Abstract
Complex I is a critical site of O2•− production and the major host of reactive protein thiols in mitochondria. In response to oxidative stress, Complex I protein thiols at the 51 kDa and 75 kDa subunits are reversibly S-glutathionylated. The mechanism of Complex I S-glutathionylation is mainly obtained from insight into GSSG-mediated thiol-disulfide exchange, which would require a dramatic decline in the GSH/GSSG ratio. Intrinsic Complex I S-glutathionylation can be detected in the rat heart at a relatively high GSH/GSSG ratio (Chen, J., et al. J. Biol. Chem 285: 3168–3180, 2010). Thus, we hypothesized that reactive thiyl radical is more likely to mediate protein S-glutathionylation of Complex I. Here we employed immuno-spin trapping and tandem mass spectrometry (LC/MS/MS) to test the hypothesis in the 75 kDa subunit from S-glutathionylated Complex I. Under the conditions of O2•− production in the presence of GSH, we detected Complex I S-glutathionylation at the Cys-226, Cys-367, and Cys-727 of the 75 kDa subunit. Addition of a radical trap, 5, 5-dimethyl-1-Pyroline-N-Oxide (DMPO), significantly decreased Complex I S-glutathionylation, and subsequently increased the protein radical adduct of Complex I-DMPO as detected by immunoblotting using an anti-DMPO antibody. LC/MS/MS analysis indicated that Cys-226, Cys-554, and Cys-727 were involved in DMPO-binding, confirming that formation of the Complex I thiyl radical mediates S-glutathionylation. LC/MS/MS analysis also showed that Cys-554 and Cys-727 were S-sulfonated under conditions of O2•− generation in the absence of DMPO. In myocytes (HL-1 cell line) treated with menadione to trigger mitochondrial O2•− generation, Complex I protein radical and S-glutathionylation were increased. Thus mediation of Complex I S-glutathionylation by the protein thiyl radical provides a unique pathway for the redox regulation of mitochondrial function.
Keywords: Complex I, S-glutathionylation, Protein Thiyl Radical, Immuno-spin Trapping, Myocytes
INTRODUCTION
Mitochondrial Complex I (EC 1.6.5.3. NADH:ubiquinone oxidoreductase, NQR2) is the first energy-conserving segment of the electron transport chain (ETC) [1–3]. Purified bovine heart Complex I contains up to 45 different subunits with a total molecular mass approaching 1 million Da (see Supplementary Fig. 1S). The enzyme catalyzes electron transfer from NADH to ubiquinone coupled with the translocation of four protons across the membrane. In addition to its functions of electron transfer and energy transduction, the catalysis of Complex I provides the major source of O2•− in mitochondria [4–6]. Two regions of the enzyme complex are hypothesized to be responsible for generating the superoxide anion radical (O2•−). One is located on the FMN cofactor and is modulated by its binding protein moiety [4, 5, 7], while the other is likely located on the ubiquinone-binding site and probably acts in the mediation of ubiquinone reduction [8–10].
In mitochondria, the generation of O2•− and the oxidants derived from it can act as a redox signal in triggering cellular events such as apoptosis, proliferation, and senescence. The mitochondrial redox pool is enriched in glutathione (GSH) with a high physiological concentration (in the mM range) [11]; overproduction of O2•− and O2•−-derived oxidants increases the ratio of GSSG (oxidized glutathione) to GSH. Complex I is the major component of the ETC to host protein thiols, which comprise structural thiols involved in the ligands of iron sulfur clusters and the reactive/regulatory thiols that are thought to function in antioxidant defense and redox signaling [12, 13]. Physiologically, the Complex I-derived regulatory thiols have been implicated in the regulation of respiration, nitric oxide utilization [14, 15], and redox status of mitochondria [11, 16, 17]. It has been documented that the subunits of 51 kDa and 75 kDa from the hydrophilic domain of Complex I are two major polypeptides that host reactive thiols in mitochondria [12, 13, 18].
We and others have reported that both the 51 kDa and 75 kDa subunits are involved in the redox modification of S-glutathionylation in vitro and in vivo [10, 12, 13, 18]. In the animal disease model of myocardial ischemia and reperfusion injury, we have demonstrated that intrinsic S-glutathionylation of Complex I can be detected in the rat myocardium and isolated mitochondria, and enhanced in the post-ischemic heart [10]. In vitro studies using isolated mitochondria also indicate that increasing formation of Complex I S-glutathionylation is favored by the conditions of oxidative stress such as exposure to organic peroxide [13], excess NO [13], or the thiol oxidant diamide [19]. In vitro oxidant exposure of mitochondria also resulted in impaired Complex I activity, but reversal of glutathionylation by DTT failed to restore Complex I activity, suggesting a non-essential role for glutathionylation in oxidant-induced damage to Complex I activity [19]. In vitro S-glutathionylation of Complex I with a low dose of GSSG leads to marginal enhancement of the electron transfer efficiency and a decrease in the electron leakage [18]. Therefore, an increase in Complex I S-glutathionylation in vivo is likely important in preventing oxidative damage to the enzyme.
The redox signal in the regulation of Complex I-derived S-glutathionylation is thus hypothesized to involve reactive oxygen species (ROS) production and homeostasis of the GSH pool in mitochondria. S-glutathionylation of Complex I may proceed spontaneously by thiol-disulfide exchange [12, 18]. However, achieving this thermodynamic equilibrium would require a marked decline in the intracellular GSH/GSSG ratio [20, 21]. Therefore, the mechanism of thiol-disulfide exchange leading to S-glutathionylation can only occur under extreme conditions, which is not a likely mechanism in vivo. A kinetic mechanism, a reaction of protein reactive sulfhydryl intermediate with GSH or vice versa, is more likely to mediate S-glutathionylation of Complex I [20, 21]. We have reported previously that in oxidative damage to Complex I and its flavin subcomplex (NADH dehydrogenase), the C206 moiety of the 51 kDa subunit plays a unique role as a reactive protein thiyl radical intermediate, based on the evidence of immuno-spin trapping with 5,5-dimethyl pyrroline N-oxide (DMPO) and mass spectrometry (MS) [7]. With a proteomic approach, we further determined that the C206 moiety of the 51 kDa subunit is involved in site-specific S-glutathionylation [18], thus lending support to the idea that Complex I-derived thiyl radical formation induced by O2•− mediates protein S-glutathionylation in the presence of GSH.
Although there is strong evidence indicating that thiol-disulfide exchange is involved in directly modulating S-glutathionylation of Complex I, there is a lack of prior investigation directed toward understanding how the protein reactive sulfhydryl intermediate mediates S-glutathionylation of Complex I. Determination of the molecular mechanism of Complex I S-glutathionylation is of particular importance because of the implications of this regulation in cardiovascular disease and the physiological settings of mitochondrial redox. Therefore, studies were performed to gain insights into the novel mechanism of Complex I S-glutathionylation with a focus on the 75 kDa subunit. By immuno-spin trapping and subsequent LC/MS/MS analysis, we report the evidence of protein thiyl radicals participating in reversible S-glutathionylation of Complex I in the levels of isolated enzyme and myocytes. We have further detected Complex I-derived irreversible S-sulfonation that is mediated by protein thiyl radical intermediates under conditions of oxidant stress in vitro.
MATERIAL AND METHODS
Reagents
Glutathione (GSH), ammonium sulfate, diethylenetriaminepentaacetic acid (DTPA), ubiquinone-1 (Q1), decyl ubiquinone (DBQ), sodium cholate, deoxycholic acid, rotenone, PEGSOD (polyethylene glycol-linked superoxide dismutase), and β-nicotinamide adenine dinucleotide (reduced form, NADH) were purchased from Sigma Chemical Company (St. Louis, MO) and used as received. The anti-DMPO polyclonal antibody was purchased from ENZO (San Diego, CA). The DMPO spin trap from Sigma-Aldrich was vacuum-distilled twice and stored under nitrogen at −80 °C until needed.
Preparations of Mitochondrial Complex I
Bovine heart mitochondrial Complex I was prepared under non-reducing conditions according to the published method by Hatefi et al. with minor modifications [22] detailed in previous publications [10, 18]. Preparation of Complex I contains 4.2–4.5 nmol ubiquinone-10 and 0.2 mg of phospholipid per mg protein. The contamination of Complex III in this preparation is less than 0.5% (~0.05–0.08 nmol of heme b plus heme c1 per mg of Complex I preparation), and exhibits adequate activity to generate O2•− (determined by EPR spin-trapping with DEPMPO) under the conditions of enzyme turnover [10, 18]. The SDS-PAGE of the isolated Complex I is indicated in the supplementary Fig. S1.
