Summary
Over-expression of the Candida albicans ATP-binding cassette transporter CaCdr1p causes clinically significant resistance to azole drugs including fluconazole (FLC). Screening of a ~1.89 × 106 member d-octapeptide combinatorial library that concentrates library members at the yeast cell surface identified RC21v3, a 4-methoxy-2,3,6-trimethylbenzenesulphonyl derivative of the d-octapeptide d-NH2-FFKWQRRR-CONH2, as a potent and stereospecific inhibitor of CaCdr1p. RC21v3 chemosensitized Saccharomyces cerevisiae strains over-expressing CaCdr1p but not other fungal ABC transporters, the C. albicans MFS transporter CaMdr1p or the azole target enzyme CaErg11p, to FLC. RC21v3 also chemosensitized clinical C. albicans isolates over-expressing CaCDR1 to FLC, even when CaCDR2 was over-expressed. Specific targeting of CaCdr1p by RC21v3 was confirmed by spontaneous RC21v3 chemosensitization resistant suppressor mutants of S. cerevisiae expressing CaCdr1p. The suppressor mutations introduced a positive charge beside, or within, extracellular loops 1, 3, 4 and 6 of CaCdr1p or an aromatic residue near the extracytoplasmic end of transmembrane segment 5. The mutations did not affect CaCdr1p localization or Cdr1p ATPase activity but some increased susceptibility to the CaCdr1p substrates FLC, rhodamine 6G, rhodamine 123 and cycloheximide. The suppressor mutations showed that the drug-like CaCdr1p inhibitors FK506, enniatin, milbemycin α11 and milbemycin β9 have modes of action similar to RC21v3.
Keywords: Candida albicans ABC transporter, d-octapeptide, fluconazole, chemosensitization
Introduction
The pathogenic fungus Candida albicans still dominates the incidence of potentially fatal disseminated fungal disease and the more readily treated superficial infections such as oral thrush in the immune deficient (the very young and the elderly) and the immunocompromised (AIDS patients, transplant recipients, and cancer patients). In C. albicans, and some other pathogenic fungi, multidrug efflux mediated by ATP-binding cassette (ABC) transporters is a major cause of clinically significant resistance to the widely-used and well-tolerated azole drug fluconazole (FLC) (Cannon et al., 2009). The fungal efflux pumps responsible for this activity belong to the Pleiotropic Drug Resistance A (PDR A) subfamily (Cannon et al., 2009, Lamping et al., 2010). The founding member is the extensively characterized but yet to be structurally resolved Pdr5p from Saccharomyces cerevisiae (Balzi & Goffeau, 1994, Balzi & Goffeau, 1995). In C. albicans the Pdr5p homologue CaCdr1p, which was also first cloned in Goffeau’s laboratory (Prasad et al., 1995), appears to be responsible for the bulk of the drug resistance detected in clinical isolates showing high-level fluconazole resistance (Holmes et al., 2008, Tsao et al., 2009, Sanglard et al., 1996). Although a broad range of PDR efflux pump substrates have been identified (Kolaczkowski et al., 1996), and some features that determine substrate specificity investigated (Egner et al., 2000, Egner et al., 1998, Hiraga et al., 2001, Tanabe et al., 2011), the discovery and/or characterization of compounds (chemosensitizers) that specifically inhibit fungal drug efflux (Watkins et al., 2004, Schuetzer-Muehlbauer et al., 2003, Lemoine et al., 2004, Hiraga et al., 2001, Egner et al., 2000, Tanabe et al., 2007, Lee et al., 2001) has not had a commercial outcome and remains limited by knowledge of relevant efflux pump structure.
The PDR A drug efflux pumps are thought to comprise 170 kDa subunits composed of two cytoplasmic nucleotide binding domains (NBDs) and two transmembrane domains (TMDs) in the order Nterminus-NBD1-TMD1-NBD2-TMD2-Cterminus. This domain order is mimicked in part by the Nterminus-NBD1-TMD1-Cterminus order of the half-sized ABC transporter HsABCG2 and contrasts with the Nterminus-TMD1-NBD1-TMD2-NBD2-Cterminus order found in comparable full-sized mammalian ABC transporters like P-glycoprotein (HsABCB1) (Cannon et al., 2009). P-glycoprotein and ABCG2 homologues are major contributors to multidrug efflux in higher eukaryotes. These pumps help define normal drug distribution and are highly expressed in several cancers that are refractory to chemotherapy. Structures obtained by X-ray crystallography over the last decade, including the prokaryotic BtuCD vitamin B12 uptake protein (Locher et al., 2002) and Sav1866 multidrug efflux transporters (Dawson et al., 2007, Dawson & Locher, 2006, Dawson & Locher, 2007), together with mouse ABCB1 (Aller et al., 2009), have provided important insights into the structure and function of ABC transporters. Conformational change between open and closed states involve adenine nucleotide-mediated interactions between the NBDs during the catalytic cycle. This affects the affinity of the TMDs for transport substrates and enables transport across the membrane. Homology between fungal PDR transporters and their prokaryotic and higher eukaryotic orthologs is sufficient to suggest that their NBDs may operate in a similar, but not necessarily identical, manner (Gupta et al., 2011). However, the low homology between the TMDs of the PDR drug efflux pumps with their structurally resolved eukaryotic and prokaryotic counterparts means that homology modeling of the regions that confer efflux substrate specificity may be premature. This problem limits understanding of the structural features of the TMDs that confer promiscuous overlapping specificity on fungal drug efflux pumps, such as that described for S. cerevisiae Pdr5p (Golin et al., 2003, Golin et al., 2007, Hanson et al., 2005). We recently used chimeras of the C. albicans homologs Cdr1p and Cdr2p over-expressed in S. cerevisiae to identify segments of TMDs that conferred specificity for pump substrates including FK506 and monensin and the pump inhibitor enniatin (Tanabe et al., 2011). Clearly, the identification of the binding sites that face the cytoplasm and the periplasm during the drug binding and release phases of the reaction cycle, respectively, would be a considerable advance (Ernst et al., 2010, Ernst et al., 2005). Hydropathy plots and consensus secondary structure profiles suggest that each fungal TMD comprises 6 transmembrane helices, two relatively short helix-rich cytoplasmic sequences, plus two very short and one larger extracytoplasmic loop (Cannon et al., 2009, Lamping et al., 2010). In addition to the primary structure linking NBD1 with TMD1 and NBD2 with TMD2, highly charged regions of 4 amino acids bordered by helices of the short intracellular loops connecting transmembrane segment (TMS) 2 and 3 and connecting TMS 8 and 9 may be analogous to the coupling sequences that interact with the NBDs of the prokaryotic ABC transporters (Dawson & Locher, 2006). The extracellular loops contain conserved sequences that may contribute to pathways used during xenobiotic efflux in fungi, but their form and function is poorly understood (Lamping et al., 2010). Rutledge and colleagues (Rutledge et al., 2010) recently used both homology and ab initio modeling to produce structural models of S. cerevisiae Pdr5p in both the open and closed conformations by selectively incorporating similarities and differences between the fungal PDR efflux pumps and the resolved structures of bacterial ABC transporters and rat ABCB1. Despite their speculative nature, these models may help visualize the mode of action of efflux pump inhibitors. In addition, unlike the major higher eukaryotic drug efflux pumps, the ATPase activity of fungal efflux pumps is not significantly stimulated by the presence of xenobiotic efflux substrates (Shukla et al., 2006) and enzyme activity is lost when endogenous lipids are displaced with solubilizing detergents. These findings suggest that xenobiotics are unnatural substrates of the fungal efflux pumps, the physiological role of the pumps may be as floppases that help maintain transmembrane lipid asymmetry (Mishra et al., 2007), xenobiotic release into the periplasm may be the rate limiting step in efflux activity (Ernst et al., 2010) and that inhibitors targeting the ectodomain of fungal efflux pumps can be identified.
We have probed transporter function and overcome the resistance-conferring efflux of antifungal agents by the most prominent efflux pumps in multidrug resistant fungal clinical isolates of C. albicans by identifying and characterizing a highly specific surface-active inhibitor of the CaCdr1p drug efflux pump that chemosensitizes cells expressing CaCdr1p to FLC. The chemosensitizer was identified by screening a 1.89 × 106-member d-octapeptide combinatorial library for surface-active efflux inhibitors that are not substrates of multidrug efflux pumps. The screen used strains of S. cerevisiae that hyperexpressed the fully-functional drug efflux pumps CaCdr1p and CaMdr1p in a background that is hyper-sensitive to xenobiotics (Lamping et al., 2007). Each member of the combinatorial peptide library contains a C-terminal tri-arginine motif that confers membrane impermeability and concentrates library members at the cell surface of fungal but not host cells (Monk et al., 2005, Niimi et al., 2004). We have identified RC21v3, a potent, stereospecific, surface-active, high-affinity inhibitor that interacts with the extracellular surface of CaCdr1p. Inhibition of Cdr1p by RC21v3 chemosensitized azole-resistant C. albicans clinical isolates to FLC and other azole drugs. Genetic analysis of suppressors of RC21-mediated chemosensitization to FLC and the detection of altered susceptibilities to a range of transport substrates and inhibitors were used to map amino acids affecting functional interactions between RC21v3 and other CaCdr1p inhibitors to within the membrane sector and cell surface of CaCdr1p.
Results
Discovery of RC21
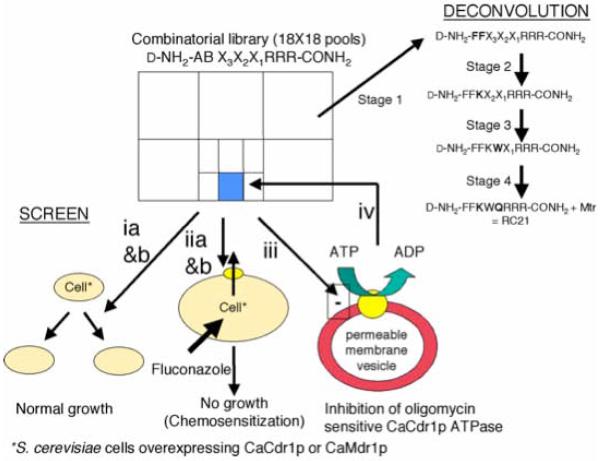
A system that hyper-expresses functional fungal drug efflux pumps in S. cerevisiae was used to screen a surface-targeting d-octapeptide combinatorial library for chemosensitizers of multidrug efflux mediated by CaCdr1p (Lamping et al., 2007, Niimi et al., 2004). The screening system (Fig. 1) used a yeast strain (AD/CDR1B, Table 1) that over-expressed functional CaCdr1p at ~30% of plasma membrane protein (Lamping et al., 2007) to identify a specific chemosensitizer of FLC resistance caused by this efflux pump. The primary screen (Fig. 1, ia & iia) visualized zones of strong growth inhibition in agarose diffusion assays caused by about 10% of the 324 library pools in the presence of FLC (Fig. 1, iia) that were considerably larger than in the absence of FLC (Fig. 1, ia). These pools included the d-NH2-FFX3X2X1RRR-CONH2 pool which give a barely detectable zone of growth inhibition in the absence of FLC (Fig. 2A). During deconvolution of the d-NH2-FFX3X2X1RRR-CONH2 pool, peptides with the most potent amino acids in the X3, X2 and X1 positions gave negligible zones of growth inhibition in the absence of FLC and even larger zones of growth inhibition in the presence FLC, despite the use of less d-octapeptide (Fig 2A).
Fig. 1.
Screens and counterscreens for ABC transporter chemosensitizer discovery in a D-octapeptide combinatorial library. Strain AD/CaCDR1 was used in cell-based primary screens (ia, growth inhibition screen in the absence of FLC; iia, chemosensitization screen in the presence of 40 μg ml−1 FLC) and to provide an enriched plasma membrane fraction containing CaCdr1p for the in vitro secondary screen (iii, oligomycin sensitive CaCdr1p ATPase activity screen). Strain AD/CaMDR1 was used in cell-based counterscreens (ib, a growth inhibition screen in the absence of FLC; iib, a chemosensitization screen in the presence of 10 μg ml−1 FLC). Screening was carried out using the following steps. 1) The combinatorial library pool d-NH2-FFX3X2X1RRR-CONH2 was identified as giving strong chemosensitization of AD/CDR1 and the lowest CaMDR1/CaCDR1 chemosensitization ratio based on the relative sizes of the growth inhibition zones in agarose diffusion assays for chemosensitization of AD/CaCDR1 to FLC (40 μg ml−1) and AD/CaMDR1 to FLC (10 μg ml−1). 2) This pool also gave strong inhibition the in vitro oligomycin sensitive ATPase activity of CaCdr1p. 3) The process was repeated (iv) with resynthesized subpools (stages 2-4). HPLC purified peptides, quality assured using mass spectrometry analysis, were used to ensure that corrected lead had been identified in stage 4.
