Abstract
Deficiency of UDP-galactose 4′-epimerase is implicated in type III galactosemia. Two variants, p.K161N-hGALE and p.D175N-hGALE, have been previously found in combination with other alleles in patients with a mild form of the disease. Both variants were studied in vivo and in vitro and showed different levels of impairment. p.K161N-hGALE was severely impaired with substantially reduced enzymatic activity, increased thermal stability, reduced cofactor binding and inability to rescue the galactose-sensitivity of gal10-null yeast. Interestingly p.K161N-hGALE showed less impairment of activity with UDP-N-acetylgalactosamine in comparison to UDP-galactose. Differential scanning fluorimetry revealed that p.K161N-hGALE was more stable than the wild-type protein and only changed stability in the presence of UDP-N-acetylglucosamine and NAD+. p.D175N-hGALE essentially rescued the galactose-sensitivity of gal10-null yeast, was less stable than the wild-type protein but showed increased stability in the presence of substrates and cofactor. We postulate that p.K161N-hGALE causes its effects by abolishing an important interaction between the protein and the cofactor, whereas p.D175N-hGALE is predicted to remove a stabilizing salt bridge between the ends of two α-helices that contain residues that interact with NAD+. These results suggest that the cofactor binding is dynamic and that its loss results in significant structural changes that may be important in disease causation.
Keywords: Type III galactosemia, yeast model, GALE, disease-associated mutation, UDP-galactose 4′-epimerase, Differential scanning fluorimetry
1. Introduction
Galactosemia is an autosomal recessive disease that results from impaired ability to metabolise the sugar galactose due to compromised expression or activity of the enzymes of the Leloir pathway [1–4]. Patients may develop a pathology that includes failure to thrive, vomiting, jaundice and sometimes bacterial infection in the most severe cases [4]. In the long term, disabilities in learning, speech, ovarian function, and movement can manifest [5–7]. The disease is classified into three types that vary in severity, pathology and occurrence and depend on which enzyme is affected. Type I is associated with galactose-1-phosphate uridylyltransferase deficiency (E.C. 2.7.7.12; OMIM#230400) and is the most commonly detected clinically severe form [8]. The occurrence of this type is geographically dependent and patients can show the most severe pathology of the three types. Over 230 different mutations have been detected by genetic screening, each associated with varying degrees of impairment. These mutations cause structural changes in the corresponding enzyme resulting in less efficient catalysis and/or reduced protein stability (for reviews see [9–11]). Galactokinase deficiency (EC 2.7.1.6; OMIM# 230200) is implicated in type II galactosemia [12] and this type is the least clinically severe of the three. Patients tend to develop galactose-dependent early onset cataracts and do not appear to suffer any other acute or long-term complications [5;12].
Human UDP-galactose 4′-epimerase (E.C. 5.1.3.2; hGALE) catalyses the interconversion of both UDP-galactose and UDP-N-acetylgalactosamine to their glucose counterparts and contains a bound NAD+ as an essential cofactor [3;13–16]. Deficiency of this enzyme is implicated in type III galactosemia (OMIM #230350) [17;18] and this is the least understood form of galactosemia [2;19]. Only one nonsense and 21 missense mutations of hGALE are known [20]. Originally, epimerase deficiency was identified as a biochemical oddity that impacted only red and white blood cells in apparently healthy individuals ([21], and reviewed in [22]); this condition was termed “peripheral” epimerase deficiency because it impacted only cells in peripheral circulation. Other cell types tested, including fibroblasts, liver, and even stimulated or EBV-transformed lymphoblasts, were not affected (reviewed in [22]). Subsequently, rare patients were identified who were severely symptomatic and demonstrated epimerase deficiency in all cell types tested; these patients were said to have “generalized” epimerase deficiency galactosemia [7]. Further studies of infants identified by newborn screening clearly established that there is a continuum of biochemical phenotypes [23].
The GALE enzymes of Aeromonas hydrophila and Trypanosoma sp. [24–26] are important in the viability of these pathogens, and GALE is also essential for survival of the “model” multicellular eukaryote Drosophila melanogaster [27]. The crystal structures of GALE from a variety of different species interacting with various substrates and substrate analogues are also known providing insight into how the enzyme functions [15;16;28–31]. The human GALE structure has been an invaluable tool in understanding how mutations can affect the enzyme and the structure of the variant associated with the most common severe phenotype, p.V94M, has been solved [32]. Coupled with a yeast model and biochemical work on the recombinant p.V94M protein, it was revealed that this variant’s lower activity was due to improper binding of both UDP- and UDP-N-acetyl-sugars and this caused elevated galactose 1-phosphate levels in vivo [32;33]. Both the yeast model and recombinant proteins have been used to dissect how each mutation causes its effects. From these studies it has been shown that there is a correlation between enzyme activity and the ability to rescue growth in gal10-null yeast cells, which lack endogenous GALE [34–36]. Enzyme activity also correlates inversely with cellular Gal-1P accumulation [34;35], and Gal-1P and UDP-Gal accumulation correlate inversely with growth of gal10-null yeast [37]. The enzyme’s turnover number (kcat) is reduced in variant enzymes associated with type III galactosemia [20;38;39]. Biochemically, the stability of the enzyme is also important: decreased stability and protein aggregation in vivo have been reported for some of the disease-associated variants [20;34;38;40;41].
Here, both in vivo and in vitro approaches have been used to investigate two previously uncharacterized variants of hGALE, p.K161N and p.D175N. Each corresponding allele was originally detected in the heterozygous state in patients who had been diagnosed biochemically with the clinically benign form of epimerase deficiency [23]. Of note, the patient who is heterozygous for hGALE.K161N is also heterozygous for two intronic variants of unknown significance [23], and the patient who is heterozygous for hGALE.D175N is also heterozygous for two silent substitutions of unknown significance [23]. Parental DNAs were unavailable, leaving linkage of the different variants in each patient unclear. The relationship between functional significance of the K161N and D175N substitutions and the biochemical and clinical outcomes of the patients who carry them is considered in the Conclusions below.
Both the K161N and the D175N variants of hGALE were investigated for their ability to rescue gal10-null yeast, for the levels of internal metabolites that accumulated in these yeast strains, and for enzyme structure, stability, activity, and ability to bind substrates and the NAD+ cofactor. p.K161N-hGALE was found to be severely impaired while p.D175N-hGALE was found to be only mildly affected. That the patient carrying the K161N variant was nonetheless clinically mild may imply that the other allele present in that patient remained functional, as is discussed below. Results of stability studies and investigation of each residue’s location in the overall enzyme structure suggest altered conformations affect substrate and cofactor binding as well as stability.
2. Materials & Methods
2.1 Expression and purification of recombinant proteins in Escherichia coli
Recombinant wild-type GALE proteins were expressed in E. coli and purified as previously described [36;38]. The “QuikChange” protocol [42] was used to change the appropriate codons in the expression vector. The following primers were used: hGALE.K161N for (5’-CCTTACGGCAAGTCCAATTTCTTCATCGAGGAA-3’) with hGALE.K161N rev (5’-TTCCTCGATGAAGAAATTGGACTTGCCGTAAGG-3’) and hGALE.D175N for (5’-GACCTGTGCCAGGCAAACAAGACTTGGAACGTA-3’) with hGALE.D175N rev (5’-TACGTTCCAAGTCTTGTTTGCCTGGCACAGGTC-3’). The sequences were verified (MWG-Biotech, Ebersburg, Germany) and the mutated plasmids were used to express p.K161N-hGALE and p.D175N-hGALE using essentially the same protocol as used with the wild-type protein.
Recombinant human UDP-glucose dehydrogenase was expressed and purified as previously described [36]. All protein concentrations were determined using the Bradford assay [43] with bovine serum albumin as a standard.
2.2 Measurement of the steady state kinetic parameters for UDP-galactose 4′-epimerase
The activity of each purified variant protein was determined as previously described [36]. Here the conversion of UDP-galactose to UDP-glucose was measured by the UDP-glucose dehydrogenase catalyzed oxidation of UDP-glucose by NAD+ [44]. Since the oxidation of one molecule of UDP-glucose consumes two molecules of NAD+, the rate of production of NADH, measured by absorbance at 340 nm, corresponds to twice the rate of production of UDP-glucose.
Initial rates of NADH production were determined from the linear segment of each progress curve. Each rate was halved and then plotted against the corresponding substrate concentration. The data was fitted to equation (1) using non-linear curve fitting on GraphPad Prism (GraphPad Software, CA, USA) with all points weighted equally.
| (1) |
where Vmax is the maximum, limiting rate and Km is the Michaelis constant.
2.3 Biophysical characterisation of variant proteins
Chemical cross-linking with BS3 was carried out as previously described [38;45]. Limited proteolysis was carried out as described [36] with a few modifications. In addition to incubating with UDP-galactose, protein samples were also incubated with 1 mM and 10 mM NAD+. The amount of protection from limited proteolysis was analysed by 15 % SDS-PAGE. The ANS unfolding assay was carried out as previously described [36].
2.4 Fluoresence spectra
Each hGALE variant (200 µl of 20 µM), dissolved in 10 mM HEPES, pH 8.8 was aliquoted in triplicate into a black 96 well plate. Samples were excited at 280 nm or 350 nm and emission was measured from 300 to 510 nm or 400 to 500 nm respectively. All measurements were done at room temperature using a Spectra Max Gemini X U.V. plate reader fluorimeter.
2.5 Plasmids and Yeast Strains
The p.K165N and p.D171N hGALE alleles were each re-created by site-directed mutagenesis (Quikchange, Stratagene, Inc) of a wild-type hGALE coding sequence within a low-copy-number yeast expression plasmid, pMM33, as described previously [34]. The corresponding positive (wild-type hGALE) and negative (plasmid backbone only) controls have been reported previously [34].
All yeast strains used in this study were derived by transformation of JFy3835, a gal10-null haploid yeast strain that lacks endogenous GALE, and that has been reported previously [35]. For enzyme assays, yeast were cultured at 28°C in synthetic medium containing 2% dextrose (SD) prior to lysis. For growth curves, yeast were cultured at 28°C in synthetic medium containing 2% glycerol and 2% ethanol (SGE), with or without the addition of galactose, as indicated.