Alkylation of Complex I with Iodoacetamide
DTT-treated Complex I (0.2 mg/ml) in PBS was incubated with iodoacetamide (1 mM) at room temperature. After 1 h incubation, more iodoacetamide was added to the final concentration of 1.5 mM and the mixture was incubated at 4 °C for 8h. The protein band of 75 kDa subunit in the SDS-PAGE was subjected to in-gel digestion with trypsin or chymotrypsin or both, and followed by nano-LC/MS/MS analysis.
Analytical Methods
Optical spectra were measured on a Shimadzu 2401 UV/VIS recording spectrophotometer. The protein concentrations of SMP and Complex I were determined by the Biuret method using BSA as a standard. The concentration of Q1 or DBQ was determined by absorbance spectra from NaBH4 reduction using a millimolar extinction coefficient ε(275nm–290nm) = 12.25 mM−1cm−1 [23]. The electron transfer activity of Complex I was assayed by measuring rotenone-sensitive NADH oxidation by Q1 as developed by Hatefi et al. [24], where an appropriate amount of Complex I was added to an assay mixture (1 ml) containing 20 mM potassium phosphate buffer, pH 8.0, 2mM NaN3, and 0.1 mM Q1, and 0.15 mM NADH. The Complex I activity was determined by measuring the decrease in absorbance at 340nm. The specific activity of Complex I was calculated using a molar extinction coefficient of ε340nm = 6.22 mM−1cm−1. The isolated Complex I using the above Hatefi’s method exhibited a specific activity of ~1.0 μmol NADH oxidized min−1 mg−1 at 23° C.
Electron Paramagnetic Resonance Experiments
EPR measurements were carried out on a Bruker EMX Micro spectrometer operating at 9.43 GHz with 100 kHz modulation frequency at room temperature. The reaction mixture was transferred to a 50 μl capillary, which was then positioned in the HS cavity (Bruker Instrument, Billerica, MA). The sample was scanned using the instrumental parameters described in a previous publication [10]. The spectral simulations were performed using the WinSim program developed at NIEHS by Duling [25].
Immunoblotting Analysis
Reaction mixture was mixed with the Laemmli sample buffer at a ratio of 4:1 (v/v), incubated at 70 °C for 10 min, and then immediately loaded onto a 4–12 % Bis-Tris polyacrylamide gradient gel. Samples were run at room temperature for 55 min at 190 V. Protein bands were electrophoretically transferred to a nitrocellulose membrane in 25 mM Bis-Tris, 25 mM Bicine, 0.029% (w/v) EDTA, and 10% methanol. Membranes were blocked for 1 h at room temperature (R.T.) in Tris-buffered saline (TBS) containing 0.1% Tween-20 (TTBS) and 5% dry milk (BioRad, Hercules, CA). The blots were then incubated overnight with anti-GSH monoclonal antibody or anti-DMPO polyclonal antibody at 4 °C. Blots were then washed 3 times in TTBS, and incubated for 1 h with horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG in TTBS at R.T. The blots were again washed twice in TTBS and twice in TBS, and then visualized using ECL Western Blotting Detection Reagents (GE Healthcare Life Sciences, Fairfield, CT).
Trypsin, Chymotrypsin and Trypsin/Chymotrypsin Digestion
The protein separated by SDS-PAGE gels was digested with sequencing grade trypsin from Promega (Madison, WI) or sequencing grade chymotrypsin from Roche (Indianapolis, IN) using the Montage In-Gel Digestion Kit from Millipore (Bedford, MA) following the manufacturer’s recommended protocols detailed in a previous publication [26].
Nano-Liquid Chromatography-Nanospray Tandem Mass Spectrometry (Nano-LC/MS/MS)
Nano LC/MSMS was performed on a Thermo Finnigan LTQ mass spectrometer equipped with a nanospray source operated in positive ion mode. The LC system was an UltiMate™ Plus system from LC-Packings A Dionex Co (Sunnyvale, CA). Solvent A was water containing 50 mM acetic acid and solvent B was acetonitrile. Each sample was first injected onto the trapping column (LC-Packings A Dionex Co, Sunnyvale, CA), and washed with 50 mM acetic acid before being eluted off the trap onto a 5 cm 75 μm ID ProteoPep II C18 column (New Objective, Inc. Woburn, MA) for chromatographic separation. Peptides were eluted directly off the column into the LTQ system using a gradient of 2–80% B over 30 minutes, with a flow rate of 300 nL/min. The total run time was 58 minutes. The MS/MS was acquired according to standard conditions established in the laboratory [27].
The RAW data files collected on the mass spectrometer were converted to mzXML and MGF files by use of MassMatrix data conversion tools (http://www.massmatrix.net/download). Isotope distributions for the precursor ions of the MS/MS spectra were deconvoluted to obtain the charge states and monoisotopic m/z values of the precursor ions during the data conversion. Database searching was performed against the NCBInr database using the MASCOT 2.0 (Matrix Science, Boston, MA) for the identification of carbamidomethylated cysteines. The mass tolerance of the precursor ions was set to 1.5 Da to accommodate accidental selection of the C13 ion and the fragment mass tolerance was set to 0.5 Da. The number of missed cleavages permitted in the search was 2 for both tryptic and chymotryptic digestions. The variable modifications were methionine oxidation and cysteine carbamidomethylation.
Cell Culture and Fluorescence Microscopy
Mouse myocytes (HL-1 cell line) were grown and maintained in Claycomb medium (Sigma-Aldrich, St. Louise, MO) supplemented with 10% fetal bovine serum, 0.01% penicillin/streptomycin antibiotic, 2 mM glutamine, and 100 μM norepinephyrine in 35 mm polystyrene tissue culture flasks at 37°C in the presence of 5% CO2 as described (28). Confluent cells with > 60% viability were used to conduct immunofluorescent staining with anti-GSH or anti-DMPO antibodies.
HL-1 myocytes were then cultured on sterile coverslips in 35-mm dishes and subjected to menadione (50 μM) treatment for 20 min at 37 °C. For the immunostaining experiment, HL-1 cells (menadione treatment with or without DMPO addition) on coverslips were washed with PBS and fixed with 3.7% paraformaldehyde for 20 min. After permeabilization and blocking with 0.3% Triton X-100 and 5% goat serum in PBS for 30 min, cells were incubated with anti-GSH monoclonal antibody for detection of glutathionylation or anti-DMPO monoclonal antibody for detecting protein radical adduct of DMPO, or Ab51/Ab75 for the visualization of 51 kDa/75 kDa subunit of Complex I. After treatment of cells with the chosen primary antibodies, they were incubated with secondary anti-rabbit AlexaFluor 488-conjugated and secondary anti-mouse AlexaFluor 594-conjugated antibodies for 1h at R.T. The coverslips with cells were mounted on a glass slide with antifade mounting medium, VECTA SHIELD (Vector Laboratories, Burlingame, CA), viewed with fluorescence microscopy (OLYMPUS American Cooperate, model IX-71, Center Valley, PA) with a 40× objective, and overlaid with QCapture Pro Software (QImaging, Surrey, BC, Canada), generating a merged image for each co-stained specimen.
