Cpf1 is a conserved subunit of the Set1 methyltransferase. Bird and colleagues investigate Cpf1 activity in controlling H3K4me3 deposition. Interestingly, Cpf1 is found to have two opposing roles: a DNA-binding-independent role in targeting Set1 to active promoters with CpG islands, and a DNA-binding-dependent role in preventing Set-mediated H3K4me3 deposition at non-CpG island regulatory regions. Cpf1 is thus recognized as a specificity factor that integrates information on promoter content and transcriptional activity to modulate H3K4me3 deposition genome-wide.
Keywords: epigenetics, chromatin, H3K4me3, Cfp1, CpG islands
Abstract
Trimethylation of histone H3 Lys 4 (H3K4me3) is a mark of active and poised promoters. The Set1 complex is responsible for most somatic H3K4me3 and contains the conserved subunit CxxC finger protein 1 (Cfp1), which binds to unmethylated CpGs and links H3K4me3 with CpG islands (CGIs). Here we report that Cfp1 plays unanticipated roles in organizing genome-wide H3K4me3 in embryonic stem cells. Cfp1 deficiency caused two contrasting phenotypes: drastic loss of H3K4me3 at expressed CGI-associated genes, with minimal consequences for transcription, and creation of “ectopic” H3K4me3 peaks at numerous regulatory regions. DNA binding by Cfp1 was dispensable for targeting H3K4me3 to active genes but was required to prevent ectopic H3K4me3 peaks. The presence of ectopic peaks at enhancers often coincided with increased expression of nearby genes. This suggests that CpG targeting prevents “leakage” of H3K4me3 to inappropriate chromatin compartments. Our results demonstrate that Cfp1 is a specificity factor that integrates multiple signals, including promoter CpG content and gene activity, to regulate genome-wide patterns of H3K4me3.
Genomic DNA of eukaryotes is organized into chromatin, a structure involved in all aspects of DNA metabolism, including transcription, replication, or repair. These roles are mediated in part by diverse post-translational modifications of histone residues (Kouzarides 2007). Recent systematic profiling of histone modifications and chromatin-associated factors revealed that chromatin can be subdivided into discrete states with distinct functional properties (Filion et al. 2010; Kharchenko et al. 2010; Ernst et al. 2011). The new classifications recapitulate known distinctions between, for example, euchromatin and heterochromatin, but recognize more subtle states relating to active and poised promoters or strong and weak enhancers (Ernst et al. 2011). Therefore, histone modification patterns can have predictive value regarding the status of the underlying DNA sequence, even though the role for each individual modification is not yet established. To fully appreciate the significance of structural adaptations of chromatin, it is important to understand their origin. Are they a secondary response to transcriptional activity per se? Or do cis-acting factors set up appropriate chromatin conformations independently regardless of transcriptional status?
These questions can be posed specifically for trimethylation of histone H3 Lys 4 (H3K4me3), a modification found at gene promoters. In yeast, it is a feature of active gene expression (Santos-Rosa et al. 2002; Ng et al. 2003), but in mammals, H3K4me3 is found at active and inactive promoters at a level dependent on gene activity (Barski et al. 2007; Guenther et al. 2007). There is an intriguingly close correlation between sites of H3K4me3 and another feature of genomic DNA: CpG islands (CGIs). CGIs are 1 kb on average, show an elevated G+C base composition and high CpG content, and are usually free of DNA methylation (Illingworth and Bird 2009; Deaton and Bird 2010). The majority of annotated gene promoters in mice and humans are associated with a CGI and tend to be marked by H3K4me3 independent of gene activity (Guenther et al. 2007; Mikkelsen et al. 2007). A significant fraction of all CGIs in both humans and mice are not associated with annotated transcription start sites (TSSs) but nevertheless exhibit characteristics of promoters, including H3K4me3 (Illingworth et al. 2010). The idea that DNA sequence itself contributes to recruitment of histone methyltransferases has been strongly supported by the finding that insertion of G+C-rich DNA lacking a promoter into the genome is sufficient to create a novel domain of H3K4me3 (Mendenhall et al. 2010; Thomson et al. 2010). These findings suggest that a key function of CGIs is to recruit H3K4me3, which in turn favors transcriptional competence.
