder et al. [102] used the ChIP-chip approach for the same purpose using promoter arrays of mESCs that covered 28000 promoter regions in the genome. From this study they identified 948 putative target genes for STAT3, with 29 of these genes also being bound by both Oct4 and Nanog. The target gene list contains both transcriptionally active and inactive genes. The inactive genes include developmentally regulated tissue-specific genes, such as Gata3 (ectoderm-specific), Foxa2 (forkhead box A2), Gata4 (both endoderm), T (brachyury) and Lhx1 (LIM homeobox protein 1; mesoderm), and Eomes (trophectoderm). Although it is not known whether STAT3 suppresses these genes, it is possible that STAT3 mediates suppression of differentiation genes and that this mechanism could be one way in which LIF prevents differentiation of mESCs into endoderm and mesoderm lineages [103]. In a previous study, it was found that STAT3 was bound to 113 genes that were also bound by Suz12 and Eed, which are subunits of the PRC2 (polycomb repressive complex 2), although the authors did not report whether any of these 113 suppressed genes included the developmentally regulated genes noted above [102]. Although STAT proteins generally function as transcriptional activators, STAT1 is known to suppress transcription of matrix metalloproteinases and cell-cycle genes (c-Myc, cyclin D and cyclin A) [104] and therefore STAT3 may also function as a suppressor. Along a related line of inquiry, Bourillot et al. [105] knocked down 22 STAT3 target genes and found that 16 induced activation of endodermal genes and one activated mesodermal genes, supporting the conclusion that STAT3 contributes to the prevention of mESC differentiation by suppressing lineage-specific genes.
Table 2. Genome-wide analyses of STAT3-binding sites.
| Cells | Method | Number of binding sites | Co-occupied sites with Oct4, Sox2 and Nanog |
Reference |
|---|---|---|---|---|
| mESCs | ChIP sequencing | 2546 | Oct4, Sox2 and Nanog 718 | [40] |
| mESCs | ChIP-chip with promoter arrays | 948 | Oct4 58 Nanog 114 Oct4 and Nanog 29 |
[102] |
| NIH 3T3 cells treated with oncostain M to activate the Stat3 gene |
ChIP subcloning for sequencing | Approximately 400 | Not tested | [156] |
| mESCs | DNA microarray | 58 genes activated by STAT3 overexpression |
Not tested | [105] |
STAT3-binding proteins and gene activation
Another approach to understanding how STAT3 regulates gene expression is to identify proteins that interact with STAT3. The list of interacting proteins includes the transcription factors NF-κB (nuclear factor κB) [106] and c-Jun [107], co-activators NcoA/SRC1a [108] and Ctr9 [109], and the chromatin remodelling ATPase Brg1 (Brahma-related gene 1) [110,111], in addition to p300/CBP discussed above. Although these interactions have been studied outside the field of stem cell biology, similar interactions are probably taking place in mESCs since some of the interacting partners are co-localized with STAT3 on the mESC genome. The transactivation domain of STAT3 at the C-terminus interacts with many chromatin proteins such as p300/CBP [27] and not surprisingly it is co-localized with STAT3 on many pluripotency genes [40].
Of particular interest is Ctr9, a subunit of the Paf1 complex which indirectly induces multiple histone modifications including trimethylation of Lys4 and Lys36, dimethylation of Lys79 on histone H3 and ubiquitination of histone H2B, all of which are important for gene activation [112]. Consistent with this, the level of trimethylation of Lys4 on histone H3 on some of the IL-6-inducible genes is dependent on the presence of Ctr9 [109]. It appears that the interaction between STAT3 and Ctr9 is important for the recruitment of STAT3 to its target genes in this case. Although it is not known whether LIF also induces the interaction between STAT3 and Ctr9 in mESCs, if confirmed it would provide the first molecular link between LIF and epigenetic modifications in ESCs. The Paf1 complex also binds to Oct4 in mESCs [113,114], but it is unknown whether STAT3 is relevant to this binding.
The STAT3–Brg1 interaction is highly significant for pluripotency of mESCs because Brg1 is linked with pluripotency at several levels. Brg1 is a catalytic subunit of the SWI/SNF (switch/sucrose non-fermentable) ATPase complex which is involved in chromatin relaxation [115]. However, the role of Brg1 in mESCs is not limited to chromatin relaxation. Recently, it was demonstrated that the ESC-specific chromatin remodelling esBAF complex contains Brg1 [116,117]. esBAF is co-localized with STAT3 throughout the genome including on pluripotency genes. Although esBAF binds to many pluripotency genes in ESCs, it maintains pluripotency primarily by suppressing differentiation-specific genes [115]. Additionally, Brg1 facilitates binding of Oct4 to its target genes and increases the efficiency of dedifferentiation of fibroblasts to a pluripotent state [118]. Although little is known about the details of the interaction between Brg1 and STAT3 during LIF stimulation, recruitment of Brg1 can happen before or after STAT3 binding to its target genes, depending on the particular IL-6 target genes. For some IL-6 target genes, Brg1 is constitutively bound and its presence is necessary for the recruitment of STAT3 [110]. For other IL-6 target genes, STAT3 binds first to its DNA and then Brg1 is recruited depending of the presence of STAT3 [111].
LIF IN ESC BIOLOGY
Two findings have further extended our understanding of the role of LIF in self-renewal and pluripotency of ESCs. First, mESCs can be maintained undifferentiated in the absence of LIF as long as the FGF receptor, MEK and GSK3 are inhibited by chemicals [73]. Secondly, there are two stages of pluripotency, and LIF is important for only the more undifferentiated ‘näive state’ of pluripotency [119,120]. This finding may explain why hESCs do not require LIF to remain undifferentiated unlike mESCs. These two developments will be discussed in the next sections.
Self-renewal and pluripotency of mESCs without LIF
mESCs are usually maintained in an undifferentiated state using a combination of LIF and FBS or a combination of LIF and BMP (bone morphogenetic protein) 2 or BMP4 [103]. Previously, mESCs were thought to be programmed to spontaneously differentiate unless blocked by these cytokines, with LIF inhibiting endodermal and mesodermal differentiation and BMP inhibiting neural differentiation [103] (Figure 6a). Although the inhibitory mechanism of LIF for these two germ-layer lineages remains unknown, BMP is known to prevent neural differentiation of mESCs by activating the inhibitor of differentiation proteins (Id1 and Id3) and inhibiting ERK and p38 [121]. However, previous reports indicate that LIF and BMPs can be replaced by chemical inhibitors of cytokine signalling pathways (see [4,7-9,122,123] for reviews). The combination of three inhibitors called 3i, which includes SU5402 (inhibiting FGF receptor tyrosine kinase), PD184352 (inhibiting MEK which activates ERK by phosphorylation) and CHIR99021 (inhibiting GSK3), is sufficient to derive and maintain mESCs in the absence of LIF and FBS (Figure 6b) [73]. Independence of LIF for self-renewal was shown by the lack of STAT3 phosphorylation when mESCs were cultured with 3i. Additionally, ESCs could be established from Stat3−/− mouse blastocysts with the 3i protocol [73]. When the more potent MEK inhibitor PD0325901 is used, this and CHIR99021 (called 2i) is sufficient for self-renewal of mESCs without LIF and FBS [73] (Figure 6c). Although SU5402, PD184352 and PD0325901 maintain self-renewal via blocking the autocrine FGF4 signalling which promotes differentiation, the precise role for GSK3 inhibition remains unclear. Because GSK3 inhibits Nanog expression [63] and overexpression of Nanog supports self-renewal in the absence of LIF [124,125], release of Nanog suppression by the GKS3 inhibitor might explain the role of GSK3 inhibition. However, Ying et al. [73] showed that GSK3 inhibition does not induce Nanog. Instead, they suggested that the role of GSK3 inhibition is beyond the issue of self-renewal and is more widely related to overall cell metabolism and viability. Given the rapid cell cycle in mESCs [126], this is certainly a possibility for the future study.