2.6 Yeast Protein Lysates and Enzyme Assays
Yeast proteins were extracted by standard methods as previously described [35;46] from 5 ml cultures grown to an OD600 between 0.8 and 1.2 in SD-Ura medium. Soluble protein extracts were placed over P-6 Bio-Spin columns (Bio-Rad) to remove small metabolites prior to further analysis. Protein determinations were made using the Bio-Rad protein assay reagent, as recommended by the manufacturer, and quantified using a standard curve of bovine serum albumin.
GALE enzyme activity was measured in soluble yeast lysates essentially as previously described [33] with quantification of reactants and products by HPLC. For assays using UDP-Gal as substrate, initial reaction conditions included 0.8 mM UDP-Gal, 40 mM glycine, 0.5 mM NAD+ and 0.05 µg total soluble protein in a final volume of 12.5 µl. The reaction was incubated at 30°C for 30min and stopped by addition of 187.5 µl of chilled water. For assays using UDP-GalNAc as substrate, initial reaction conditions included 40 mM glycine, 0.8 mM UDP-GalNAc, and 0.5 mM NAD+ with 0.3 µg soluble protein in a final volume of 12.5 µl. Reactions were carried out at 30°C for 30min and stopped by addition of 187.5 µl chilled water. Samples were filtered through 0.2 µm nylon filters (Alltech) before HPLC analysis.
To test the relative dependence of p.K165N-hGALE and p.D171N-hGALE on exogenous NAD+, analyses of soluble proteins from yeast expressing either wild-type hGALE, p.K165N-hGALE, or p.D171N-hGALE were performed in triplicate over a range of six different NAD+ concentrations, as indicated; UDP-Gal and UDP-GalNAc were each held constant at 0.8 mM.
The following procedures were used for separation and quantification of enzymatic substrates and products on a DX600 HPLC system (Dionex, Sunnyvale, CA) consisting of a Dionex AS50 autosampler, a Dionex GP50 gradient pump, and a Dionex ED50 electrochemical detector, as previously described [47]. Samples were maintained at 4°C in the autosampler tray and the chromatography was performed at room temperature. Substrates and products was separated from other metabolites on a CarboPac PA10 column (250 × 4mm) with an amino-trap (50 × 4mm) placed before the analysis column and a borate-trap (50 × 4mm) placed before the injector port to remove trace amounts of borate from the mobile phase. Elution programs were as follows: For GALE (UDP-Gal), Solvent A: 10 mM NaOH; Solvent B: 40 mM NaOH/200mM NaAc. Solvents were degased and maintained under a helium atmosphere. An isocratic method was used with 35% A and 65% B for 25 min at 0.8 ml/min flow rate. UDP-Gal and UDP-Glc were eluted at 17.6 and 20.5 min respectively. For GALE (UDP-GalNAc), a similar isocratic method with the same solvent system was applied. The method used 45% A and 55% B for 40 min at 0.75 ml/min flow rate. UDP-GalNAc and UDP-GlcNAc were eluted at 20.2 and 22.3 min respectively.
2.7 Determination of growth in galactose
Yeast growth curves were performed as described previously [39;45]. All yeast cultures were grown in SGE-Ura medium to mid-logarithmic phase, at which point they were diluted to OD600 = 0.05 and allowed one more doubling. Finally, cells were inoculated into the wells of a 96-well flat-bottom plate (NalgeNunc International, Rochester, NY) at a density of 5 × 105 cells/ml in 100 µl/well of SGE-Ura medium supplemented with the indicated concentration(s) of galactose. The plate was incubated for 40 h at 30 °C with continuous shaking between OD630 absorbance readings, which occurred every 30 min. Incubation, shaking, and absorbance readings were performed using a microplate reader (Bio-Tek, model #EL808UI) run by KC junior software (Bio-Tek Instruments, Winooski, VT).
2.8 Measurement of intracellular metabolites in yeast
Yeast JFy3835 expressing either WT-hGALE, no hGALE, p.K165N-hGALE, or p.D171N-hGALE, each from a centromeric expression plasmid, were grown in SGE-Ura medium at 28°C to OD600 between 0.8 and 1, then diluted to OD600=0.05 in parallel sets of cultures and allowed to double to OD600 = 0.1. Galactose was then added (t=0) to a final concentration of 0.01% to one culture from each pair. 10 ml of cell suspension from each culture were collected for analysis of intracellular metabolites at t=0h, 6h, and 24h. Extracts were prepared as described previously [47]. In brief, 10 ml of yeast culture were quenched in 20 ml of ice-cold 60% (v/v) methanol. Cells were collected by centrifugation at 2000rpm for 20min at 4 °C, then transferred to microfuge tubes and washed once with 1ml of sterile water. Intracellular metabolites were extracted by vigorous agitation of the cells for 45min at 4°C in chloroform: methanol: water, 4:2:1, with a final volume of 875 µl. The aqueous layer was collected after centrifugation at 2000rpm in a microfuge for 20min at 4°C. The remaining organic phase was back-extracted with 125 µl methanol and 125µl water. Corresponding aqueous layers were combined and dried under vacuum without heat. Finally, dried metabolites were rehydrated with sterile water to a concentration of 5 mg cell dry mass/ml and filtered through 0.2 µm nylon filters (Alltech) before HPLC analysis.
Gal-1P and Glc-1P were quantified using the same DX600 HPLC system (Dionex, Sunnyvale, CA) with a CarboPac PA10 column (250 × 4mm). 20 µl of each sample were injected. The following mobile phase solvents were used: Solvent A, 10 mM NaOH, and Solvent B, 40mM NaOH/200mM NaAc. Flow rate was maintained at1 ml/min. Gal-1P and Glc-1P were detected using a low salt gradient procedure: 98 % A and 2 % B (−10 to 8min), a linear increase of B to 30% (8–15 min), a linear increase of B to 50% (15–25 min), hold for 5min (25–30 min), a linear decrease of B to 2% (30 to 35min).
UDP-Gal and UDP-Glc were quantified using the same HPLC and solvent system with a high salt gradient procedure: 50 % A and 50 % B (−5 to 5 min), a linear increase of B to 70 % (5–22 min), hold for 5 min (22–27 min), a linear decrease of B to 50 % (27 to 40 min).
2.9 Differential scanning fluorimetry
Sypro orange (Sigma, Poole, UK), a fluorescent protein dye, was diluted from a 5000× solution (manufacturer’s concentration definition) into a 50× solution with 10 mM HEPES, pH 8.8. This stock solution was mixed well prior to each use. Recombinant protein samples were diluted in 10 mM HEPES, pH 8.8 to a final concentration of 5 µM and 1 µl of 50× sypro orange was added. Any ligands used were diluted in 10 mM HEPES, pH 8.8 and added to a final concentration of 1 mM. Reactions were set up in a total volume of 20 µl in 0.2 ml PCR tubes and controls with no protein were always run alongside experimental samples.
Samples were loaded into a Rotor-Gene Q cycler (Qiagen) and the following protocol was used: High resolution melt run (460 nm source, 510 nm detector), 25 °C to 95 °C ramp with a 1 °C rise for each step and no gain optimisation. The melting temperatures, (Tm), were calculated using the inbuilt analysis software.
One way ANOVA with Dunnett comparison test was used to determine the significance of the difference in Tm due to substrate binding.
2.10 Computational analysis of mutant proteins
The cupsat server (http://cupsat.tu-bs.de/) was used to estimate the predicted energy of unfolding [48] of both variants to add further insight into any destabilising affects the two mutations have on the structure. The crystal structure 1EK6 was used through this server and the effects of each substitution on the structure were determined by the thermal option.
3. Results
3.1 Steady-state kinetics of recombinant proteins: both variants have impaired activity
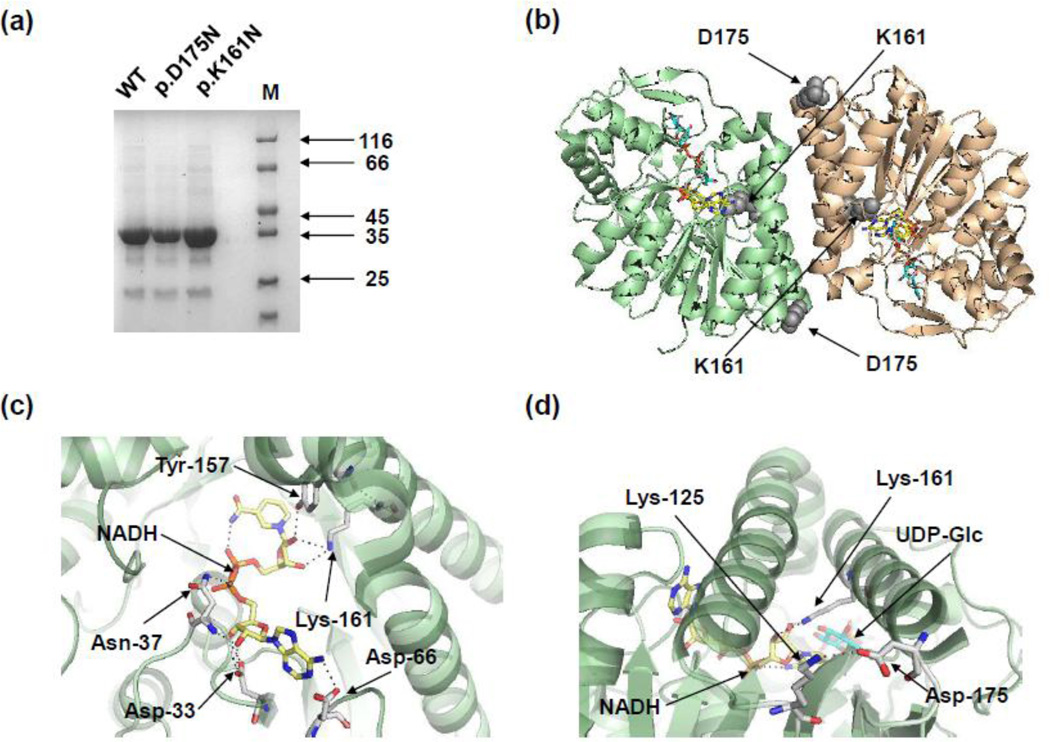
To understand the consequences of these substitutions, p.K161N and p.D175N variants of hGALE were expressed and purified using a bacterial expression system. Each substituted protein was produced to a similar yield (approximately 7.0 mg of recombinant protein per litre of culture) and quality as the wild-type protein (Figure 1a).