Set1 is the sole H3K4 histone methyltransferase in budding yeast (Briggs et al. 2001; Krogan et al. 2002; Nagy et al. 2002), and in Drosophila melanogaster, dSet1 is responsible for the bulk of H3K4me2 and H3K4me3 (Ardehali et al. 2011; Mohan et al. 2011). In mammals, there are at least six Set1-related proteins, including two homologs of Set1—Set1A and Set1B—and the mixed-lineage leukemia (Mll) family (Mll1–4), which comprises orthologs of the Drosophila trithorax gene (Eissenberg and Shilatifard 2010). All of these proteins form complexes that are related to the yeast Set1 complex (or COMPASS) and include the common subunits Wdr5, Rbbp5, Ash2L, and Dpy-30 (Miller et al. 2001; Roguev et al. 2001). Of particular relevance regarding CGIs is CxxC finger protein 1 (Cfp1, CXXC1, or CGBP), which is a component of both Set1A and Set1B complexes (Lee and Skalnik 2005; JH Lee et al. 2007). Cfp1 binds unmethylated CpGs in vitro via its CxxC zinc finger domain (Lee et al. 2001). Mll1 and Mll2 also have CxxC domains that bind unmethylated CpGs in vitro (Birke et al. 2002; Bach et al. 2009). We previously reported that Cfp1 colocalizes with CGIs in mouse brains, and its depletion in fibroblasts leads to decreased H3K4me3 at tested CGIs (Thomson et al. 2010). The results suggest that Cfp1 targets the Set1 complex to CGIs regardless of their transcriptional activity. Similarly, the CxxC protein KDM2A, which demethylates H3K36, depletes H3K36me2 at CGIs (Blackledge et al. 2010).
In the present study, we examined how genome-wide trimethylation of H3K4 in mouse embryonic stem (ES) cells is dependent on Cfp1. Targeted disruption of the Cfp1 gene results in peri-implantation embryonic lethality in mice (Carlone and Skalnik 2001). Also, RNAi-mediated knockdown of Cfp1 in somatic cells is severely detrimental (Young and Skalnik 2007; Thomson et al. 2010). Mouse ES cells lacking Cfp1, however, are viable and can self-renew, although they are unable to differentiate (Carlone et al. 2005). We set out to understand the role of Cfp1 in dictating H3K4me3 patterns using Cfp1−/− ES cells. Our findings demonstrate that Cfp1 plays a key role in organizing genome-wide H3K4me3 in mouse ES cells. We found that Cfp1 is essential for proper H3K4me3 accumulation at a large number of promoters, with a strong bias toward CGIs and active genes. We also found that decreased H3K4me3 at active promoters did not consistently affect transcription of the associated gene, raising questions about the relationship between this histone modification and gene expression. Unexpectedly, reduced H3K4me3 was rescued by a DNA-binding-deficient version of Cfp1, indicating that recruitment of H3K4me3 to CGIs can occur independent of CpG binding by Cfp1. In contrast, the CpG-binding mutant of Cfp1 was unable to rescue a second prominent phenotype of Cfp1−/− ES cells; namely, the aberrant accumulation of H3K4me3 at ectopic sites. Many of these ectopic H3K4me3 sites corresponded to enhancers and sites bound by CTCF and cohesin. Furthermore, aberrant H3K4me3 accumulation is linked to increased expression of the proximal gene. Our results reveal roles for Cfp1 as a specificity factor that regulates genome-wide H3K4me3 by integrating both CpG content at promoters and gene activity. They also establish that the ability of Cfp1 to bind unmethylated CpGs is essential to restrict the activity of the Set1 complex to promoters and avoid “leakage” of this modification to inappropriate chromatin compartments with consequent effects on local gene expression.