Table 1. Yeast strains used in this study.
| Species | Strain | Source of cloned gene |
Genotype/phenotype | Reference/source |
|---|---|---|---|---|
|
Saccharomyces
cerevisiae |
AD12345678u− (AD1-8u−) |
Matαpdr1-3 ura3 his1 yor1Δ::hisG
snq2Δ::hisGpdr10Δ::hisG pdruΔ:: hisGycf1Δ::hisG pdr3Δ::hisG pdr15 Δ::hisG pdr5Δ::hisG |
Lamping et al., 2007) | |
| AD/pABC3 | plasmid vector | AD1-8u−, pdr5Δ::URA3 | Lamping et al., 2007) | |
| AD/CaMDR1 | CaATCC10261 | AD1-8u−, pdr5Δ::MDR1-URA3 | (Lamping et al., 2007) | |
| AD/CaCDR1A a (7D) |
CaATCC10261 | AD1-8u−, pdr5Δ::CDR1A-URA3 | Lamping et al., 2007) | |
| AD/CaCDR1Ba (6C) |
CaATCC10261 | AD1-8u−, pdr5Δ::CDR1B-URA3 | Lamping et al., 2007) | |
| AD1002 | CaATCC10261 | AD1-8u−, pdr5Δ::CDR1A-URA3 | Lamping et al., 2007) | |
| AD/CaCDR2Ba | CaATCC10261 | AD1-8u−, pdr5Δ::CDR2B-URA3 | Lamping et al., 2007) | |
| AD/ERG11 | CaATCC10261 | AD1-8u−, pdr5Δ::ERG11-URA3 | Lamping et al., 2007) | |
| AD/CgCDR1 | CgCBS138 | AD1-8u−, pdr5Δ::CgCDR1-URA3 | Lamping et al., 2007) | |
| AD/CgCDR2 | CgCBS138 | AD1-8u−, pdr5Δ::CgPDH1-URA3 | Lamping et al., 2007) | |
| AD/CnMDR1 | CnCDC551 | AD1-8u−, pdr5Δ:: CnMDR1-URA3 | Lamping et al., 2007) | |
| AD/CkABC1 | CkB2399 | AD1-8u−, pdr5Δ:: CkABC1-URA3 | Lamping et al., 2007) | |
| AD124567 |
Matαpdr1-3 ura3 his1 yor1Δ::hisG
snq2Δ::hisG pdr10 Δ::hisG pdru Δ::hisG ycf1 Δ::hisG pdr3Δ::hisG |
(Decottignies et al., 1998) | ||
|
Candida
albicans |
ATCC 10261 | American Type Culture Collection | ||
| CIA4 | Δ ura3::imm434/Δura3::imm434 | (Fonzi and Irwin, 1993) | ||
| MML33 |
Δ
ura3::imm434/
Δ
ura3::imm434,
Δ cdr1::hisG/ Δ cdr1::hisG |
Nippon Roche (Kanagawa, Japan) |
||
| TIMM3163 | Azole-resistant | (Maebashi et al., 2001) Teikyo University (Tokyo, Japan) |
||
| TIMM3165 | Azole-resistant | (Maebashi et al., 2001) Teikyo University (Tokyo, Japan) |
||
| B59630 | Azole-resistant | (Albertson et al., 1996) F. Odds (Aberdeen, UK) |
||
| IFO1060 | Azole-susceptible | Institute for Fermentation (Osaka, Japan) |
||
| KB | Azole-resistant | (Albertson et al., 1996) Pfizer (Sandwich, UK) |
||
| MML610 | Azole-susceptible TL1 | (Holmes et al., 2008) T. White (Seattle, WA) |
||
| MML611 | Azole-resistant TL3 | (Holmes et al., 2008) T. White (Seattle, WA) |
||
| C. glabrata | CBS138 | Centraalbureau voor Schimmelcultures (Utrecht, The Netherlands) |
||
| C. krusei | B2399 | Centre for Disease Control (Atlanta, GA) |
Separate alleles of each transporter gene from C. albicans ATCC 10261 are designated A or B (Holmes et al., 2006).
Fig. 2.
Discovery of RC21 by deconvolution of a d-octapeptide combinatorial library. A, Agarose diffusion assays used to screen for chemosensitization of AD/CaCDR1B to FLC. B, Agarose diffusion assay used as a counterscreen by comparing chemosensitization to FLC of AD/CaMDR1 with AD/CaCDR1B.
A counterscreen (Fig. 1, ib and iib), which employed a strain over-expressing the Major Facilitator Superfamily transporter Mdr1p from C. albicans (Lamping et al., 2007), was used to eliminate compounds that gave increased susceptibility to FLC through effects on energy metabolism or mechanisms of drug efflux dependent on the generation of a plasma membrane electrochemical gradient. All library pools which chemosensitized yeast over-expressing CaCdr1p to FLC also chemosensitized yeast over-expressing Mdr1p. Of the 30 pools that most strongly chemosensitized CaCdr1p, the d-NH2-FFX3X2X1RRR-CONH2 pool gave the lowest CaMdr1p/CaCdr1p chemosensitization ratio based on the relative sizes of the growth inhibition zones. The counterscreen selected against this undesirable activity (i.e growth inhibition of CaMdr1p expressing cells in the presence of FLC) in subpools of the d-NH2-FFX3X2X1RRR-CONH2 pool (Fig. 2B). A secondary screen (Fig. 1, iii) used inhibition of the oligomycin-sensitive ATPase activity of CaCdr1p in isolated plasma membrane preparations from strain AD/CDR1B to confirm target-directed activity in selected library pools, subpools or isolated compounds (Table 2) (Niimi et al., 2004). Of the four candidate stage 1 pools assessed, the d-NH2-FFX3X2X1RRR-CONH2 pool gave the strongest inhibitory effect on CaCdr1p ATPase activity (data not shown).
Table 2. In vitro inhibition of Cdr1p and Pma1p ATPase activity.
| IC50 (μM) |
||||
|---|---|---|---|---|
| Stage of deconvolution |
Formula | Cdr1p ATPase |
Pma1p ATPase |
|
| 1 | d-NH2-FFX3X2X1RRR-CONH2 | 20 | 5 | |
| 2 | d-NH2-FFKX2X1RRR-CONH2 | 10 | 5 | |
| 3 | d-NH2-FFKWX1RRR -CONH2 | 4 | 5 | |
| 4 | d-NH2-FFKWQRRR-CONH2 | 2 | 5 | |
| 5 | RC1 | d-NH2-FFKWQRRR-CONH2 | >160 | 5 |
| 5 | RC21 | d-NH2-FFKWQRRR-CONH2 + Mtr* | 1.3 | 5 |
Cdr1p and Pma1p activities were measured in the presence of Stage 1 to Stage 5 peptide pools, subpools and purified peptides as described under Experimental Procedures. Values are the mean of two separate measurements which did not vary by more than 5%.
Mtr: 4-Methoxy-2,3,6-trimethylbenzensulfonyl.
The cycle of screen, counterscreen and in vitro ATPase assay was repeated three more times on resynthesized subpools to deconvolute the library and sequentially identify the primary sequence of the lead peptide RC21 as d-NH2-FFKWQRRR-CONH2 (Fig. 1, Stages 1 - 4). RC21 chemosensitized CaCdr1p-overpressing S. cerevisiae to FLC but did not inhibit fungal growth in the absence of FLC (Fig. 2A). The deconvolution also found that a 4-methoxy-2,3,6-trimethylbenzenesulphonyl (Mtr) derivative, but not the naked peptide RC1 (Table 3), was the active principal (Fig. 2A). The requirement for the Mtr group, an arginine sidechain blocking agent used in solid phase peptide synthesis (Niimi et al., 2004), was identified using HPLC purified peptides that were characterized by mass spectrometry after the 4th stage of the combinatorial library deconvolution (Fig. 2A).
Table 3. Peptides and peptide derivatives used in the present study.
| Name | Structures |
|---|---|
| RC1 | d-NH2-FFKWQRRR-CONH2 |
| RC21 | d-NH2-FFKW(Mtr)QRRR-CONH2 |
| d-NH2-FFKWQR(Mtr)RR-CONH2 | |
| d-NH2-FFKWQRR(Mtr)R-CONH2 | |
| d-NH2-FFKWQRRR(Mtr)-CONH2 | |
| RC21v1 | d-NH2-FFKW(Mtr)QRRR-CONH2 |
| RC21v2 | d-NH2-FFKWQR(Mtr)RR-CONH2 |
| d-NH2-FFKWQRR(Mtr)R-CONH2 | |
| d-NH2-FFKWQRRR(Mtr)-CONH2 | |
| RC21v3 | d-NH2-FFKWQR(Mtr)RR-CONH2 |
| RC22 | d-Acetyl-NH-FFKW(Mtr)QRRR-CONH2 |
| d-Acetyl-NH-FFKWQR(Mtr)RR-CONH2 | |
| d-Acetyl-NH-FFKWQRR(Mtr)R-CONH2 | |
| d-Acetyl-NH-FFKWQRRR(Mtr)-CONH2 | |
| RC23 | d-NH2-FFHWQR(Mtr)RR-CONH2 |
| d-NH2-FFHWQRR(Mtr)R-CONH2 | |
| d-NH2-FFHWQRRR(Mtr)-CONH2 | |
| RC24 | d-NH2-FFHWQR(Mtr)R(Mtr)R-CONH2 |
| d-NH2-FFHWQRR(Mtr)R(Mtr)-CONH2 | |
| d-NH2-FFHWQR(Mtr)RR(Mtr)-CONH2 |
The surface active d-octapeptide pools and subpools identified as inhibitors of the ATPase activity of CaCdr1p were also tested for their effects on the in vitro enzyme activity of the plasma membrane proton pump (Pma1p), a key player in yeast energy supply (Table 2). The IC50 for inhibition of Pma1p ATPase activity (5 μM) was unaffected by the stage of library deconvolution. In contrast, there was a 16-fold increase in potency against CaCdr1p ATPase activity (IC50 = 20 → 1.3 μM) during the deconvolution used to identify RC21 in the d-NH2-FFX3X2X1RRR-CONH2 pool. Furthermore, the naked peptide RC1 inhibited Pma1p, but inhibition of CaCdr1p required the Mtr-derivatized d-octapeptide RC21. Inhibition of the oligomycin-sensitive ATPase activity of CaCdr1p was unaffected by preincubation of the membranes in the presence of ATP (data not shown), indicating that RC21 is a not a competitive inhibitor of CaCdr1p.
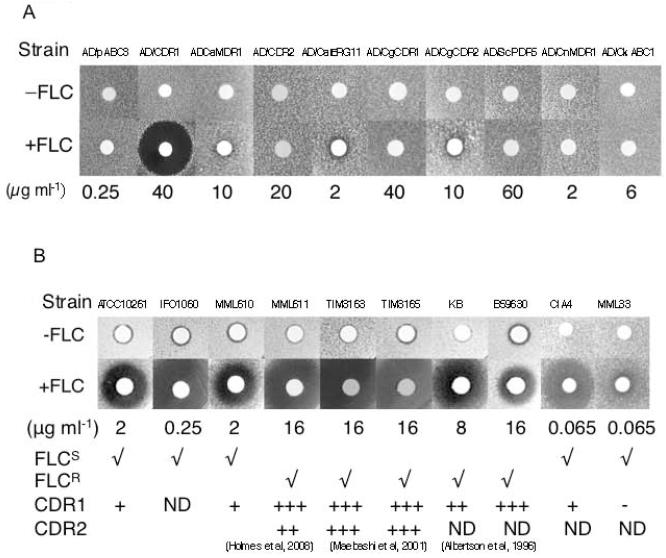
RC21 inhibited FLC resistance mediated by the over-expression in S. cerevisiae of CaCdr1p but not by the over-expression of the following ABC proteins: S. cerevisiae Pdr5p, C. albicans Cdr2p, Mdr1p or Erg11p, Candida glabrata Cdr1p and Pdh1p, Candida krusei Abc1p or Cryptococcus neoformans Mdr1p (agarose diffusion assays in Fig. 3A and selected checkerboard assays in Fig 1S). As expected, RC21 had no effect on the growth of the strain AD/pABC3 in the presence or absence of FLC at 1/4 MIC (Fig 3A). RC21 conferred increased FLC susceptibility on the FLC-susceptible C. albicans strain CIA4 but had a minor effect on the derivative strain MML33 (from Dr M. Arisawa, Nippon Roche), which has been selectively deleted of both genomic copies of CDR1 using the ura-blaster method (Alani et al., 1987). This was shown using both agarose diffusion assays (Fig 3B) and checkerboard assays (Fig S2). RC21 increased the FLC susceptibility of both FLC-susceptible (CIA4, ATCC10261, IFO1060, MML610) and FLC-resistant (MML611, TIMM3163, TIMM3165, KB and B59630) clinical isolates of C. albicans (Fig. 3B), including isolates known to over-express CaCdr1p alone or both CaCdr1p and CaCdr2p. MML611 and its parental FLC-sensitive strain MML610 were supplied by Dr T.H. White, Seattle (Holmes et al., 2008). These experiments suggested that RC21 in low nanomole quantities is a specific inhibitor CaCdr1p with potential clinical relevance.
