Natural chromosome ends are protected in order to prevent the DNA damage response that is usually triggered by DNA breaks. In this study, Riha and colleagues make an unexpected discovery: They find that a significant proportion of plant telomeres lack G-overhangs and are instead blunt-ended and retained as such in post-mitotic tissue. The Ku complex maintains these blunt ends, protecting them from Exol degradation. This study provides intriguing and novel insights into telomere biology and mechanisms of chromosome end protection.
Keywords: telomere, telomerase, t-circle, Arabidopsis, recombination, Ku
Abstract
Single-stranded telomeric DNA protrusions are considered to be evolutionarily conserved structural elements essential for chromosome end protection. Their formation at telomeres replicated by the leading strand mechanism is thought to involve poorly understood post-replicative processing of blunt ends. Unexpectedly, we found that angiosperm plants contain blunt-ended and short (1- to 3-nucleotide) G-overhang-containing telomeres that are stably retained in post-mitotic tissues, revealing a novel mechanism of chromosome end protection. The integrity of blunt-ended telomeres depends on the Ku70/80 heterodimer but not on another telomere capping protein, STN1. Curiously, Ku-depleted telomeres are fully functional. They are resected by exonuclease 1, promoting intrachromatid recombination, which may facilitate formation of an alternative capping structure. These data challenge the view that telomeres require ssDNA protrusions for forming a functional capping structure and demonstrate flexibility in solutions to the chromosome end protection problem.
Telomeres form specialized chromatin that protects natural chromosome ends from being recognized and processed as DNA double-strand breaks (DSBs). Furthermore, a unique mode of telomere replication that involves telomerase, a reverse transcriptase that adds telomeric repeats to 3′ chromosome ends, counteracts the end replication problem and prevents chromosome erosion. Telomeres are evolutionarily highly conserved, and one of their key structural hallmarks is single-stranded 3′ DNA protrusions (G-overhangs) that range, depending on the species, from 16 to ∼200 nucleotides (nt). G-overhangs are important for telomere maintenance by acting as a primer for telomerase (Lingner and Cech 1996). They are also thought to be required for chromosome end protection by either serving as a substrate for specific DNA-binding protein complexes or forming secondary DNA structures such as t-loops (Horvath et al. 1998; Griffith et al. 1999; Baumann and Cech 2001; Gao et al. 2007; Raices et al. 2008). Thus, G-overhangs are considered to be essential functional elements of telomeres.
Although the primary function of telomeres is to inhibit DNA repair, the post-replicative activation of DNA damage responses appears to be a prerequisite for telomere cap assembly (Verdun et al. 2005; Verdun and Karlseder 2006). The emerging models suggest that temporal recognition of newly replicated chromosome ends by DNA damage checkpoint and repair factors may facilitate telomerase recruitment, G-overhang formation, and t-loop assembly (Gilson and Geli 2007; Verdun and Karlseder 2007; Bianchi and Shore 2008). It is assumed that all telomeres in a cell are functionally and structurally identical. However, the processes that lead to the formation of telomere caps at opposite ends of the same chromosome likely differ. The inherent polarity of DNA strand synthesis postulates that one end of a chromosome is a product of leading strand replication, whereas the opposite end is synthesized by the lagging strand replication mechanism. While removal of the RNA primer from the most terminal Okazaki fragment results in a 3′ G-overhang at the chromosome end replicated by the lagging strand mechanism, synthesis of the telomeric C-rich strand by the leading strand mechanism is expected to produce a blunt end (Lingner et al. 1995). Telomeres replicated by the leading mechanism are thought to undergo nucleolytic processing to generate 3′ G-overhangs at both chromosome ends. At least three lines of evidence support this model. First, G-overhangs were detected at the majority of telomeres in yeast and humans (Wellinger et al. 1996; Makarov et al. 1997; Chai et al. 2006a). Second, G-overhang formation in Saccharomyces cerevisiae and mammals is partially dependent on Mre11 and Apollo nucleases (Larrivee et al. 2004; Chai et al. 2006b; Bonetti et al. 2009; Lam et al. 2010; Wu et al. 2010), and, finally, these nucleases appear to be important for protecting telomeres synthesized by the leading strand mechanism (Deng et al. 2009; Faure et al. 2010; Lam et al. 2010; Wu et al. 2010). However, post-replicative processes that govern the resection of telomeres and formation of a functional cap are still elusive, partially due to the transient nature of these events and the technical challenges associated with detailed structural analysis of chromosome ends.
Here we describe several methods that we developed for detailed and quantitative analysis of the terminal structure of telomeric DNA. We discovered that a substantial portion of telomeres in Arabidopsis thaliana does not undergo nucleolytic resection and remains blunt-ended. The presence of stable blunt-ended telomeres in plant post-replicative tissues demonstrates the existence of a novel mechanism of chromosome end protection that does not rely on single-stranded G-overhangs. We further show that inactivation of the conserved DNA repair factor Ku leads to resection of blunt-ended telomeres by exonuclease 1, which in turn promotes intrachromatid recombination. This may facilitate stabilization of the resected chromosome ends through t-loop formation and to some extent recapitulate events that occur during post-replicative processing of leading strand telomeres in humans and other organisms that do not retain blunt-ended telomeres.
Results
A portion of Arabidopsis telomeres lacks ssDNA protrusions and is blunt-ended
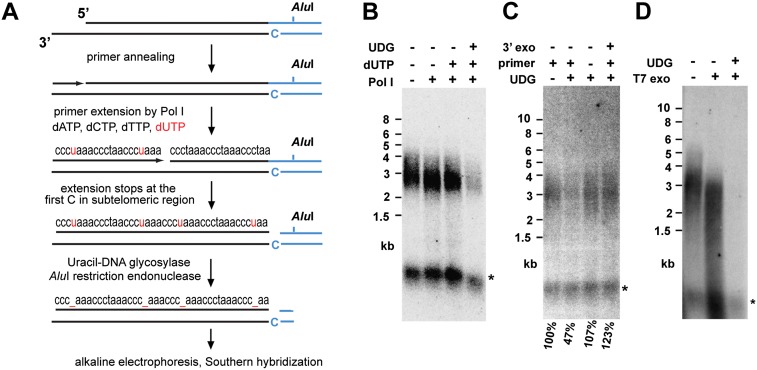
Our experiments using the primer extension nick translation (PENT) protocol indicated that only a portion of telomeres in the angiosperm plants Silene latifolia and A. thaliana is amenable to the PENT reaction, contradicting the view that G-overhangs are uniformly present at all telomeres (Supplemental Fig. S1; Riha et al. 2000). The refractory PENT reaction signal could be derived from telomeres with no G-overhangs or from intrachromosomal telomeric sequences that are particularly abundant in the Arabidopsis genome. To distinguish these possibilities, we modified the assay by incorporating dUTP in the nascent telomeric C strand synthesized by PENT (Fig. 1A). Subsequent treatment with uracil DNA glycosylase (UDG) converts uracils into abasic sites that render DNA strands fragile under alkaline conditions. Hence, separation of terminal restriction fragments (TRFs) from intrachromosomal telomeric DNA by alkaline gel electrophoresis should lead to preferential degradation of C strands at telomeres with a G-overhang. Indeed, dUTP-PENT with Arabidopsis DNA resulted in a decline of the TRF signal, which was suppressed by the pretreatment of the DNA with a nuclease that specifically degrades G-overhangs (Fig. 1B,C). Interestingly, ∼40%–50% of telomeres appear to be refractory to the PENT reaction and were detected as intact TRFs after alkaline electrophoresis. This signal diminishes when 3′ single-stranded protrusions are generated at all DNA ends by T7 gene 6 exonuclease (Fig. 1D), demonstrating that the dUTP-PENT-resistant telomeres do not provide a binding site sufficient for primer annealing.