Results
Cfp1 is required for H3K4me3 at many gene promoters
Previous work suggested that Cfp1 dictates H3K4me3 patterns by targeting the Set1 complex to CGI promoters (Thomson et al. 2010). To investigate this hypothesis further, we analyzed H3K4me3 distribution in Cfp1−/− ES cells, which show normal global levels of H3K4me3 (Supplemental Fig. S1A; Lee and Skalnik 2005). We carried out H3K4me3 chromatin immunoprecipitation (ChIP) coupled to massively parallel DNA sequencing (ChIP-seq) in wild-type and Cfp1−/− ES cells. We identified 18,244 Ensembl gene promoters in wild-type ES cells showing a detectable H3K4me3 peak within 2 kb of their TSSs—a number similar to that observed in human ES cells (Guenther et al. 2007). Consistent with previous observations, the vast majority of H3K4me3-positive promoters (15,441, or 85%) were associated with a CGI (Supplemental Fig. S1B; Barski et al. 2007; Guenther et al. 2007; Mikkelsen et al. 2007).
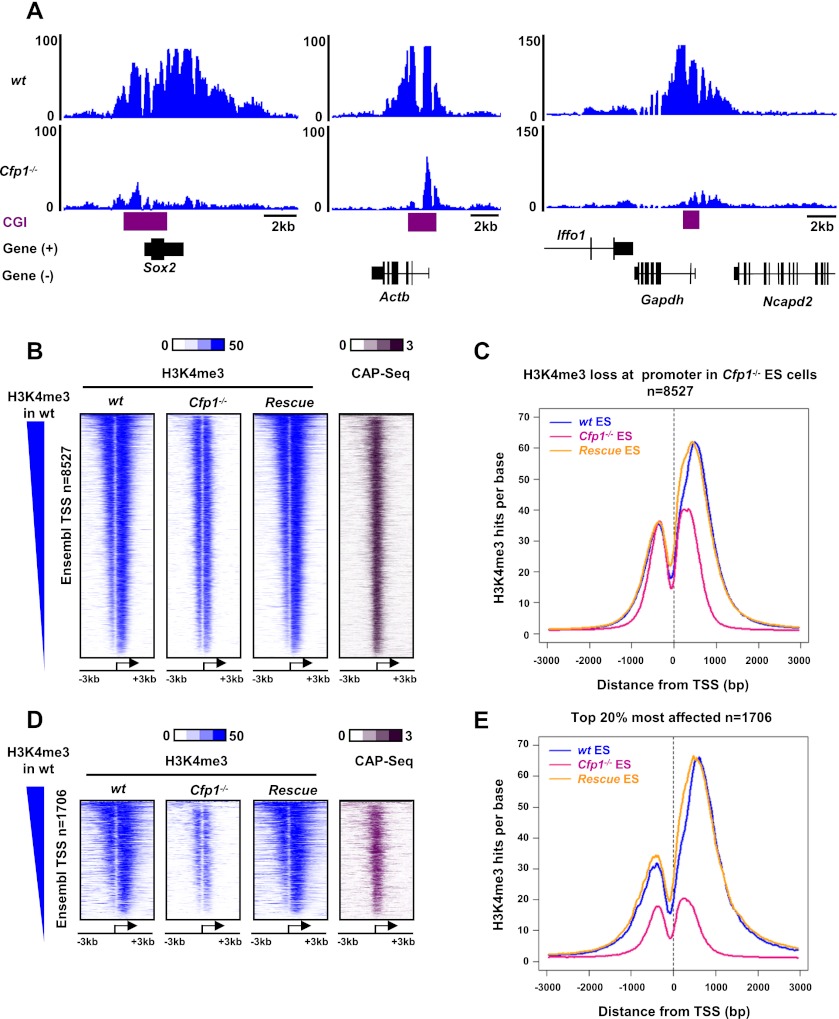
Cfp1−/− ES cells showed loss of H3K4me3 at many loci, ranging from severe loss (exemplified by Sox2, Actb, and Gapdh loci) (Fig. 1A) to no discernable effect (Supplemental Fig. S1C). The magnitude of these changes was confirmed at candidate loci (Actb and Sox2) in three independent Cfp1−/− ES cell clones using ChIP followed by quantitative PCR (qPCR) (Supplemental Fig. S1D,E). Overall, we identified 8527 Ensembl gene promoters showing a detectable reduction in H3K4me3 in Cfp1−/− ES cells (Fig. 1B) as well as 1799 promoters with increased H3K4me3, leaving 8663 that are weakly or not affected (Supplemental Fig. S1F; Supplemental Table S1). Overall, about half of all H3K4me3-positive promoters are affected by loss of Cfp1 in ES cells, 95% of which are associated with a CGI (Fig. 1B). Composite profiles revealed that the loss of H3K4me3 is mainly located downstream from the TSS (Fig. 1C). A similar composite profile for the 20% most affected promoters showed a more drastic depletion of H3K4me3 both upstream of and downstream from the TSS (Fig. 1D,E). To confirm that the absence of Cfp1 was primarily responsible for the observed decrease, we performed H3K4me3 ChIP-seq in a rescue cell line obtained by stable transfection of a human Cfp1 cDNA into Cfp1−/− ES cells (Carlone et al. 2005; Tate et al. 2009a). These wild-type rescue cells, which express Cfp1 at near endogenous levels (Supplemental Fig. S1G), generally regained appropriate H3K4me3 patterns at affected promoters (Fig. 1B–E).