Fig. 3.
Specificity of RC21 as a chemosensitizer. The drug efflux pump substrate FLC was used at concentrations that were 4x lower than the FLC MIC for each strain. A, Agarose diffusion assays showing RC21 chemosensitization to FLC of S. cerevisiae cells expressing drug efflux pumps or CaErg11p. Crude RC21 (not HPLC purified) was applied at 6 nmole/disk in the presence or absence of the indicated concentration of FLC. B, Agarose diffusion assays showing RC21 chemosensitization to FLC of C. albicans clinical isolates that overexpress CaCdr1p. Crude RC21 was applied at 48 nmole/disk in the presence or absence of the indicated concentration of FLC for all strains except CIA4 and MML33, for which 7.5 nmole of RC21 was applied. Strains ATC10261, IFO1060, MML610 and CIA4 are FLC susceptible strains that express CaCdr1p at low levels (Holmes et al., 2008). MML31 is derived from CIA4 by deletion of both alleles of CaCdr1p.
Mode of action of RC21
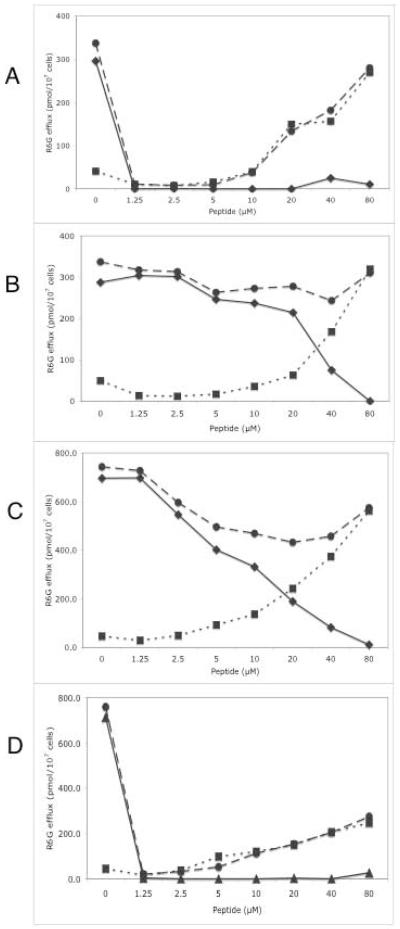
Chemical properties of RC21 that contribute to its biological activity are illustrated in Fig. 4A,B. Two HPLC-purified Mtr derivatives of RC21 were characterized by mass spectrometry and spectrophotometry. One derivative (RC21v1, MW = 1437 Da) had an absorbance peak at 300 nm characteristic of Mtr modified tryptophan (W). The other derivative (RC21v2, MW = 1437 Da, with an absorbance peak at 280 nm characteristic of unmodified W) has a single Mtr group attached to one of its three C-terminal arginine residues. Of these two derivative classes, only RC21v2 at ~5.5 nmol per disk chemosensitized yeast over-expressing CaCdr1p to FLC (Fig. 4A). No additive or synergistic sensitization activity was observed when RC21v2 was supplemented with RC21v1, confirming that RC21v1 is inactive. Cells overexpressing CaMdr1p (Fig. 4A) were not chemosensitized by any of these derivatives. The comparable l-derivatives of RC21 (Fig. 4B), either alone or together with RC21v1, did not chemosensitize cells overexpressing CaCdr1p to FLC or increase the effect of RC21v2. That is, d-RC21v2 (~6 nmole/disk), but not its stereoisomer l-RC21v2 (~28 nmole/disk), chemosensitized cells overexpressing CaCdr1p to FLC. The stereospecificity of RC21v2 is consistent with the active principal in RC21 targeting a protein rather than a general component of the plasma membrane. In contrast, high concentrations of the l-enantiomers of RC21 weakly chemosensitized yeast over-expressing CaMdr1p to FLC (Fig. 4B).
Fig. 4.
Stereospecificity of RC21 and localization of the Mtr-group in RC21 required for chemosensitization. Crude D-RC21 was compared with the HPLC purified peptides d- and l-RC21v1 and v2 quantitated spectrophotometrically. A, each peptide was used at 6 nmole/disk in the presence and absence of the indicated concentrations of FLC. The Mtr group required for AD/CaCDR1B chemosensitization is associated with an arginine residue (R6, R7 or R8) and not the tryptophan residue (W4). B, d-RC21 was used at 6 nmol per disk and the HPLC purified L-peptides were used at 28 nmole/disk. D-RC21 but not L-RC21 chemosensitized AD/CaCDR1B to FLC.
At concentrations <20 μM RC1 and RC21 had no significant effect on the rate of uptake of R6G in de-energised control AD/pABC3 or AD/CaCDR1 cells (data not shown). RC21 or RC21v2 at 1.25 - <20 μM fully and specifically inhibited glucose-dependent CaCdr1p-mediated rhodamine 6G efflux in whole AD/CaCDR1 cells (Fig. 5A, D). Higher concentrations of RC21 and RC21v2 caused non-specific energy-independent release of R6G. At least 40 μM RC1 or RC21v1 was required to obtain a significant effect on R6G efflux in the same assay (Fig. 5B,C). The effects of these higher RC1 and RC21v1 concentrations on R6G efflux were independent of glucose energization of efflux and therefore indicated non-specific membrane permeabilization. The whole cell effects of RC1 at >40 μM and RC21 at <20 μM therefore involve different processes. This result, plus the absence of growth inhibition of yeast by RC21 and RC21v2, imply that the chemosensitizing activity of these two compounds on intact cells was unrelated to the in vitro inhibition of Pma1p by RC1 and RC21 (IC50 = 5 μM, Table 2). We infer that the site on Pma1p affected by RC1 and RC21 in vitro is not accessible in whole cells treated with these peptides.
Fig. 5.
Localization of Mtr group on RC21 required for inhibition of R6G pumping by CaCdr1p. R6G efflux in the presence and absence of glucose was used to identify the glucose-dependent efflux of R6G from AD/CDR1 cells exposed to the indicated concentrations of: A, the mixed Mtr derivative crude RC21; B, the HPLC-purified peptide RC1; C, the HPLC-purified derivative RC21v1 (W4-Mtr); D, the HPLC-purified derivatives RC21v2 (R6-R8-Mtr). The efflux of R6G from preloaded cells into the supernatant fraction was measured as described under Experimental Procedures in the presence  or absence
or absence  of glucose. The glucose-sensitive efflux remaining after RC21 treatment is shown
of glucose. The glucose-sensitive efflux remaining after RC21 treatment is shown  .
.
Other experiments have shown that RC21 at 1.25 μM inhibited glucose-dependent R6G efflux of a S. cerevisiae strain overexpressing CaCdr1p by 90% but had no effect on a strain overexpressing CaCdr2p (Holmes et al., 2008). Furthermore, RC21 at 0.62 μM inhibited glucose-dependent R6G efflux from the FLC-resistant C. albicans MML611 clinical isolate by 90% and detectably inhibited the 20-fold lower levels of efflux from its FLC-susceptible parental strain MML610 (Holmes et al., 2008). The specificity of RC21 inhibition of drug efflux mediated by CaCdr1p in this strain confirms the Cdr1p-specific chemosensitization to FLC obtained using drug diffusion assays (Fig. 3B). Despite evidence of significant expression of CaCdr2p in strain MML611, CaCdr1p contributes much more to drug efflux than CaCdr2p (Holmes et al., 2008).
RC21 structure and function
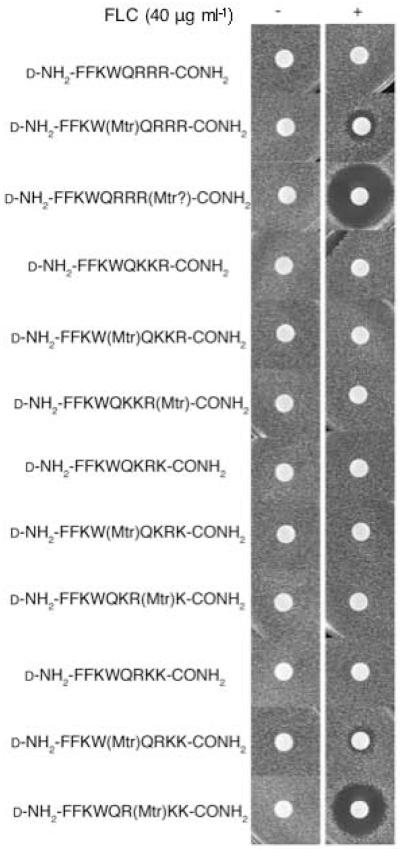
The location of the functional Mtr group in RC21 was explored using purified derivatives in which pairs of residues in the C-terminal tri-arginine motif were changed to lysine (Table 3). Each derivative contained a single arginine sidechain within the tri-arginine motif that could retain an Mtr group after manual solid phase synthesis. Agarose diffusion assays using strain AD/CaCDR1 showed that neither naked peptides nor peptides Mtr-derivatized at W4 had chemosensitizing activity (Fig. 6). Only d-NH2-FFKWQR(Mtr)KK-CONH2 with Mtr derivitization of the most N-terminal arginine (R6) in the C-terminal tri-arginine motif was found to have chemosensitizing activity (Fig. 6). A combination of peptide synthesis chemistries (Fmoc and Pdf) that employ different arginine side chain blocking agents was subsequently used to obtain RC21v3 (d-NH2-FFHWQR(Mtr)RR-CONH2) which has a chemosensitizing activity identical to RC21v2 (data not shown).
Fig. 6.
Chemosensitization of AD/CaCDR1 requires an Mtr group on the R6 sidechain of RC21. Agarose diffusion assays were used to measure the potency of HPLC-purified congeners of RC21 and their derivatives. Chemosensitization in agarose diffusion assays was carried out as described under Experimental Procedures using strain AD/CaCDR1. Crude RC21 or HPLC purified peptide was applied at 6 nmole/disk in the presence or absence of FLC (40 μg ml−1).
The structure of RC21 and its modification by the Mtr group provided significant challenges in determining structure-activity relations. Tetramethylrhodamine isothiocyanate (TRITC) labeling inactivated RC21, either because the amino terminus or the lysine side chain was essential for activity, or because introduction of TRITC blocked binding to CaCdr1p. Additional derivatives of RC21 were therefore synthesized and tested (Table 3). Acetylation of the RC21 amino terminus (RC22) or conversion of RC21’s K3 residue to histidine (RC23) did not affect chemosensitization of CaCdr1p-dependent FLC resistance (data not shown). An amino-terminal positive charge was therefore not necessary for RC21 activity and the ε-amino group of lysine could be replaced with an imidazole that is not susceptible to TRITC derivitization. One Mtr group (RC23) was required for chemosensitizing activity but two Mtr groups (RC24) ablated activity (data not shown). Two positive charges were therefore required within the C-terminal tri-arginine motif of RC23 for activity. N-terminal derivatization of RC23 with TRITC gave an inactive adduct that was shown by confocal microscopy to be excluded from cells, as were TRITC-labelled RC21 and RC22 (Fig. S3). Either the size and/or charge of these N-terminal and/or sidechain modifications ablated chemosensitizer activity i.e. TRITC contains three aromatic rings and introduces four additional negative charges into RC21 and two additional negative charges into RC22 and RC23.
Toxicity and efficacy in a mouse model of oral candidiasis
RC1, RC21 and RC21v2 (≤100 μM) did not lyse red blood cells, in contrast to amphotericin B (AMB) which completely lysed these cells at 3 μM (Fig. 7A). RC21v2 at 100 μM did not affect the growth or viability of a human epithelial cell line (Fig. 7B). AMB rendered these cells inviable at 15 μM. We also believe that RC21v3 is likely to be stable to endogenous proteases because it is a C-terminal amidated D-peptide and may therefore be more suitable for animal studies than conventional l-peptides. RC21v3 was found to chemosensitize C. albicans strains MML610 and MML611 to FLC in a mouse model of oral candidiasis (Hayama et al., 2012). Tissue burden and symptom scores were reduced signficantly at concentrations of RC21v3 that were found to be non-toxic when administered orally.