RESULTS AND DISCUSSION
O2•− Mediates S-Glutathionylation of Complex I in the Presence of GSH
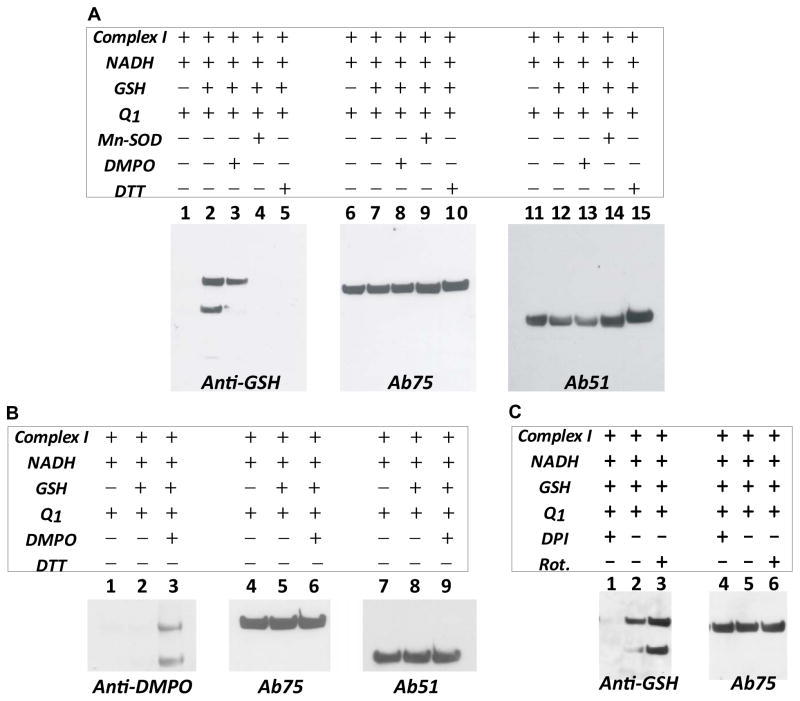
Mitochondrial Complex I is one of the major sources of oxygen free radical generation in mitochondria. O2•− production mediated by isolated Complex I has been unambiguously detected by EPR spin trapping with DEPMPO (5-diethoxyphosphoryl-5-methyl-1-pyrroline N-oxide) under conditions of enzyme turnover [10, 18]. We hypothesize that O2•− may induce protein-derived thiyl radical(s) on the surface of Complex I, and these nascent protein thiyl radical(s) may react with GSH, leading to Complex I S-glutathionylation. To test this hypothesis, as-isolated enzyme (5 mg/ml) was incubated with dithiothretol (DTT, 1 mM) to ensure fully cysteinyl sulfhydryl form. The DTT-treated Complex I was then precipitated by ammonium sulfate (36% saturation) to remove excess reducing agent. DTT-treated Complex I (0.5 mg/ml–2.0 mg/ml, ~0.5 μM–2 μM) was pre-incubated with Q1 (or decylubiquinone, 100 μM) and GSH (2 mM) in the phosphate buffered saline (PBS, pH 7.4), and the reaction was then initiated by the addition of NADH (1.0 mM) at 37 °C. After 1h of incubation, the reaction mixture was subjected to SDS polyacrylamide gel electrophoresis under non-reducing conditions and then immunoblotted with an anti-GSH monoclonal antibody (Fig. 1A, lane 1–5). As indicated in Fig. 1A (lane 2), both 75 kDa and 51 kDa subunits of Complex I were detected to be S-glutathionylated, as verified by re-probing the blotting with the polyclonal antibodies against the Complex I 75 kDa (Fig. 1A, lane 6–10) and 51 kDa (Fig. 1A, lane 11–15) subunits respectively [10, 29]. The detected Western signal of S-glutathionylation depends on the presence of GSH (Fig. 1A, lane 1 versus lane 2). Addition of Mn-SOD (Fig. 1A, lane 4) can eliminate this signal, indicating that the presence of O2•− is essential for the production of this S-glutathionylated band. These results support the hypothesis that protein S-glutathionylation is enhanced by conditions of oxidant stress, and that both the reactive thiol intermediate and GSH are required for this process.
FIGURE 1.
O2•− generated from Complex I induced Complex I S-glutathionylation in the presence of GSH. A, Isolated Complex I (2.0 mg/ml) in PBS containing DTPA (1 mM) was incubated with Q1 (0.1 mM) and GSH (2 mM), and the reaction was initiated by the addition of NADH (1.0 mM) at 37 °C for 60 min. An aliquot (100 μg protein) of reaction mixture was subjected to SDS-PAGE under non-reducing conditions, and then immunoblotted with anti-GSH monoclonal antibody (lane 2). Lane 3, same as lane 2, except that the radical trap DMPO (200 mM) was included in the reaction system. Lane 4, same as lane 2, except that Mn-SOD (1 unit/μl) was included in the system. Lane 5, same as lane 2, except that DTT (1 mM) was included in the system. Lanes 6–10, blotting was probed with antibodies against the 75 kDa subunit (Ab75). Lanes 11–15, blotting was probed with antibodies against the 51 kDa subunit (Ab51). B, samples from lanes 1–3 in A were probed with anti-DMPO polyclonal antibody, Ab 75, and Ab51. C, Effect of DPI or rotenone on the S-glutathionylation of Complex I. System is the same as lane 2 in A, except that DPI (50 μM) or rotenone (20 μM) was included in the system.
Detection of Protein Radical Intermediate for S-Glutathionylation of Complex I
To test the hypothesis that O2•− can induce the formation of protein radical(s) on Complex I, we included a radical trap DMPO (200 mM) in the complete system prior to the addition of NADH (Fig. 1A, lane 3). This resulted in a significant decrease in the 75 kDa-derived S-glutathionylation and nearly complete elimination of the 51 kDa-derived S-glutathionylation, suggesting that (i) the high dosage of DMPO competed with GSH in binding to the protein thiyl radical(s) of Complex I, and (ii) DMPO trapping of Complex I-derived thiyl radical out-competed the rate of S-glutathionylation. DMPO traps the protein radical, forming the DMPO-protein radical adduct which can be detected by immunoblotting using anti-DMPO antibody (Reaction 2 of the Fig. 7). To determine whether the protein radical formed was trapped by DMPO or not, the membrane (Fig. 1A, lanes 1–3) was re-probed with anti-DMPO polyclonal antibodies to detect DMPO-protein radical adduct. As indicated in Fig. 1B (lanes 1–3), both the 51 kDa and 75 kDa subunits reacted specifically with anti-DMPO antibodies, thus confirming the formation of DMPO-protein radical adducts of Complex I at the 51 kDa and 75 kDa subunits (Fig. 1B, lanes 4–6 and lanes 7–9).
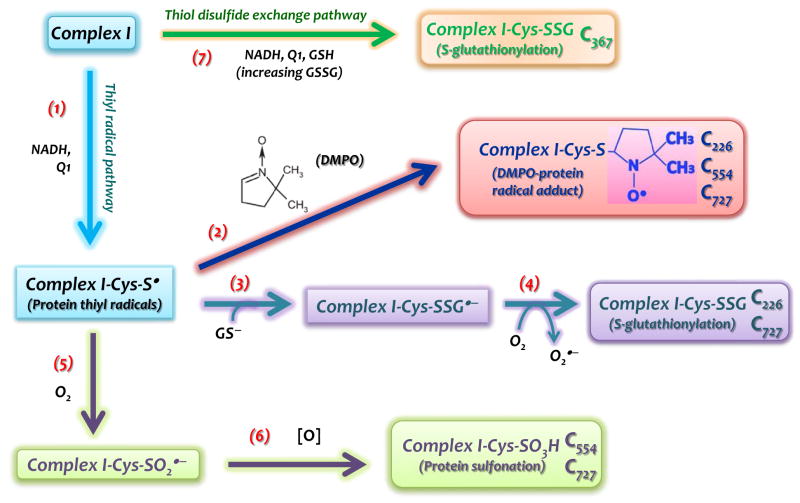
FIGURE 7.
Schematic delineation in which protein thiyl radical mediates Complex I S-glutathionylation and other oxidative modifications. The formation of Complex I thiyl radical intermediate (Complex I-Cys-S•) at the 75 kDa subunit is induced by oxidative attack of O2•− under the enzyme turnover conditions (Reaction 1). Immuno-spin trapping with DMPO (Reaction 2) and MS analysis reveal the residues of C226, C554, and C727 to be involved in the Complex I-Cys-S•. Intrinsic protein S-glutathionylation (Complex I-Cys-SSG) of complex I at C226 and C727 is facilitated by the reaction of Complex I-Cys-S• with glutathione thiolate ion (GS−) (Reaction 3) and subsequent production of O2•− (Reaction 4).. Complex I-Cys-S• can also mediate complex I S-sulfonation (Complex I-Cys-SO3H) at C554 and C727 via cysteine sulfinyl radical (Complex I-Cys-SO2•) and further reaction with oxidants (Reactions 5&6). Complex I-Cys-SSG can also occur independently of protein thiyl radical. GSSG, a product accumulated by the reaction system of complex I/NADH/Q1/GSH, may directly mediate Complex I-Cys-SSG at the C367 (Reaction 7) via thiol disulfide exchange.
Several protein-derived reactive thiol intermediates are likely to mediate S-glutathionylation in cells [20, 21]. Protein thiyl radicals (Pr-S•), among the shortest-lived sulfhydryl intermediates, readily mediate protein S-glutathionylation via reaction with glutathione thiolate (GS−) to form protein-GSH mixed disulfide anion radicals (Pr-SSG•− in eq. 1) [20, 21]. Pr-SSG•− is a source of O2•− (eq. 2), which increases cellular oxidant stress. Therefore, an increasing marker of Complex I-derived S-glutathionylation can serve as an indicator of overproduction of O2•− by mitochondria under disease conditions such as myocardial infarction [10]. A similar case is also observed in the enhanced eNOS S-glutathionylation in the vascular system of the hypertensive rat [30].
| (eq. 1) |
| (eq. 2) |
Effect of DPI and Rotenone on the Protein S-Glutathionylation of Complex I
Diphenyleneiodonium (DPI) is a flavoprotein inhibitor which was reported to abolish O2•− generation by Complex I via directly modifying the reduced form of FMN of the 51 kDa subunit [5, 7]. Addition of DPI (50 μM) to the complete system (Fig. 1A, lane 2) inhibited Complex I-derived S-glutathionylation, as indicated in Fig. 1C (lane 1), confirming that FMN and its binding moiety control protein S-glutathionylation of Complex I. Rotenone inhibits the O2•− generation mediated by unstable semiquinone radical via binding to the ubiquinone-binding site of Complex I, but does not affect the O2•− production mediated by FMN and its binding moiety [10]. Inclusion of rotenone (20 μM) in the mixture of the complete system resulted in the enhancement of protein S-glutathionylation of Complex I, implying that electron leakage for O2•− generation accumulated at the domain of the flavoprotein and subsequently increased protein thiyl radical intermediates for Complex I S-glutathionylation (Fig. 1C, lane 3).