Figure 1.
Cfp1 regulates H3K4me3 at many gene promoters. (A) Genome browser screenshots representing H3K4me3 signal (as read coverage) in wild-type (wt) and Cfp1−/− ES cells at selected gene loci (Sox2, Actb, and Gapdh). CGIs are represented in purple, and RefSeq genes are in black. (B) Heat map representing ChIP-seq signal for H3K4me3 (blue) in wild-type, Cfp1−/−, and wild-type rescue ES cells for Ensembl promoters showing decreased H3K4me3 signal in Cfp1−/− ES cells (n = 8527). CxxC affinity purification (CAP)-seq enrichment of CGIs in wild-type ES cells is also shown (purple). CAP-seq identifies CGIs using biochemical affinity for an immobilized CxxC domain. Genes are rank-ordered from the highest to the lowest H3K4me3 signal in wild-type ES cells. Signal is displayed from −3 kb to +3 kb surrounding each annotated TSS. (C) Composite profile showing H3K4me3 signal for wild-type (blue), Cfp1−/− (pink), and wild-type rescue (orange) ES cells at 8527 Ensembl promoters showing decreased H3K4me3 in Cfp1−/− ES cells. D and E are equivalent to B and C, respectively, but represent only the 20% most affected promoters with respect to H3K4me3 loss in Cfp1−/− ES cells (n = 1706).
H3K4me3 at highly expressed genes is most affected by Cfp1 deficiency
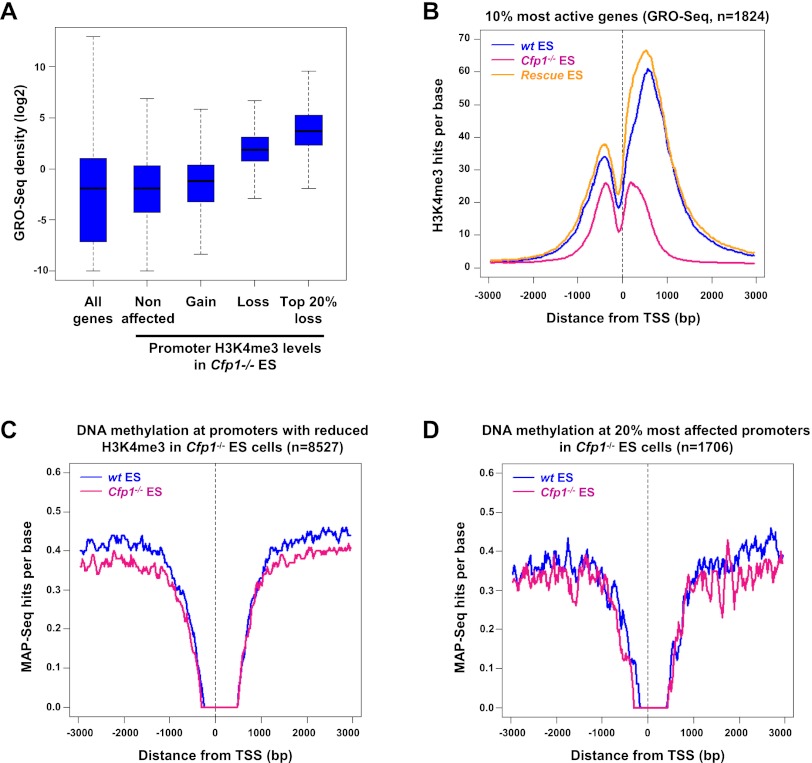
Since only a fraction of H3K4me3-positive promoters are significantly affected by the absence of Cfp1, we hypothesized that transcriptional status could distinguish Cfp1-sensitive from Cfp1-insensitive loci. To measure transcription rates, we performed global run-on (GRO)-seq, which provides a snapshot of genome-wide RNA polymerase activity (Core et al. 2008). GRO-seq read density across Ensembl genes showed that genes with decreased H3K4me3 in Cfp1−/− ES cells are the most transcriptionally active in wild-type ES cells (Fig. 2A). Within this subset, the genes whose H3K4me3 was most severely affected by the absence of Cfp1 (top 20%) had an even higher level of transcription (Fig. 2A). This trend was confirmed using published expression microarray data (Mikkelsen et al. 2007) and by RNA polymerase II (RNA Pol II) ChIP-seq for mouse ES cells (Supplemental Fig. S2A,B). Specifically, the H3K4me3 status of the top 10% most expressed genes in wild-type cells was highly sensitive to loss of