Fig. 7.
Toxicity of RC21. A, Crude RC21, and its HPLC purified peptide backbone RC1 and HPLC-purified derivatives RC21v2 (≤100 μM) did not hemolyse human red blood cells unlike amphotericin B (AMB) which caused complete hemolysis at 3 μM. B, RC21v2 (≤100 μM) did not kill cultured HEp2 cells.
Intragenic suppressors of RC21-mediated chemosensitization
The interaction between RC21 and CaCdr1p was explored using molecular genetic analysis of spontaneous yeast suppressor mutants that were chemosensitization-resistant to either RC21 or RC21v2 (Table 4). These mutants appeared after 7 days within the growth inhibitory zones obtained in agarose diffusion chemosensitization tests of the AD/CaCDR1 strain in the presence of 40 μg ml−1 FLC (Fig. 8A). Each isolate appeared at a low frequency consistent with spontaneous mutation, possessed a specific level of resistance to RC21 or RC21v2, and their phenotypes were stable both in agar diffusion assays and checkerboard liquid susceptibility assays (Fig. 8A, B). The phenotype of resistance to RC21 was maintained in all isolates after ten passages on non-selective YPD agar plates (data not shown).
Table 4. S. cerevisiae AD/CaCDR1 variants resistant to FLC chemosensitization by RC21.
| Variant | Selected using |
Resistance | Amino acid change |
Predicted location | Strain name (Tables 5 & 6) |
|---|---|---|---|---|---|
| C1 | RC21 | High | Q1226K | External loop 4 | EL4-1 |
| C2 | RC21 | High | G547R | TMD1, TMS2 | EL1/TM2 |
| R1 | RC21v2 | High | Q714K | External loop 3 | EL3 |
| R2 | RC21v2 | High | Q1445K | External loop 6 | EL6 |
| R3 | RC21v2 | High | Q1226K | External loop 4 | |
| R4 | RC21v2 | High | Q1226K | External loop 4 | |
| F1 | RC21 | High | Q1226K | External loop 4 | |
| F2 | RC21 | High | Q1226K | External loop 4 | |
| F3 | RC21 | High | Q714K | External loop 3 | |
| HE2 | RC21 | High | Q1226R | External loop 4 | EL4-2 |
| E1 | RC21 | Moderate | V674F | TMD1, TMS5 | TM5 |
| E2 | RC21 | Moderate | V674F | TMD1, TMS5 |
Chemosensitization resistant variants with different capital letters were isolated in separate experiments. TMD, transmembrane domain; TMS, transmembrane segme
Fig. 8.
Molecular genetic analysis of RC21-resistant variants. A, Selection and stability of RC21 chemosensitization-resistant variants. Examples of RC21 variants (strongly resistant C1 and C2 and weakly resistant E1 and E2) selected from within the zone of inhibition and retested in an agarose diffusion assays are shown. B, Checkerboard assay showing suppression of synergy between RC21 chemosensitization of AD/CaCDR1B to FLC by the RC21 chemosensitization-resistant variant AD/CDR1B-C2. C, Mutations in RC21 chemosensitization-resistant variants occur within the CaCDR1B-URA3 transformation cassette. The CaCDR1B-Ura3 transformation cassette was obtained by PCR of the genomic DNA from strain AD/CDR1-C2 and used to transform the null parental strain AD1-8u− to Ura+. Selection on CSM-ura medium gave the strain AD/CDR1-C2.1 which showed RC21 chemosensitization-resistance identical to AD/CDR1-C2.
The PDR5 region, containing the CaCDR1 ORF and URA3 gene, was prepared by PCR amplification of the genomic DNA from 12 RC21 or RC21v2 resistant variants and used to transform the host strain AD1-8u− to Ura+ (Fig. 8C) (Wada et al., 2005). The ORF from each variant (e.g. AD/CaCDR1-C2) gave Ura+ transformants that were FLCR. Each transformant (e.g. AD/CaCDR1-C2.1) had a phenotype identical to the original mutant and showed resistance to both RC21 and RC21v2 in the presence of FLC. These properties suggested that the resistant variants were the result of suppressor mutations found within the CaCDR1 ORF. DNA sequence analysis of each CaCDR1 ORF cloned identified a single nucleotide mutation that modified the amino acid sequence of CaCdr1p within a predicted extracellular domain or a predicted transmembrane domain near the extracellular surface (Table 4 and Fig. 9). Five separate mutations were found within or adjacent to the predicted extracellular loops 1, 3, 4 and 6 of CaCdr1p. Each of these mutations replaced the native amino acid in the CaCdr1p ectodomain with a positively charged amino acid i.e. G547R at the N-terminal end of TMS2, Q714K in Extracellular Loop 3 (EL3), Q1226K and Q1226R in EL4, and Q1445K in EL6 according a model of CaCdr1p based on the Pdr5p model of Rutledge et al (Rutledge et al., 2010). The active principal of RC21 should exhibit four positive charges at physiological pH (N-terminal amino group, K3 amino sidechain and the R7 plus R8 guanidino sidechains), yet the introduction of a single positive charge into CaCdr1p overcame the inhibitory interaction between RC21 and the CaCdr1p ectodomain. The other two suppressor mutants gave the identical amino acid change V674F in CaCdr1p. This mutation was adjacent to the predicted ectodomain end of TMS5 in TMD1. Although the V674F mutation was the weakest of the suppressor mutations analyzed (Table 4) the introduction of the aromatic F residue in the TMD1 significantly reduced the effects of the RC21v2 on CaCdr1p. We note that the suppressor mutations Q1226K, Q714K and V674F were recovered at least twice in separate experiments. This suggests that saturation of spontaneous RC21suppressor mutations was being approached and that the bulk of the mutations affecting peptide binding may have been identified.
Fig. 9.
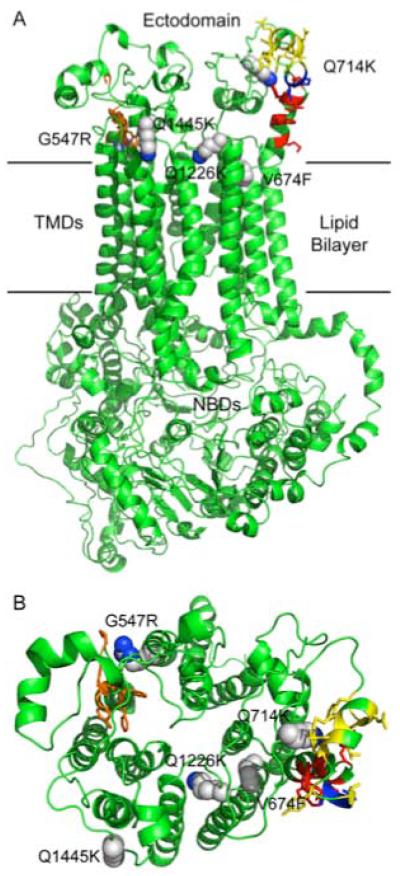
A map of RC21 suppressor mutations using a CaCdr1p homology model. The open (no ATP bound) CaCdr1p model, viewed from the side (A) and from the periplasm (B), was generated in Modeller 9v7 (Eswar et al., 2006) using the S. cerevisiae Pdr5p model of Rutledge and colleagues (Rutledge et al., 2010). A sequence alignment was generated for CaCdr1p and ScPdr5p using clustalW. Coordinates for the Pdr5p model were kindly provided by Robert Rutledge. Conserved amino acid sidechains of the PDR motif A are shown in red, the connecting sequence in blue and the PDR motif B in yellow. Conserved amino acid sidechains of the EL6 hydrophobic motif are shown in orange. Amino acid side chains of 5 of the suppressor mutants are shown as spheres, with carbons in grey and nitrogens in blue.
We considered the possibility that the Mtr group of RC21 is a pseudosubstrate of Cdr1p that cannot flip between the inner and outer layers of the lipid bilayer because of the attached positively charged peptide. Since the V674F mutation blocked the activity of RC21 while modestly diminishing the conferral of resistance to FLC in overexpressed CaCdr1p (Table 5), we first envisaged that the mutant phenylalanine might directly block insertion of the RC21 Mtr into the substrate exit site for FLC. However, the naked peptide RC1 failed to chemosensitize the V674F mutant to FLC. It is therefore more likely that the V674F mutation indirectly blocked RC21 binding to the pump ectodomain by altering the conformation of CaCdr1p TMD1. Our homology model of Cdr1p (Fig. 9) shows CaCdr1p V674 to be located immediately behind TMS6 which borders the efflux channel. The insertion of the bulky F residue could position the extracytoplasmic end of TMS6 slightly deeper into the efflux channel and restrict the closed conformation of the enzyme.
Table 5. Effects of xenobiotics on chemosensitization-resistant variant strains.
| MIC* |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Strain |
AD/pABC3 | AD/CDR1 | EL1/TM2 | EL3 | EL4-1 | EL4-2 | EL6 | TM5 | |
| Compound | Control | Wild type | G547R | Q714K | Q1226K | Q1226R | Q1445R | V674F | |
| Azole substrates | Fluconazole | 0.5 | 240 | 100 | 240 | 80 | 240 | 240 | 160 |
| Voriconazole | 0.156 | 8 | 2.5 | 8 | 2.5 | 6 | 10 | 4 | |
| Ketoconazole | 0.156 | 4 | 1 | 4 | 1 | 4 | 4 | 3 | |
| Miconazole | 0.156 | 2.5 | 1 | 2.5 | 1 | 3 | 3 | 2 | |
| Fluorescent dye substrates | Rhodamine 6G | 1 | 75 | 50 | 75 | 50 | 50 | 75 | 50 |
| Rhodamine 123 | 8 | 300 | 150 | 200 | 150 | 200 | 200 | 150 | |
| Xenobiotic substrates | Cycloheximide | 0.008 | 1 | 0.5 | 1 | 0.5 | 1 | 2 | 1 |
| Trifluoperazine | 5 | 40 | 40 | 40 | 40 | 40 | 80 | 20 | |
| Xenobotic substrate/inhibitor | FK506 | 0.313 | 40 | 40 | 40 | 40 | 40 | 40 | 40 |
| Pump inhibitors | RC21 | >20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 |
| Milbemycin α11 | 4 | 4 | 8 | 4 | 8 | 4 | 4 | >8 | |
| Milbemycin α20 | >8 | >8 | >8 | >8 | >8 | >8 | >8 | >8 | |
| Milbemycin α25 | 8 | 8 | 8 | >8 | 8 | >8 | >8 | >8 | |
| Milbemycin α26 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | |
| Milbemycin β9 | 8 | >8 | >8 | 8 | >8 | >8 | >8 | >8 | |
| Milbemycin β11 | >8 | >8 | >8 | >8 | >8 | >8 | >8 | >8 | |
| Enniatin | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | |
| Other | Micafungin | 0.25 | 0.125 | 0.125 | 0.25 | 0.125 | 0.125 | 0.25 | 0.125 |
MIC was determined using CSM medium (pH 7.0) after incubation at 30°C for 48 h with shaking.
MIC: Azoles, μg ml−1 (I80); RC21 and rhodamines, μM; milbemycins and micafungin, μg ml−1 (I95); cycloheximide, enniatin and FK506, μg ml−1; trifluoperazine, μM (I95).