In our previous studies, we employed EPR spin trapping with DEPMPO to demonstrate that ~50% of the Complex I-mediated O2•− generation was derived from a source of unstable ubisemiquinone-1 (Q1•−) under conditions of enzyme turnover [10]. ~80% of O2•− generation mediated by semiquinone can be inhibited by rotenone [10]. Presumably, rotenone marginally stimulates the Fp-mediated O2•− generation by ~20%, which in turn enhances Complex I S-glutathionylation via increasing protein radical formation.
Identification of the Glutathione (GS)–binding Sites from Complex I by Mass Spectrometry
To understand how O2•−-induced protein S-glutathionylation affects Complex I function, it is important to determine the precise sites of GS-binding. Therefore, the reaction mixture of the complete system was subjected to SDS-PAGE separation (Fig. 1A, lane 2), and the protein band corresponding to the glutathionylated 75 kDa polypeptide was cut and subjected to in-gel digestion with trypsin, chymotrypsin or both. The digested peptide fragments were analyzed by nano-LC/MS/MS analysis as described under “Experimental Procedure.” Table I summarizes the peptide sequences and their corresponding oxidative modification of cysteine based on the MS analysis.
Table I.
Summary of the peptide sequence and corresponding oxidative post-translational modification (OPTM) obtained from MS/MS analysis
| Amnio Acid Residue in the 75 kDa Subunit | Ion and m/z | Peptide Sequence and OPTM | Remarks |
|---|---|---|---|
| C226 | 788.822+ | 219SGNIIDIC226(SG)PVGAL231 | S-glutathionylation |
| C226 | 692.693+ | 219SGNIIDIC226(DMPO)PVGAL231 | thiyl radical adduct |
| C367 | 1301.852+ | 361VDSDTLC367(SG)TEEVFPTAGAGTDLR382 | S-glutathionylation |
| C554 | 527.033+ | 544MLFLLGADGGC554(DMPO)ITR557 | thiyl radical adduct |
| C554 | 780.832+ | 544M(ox)LFLLGADGGC554(SO3)ITR557 | S-sulfonation |
| C727 | 607.013+ | 713AVTEGAHAVEEPSIC727(SG) | S-glutathionylation |
| C727 | 542.183+ | 713AVTEGAHAVEEPSIC727(DMPO) | thiyl radical adduct |
| C727 | 766.182+ | 713AVTEGAHAVEEPSIC727(SO3) | S-sulfonation |
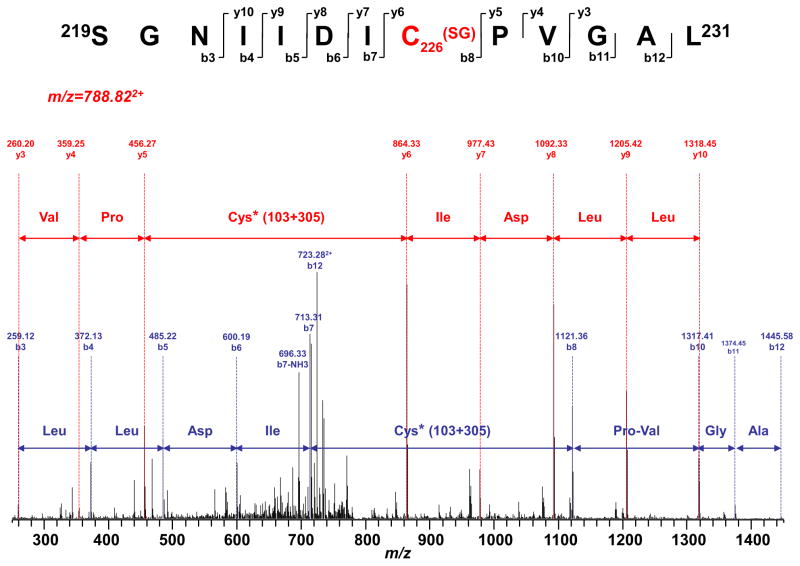
Whether digested with trypsin, chymotrypsin or both, the LC/MS/MS results from the 75 kDa subunit showed that 72.6% of the polypeptide amino acid sequence was identified (Supplementary Fig. S2). The spectra of the digests were also examined for S-glutathionylation, which causes a mass shift of 305 Da to the native intact peptide. Three peptides were identified with the mass shift, and MS/MS analysis of these three peptides further confirmed S-glutathionylation at C367 (361VDSDTLC367TEEVFPTAGAGTDLR382, m/z 1301.852+, Table I and Supplementary Fig. S3a), C226 (219SGNIIDIC226PVGAL231, m/z 788.822+, Fig. 2 and Table I), and C727 (713AVTEGAHAVEEPSIC727, m/z 607.013+, Table I and Supplementary Fig. S3b).
FIGURE 2.
MS/MS spectra of the protonated molecular ions of S-glutathionylated peptides 219SGNIIDIC226PVDAL231. The sequence-specific ions are labeled as y and b ions. The amino acid residues involved in S-glutathionylation are labeled at C226.
S-glutathionylation of C367 was likely mediated by GSSG via thiol disulfide exchange (or GSH-mixed disulfide exchange) as reported previously [18] and illustrated in the scheme of Fig. 7 (Reaction 7). This hypothesis was further supported by the detection of an increasing GSSG/GSH ratio from the reaction mixture. Quantitative determination of GSH and GSSG levels in the reaction mixture of the complete system by an enzymatic recycling method 60 min after NADH addition indicated a dramatic elevation in the GSSG level (from 0.032±0.001 mM to 0.54±0.02 mM, n=6) and reduction of the GSH/GSSG ratio (decreasing from 66.67 to 1.95±0.11) as indicated in Supplementary Fig. S4A.
C727 is located at the C-terminus. GSSG-mediated S-glutathionylation of C727 can only occur at a higher dosage (5 mM) of GSSG used [19], which is not likely physiologically relevant. However, C727 glutathionylation was also detected from diamide-treated mitochondria, or induced by endogenous oxidative stress (antimycin A, rotenone, or H2O2) within mitochondria [19]. An additional mechanism for C727 glutathionylation independent of GSSG should thus be considered. Formation of an oxidant-induced protein-derived reactive thiol intermediate is likely involved in C727 S-glutathionylation.
C226 is involved with iron-sulfur center ligation (the N4 center of the 75 kDa subunit). GSSG-mediated C226 glutathionylation can only be detected using a very high and non-physiologically relevant concentration of GSSG (20 mM) or SDS-denatured Complex I [19]. Therefore, C226 glutathionylation in this study is independent of GSSG, and to account for the O2•− generation-inducing self-inactivation of Complex I. To support this conclusion, we assayed the electron transfer activity of Complex I from the reaction mixture by Q1-mediated NADH oxidation after a 60 min incubation. We observed that 31% of the Complex I activity was irreversibly impaired (Supplementary Fig. S4B), presumably due to C226 S-glutathionylation, leading to destruction of the N4 center.