Figure 6. Self-renewal using cytokines (LIF and BMP) or chemical inhibitors.
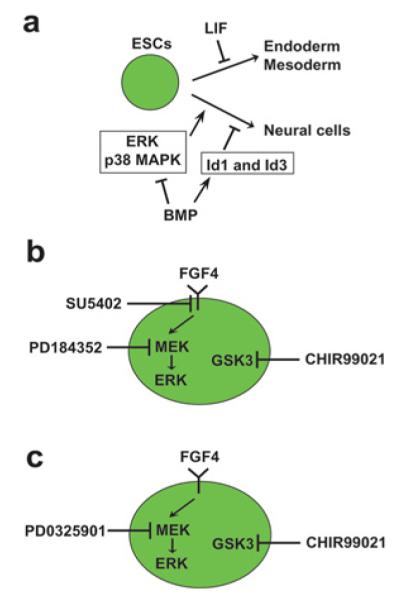
(A) Inhibition of differentiation by LIF and BMP. (B) Inhibition of differentiation by 3i. (C) Inhibition of differentiation by 2i.
Although the LIF/JAK/STAT3 pathway is critical for self-renewal of mESCs, mouse blastocysts can develop normally in the absence of key components of this pathway, with the exception of failed recovery from diapause in the absence of gp130. Mouse embryogenesis can be temporarily arrested at the blastocyst stage in the uterus without implantation for up to several weeks (a phenomenon called diapause) when the maternal physical condition is suboptimal (see [127] for details). The arrested blastocysts can still implant into the uterus and resume normal development when the maternal condition improves. However, pluripotent cells in the inner cell mass of gp130−/− blastocysts undergo apoptosis after 6 days of diapause, resulting in failed development [127]. This result indicates the necessity of gp130 for the prolonged maintenance of pluripotent cells in the inner cell mass. Other than this diapause problem, each of the homozygous knockout mice for the genes encoding LIF, LIF receptor, gp130, JAK1 and STAT3 can develop beyond the blastocyst stage (Table 3). The most severe phenotype is observed with Stat3−/−, which results in embryonic lethality between 6.5 and 7.5 dpc (days post-coitum) [128]. Although smaller than the wild-type embryos, they can develop to the egg cylinder stage (6 dpc) which comes after the blastocyst stage (4–5 dpc). Female sterility in the Lif−/− mouse is due to a failure of the uterus to support implantation of blastocysts, not the developmental potential of blastocysts themselves [129]. These blastocysts develop normally when implanted into the uterus of a pseudopregnant wild-type female. There are several possibilities that might explain why LIF-dependency is different between mESCs and blastocysts. First, ESCs might represent an artificial state created by culture conditions that do not exist in vivo. For instance, ESCs are commonly cultured in the presence of 21% oxygen, instead of approximately 0.7–7% oxygen in vivo. Oxygen concentration is known to influence a variety of cell functions [130]. Secondly, some unknown aspects of the complex cellular environment in vivo might compensate for the disrupted LIF/STAT3 pathway in vivo.
Table 3. Phenotypes of knockout mice homozygous for key genes involved in the LIF/JAK/STAT3 signalling pathway.
| Gene | Major phenotype | Reference(s) |
|---|---|---|
| Lif | Mice grow normally to adulthood A homozygous male is fertile when mated with a wild-type female A homozygous female is sterile when mated with a wild-type male because blastocysts do not implant in the uterus. The blastocysts can implant in the wild-type uterus Several types of blood stem/progenitor cells are severely decreased in the spleen and bone marrow |
[129,157] |
| Lifr | Newborn mice die within 24 h after birth Placental architecture is disrupted Bone volume is lost by two-thirds Motor neurons are lost in the brainstem, spinal cord and face |
[158,159] |
| gp130 | Embryos die between 12.5 dpc and term Multiple developmental failures occur in cardiac, haemapoietic and neural tissues Inner cell mass cells undergo apoptosis after 6 days of diapause |
[127,160,161] |
| Jak1 | Newborn mice die within 24 h after birth Lymphocyte development is severely impaired |
[162] |
| Stat3 | Embryos develop to the egg cylinder stage, but die between 6.5 and 7.5 dpc |
[128] |
LIF-independent self-renewal of mESCs with overexpressed pluripotency genes
Overexpression of some of the key genes for pluripotency, such as Nanog [124,125], Klf2 (Krüppel-like transcription factor 2) [131] and a mutant form of Myc (c-Myc) [62], also allows self-renewal of mESCs in the absence of the LIF signalling system. Before describing these genes in detail, it should be noted that mESCs secrete LIF [50] and the level of LIF secreted at a low cell density could be sufficient for self-renewal of mESCs in some cases [131]. Therefore, even if a given mESC population can continue self-renewal in the absence of exogenous LIF, it does not prove LIF-independence of self-renewal. LIF-independence is commonly tested through inhibiting multiple LIF signalling components, such as by adding a JAK inhibitor, a LIF antagonist (hLIF-05) and a dominant-negative STAT3, and by using mutant ESCs that lack one of the LIF signalling components, such as Lifr−/− cells [62,124,131].
Among the three master transcription factors for self-renewal, only Nanog has been shown to be sufficient for self-renewal of mESCs in the absence of LIF [124,125]. Because activation of STAT3 is essential for self-renewal of mESCs [49], overexpressed Nanog probably plays a function equivalent to STAT3 activation, but this link has not been well characterized. Although STAT3 binds to the regulatory region of Nanog as described above [40], the expression of Nanog is not regulated by the LIF/STAT3 pathway as shown by blocking this pathway using several approaches [124]. Since at least 18 genes are bound by both Nanog and STAT3 and co-regulated by these two proteins [105], overexpressed Nanog might compensates for the lack of LIF at least for some common target genes. However, the details of this possibility have not been investigated.
At least three members of the Klf genes, Klf2, Klf4 and Klf5, are expressed in mESCs and only overexpressed Klf2 can sustain self-renewal independently of LIF [131]. Although the expression levels of Klf4 and Klf5 are increased by LIF, the level of Klf2 is not altered by LIF, consistent with the observation that only Klf4 and Klf5 are direct targets of STAT3 [105]. Additionally, Klf2 does not require Nanog to support LIF-independent self-renewal [131], but it is not known whether Nanog can sustain self-renewal in the absence of Klf2 and LIF. Ema et al. [132] showed that Klf5 also supports self-renewal in the absence of exogenous LIF when overexpressed in mESCs, but Hall et al. [131] demonstrated the inability of Klf5 to sustain self-renewal of Lifr−/− mESCs. Since mESCs secrete LIF [50], the result based on Lifr−/− cells appears to more accurately indicate the LIF-dependence of overexpressed Klf5 for self-renewal. It is possible that LIF secreted by the feeder cells contributed to the maintenance of self-renewal in the study by Ema et al. [132].