Figure 1. Purified recombinant hGALE variants from E. coli with the location and interactions of Lys-161 and Asp-175 within the hGALE protein structure.
(a) Proteins were resolved by SDS-PAGE (10%) and seen by staining with Coomassie blue. M = molecular mass markers (kDa). (b) The structure of the hGALE dimer showing the locations of the amino acids altered by the disease-associated mutations. Altered residues are shown as sphere models in grey. (c) Lys-161 interacts with the 2’- and 3’- hydroxyl groups of the riboside moiety of the nicotinamide adenine dinucleotide cofactor and is one of five key residues which interact with the cofactor. (d) Asp-175 forms a salt bridge with Asp-125 near the ends of two α-helices at the dimer interface. The helix that contains Asp-175 also contains Lys-161which interacts with the cofactor. Selected residues are shown as stick models in white with nitrogen, oxygen and phosphate atoms in blue, red and orange respectively. Hydrogen bonds are shown as black dotted lines. NAD+ is coloured yellow while UDP-glucose is cyan in all figures. Structures were made in PyMol (www.pymol.org) using PDB entry 1EK6 [15]. Readers are directed to the online version of this article for the colour version of this figure.
The ability of each variant to catalyse the conversion of UDP-Gal to UDP-Glc was determined using a coupled enzyme assay (Table 1). Both p.K161N- and p.D175N-hGALE had reduced overall activity in comparison to the wild-type protein (WT). p.K161N-hGALE was most severely affected and had only 0.5% of the turnover number (kcat) of the WT, whereas p.D175N-hGALE had a kcat that was 43% of the wild-type’s value. The Km of p.D175N was essentially unaltered compared to the wild-type enzyme, but for p.K161N-hGALE the value was increased almost 10-fold.
Table 1.
Kinetic constants of recombinant wild-type and variant hGALE enzymes acting on UDP-Gal.
| Protein |
Km µM |
kcat s−1 |
kcat/Km l.mol−1.s−1 |
|---|---|---|---|
| WTa | 48 ± 16 | 14 ± 1 | 3.0 ± 1.1 × 105 |
| p.K161N | 462 ± 75 | 0.07 ± 0.01 | 1.6 ± 0.3 × 102 |
| p.D175N | 86 ± 34 | 6.1 ± 0.6 | 7.1 ± 3.0 × 104 |
The values of the wild-type enzyme were determined previously [36] and are included here for comparison. Values are the means ± SD; n = 3.
3.2 Both variants retain the ability to form dimers
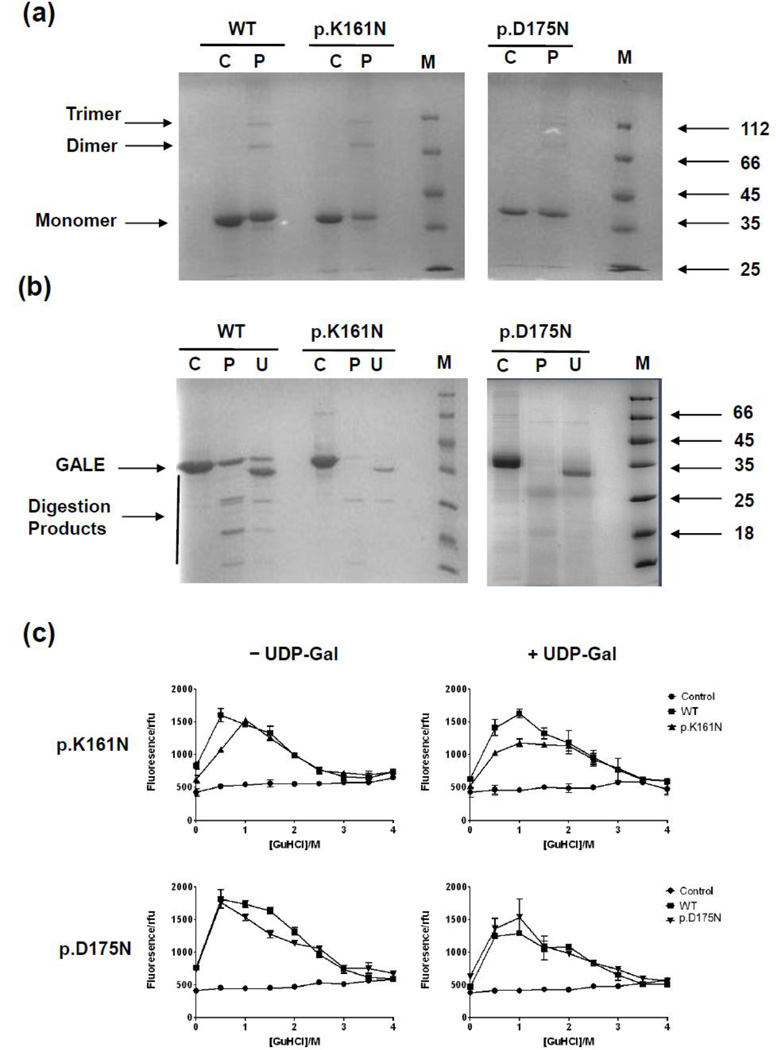
Since hGALE is a homodimer in solution [3] it was thought that the effects on enzyme activity might be due to disruption of the ability to dimerise. The chemical cross-linker BS3 has been used previously to determine if disease-associated variants of hGALE could dimerise [38;45] and was employed here. Both p.K161N-hGALE and p.D175N-hGALE formed detectable dimers (M.W. = 76 kDa) and higher order oligomers (that have been observed before [38;45]), as judged by SDS-PAGE (Figure 2a). Subtle affects on dimerisation, however, cannot be ruled out but this method but these results demonstrate that these variants have the potential to form dimers in a manner similar to the wild-type.
Figure 2. Chemical cross-linking, limited proteolysis and chemical denaturation of hGALE variants in comparison to wild-type hGALE.
(a) The hGALE variants showed no detectable difference in their ability to form dimers in comparison to the wild-type protein. C, control, protein with no cross-linker (10 µM); P, protein with 100 µM of the cross-linker BS3. (b) UDP-galactose partially protected the variants from proteolysis, to a similar extent to the wild-type protein. C, control, undigested protein (10 µM); P, protein digested with 600 nM trypsin; U, protein digested with 600 nM trypsin in the presence of 1 mM UDP-galactose. (c) Guanidine hydrochloride denaturation of the wild-type protein followed by ANS fluorescence. In the absence of UDP-galactose, a peak occurred at approximately 0.5 M GuHCl. This peak shifted to approximately 1.0 M GuHCl in the presence of 50 µM UDP-galactose. A similar shift was seen with the p.D175N-hGALE protein but not with p.K161N-hGALE. The control data represent the fluorescence of ANS in increasing concentrations of GuHCl in the absence of protein. Each point represents the mean ± SD; n = 3.
3.3 Both variants show altered susceptibility to denaturation by GuHCl and protease digestion
Since both variants showed decreased activity it was thought that instability might be a contributing factor. A number of mutants of hGALE have been previously shown to have an increased susceptibility to protease degradation [38;41;45].
An ANS unfolding assay and limited proteolytic degradation by trypsin were used to gain insight into what effects each alteration had on the overall structure (Figure 2b, c). p.K161N and p.D175N were both more susceptible than the wild-type protein to proteolytic digestion but gained some protection in the presence of UDP-Gal. p.D175N-hGALE showed little difference in response to denaturation by GuHCl compared to WT. Interestingly, p.K161N appeared to be more stable than the WT showing a greatest fluorescence emission at 1.0 M GuHCl. This variant also showed no shift in the concentration of GuHCl corresponding to the maximum fluorescence in the presence of UDP-Gal, whereas both the WT and p.D175N-hGALE showed an upward shift from 0.5 M to 1.0 M GuHCl (Figure 2c). Comparing the fluorescence signals at 0.5 M GuHCl using the student’s T-test confirmed these shifts to be statistically significant for both WT and p.D175N-hGALE (WT: p=0.0026; D175N: p=0.0152) and the increased stability of K161N compared to WT (p=0.0010).
3.4 p.K161N has an altered ability to bind the NAD+ cofactor
Since both variants had decreased activity and altered stabilities it was suggested that this may be, in part, due to altered cofactor binding. This was supported by the location of Lys-161 in the cofactor-binding site (Figure 1c) [15].