Identification of the DMPO- binding Sites from the Complex I 75 kDa subunit by Mass Spectrometry
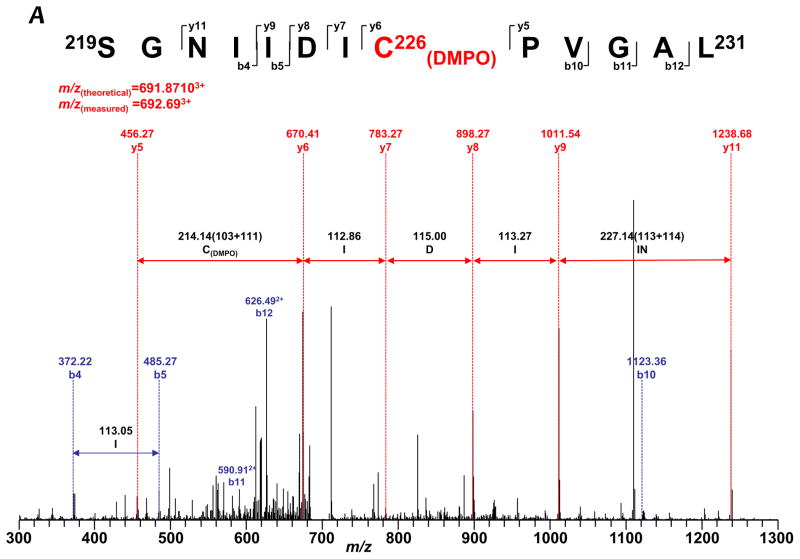
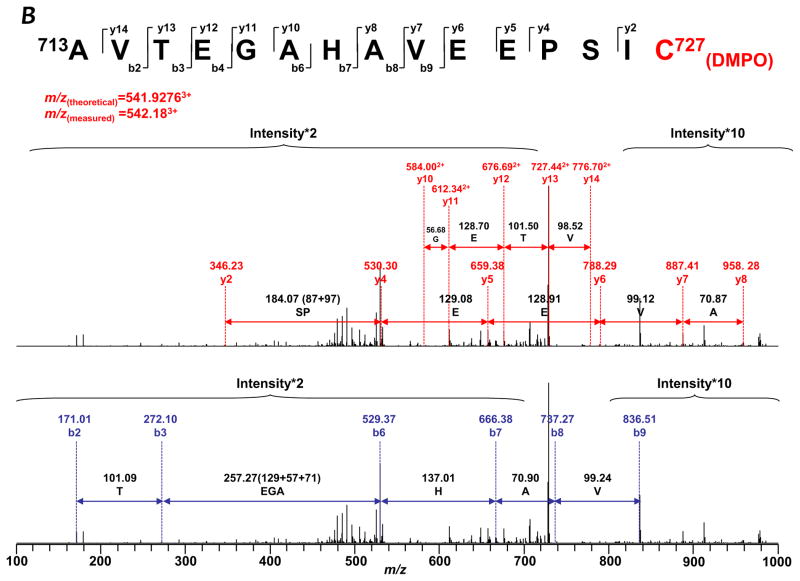
To gain a deeper insight into how protein radicals mediate S-glutathionylation, it is imperative to determine the location of DMPO binding. Therefore, the reaction mixture of the complete system with DMPO (Fig. 1B, lane 2) was subjected to SDS-PAGE separation under reducing conditions. The protein band of 75 kDa subunit was subjected to in-gel digestion with trypsin, chymotrypsin, or both, followed by nano-LC/MS/MS analysis. 96.4% of the amino acid sequence was identified in the MS/MS spectra (Supplementary Fig. S5), and the DMPO was exclusively bound to the cysteinyl residue of the 75 kDa subunit as revealed by MS/MS analysis. The addition of DMPO to cysteine residues causes a mass increase of 111 Da to the native intact peptide. This mass difference was observed for one chymotryptic peptide (229SGNIIDIC236PVGAL241, observed m/z 692.693+), and two tryptic peptides (713AVTEGAHAVEEPSIC727, observed m/z 542.183+, and 544MLFLLGADGGC554ITR557, m/z 527.033+). This mass shift was further observed in MS/MS fragmentation ions y6–y9, y11, and b10–b12 for peptide 229SGNIIDIC(DMPO)236PVGAL241 (Table I and Fig. 3A), which indicates that one DMPO molecule is covalently bound to C226. Similarly, the mass shift was detected for fragmentation ions y2, y4–y8, y10–y14 from peptide 713AVTEGAHAVEEPSIC(DMPO)727 (Table I and Fig. 3B), and y6–y13 from peptide 544MLFLLGADGGC(DMPO)554ITR557 (Table I and Supplementary Fig. S6). This observation suggests that DMPO adducts are formed on C226, C554, and C727.
FIGURE 3.
MS/MS spectra of the triply protonated molecular ions of DMPO-binding peptides. A, 219SGNIIDIC226PVGAL231. B, 713AVTEGAHAVEEPSIC727. The amino acid residues involved in DMPO binding are identified as C226 and C727.
Detection of DMPO-binding at the residues of C226 and C727 reinforces the proposed mechanism of Complex I S-glutathionylation as driven by the reaction of specific protein thiyl radicals with glutathione thiolate (Eq. 1, and Reaction 3 of the Fig. 7), which renders glutathionylation of C226 and C727 as potential sources of O2•− (Eq. 2, and Reaction 4 of the Fig. 7). Identification of DMPO-binding at the residue of C554 further supports the C554 as the specific site of the protein thiyl radical formation. C554 glutathionylation can be induced by a higher dosage of GSSG (5 mM), and detected in diamide-treated mitochondria [19]. In the diamide-treated mitochondria, a higher concentration of GSSG (GSH/GSSG ~ 0.1 [19]) is built up with partial depletion of GSH, thus facilitating the GSSG-mediating pathway. Failure to detect C554 glutathionylation in this study is likely due to a lower amount of GSSG produced (~0.5 mM and GSH/GSSG ratio ~ 2, vide ante) in the reaction mixture of the complete system. Presumably, the redox of the C554 thiol was controlled by enzyme-mediated O2•− generation, forming a reactive cysteine thiyl radical intermediate and then trapped by DMPO or oxidized to sulfinic acid or sulfonic acid (vide infra).
It should be noted that, although O2•− may not directly react with GSH and cysteine for the formation of protein thiyl radicals, an indirect pathway, the Haber-Weiss reaction (O2•−+H2O2 → HO•+O2+HO3), should be considered as a possible mechanism. In this reaction, O2•−-dependent hydroxyl radical could be generated in the presence of iron, O2•− and H2O2, and the hydroxyl radical could subsequently induce the formation of the Complex I protein thiyl radicals. This mechanism favors the formation of C226-derived thiyl radical due to involving of the 4Fe-4S ligand.
Cysteine S-sulfonation of the Complex I 75 kDa subunit
To better understand the Complex I-derived cysteine oxidation mediated by O2•−, we further examined the oxidative modification with cysteine S-sulfonation (conversion of SH to SO3H) from the system of Complex I/NADH/Q1 (complete system in the absence of GSH) using nano-LC/MS/MS as described previously. The mass spectra were scrutinized for a mass shift of 48 Da caused by S-sulfonation. Mass spectrometric analysis showed a 48 Da mass increase in the tryptic peptide 713AVTEGAHAVEEPSIC(SO3)727. Further MS/MS analysis of this peptide revealed that the S-sulfonation was located on C727 (Table I and Supplementary Fig. S7a). Likewise, another S-sulfonation site was identified at C554 (544M(ox)LFLLGADGGC(SO3)554ITR557, m/z 780.832+, Table I and Supplementary Fig. S7b). Specific S-sulfonation of C727 and C554 represents the major mechanism of irreversible cysteine oxidation under the conditions of oxidant stress.
In the presence of DMPO (100 mM, the reaction system of Complex I/NADH/Q1/DMPO), S-sulfonation at C554 and C727 was not detected in the MS/MS analysis. Instead, cysteine sulfination (conversion of SH to SO2H, mass shift 32 Da) and DMPO adducts (in Table I) were detected at C554 (LLGADGGC554 (SO2), m/z 554.682+) and C727 (AVTEGAHAVEEPSIC727 (SO2), m/z 772.692+) as indicated in the Table II and Supplementary Table, presumably due to incomplete oxidation of cysteine residue. It is likely that the formation of protein thiyl radicals at C727 and C554 accelerates the corresponding cysteine oxidation to form protein cysteinyl sulfinyl radical (cys-SOO•) (Reaction 5 of the Fig. 7), and protein sulfonation is a byproduct derived from further reactions with oxidants (e.g., H2O2). Presumably, the presence of DMPO can prevent protein cysteine sulfonation via the trapping of O2•− and the diminishing of the O2•−-derived oxidants. It is worth noting that the observed protein sulfonation could be a byproduct of H2O2-mediated thiolate anion hyperoxidation [31], because this possibility could not be ruled out under our current experimental settings. Although the protein cysteine oxidation can also be mediated by peroxynitrite as reported in our earlier studies of Complex II [26], MS/MS results of earlier studies do not support this mechanism with a Complex II-derived thiyl radical intermediate involved [26].
Table II.
Effect of DMPO on the cysteine sulfonation at the C554 and C727 of the Complex I from the reaction mixture containing Complex I, NADH, and Q1
| Treatment | m/z (Theoretical) | m/z (Observed) | Peptide Sequence and OPTM |
|---|---|---|---|
| −DMPO (Ta) | 765.86822+ | 766.182+ | 544M(ox)LFLLGADGGC554(SO3)ITR557 |
| +DMPO (T) | 757.87072+ | 758.172+ | 544M(ox)LFLLGADGGC554(SO2)ITR557 |
| −DMPO (TCb) | 562.27422+ | 562.372+ | 547LLGADGGC554(SO3)ITR557 |
| +DMPO (TC) | 554.27682+ | 554.682+ | 547LLGADGGC554(SO2)ITR557 |
| −DMPO (T) | 780.84602+ | 780.932+ | 713AVTEGAHAVEEPSIC727(SO3) |
| +DMPO (T) | 772.84852+ | 772.692+ | 713AVTEGAHAVEEPSIC727(SO2) |
T: Trypsin digestion of the sample
TC: Trypsin+Chymotrypsin digestion of the sample
To further confirm the protein thiyl radical mediating S-glutathionylation and S-sulfonation of the Complex I, redox status of control Complex I 75 kDa was analyzed by MS/MS using the enzyme treated with DTT and then iodoacetamide alkylation as a control of in vitro reaction. 90.1% of the amino acid sequence was identified in the MS/MS spectra (Supplementary Fig. S10). MS/MS analysis revealed no cysteine S-glutathionylation and cysteine S-sulfination/S-sulfonation detected from the 75 kDa subunit of as-isolated Complex I.