Overexpression of a stabilized mutant of Myc, one of the key target genes of LIF, also maintains self-renewal of mESC in the absence of exogenous LIF [62]. The mRNA level of Myc is regulated by LIF through the binding of phosphorylated STAT3 to the Myc promoter. When LIF is removed from the culture medium of mESCs, the level of Myc mRNA is rapidly down-regulated. In addition, LIF withdrawal triggers GSK3β-dependent phosphorylation of Thr58 of c-Myc, resulting in ubiquitin-mediated degradation of c-Myc. However, if the c-Myc T58A mutant, in which Thr58 is replaced with an alanine residue [133], is overexpressed, it can sustain self-renewal of mESCs in the absence of LIF or in the presence of a dominant-negative STAT3. The contribution of c-Myc to self-renewal is partially explained by its inhibition of endoderm differentiation through suppression of Gata6 (GATA-binding protein 6), a key regulator of endoderm differentiation [134]. Potential roles of c-Myc in ectoderm and endoderm differentiation remain to be investigated. Another approach to investigate how the c-Myc T58A mutation compensates for the lack of LIF signalling is to compare DNA-binding sites between c-Myc and STAT3. A ChIP-chip study with promoter arrays detected only 218 genes bound by both c-Myc and STAT3 among the 1459 genes bound by c-Myc and the 948 genes bound by STAT3 [102]. Thus it is highly unlikely that overexpressed c-Myc T58A is sufficient to regulate all of those STAT3 target genes. Additionally, Nanog does not seem to be involved in c-Myc-dependent self-renewal [134]. Therefore, as with Nanog and Klf2, it is still unclear how the c-Myc T58A mutant maintains self-renewal of mESCs.
EpiSCs (epiblast stem cells) and LIF
It is well known that mouse and human ESCs are biologically different despite sharing a core genetic regulatory network for pluripotency (see [4,7-9,135,136] for reviews) (Table 4). One important difference is that, whereas mESCs rely on LIF and BMP for self-renewal and pluripotency, LIF is dispensable for hESCs. Instead, FGF2 and activin A are the primary determinants of hESC self-renewal and pluripotency. This is not because hESCs do not respond to LIF. LIF does induce phosphorylation and nuclear translocation of STAT3 in hESCs as in mESCs [42,84,137]; however, LIF fails to have any effect on self-renewal of hESCs for unknown reasons. Another noticeable difference is that mESCs express the cell-surface antigen SSEA1 (stage-specific embryonic antigen1), but hESCs express SSEA3, SSEA4, TRA1-60 and TRA1-81 [4,7-9,135,136]. A third difference is that mESCs can be dissociated to a single cell by trypsin without compromising cell viability, but hESCs are prone to enter apoptosis during this procedure due to actin- and myosin-based cell contraction [138,139]. Colony morphologies are also different: mESC colonies are dome-shaped and hESC colonies display a flatter appearance. The differences between mESCs and hESCs are also applicable to mouse and human iPSCs. iPSCs are prepared from differentiated somatic cells, most commonly fibroblasts, by introducing up to four genes, such as Oct4, Sox2, Klf4 and Myc [140-143]. Mouse iPSCs are generally prepared with LIF and FBS, and human iPSCs are established with FGF2 and FBS. These differences did not have an adequate mechanistic explanation until the recent discovery of a new type of pluripotent stem cell EpiSCs.
Table 4. Comparison between naïve and primed pluripotent stem cells.
| Mouse ESCs | Mouse EpiSCs | Human ESCs | |
|---|---|---|---|
| Type of pluripotency | Naïve | Primed | Probably primed |
| Colony morphology | Dome | Flat | Flat |
| Cloning efficiency after single-cell dissociation | Good | Poor | Poor |
| Cell-surface antigen | SSEA1 | SSEA1 | SSEA3, SSEA4, TRA1-60 and TRA1-81 |
| Expression of Oct4 , Sox2 and Nanog (core genes for pluripotency) | Yes | Yes | Yes |
| Expression of Rex1 (Zfp42), Stella (Dppa3), Gbx2 , Fgf4 and Klf4 | Yes | No | Yes |
| Expression of Fgf5 and T (brachyury) | No | Yes | No |
| X chromosome inactivation | Both X chromosomes active | One X chromosome inactive | Tend to inactivate one X chromosome |
| Teratoma formation | Yes | Yes | Yes |
| Chimaera contribution | High | Extremely low or none | Not tested |
| Cytokine requirement | LIF with BMP2 or BMP4 | FGF2 and activin A | FGF2 and activin A |
In 2007, two groups independently reported the establishment of EpiSCs from mouse epiblasts [119,120]. Epiblast is a tissue in a post-implantation embryo destined to become the embryo proper, thus a descendant of the inner cell mass from which mESCs are derived (Figure 7; see [144,145] for reviews of early mouse embryogenesis). More recently, EpiSCs were also established from mouse pre-implantation blastocysts, the same as is the case for ESCs, by changing the culture condition [146]. mEpiSCs (mouse EpiSCs) closely resemble hESCs in several respects including colony morphology and cytokine requirements (Table 4) (see [4,7-9,135,136] for reviews). These similarities suggest that hESCs are in fact more closely related to a later stage of development than mESCs. Subsequent studies by several research groups have confirmed this notion and have established that there are at least two states of pluripotency: the ‘näive’ state (or ground state) represented by mESCs and the ‘primed’ state represented by mEpiSCs and hESCs (Table 4). The most striking functional difference between these two states is that cells in the primed state have very limited or no capability of contributing to a chimaeric animal, although for an ethical reason this can not be tested for hESCs. Although both mESCs and mEpiSCs form teratomas upon injection into immunocompromized mice, mEpiSCs cannot contribute to chimaeric mice. Thus mEpiSCs have more restricted pluripotency than mESCs. Both mESCs and mEpiSCs express core pluripotency genes such as Oct4, Sox2 and Nanog; however, expression of other pluripotency genes, such as Rex1 [Zfp42 (zinc finger protein 42)], Stella [Dppa3 (development pluripotency-associated 3)], Gbx2 (gastrulation brain homeobox 2), Fgf4 and Klf4, is limited to mESCs, and Fgf5 is specifically expressed in mEpiSCs [4,7,119,120]. In addition, only one X chromosome is active in female mEpiSCs unlike in mESCs. Most importantly for the current discussion, mEpiSCs require FGF2 and activin A, like hESCs, in order to maintain pluripotency and self-renewal.
Figure 7. Embryonic sources of mESCs and mEpiSCs.
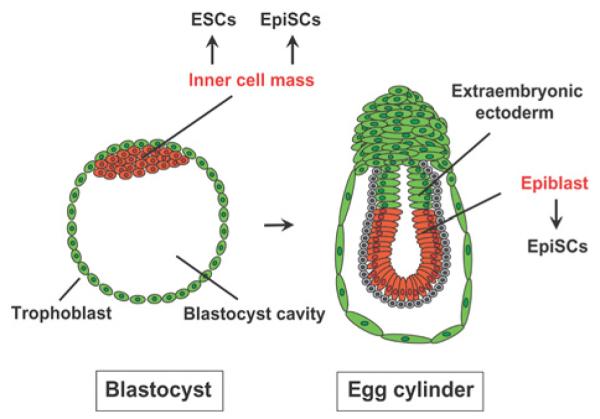
mEpiSCs and mESCs can be bidirectionally converted by changing the cytokines in the culture medium [7,136]. Differentiation from the näive state to the primed state is readily achieved as shown by Guo et al. [147], who created mEpiSC-like cells from mESCs by replacing LIF plus 2i with the combination of FGF2 and activin A. In contrast, dedifferentiation of primed pluripotent cells to näive pluripotent cells is more difficult and is accomplished at a frequency of 0.1–10% (Table 5). This process usually involves the replacement of FGF2 and activin A with LIF and 2i or other chemical inhibitors of signalling proteins. For instance, transduction of the Klf4 gene combined with LIF and 2i reverses mEpiSC to mESCs at the frequency of 0.1–1% [147]. However, it is important to note that the dedifferentiation from the primed state to the näive state can be achieved without LIF or in the presence of a JAK inhibitor, although it becomes less efficient [148]. This is consistent with mouse iPSC production, another example of the acquisition of a näive state. Mouse iPSCs can be prepared from neural stem cells with 2i in the absence of LIF at a 3–4-fold reduced efficiency compared with the efficiency in the presence of LIF [149]. The current understanding is that LIF is not essential for acquisition of the näive state, but the activation of the JAK/STAT3 pathway does accelerate this process [148].