Figure 1.
G-overhangs are present at 50% of Arabidopsis telomeres. (A) Schematic representation of the dUTP-PENT reaction. (Black lines) Telomeric DNA; (blue lines) subtelomeric DNA. TRFs are released by AluI and separated by alkaline electrophoresis, and telomeres are detected by Southern hybridization with a (TTTAGGG)4 probe. (B) Arabidopsis DNA assayed by dUTP-PENT. Treatment with UDG reduces a signal from AluI-generated TRFs to 55% relative to the signal from intrachromosomal telomeric DNA (indicated by an asterisk). (C) Omission of the primer or removal of the G-overhangs by 3′–5′ exonuclease does not lead to a reduction of the TRF signal after UDG treatment. Relative intensity of the TRF signal in comparison with the reaction without UDG treatment is indicated below the autoradiogram. TRF signals were normalized in each lane using the signal from intrachromosomal telomeric DNA. (D) dUTP-PENT completely eliminates the TRF signal after DNA pretreatment with the 5′–3′ T7 gene 6 exonuclease.
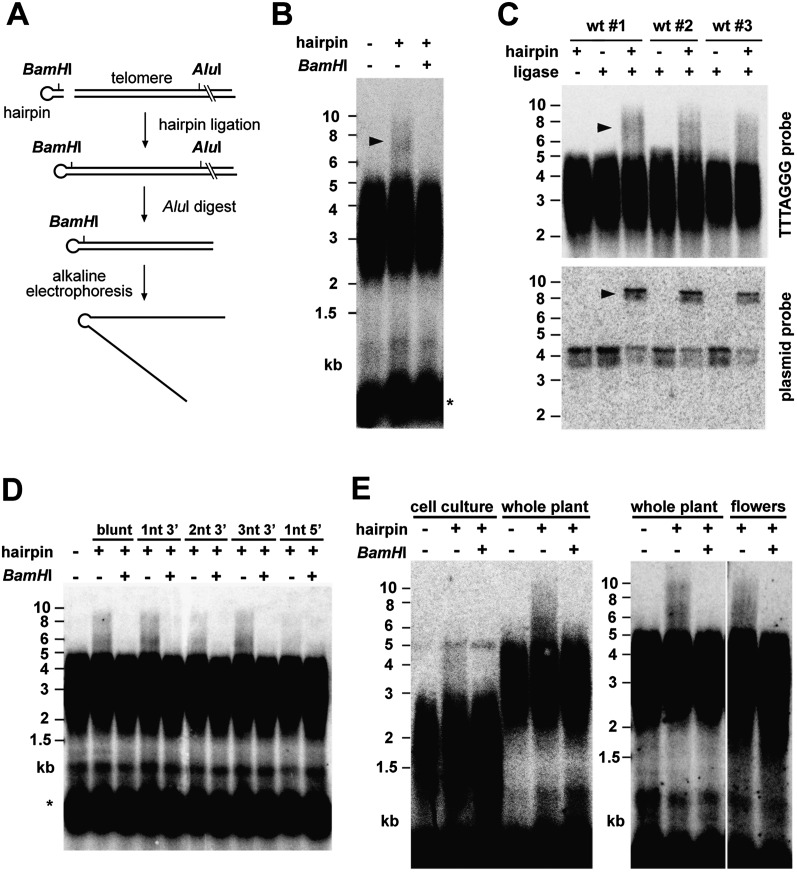
Such telomeres may represent unprocessed chromosome ends replicated by the leading strand mechanism that are expected to carry blunt ends (Ohki et al. 2001). We assayed for the presence of telomeric blunt ends by ligating a hairpin that covalently links both C-rich and G-rich complementary strands, thereby shifting the electrophoretic mobility of TRFs to double-sized fragments under denaturing conditions (Fig. 2A). Arabidopsis TRFs generated by AluI range from 2 to 5 kb, and hairpin ligation indeed slowed the mobility of a fraction of the TRFs to 5–10 kb (Fig. 2B). Ligation products were sensitive to cleavage of the hairpin with BamHI, demonstrating that the mobility shift is caused by a covalent linkage of complementary strands at some telomeres. A series of control experiments demonstrated that the hairpin specifically ligates to blunt ends and not to telomeres with G-overhangs (Supplemental Fig. S2). Furthermore, we verified that ligation occurs at natural chromosome ends and not in telomere internal sites that may arise from rare DSBs or t-loop resolution events (Supplemental Fig. S3). To estimate the fraction of telomeres with blunt ends, we quantified the proportion of TRFs in the ligation-dependent shift and divided it by the efficiency of the hairpin ligation, which was determined for each reaction individually by using an internal control (Fig. 2C). This quantification showed that ∼11% of telomeres terminate with a blunt end. Considering that approximately half of the telomeres have G-overhangs detectable by dUTP-PENT, we next asked whether the remaining fraction of telomeres contains ssDNA protrusions too short for efficient primer annealing. To assay for such structures, we performed hairpin ligations with mixtures of hairpins containing 1-, 2-, or 3-nt long 3′ protrusions with sequences corresponding to all possible permutations of plant telomeric DNA. Ligation of these hairpins to Arabidopsis genomic DNA yielded a BamHI-sensitive shift similar to the one obtained with a blunt-ended hairpin (Fig. 2D). In contrast, a hairpin with a single-nucleotide 5′ protrusion produced no or only a barely detectable signal in the shift. The amount of signal in the shift after ligation of 1- to 3-nt hairpins is in the range of 3%–6%. Assuming that the ligation efficiency is similar to the ligation efficiency of the blunt-ended hairpin (∼60%), telomeres with 1- to 3-nt G-overhangs may form up to 30% of all telomeres. This is consistent with the prediction that the dUTP-PENT-resistant signal is derived from telomeres containing either no or 1- to 3-nt long G-overhangs.
Figure 2.
Detection of blunt-ended telomeres in Arabidopsis. (A) Diagram of the hairpin ligation assay for detecting blunt-ended telomeres. The hairpin containing the BamHI site is indicated in the diagram. (B) TRFs of wild-type plants that were ligated to the hairpin prior to digestion with AluI. (Arrowhead) TRF mobility shift caused by hairpin ligation. (C) Quantification of blunt-ended telomeres. Efficiency of hairpin ligation was determined in each reaction by mixing linearized blunt-ended plasmid with genomic DNA prior to hairpin ligation. (Bottom panel) Plasmid dimers and monomers were detected in one-half of the reaction by Southern hybridization with a plasmid probe (ligation products are indicated by an arrowhead). (Top panel) The second half of the reaction was analyzed for hairpin ligation to telomeres. Ligation efficiency was determined as the portion of plasmid dimers ranging from 61% to 78%. The fraction of telomeres carrying blunt ends (11.0% ± 1.5%; N = 6) was estimated as the portion of telomeres in the shift divided by the efficiency of ligation. (D) Detection of telomeres with 1- to 3-nt long 3′ ssDNA protrusions by hairpin ligation assay. (E) Detection of blunt-ended telomeres in a rapidly proliferating cell culture and flowers.
From these data, we conclude that Arabidopsis contains at least two populations of telomeres. Approximately half of the telomeres have G-overhangs detectable by dUTP-PENT. In our previous study, we showed that PENT is still quantitative at 65°C, indicating that the minimum length of G-overhangs to provide a stable substrate for efficient primer annealing should be at least 15–20 nt (Riha et al. 2000). The remaining telomeres appear to contain blunt ends or up to several-nucleotide long 3′ protrusions. Formation of G-overhangs at both chromosome termini requires post-replicative processing of presumably blunt-ended telomeres synthesized by the leading strand replication. The DNA used in described experiments was extracted from adult plants that consist almost exclusively of post-mitotic nonproliferating cells. Nevertheless, we also detected telomeres with blunt ends in flowers and cell culture that contain a high proportion of rapidly proliferating cells (Fig. 2E). The presence of blunt-ended telomeres in dividing as well as quiescent cells argues that the blunt ends do not represent temporary preprocessing intermediates, but rather mature fully functional chromosome ends. Interestingly, the blunt-ended telomeres appear to be present only in angiosperm plants, as they were detected in Arabidopsis, Silene, and maize but not in moss, yeasts, and humans (Supplemental Fig. S4).