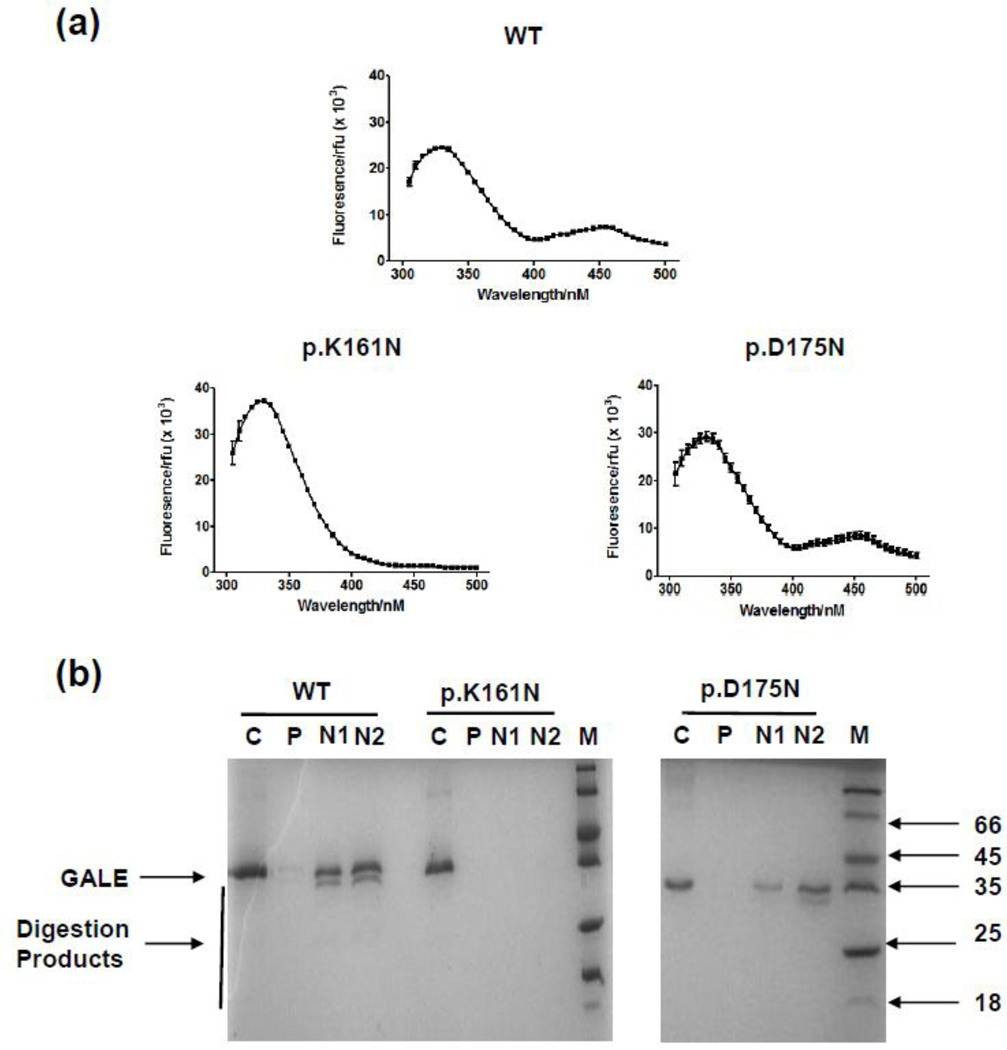
NAD(H)-containing epimerases have a unique fluorescence spectrum where excitement at 280 nm results in two emission peaks: a peak at 340 nm due to tryptophan emission and a much smaller peak at 450 nm due to FRET from tryptophan to the bound cofactor [25]. The resulting fluorescence spectra (Figure 3a) showed that while the wild-type and p.D175N show this expected pattern, the p.K161N variant demonstrated a greatly diminished peak at 450 nm. Direct excitation of the bound cofactor at 340 nm was also carried out. Here WT hGALE showed a concentration dependent increase in the 450 nm signal from the cofactor, validating this technique in determining cofactor content (Figure S1a). Comparison of the hGALE variants showed similar findings to the FRET data with p.K161N having a greatly reduced signal at 450 nm (Figure S1b). Additionally, unlike the WT and p.D175N-hGALE proteins, p.K161N-hGALE gained no protection from limited proteolysis in the presence of 1 and 10 mM NAD+ (Figure 3b).
Figure 3. p.K161N-hGALE contained no detectable endogenous cofactor and is incapable of binding exogenous NAD+.
(a) Fluorescence spectra of wild-type and variant hGALEs (20 µM) in 10 mM HEPES, pH 8.8. Excitation at 280 nm results in an emission peak at 340 nm due to tryptophan fluorescence and a smaller peak at 450 nm due to FRET between the protein’s tryptophan residues and the endogenous NAD(H) cofactor. Each point represents mean ± SD; n = 3. (b) NAD+ partially protected wild-type GALE and p.D175N-hGALE proteins from proteolysis, whereas p.K161N-hGALE gained no such protection. C, control, undigested protein (10 µM); P, protein digested with 600 nM trypsin; N1, protein digested with 600 nM trypsin in the presence of 1 mM NAD+. N2, protein digested with 600 nM trypsin in the presence of 10 mM NAD+.
3.5 Impact of the p.K161N and p.D175N substitutions on hGALE activity measured in soluble yeast lysates
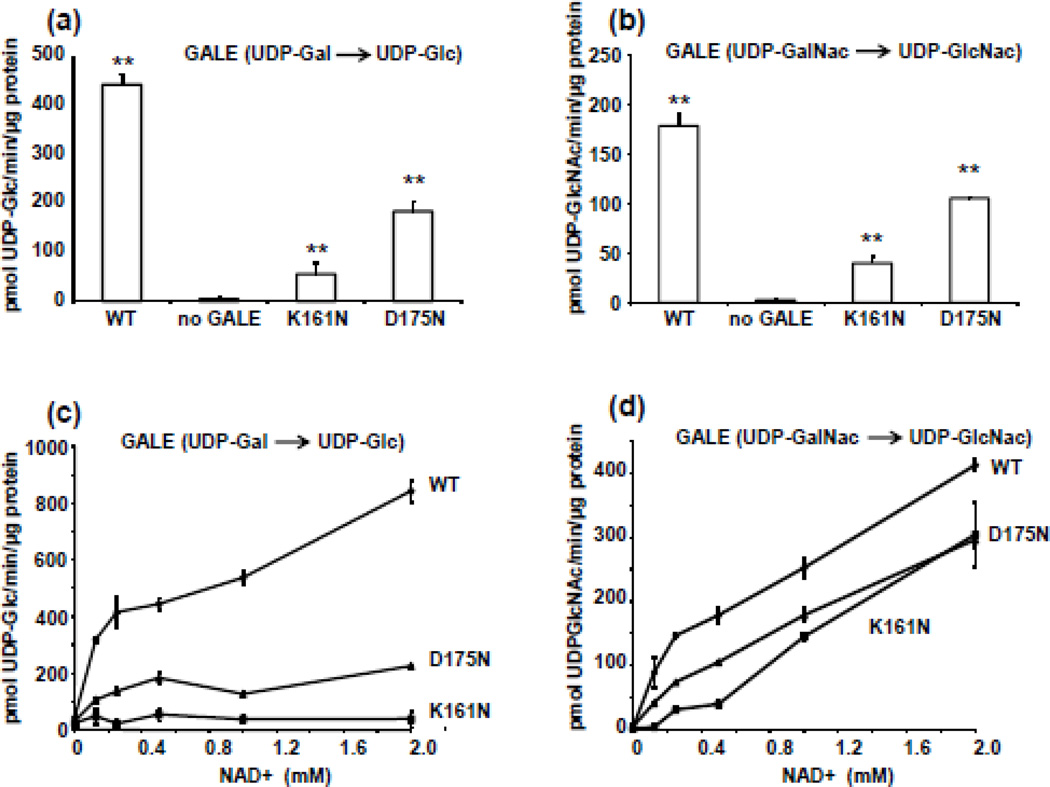
In order to further understand the different behaviours of the mutants in vitro and in vivo, both p.K161N- and p.D175N-hGALE alleles were expressed in a yeast model. GALE enzyme activity assays performed on soluble lysates of yeast expressing individual human GALE alleles in a null background demonstrated that p.K161N-hGALE has substantially reduced enzyme activity relative to p.D175N-hGALE, and both alleles demonstrated significantly reduced GALE activity relative to the positive control (Figure 4a, b). This same result was obtained using both UDP-Gal and UDP-GalNAc as substrates.
Figure 4. Effects of p.K161N and p.D175N substitutions on hGALE activity measured in soluble yeast lysates.
Soluble protein lysates were prepared from gal10-null yeast expressing no human GALE, wild-type hGALE, p.K161N-hGALE, or p.D175N-hGALE. GALE activity assays were performed using either (a) UDP-Gal or (b) UDP-GalNAc. For assays using UDP-Gal as substrate, initial reaction conditions included 0.8 mM UDP-Gal, 40 mM glycine, 0.5 mM NAD+ and 0.05 µg total soluble protein in a final volume of 12.5 µl. For assays using UDP-GalNAc as substrate, initial reaction conditions included 40 mM glycine, 0.8 mM UDP-GalNAc, and 0.5 mM NAD+ with 0.3 µg soluble protein in a final volume of 12.5 µl. To test the potential role of NAD+ affinity (c, d), enzyme assays were repeated using a series of NAD+ concentrations, but UDP-Gal and UDP-GalNAc were each maintained at a starting concentration of 0.8 mM. Following each reaction, substrates and products were separated and quantified by HPLC. The enzyme activity is expressed as mean ± SD pmol product/min/µg soluble protein; n = 3. ** indicates p < 0.01, by the Students’ two tailed T test.
To test whether the apparent activity impairment might reflect reduced affinity for NAD+, we repeated the enzyme activity assays holding substrate concentrations constant and varying the level of exogenous NAD+. As illustrated (Figure 4c, d), the p.K165N-hGALE enzyme was unresponsive to increased NAD+ when the substrate was UDP-Gal, but demonstrated a striking NAD+-dependent rise in activity when the substrate was UDP-GalNAc. In contrast, the p.D175N-hGALE enzyme was marginally responsive to increased NAD+ when the substrate was UDP-Gal, and strikingly responsive to increased NAD+ when the substrate was UDP-GalNAc.