Molecular Modeling of the Bovine Complex I 75 kDa Subunit
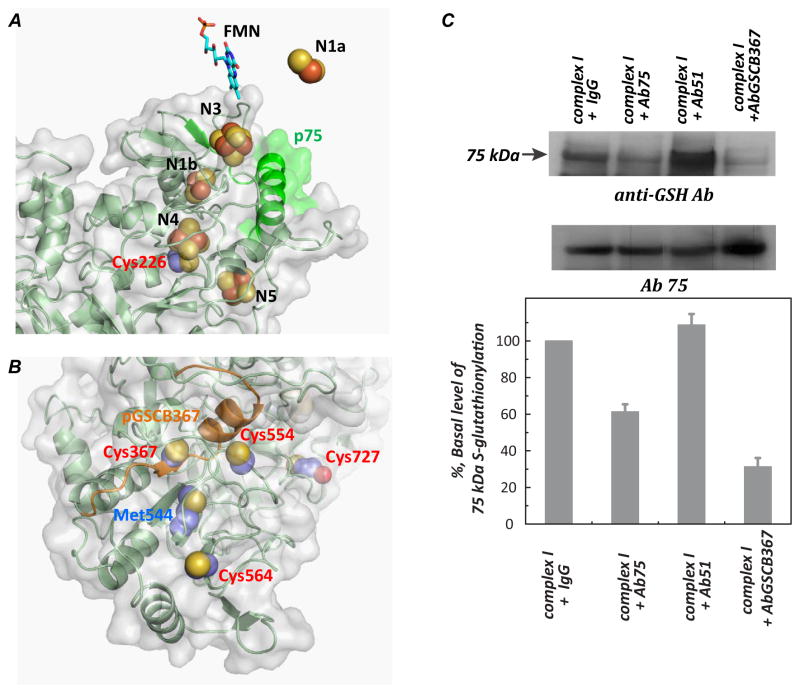
The homology model of the Complex I 75 kDa subunit was built and generated from SWISS-MODEL as detailed in a previous publication [29]. As indicated in Fig. 4A, C226 is one of the ligands involved in the [4Fe-4S] binding of the N4 center. The S-glutathionylation or binding of DMPO at C226 would impair Complex I-derived electron transfer activity due to the interruption of electron flow from the upstream cofactors [32]. The detection of C226 thiyl radical under conditions of enzyme turnover strongly suggests the N4 center as a potential site of electron leakage for O2•−. This hypothesis is further supported by previous studies of immunoinhibition [10] using an antibody (Ab75) against the p75 peptide. In previous studies, we demonstrated that binding of the antibody (Ab75) against the p75 domain (aa100–120, β-sheet-α-helix in green from Fig. 4A) to Complex I stabilizes the conformation of the protein matrix surrounding iron sulfur clusters (N1b, N4, N5), preventing molecular oxygen’s accessing the iron-sulfur clusters, resulting in the inhibition of O2•− generation and enhancing electron transfer efficiency [10]. We have further observed that binding of Ab75 to Complex I modestly inhibited (by 38.7±4.1 %, n=3) S-glutathionylation of the 75 kDa subunit in the presence of GSH (Fig. 4C). Therefore, transient formation of the C226 thiyl radical is likely a reversible process under conditions of enzyme turnover, and subsequently becomes irreversible after oxidative modification with GS-binding (Fig. 2) or DMPO-binding (Fig. 3A).
FIGURE 4.
A and B Molecular Modeling of the Complex I 75 kDa subunit. The three-dimensional structure of bovine Complex I 75 kDa was generated using the respiratory Complex I of T. thermophiles as a template (protein data bank code: 3i9vC (3.1Å)) by Swiss molecular modeling as detailed in a previous publication [29]. C: Effect of the antibodies of Ab75 and AbGSCB367 (10, 29) on the S-glutathionylation of the Complex I 75 kDa subunit. Isolated Complex I (1 mg/ml) in 50 mM sodium phosphate buffer with 0.1 % lauryl maltoside (pH 7.4) was incubated with the antibody (0.36 mg/ml), Ab75 or AbBGSC367 or preimmune IgG (as a non-binding control) or Ab51 ([10, 29], as a binding control) at 4 °C for 8 h. S-glutathionylation of Complex I was initiated by the addition of NADH (1 mM), Q1 (0.1 mM), and GSH (2 mM) as described in the Fig. 1 caption. 33.6 μg of Complex I was subjected to SDS-PAGE and immunoblotted with anti-GSH and Ab75 antibodies respectively.
C727 is located at the un-structural c-terminus of the 75 kDa subunit. The peptide of the c-terminal domain hosting C727 is likely wobbling based on the homology model, therefore C727 is surface-exposed [19] (Fig. 4B and Supplementary Fig. S8), and involved in producing an O2•−-induced thiyl radical intermediate for subsequent S-glutathionylation in the presence of GSH (Reaction 3 of the Fig. 7) or further irreversible S-sulfonation in the absence of GSH or DMPO (Reactions 5–6 of the Fig. 7).
On the basis of the homology model (Supplementary Fig. S8), the residue of C554 is also located on the surface of the 75 kDa subunit without the hindrance of other associated subunits. A positive amino acid (K562) neighbor C554, which can increase the thiolate formed at C554. The above two factors may render C554 susceptible to oxidative attack, forming protein thiyl radical for subsequent protein sulfonation (Reactions 5–6 of the Fig. 7).
The homology model also reveals that C367, C554, C564, and C727 are close to each other (Fig. 4B). In our previous studies (10), we demonstrated that binding of an antibody (AbGSCB367) against the pGSCB367 peptide (aa354–372, β-sheet-α-helix in orange ribbon from the Fig. 4B) significantly inhibits Complex I-mediated O2•− production. Likewise, a similar result indicates that the binding of AbGSCB367 significantly diminishes (by 70±8 %, n=3) protein S-glutathionylation of the Complex I 75 kDa subunit (Fig. 4C), which is likely due to (i) deceasing C727 glutathionylation via weakening the O2•−-induced thiyl radical at C727, and (ii) deceasing C367 glutathionylation by reducing GSSG access to C367 or by lessening Complex I denaturation by oxygen radicals [19].
Formation and Trapping of Oxygen Free Radicals in Myocytes and Mitochondria under Oxidative Stress
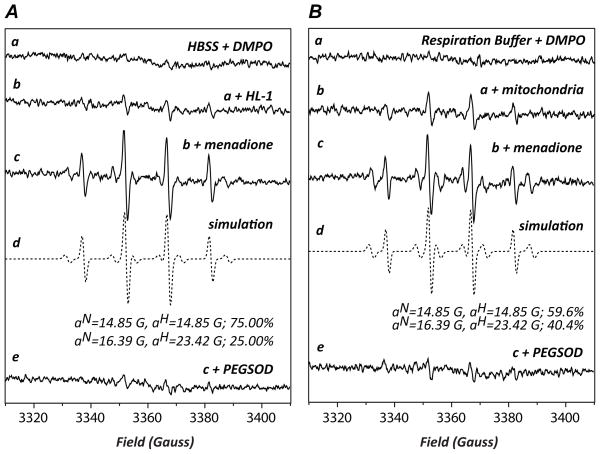
Experiments were performed to determine whether protein S-glutathionylation of Complex I occurs in myocytes under oxidative stress using the HL-1 cell line [28]. DMPO EPR spin trapping was first used to determine the extent of O2•− generation from myocytes induced by menadione, which can uncouple the mitochondrial flavin-containing enzyme in mammalian cells [33]. Menadione-treated HL-1 exhibited a prominent four-line EPR spectrum of DMPO/•OH (aN= aH=14.89G, 75%) and a minor six-line spectrum composed of the carbon centered radical adduct of DMPO (DMPO-R, aN=16.39G and aH=23.42G, 25% indicated in the Fig. 5A, c–d) (34). The detected EPR spectrum was inhibited by PEGSOD, indicating that menadione activates O2•− generation in myocytes [33] (Fig. 5A, e). It was further observed that menadione-treated isolated mitochondria (from the rat heart) resulted in a similar and SOD-sensitive EPR spectrum composed of DMPO/•OH (59.6%) and a smaller DMPO-R adduct (40.4%) under the energized conditions driven by malate/glutamate (NADH dependent, Fig. 5B, spectra c–e), thus confirming that menadione-mediated mitochondrial uncoupling contributed to a fraction of the O2•− –derived radical generation from HL-1 (Fig. 5A).
FIGURE 5.