STN1 affects sequence composition of blunt-ended telomeres
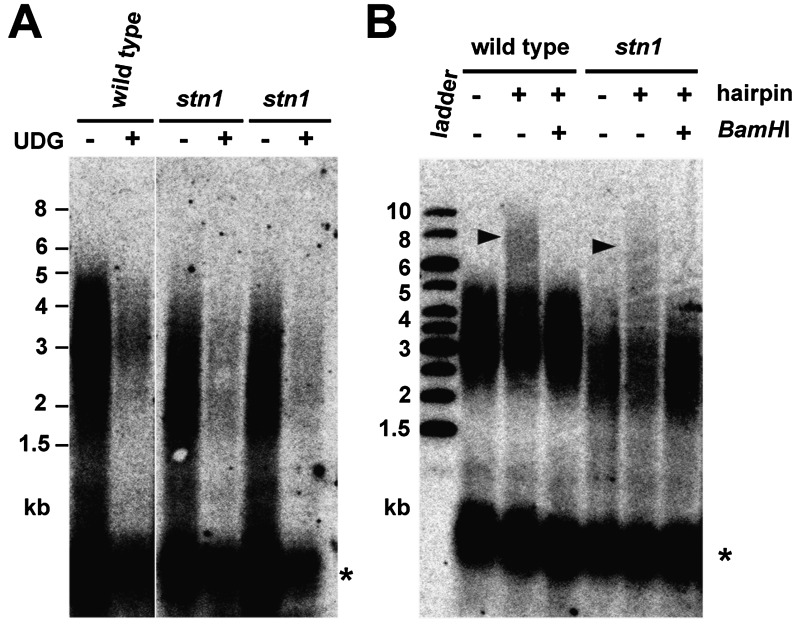
Our data demonstrate that Arabidopsis contains at least two fundamentally distinct populations of telomeres: Approximately half of the telomeres have long G-overhangs detectable by dUTP-PENT, while the remaining telomeres are blunt- or nearly blunt-ended. This may reflect different modes of replication of opposite chromosome ends by lagging and leading strand mechanisms. To gain support for this hypothesis, we investigated the telomere structure of STN1-deficient plants. STN1 is an evolutionarily conserved component of the CST complex that, in budding yeast, is required for chromosome end protection and telomere maintenance (Gao et al. 2007; Giraud-Panis et al. 2010). STN1 function at telomeres may be linked to the replication of lagging strand telomeres, as STN1 associates with and acts as a stimulatory factor of polymerase α (Grossi et al. 2004; Casteel et al. 2009). Inactivation of STN1 in Arabidopsis has been reported to result in telomere deprotection that is manifested by shorter heterogeneous telomeres, chromosome end-to-end fusions, and longer G-overhangs (Song et al. 2008). Intriguingly, despite the increased amount of telomeric ssDNA, we found G-overhangs on only 52% of telomeres in stn1 mutants by dUTP-PENT (52.4% ± 12.4%; N = 9) (Fig. 3A). In addition, telomeres with blunt ends were readily detected by the hairpin ligation assay in stn1 mutants (Fig. 3B).
Figure 3.
Blunt-ended telomeres remain intact in stn1 mutants. dUTP-PENT (A) and hairpin ligation (B) assays with DNA from Arabidopsis stn1 mutants. The signal from intrachromosomal telomeric DNA is indicated by an asterisk.
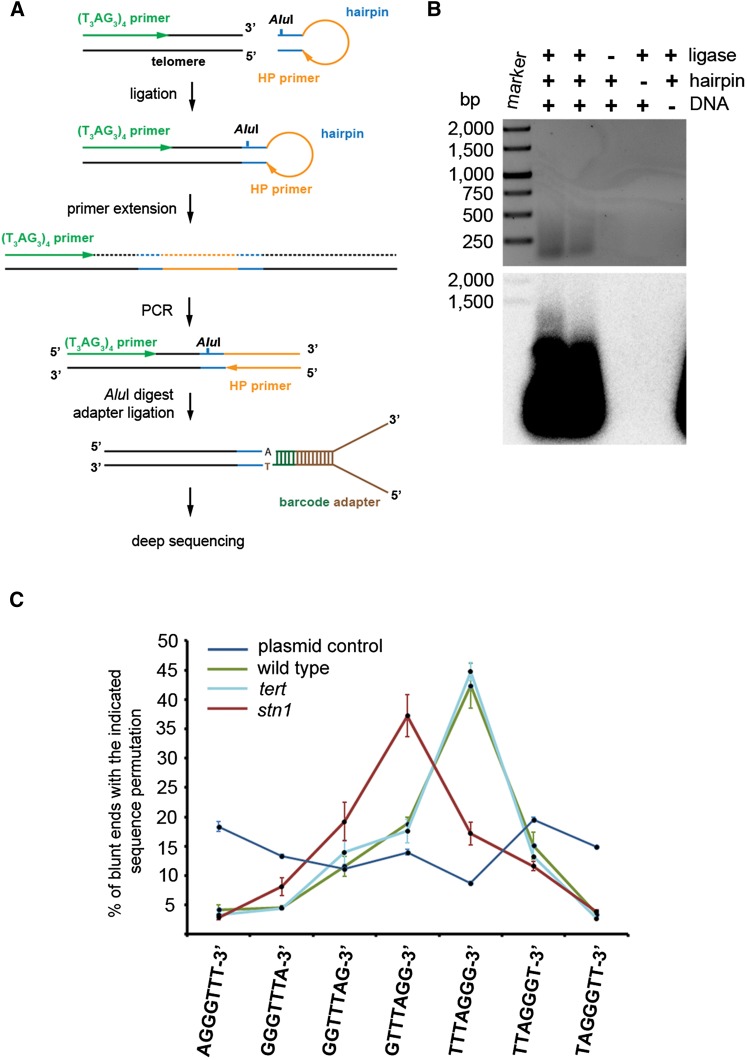
These data show that STN1 dysfunction affects only a subset of telomeres, presumably the ones that carry G-overhangs, while it is not required for the integrity of blunt-ended telomeres. A recent study with Xenopus egg extracts indicated that STN1 may be involved in the priming step during lagging strand replication (Nakaoka et al. 2012). The 5′ end of the telomeric strand synthesized by the lagging mechanism serves as a template for replication of telomeric 3′ ends by the leading strand mechanism in the following S phase. Hence, we predicted that aberrant priming or processing of the lagging strand in the absence of STN1 may result in alteration of the terminal sequence of the blunt-ended telomeres. To test this scenario, we developed a methodology for assessing the frequency of telomeric permutations at the termini of blunt-ended telomeres (Fig. 4A,B; Supplemental Fig. S5). This technique is based on adapter ligation-coupled PCR and quantitative analysis of PCR products by Illumina sequencing. We found that blunt-ended telomeres in wild type and tert (telomerase reverse transcriptase) mutants preferentially terminate with the –TTTAGGG-3′ sequence (∼45%) (Fig. 4C); such bias was not detected in a control reaction containing an equimolar mixture of seven blunt-ended plasmid constructs, each carrying a different permutation of plant telomeric DNA at its end (Fig. 4C; Supplemental Fig. S6). Remarkably, the predominant terminal sequence was shifted to the –GTTTAGG-3′ permutation in stn1 mutants (Fig. 4C). These data support the model in which STN1 influences processing/maintenance of the 5′ chromosome end synthesized by the lagging strand mechanism, which in the following replication cycle serves as a template for synthesis of the blunt-ended 3′ chromosome end by the leading strand mechanism.