Table 5. Dedifferentiation of primed pluripotent stem cells to naïve pluripotent stem cells in mouse and human CDK1, cyclin-dependent kinase 1.
| Parent cells (primed pluripotency) | Inducer of reversal to a naïve state | Success rate | Reference |
|---|---|---|---|
| Mouse EpiSCs | Transfection of Klf4 and addition of LIF and 2i (the ERK inhibitor PD0325901 and the GSK3 inhibitor CHIR99021) |
0.1–1% | [147] |
| Mouse EpiSCs | Transfection of Klf2 and addition of LIF and 2i | 0.11% | [131] |
| Mouse EpiSCs | Transfection of Nanog and addition of LIF and 2i Transfection of Nanog and addition of LIF and BMP4 |
1–10% Not described 2% |
[163] |
| Mouse EpiSCs | One of the following five options in the presence of LIF: transfection of Klf4 transfection of Myc addition of Kenpaullone (GSK3b and CDK1/cyclin B inhibitor) addition of Kenpaullone and CHIR99021 addition of PD184352 (MEK inhibitor) and CHIR99021 |
[150] | |
| Mouse EpiSCs | Transfection of the nuclear receptor Nr5a1 or Nr5a2 gene with the addition of LIF and 2i |
1% | [152] |
| Mouse EpiSCs | 2i and activation of the JAK/STAT3 pathway with a chimaeric receptor |
0.5–1.5% | [148] |
| Human ESCs | LIF, PD0325901 and SB203580 (p38 inhibitor) | Not described | [164] |
| Human iPSCs | Reversal to the naïve state by LIF and 2i in addition to the continued expression of the transgenes OCT4 , SOX2 and KLF4 Maintenance of the reversed state by LIF, 2i and forskolin (activator of adenylate cyclase) without the three transgenes |
Not described | [165] |
| Human ESCs | Transient expression of OCT4 plus KLF4, or KLF2 plus KLF4, followed by addition of LIF, 2i and forskolin. |
Not described | [165] |
FUTURE DIRECTIONS
The biochemical analysis of the intracellular signalling mechanism of LIF appears to have already matured, due in part to the extensively overlapping mechanisms of the IL-6 signalling system. We expect that future development of the study of LIF primarily lies in working out the functions of LIF in cell and developmental biology. In particular, LIF can be used as a tool to dissect molecular differences between the näive and primed states of pluripotency. Although many target genes of STAT3 have been identified, the roles of these genes in distinguishing between these two pluripotent states have not yet been elucidated. The functions of the binding proteins of STAT3 and the potential contribution of microRNAs require further investigation as well. It is also possible that LIF/PI3K/AKT, LIF/SHP2/MAPK and other LIF signalling pathways may be involved in the distinction between the näive and primed states of pluripotency. Additional study of these molecular mechanisms is expected to further expand our understanding of pluripotency, which will in turn greatly benefit medical applications of pluripotent stem cells.
ACKNOWLEDGEMENT
We thank Michael Franklin (University of Minnesota) for his critical reading of the manuscript prior to publication.
FUNDING
N.K is supported by the Richard M. Schulze Family Foundation and the National Institutes of Health [grant number R01 DK082430].
Abbreviations used
- BMP
bone morphogenetic protein
- Brg1
Brahma-related gene 1
- CBP
CREB (cAMP-response-element-binding protein)-binding protein
- ChIP
chromatin immunoprecipitation
- DIA
differentiation inhibiting activity
- dpc
days post coitum
- Dppa3
development pluripotency-associated 3
- EpiSC
epiblast stem cell
- ERK
extracellular-signal-regulated kinase
- ESC
embryonic stem cell
- FBS
fetal bovine serum
- FGF
fibroblast growth factor
- Gbx2
gastrulation brain homeobox 2
- gp130
glycoprotein 130
- GSK3
glycogen synthase kinases 3
- Hck
hemopoietic cell kinase
- hESC
human ESC
- HILDA
human interleukin for DA cells
- Id
inhibitor of differentiation protein
- IL
interleukin
- iPSC
induced pluripotent stem cell
- JAK
Janus kinase
- JH
JAK homology
- Klf
Krüppel-like transcription factor
- LIF
leukaemia inhibitory factor
- LIFr
LIF receptor
- MAPK
mitogen-activated protein kinase
- MEK
MAPK/ERK kinase
- mEpiSC
mouse EpiSC
- mESC
mouse ESC
- Oct4
octamer-binding protein 4
- PI3K
phosphoinositide 3-kinase
- PTP1B
protein tyrosine phosphatase 1B
- PTP-BL
protein tyrosine phosphatase basophil-like
- SH2
Src homology 2
- SHP
SH2 domain-containing tyrosine phosphatase
- siRNA
short interfering RNA
- SOCS
suppressors of cytokine signalling
- STAT
signal transducer and activator of transcription
- PIAS
protein inhibitor of activated STAT
- Sox2
SRY-box-containing gene 2
- SSEA
stage-specific embryonic antigen
- Tbx3
T-box 3
- TCF/LEF
T-cell factor/lymphoid-enhancer-binding factor
- Zfp42
zinc finger protein 42
REFERENCES
- 1.Smith AG. Embryo-derived stem cells: of mice and men. Annu. Rev. Cell Dev. Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- 2.Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–7160. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- 3.Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- 4.Pera MF, Tam PP. Extrinsic regulation of pluripotent stem cells. Nature. 2010;465:713–720. doi: 10.1038/nature09228. [DOI] [PubMed] [Google Scholar]
- 5.Nichols J, Smith A. The origin and identity of embryonic stem cells. Development. 2011;138:3–8. doi: 10.1242/dev.050831. [DOI] [PubMed] [Google Scholar]
- 6.Yu J, Thomson JA. Pluripotent stem cell lines. Genes Dev. 2008;22:1987–1997. doi: 10.1101/gad.1689808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Buecker C, Geijsen N. Different flavors of pluripotency, molecular mechanisms, and practical implications. Cell Stem Cell. 2010;7:559–564. doi: 10.1016/j.stem.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith AG, Nichols J, Robertson M, Rathjen PD. Differentiation inhibiting activity (DIA/LIF) and mouse development. Dev. Biol. 1992;151:339–351. doi: 10.1016/0012-1606(92)90174-f. [DOI] [PubMed] [Google Scholar]
- 11.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 12.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U.S.A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith AG, Hooper ML. Buffalo rat liver cells produce a diffusible activity which inhibits the differentiation of murine embryonal carcinoma and embryonic stem cells. Dev. Biol. 1987;121:1–9. doi: 10.1016/0012-1606(87)90132-1. [DOI] [PubMed] [Google Scholar]
- 14.Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 15.Tomida M, Yamamoto-Yamaguchi Y, Hozumi M. Purification of a factor inducing differentiation of mouse myeloid leukemic M1 cells from conditioned medium of mouse fibroblast L929 cells. J. Biol. Chem. 1984;259:10978–10982. [PubMed] [Google Scholar]
- 16.Hilton DJ, Nicola NA, Gough NM, Metcalf D. Resolution and purification of three distinct factors produced by Krebs ascites cells which have differentiation-inducing activity on murine myeloid leukemic cell lines. J. Biol. Chem. 1988;263:9238–9243. [PubMed] [Google Scholar]