DMPO EPR spin trapping of O2•−-dependent free radical generated from HL-1 myocytes (in A) and isolated mitochondria (in B) as induced by menadione. A, Spectrum a is the EPR signal of Hank’s balanced salt solution (HBSS) with DMPO (90 mM). Spectrum b is the signal of HL-1 (2×105 cells) with DMPO. Spectrum c is the EPR signal of HL-1 with DMPO activated by the addition of menadione (20 μM). Spectrum d is a computer simulation (dashed line) of the experimental EPR spectrum (c, solid line). The spectrum e is the same as c except that polyethylene glycol-linked superoxide dismutase (PEGSOD, 40 mU/μl) is included in the system. B, Mitochondria were isolated from the rat heart according to the published procedure [36]. Spectrum a: DMPO (90 mM) was mixed with the respiration buffer containing the following reagents (in mM): K-glutamate 140, NaCl 10, mannitol 10, MgCl2 1, EGTA 1, malate 5, Trizma 1, phosphate 2.5, adjusted to pH 7.4. Spectrum b: the reaction mixture containing the respiration buffer supplemented with 1 mM of DTPA, 90 mM of DMPO, was mixed with isolated mitochondria (final 0.5 mg/mL) at 30° C for 5 min. The reaction mixture was subjected to EPR measurement. Spectrum c: the same as b except that menadione (20 μM) was included in the reaction mixture. Spectrum d: computer simulation of spectrum c. Spectrum e: the same as c except that PEGSOD, 40 mU/μl was included in the system. Instrumental settings: center field 3360 G, sweep width 100 G, power 20 mW, receiver gain 1 × 105, modulation frequency 100 kHz, modulation amplitude 1 G, conversion time 40.96 ms, time constant 163.84 ms, 5 scans.
Protein Thiyl Radical Mediates S-glutathionylation of Complex I in Myocytes
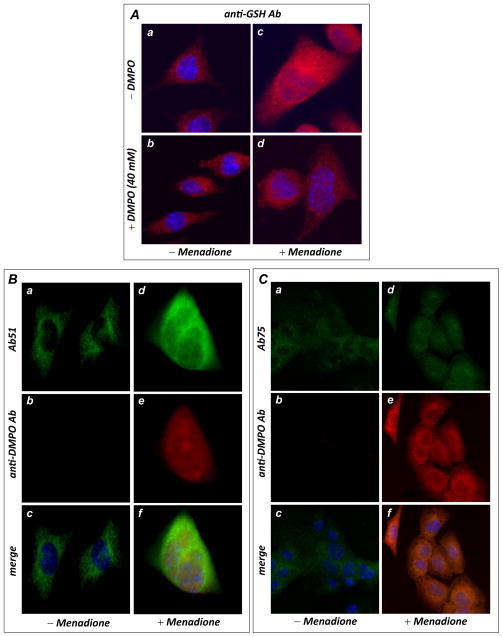
In HL-1 cells treated with menadione (0–100 μM), fluorescence microscopy with anti-GSH antibody illustrated a dosage-dependent (shown in Supplementary Fig. S9A) and marked increase in cellular S-glutathionylation (by 149.3 ± 11.4 % at a dosage of 50 μM menadione as indicated in Supplementary Fig. S9A and in Fig. 6B, a & c) that is subsequently decreased by the addition of DMPO (38.2 ± 4.2 % reduction at a DMPO dosage of 40 mM as indicated in Fig. 6B, b & d), suggesting that DMPO competes with cellular GSH in binding to protein thiyl radicals induced by menadione. Fluorescence microscopy with an anti-DMPO monoclonal antibody further demonstrated a marked increase in cellular DMPO protein radical adducts that colocalized with Complex I from the menadione-treated HL-1 (Fig. 6C for the Complex I 51 kDa subunit and Fig. 6D for the Complex I 75 kDa subunit) Note, immunomicroscopic fluorescence using anti-DMPO antibody also detected a dosage-dependent and marked increase in cellular protein radical adduct of DMPO from menadione-treated HL-1 (Supplementary Fig. S9B).
FIGURE 6.
In vivo protein radical formation and consequent S-glutathionylation from HL-1 myocytes under oxidative stress induced by menadione. A, immunomicroscopic analysis of S-glutathionylation in HL-1 using an anti-GSH monoclonal antibody. The upper panels (images a & c) are control HL-1 with or without menadione (40 μM) treatment, demonstrating enhancement of cellular S-glutathionylation by menadione. The lower panels (images b & d) are HL-1 with or without menadione treatment in the presence of DMPO (40 mM), demonstrating that DMPO outcompeted endogenous GSH for the protein radicals, decreasing cellular S-glutathionylation. B, Immunomicroscopic detection of Complex I protein radical formation using an anti-DMPO monoclonal antibody (red) and Ab51 (green). The left panels (images a, b & c) are control HL-1 with the addition of 40 mM DMPO trap, demonstrating that no detectable protein radical was formed. The right panels (images d, e, & f) are HL-1 treated with 40 μM of menadione with addition of the spin trap DMPO, demonstrating marked protein radical formation that colocalized with the Complex I 51 kDa subunit (green). C, same as B, except that cellular proteins were probed with Ab75 to show marked protein radical formation that colocalized with the 75 kDa subunit of Complex I.
CONCLUSION
Mitochondrial thiols are composed of protein thiols and the GSH pool. The proteins of the mitochondrial electron transport chain are rich in protein thiols. The protein thiols of the Complex I appear to play an important physiological role in controlling the redox status of mitochondria. For example, the physiological role of Complex I-derived thiols has been related to S-glutathionylation during myocardial ischemia and reperfusion [10]. Our data obtained from the in vitro isolated enzyme and ex vivo myocyte models should provide a deeper insight into the major mechanism of S-glutathionylation as mediated by the reactive thiol intermediate of protein radical.
The overall mechanism in which protein thiyl radical mediates Complex I S-glutathionylation and other oxidative modifications is summarized in the Fig. 7. The current studies revealed C226 and C727 to be involved in protein thiyl radical-mediated S-glutathionylation (Reactions 3–4). C226 S-glutathionylation is the primary mechanism for Complex I inactivation caused by S-glutathionylation, and S-glutathionylation of C727 should protect the residue from further S-sulfination and S-sulfonation (Reactions 5–6). Current studies also indicate C554 to be involved in a protein thiyl radical-mediated S-sulfination/S-sulfonation under the oxidant stress of O2•− (Reactions 5–6). Physiological conditions of GSH depletion or low GSH/GSSG ratios favor the mechanism of protein thiyl radical-mediated S-sulfonation, which should be relevant to the disease pathogenesis. Furthermore, the antibodies against the specific domains of p75 and pGSCB367 significantly abolished O2•− generation and protein thiyl radical-mediated Complex I S-glutathionylation (Fig. 4 and [10]). The result suggests agent or drug design that reset this redox switch, thereby restoring normal Complex I function, may have therapeutic potential.
The milieu of the mitochondrial matrix is anoxic in the presence of the GSH/GSSG pool under normal physiological conditions (35). Analysis of redox compartmentation indicates that the relative redox states from most reductive to most oxidative are as follows: mitochondria > nuclei > endoplasmic reticulum > extracellular space [35]. Thus, it is expected that low oxygen tension in the mitochondrial environment should facilitate the free thiol state for most cysteines of the Complex I 75 kDa subunit. The specific thiols subsequently become the targets of excess reactive oxygen species produced by pathological conditions. They are vulnerable to oxidation such as protein-thiyl radical formation, and even S-sulfonation. Recognition of this molecular event is valuable in understanding the fundamental basis of disease pathogenesis by redox alterations.
Supplementary Material
Highlights.
S-glutathionylation of Complex I is a redox modification induced by oxidant stress.
Protein thiyl radical intermediate is proposed to mediate S-glutathionylation.
The mechanism of S-glutathionylation strictly depends on oxidant stress and GSH.
The mechanism explains the physiological relevance of S-glutathionylation.
Acknowledgments
This work was supported by National Institutes of Health Grant HL83237 (YRC).