RC21/RC21v2 suppressor mutations allowed preliminary genetic and biochemical mapping of a putative RC21v3 binding site, the identification of substrates and other chemosensitizers that interact with this site, and probing of the peptide’s mode of action. First, the possibility that CaCdr1p in the RC21 suppressor mutants was mislocalized or severely inactivated was eliminated. Plasma membranes were isolated from representative suppressor strains and their polypeptide composition and the ATPase activity of Cdr1p compared with the parental strain AD/CaCDR1 (Fig. 10 A,B). The polypeptide composition of these plasma membrane preparations, as revealed by SDS-PAGE and staining with coomassie blue R250, appeared identical. The vanadate-, oligomycin- and FK506-sensitive ATPase activities of the CaCdr1p overexpressing preparations were essentially unaffected or, in one case only, reduced <40% due to the suppressor mutations. RC21v3 gave extensive monophasic inhibition (≥68% - 100% at 40 μM) of the ATPase activity of the wild type enzyme and each of the RC21 chemosensitization suppressor mutants tested, but the IC50 for each suppressor mutant was several-fold higher (3.6 – 9.4 μM) than the IC50 (1.1 μM) determined for wild type CaCdr1p (Fig. S5 and Table 1S). In addition, treatment of cells expressing CaCdr1p-EGFP with 2.5 μM RC21v3 did not modify cell growth rate, or the biosynthesis, folding and delivery of the enzyme to the plasma membrane as assessed by confocal microscopy (Fig. S4). The simplest model for RC21v3 action assumes that the peptide derivative interacts with a single surface accessible site in CaCdr1p at the plasma membrane and in a manner that indirectly affects the cytoplasmic ATPase activity of the NBDs. By occupying or modifying a low affinity binding site required for the efflux of substrates from the enzyme’s extracytoplasmic surface, the pathway for drug efflux would be blocked and ATP hydrolysis, the step completing the reaction cycle, prevented. Some suppressor strains showed slightly increased susceptibility to CaCdr1p xenobiotic substrates (Table 5). These substrates, which include a range of azole drugs (FLC, voriconazole (VCZ), ketoconazole (KTZ) and miconazole (MCZ), two rhodamine dyes (R6G and R123) and CHX, gave the expected reduced susceptibility when wild type CaCdr1p was overexpressed and slightly increased susceptibility in some suppressor mutants (G547R, Q1226K and V674F). This result appears consistent with deleterious interactions between suppressor mutations and the efflux pathway for the azoles, rhodamines and CHX. In contrast, susceptibility to the CaCdr1p substrates FK506 and trifluoperazine were unaltered by the RC21 chemosensitization suppressor mutations. This observation appears consistent with the presence of multiple efflux pathways within CaCdr1p, like its S. cerevisiae homologue Pdr5p (Golin et al., 2003). We also found that FK506, enniatin and two (α11 and β9) of the five milbemycins tested chemosensitized the parental AD/CaCDR1B strain to FLC more effectively than particular RC21 suppressor mutants (Table 6). Like FK506, α11 milbemycin, β9 milbemycin and enniatin inhibited FLC efflux by CaCdr1p. Unlike FK506, none of these xenobiotics appeared to be substrates of the efflux pump because CaCdr1p over-expression did not confer detectable resistance (Table 5). These observations identify specific amino acid residues on CaCdr1p, the modification of which affect the transport of pump substrates and the binding of the inhibitors FK506, enniatin and the milbemycins α11 and β9 (Fig. 9).
Fig. 10.
CaCdr1ps in RC21 chemosensitization-resistant variants have functional ATPase activities localized to the plasma membrane. A, SDS-PAGE of plasma membrane preparations obtained from AD1-8u−, AD/CaCDR1 and representative RC21 and RC21v2 suppressor mutants. B, CaCdr1p ATPase activity of plasma membrane preparations described in A. ATPase activity was measured as described under Experimental Procedures. The CaCdr1p ATPase activity measurements shown are for a representative of two independent experiments, with duplicate measurements of ATPase activity sensitive to vanadate (100 μM), oligomycin (20 μM) and FK506 (10 μg/ml) which did not vary by more than 5%.
Table 6. Chemosensitization of RC21 resistant variants to FLC.
| Strain | AD/pABC3 | AD/CDR1 | EL1/TM2 | EL4-1 | EI4-2 | EL3 | EL6 | TM5 |
|---|---|---|---|---|---|---|---|---|
| Compound (per disk) | Control | Wild type | G547R | Q1226K | Q1226R | Q714K | Q1445K | V674F |
| RC21 (6 nmoles) | − | ++++ | − | − | − | − | − | − |
| Milbemycin α11 (5 μg) | − | ++++ | + | + | + | +++ | +++ | ± |
| Milbemycin α20 (5 μg) | − | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| Milbemycin α25 (5 μg) | − | +++++ | ++++ | ++++ | ++++ | +++++ | +++++ | ++++ |
| Milbemycin β9 (5 μg) | − | ++ | − | ± | ± | +++ | + | − |
| Milbemycin β11 (5 μg) | − | +++ | ++ | ++ | ++ | +++ | +++ | ++ |
| FK506 (10 μg) | − | + | − | ± | ± | + | + | + |
| Enniatin (0.5 μg) | − | ++ | ± | + | ++ | + | + | ++ |
| Amphotericin B (20 μg) | − | − | − | − | − | − | − | − |
| DMSO (5 μl) | − | − | − | − | − | − | − | − |
Chemosensitization activies of the compounds were measured using the agarose diffusion assay as described under Experimental Procedures in the presence of 40 μg ml−1 FLC. Chemosensitization, + to +++++; marginal, ±; none, −.
Discussion
Screening strategy for a transporter-specific chemosensitizer
The d-NH2-FFKWQRRR-CONH2 amino acid sequence of RC21v3 was identified by deconvolution of a ~2 million-member d-octapeptide library for chemosensitizers that targeted specifically the ABC transporter CaCdr1p functionally overexpressed in S. cerevisiae. The specificity of the screen was enhanced using a counterscreen that selected against inhibitors of the MFS transporter CaMdr1p or other properties of the system that might modify yeast growth in the presence of FLC. The specificity of RC21v3 for its target but not other efflux pumps contrasts with the modest specificity obtained in our previous effort to identify an inhibitor of yeast multidrug efflux that targeted S. cerevisiae Pdr5p (Niimi et al., 2004). That study identified the d-octapeptide derivative KN20 (d-NH2-NWWKVRRR-CONH2 + Mtr) which includes an Mtr group attached to one of the arginine or tryptophan residues. Inhibition of ScPdr5p by KN20 was observed against a significant background of non-specific effects that included drug efflux mediated by MFS transporters in the presence of FLC. This lack of target specificity prompted the use of the CaMdr1p-based counterscreen in the present study.
Biochemical and physiological analysis of the mode of action of RC21 derivatives
Several classes of compound, including the immunosuppressant FK506, enniatin, ATP analogs and some milbemycins, inhibit drug efflux by the fungal ABC transporter CaCdr1p. As far as we are aware, the d-octapeptide derivative RC21 is the first compound shown to specifically inactivate both the ATPase and drug efflux activity of CaCdr1p via an inhibitory site likely to be at the extracytoplasmic face of the plasma membrane. The active principal of RC21, the d-octapeptide derivative RC21v3 (d-NH2-FFKWQR(Mtr)RR-CONH2), has three key components: (1) a tri-arginine motif which we have previously shown renders model peptides membrane impermeant and concentrates them at the fungal cell surface in association with phosphomannan (Niimi et al., 2004), (2) a targeting specificity for CaCdr1p in vivo that is resident in the peptide’s overall amino acid sequence and stereochemistry, plus (3) an essential Mtr group attached to the R6 guanidino group that was adventitiously retained by the peptide despite the deprotection strategy used during manual synthesis. We hypothesize that RC21v3 is guided to its inhibitory site in CaCdr1p because of the combined cationic and aromatic nature of the RC21v3 sequence. It appears to act on the drug efflux pathway for azole drugs, rhodamines and CHX as a surface-active, pseudosubstrate of the pump. The compound’s binding site appears to be shared with inhibitory sites for FK506, enniatin and some milbemycins.
Chemosensitization of the CaCdr1p overexpressing yeast strains to FLC by the fractions containing RC21 or the RC21v2 subfraction required an Mtr group attatched to a side chain guanidino group because the HPLC purified peptides RC1 (d-NH2-FFKWQRRR-CONH2) and RC21v1 (d-NH2-FFKW(Mtr)QRRR-CONH2) were both ineffective in agarose diffusion assays. Although RC21v2 and the naked peptide RC1 both inhibited in vitro the dominant ATPase (Pma1p) of yeast plasma membrane preparations (IC50 = 5 μM), RC21 strongly inhibited the ATPase activity of CaCdr1p (IC50 = 1.3 μM) while RC1 had little or no effect (IC50 >200μM). Since concentrations of RC21v2 that completely inhibited both CaCdr1p ATPase activity in vitro and the CaCdr1p-dependent R6G efflux activity of whole cells had no effect on the membrane electrochemical gradient-dependent (i.e. Pma1p-dependent) activity of CaMdr1p over-expressed in S. cerevisiae, it is unlikely that the peptide derivative crossed the plasma membrane and affected Pma1p in whole cells. These results, and the finding that TRITC-derivatives of RC21, RC22 and RC23 were all excluded from cells, support the idea that RC21 is a surface-active inhibitor of CaCdr1p. Comparison of the potency of HPLC-purified RC21 derivatives with the Mtr attached to specific residues in the C-terminal triarginine motif (R6, R7 or R8, and with the other two arginines replaced with lysine) showed that Mtr protection at R6 was required for chemosensitization of CaCdr1p-overexpressing cells to FLC. This was confirmed with a purifed RC21v3 (d-NH2-FFKWQR(Mtr)RR-CONH2) preparation obtained using an alternative peptide synthesis and protection/deprotection strategy. In addition, 24 derivatives of the K6 side chain amino group for the peptide d-NH2-FFHWQKRR-CONH2 were prepared. This peptide and all 24 derivatives were found to be inactive as chemosensitizers of cells over-expressing CaCdr1p (data not shown). Thus the R6 side chain and its Mtr substituant confer properties required for chemosensitization of FLC efflux mediated by CaCdr1p. These properties are not retained when a K6 amino side chain is used instead of an R6 guanidino side chain. Other studies showed that K3 can be replaced with H (RC23), the N-terminus of RC21 can be acetylated without affecting activity in agarose diffusion assays, while N-terminal derivatization of RC23 with TRITC caused complete loss of activity. Thus, the N-terminal amino group is not essential for activity, but peptide derivative bulk and charge are likely to be key in RC21 accessing its target. We have also found that the combinatorial library d-NH2-FFHX2X3RRR-CONH2 peptide subpool contains species that inhibit CaCdr1p, CgCdr1p, CgPdh1p and CkAbc1p drug efflux pumps but not CaCdr2p (data not shown). This finding has provided an entry point for the identification of d-octapeptide inhibitors of fungal drug efflux with broader target specificity than RC21 (Monk and Keniya, unpublished observations). Preliminary studies indicate that these compounds may have lower affinity for their targets than RC21 has for CaCdr1p.
Genetic analysis of the specificity and mode of action of RC21v3
The stereospecificity of RC21 and its derivatives as inhibitors of CaCdr1p activity in both whole cells and in vitro in plasma membrane preparations clearly indicated CaCdr1p as the target. Yeast cells over-expressing CaCdr1p-EGFP grew equally well and presented normal levels of the fluorescent chimera at the plasma membrane in the presence or absence of 2.5 μM RC21v3. We conclude, by extension, that RC21v3 does not affect the biosynthesis, folding or localization of CaCdr1p. Finally, RC21v2 at 1.25 μM fully inhibited glucose-dependent R6G efflux from cells over-expressing CaCdr1p after 5 minutes of preincubation, consistent with a direct interaction between the drug and the fully-folded efflux pump.
The isolation of stable, spontaneous RC21 and RC21v2 chemosensitization suppressor mutants confirmed that CaCdr1p is the target of its active principal, provided information about the mode of action of the inhibitory peptide and identified additional compounds that appear to operate in a similar way. Of the 12 suppressor mutants tested, complementation analysis using a parental host strain demonstrated that all mutations occurred within the PDR5::CaCDR1-URA3 transformation cassette of the AD/CaCDR1B strain (Wada et al., 2005). DNA sequence analysis of the genomic DNA from the transformants found that each suppressor mutation involved a single nucleotide change within the CaCDR1 ORF at one of 5 sites predicted to encode amino acids that are at or near the boundary between the membrane sector and the ectodomain (G547R, extracellular end of TM2; V674F, transmembrane segment 5), or within the ectodomain itself (Q714K, extracellular loop 3; Q1226K/R, extracellular loop 4; Q1445K, extracellular loop 6). None of the mutations appeared to affect significantly the biosynthesis and correct localization of CaCdr1p, and only modestly affected the in vitro ATPase activity of the enzyme at pH 7.5 or its inhibition by vanadate (100 μM), oligomycin (20 μM) or FK506 (20 μg ml−1). Furthermore, the CaCdr1p ATPase activity of each preparation showed a monophasic inhibition profile (Fig. S5), the IC50 of RC21v3 for each CaCdr1p ATPase tested was several fold greater than for wild type enzyme (Table 1S) and each enzyme was extensively inhibited by excess RC21v3. Thus RC21v3 appears to bind at a single site that blocks the complete reaction cycle, including drug efflux (FLC and R6G) and the ATP hydrolysis that resets the enzyme for drug binding. This was despite the suppressor mutations occurring distant from the NBDs and within or near the enzyme’s ectodomain in regions of relatively well conserved primary sequence.