- 17.Gearing DP, Gough NM, King JA, Hilton DJ, Nicola NA, Simpson RJ, Nice EC, Kelso A, Metcalf D. Molecular cloning and expression of cDNA encoding a murine myeloid leukaemia inhibitory factor (LIF) EMBO J. 1987;6:3995–4002. doi: 10.1002/j.1460-2075.1987.tb02742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godard A, Gascan H, Naulet J, Peyrat MA, Jacques Y, Soulillou JP, Moreau JF. Biochemical characterization and purification of HILDA, a human lymphokine active on eosinophils and bone marrow cells. Blood. 1988;71:1618–1623. [PubMed] [Google Scholar]
- 19.Moreau JF, Bonneville M, Godard A, Gascan H, Gruart V, Moore MA, Soulillou JP. Characterization of a factor produced by human T cell clones exhibiting eosinophil-activating and burst-promoting activities. J. Immunol. 1987;138:3844–3849. [PubMed] [Google Scholar]
- 20.Moreau JF, Donaldson DD, Bennett F, Witek-Giannotti J, Clark SC, Wong GG. Leukaemia inhibitory factor is identical to the myeloid growth factor human interleukin for DA cells. Nature. 1988;336:690–692. doi: 10.1038/336690a0. [DOI] [PubMed] [Google Scholar]
- 21.Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 22.Murray PJ. The JAK/STAT signaling pathway: input and output integration. J. Immunol. 2007;178:2623–2629. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- 23.O’Shea JJ, Gadina M, Schreiber RD. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell. 2002;109:S121–S131. doi: 10.1016/s0092-8674(02)00701-8. [DOI] [PubMed] [Google Scholar]
- 24.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 25.Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinrich PC, Behrmann I, Muller-Newen G, Schaper F, Graeve L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem. J. 1998;334:297–314. doi: 10.1042/bj3340297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J. Biol. Chem. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 28.Leonard WJ, O’Shea JJ. Jaks and STATs: biological implications. Annu. Rev. Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 29.Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene. 2002;285:1–24. doi: 10.1016/s0378-1119(02)00398-0. [DOI] [PubMed] [Google Scholar]
- 30.Kristensen DM, Kalisz M, Nielsen JH. Cytokine signalling in embryonic stem cells. APMIS. 2005;113:756–772. doi: 10.1111/j.1600-0463.2005.apm_391.x. [DOI] [PubMed] [Google Scholar]
- 31.Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T, Yokota T. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy J, Tannahill G, Hilton D, Greenhalgh C. The negative regulation of JAK/STAT signaling. In: Bradshaw R, Dennis E, editors. Handbook of Cell Signaling. Elsevier; San Diego: 2010. pp. 467–480. [Google Scholar]
- 33.Ernst M, Oates A, Dunn AR. Gp130-mediated signal transduction in embryonic stem cells involves activation of Jak and Ras/mitogen-activated protein kinase pathways. J. Biol. Chem. 1996;271:30136–30143. doi: 10.1074/jbc.271.47.30136. [DOI] [PubMed] [Google Scholar]
- 34.Stahl N, Farruggella TJ, Boulton TG, Zhong Z, Darnell JE, Jr, Yancopoulos GD. Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science. 1995;267:1349–1353. doi: 10.1126/science.7871433. [DOI] [PubMed] [Google Scholar]
- 35.Gerhartz C, Heesel B, Sasse J, Hemmann U, Landgraf C, Schneider-Mergener J, Horn F, Heinrich PC, Graeve L. Differential activation of acute phase response factor/STAT3 and STAT1 via the cytoplasmic domain of the interleukin 6 signal transducer gp130. I. Definition of a novel phosphotyrosine motif mediating STAT1 activation. J. Biol. Chem. 1996;271:12991–12998. doi: 10.1074/jbc.271.22.12991. [DOI] [PubMed] [Google Scholar]
- 36.Ihle JN, Kerr IM. Jaks and STATs in signaling by the cytokine receptor superfamily. Trends Genet. 1995;11:69–74. doi: 10.1016/s0168-9525(00)89000-9. [DOI] [PubMed] [Google Scholar]
- 37.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 38.Liu L, McBride KM, Reich NC. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-α3. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8150–8155. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reich NC, Liu L. Tracking STAT nuclear traffic. Nat. Rev. Immunol. 2006;6:602–612. doi: 10.1038/nri1885. [DOI] [PubMed] [Google Scholar]
- 40.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 41.Leonard W. Fundamental Immunology. Wolters Kluwer Lippincott Williams & Wilkins; Philadelphia: 2008. Type I cytokines and interferons and their receptors; pp. 706–749. [Google Scholar]
- 42.Daheron L, Opitz SL, Zaehres H, Lensch MW, Andrews PW, Itskovitz-Eldor J, Daley GQ. LIF/STAT3 signaling fails to maintain self-renewal of human embryonic stem cells. Stem Cells. 2004;22:770–778. doi: 10.1634/stemcells.22-5-770. [DOI] [PubMed] [Google Scholar]
- 43.Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–2637. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, Blenis J, Li HC, Schindler C, Chen-Kiang S. Requirement of serine phosphorylation for formation of STAT-promoter complexes. Science. 1995;267:1990–1994. doi: 10.1126/science.7701321. [DOI] [PubMed] [Google Scholar]
- 45.Wen Z, Zhong Z, Darnell JE., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 46.Rose TM, Weiford DM, Gunderson NL, Bruce AG. Oncostatin M (OSM) inhibits the differentiation of pluripotent embryonic stem cells in vitro. Cytokine. 1994;6:48–54. doi: 10.1016/1043-4666(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 47.Pennica D, Shaw KJ, Swanson TA, Moore MW, Shelton DL, Zioncheck KA, Rosenthal A, Taga T, Paoni NF, Wood WI. Cardiotrophin-1. Biological activities and binding to the leukaemia inhibitory factor receptor/gp130 signaling complex. J. Biol. Chem. 1995;270:10915–10922. doi: 10.1074/jbc.270.18.10915. [DOI] [PubMed] [Google Scholar]
- 48.Conover JC, Ip NY, Poueymirou WT, Bates B, Goldfarb MP, DeChiara TM, Yancopoulos GD. Ciliary neurotrophic factor maintains the pluripotentiality of embryonic stem cells. Development. 1993;119:559–565. doi: 10.1242/dev.119.3.559. [DOI] [PubMed] [Google Scholar]
- 49.Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rathjen PD, Nichols J, Toth S, Edwards DR, Heath JK, Smith AG. Developmentally programmed induction of differentiation inhibiting activity and the control of stem cell populations. Genes Dev. 1990;4:2308–2318. doi: 10.1101/gad.4.12b.2308. [DOI] [PubMed] [Google Scholar]
- 51.Shuai K, Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat. Rev. Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 52.Larsen L, Ropke C. Suppressors of cytokine signalling: SOCS. APMIS. 2002;110:833–844. doi: 10.1034/j.1600-0463.2002.1101201.x. [DOI] [PubMed] [Google Scholar]
- 53.Alexander WS. Suppressors of cytokine signalling (SOCS) in the immune system. Nat. Rev. Immunol. 2002;2:410–416. doi: 10.1038/nri818. [DOI] [PubMed] [Google Scholar]
- 54.Alexander WS, Hilton DJ. The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu. Rev. Immunol. 2004;22:503–529. doi: 10.1146/annurev.immunol.22.091003.090312. [DOI] [PubMed] [Google Scholar]