3.6 Differential impact of the p.K161N and p.D175N substitutions on hGALE function in living yeast
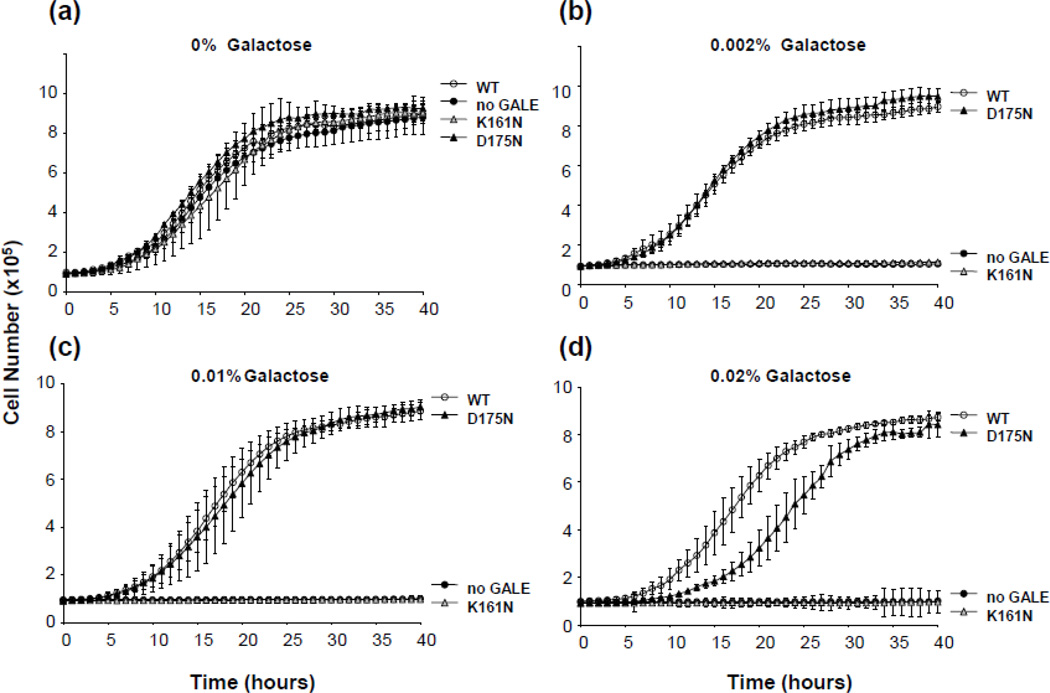
We have previously demonstrated a relationship between growth rate of yeast exposed to galactose and function of the GALE allele(s) they express (e.g. [34–36;39]). To explore the potential functional impairment of the p.K161N- and p.D175N-hGALE alleles in living yeast we applied this “growth test” (Figure 5) and found that, indeed, both substituted alleles demonstrated impaired function, and of the two, p.K161N-hGALE was, by far, the more severely impaired. These results confirm that the GALE impairment observed in soluble lysates (Figure 4) and purified enzyme (Table 1) is not a vagary of the in vitro system, but reflects a true enzyme impairment that is also evident in vivo.
Figure 5. Impact of p.K161N and p.D175N substitutions on hGALE function in living yeast.
Cultures of gal10-null yeast expressing either WT, p.K161N-hGALE, p.D175N-hGALE, or no hGALE were inoculated into 96-wellplates at a density of 5 × 105cells/ml in SGE-Ura medium with the indicated amount of galactose added at t = 0. Growth of each culture was monitored as described in Materials and Methods. Values represent means ± SD; n ≥ 6. Resulting growth profiles are demonstrated for all four strains cultured in the presence of medium containing (a) 0 %, (b) 0.002%, (c) 0.01 % and (d) 0.02 % galactose.
3.7 Differential accumulation of intracellular metabolites in galactose-exposed yeast expressing p.K161N-hGALE and p.D175N-hGALE
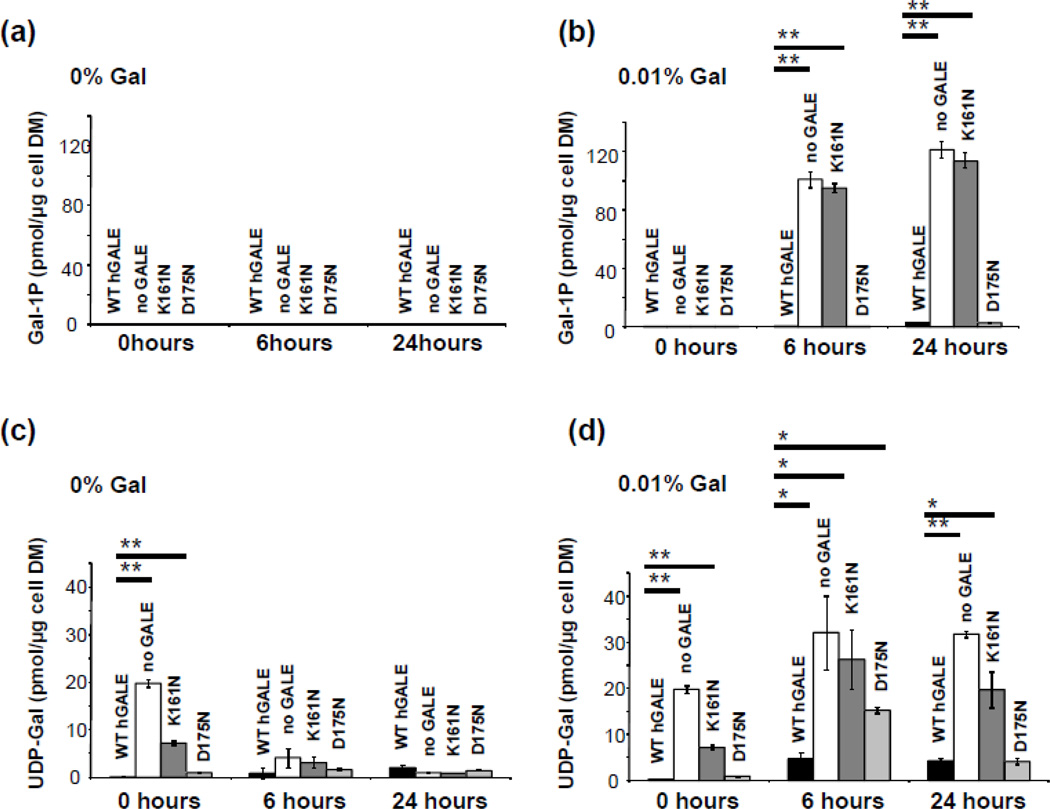
As a final in vivo test of the p.K161N- and p.D175N-hGALE alleles, we asked whether yeast expressing either of these alleles accumulate abnormal levels of galactose metabolites when exposed to 0.01% (w/v) galactose in culture (Figure 6). As predicted from their growth (Figure 5a), none of the strains accumulated Gal-1P (Figure 6a) when cultured in the absence of galactose, and yeast expressing normal hGALE or p.D175N-hGALE also demonstrated little, if any, Gal-1P accumulation even when exposed to galactose (Figure 6b). In contrast, yeast expressing p.K161N-hGALE accumulated Gal-1P levels comparable to those seen in GALE-deficient yeast.
Figure 6. Accumulation of intracellular metabolites in yeast expressing p.K161N- and p.D175N-hGALE.
Cultures of yeast expressing the indicated alleles of hGALE were grown in SGE-Ura medium supplemented with 0 or 0.01% galactose at t=0; samples were harvested for analysis of intracellular metabolites as described in Materials and Methods at t= 0, t=6h, and t=24h. Values plotted represent mean ± SEM; n = 3. (* indicates p < 0.05, ** indicates p < 0.01, by the Students’ two tailed T test). (a) Intracellular Gal-1P levels in yeast cultured in SGE-Ura. (b) Intracellular Gal-1P levels in yeast cultured in SGE-Ura plus 0.01 % of galactose. (c) Intracellular UDP-Gal levels in yeast cultured in SGE-Ura. (d) Intracellular UDP-Gal levels in yeast cultured in SGE-Ura plus 0.01% of galactose.
Cultured in synthetic glycerol-ethanol medium (SGE-Ura) lacking galactose, yeast expressing normal hGALE or p.D175N-hGALE demonstrated almost no accumulation of UDP-Gal (Figure 6c), while yeast expressing no GALE or p.K161N-hGALE showed some accumulation of UDP-Gal at the 0 hour time point which decreased to baseline after dilution into fresh SGE-Ura medium. Cultured in SGE-Ura medium spiked to 0.01 % (w/v) galactose, yeast expressing normal hGALE or p.D175N-hGALE demonstrated only a slight or transient UDP-Gal accumulation (Figure 6d). In contrast, yeast expressing no GALE or p.K161N-hGALE accumulated substantial and persistent levels of UDP-Gal. Intracellular levels of Glc-1P and UDP-Glc appeared insensitive to galactose exposure in all four yeast strains (data not shown).
3.8 A thermal shift assay confirms the different impairments of substrate and cofactor binding for both hGALE variants
To further investigate these variants, differential scanning fluorimetry (DSF) was carried out on the recombinant proteins. This is a technique that uses an extrinsic hydrophobic fluorophore to detect conformational changes due to increasing temperature [49]. Here a dye that fluoresces strongly when bound to hydrophobic residues is mixed with a purified protein sample and the mixture is heated in a real-time thermal cycler. The fluorescence initially increases resulting from the unfolding of the protein as more hydrophobic patches appear and then decreases when the protein unfolds into a random coil. The melting temperature, Tm, is then calculated as the midpoint between the maximum and initial minimum fluorescence. When a protein is in the presence of a ligand, additional interactions and induced conformational changes due to binding can then be inferred from the change in Tm (ΔTm) [50]. In addition, there is good agreement between the results from this technique and differential scanning calorimetry [49]. However, when using this assay it is assumed that the unfolding of the protein follows a two state process, that the unfolded state does not bind to the ligands, and that the unfolding process is irreversible under the conditions of the experiment [49;50]. Previously we have used this technique to assess the binding of putative activators to hGALE [51;52].
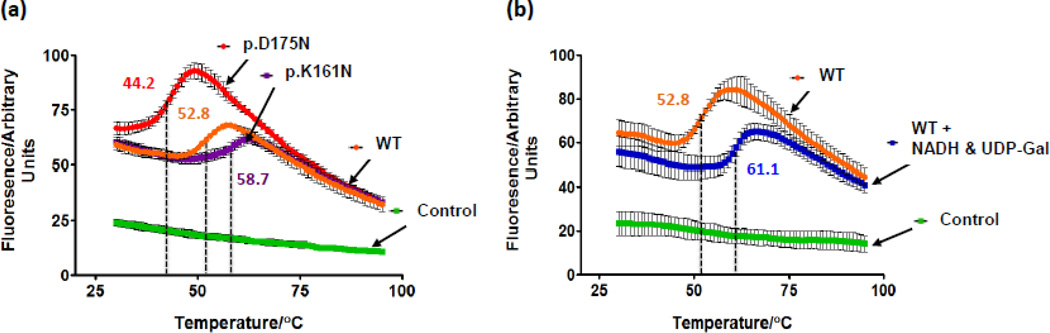
Initial computational analysis using the cupsat server suggested that p.K161N-hGALE was a stabilising mutation with an estimated free energy change of unfolding of 5.85 kJ/mol. p.D175N-hGALE, however, was predicted to be destabilising with an estimated free energy change of unfolding of −6.53 kJ/mol. DSF was then used to investigate the effects each alteration has on the stability of the protein and the ability to bind substrates and cofactors. The resulting melting curves for each variant are shown in figure 7a, Tm values are presented in table S1 and ΔTm data are shown in table 2. An example of the shift in melting temperature of wild-type hGALE due to substrate binding is shown in figure 7b. The melting temperatures were found to be in good agreement with the computationally predicted results. p.K161N-hGALE was more stable with Tm increased by 5.9 K compared to the wild-type protein (Figure 7a). The only significant change in stability was detected when this variant was in the presence of UDP-GlcNAc and NAD+, which reduced the Tm by 4.8 K (Table 2). p.D175N-hGALE was less stable with Tm reduced by 8.6 K compared to the wild-type protein without ligands (Figure 7a). However, this variant showed significant changes in Tm when in the presence of all substrates and cofactors individually, whereas the wild-type protein did not (Table 2).