The fungal ABC transporters most closely associated with antifungal drug resistance belong to the 42 member yeast PDR A subfamily of efflux pumps (Lamping et al., 2010). The residue corresponding to G547 in CaCdr1p TM2 is strongly conserved in the PDR A subfamily. It is either S, T, A or G, apart from one case of a C. Similarly, the residue corresponding to Q1226 in EL4 in CaCdr1p is strongly conserved but is S, T, N or K at low frequency. The residue equivalent to V674 in CaCdr1p TM5 is primarily a small hydrophobic residue (V, A, I) and rarely S, and so is also well conserved. It is C-terminal of a conserved F and is adjacent to a variably-charged, proline-containing sequence of 5 amino acids (ie uncharged FVIPTPS in CaCdr1p, FVLPIPY in CaCdr2p and FVLPTPS in CkABC1 and charged FAIPKKK in ScPdr5p, FAIPKGN in CgCdr1p, and FAIPRTK in CgCdr2p also known as CgPdh1p) that may change conformation during the reaction cycle (Rutledge et al., 2010). It is also immediately N-terminal of the PDR motif A and PDR motif B recently identified in EL3 of PDR efflux pumps (Lamping et al., 2010). CaCdr1p Q714 is just C-terminal of the same PDR A and B motifs. Within the PDR A subfamily of transporters, the residues that align with CaCdr1p Q714 are almost exclusively polar or charged (Q, D, N, E, R, K, H), with a single instance of an A. Q714 is 2 residues C-terminal of a conserved C, and is 3 and 5 residues amino-terminal of a conserved P and a conserved G, respectively. CaCdr1p Q1445 is beside a conserved C but the residue itself varies considerably among PDR A subfamily members with hydrophobic, polar and positively charged residues dominating. Collectively, these observations suggest but do not prove that RC21 binding involves structurally conserved features at or near the ectodomain of CaCdr1p. While RC21 appears specific for CaCdr1p, we speculate that conserved structures involved in RC21 binding might provide a broader spectrum target for chemosensitization.
Modeling of suppressor mutant interactions
We used an homology model of CaCdr1p (Fig. 9) based on the the open (no ATP bound) conformation model of Pdr5p (Rutledge et al., 2010) to identify possible interactions between the RC21v3 suppressor mutations and conserved structures in extracellular loops. Although the Pdr5p and CaCdr1p homology models have yet to be supported by direct structural analysis, they are based on resolved bacterial and eukaryotic structures and provide the most informative visualisations currently available for the yeast efflux pumps. They lead us to suggest that G547R may be adjacent the EL6 hydrophobic motif (coloured in orange). Conformers of the Q1226K/Q1226R mutant may either project into the egress channel or towards the PDR motif A (red). The new interactions between extracellular loops (EL1-EL6 and EL4-EL3) in different halves of CaCdr1p could modify the accessibility of RC21v3 to two lateral substrate egress pathways that are lined by the extracellular loops EL3 plus EL6 and the hydrophilic EL1 or EL4. New interactions also occur between suppressor mutations and conserved structures in the same half of CaCdr1p. Q714K in EL3 is adjacent to the conserved PDR motif B (yellow), while the nearby V674F mutation in TM5 may perturb TMS6. Finally Q1445K in EL6 is modelled on the same face as PDR motif A and the Q1226 mutations. These three mutations may lie beside an opening into the substrate egress pathway.
The single amino acid suppressor mutations encoded in CaCDR1 and their predicted spatial locations within CaCdr1p suggest RC21v3 could behave as a strongly cationic surface-active inhibitor/pseudosubstrate that need not cross the plasma membrane to block drug efflux. For example, the five suppressor mutations that occur within an extracellular loop or at the transmembrane-extracellular loop boundary all introduce a large positive charge (K or R) into CaCdr1p by modifying the small uncharged (G) or larger polar (Q) amino acid side chain. Each of these mutations has similar efficacy in suppressing chemosensitization of FLC efflux by the active principal of RC21. Their phenotypes appear consistent with the introduction, at or near lateral efflux channels, of a positive charge that repels the positively charged RC21v3 molecule, preventing it from reaching its target. For CaCdr2p, all the amino acid residues affected by the suppressor mutations in Cdr1p are homologous to those found in wild type CaCdr1p, apart from R1443 (Sanglard et al., 1997) which is similar to the EL6 Q1445K mutation of a CaCdr1p suppressor mutant. The R1443 residue may be sufficient to block inhibition of CaCdr2p function by RC21v3 and predicts that an R1443Q mutation in CaCdr2p might render the enzyme sensitive to RC21v3. In other PDR A subfamily pumps, neighboring non-conserved positively charged residues may suppress the ability of RC21v3 to bind to ScPdr5p, CgCdr1p, and CgCdr2p and explain why these efflux pumps are not susceptible to the inhibitor. For example, ScPdr5p has an arginine located 4 residues C-terminal of E724 (which aligns with CaCdr1p Q714) and a unique patch of 3 lysine residues within the 5 residues C-terminal of A684 (which aligns with CaCdr1p V674). CgCdr1p and CgCdr2p have one and two positively charged residues, respectively, within 5 residues C-terminal of the A that aligns with CaCdr1p V674. The CaCdr1p residues that align with these charged residues are not charged.
Suppression of R21v3 chemosensitizing activity by the V674F mutation within CaCdr1p TMD1 is consistent with RC21v3 penetrating the egress channel(s) to occupy or modify the conformation of sites that might normally bind efflux substrates. The V674F mutation introduced an uncharged aromatic side chain in place of a small non-polar side chain at the extracytoplasmic end of predicted TMS5 without significantly modifying cellular localization or the ATPase activity of CaCdr1p. Suppression of RC21v3 inhibition of CaCdr1p may involve amino acids in the vicinity of the V674F mutation. Unlike the V674A mutation and other mutations that were introduced into TMS5 of CaCdr1p by alanine scanning mutagenesis (Puri et al., 2009), the V674F mutation modestly uncoupled the ATP hydrolysis and substrate transport activities of the efflux pump i.e. R6G, R123 and FLC efflux were <2-fold reduced by the mutation. RC21v3 is rich in aromatic groups (two phenylalanines, a tryptophan and the Mtr group). The space occupied by the benzene ring of the V674F suppressor mutation in CaCdr1p might therefore be construed to indicate a site required for RC21v3 function. Because RC1, which is identical to RC21 but lacks the Mtr group, failed to chemosensitize the V674F suppressor mutant, the Mtr group of RC21 is unlikely to occupy the equivalent location in wild type CaCdr1p. The V674F mutation may instead indirectly affect a more distal RC21v3 binding site. The homology model of CaCdr1p (Fig. 9) suggests that the V674F mutation located in TMS5 may indirectly affect substrate transport, perhaps via interaction with TMS6 which lines the efflux channel.
In vitro kinetic studies indicate that membrane impermeant RC21v3 binds at a single inhibitory site and is not a competitive inhibitor of CaCdr1p ATPase activity, while whole cell studies suggest that RC21v3 inhibits the efflux pump activity of CaCdr1p. The resultant high affinity interaction between RC21v3 and structural elements at the extracellular surface of CaCdr1p would then be expected to block pump conformational change, locking the enzyme in a closed state. This would make cytoplasmically-oriented high affinity substrate binding sites unavailable and block recognition of efflux substrates in the cytoplasm or inner leaflet of the plasma membrane. Alternatively, the occupation of the egress channel or a periplasmically-oriented efflux substrate binding site by RC21 may physically block substrate transfer across the lipid bilayer. In both scenarios in the absence of chemosensitizer, the charged suppressor mutations that inhibit RC21v3 binding to CaCdr1p are unlikely to affect the high affinity pump substrate binding sites in the enzyme’s open conformation but could cause lower rates of substrate efflux by placing a positive charge barrier in the efflux channel or restricting the channel diameter. Slightly reduced (2-4 fold) resistance to several azole drugs (FLC, VCZ, KTZ, and MCZ), the rhodamines (R6G and R123) and CHX was conferred by particular RC21 suppressor mutations i.e. most strongly by the G547R and Q1226K and to a lesser extent by V674F. Thus, mutations near the membrane-ectodomain interface interfered significantly with the efflux of multiple pump substrates, while mutations more distant from the membrane surface (ie in extracellular loops 3 and 6) did not. These observations suggest that pump substrates acquired from the cytoplasm or lipid bilayer enter into a spatially confined externally facing egress channel near the surface of the pump. Chemical-crossing linking experiments and in vitro studies of pump function are now needed to determine whether RC21v3 binding to suppressor mutant or wild type enzyme CaCdr1p interferes with substrate binding either directly or indirectly. Finally, suppressor mutations that screen RC21v3 from entering the egress channel might be expected to block the action of other chemosensitizers that act in a similar manner. Agarose diffusion assays demonstrated that some FLC chemosensitization suppressor mutants, in comparison with the control strain overexpressing CaCdr1p, were less responsive to the CaCdr1p inhibitors FK506, enniatin and the milbemycins α11 and β9 but not the remainder of the milbemycins tested. This pattern of response suggests that the RC21v3 binding site is accessed from outside the cell by RC21v3, enniatin and the milbemycins, and perhaps from both sides of the membrane by the pump substrate FK506. The impact of the suppressor mutations on chemosensitization and pump substrate efflux suggests the binding sites for RC21v3, enniatin, FK506 and the two milbemycins overlap, and that the charge changes/conformational modifications introduced by these mutations can inhibit significantly the access of these compounds and possibly modify the binding affinity of CaCdr1p for particular chemosensitizers and pump substrates.
Drug-like congeners and the clinical relevance of RC21v3
The screening system and panel of suppressor mutants provide a strategy to discover novel, drug-like chemosensitizers with modes of action similar to RC21, enniatin and the milbemycins α11 and β9. Unfortunately enniatin is toxic for humans, probably because it is an ionophore. The milbemycins and related compounds are more promising candidate antifungal chemosensitizers because this class of compounds is less toxic, with members used as anthelminthics and miticides in animals and humans. They also differ from the α9 class of milbemycins developed by Microcide Pharmaceuticals Inc as inhibitors of fungal drug efflux. Furthermore, we recently found that the combination of RC21v3 and FLC dramatically reduced oral infection by FLC-resistant C. albicans MML611 in a mouse model (Hayama et al., 2012), illustrating the clinical relevance of the compound. Non-toxic drugs with a mode of action similar to RC21v3 are desirable because they need not penetrate cells and are thus less likely to be degraded by intracellular detoxification pathways. Their potential to dramatically increase efficacy could extend the commercial life of the inexpensive but efflux-susceptible azole drugs.
Experimental Procedures
Strains and culture condition
Strains of S. cerevisiae and Candida species used in this study are listed in Table 1. The S. cerevisiae strain AD/CaCDR1B that hyper-expresses the C. albicans ABC transporter CaCDR1 (Lamping et al., 2007) was used throughout the screening (stage 1 through 5) in both primary cell-based screens and for in vitro secondary screens. The S. cerevisiae strain AD/CaMDR1 which over-expresses the C. albicans MFS transporter CaMDR1 (BENr) gene (Lamping et al., 2007) provided a counterscreen that helped eliminate peptide pools that affected both the CaCdr1p ABC transporter and other cellular activities. CaMdr1p uses the membrane potential generated by the plasma membrane proton pump (Pma1p) to efflux toxic substances such as FLC. The strains hyper-expressing CaCDR1 and CaMDR1 had FLC MICs of 320 μg/ml and 64 μg/ml, respectively, in liquid complete synthetic medium without uracil (CSM-ura) buffered with 10 mM morpholinoethanesulfonic acid and 20 mM HEPES at pH 7.0. Strains functionally over-expressing the ABC and MFS transporters or ERG11 were constructed using a transformation cassette from plasmid pABC3 and the AD1-8u− strain which is deleted in seven ABC transporters (Lamping et al., 2007). S. cerevisiae strains were routinely maintained on CSM-ura, which contained 0.67% (wt/vol) yeast nitrogen base (Difco, Becton Dickinson, Sparks, MD), 0.077% (wt/vol) CSM-ura (Bio 101, Vista, CA), 2% (wt/vol) glucose, and 2% (wt/vol) agar (pH 5.5). Candida species were maintained on YPD agar, which contained 1% (wt/vol) yeast extract (Difco), 2% (wt/vol) Bacto Peptone (Difco), 2% (wt/vol) glucose, and 2% (wt/vol) agar (pH 5.5). For growth inhibition and checkerboard assays, liquid CSM-ura (pH 7.0) was used. Uridine (50 μg/ml) was added to CSM-ura medium for growth of the AD1-8u− host. Cells were grown at 30°C with shaking (150 rpm).