- 55.Boyle K, Zhang JG, Nicholson SE, Trounson E, Babon JJ, McManus EJ, Nicola NA, Robb L. Deletion of the SOCS box of suppressor of cytokine signaling 3 (SOCS3) in embryonic stem cells reveals SOCS box-dependent regulation of JAK but not STAT phosphorylation. Cell. Signalling. 2009;21:394–404. doi: 10.1016/j.cellsig.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forrai A, Boyle K, Hart AH, Hartley L, Rakar S, Willson TA, Simpson KM, Roberts AW, Alexander WS, Voss AK, Robb L. Absence of suppressor of cytokine signalling 3 reduces self-renewal and promotes differentiation in murine embryonic stem cells. Stem Cells. 2006;24:604–614. doi: 10.1634/stemcells.2005-0323. [DOI] [PubMed] [Google Scholar]
- 57.Li Y, McClintick J, Zhong L, Edenberg HJ, Yoder MC, Chan RJ. Murine embryonic stem cell differentiation is promoted by SOCS-3 and inhibited by the zinc finger transcription factor Klf4. Blood. 2005;105:635–637. doi: 10.1182/blood-2004-07-2681. [DOI] [PubMed] [Google Scholar]
- 58.Migone TS, Rodig S, Cacalano NA, Berg M, Schreiber RD, Leonard WJ. Functional cooperation of the interleukin-2 receptor β chain and Jak1 in phosphatidylinositol 3-kinase recruitment and phosphorylation. Mol. Cell Biol. 1998;18:6416–6422. doi: 10.1128/mcb.18.11.6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paling NR, Wheadon H, Bone HK, Welham MJ. Regulation of embryonic stem cell self-renewal by phosphoinositide 3-kinase-dependent signaling. J. Biol. Chem. 2004;279:48063–48070. doi: 10.1074/jbc.M406467200. [DOI] [PubMed] [Google Scholar]
- 60.Bechard M, Dalton S. Subcellular localization of glycogen synthase kinase 3β controls embryonic stem cell self-renewal. Mol. Cell Biol. 2009;29:2092–2104. doi: 10.1128/MCB.01405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell Sci. 2003;116:1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 63.Storm MP, Bone HK, Beck CG, Bourillot PY, Schreiber V, Damiano T, Nelson A, Savatier P, Welham MJ. Regulation of Nanog expression by phosphoinositide 3-kinase-dependent signaling in murine embryonic stem cells. J. Biol. Chem. 2007;282:6265–6273. doi: 10.1074/jbc.M610906200. [DOI] [PubMed] [Google Scholar]
- 64.Hoeflich KP, Luo J, Rubie EA, Tsao MS, Jin O, Woodgett JR. Requirement for glycogen synthase kinase-3β in cell survival and NF-κB activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- 65.Doble BW, Patel S, Wood GA, Kockeritz LK, Woodgett JR. Functional redundancy of GSK-3αand GSK-3βin Wnt/β-catenin signaling shown by using an allelic series of embryonic stem cell lines. Dev. Cell. 2007;12:957–971. doi: 10.1016/j.devcel.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- 67.Wang R, Cherukuri P, Luo J. Activation of Stat3 sequence-specific DNA binding and transcription by p300/CREB-binding protein-mediated acetylation. J. Biol. Chem. 2005;280:11528–11534. doi: 10.1074/jbc.M413930200. [DOI] [PubMed] [Google Scholar]
- 68.Ohbayashi N, Ikeda O, Taira N, Yamamoto Y, Muromoto R, Sekine Y, Sugiyama K, Honjoh T, Matsuda T. LIF- and IL-6-induced acetylation of STAT3 at Lys-685 through PI3K/Akt activation. Biol. Pharm. Bull. 2007;30:1860–1864. doi: 10.1248/bpb.30.1860. [DOI] [PubMed] [Google Scholar]
- 69.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 70.Braunstein J, Brutsaert S, Olson R, Schindler C. STATs dimerize in the absence of phosphorylation. J. Biol. Chem. 2003;278:34133–34140. doi: 10.1074/jbc.M304531200. [DOI] [PubMed] [Google Scholar]
- 71.Hou T, Ray S, Lee C, Brasier AR. The STAT3 NH2-terminal domain stabilizes enhanceosome assembly by interacting with the p300 bromodomain. J. Biol. Chem. 2008;283:30725–30734. doi: 10.1074/jbc.M805941200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watanabe S, Umehara H, Murayama K, Okabe M, Kimura T, Nakano T. Activation of Akt signaling is sufficient to maintain pluripotency in mouse and primate embryonic stem cells. Oncogene. 2006;25:2697–2707. doi: 10.1038/sj.onc.1209307. [DOI] [PubMed] [Google Scholar]
- 73.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 75.Rubin R, Arzumanyan A, Soliera AR, Ross B, Peruzzi F, Prisco M. Insulin receptor substrate (IRS)-1 regulates murine embryonic stem (mES) cells self-renewal. J. Cell Physiol. 2007;213:445–453. doi: 10.1002/jcp.21185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen L, Khillan JS. A novel signaling by vitamin A/retinol promotes self renewal of mouse embryonic stem cells by activating PI3K/Akt signaling pathway via insulin-like growth factor-1 receptor. Stem Cells. 2010;28:57–63. doi: 10.1002/stem.251. [DOI] [PubMed] [Google Scholar]
- 77.Takahashi K, Mitsui K, Yamanaka S. Role of ERas in promoting tumour-like properties in mouse embryonic stem cells. Nature. 2003;423:541–545. doi: 10.1038/nature01646. [DOI] [PubMed] [Google Scholar]
- 78.Takahashi K, Murakami M, Yamanaka S. Role of the phosphoinositide 3-kinase pathway in mouse embryonic stem (ES) cells. Biochem. Soc. Trans. 2005;33:1522–1525. doi: 10.1042/BST0331522. [DOI] [PubMed] [Google Scholar]
- 79.Sun H, Lesche R, Li DM, Liliental J, Zhang H, Gao J, Gavrilova N, Mueller B, Liu X, Wu H. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5,-trisphosphate and Akt/protein kinase B signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6199–6204. doi: 10.1073/pnas.96.11.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nichols J, Silva J, Roode M, Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 82.Willert K, Jones KA. Wnt signaling: is the party in the nucleus? Genes Dev. 2006;20:1394–1404. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
- 83.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 84.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 85.Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ogawa K, Nishinakamura R, Iwamatsu Y, Shimosato D, Niwa H. Synergistic action of Wnt and LIF in maintaining pluripotency of mouse ES cells. Biochem. Biophys. Res. Commun. 2006;343:159–166. doi: 10.1016/j.bbrc.2006.02.127. [DOI] [PubMed] [Google Scholar]
- 87.Takao Y, Yokota T, Koide H. β-catenin up-regulates Nanog expression through interaction with Oct-3/4 in embryonic stem cells. Biochem. Biophys. Res. Commun. 2007;353:699–705. doi: 10.1016/j.bbrc.2006.12.072. [DOI] [PubMed] [Google Scholar]
- 88.Kielman MF, Rindapaa M, Gaspar C, van Poppel N, Breukel C, van Leeuwen S, Taketo MM, Roberts S, Smits R, Fodde R. Apc modulates embryonic stem-cell differentiation by controlling the dosage of β-catenin signaling. Nat. Genet. 2002;32:594–605. doi: 10.1038/ng1045. [DOI] [PubMed] [Google Scholar]
- 89.Hao J, Li TG, Qi X, Zhao DF, Zhao GQ. WNT/β-catenin pathway up-regulates Stat3 and converges on LIF to prevent differentiation of mouse embryonicstem cells. Dev. Biol. 2006;290:81–91. doi: 10.1016/j.ydbio.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 90.Marson A, Foreman R, Chevalier B, Bilodeau S, Kahn M, Young RA, Jaenisch R. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3:132–135. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wray J, Kalkan T, Smith AG. The ground state of pluripotency. Biochem. Soc. Trans. 2010;38:1027–1032. doi: 10.1042/BST0381027. [DOI] [PubMed] [Google Scholar]