Figure 7. p.K161N-hGALE showed a higher Tm than the wild-type protein, whereas p.D175N-hGALE showed a lower Tm.
(a) Melting profiles and melting temperatures of WT, p.K161N-hGALE, and p.D175N-hGALE in the absence of substrates and cofactors. (b) Melting profiles and melting temperatures of WT hGALE with and without 1 mM NADH and 1 mM UDP-gal. Reaction mixtures contained 5 µM protein and 2.5 × Sypro orange (manufacturer’s concentration definition) dissolved in 10 mM HEPES pH 8.8. Controls contained no protein. Values represent mean ± SD; n = 3. Readers are directed to the online version of this article for the colour version of this figure.
Table 2.
Change of melting temperatures (ΔTm) of recombinant wild-type and variant hGALE enzymes in the presence of various ligands.
| ΔTm (K) Variant |
Substrate UDP-Gal |
UDP-Glc | UDP-GlcNAc | Cofactor NAD+ |
NADH |
|---|---|---|---|---|---|
| WT | −0.3 ± 1.2 | −0.1 ± 0.5 | 2.2 ± 0.3*** | 1.1 ± 0.4** | 0.5 ± 0.5 |
| p.K161N | 0.7 ± 0.5 | − 0.4 ± 1.1 | − 0.9 ± 0.3 | 0.6 ± 0.3 | 0.4 ± 0.4 |
| p.D175N | 3.0 ± 0.5*** | 3.0 ± 0.5*** | 6.5 ± 0.3*** | 5.5 ± 0.5*** | 4.2 ± 0.4*** |
| ΔTm (K) | Substrate & Cofactor | |||||
|---|---|---|---|---|---|---|
| Variant | NAD+and UDP-Gal |
NAD+and UDP-Glc |
NAD+and UDP-GlcNAc |
NADH and UDP-Gal |
NADH and UDP-Glc |
NADH and UDP-GlcNAc |
| WT | 3.4 ± 0.4*** | 3.4 ± 0.4*** | 6.7 ± 0.5*** | 8.3 ± 0.5*** | 8.3 ± 0.4*** | 12.0 ± 0.3*** |
| p.K161N | −0.8 ± 1.9 | −0.6 ± 0.6 | −4.8 ± 0.9*** | −0.6 ± 0.5 | −0.3 ± 0.6 | −1.1 ± 0.5 |
| p.D175N | 9.1 ± 0.6*** | 9.2 ± 1.9*** | 15.3 ± 0.5*** | 15.6 ± 0.4*** | 15.7 ± 0.6*** | 19.8 ± 0.4*** |
All ligands were added to a final concentration of 1 mM in 10 mM HEPES, pH 8.8. The change of melting temperature, ΔTm, for mutant stability and ligand binding were calculated using equations (2) and (3) respectively.
| (2) |
| (3) |
Tmand ΔTm values are the means ± SD; n = 3.
p< 0.001,
p < 0.0001 (One way ANOVA with Dunnett comparison test).
4. Discussion
p.K161N is the more severe substitution due to altered cofactor binding
The p.K161N variant showed a substantially decreased activity for UDP-Gal determined from yeast lysates (Figure 4) and the purified protein (Table 1). Coupled with the inability to rescue gal10-null yeast (Figure 5) and accumulation of the metabolites Gal-1P (Figure 6b) and UDP-Gal (Figure 6d), these data demonstrate that p.K161N-hGALE is a severely impaired variant. The altered Km for UDP-Gal and increased protease degradation indicate this mutation may cause conformational changes and this is quite similar to variants associated with the more severe disease phenotypes [38]. However, this mutant also showed greater resistance to chemical denaturation and showed some activity with UDP-GalNAc suggesting a more complex situation (Figure 2). This is likely to be due to altered cofactor binding as demonstrated by a much lower FRET signal, lack of protection from protease by NAD+ (Figure 3) and the NAD+-dependent rise in activity with UDP-GalNAc but not with UDP-Gal (Figure 4c, d). The decreased FRET and directly excited cofactor signals (Figures 3, S1b) considered together with the limited proteolysis data suggest that this altered cofactor binding decreases the cofactor content of this variant under the conditions investigated.
In the DSF assay, when in the presence of each substrate and cofactor alone, the wild-type hGALE protein showed only a significant change of stability with NAD+ and UDP-GlcNAc. However when both the cofactor and a substrate were present together, a significant increase in stability was detected. Most likely, this increase results from the structural change that has been postulated based on the crystal structures in which the N- and C-terminal domains clamp over the active site when substrate is present [15]. In addition, these stability changes were more than additive when compared to the changes resulting from the addition of substrate or cofactor alone. This demonstrates that substrate and cofactor binding at an active site is synergistic. We have previously observed that hGALE exhibits sigmoidal kinetic behaviour at 24 °C, suggesting cooperative interactions between, as well as within, the subunits [36]. Furthermore, the higher stability of the enzyme with NADH and substrate compared to NAD+ with substrate reflects previous studies where the NADH-bound enzyme has a higher affinity for substrates than the NAD+-bound enzyme [28]. Even larger changes in stability of the wild-type protein were detected when UDP-GlcNAc was present, suggesting additional stabilising interactions. This may be due to the small conformational change that was seen in the crystal structure of the enzyme bound to UDP-GlcNAc [16]. p.K161N-hGALE was more stable and, unlike the wild-type protein, showed no significant changes in stability in the presence of both substrate and cofactor except with UDP-GlcNAc and NAD+ together (Table 2).
Lys-161 is located in the cofactor binding site (Figure 1c) forming hydrogen bonds with the 2′- and 3′-hydroxyl groups of the riboside moiety of the NAD+ cofactor [15]. The corresponding residue has been associated with maintaining enzyme activity [53] and is conserved in the GALE proteins of yeast, E. coli, rat, and human, highlighting its importance [23]. It is thought that this lysine increases the reactivity of the cofactor by destabilising the nicotinamide ring [53]. Mutational studies at this site of the enzymes from E. coli and A. hydrophila show similar decreases in activity with differences in cofactor binding. The E. coli mutants, K153M and K153A presented no changes in binding while the A. hydrophila mutant K153N showed a decreased affinity [26;53]. These differences are due to the different number of interactions that the bacterial and human epimerases have between the protein and cofactor. The E. coli enzyme has 19 hydrogen bonds whereas the human enzyme has only 11 [15] and this is further supported by the observation that hGALE requires external NAD+ for optimal activity unlike the bacterial enzyme [40]. Additionally removal of the cofactor causes aggregation of the E. coli GALE whereas this does not occur in the human enzyme [15]. Taken together, these observations suggest that NAD+ is less tightly bound to the human enzyme than to the bacterial ones and is exchangeable under the conditions of these experiments. The higher stability of p.K161N-hGALE towards thermal and chemical denaturation combined with the fluorescence and proteolysis data suggests that this variant has a low cofactor content and most likely additional stabilising interactions occurring in the binding pocket perhaps because the binding pocket has partially collapsed. These structural changes affect the UDP-Gal binding site, decreasing UDP-sugar affinity (Figure 2c, Table 1) and increase overall sensitivity to proteases (Figure 2b, 3b). This possible collapse of the cofactor binding site appears not to be reversible under the conditions of these experiments, except in the presence of UDP-GlcNAc and NAD+. The observation that UDP-GalNAc activity is dependent on NAD+ concentration (Figure 4b, d) and the stability changes of this mutant when UDP-GlcNAc is bound together with NAD+ (Table 2) suggests that conformational changes at the substrate binding site reciprocally affect the cofactor binding pocket. This observation further supports the hypothesis that additional interactions with UDP-GlcNAc or UDP-GalNAc cause different conformational changes compared to the binding of UDP-Glc or UDP-Gal. In the case of p.K161NhGALE, these additional interactions appear to be able to open the active site, permit partial repopulation of the cofactor binding site and, thus, reduce the overall rigidity of the enzyme.
p.D175N is a mild variant due to the stability of the protein
The less severe enzymological impairment of p.D175N-hGALE, compared to p.K161N-hGALE, suggests that less radical conformational changes have occurred. This variant showed no significant differences from the wild-type protein in terms of chemical denaturation (Figure 2c) and cofactor binding (Figure 3, S1b). Additionally, in vivo, yeast expressing p.D175N-hGALE showed only a slight impairment of growth when challenged with galactose (Figure 5) and only a transient UDP-Gal accumulation (Figure 6) and intermediate levels of activity for both substrates (Figure 4). These results are consistent with p.D175N-hGALE being an intermediately impaired enzyme.
However this variant appeared to be more prone to digestion (Figure 2b) and, in the more sensitive DSF assay, p.D175N-hGALE was found to be less stable than the WT protein by 8.6 K (Figure 7a, Table 2). Also, significant changes in stability in the presence of all substrates and cofactors were detected and even larger changes in stability occurred in the presence of both substrate and cofactor pairs (Table 2). These results indicate a stabilising interaction being disrupted but the resulting structural changes can be compensated by additional interactions caused by cofactor and substrate binding. Additionally, the finding that the stability of this variant is quite close to that of the wild-type protein when cofactor and substrate are both bound (Table S1) supports the hypothesis that this protein is only slightly different from the wild-type protein in conformation.
Asp-175 is located towards the end of an α-helix in the N-terminal domain (Figure 1d) and this forms a salt bridge with Lys-125 that is close to the end of an adjacent α-helix [15]. Interestingly, Lys-161 is located in one of these helices (Figure 1d). Changing an aspartate to asparagine will remove this salt bridge, most likely resulting in a reduction in stability. Our results support this hypothesis but also suggest that this salt-bridge is only marginally important in maintaining overall structure, possibly helping maintain these α-helices in position.