Chemicals and antifungal agents
The chemicals and antifungal agents used in this study were obtained from the following sources: FLC (Diflucan; Pfizer Laboratories Limited, Auckland, New Zealand); itraconazole (ITC; Janssen Research Foundation, Beerse, Belgium); ketoconazole (KTC; Janssen Pharmaceutica Limited, Sydney, Australia); miconazole (MCZ), cycloheximide (CHX), oligomycin, aurovertin B, sodium metavanadate, sodium azide, ATP, bicinchoninic (BCA) protein assay kit (Sigma, St. Louis, Mo.); phenylmethylsulfonyl fluoride (Roche Diagnostics NZ Limited, Auckland, New Zealand); and L-ascorbic acid, ammonium molybdate, and dimethyl sulfoxide (DMSO; BDH, Poole, United Kingdom). ITC, KTC, oligomycin, and the peptide combinatorial library pools and subpools were prepared as stock solutions dissolved in DMSO. Agarose was obtained from Gibco (Invitrogen Corporation, Auckland, New Zealand).
Combinatorial library
The d-octapeptide combinatorial library was synthesized manually by using solid-phase 9-fluorenylmethoxy carbonyl (Fmoc) chemistry as described previously (Monk et al., 2005, Niimi et al., 2004). The combinatorial library is comprised of 324 peptide pools (each theoretically containing 5,832 separate peptides), with all possible peptides being present in approximately equimolar amounts (Fig. 1). Each peptide can be represented by the formula d-NH2-ABX3X2X1RRR-CONH2 where, for each pool, A and B are known and X represents any of 18 amino acids (cysteine was excluded because a cysteine-targeting sub-library was also constructed, and glycine was excluded because it lacks a side chain). The most active peptide library pools were identified by bioassay (Fig. 1, stage 1). The most suitable pools were then deconvoluted by cycles of resynthesis and bioassay to sequentially determine the optimal amino acids present at positions X3 (stage 2), X2 (stage 3), and X1 (stage 4). Finally, the most potent peptides identified in stage 4 were manually resynthesized and purified by high-pressure liquid chromatography (HPLC) with a Shimadzu LC-6 system equipped with 15μm, 300-A° Phenomenex guard and separation (250 by 21.20 mm) columns. The HPLC buffers used were as follows: buffer A, 2% acetonitrile and 0.1% trifluoroacetic acid; and buffer B, 90% acetonitrile and 0.1% trifluoroacetic acid. As an example, RC21 was purified as follows. The crude peptide was loaded at 5% acetonitrile (ACN) in 0.1% trifluoracetic acid (TFA), and the column was run at 5 to 15% ACN in 0.1% TFA over 10 min and then at 15 to 45% ACN in 0.1% TFA over 2 h. The recovered peptides were analyzed by HPLC with a Waters Alliance HT 2790 system coupled to a Micromass ZMD 4000 electrospray mass spectrometer. The purification separated the naked peptide and monosubstituted 4-methoxy-2,3,6-trimethylbenzensulfonyl (Mtr) derivatives.
Determination of the concentrations of peptide pools, subpools and peptides
The concentration of stage 1, 2, 3 and 4 d-octapeptide pools, and crude peptides dissolved in DMSO were determined using the BCA protein assay kit (Sigma) in comparison with a standard peptide, the concentration of which was estimated using a molar extinction coefficient that took into account its composition of aromatic amino acids. The concentrations of stage 5 (Fig. 1) purified peptides and peptide derivatives were determined by measuring their absorbance at 280 nm as they each contained a single tryptophan (E280nm = 5600 M−1 cm−1).
In vivo FLC chemosensitization assay
(1) Agarose diffusion assay
Twenty-five ml CSM–ura containing the indicated amount of FLC was solidified with 0.6% agarose in an Omnitray (126 by 86 by 19 mm; Nunc, Roskilde, Denmark) or a plastic Petri dish. Cells hyper-expressing CaCdr1p or CaMdr1p were grown in CSM-ura liquid medium (pH 7.0) until OD600nm reached ~2.0, and diluted to OD600nm = 0.2 in the same medium. Cells (1-2 × 105) in 5 ml of melted overlay CSM–ura containing 0.4% agarose and FLC were placed over the base medium. An appropriate volume of each pool, crude peptide or purified peptide (5 μl unless otherwise stated) was loaded on a sterile paper disk (for stage 1, 3MM Whatman, UK; for stages 2-5, Becton, Dickinson and Co., Sparks, MD), the disk dried at 37 °C for 1-2 h and applied to the solidified overlay. The Omnitray or Petri dish was incubated at 30°C for 48 h. FLC-free medium was also used in the agarose diffusion assay to assess the effects of test peptides alone on the growth of cells. The diameters of growth inhibitory zones were compared between the presence and the absence of FLC. Peptide pools that inhibited cell growth of a strain overexpressing CaCdr1p in the absence of FLC were excluded from further assay.
(2) Checkerboard chemosensitization assay
(i) Peptide MIC as a measure of toxicity against S. cerevisiae
The MIC of RC21 for S. cerevisiae strains hyper-expressing fungal drug efflux pumps was determined using a microdilution method in 96-well microplates. Cells were inoculated at 4 × 103 per well in CSM-ura (200 μl) at pH 7.0 and incubated at 30°C for 48 h with shaking (150 rpm) in the presence of a twofold dilution series of the peptide. Cell growth was monitored at 590 nm using an EL340 Bio Kinetics reader (BioTek Instruments).
(ii) FLC chemosensitization assay
Checkerboard chemosensitization assays were performed with RC21 to evaluate the effects on cell growth of both peptide and FLC concentrations (Monk et al., 2005, Niimi et al., 2004). In brief, six rows each comprising ten samples of twofold dilutions of the peptide were prepared in a microplate. In a separate microplate, ten columns each comprising six samples of twofold dilutions of FLC were prepared to yield final concentrations in the test plate according to the individual FLC MIC for test strains. The FLC dilutions were added to wells containing the peptide dilutions, and 4 × 103 cells in a final volume of 200 μl were inoculated per well. The microplates were incubated at 30°C with shaking (150 rpm), and cell growth was monitored at 24 and 48 h. After 48 h, 5 μl samples from wells with no visible growth were spotted on YPD agar and incubated at 30°C for 48 h to test for cell viability.
In vitro assays
(i) Isolation of plasma membranes
Plasma membranes from strains AD1-8u− and AD/CaCDR1B were used for in vitro CaCdr1p ATPase assays. S. cerevisiae strain T48 (Monk et al., 2005) was used as a source of membranes for plasma membrane ATPase (Pma1p) assays. Cells were grown in YPD liquid medium at 30°C with shaking (200 rpm) and harvested in the diauxic phase of growth (OD600nm = ~6). AD1-8u− and AD/CaCDR1B cells were washed twice and starved for glucose on ice for 30 min in distilled water, while T48 cells were harvested without the washing steps. The glucose starvation step minimized Pma1p activity in the AD/CaCDR1B cells, while the T48 cells were harvested to maximize the ATPase activity of Pma1p. Enriched plasma membrane fractions were prepared as described previously (Niimi et al., 2004), resuspended in 10 mM Tris (pH 7.0), 0.5 mM EDTA, 20% (vol/vol) glycerol, and stored at −80°C. Protein concentration were determined by a micro-Bradford assay (Bio-Rad Laboratories, Hercules, CA.) with bovine gamma globulin as the standard. Protein profiles of membrane samples were otained by electrophoresis in sodium dodecyl sulfate (SDS)–8% polyacrylamide gels and staining with Coomassie blue R250.
(ii) CaCdr1p ATPase inhibition assay
CaCdr1p ATPase assays were optimized according to the standard method described previously (Niimi et al., 2004). Cdr1p has nucleoside triphosphatase activity over a broad pH range, with optimal vanadate- and oligomycin-sensitive activities between pH 7 and 8. Peptides that sensitized strain AD/CaCDR1B to FLC were assessed for their inhibition of oligomycin-sensitive ATPase activity at pH 7.5. CaCdr1p ATPase activity was measured in the presence or absence of test peptides by incubating plasma membrane fractions (5-10 μg) at 30°C for 30 min in a final volume of 120 μl containing 6 mM ATP and 7 mM MgSO4 in 59 mM morpholinoethanesulfonic acid-Tris as previously described (Niimi et al., 2004). In ATP protection assays, 6 mM ATP was added 5 min prior to the addition of peptides, and the reaction was initiated by the addition of 7 mM MgSO4 after an additional 5 min of incubation at 30°C. The ATPase specific activity of hyperexpressed efflux pumps was corrected for the activities of other ATPases by subtraction of the background oligomycin-sensitive specific activity detected in plasma membranes from the isogenic null strain AD1-8u−.
(iii) Pma1p assay
Plasma membrane fractions were prepared from S. cerevisiae strain T48 by the method described above. ATPase assays were carried out at 30°C and pH 7.0 for 20 min with 2 μg of membrane protein per assay in the presence of 10 μM oligomycin (Monk et al., 2005).
Whole cell assays
(i) Rhodamine uptake and efflux
Log-phase (OD600 = 1.5) AD/CaCDR1B or AD1-8u− cells were stored overnight on ice. The cells were harvested by centrifugation, washed twice with distilled water, resuspended in 50 mM HEPES-NaOH pH 7.0 (HEPES buffer), and incubated with 5 mM 2-deoxyglucose at 30°C for 30 min to deplete intracellular energy levels and then with 15 μM rhodamine 6G (R6G). For uptake experiments, 1-ml samples of cells (OD600nm = 1.0) were harvested by centrifugation at various time intervals after the addition of R6G with or without test peptides and lysed by boiling for 10 min with 250 μl of 2% (w/v) SDS, 2% (v/v) mercaptoethanol, 125 mM Tris-HCl pH 6.8 to release cell-associated R6G (Niimi et al., 2004). After centrifugation, the amount of R6G in each supernatant was quantitated fluorimetrically by using a standard curve generated from known amounts of R6G in the same lysis buffer. For R6G efflux experiments, cells preloaded with 15 μM R6G for 30 min were washed twice and resuspended in HEPES buffer at an OD600nm of 10-15. Cells (100 μl) were distributed to wells of a 96-well microplate, pre-warmed at 30°C for 5, then glucose (100 μl) added to the cells (Niimi et al., 2004). Cells were incubated for an appropriate period (usually 6 min) and 80 μl of the suspension transferred to wells of Multi-well Filter plate (Acro Prep, Pall Corporation, USA) placed on a Multiscreen resist vacuum manifold (Millipore) over a 96-well black flat-bottom microtitre plate (BMG Labtechnologies GmbH, Offenburg, Germany). The supernatants recovered in the black plate after passage through a glass fibre filter were combined with two washes of 80 μl of ice-cold HEPES buffer to a final volume of 240 μl. The R6G fluorescence of 100 μl samples of the filtrate were measured using a POLARstar OPTIMA (BMG Labtechnologies) with a filter number 3 (the excitation and the emission wavelengths were 485 and 520 nm, respectively) and analysed using Fluostar Optima software. The fluorescence of a 100 μl dilution series of R6G in the same buffer gave a linear standard curve between 0.1 and 1 nmole.
Toxicity for human cells
(i) Hemolysis
The lytic effects of peptides on human erythrocytes were assessed by the method of Helmerhorst (Helmerhorst et al., 1997).
(ii) Toxicity for cultured human epithelial cells
Assays for determination of the toxicity of peptides for human epithelial (HEp2) cells were conducted as previously described (Niimi et al., 2004).
Confocal microscopy
S. cerevisiae strains expressing enhanced GFP (EGFP)-tagged CaCdr1p proteins were examined using a Zeiss 510 Axiovert 200 M inverted confocal laser scanning microscope.
Supplementary Material
Acknowledgements
This research was supported by funding from the Health Research Council of New Zealand (BCM and RDC), the National Institutes of Health (DE015075, DE016885, RDC) and the Japan Health Sciences Foundation (BCM, MN and RDC). Dr Mikhail Keniya is acknowledged for asistance with confocal microscopy experiments reported in the Supplementary Data section. We also wish to acknowledge the provision of some peptides and peptide derivatives by Dr Edmond Fleischer and Dr Anette Klinger of MicroCombiChem (Wiesbaden, Germany).
Footnotes
Dr K Niimi and Dr BC Monk contributed equally to this research.