- 92.Price MA. CKI, there’s more than one: casein kinase I family members in Wnt and Hedgehog signaling. Genes Dev. 2006;20:399–410. doi: 10.1101/gad.1394306. [DOI] [PubMed] [Google Scholar]
- 93.Pereira L, Yi F, Merrill BJ. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol. Cell Biol. 2006;26:7479–7491. doi: 10.1128/MCB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schiemann WP, Bartoe JL, Nathanson NM. Box 3-independent signaling mechanisms are involved in leukaemia inhibitory factor receptor α- and gp130-mediated stimulation of mitogen-activated protein kinase. Evidence for participation of multiple signaling pathways which converge at Ras. J. Biol. Chem. 1997;272:16631–16636. doi: 10.1074/jbc.272.26.16631. [DOI] [PubMed] [Google Scholar]
- 95.Schaper F, Gendo C, Eck M, Schmitz J, Grimm C, Anhuf D, Kerr IM, Heinrich PC. Activation of the protein tyrosine phosphatase SHP2 via the interleukin-6 signal transducing receptor protein gp130 requires tyrosine kinase Jak1 and limits acute-phase protein expression. Biochem. J. 1998;335:557–565. doi: 10.1042/bj3350557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fukada T, Hibi M, Yamanaka Y, Takahashi-Tezuka M, Fujitani Y, Yamaguchi T, Nakajima K, Hirano T. Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: involvement of STAT3 in anti-apoptosis. Immunity. 1996;5:449–460. doi: 10.1016/s1074-7613(00)80501-4. [DOI] [PubMed] [Google Scholar]
- 97.Hermanns HM, Radtke S, Schaper F, Heinrich PC, Behrmann I. Non-redundant signal transduction of interleukin-6-type cytokines. The adapter protein Shc is specifically recruited to rhe oncostatin M receptor. J. Biol. Chem. 2000;275:40742–40748. doi: 10.1074/jbc.M005408200. [DOI] [PubMed] [Google Scholar]
- 98.Hamazaki T, Kehoe SM, Nakano T, Terada N. The Grb2/Mek pathway represses Nanog in murine embryonic stem cells. Mol. Cell Biol. 2006;26:7539–7549. doi: 10.1128/MCB.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Burdon T, Stracey C, Chambers I, Nichols J, Smith A. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev. Biol. 1999;210:30–43. doi: 10.1006/dbio.1999.9265. [DOI] [PubMed] [Google Scholar]
- 100.Anneren C, Cowan CA, Melton DA. The Src family of tyrosine kinases is important for embryonic stem cell self-renewal. J. Biol. Chem. 2004;279:31590–31598. doi: 10.1074/jbc.M403547200. [DOI] [PubMed] [Google Scholar]
- 101.Ernst M, Gearing DP, Dunn AR. Functional and biochemical association of Hck with the LIF/IL-6 receptor signal transducing subunit gp130 in embryonic stem cells. EMBO J. 1994;13:1574–1584. doi: 10.1002/j.1460-2075.1994.tb06420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kidder BL, Yang J, Palmer S. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS ONE. 2008;3:e3932. doi: 10.1371/journal.pone.0003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 104.Ramana CV, Chatterjee-Kishore M, Nguyen H, Stark GR. Complex roles of Stat1 in regulating gene expression. Oncogene. 2000;19:2619–2627. doi: 10.1038/sj.onc.1203525. [DOI] [PubMed] [Google Scholar]
- 105.Bourillot PY, Aksoy I, Schreiber V, Wianny F, Schulz H, Hummel O, Hubner N, Savatier P. Novel STAT3 target genes exert distinct roles in the inhibition of mesoderm and endoderm differentiation in cooperation with Nanog. Stem Cells. 2009;27:1760–1771. doi: 10.1002/stem.110. [DOI] [PubMed] [Google Scholar]
- 106.Yu Z, Zhang W, Kone BC. Signal transducers and activators of transcription 3 (STAT3) inhibits transcription of the inducible nitric oxide synthase gene by interacting with nuclear factor κB. Biochem. J. 2002;367:97–105. doi: 10.1042/BJ20020588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang X, Wrzeszczynska MH, Horvath CM, Darnell JE., Jr Interacting regions in Stat3 and c-Jun that participate in cooperative transcriptional activation. Mol. Cell Biol. 1999;19:7138–7146. doi: 10.1128/mcb.19.10.7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Giraud S, Bienvenu F, Avril S, Gascan H, Heery DM, Coqueret O. Functional interaction of STAT3 transcription factor with the coactivator NcoA/SRC1a. J. Biol. Chem. 2002;277:8004–8011. doi: 10.1074/jbc.M111486200. [DOI] [PubMed] [Google Scholar]
- 109.Youn MY, Yoo HS, Kim MJ, Hwang SY, Choi Y, Desiderio SV, Yoo JY. hCTR9, a component of Paf1 complex, participates in the transcription of interleukin 6-responsive genes through regulation of STAT3-DNA interactions. J. Biol. Chem. 2007;282:34727–34734. doi: 10.1074/jbc.M705411200. [DOI] [PubMed] [Google Scholar]
- 110.Ni Z, Bremner R. Brahma-related gene 1-dependent STAT3 recruitment at IL-6-inducible genes. J. Immunol. 2007;178:345–351. doi: 10.4049/jimmunol.178.1.345. [DOI] [PubMed] [Google Scholar]
- 111.Giraud S, Hurlstone A, Avril S, Coqueret O. Implication of BRG1 and cdk9 in the STAT3-mediated activation of the p21waf1 gene. Oncogene. 2004;23:7391–7398. doi: 10.1038/sj.onc.1207972. [DOI] [PubMed] [Google Scholar]
- 112.Gerber M, Shilatifard A. Transcriptional elongation by RNA polymerase II and histone methylation. J. Biol. Chem. 2003;278:26303–26306. doi: 10.1074/jbc.R300014200. [DOI] [PubMed] [Google Scholar]
- 113.Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 114.Ponnusamy MP, Deb S, Dey P, Chakraborty S, Rachagani S, Senapati S, Batra SK. RNA polymerase II associated factor 1/PD2 maintains self-renewal by its interaction with Oct3/4 in mouse embryonic stem cells. Stem Cells. 2009;27:3001–3011. doi: 10.1002/stem.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lessard JA, Crabtree GR. Chromatin regulatory mechanisms in pluripotency. Annu. Rev. Cell Dev. Biol. 2010;26:503–532. doi: 10.1146/annurev-cellbio-051809-102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Singhal N, Graumann J, Wu G, Arauzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Scholer HR. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 119.Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 120.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 121.Qi X, Li TG, Hao J, Hu J, Wang J, Simmons H, Miura S, Mishina Y, Zhao GQ. BMP4 supports self-renewal of embryonic stem cells by inhibiting mitogen-activated protein kinase pathways. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6027–6032. doi: 10.1073/pnas.0401367101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lanner F, Rossant J. The role of FGF/Erk signaling in pluripotent cells. Development. 2010;137:3351–3360. doi: 10.1242/dev.050146. [DOI] [PubMed] [Google Scholar]
- 124.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 125.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 126.White J, Dalton S. Cell cycle control of embryonic stem cells. Stem Cell Rev. 2005;1:131–138. doi: 10.1385/SCR:1:2:131. [DOI] [PubMed] [Google Scholar]