Conclusions
The p.K161N and p.D175N mutations were found in two unrelated patients, each with the peripheral form of epimerase-deficient galactosemia [23]. Detailed biochemical analyses in vivo and in vitro have revealed that the K161N and D175N substitutions lead to very different levels of hGALE impairment. It is important to point out that both alleles were found in the heterozygous state, not the homozygous state, in their respective patients [23]. The patient who is heterozygous for hGALE.K161N is also heterozygous for two intronic variants of unknown significance, and the patient who is heterozygous for hGALE.D175N is also heterozygous for two silent substitutions of unknown significance [23]. Parental DNAs were unavailable for both patients, leaving linkage of the different variants in each patient unclear. Nonetheless, given the compound heterozygous nature of the alleles in both patients, that an apparently “severe” substitution (K161N) could be found in a mildly affected patient suggests that the other allele in that patient is likely to retain significant function.
In this way, p.K161N is, functionally, quite similar to the p.G90E variant [38]. p.G90E was also detected in a patient who was a compound heterozygote and presented biochemically and clinically with the benign, peripheral form of epimerase deficiency [1]. Glycine 90 is near the cofactor binding site and its mutation has been proposed to perturb NAD+ binding [38]. Altered cofactor binding has also been found before for one variant, p.N34S, but not for others [40;46]. We cannot rule out the possibility that the mutations studied here cause increased aggregation in vivo as previously observed with some mutants expressed in mammalian GALE-null cells [41]. Coupled with these findings, this suggests the possibility of developing a molecular chaperone treatment similar to that for phenylketonuria and other misfolding diseases [54–57].
In summary we have determined the biochemical basis of impairment of hGALE due to the two substitution mutations, p.K161N and p.D175N. These studies have confirmed that the impairment of the two activities of hGALE, toward UDP-Gal and UDP-GalNAc, may not be strictly correlated and that altered cofactor binding can affect stability as well as activity. They also demonstrate that UDP-galactose 4’-epimerase requires the correct level of flexibility for proper function. An increase or decrease in flexibility, as seen here with p.K161N and p.D175N respectively, results in loss of function. It also cannot be ruled out that more subtle affects are occurring in terms of dimerisation and that these may be important in the enzyme’s possible communication between the two active sites. We recommend that future research on all known and new variants should consider the role of cofactor binding in the stability and structure and flexibility of the protein in the presence of both substrates. This will add further insight into the relationship between specific hGALE mutations and different forms of the disease epimerase-deficiency galactosemia.
Supplementary Material
Highlights.
The K161N variant of human GALE is more resistant to denaturation
K161N has very low detectable NAD+ and greatly reduced enzymatic activity
The D175N variant is less stable and slightly less active than the wild type
GALE’s activities with UDP-Gal and UDP-GalNAc are not necessarily correlated
Acknowledgements
TJM thanks the Department of Employment and Learning, Northern Ireland for a PhD studentship. We wish to thank Prof Aaron Maule (Queen’s University, Belfast) for use of a thermal cycler for the thermal scanning fluorimetry assay and Dr. Kostya Panov (Queen’s University, Belfast) for suggesting this assay. Work conducted in the Fridovich-Keil lab was supported, in part, by funds from the National Institutes of Health (USA) grant R01 DK059904 (to JLFK).
Abbreviations
- ANS
1-anilinonaphthalene-8-sulphonic acid
- BS3
suberic acid bis(3-sulfo-N-hydroxysuccinimide ester)
- Gal-1P
galactose 1-phosphate
- Glc-1P
glucose 1-phopshate
- GALE
UDP-galactose 4′-epimerase
- GuHCl
guanidine hydrochloride
- hGALE
human GALE
- FRET
Förster resonance energy transfer
- NAD+
oxidized nicotinamide adenine dinucleotide
- NADH
reduced nicotinamide adenine dinucleotide
- NAD(H)
nicotinamide adenine dinucleotide (both oxidized and reduced)
- UDP-Gal
uridine diphosphate galactose
- UDP-Glc
uridine diphosphate glucose
- UDP-GalNAc
uridine diphosphate-N-acetylgalactosamine
- UDP-GlcNAc
uridinediphosphate-N-acetylglucosamine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflicts of interest to declare.
References
- 1.Maceratesi P, Daude N, Dallapiccola B, Novelli G, Allen R, Okano Y, Reichardt J. Human UDP-galactose 4' epimerase (GALE) gene and identification of five missense mutations in patients with epimerase-deficiency galactosemia. Mol.Genet.Metab. 1998;63:26–30. doi: 10.1006/mgme.1997.2645. [DOI] [PubMed] [Google Scholar]
- 2.Holton JB, Walter JH, Tyfield LA. Galactosemia. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill, Inc; 2002. [Google Scholar]
- 3.Holden HM, Rayment I, Thoden JB. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J.Biol.Chem. 2003;278:43885–43888. doi: 10.1074/jbc.R300025200. [DOI] [PubMed] [Google Scholar]
- 4.Berry GT, Segal S, Gitzelmann R. Disorders of galactose metabolism. In: Fernandes J, Saudubray JM, van den Berghe G, Walter JH, editors. Inborn Metabolic Diseases: Diagnosis and Treatment. Chapter 7. New York: Springer; 2006. [Google Scholar]
- 5.Bosch AM, Bakker HD, van Gennip AH, van Kempen JV, Wanders RJ, Wijburg FA. Clinical features of galactokinase deficiency: a review of the literature. J.Inherit.Metab.Dis. 2002;25:629–634. doi: 10.1023/a:1022875629436. [DOI] [PubMed] [Google Scholar]
- 6.Waggoner DD, Buist NR, Donnell GN. Long-term prognosis in galactosaemia: results of a survey of 350 cases. J.Inherit.Metab.Dis. 1990;13:802–818. doi: 10.1007/BF01800204. [DOI] [PubMed] [Google Scholar]
- 7.Walter JH, Roberts RE, Besley GT, Wraith JE, Cleary MA, Holton JB, MacFaul R. Generalised uridine diphosphate galactose-4-epimerase deficiency. Arch.Dis.Child. 1999;80:374–376. doi: 10.1136/adc.80.4.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fridovich-Keil JL, Walter JH. Galactosemia. In: Valle D, Beaudet AL, Vogelstein B, Kinzler KW, Antonarakis SE, Ballabio A, editors. The Online Metabolic and Molecular Bases of Inherited Diseases. New York: McGraw-Hill; 2008. [Google Scholar]
- 9.Lai K, Elsas LJ, Wierenga KJ. Galactose toxicity in animals. IUBMB Life. 2009;61:1063–1074. doi: 10.1002/iub.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCorvie TJ, Timson DJ. The structural and molecular biology of type I galactosemia: Enzymology of galactose 1-phosphate uridylyltransferase. IUBMB Life. 2011;63:694–700. doi: 10.1002/iub.511. [DOI] [PubMed] [Google Scholar]
- 11.McCorvie TJ, Timson DJ. Structural and molecular biology of type I galactosemia: disease-associated mutations. IUBMB Life. 2011;63:949–954. doi: 10.1002/iub.510. [DOI] [PubMed] [Google Scholar]
- 12.Holden HM, Thoden JB, Timson DJ, Reece RJ. Galactokinase: structure, function and role in type II galactosemia. Cell Mol.Life Sci. 2004;61:2471–2484. doi: 10.1007/s00018-004-4160-6. [DOI] [PubMed] [Google Scholar]
- 13.Leloir LF. The enzymatic transformation of uridine diphosphate glucose into a galactose derivative. Arch.Biochem. 1951;33:186–190. doi: 10.1016/0003-9861(51)90096-3. [DOI] [PubMed] [Google Scholar]
- 14.Maxwell ES. The enzymic interconversion of uridine diphosphogalactose and uridine diphosphoglucose. J.Biol.Chem. 1957;229:139–151. [PubMed] [Google Scholar]
- 15.Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase. Biochemistry. 2000;39:5691–5701. doi: 10.1021/bi000215l. [DOI] [PubMed] [Google Scholar]
- 16.Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Human UDP-galactose 4-epimerase. Accommodation of UDP-N-acetylglucosamine within the active site. J.Biol.Chem. 2001;276:15131–15136. doi: 10.1074/jbc.M100220200. [DOI] [PubMed] [Google Scholar]
- 17.Holton JB, Gillett MG, MacFaul R, Young R. Galactosaemia: a new severe variant due to uridine diphosphate galactose-4-epimerase deficiency. Arch.Dis.Child. 1981;56:885–887. doi: 10.1136/adc.56.11.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fridovich-Keil JL. Galactosemia: the good, the bad, and the unknown. J.Cell.Physiol. 2006;209:701–705. doi: 10.1002/jcp.20820. [DOI] [PubMed] [Google Scholar]
- 19.Alano A, Almashanu S, Chinsky JM, Costeas P, Blitzer MG, Wulfsberg EA, Cowan TM. Molecular characterization of a unique patient with epimerase-deficiency galactosaemia. J.Inherit.Metab.Dis. 1998;21:341–350. doi: 10.1023/a:1005342306080. [DOI] [PubMed] [Google Scholar]
- 20.Timson DJ. The structural and molecular biology of type III galactosemia. IUBMB Life. 2006;58:83–89. doi: 10.1080/15216540600644846. [DOI] [PubMed] [Google Scholar]
- 21.Gitzelmann R, Steinmann B, Mitchell B, Haigis E. Uridine diphosphate galactose 4'-epimerase deficiency. IV. Report of eight cases in three families. Helv.Paediatr.Acta. 1977;31:441–452. [PubMed] [Google Scholar]
- 22.Fridovich-Keil J, Bean L, He M, Schroer R. Epimerase Deficiency Galactosemia. In: Pagon RA, Bird TD, Dolan CR, Stephens K, Adam MP, editors. GeneReviews™. Seattle (WA): University of Washington, Seattle; 2011. http://www.ncbi.nlm.nih.gov/books/NBK51671/ [Google Scholar]