Cfp1 (Fig. 2B). Similarly, promoters of active genes (RNA Pol II+ and H3K79me2+) (Kagey et al. 2010; Rahl et al. 2010) again showed reduced H3K4me3 in Cfp1−/− ES cells compared with wild-type and the wild-type rescue cell lines (Supplemental Fig. 2C). In contrast, little change in H3K4me3 profile was detected at promoters of nonproductive (RNA Pol II+ and H3K79me2−) or inactive (RNA Pol II−) genes (Supplemental Fig. S2C). Consistent with these observations, we found that H3K4me3 levels at bivalent promoters, which are silent promoters enriched for both H3K4me3 and H3K27me3 (Azuara et al. 2006; Bernstein et al. 2006), were not significantly altered in Cfp1−/− ES cells (Supplemental Fig. S2D,E). We conclude that H3K4me3 at promoters of highly expressed genes is most sensitive to the lack of Cfp1. Finally, we could confirm by ChIP-qPCR that the absence of Cfp1 leads to decreased Set1A binding at Actb, Gapdh, and Rpl3 gene loci (Supplemental Fig. S2F,G). We conclude that Cfp1 is required for proper Set1-dependent H3K4me3 at active gene promoters.
Figure 2.
Cfp1 deficiency preferentially affects H3K4me3 at highly expressed genes without altering their DNA methylation. (A) GRO-seq density distribution in wild-type (wt) ES cells for all Ensembl genes (All genes); for genes whose H3K4me3 is not affected (Nonaffected, n = 8663), increased (Gain, n = 1799), or decreased (Loss, n = 8527); or for the 20% most affected (Top 20% loss, n = 1706) in Cfp1−/− ES cells. Box plots represent the central 50% of the data (filled box), the median value (central bisecting line), and 1.5× the interquartile range (whiskers). (B) Composite profile showing H3K4me3 signal for wild-type (blue), Cfp1−/− (pink), and wild-type rescue (orange) ES cells for the 10% most active of all H3K4me3-positive Ensembl genes in wild-type ES cells, as judged by GRO-seq read density (n = 1824). (C) Composite profile showing MAP-seq signal for wild-type (blue) and Cfp1−/− (pink) ES cells at 8527 Ensembl promoters showing decreased H3K4me3 in Cfp1−/− ES cells. MAP-seq identifies clusters of methylated CpGs using biochemical affinity for an immobilized methyl-CpG-binding domain (MBD). (D) The same as C but representing only the 20% most affected promoters with respect to H3K4me3 loss in Cfp1−/− ES cells (n = 1706).