Footnotes
Abbreviations: NQR, NADH ubiquinone reductase, or mitochondrial Complex I; O2•−, superoxide anion radical; ETC, electron transport chain; SMP, submitochondrial particles; Q1, ubiquinone-1; DMPO, 5,5-dimethyl pyrroline N-oxide; FMN, flavin mononucleotide; GSH, glutathione; SDS-PAGE, SDS polyacrylamide gel electrophoresis; EPR, electron paramagnetic resonance; MS, mass spectrometry; MS/MS, tandem mass spectrometry; Ab, antibody; PBS, phosphate buffered saline; HBSS, Hank’s balanced salt solution.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hirst J, Carroll J, Fearnley IM, Shannon RJ, Walker JE. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim Biophys Acta. 2003;1604:135–150. doi: 10.1016/s0005-2728(03)00059-8. [DOI] [PubMed] [Google Scholar]
- 2.Carroll J, Fearnley IM, Shannon RJ, Hirst J, Walker JE. Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol Cell Proteomics. 2003;2:117–126. doi: 10.1074/mcp.M300014-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Walker JE. The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Q Rev Biophys. 1992;25:253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
- 4.Galkin A, Brandt U. Superoxide radical formation by pure complex I (NADH:ubiquinone oxidoreductase) from Yarrowia lipolytica. J Biol Chem. 2005;280:30129–30135. doi: 10.1074/jbc.M504709200. [DOI] [PubMed] [Google Scholar]
- 5.Kudin AP, Bimpong-Buta NY, Vielhaber S, Elger CE, Kunz WS. Characterization of superoxide-producing sites in isolated brain mitochondria. J Biol Chem. 2004;279:4127–4135. doi: 10.1074/jbc.M310341200. [DOI] [PubMed] [Google Scholar]
- 6.Turrens JF, Boveris A. Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem J. 1980;191:421–427. doi: 10.1042/bj1910421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen YR, Chen CL, Zhang L, Green-Church KB, Zweier JL. Superoxide generation from mitochondrial NADH dehydrogenase induces self-inactivation with specific protein radical formation. J Biol Chem. 2005;280:37339–37348. doi: 10.1074/jbc.M503936200. [DOI] [PubMed] [Google Scholar]
- 8.Lambert AJ, Brand MD. Inhibitors of the quinone-binding site allow rapid superoxide production from mitochondrial NADH:ubiquinone oxidoreductase (complex I) J Biol Chem. 2004;279:39414–39420. doi: 10.1074/jbc.M406576200. [DOI] [PubMed] [Google Scholar]
- 9.Ohnishi ST, Ohnishi T, Muranaka S, Fujita H, Kimura H, Uemura K, Yoshida K, Utsumi K. A possible site of superoxide generation in the complex I segment of rat heart mitochondria. J Bioenerg Biomembr. 2005;37:1–15. doi: 10.1007/s10863-005-4117-y. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Chen C, Rawale S, Chen C, Zweier J, Kaumaya P, Chen Y. Peptide-based antibodies against glutathione-binding domains suppress superoxide production mediated by mitochondrial complex I. J Biol Chem. 2010;285:3168–3180. doi: 10.1074/jbc.M109.056846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa NJ, Dahm CC, Hurrell F, Taylor ER, Murphy MP. Interactions of mitochondrial thiols with nitric oxide. Antioxid Redox Signal. 2003;5:291–305. doi: 10.1089/152308603322110878. [DOI] [PubMed] [Google Scholar]
- 12.Beer SM, Taylor ER, Brown SE, Dahm CC, Costa NJ, Runswick MJ, Murphy MP. Glutaredoxin 2 catalyzes the reversible oxidation and glutathionylation of mitochondrial membrane thiol proteins: implications for mitochondrial redox regulation and antioxidant DEFENSE. J Biol Chem. 2004;279:47939–47951. doi: 10.1074/jbc.M408011200. [DOI] [PubMed] [Google Scholar]
- 13.Taylor ER, Hurrell F, Shannon RJ, Lin TK, Hirst J, Murphy MP. Reversible glutathionylation of complex I increases mitochondrial superoxide formation. J Biol Chem. 2003;278:19603–19610. doi: 10.1074/jbc.M209359200. [DOI] [PubMed] [Google Scholar]
- 14.Beltran B, Orsi A, Clementi E, Moncada S. Oxidative stress and S-nitrosylation of proteins in cells. Br J Pharmacol. 2000;129:953–960. doi: 10.1038/sj.bjp.0703147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clementi E, Brown GC, Feelisch M, Moncada S. Persistent inhibition of cell respiration by nitric oxide: crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proc Natl Acad Sci U S A. 1998;95:7631–7636. doi: 10.1073/pnas.95.13.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurd TR, Filipovska A, Costa NJ, Dahm CC, Murphy MP. Disulphide formation on mitochondrial protein thiols. Biochem Soc Trans. 2005;33:1390–1393. doi: 10.1042/BST0331390. [DOI] [PubMed] [Google Scholar]
- 17.Hurd TR, Costa NJ, Dahm CC, Beer SM, Brown SE, Filipovska A, Murphy MP. Glutathionylation of mitochondrial proteins. Antioxid Redox Signal. 2005;7:999–1010. doi: 10.1089/ars.2005.7.999. [DOI] [PubMed] [Google Scholar]
- 18.Chen CL, Zhang L, Yeh A, Chen CA, Green-Church KB, Zweier JL, Chen YR. Site-specific S-glutathiolation of mitochondrial NADH ubiquinone reductase. Biochemistry. 2007;46:5754–5765. doi: 10.1021/bi602580c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurd TR, Requejo R, Filipovska A, Brown S, Prime TA, Robinson AJ, Fearnley IM, Murphy MP. Complex I within oxidatively stressed bovine heart mitochondria is glutathionylated on Cys-531 and Cys-704 of the 75-kDa subunit: potential role of CYS residues in decreasing oxidative damage. J Biol Chem. 2008;283:24801–24815. doi: 10.1074/jbc.M803432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mieyal JJ, Gallogly MM, Qanungo S, Sabens EA, Shelton MD. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid Redox Signal. 2008;10:1941–1988. doi: 10.1089/ars.2008.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallogly MM, Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr Opin Pharmacol. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Galante YM, Hatefi Y. Purification and molecular and enzymic properties of mitochondrial NADH dehydrogenase. Arch Biochem Biophys. 1979;192:559–568. doi: 10.1016/0003-9861(79)90126-7. [DOI] [PubMed] [Google Scholar]
- 23.Redfearn ER, Whittaker PA. The determination of the oxidation-reduction states of ubiquinone (coenzyme Q) in rat-liver mitochondria. Biochim Biophys Acta. 1966;118:413–418. doi: 10.1016/s0926-6593(66)80050-4. [DOI] [PubMed] [Google Scholar]
- 24.Hatefi Y. Preparation and properties of NADH: ubiquinone oxidoreductase (complexI), EC 1.6.5.3. Methods Enzymol. 1978;53:11–14. doi: 10.1016/s0076-6879(78)53006-1. [DOI] [PubMed] [Google Scholar]
- 25.Duling DR. Simulation of multiple isotropic spin-trap EPR spectra. J Magn Reson B. 1994;104:105–110. doi: 10.1006/jmrb.1994.1062. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L, Chen C, Kang P, Garg V, Hu K, Green-Church K, Chen Y. Peroxynitrite-mediated oxidative modifications of complex II: relevance in myocardial infarction. Biochemistry. 2010;49:2529–2539. doi: 10.1021/bi9018237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu H, Freitas MA. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics. 2007;8:133. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White S, Constantin P, Claycomb W. Cardiac physiology at the cellular level: use of cultured HL-1 cardiomyocytes for studies of cardiac muscle cell structure and function. Am J Physiol Heart Circ Physiol. 2004;286:H823–829. doi: 10.1152/ajpheart.00986.2003. [DOI] [PubMed] [Google Scholar]
- 29.Kang P, Yun J, Kaumaya P, Chen Y. Design and use of peptide-based antibodies decreasing superoxide production by mitochondrial complex I and complex II. Biopolymer: Peptide Science. 2011;96:207–220. doi: 10.1002/bip.21457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C, Wang T, Varadharaj S, Reyes L, Hemann C, Talukder M, Chen Y, Druhan L, Zweier J. S-glutathionylation uncouples eNOS and regulates its cellular and vascular function. Nature. 2010;468:1115–1118. doi: 10.1038/nature09599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roos G, Messens J. Protein sulfenic acid formation: from cellular damage to redox regulation. Free Radic Biol Med. 2011;51:314–326. doi: 10.1016/j.freeradbiomed.2011.04.031. [DOI] [PubMed] [Google Scholar]
- 32.Sazanov LA, Hinchliffe P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 33.Rosen G, Freeman B. Detection of superoxide generated by endothelial cells. Proc Natl Acad Sci U S A. 1984;81:7269–7273. doi: 10.1073/pnas.81.23.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zweier JL. Measurement of superoxide-derived free radicals in the reperfused heart. Evidence for a free radical mechanism of reperfusion injury. J Biol Chem. 1988;263:1353–1357. [PubMed] [Google Scholar]
- 35.Hansen JM, Go YM, Jones DP. Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annu Rev Pharmacol Toxicol. 2006;46:215–234. doi: 10.1146/annurev.pharmtox.46.120604.141122. [DOI] [PubMed] [Google Scholar]
- 36.Lee H, Chen C, Yeh S, Zweier J, Chen Y. Biphasic Modulation of the Mitochondrial Electron Transport Chain in Myocardial Ischemia and Reperfusion. Am J Physiol Heart Circ Physiol. 2012;302:H1410–1422. doi: 10.1152/ajpheart.00731.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.