Figure 4.
STN1 deficiency alters the sequence of termini of blunt-ended telomeres. (A) Diagram illustrating the strategy to determine the terminal sequence permutation by adapter-guided PCR and deep sequencing. Genomic DNA was ligated with a blunt-ended hairpin containing an AluI restriction site. Primer extension from a ligated telomere opens the hairpin and creates an annealing site for a HP primer, whose 3′ sequence is identical with the sequence within bulge of the hairpin. Ligated products were PCR-amplified from the telomeric (T3AG3)4 primer and HP primer. This experimental design minimizes the formation of PCR products from two ligated HP primers. A maximum of 20 cycles were performed to ensure that PCR amplification is exponential. PCR products were digested with AluI, ligated to adapters used for generating deep-sequencing libraries, and subjected to single-end sequencing. The deep-sequencing adapters include bar codes to allow sequencing of multiple samples in a single run. As we demonstrated that ligation of a blunt-ended hairpin primarily occurs at telomere ends (Supplemental Fig. S3), the sequence at the adapter telomere junction represents natural chromosome termini. (B) Products obtained by PCR amplification of ligated products prior to the AluI digest. Ethidium bromide-stained gel is shown in the top panel. (Bottom panel) The PCR products were then analyzed by Southern hybridization with a telomeric probe. The expected smear consisting of amplified telomeric DNA as verified by Southern hybridization was obtained only in the reaction containing genomic DNA, ligase, and hairpin. Reactions in which any of these components were omitted did not produce the smear, demonstrating the specificity of the amplification. (C) Frequency of telomeric sequence permutations at blunt-end chromosome termini. Error bars indicate SD from five wild-type and tert and three stn1 biological replicas. Error bars for the control plasmid constructs represent SD from three technical replicas (the same mixture was used as a template in three independent experiments).
Integrity of blunt-ended telomeres requires Ku
Blunt ends and 1- to 3-nt short 3′ protrusions do not allow t-loop formation and binding of specific ssDNA-binding complexes. Therefore, protection of such chromosome ends must be mediated by a novel mechanism that is different from the mechanisms used at telomeres with G-overhangs and may involve proteins capable of binding to blunt-ended DNA termini. A strong candidate with such activity is the evolutionarily conserved Ku complex that is involved in the early steps of nonhomologous end-joining (NHEJ) DNA repair. The Ku complex binds to broken DNA ends in a sequence-independent manner and stabilizes them by preventing excessive degradation prior to ligation (Lee et al. 1998; Mimitou and Symington 2010). The Ku complex also plays a role at telomeres in a broad variety of organisms, where it protects chromosome ends from nucleolytic degradation and recombination (Gravel et al. 1998; Maringele and Lydall 2002; Celli et al. 2006; Wang et al. 2009).
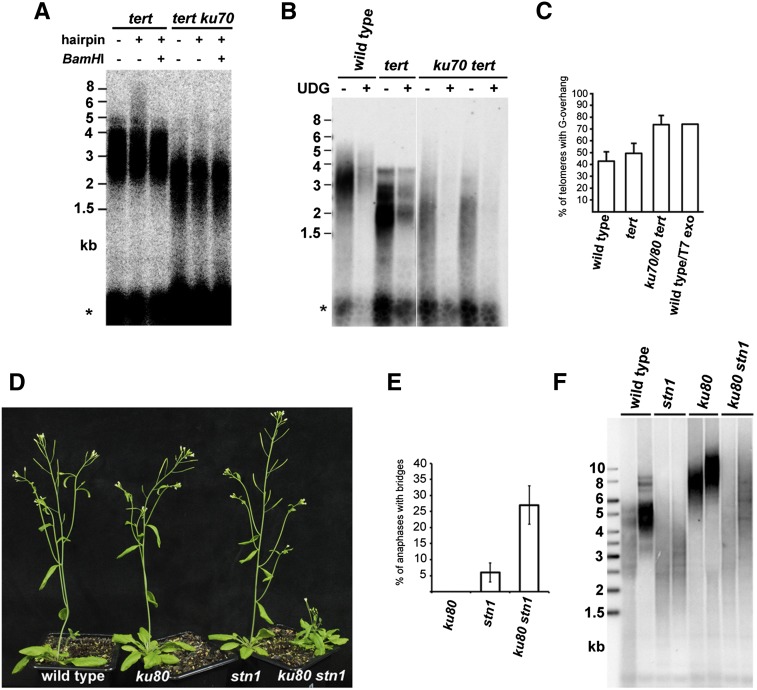
In Arabidopsis, the Ku complex acts as a negative regulator of telomerase and protects chromosome termini from 5′-to-3′ resection and aberrant recombination (Riha and Shippen 2003; Zellinger et al. 2007). Because Ku-deficient plants have long telomeres difficult to resolve by alkaline electrophoresis, we tested the role of Ku at blunt-ended telomeres by analyzing Arabidopsis mutants deficient in Ku and tert. Although a characteristic mobility shift was observed after hairpin ligation in tert mutants, we failed to detect blunt ends in tert ku70 mutants (Fig. 5A). Furthermore, by dUTP-PENT, we determined that the majority of tert ku telomeres possess G-overhangs, whereas these structures were detected on only ∼43% and 49% of telomeres in wild-type and tert plants, respectively (Fig. 5B,C). These data imply that, in the absence of Ku, blunt-ended telomeres are converted into 3′ G-overhangs, likely through 5′-to-3′ nucleolytic resection.
Figure 5.
The integrity of blunt-ended telomeres depends on Ku. Hairpin ligation (A) and dUTP-PENT (B) assays with DNA from tert and ku70 tert mutants. Signals from intrachromosomal telomeric sequences are indicated by an asterisk. (C) The proportion of telomeres carrying G-overhangs detectable by the dUTP-PENT assay in wild type and indicated mutants. Error bars indicate SD from nine to 15 independent samples. (D) Ku and STN1 act in parallel chromosome end protection pathways. F2 siblings carrying indicated mutations outsegregating from a STN1+/−KU80+/− plant. Frequency of anaphase bridges (E) and TRF analysis (F) in the G1/F2 generation of ku80, stn1, and ku80 stn1 mutants.
These data established that Ku is important for the protection of blunt-ended telomeres, yet neither ku nor early-generation ku tert mutants exhibit any obvious hallmarks of telomere dysfunction, such as chromosome end-to-end fusions and growth retardation. This implies that telomeres that lose their blunt ends are still functional. We propose that their nucleolytic processing (and the action of telomerase in the case of ku mutants) generates G-overhangs, which may permit formation of a canonical capping structure. If STN1 acts in this “G-overhang” capping pathway, concomitant inactivation of both Ku and STN1 should have a synergistic effect on telomere protection. To test this prediction, we crossed plants heterozygous for the ku80 and stn1 mutations and examined mutant and control plants outsegregating from a single F1 parent (Supplemental Fig. S7A). While the first outsegregating generation (G1) stn1 mutants have a wild-type appearance and low frequency of anaphase bridges (Fig. 5D,E; Supplemental Fig. S7C,D), telomere length progressively shortens, and G2 plants exhibit phenotypes consistent with massive telomere dysfunction (Supplemental Fig. S7B). This observation indicates a role for STN1 in telomere maintenance. The G1 ku80 stn1 double mutants were slightly underrepresented in the F2 population and showed a high frequency of chromosome end-to-end fusions and severe developmental defects (Fig. 5D,E). Despite telomere dysfunction, telomeres in G1 ku80 stn1 mutants were longer than in G1 stn1 siblings, arguing that chromosome end capping, rather than telomere maintenance, is strongly affected (Fig. 5F). These data support the idea that STN1 and Ku act in parallel end protection/maintenance pathways and that the function of STN1 becomes more critical in the absence of Ku.