H3K4me3 depletion at active CGIs does not lead to increased DNA methylation
Methylation at H3K4 potentially inhibits DNA methylation by preventing recruitment of de novo DNA methyltransferases (Ooi et al. 2007; Zhang et al. 2010). We tested whether decreased H3K4me3 at active CGI promoters influenced their DNA methylation status by methyl-CpG affinity purification (MAP) coupled with high-throughput DNA sequencing (MAP-seq) (Illingworth et al. 2010) in wild-type and Cfp1−/− ES cells. No increase in CGI methylation was detected in Cfp1−/− ES cells at either gene promoters showing reduced H3K4me3 (Fig. 2C,D) or promoters with increased or unaffected H3K4me3 (Supplemental Fig. S3A). Cfp1−/− ES cells are reported to display a global decrease in genomic DNA methylation (Carlone et al. 2005). Accordingly, our MAP-seq data confirmed reduced DNA methylation at promoters that are normally methylated in wild-type ES cells (Supplemental Fig. S3B,C). The absence of increased methylation at H3K4me3-deficient promoters may be related to their persistent transcription (see below) or the compromised DNA methylation system in Cfp1−/− ES cells.
Loss of H3K4me3 at active genes does not affect gene expression
If H3K4me3 is an important contributor to the transcription process at highly expressed genes, then gene expression should be disturbed in Cfp1−/− ES cells. Remarkably, gene expression microarrays showed only subtle changes, with few probes changing twofold or more (P < 0.05) between wild-type with Cfp1−/− ES cells (Fig. 3A). In fact, stable RNA from genes with decreased H3K4me3 in Cfp1−/− ES cells (Fig. 3A, highlighted in red) exhibited mild up-regulation or down-regulation in a manner similar to genes whose H3K4me3 was not altered by the mutation (Fig. 3A, highlighted in blue). Focusing on the 20% of promoters with the most severe H3K4me3 depletion (Fig. 3A, orange) revealed a weak tendency to accumulate toward the right quadrant, suggesting somewhat reduced expression. A reciprocal weak tendency toward increased expression was observed at the few genes whose promoters gain H3K4me3 in Cfp1−/− ES cells (Fig. 3A, green). Both of these effects are modest. Equivalent results were obtained when comparing Cfp1−/− ES cells with the wild-type rescue line (Supplemental Fig. S4A).
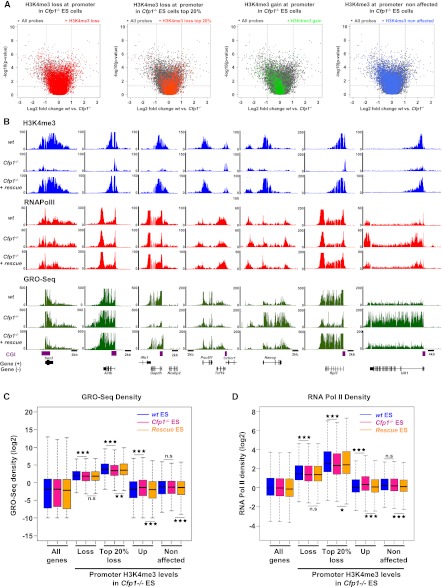
Figure 3.
Transcription is weakly affected by decreased H3K4me3 at active gene promoters. (A) Volcano plots comparing expression values between wild-type and Cfp1−/− ES cells. Fold change in expression values (wild-type vs. Cfp1−/− ES cells) and statistical significance of the differences (two-sided t-test) were computed for each probe on the array (gray dots). Genes differentially expressed twofold or more lie above the horizontal threshold (P = 0.05) and outside the two vertical thresholds (plus or minus twofold differential expression). Colors denote genes in Cfp1−/− ES cells with decreased H3K4me3 (red), with the 20% most H3K4me3-depleted (orange), with increased H3K4me3 (green), and unaffected (blue). (B) Screenshots representing read coverage of H3K4me3 ChIP-seq signal (blue), RNA Pol II ChIP-seq signal (red), or GRO-seq signal (green) in wild-type, Cfp1−/− and wild-type rescue ES cells at selected gene loci (Sox2, Actb, Gapdh, Pou5f1, Nanog, Rpl3, and Ulk1). (C) Comparison of GRO-seq read density for wild-type (blue), Cfp1−/− (pink), and wild-type rescue (orange) at all Ensembl genes (All), genes whose H3K4me3 is decreased (Loss, n = 8527), the 20% most affected (Top 20% loss, n = 1706), genes whose H3K4me3 is increased (Gain, n = 1799), or genes whose H3K4me3 is unaffected (n = 8663) in Cfp1−/− ES cells. Box plots show the central 50% of the data (filled box), the median (central bisecting line), and 1.5× the interquartile range (whiskers). P-values were calculated using the nonpaired Wilcoxon test; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (D) As C, but showing the density distribution for RNA Pol II ChIP-seq reads.