References
- Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson GD, Niimi M, Cannon RD, Jenkinson HF. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob Agents Chemother. 1996;40:2835–2841. doi: 10.1128/aac.40.12.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzi E, Goffeau A. Genetics and biochemistry of yeast multidrug resistance. Biochim Biophys Acta. 1994;1187:152–162. doi: 10.1016/0005-2728(94)90102-3. [DOI] [PubMed] [Google Scholar]
- Balzi E, Goffeau A. Yeast multidrug resistance: the PDR network. J Bioenerg Biomembr. 1995;27:71–76. doi: 10.1007/BF02110333. [DOI] [PubMed] [Google Scholar]
- Cannon RD, Lamping E, Holmes AR, Niimi K, Baret PV, Keniya MV, Tanabe K, Niimi M, Goffeau A, Monk BC. Efflux-mediated antifungal drug resistance. Clin Microbiol Rev. 2009;22:291–321. doi: 10.1128/CMR.00051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson RJ, Hollenstein K, Locher KP. Uptake or extrusion: crystal structures of full ABC transporters suggest a common mechanism. Molec Microbiol. 2007;65:250–257. doi: 10.1111/j.1365-2958.2007.05792.x. [DOI] [PubMed] [Google Scholar]
- Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- Dawson RJ, Locher KP. Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 2007;581:935–938. doi: 10.1016/j.febslet.2007.01.073. [DOI] [PubMed] [Google Scholar]
- Decottignies A, Grant AM, Nichols JW, de Wet H, McIntosh DB, Goffeau A. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J Biol Chem. 1998;273:12612–12622. doi: 10.1074/jbc.273.20.12612. [DOI] [PubMed] [Google Scholar]
- Egner R, Bauer BE, Kuchler K. The transmembrane domain 10 of the yeast Pdr5p ABC antifungal efflux pump determines both substrate specificity and inhibitor susceptibility. Molec Microbiol. 2000;35:1255–1263. doi: 10.1046/j.1365-2958.2000.01798.x. [DOI] [PubMed] [Google Scholar]
- Egner R, Rosenthal FE, Kralli A, Sanglard D, Kuchler K. Genetic separation of FK506 susceptibility and drug transport in the yeast Pdr5 ATP-binding cassette multidrug resistance transporter. Mol Biol Cell. 1998;9:523–543. doi: 10.1091/mbc.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst R, Klemm R, Schmitt L, Kuchler K. Yeast ATP-binding cassette transporters: cellular cleaning pumps. Methods Enzymol. 2005;400:460–484. doi: 10.1016/S0076-6879(05)00026-1. [DOI] [PubMed] [Google Scholar]
- Ernst R, Kueppers P, Stindt J, Kuchler K, Schmitt L. Multidrug efflux pumps: substrate selection in ATP-binding cassette multidrug efflux pumps--first come, first served? FEBS J. 2010;277:540–549. doi: 10.1111/j.1742-4658.2009.07485.x. [DOI] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006 doi: 10.1002/0471250953.bi0506s15. Chapter 5: Unit 5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golin J, Ambudkar SV, Gottesman MM, Habib AD, Sczepanski J, Ziccardi W, May L. Studies with novel Pdr5p substrates demonstrate a strong size dependence for xenobiotic efflux. J Biol Chem. 2003;278:5963–5969. doi: 10.1074/jbc.M210908200. [DOI] [PubMed] [Google Scholar]
- Golin J, Ambudkar SV, May L. The yeast Pdr5p multidrug transporter: how does it recognize so many substrates? Biochem Biophys Res Commun. 2007;356:1–5. doi: 10.1016/j.bbrc.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Gupta RP, Kueppers P, Schmitt L, Ernst R. The multidrug transporter Pdr5: a molecular diode? Biol Chem. 2011;392:53–60. doi: 10.1515/BC.2011.011. [DOI] [PubMed] [Google Scholar]
- Hanson L, May L, Tuma P, Keeven J, Mehl P, Ferenz M, Ambudkar SV, Golin J. The role of hydrogen bond acceptor groups in the interaction of substrates with Pdr5p, a major yeast drug transporter. Biochemistry. 2005;44:9703–9713. doi: 10.1021/bi0502994. [DOI] [PubMed] [Google Scholar]
- Hayama K, Ishibashi H, Ishijima SA, Niimi K, Tansho S, Ono Y, Monk BC, Holmes AR, Harding DR, Cannon RD, Abe S. A D-octapeptide drug efflux pump inhibitor acts synergistically with azoles in a murine oral candidiasis infection model. FEMS Microbiol Lett. 2012;328:130–137. doi: 10.1111/j.1574-6968.2011.02490.x. [DOI] [PubMed] [Google Scholar]
- Helmerhorst EJ, Van’t Hof W, Veerman EC, Simoons-Smit I, Nieuw Amerongen AV. Synthetic histatin analogues with broad-spectrum antimicrobial activity. Biochem J. 1997;326:39–45. doi: 10.1042/bj3260039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga K, Wanigasekera A, Sugi H, Hamanaka N, Oda K. A novel screening for inhibitors of a pleiotropic drug resistant pump, Pdr5, in Saccharomyces cerevisiae. Biosci Biotechnol Biochem. 2001;65:1589–1595. doi: 10.1271/bbb.65.1589. [DOI] [PubMed] [Google Scholar]
- Holmes AR, Lin YH, Niimi K, Lamping E, Keniya M, Niimi M, Tanabe K, Monk BC, Cannon RD. ABC transporter Cdr1p contributes more than Cdr2p does to fluconazole efflux in fluconazole-resistant Candida albicans clinical isolates. Antimicrob Agents Chemother. 2008;52:3851–3862. doi: 10.1128/AAC.00463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AR, Tsao S, Ong SW, Lamping E, Niimi K, Monk BC, Niimi M, Kaneko A, Holland BR, Schmid J, Cannon RD. Heterozygosity and functional allelic variation in the Candida albicans efflux pump genes CDR1 and CDR2. Molec Microbiol. 2006;62:170–186. doi: 10.1111/j.1365-2958.2006.05357.x. [DOI] [PubMed] [Google Scholar]
- Kolaczkowski M, van der Rest M, Cybularz-Kolaczkowska A, Soumillion JP, Konings WN, Goffeau A. Anticancer drugs, ionophoric peptides, and steroids as substrates of the yeast multidrug transporter Pdr5p. The J Biol Chem. 1996;271:31543–31548. doi: 10.1074/jbc.271.49.31543. [DOI] [PubMed] [Google Scholar]
- Lamping E, Baret PV, Holmes AR, Monk BC, Goffeau A, Cannon RD. Fungal PDR transporters: Phylogeny, topology, motifs and function. Fungal Genet Biol. 2010;47:127–142. doi: 10.1016/j.fgb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamping E, Monk BC, Niimi K, Holmes AR, Tsao S, Tanabe K, Niimi M, Uehara Y, Cannon RD. Characterization of three classes of membrane proteins involved in fungal azole resistance by functional hyperexpression in Saccharomyces cerevisiae. Eukaryot Cell. 2007;6:1150–1165. doi: 10.1128/EC.00091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MD, Galazzo JL, Staley AL, Lee JC, Warren MS, Fuernkranz H, Chamberland S, Lomovskaya O, Miller GH. Microbial fermentation-derived inhibitors of efflux-pump-mediated drug resistance. Farmaco. 2001;56:81–85. doi: 10.1016/s0014-827x(01)01002-3. [DOI] [PubMed] [Google Scholar]
- Lemoine RC, Glinka TW, Watkins WJ, Cho A, Yang J, Iqbal N, Singh R, Madsen D, Lolans K, Lomovskaya O, Oza U, Dudley MN. Quinazolinone-based fungal efflux pump inhibitors. Part 1: Discovery of an (N-methylpiperazine)-containing derivative with activity in clinically relevant Candida spp. Bioorg Med Chem Lett. 2004;14:5127–5131. doi: 10.1016/j.bmcl.2004.07.070. [DOI] [PubMed] [Google Scholar]
- Locher KP, Lee AT, Rees DC. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- Maebashi K, Niimi M, Kudoh M, Fischer FJ, Makimura K, Niimi K, Piper RJ, Uchida K, Arisawa M, Cannon RD, Yamaguchi H. Mechanisms of fluconazole resistance in Candida albicans isolates from Japanese AIDS patients. J Antimicrob Chemother. 2001;47:527–536. doi: 10.1093/jac/47.5.527. [DOI] [PubMed] [Google Scholar]
- Mishra NN, Prasad T, Sharma N, Lattif AA, Prasad R, Gupta DK. Membrane fluidity and lipid composition in clinical isolates of Candida albicans isolated from AIDS/HIV patients. Acta Microbiol Immunol Hung. 2007;54:367–377. doi: 10.1556/AMicr.54.2007.4.4. [DOI] [PubMed] [Google Scholar]
- Monk BC, Niimi K, Lin S, Knight A, Kardos TB, Cannon RD, Parshot R, King A, Lun D, Harding DR. Surface-active fungicidal D-peptide inhibitors of the plasma membrane proton pump that block azole resistance. Antimicrob Agents Chemother. 2005;49:57–70. doi: 10.1128/AAC.49.1.57-70.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi K, Harding DR, Parshot R, King A, Lun DJ, Decottignies A, Niimi M, Lin S, Cannon RD, Goffeau A, Monk BC. Chemosensitization of fluconazole resistance in Saccharomyces cerevisiae and pathogenic fungi by a D-octapeptide derivative. Antimicrob Agents Chemother. 2004;48:1256–1271. doi: 10.1128/AAC.48.4.1256-1271.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, De Wergifosse P, Goffeau A, Balzi E. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr Genet. 1995;27:320–329. doi: 10.1007/BF00352101. [DOI] [PubMed] [Google Scholar]
- Puri N, Gaur M, Sharma M, Shukla S, Ambudkar SV, Prasad R. The amino acid residues of transmembrane helix 5 of multidrug resistance protein CaCdr1p of Candida albicans are involved in substrate specificity and drug transport. Biochim Biophys Acta. 2009;1788:1752–1761. doi: 10.1016/j.bbamem.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge RM, Esser L, Ma J, Xia D. Toward understanding the mechanism of action of the yeast multidrug resistance transporter Pdr5p: A molecular modeling study. J Struct Biol. 2010;173:333–344. doi: 10.1016/j.jsb.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D, Ischer F, Monod M, Bille J. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Chemother. 1996;40:2300–2305. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- Schuetzer-Muehlbauer M, Willinger B, Egner R, Ecker G, Kuchler K. Reversal of antifungal resistance mediated by ABC efflux pumps from Candida albicans functionally expressed in yeast. Int J Antimicrob Agents. 2003;22:291–300. doi: 10.1016/s0924-8579(03)00213-9. [DOI] [PubMed] [Google Scholar]
- Shukla S, Rai V, Banerjee D, Prasad R. Characterization of Cdr1p, a major multidrug efflux protein of Candida albicans: purified protein is amenable to intrinsic fluorescence analysis. Biochemistry. 2006;45:2425–2435. doi: 10.1021/bi0519147. [DOI] [PubMed] [Google Scholar]
- Tanabe K, Lamping E, Adachi K, Takano Y, Kawabata K, Shizuri Y, Niimi M, Uehara Y. Inhibition of fungal ABC transporters by unnarmicin A and unnarmicin C, novel cyclic peptides from marine bacterium. Biochem Biophys Res Commun. 2007;364:990–995. doi: 10.1016/j.bbrc.2007.10.110. [DOI] [PubMed] [Google Scholar]
- Tanabe K, Lamping E, Nagi M, Okawada A, Holmes AR, Miyazaki Y, Cannon RD, Monk BC, Niimi M. Chimeras of Candida albicans Cdr1p and Cdr2p reveal features of pleiotropic drug resistance transporter structure and function. Molec Microbiol. 2011;82:416–433. doi: 10.1111/j.1365-2958.2011.07820.x. [DOI] [PubMed] [Google Scholar]
- Tsao S, Rahkhoodaee F, Raymond M. Relative contributions of the Candida albicans ABC transporters Cdr1p and Cdr2p to clinical azole resistance. Antimicrob Agents Chemother. 2009;53:1344–1352. doi: 10.1128/AAC.00926-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Tanabe K, Yamazaki A, Niimi M, Uehara Y, Niimi K, Lamping E, Cannon RD, Monk BC. Phosphorylation of Candida glabrata ATP-binding cassette transporter Cdr1p regulates drug efflux activity and ATPase stability. J Biol Chem. 2005;280:94–103. doi: 10.1074/jbc.M408252200. [DOI] [PubMed] [Google Scholar]
- Watkins WJ, Lemoine RC, Chong L, Cho A, Renau TE, Kuo B, Wong V, Ludwikow M, Garizi N, Iqbal N, Barnard J, Jankowska R, Singh R, Madsen D, Lolans K, Lomovskaya O, Oza U, Dudley MN, Quinazolinone fungal efflux pump inhibitors Part 2: In vitro structure-activity relationships of (N-methyl-piperazinyl)-containing derivatives. Bioorg Med Chem Lett. 2004;14:5133–5137. doi: 10.1016/j.bmcl.2004.07.071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.