Exonuclease 1 resects Ku-depleted blunt-ended telomeres and promotes homologous recombination
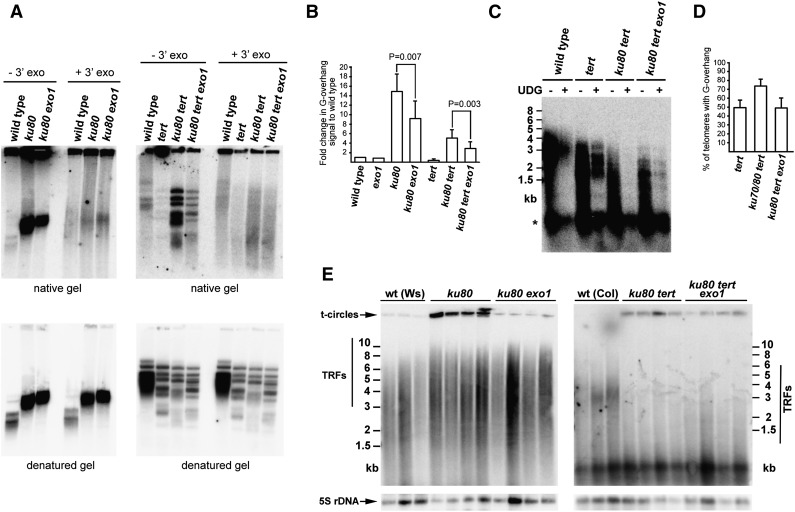
To confirm the prediction that Ku-depleted telomeres are subject to nucleolytic resection, we aimed to identify the nuclease responsible for the degradation of telomeric blunt ends. In budding yeast, Ku-depleted telomeres are mainly resected by exonuclease 1 (Maringele and Lydall 2002). Two Arabidopsis genes, At1g29630 (EXO1A) and At1g18090 (EXO1B), encode putative exonuclease 1 homologs (Supplemental Figs. S8, S9). As both genes carry the N-terminal RAD2 domain typical for exonuclease 1 and exhibit similar expression profiles, suggesting a functional redundancy, we created exo1a exo1b double mutants, referred to as exo1 mutants (Supplemental Fig. S9). Inactivation of Ku in Arabidopsis leads to an increased amount of telomeric ssDNA that can be detected by the in-gel hybridization technique (Fig. 6A). The ssDNA signal was sensitive to treatment with a 3′-to-5′ exonuclease, demonstrating that it detects terminal G-overhangs. The G-overhang signal was significantly reduced in ku80 tert in comparison with ku80 mutants (Fig. 6A,B), suggesting that uncoupling of the 3′ G strand extension by telomerase from fill-in synthesis of the complementary C strand is partially responsible for the extended G-overhangs in Ku-deficient plants. The exo1 mutations further lessened the G-overhang signal in both ku80 and ku80 tert plants, demonstrating that, in Arabidopsis, Ku-depleted telomeres are subject to 5′-to-3′ resection by EXO1 (Fig. 6A,B). However, the amount of 3′ single-stranded telomeric DNA in ku80 tert exo1 plants was still greater than in tert mutants, indicating that other nucleases act on telomeres in the absence of Ku. This notion was supported by the observation that ku80 tert exo1 mutants did not regain telomeric blunt ends as would be expected if telomeric resection was completely abolished (Supplemental Fig. S10). Nevertheless, inactivation of EXO1 in ku80 tert plants re-established dUTP-PENT-resistant telomeres (Fig. 6C,D). We interpret these data to mean that resection of blunt-ended telomeres is reduced in exo1 mutants to an extent that does not permit their efficient detection by dUTP-PENT.
Figure 6.
Ku protects blunt-ended telomeres from EXO1-mediated resection. (A) G-overhang analysis by the in-gel hybridization technique. DNA samples pretreated with Escherichia coli exonuclease 1 to remove G-overhangs are indicated (+3′ exo). The gels were first hybridized under nondenaturing conditions (top panels) and then denatured and hybridized again (bottom panels). (B) Quantification of the G-overhang signals from native gels. Signals were normalized to wild type. Error bars represent SD from three to four independent samples. P-values indicating significance of difference are indicated (one-tailed paired Student's t-test). (C) Detection of G-overhangs in ku80 tert exo1 mutants by dUTP-PENT assay. (D) Proportion of telomeres carrying G-overhangs detectable by the dUTP-PENT assay in wild type and ku tert and ku80 tert exo1 mutants. Values for wild-type and ku tert plants are from the same data as values in Figure 5C. Error bars indicate SD from nine to 15 independent samples. (E) Effect of EXO1 on t-circle formation measured by the t-circle amplification assay (TCA). Arabidopsis plant ecotypes Ws and Col-0 possessing long and short telomeres, respectively, were used for comparison. Signals from t-circles and TRFs are indicated. (Bottom panel) Detection of 5S rDNA was used as a loading control.
Inactivation of Ku in Arabidopsis is accompanied by increased excision of extrachromosomal telomeric DNA circles (t-circles) (Zellinger et al. 2007). This is likely a consequence of increased intrachromosomal recombination initiated by invasion of a G-overhang into internal arrays of a telomere. To test whether diminished chromosome end resection affects telomeric recombination, we measured the impact of exo1 mutation on the t-circle excision. We used the t-circle amplification assay (TCA), which uses highly processive ϕ29 polymerase that can generate ssDNA products in the range of 100 kb from circular templates via rolling circle replication (Zellinger et al. 2007). While TCA in ku80 mutants yielded a strong t-circle signal, inactivation of EXO1 led to, on average, an ∼10-fold reduction of the signal in comparison with EXO1-proficient plants (relative abundance 1.00 ± 0.33 in ku80 vs. 0.10 ± 0.03 in ku80 exo1; N = 4) (Fig. 6E). This supports the idea that nucleolytic resection of 5′ chromosome ends promotes telomere recombination. Interestingly, the absence of EXO1 led to only an approximately twofold reduction of the t-circle signal in ku80 tert mutants (relative abundance 1.00 ± 0.43 in ku80 tert vs. 0.48 ± 0.25 in ku80 tert exo1; N = 4) (Fig. 6E). This observation suggests that telomerase has a further inhibitory effect on telomeric recombination.
In conclusion, our data show that, in the absence of Ku, Arabidopsis blunt-ended telomeres are resected by EXO1 and other nucleases, which in turn promote intrachromatid recombination and t-circle excision.
Discussion
The prevailing assumptions in the telomere field have been that all telomeres within an organism are identical and that ssDNA protrusions are critical for their function. These paradigms are essentially based on two chromosome end-capping models, which postulate that chromosome termini are either sequestered in a t-loop or protected by oligonucleotide-binding (OB)-fold proteins with binding specificity to single-stranded telomeric DNA. In this study, we challenge these assumptions by demonstrating that Arabidopsis contains at least two distinct populations of telomeres terminating with either a long 3′ G-overhang or a blunt end. The presence of stable blunt-ended telomeres represents a new paradigm by defining a fundamentally novel mechanism of chromosome end protection that is independent of ssDNA protrusions. We also detected telomeres that possess a few-nucleotide long 3′ G-overhangs. These single-stranded protrusions are unlikely to form stable t-loops or provide a sufficient binding site for telomeric OB-fold proteins (Pot1; CST complex) (Mitton-Fry et al. 2002; Lei et al. 2004; Loayza et al. 2004), and hence we consider these telomeres to be mechanistically related to the blunt-ended telomeres.