- 23.Openo KK, Schulz JM, Vargas CA, Orton CS, Epstein MP, Schnur RE, Scaglia F, Berry GT, Gottesman GS, Ficicioglu C, Slonim AE, Schroer RJ, Yu C, Rangel VE, Keenan J, Lamance K, Fridovich-Keil JL. Epimerase-deficiency galactosemia is not a binary condition. Am.J.Hum.Genet. 2006;78:89–102. doi: 10.1086/498985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roper JR, Guther ML, Macrae JI, Prescott AR, Hallyburton I, Acosta-Serrano A, Ferguson MA. The suppression of galactose metabolism in procylic form Trypanosoma brucei causes cessation of cell growth and alters procyclin glycoprotein structure and copy number. J.Biol.Chem. 2005;280:19728–19736. doi: 10.1074/jbc.M502370200. [DOI] [PubMed] [Google Scholar]
- 25.Agarwal S, Gopal K, Upadhyaya T, Dixit A. Biochemical and functional characterization of UDP-galactose 4-epimerase from Aeromonas hydrophila. Biochim.Biophys.Acta. 2007;1774:828–837. doi: 10.1016/j.bbapap.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 26.Agarwal S, Mishra N, Agarwal S, Dixit A. Characterization of the active site and coenzyme binding pocket of the monomeric UDP- galactose 4'- epimerase of Aeromonas hydrophila. BMB Rep. 2010;43:419–426. doi: 10.5483/bmbrep.2010.43.6.419. [DOI] [PubMed] [Google Scholar]
- 27.Sanders RD, Sefton JM, Moberg KH, Fridovich-Keil JL. UDP-galactose 4' epimerase (GALE) is essential for development of Drosophila melanogaster. Dis.Model.Mech. 2010;3:628–638. doi: 10.1242/dmm.005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thoden JB, Frey PA, Holden HM. Crystal structures of the oxidized and reduced forms of UDP-galactose 4-epimerase isolated from Escherichia coli. Biochemistry. 1996;35:2557–2566. doi: 10.1021/bi952715y. [DOI] [PubMed] [Google Scholar]
- 29.Shaw MP, Bond CS, Roper JR, Gourley DG, Ferguson MA, Hunter WN. High-resolution crystal structure of Trypanosoma brucei UDP-galactose 4'-epimerase: a potential target for structure-based development of novel trypanocides. Mol.Biochem.Parasitol. 2003;126:173–180. doi: 10.1016/s0166-6851(02)00243-8. [DOI] [PubMed] [Google Scholar]
- 30.Thoden JB, Holden HM. The molecular architecture of galactose mutarotase/UDP-galactose 4-epimerase from Saccharomyces cerevisiae. J.Biol.Chem. 2005;280:21900–21907. doi: 10.1074/jbc.M502411200. [DOI] [PubMed] [Google Scholar]
- 31.Sakuraba H, Kawai T, Yoneda K, Ohshima T. Crystal structure of UDP-galactose 4-epimerase from the hyperthermophilic archaeon Pyrobaculum calidifontis. Arch.Biochem.Biophys. 2011;512:126–134. doi: 10.1016/j.abb.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 32.Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Molecular basis for severe epimerase deficiency galactosemia. X-ray structure of the human V94m-substituted UDP-galactose 4-epimerase. J.Biol.Chem. 2001;276:20617–20623. doi: 10.1074/jbc.M101304200. [DOI] [PubMed] [Google Scholar]
- 33.Wohlers TM, Fridovich-Keil JL. Studies of the V94M-substituted human UDPgalactose-4-epimerase enzyme associated with generalized epimerase-deficiency galactosaemia. J.Inherit.Metab.Dis. 2000;23:713–729. doi: 10.1023/a:1005682913784. [DOI] [PubMed] [Google Scholar]
- 34.Wohlers TM, Christacos NC, Harreman MT, Fridovich-Keil JL. Identification and characterization of a mutation, in the human UDP-galactose-4-epimerase gene, associated with generalized epimerase-deficiency galactosemia. Am.J.Hum.Genet. 1999;64:462–470. doi: 10.1086/302263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wasilenko J, Fridovich-Keil JL. Relationship between UDP-galactose 4'-epimerase activity and galactose sensitivity in yeast. J.Biol.Chem. 2006;281:8443–8449. doi: 10.1074/jbc.M600778200. [DOI] [PubMed] [Google Scholar]
- 36.McCorvie TJ, Wasilenko J, Liu Y, Fridovich-Keil JL, Timson DJ. In vivo and in vitro function of human UDP-galactose 4'-epimerase variants. Biochimie. 2011;93:1747–1754. doi: 10.1016/j.biochi.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mumma JO, Chhay JS, Ross KL, Eaton JS, Newell-Litwa KA, Fridovich-Keil JL. Distinct roles of galactose-1P in galactose-mediated growth arrest of yeast deficient in galactose-1P uridylyltransferase (GALT) and UDP-galactose 4'-epimerase (GALE) Mol.Genet.Metab. 2008;93:160–171. doi: 10.1016/j.ymgme.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timson DJ. Functional analysis of disease-causing mutations in human UDP-galactose 4-epimerase. FEBS J. 2005;272:6170–6177. doi: 10.1111/j.1742-4658.2005.05017.x. [DOI] [PubMed] [Google Scholar]
- 39.Chhay JS, Openo KK, Eaton JS, Gentile M, Fridovich-Keil JL. A yeast model reveals biochemical severity associated with each of three variant alleles of galactose-1P uridylyltransferase segregating in a single family. J.Inherit.Metab.Dis. 2008;31:97–107. doi: 10.1007/s10545-007-0786-5. [DOI] [PubMed] [Google Scholar]
- 40.Quimby BB, Alano A, Almashanu S, DeSandro AM, Cowan TM, Fridovich-Keil JL. Characterization of two mutations associated with epimerase-deficiency galactosemia, by use of a yeast expression system for human UDP-galactose-4-epimerase. Am.J.Hum.Genet. 1997;61:590–598. doi: 10.1086/515517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bang YL, Nguyen TT, Trinh TT, Kim YJ, Song J, Song YH. Functional analysis of mutations in UDP-galactose-4-epimerase (GALE) associated with galactosemia in Korean patients using mammalian GALE-null cells. FEBS J. 2009;276:1952–1961. doi: 10.1111/j.1742-4658.2009.06922.x. [DOI] [PubMed] [Google Scholar]
- 42.Wang W, Malcolm BA. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange Site-Directed Mutagenesis. BioTechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- 43.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal.Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 44.Ng WG, Donnell GN, Hodgman JE, Bergren WR. Differences in uridine diphosphate galactose-4-epimerase between haemolysates of newborns and of adults. Nature. 1967;214:283–284. doi: 10.1038/214283a0. [DOI] [PubMed] [Google Scholar]
- 45.Chhay JS, Vargas CA, McCorvie TJ, Fridovich-Keil JL, Timson DJ. Analysis of UDP-galactose 4'-epimerase mutations associated with the intermediate form of type III galactosemia. J.Inherit.Metab.Dis. 2008;31:108–116. doi: 10.1007/s10545-007-0790-9. [DOI] [PubMed] [Google Scholar]
- 46.Wasilenko J, Lucas ME, Thoden JB, Holden HM, Fridovich-Keil JL. Functional characterization of the K257R and G319E-hGALE alleles found in patients with ostensibly peripheral epimerase deficiency galactosemia. Mol.Genet.Metab. 2005;84:32–38. doi: 10.1016/j.ymgme.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 47.Ross KL, Davis CN, Fridovich-Keil JL. Differential roles of the Leloir pathway enzymes and metabolites in defining galactose sensitivity in yeast. Mol.Genet.Metab. 2004;83:103–116. doi: 10.1016/j.ymgme.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 48.Parthiban V, Gromiha MM, Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 2006;34:W239–W242. doi: 10.1093/nar/gkl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, Carver T, Asel E, Springer BA, Lane P, Salemme FR. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J.Biomol.Screen. 2001;6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 50.Ericsson UB, Hallberg BM, Detitta GT, Dekker N, Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal.Biochem. 2006;357:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 51.Durrant JD, Urbaniak MD, Ferguson MA, McCammon JA. Computer-aided identification of Trypanosoma brucei uridine diphosphate galactose 4'-epimerase inhibitors: toward the development of novel therapies for African sleeping sickness. J.Med.Chem. 2010;53:5025–5032. doi: 10.1021/jm100456a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friedman AJ, Durrant JD, Pierce LCT, McCorvie TJ, Timson DJ, McCammon JA. The molecular dynamics of Trypanosoma brucei UDP-galactose 4'-epimerase: a drug target for African sleeping sickness. Chem.Biol.Drug Des. 2012 doi: 10.1111/j.1747-0285.2012.01392.x. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swanson BA, Frey PA. Identification of lysine 153 as a functionally important residue in UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1993;32:13231–13236. doi: 10.1021/bi00211a035. [DOI] [PubMed] [Google Scholar]
- 54.Pey AL, Ying M, Cremades N, Velazquez-Campoy A, Scherer T, Thony B, Sancho J, Martinez A. Identification of pharmacological chaperones as potential therapeutic agents to treat phenylketonuria. J.Clin.Invest. 2008;118:2858–2867. doi: 10.1172/JCI34355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santos-Sierra S, Kirchmair J, Perna AM, Reiss D, Kemter K, Roschinger W, Glossmann H, Gersting SW, Muntau AC, Wolber G, Lagler FB. Novel pharmacological chaperones that correct phenylketonuria in mice. Hum.Mol.Genet. 2012;21:1877–1887. doi: 10.1093/hmg/dds001. [DOI] [PubMed] [Google Scholar]
- 56.Muntau AC, Gersting SW. Phenylketonuria as a model for protein misfolding diseases and for the development of next generation orphan drugs for patients with inborn errors of metabolism. J.Inherit.Metab.Dis. 2010;33:649–658. doi: 10.1007/s10545-010-9185-4. [DOI] [PubMed] [Google Scholar]
- 57.Sampson HM, Robert R, Liao J, Matthes E, Carlile GW, Hanrahan JW, Thomas DY. Identification of a NBD1-binding pharmacological chaperone that corrects the trafficking defect of F508del-CFTR. Chem.Biol. 2011;18:231–242. doi: 10.1016/j.chembiol.2010.11.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.