- 127.Nichols J, Chambers I, Taga T, Smith A. Physiological rationale for responsiveness of mouse embryonic stem cells to gp130 cytokines. Development. 2001;128:2333–2339. doi: 10.1242/dev.128.12.2333. [DOI] [PubMed] [Google Scholar]
- 128.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Stewart CL, Kaspar P, Brunet LJ, Bhatt H, Gadi I, Kontgen F, Abbondanzo SJ. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature. 1992;359:76–79. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- 130.Wion D, Christen T, Barbier EL, Coles JA. PO2 matters in stem cell culture. Cell Stem Cell. 2009;5:242–243. doi: 10.1016/j.stem.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 131.Hall J, Guo G, Wray J, Eyres I, Nichols J, Grotewold L, Morfopoulou S, Humphreys P, Mansfield W, Walker R, et al. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5:597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 132.Ema M, Mori D, Niwa H, Hasegawa Y, Yamanaka Y, Hitoshi S, Mimura J, Kawabe Y, Hosoya T, Morita M, et al. Kruppel-like factor 5 is essential for blastocyst development and the normal self-renewal of mouse ESCs. Cell Stem Cell. 2008;3:555–567. doi: 10.1016/j.stem.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 133.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Smith KN, Singh AM, Dalton S. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell. 2010;7:343–354. doi: 10.1016/j.stem.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kerr CL, Cheng L. Multiple, interconvertible states of human pluripotent stem cells. Cell Stem Cell. 2010;6:497–499. doi: 10.1016/j.stem.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 136.Chenoweth JG, McKay RD, Tesar PJ. Epiblast stem cells contribute new insight into pluripotency and gastrulation. Dev. Growth Differ. 2010;52:293–301. doi: 10.1111/j.1440-169X.2010.01171.x. [DOI] [PubMed] [Google Scholar]
- 137.Humphrey RK, Beattie GM, Lopez AD, Bucay N, King CC, Firpo MT, Rose-John S, Hayek A. Maintenance of pluripotency in human embryonic stem cells is STAT3 independent. Stem Cells. 2004;22:522–530. doi: 10.1634/stemcells.22-4-522. [DOI] [PubMed] [Google Scholar]
- 138.Ohgushi M, Matsumura M, Eiraku M, Murakami K, Aramaki T, Nishiyama A, Muguruma K, Nakano T, Suga H, Ueno M, et al. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell. 2010;7:225–239. doi: 10.1016/j.stem.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 139.Chen G, Hou Z, Gulbranson DR, Thomson JA. Actin-myosin contractility is responsible for the reduced viability of dissociated human embryonic stem cells. Cell Stem Cell. 2010;7:240–248. doi: 10.1016/j.stem.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 141.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 142.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 143.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 144.Rossant J, Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–713. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- 145.Gardner RL. Contributions of blastocyst micromanipulation to the study of mammalian development. BioEssays. 1998;20:168–180. doi: 10.1002/(SICI)1521-1878(199802)20:2<168::AID-BIES9>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 146.Najm FJ, Chenoweth JG, Anderson PD, Nadeau JH, Redline RW, McKay RD, Tesar PJ. Isolation of epiblast stem cells from preimplantation mouse embryos. Cell Stem Cell. 2011;8:318–325. doi: 10.1016/j.stem.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yang J, van Oosten AL, Theunissen TW, Guo G, Silva JC, Smith A. Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell. 2010;7:319–328. doi: 10.1016/j.stem.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hanna J, Markoulaki S, Mitalipova M, Cheng AW, Cassady JP, Staerk J, Carey BW, Lengner CJ, Foreman R, Love J, et al. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell. 2009;4:513–524. doi: 10.1016/j.stem.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Umehara H, Kimura T, Ohtsuka S, Nakamura T, Kitajima K, Ikawa M, Okabe M, Niwa H, Nakano T. Efficient derivation of embryonic stem cells by inhibition of glycogen synthase kinase-3. Stem Cells. 2007;25:2705–2711. doi: 10.1634/stemcells.2007-0086. [DOI] [PubMed] [Google Scholar]
- 152.Guo G, Smith A. A genome-wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development. 2010;137:3185–3192. doi: 10.1242/dev.052753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S, Smith A. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- 154.Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem. J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Snyder M, Huang XY, Zhang JJ. Identification of novel direct Stat3 target genes for control of growth and differentiation. J. Biol. Chem. 2008;283:3791–3798. doi: 10.1074/jbc.M706976200. [DOI] [PubMed] [Google Scholar]
- 157.Escary JL, Perreau J, Dumenil D, Ezine S, Brulet P. Leukaemia inhibitory factor is necessary for maintenance of haematopoietic stem cells and thymocyte stimulation. Nature. 1993;363:361–364. doi: 10.1038/363361a0. [DOI] [PubMed] [Google Scholar]
- 158.Li M, Sendtner M, Smith A. Essential function of LIF receptor in motor neurons. Nature. 1995;378:724–727. doi: 10.1038/378724a0. [DOI] [PubMed] [Google Scholar]
- 159.Ware CB, Horowitz MC, Renshaw BR, Hunt JS, Liggitt D, Koblar SA, Gliniak BC, McKenna HJ, Papayannopoulou T, Thoma B, et al. Targeted disruption of the low-affinity leukaemia inhibitory factor receptor gene causes placental, skeletal, neural and metabolic defects and results in perinatal death. Development. 1995;121:1283–1299. doi: 10.1242/dev.121.5.1283. [DOI] [PubMed] [Google Scholar]
- 160.Yoshida K, Taga T, Saito M, Suematsu S, Kumanogoh A, Tanaka T, Fujiwara H, Hirata M, Yamagami T, Nakahata T, et al. Targeted disruption of gp130, a common signal transducer for the interleukin 6 family of cytokines, leads to myocardial and hematological disorders. Proc. Natl. Acad. Sci. U.S.A. 1996;93:407–411. doi: 10.1073/pnas.93.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nakashima K, Wiese S, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Yoshida K, Kishimoto T, Sendtner M, Taga T. Developmental requirement of gp130 signaling in neuronal survival and astrocyte differentiation. J. Neurosci. 1999;19:5429–5434. doi: 10.1523/JNEUROSCI.19-13-05429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, King KL, Sheehan KC, Yin L, Pennica D, Johnson EM, Jr, Schreiber RD. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell. 1998;93:373–383. doi: 10.1016/s0092-8674(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 163.Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Xu Y, Zhu X, Hahm HS, Wei W, Hao E, Hayek A, Ding S. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc. Natl. Acad. Sci. U.S.A. 2010;107:8129–8134. doi: 10.1073/pnas.1002024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]