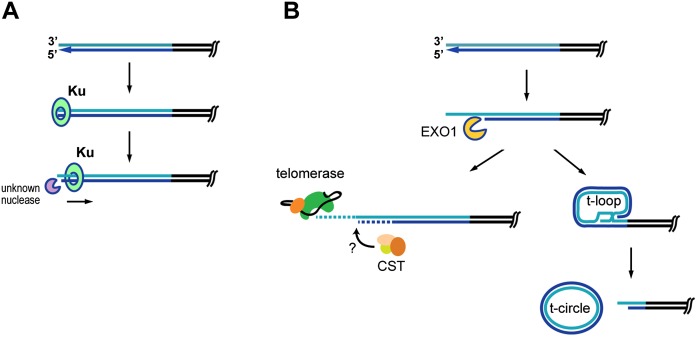
The coexistence of two types of structurally distinct telomeres terminating with either 3′ G-rich or 5′ C-rich protrusions have recently been reported in Caenorhabditis elegans and in mammalian cells that maintain telomeres by ALT (Raices et al. 2008; Oganesian and Karlseder 2011). While the 5′ C-rich overhangs were suggested to stem from telomeric recombination (Oganesian and Karlseder 2011), this mechanism does not seem to be involved in forming the blunt-ended telomeres in plants. First, telomeric recombination occurs only very rarely in wild-type Arabidopsis (Watson et al. 2005; Watson and Shippen 2007; Zellinger et al. 2007). Second, we show that telomeres carrying blunt ends have the same length distribution as remaining telomeres and hence are not subject to rapid telomere deletion or lengthening as expected from a recombination event. The most likely explanation for the origin of blunt-ended telomeres is that they represent unprocessed chromosome ends produced by the leading strand replication (Ohki et al. 2001). This would also imply that the majority of plant telomeres replicated by the leading strand mechanism are not extended by telomerase, which requires a 3′ overhang as a substrate (Lingner and Cech 1996). In most organisms, such telomeres are resected by nucleases to produce ssDNA protrusions (Larrivee et al. 2004; Chai et al. 2006b; Bonetti et al. 2009; Lam et al. 2010; Wu et al. 2010). This step is apparently important for full telomere replication in human cancer cells, where the majority of leading telomeres are extended by telomerase (Zhao et al. 2009). Interestingly, inactivation of Ku in Arabidopsis leads to telomerase-dependent telomere elongation (Riha and Shippen 2003), which may be a consequence of deregulated telomerase activity on the resected leading telomeres (Fig. 7).
Figure 7.
Model illustrating a role of Ku at blunt-ended telomeres. (A) Protection of a blunt-ended telomere in the presence of Ku. Ku loads onto the blunt end immediately after DNA replication, preventing nucleolytic resection. A slight translocation of Ku along chromosomal DNA may free the end for limited processing by unknown nucleases. (B) In the absence of Ku, the telomeric blunt end is resected by EXO1 and other nucleases involved in homologous recombination. The resulting 3′ G-overhang can be extended by telomerase followed by fill-in synthesis of the complementary C strand that may require the CST complex. Alternatively, the 3′ G-overhang may invade the duplex telomere, forming a t-loop. While this process may stabilize the majority of chromosome ends in Arabidopsis ku mutants, some t-loops undergo resolution, giving rise to extrachromosomal t-circles.
However, this resection step must be tightly regulated to prevent excessive DNA degradation that would be followed by homologous recombination or activation of a strong DNA damage response. We propose that the evolutionarily conserved function of Ku on telomeres is to control the C-strand resection in a manner further influenced by species-specific factors such as the availability of nucleases or the composition of telomeric chromatin. Resection is almost completely inhibited in angiosperm plants, resulting in telomeres with no or only a few-nucleotide long G-overhangs. Studies in budding yeast demonstrated that Ku physically associates with and protects chromosome termini in nondividing cells (Bonetti et al. 2010; Vodenicharov et al. 2010) and that telomeric function requires direct loading of Ku onto DNA through a circular channel in its structure (Lopez et al. 2011; Pfingsten et al. 2012). Blunt-ended telomeres generated by the leading strand replication are perfect substrates for Ku loading, which may be occluded after resection by proteins that bind to ssDNA (Faure et al. 2010). We suggest that in plants, Ku loads onto and remains associated with blunt-ended telomeres, forming a physical cap that protects chromosome termini from nucleolytic degradation (Fig. 7). Inactivation of Ku is accompanied by derepression of homologous recombination (Zellinger et al. 2007), which we demonstrate is promoted by EXO1-mediated 5′ chromosome end resection. Interestingly, the 1- to 3-nt protrusions detected in Ku-proficient plants are not dependent on EXO1 and MRE11 (data not shown). This indicates that Ku specifically inhibits the access of nucleases that normally promote homologous recombination (Mimitou and Symington 2008; Zhu et al. 2008) but still allows limited resection by other nucleases.
We further show that EXO1 promotes excision of telomeric circles in ku mutants, which is one of the first pieces of evidence demonstrating the role of EXO1-mediated resection in homologous recombination outside of yeast. However, despite the fact that G-overhangs are longer in ku mutants than in ku tert mutants, EXO1 activity appears to be more critical for recombination in telomerase-positive plants than in ku tert mutants. These data indicate that telomerase directly inhibits telomeric recombination and corroborate earlier findings in budding and fission yeast, where reactivation of telomerase suppresses the recombination-based mechanism of telomere maintenance (Teng and Zakian 1999; Subramanian et al. 2008). We propose that G-overhangs arising from resection of Arabidopsis blunt-ended telomeres can either undergo strand invasion and homologous recombination or serve as a substrate for telomerase (Fig. 7). Telomerase binding to a G-overhang may physically block efficient recruitment of recombinases and/or strand invasion. This notion is supported by the observations that telomere recombination or resection in Schizosaccharomyces pombe and Candida albicans, respectively, can be inhibited by catalytically inactive telomerase (Hsu et al. 2007; Subramanian et al. 2008).
Although we detected blunt-ended telomeres only in angiosperm plants, our understanding of DNA replication predicts that they arise immediately after DNA replication at one end of every chromosome, while the other end acquires a G-overhang. It appears that in the majority of organisms, the leading strand telomeres are more accessible to nucleases and form sufficiently long G-overhangs (Chai et al. 2006a). However, recent data in human cells show that the leading telomeres form G-overhangs only 1 h after replication, implying that a G-overhang-independent mechanism of chromosome end protection operates, at least temporarily, in humans as well (Chow et al. 2012). Several lines of evidence suggest that the protective function at leading telomeres is mediated by Ku. First, Ku-depleted human cells suffer from a massive loss of telomeric DNA via intrachromatid recombination (Wang et al. 2009). Second, data in mice show that DNA-PKcs, a mammalian kinase that associates with the Ku complex, is specifically involved in protecting leading telomeres from chromosome end-to-end fusions (Bailey et al. 2001). Finally, the notion that Ku acts in protecting blunt-ended telomeres is further corroborated by in vitro experiments with reconstituted human telomeres that showed an inhibitory effect of Ku- and telomere-binding proteins TRF2/RAP1 in DNA end joining (Bae and Baumann 2007; Bombarde et al. 2010).
In conclusion, our work demonstrates that telomeric blunt ends are stable in plant post-mitotic tissues and that their integrity relies on the Ku heterodimer. This challenges the view that telomeres require ssDNA protrusions for forming a functional capping structure and demonstrates flexibility in solutions to the chromosome end protection problem.
Materials and methods
Plant material
The following A. thaliana mutant lines were used in this study: ku70-1 (Riha et al. 2002), ku80-1 (T-DNA insertion line JF46-4) (Friesner and Britt 2003), tert (Fitzgerald et al. 1999); exo1a-1 (T-DNA insertion line GABI_146B05), exo1b-1 (SALK_062208), and stn1-1 (SALK_023505) (Song et al. 2008). The primers used for PCR genotyping are listed in Supplemental Table S1. Plants were grown at 22°C with 16 h/8 h light/dark periods, and whole 4- to 6-wk-old plants were usually harvested for DNA extraction.
DNA extraction
The plant tissue (1–5 g) was ground in liquid nitrogen and transferred to 30 mL of precooled DNA extraction buffer (63.77 g/L sorbitol, 100 mM Tris-HCl at pH 7.5, 5 mM EDTA at pH 8.0). Nuclei were pelleted by centrifugation and resuspended in 1.25 mL of DNA extraction buffer. The nuclei were lysed by adding 1.75 mL of nuclei lysis buffer (200 mM Tris-HCl at pH 8.0, 50 mM EDTA at pH 8.0, 2 M NaCl, 2% [w/v] cetyltrimethylammonium bromide) and 0.7% sarcosyl in the presence of 82 μg/mL RNase A. Samples were mixed and incubated for 25 min at 65°C. DNA was extracted with chloroform:isoamyl alcohol (24:1) and precipitated with isopropanol. DNA was then extracted twice with phenol:chloroform:isoamyl alcohol (50:49:1), precipitated with isopropanol, and resuspended in TE.
dUTP-PENT
Primer extension was carried out with 9 U of DNA polymerase I (Promega) in 100 μL of 1× DNA polymerase buffer (Promega) supplemented with dATP (750 μM), dCTP (750 μM), dTTP (600 μM), dUTP (150 μM), 0.1 μM primer (5′-CCCTAAACCCTAAA-3′), and ∼0.5 μg of genomic DNA for 16 h at 25°C. DNA was ethanol-precipitated and resuspended in 40 μL of dH2O. Samples were split in half and digested with 20 U of AluI (Fermentas) in 1× Tango buffer (Fermentas) for 6 h at 37°C. UDG treatment was carried out by adding 5 μL of 5× UDG buffer (New England Biolabs), 19 μL of H2O, and 2 U of UDG (New England Biolabs) to the restriction reaction and by incubating the mixture for an additional 2 h at 37°C. DNA was separated by alkaline electrophoresis and analyzed by Southern hybridization with a [γ-32P]ATP 5′-end-labeled (TTTAGGG)4 probe. Pretreatment of genomic DNA with the T7 gene 6 nuclease was carried out in 15 μL of the NEB4 buffer (New England Biolabs) with 3 U of T7 gene 6 exonuclease (New England Biolabs) for 5 min at 37°C. The reaction was stopped by adding 1 μL of 0.5 M EDTA. Exonuclease 1 treatment was performed as described previously (Riha et al. 2000). The radioactive signal was captured on Kodak screens, scanned by FX PhosphorImager (Bio-Rad), and quantified with Quantity One software (Bio-Rad). The proportion of telomeres lacking a G-overhang was calculated as the ratio of TRF signals between UDG-treated and nontreated reactions that were normalized to the signal from intrachromosomal telomeric DNA.
Hairpin ligation assay
The hairpin was prepared by heating the blunt-HP oligonucleotide (5′-P-GGATCCGACTTTTGTCGGATCC-3′; Sigma) in 10 μM concentration for 5 min at 99°C and cooling it down to 25°C. The ligation was carried out for 16 h at 16°C by T4 ligase (10 Weiss units) in 25 μL of 1× T4 ligase buffer (Fermentas) containing 1.6 μM hairpin and 1 μg of genomic DNA. T4 ligase was heat-inactivated (20 min at 65°C), and DNA was digested with AluI (Fermentas). Alkaline gel electrophoresis with 0.8% agarose gels was used to separate ligated products at 1.2 V cm−1 for 16–18 h. The separated DNA was analyzed by Southern hybridization with the (TTTAGGG)4 probe. For assessing the efficiency of hairpin ligation, 2 ng of EcoRV-linearized plasmid pCR2.1cycB12 (4.5 kb) was mixed with 2 μg of Arabidopsis genomic DNA. After hairpin ligation, the reaction was split in two. One-half of the reaction was digested with AluI and analyzed by Southern hybridization with the (TTTAGGG)4 probe. The other half of the reaction was digested with XbaI to remove any hairpin ligated to one end of the plasmid and analyzed by Southern hybridization with the plasmid-specific probe.
In-gel assay and TCA
The TCA was performed according to Zellinger et al. (2007). In-gel hybridization was performed as described by Heacock et al. (2007).
TRF analysis and cytology
TRF analysis was performed according to Riha et al. (2002). Mitotic spreads were prepared from Arabidopsis floral buds according to Riha et al. (2001).
Analysis of sequence permutations at telomeric blunt ends
Genomic DNA (1 μg) was ligated with the blunt hairpin 3 (Supplemental Table S1) at 16°C, followed by heat inactivation. The samples were digested with 20 U of Tru1I (Fermentas) overnight at 65°C, ethanol-precipitated, and dissolved in 25 μL of TE. Five microliters of the reaction was used as template for 20 cycles of PCR amplification in 1× GoTaq buffer (Promega) supplemented with 0.5 mM dNTPs, 1.25 μM (T3AG3)4 and hairpin-specific HP 3 primers, and 2.5 U of GoTaq (Promega) in a volume of 50 μL. PCR products were ethanol-precipitated, digested with AluI, and purified with Nucleospin columns (Macherey-Nagel). The samples were eluted in 30 μL of supplied elution buffer. Four-hundred nanograms of eluted DNA was sequenced by single-end read technology (36 nt) with the Genome Analyzer IIx (Illumina). A multiplexing system using bar-coded adapter primers was used, which allowed the analysis of eight different samples in one sequencing lane. The obtained sequencing reads—on average, 477.000 per sample—were analyzed by the custom-made program “TELOMERATOR” (Supplemental Fig. S5).
Acknowledgments
We thank Matt Watson for helpful discussion. This work was supported by the Austrian Science Fund (FWF; Y418-B03) and the Austrian Academy of Sciences.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.194944.112.
References
- Bae NS, Baumann P 2007. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol Cell 26: 323–334 [DOI] [PubMed] [Google Scholar]
- Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH 2001. Strand-specific postreplicative processing of mammalian telomeres. Science 293: 2462–2465 [DOI] [PubMed] [Google Scholar]
- Baumann P, Cech TR 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Bianchi A, Shore D 2008. How telomerase reaches its end: Mechanism of telomerase regulation by the telomeric complex. Mol Cell 31: 153–165 [DOI] [PubMed] [Google Scholar]
- Bombarde O, Boby C, Gomez D, Frit P, Giraud-Panis MJ, Gilson E, Salles B, Calsou P 2010. TRF2/RAP1 and DNA-PK mediate a double protection against joining at telomeric ends. EMBO J 29: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP 2009. Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol Cell 35: 70–81 [DOI] [PubMed] [Google Scholar]
- Bonetti D, Clerici M, Anbalagan S, Martina M, Lucchini G, Longhese MP 2010. Shelterin-like proteins and Yku inhibit nucleolytic processing of Saccharomyces cerevisiae telomeres. PLoS Genet 6: e1000966 doi: 10.1371/journal.pgen.1000966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB 2009. A DNA polymerase-α·primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem 284: 5807–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli GB, Denchi EL, de Lange T 2006. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol 8: 885–890 [DOI] [PubMed] [Google Scholar]
- Chai W, Du Q, Shay JW, Wright WE 2006a. Human telomeres have different overhang sizes at leading versus lagging strands. Mol Cell 21: 427–435 [DOI] [PubMed] [Google Scholar]
- Chai W, Sfeir AJ, Hoshiyama H, Shay JW, Wright WE 2006b. The involvement of the Mre11/Rad50/Nbs1 complex in the generation of G-overhangs at human telomeres. EMBO Rep 7: 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow TT, Zhao Y, Mak SS, Shay JW, Wright WE 2012. Early and late steps in telomere overhang processing in normal human cells: The position of the final RNA primer drives telomere shortening. Genes Dev 26: 1167–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Guo X, Ferguson DO, Chang S 2009. Multiple roles for MRE11 at uncapped telomeres. Nature 460: 914–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure V, Coulon S, Hardy J, Geli V 2010. Cdc13 and telomerase bind through different mechanisms at the lagging- and leading-strand telomeres. Mol Cell 38: 842–852 [DOI] [PubMed] [Google Scholar]
- Fitzgerald MS, Riha K, Gao F, Ren S, McKnight TD, Shippen DE 1999. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc Natl Acad Sci 96: 14813–14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner J, Britt AB 2003. Ku80- and DNA ligase IV-deficient plants are sensitive to ionizing radiation and defective in T-DNA integration. Plant J 34: 427–440 [DOI] [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V 2007. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol 14: 208–214 [DOI] [PubMed] [Google Scholar]
- Gilson E, Geli V 2007. How telomeres are replicated. Nat Rev Mol Cell Biol 8: 825–838 [DOI] [PubMed] [Google Scholar]
- Giraud-Panis MJ, Teixeira MT, Geli V, Gilson E 2010. CST meets shelterin to keep telomeres in check. Mol Cell 39: 665–676 [DOI] [PubMed] [Google Scholar]
- Gravel S, Larrivee M, Labrecque P, Wellinger RJ 1998. Yeast Ku as a regulator of chromosomal DNA end structure. Science 280: 741–744 [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T 1999. Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- Grossi S, Puglisi A, Dmitriev PV, Lopes M, Shore D 2004. Pol12, the B subunit of DNA polymerase α, functions in both telomere capping and length regulation. Genes Dev 18: 992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heacock ML, Idol RA, Friesner JD, Britt AB, Shippen DE 2007. Telomere dynamics and fusion of critically shortened telomeres in plants lacking DNA ligase IV. Nucleic Acids Res 35: 6490–6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC 1998. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell 95: 963–974 [DOI] [PubMed] [Google Scholar]
- Hsu M, McEachern MJ, Dandjinou AT, Tzfati Y, Orr E, Blackburn EH, Lue NF 2007. Telomerase core components protect Candida telomeres from aberrant overhang accumulation. Proc Natl Acad Sci 104: 11682–11687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YC, Akhter S, Gu P, Ye J, Poulet A, Giraud-Panis MJ, Bailey SM, Gilson E, Legerski RJ, Chang S 2010. SNMIB/Apollo protects leading-strand telomeres against NHEJ-mediated repair. EMBO J 29: 2230–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrivee M, LeBel C, Wellinger RJ 2004. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev 18: 1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94: 399–409 [DOI] [PubMed] [Google Scholar]
- Lei M, Podell ER, Cech TR 2004. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol 11: 1223–1229 [DOI] [PubMed] [Google Scholar]
- Lingner J, Cech TR 1996. Purification of telomerase from Euplotes aediculatus: Requirement of a primer 3′ overhang. Proc Natl Acad Sci 93: 10712–10717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cooper JP, Cech TR 1995. Telomerase and DNA end replication: No longer a lagging strand problem? Science 269: 1533–1534 [DOI] [PubMed] [Google Scholar]
- Loayza D, Parsons H, Donigian J, Hoke K, de Lange T 2004. DNA binding features of human POT1: A nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J Biol Chem 279: 13241–13248 [DOI] [PubMed] [Google Scholar]
- Lopez CR, Ribes-Zamora A, Indiviglio SM, Williams CL, Haricharan S, Bertuch AA 2011. Ku must load directly onto the chromosome end in order to mediate its telomeric functions. PLoS Genet 7: e1002233 doi: 10.1371/journal.pgen.1002233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov VL, Hirose Y, Langmore JP 1997. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88: 657–666 [DOI] [PubMed] [Google Scholar]
- Maringele L, Lydall D 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes Dev 16: 1919–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2010. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J 29: 3358–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS 2002. Conserved structure for single-stranded telomeric DNA recognition. Science 296: 145–147 [DOI] [PubMed] [Google Scholar]
- Nakaoka H, Nishiyama A, Saito M, Ishikawa F 2012. Xenopus laevis Ctc1–Stn1–Ten1 (xCST) protein complex is involved in priming DNA synthesis on single-stranded DNA template in Xenopus egg extract. J Biol Chem 287: 619–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesian L, Karlseder J 2011. Mammalian 5′ C-rich telomeric overhangs are a mark of recombination-dependent telomere maintenance. Mol Cell 42: 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki R, Tsurimoto T, Ishikawa F 2001. In vitro reconstitution of the end replication problem. Mol Cell Biol 21: 5753–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten JS, Goodrich KJ, Taabazuing C, Ouenzar F, Chartrand P, Cech TR 2012. Mutually exclusive binding of telomerase RNA and DNA by ku alters telomerase recruitment model. Cell 148: 922–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raices M, Verdun RE, Compton SA, Haggblom CI, Griffith JD, Dillin A, Karlseder J 2008. C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell 132: 745–757 [DOI] [PubMed] [Google Scholar]
- Riha K, Shippen DE 2003. Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc Natl Acad Sci 100: 611–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K, McKnight TD, Fajkus J, Vyskot B, Shippen DE 2000. Analysis of the G-overhang structures on plant telomeres: Evidence for two distinct telomere architectures. Plant J 23: 633–641 [DOI] [PubMed] [Google Scholar]
- Riha K, McKnight TD, Griffing LR, Shippen DE 2001. Living with genome instability: Plant responses to telomere dysfunction. Science 291: 1797–1800 [DOI] [PubMed] [Google Scholar]
- Riha K, Watson JM, Parkey J, Shippen DE 2002. Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J 21: 2819–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Leehy K, Warrington RT, Lamb JC, Surovtseva YV, Shippen DE 2008. STN1 protects chromosome ends in Arabidopsis thaliana. Proc Natl Acad Sci 105: 19815–19820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L, Moser BA, Nakamura TM 2008. Recombination-based telomere maintenance is dependent on Tel1–MRN and Rap1 and inhibited by telomerase, Taz1, and Ku in fission yeast. Mol Cell Biol 28: 1443–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng SC, Zakian VA 1999. Telomere–telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol 19: 8083–8093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J 2006. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 127: 709–720 [DOI] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J 2007. Replication and protection of telomeres. Nature 447: 924–931 [DOI] [PubMed] [Google Scholar]
- Verdun RE, Crabbe L, Haggblom C, Karlseder J 2005. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell 20: 551–561 [DOI] [PubMed] [Google Scholar]
- Vodenicharov MD, Laterreur N, Wellinger RJ 2010. Telomere capping in non-dividing yeast cells requires Yku and Rap1. EMBO J 29: 3007–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ghosh G, Hendrickson EA 2009. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci 106: 12430–12435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JM, Shippen DE 2007. Telomere rapid deletion regulates telomere length in Arabidopsis thaliana. Mol Cell Biol 27: 1706–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JM, Bulankova P, Riha K, Shippen DE, Vyskot B 2005. Telomerase-independent cell survival in Arabidopsis thaliana. Plant J 43: 662–674 [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Ethier K, Labrecque P, Zakian VA 1996. Evidence for a new step in telomere maintenance. Cell 85: 423–433 [DOI] [PubMed] [Google Scholar]
- Wu P, van Overbeek M, Rooney S, de Lange T 2010. Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol Cell 39: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellinger B, Akimcheva S, Puizina J, Schirato M, Riha K 2007. Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol Cell 27: 163–169 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE 2009. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138: 463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]