Expression arrays monitor the steady-state levels of mRNAs. To assay altered transcription in Cfp1−/− cells more directly, we examined RNA Pol II ChIP-seq and GRO-seq data. In agreement with the transcriptome data, there was no obvious link between changes in H3K4me3 and transcription or RNA Pol II occupancy. For example, Actb and Pou5f1 (encoding Oct4) showed normal RNA Pol II and GRO-seq profiles despite a strong decrease in H3K4me3 in Cfp1−/− ES cells (Fig. 3B). When altered expression was seen, this effect was not simply related to H3K4me3. For example, both GRO-seq reads and RNA Pol II density were decreased at the Gapdh or Rpl3 genes, but Nanog and Ulk1 showed increased transcriptional activity despite a similar drastic loss of H3K4me3. The aggregate distribution of GRO-seq read density over the body of genes with decreased H3K4me3 revealed a very weak, but significant (P = 1.75 × 10−7), difference due to the absence of Cfp1 (Fig. 3C). This became more noticeable at the 20% most H3K4me3-depleted promoters (Fig. 3C), but expression nevertheless remained substantially higher than at genes whose H3K4me3 was not affected by Cfp1 deficiency. Equivalent results were obtained by analyzing RNA Pol II density across the same gene set (Fig. 3D). H3K4me3 at active gene promoters has been linked to pre-mRNA splicing (Sims et al. 2007). We checked for splicing defects in pre-mRNAs produced from several candidate genes showing decreased H3K4me3 in Cfp1−/− ES cells using qRT–PCR to measure the ratio of unspliced to spliced transcripts in wild-type and Cfp1−/− ES cells (Supplemental Fig. S4B). We were unable to detect increased accumulation of unspliced pre-mRNAs (Supplemental Fig. S4C) even in cases of subtle alterations in expression (Supplemental Fig. S4D). Thus, the effect of losing H3K4me3 on ES cell transcription is variable and generally modest. However, we cannot definitively rule out that low H3K4me3 levels remaining at most genes in Cfp1−/− ES cells are sufficient to mediate proper gene expression and/or splicing.
Cfp1 DNA-binding activity is not required for H3K4me3 at promoters
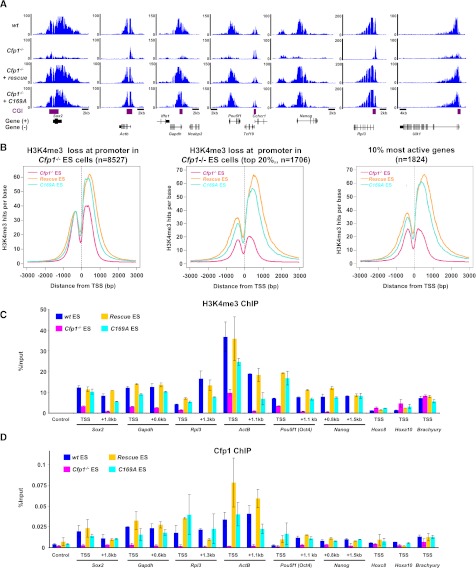
Cfp1 can bind unmethylated CpGs in vitro and in vivo via its CxxC DNA-binding domain (Lee et al. 2001; Thomson et al. 2010). Since 95% of promoters where H3K4me3 decreased in Cfp1−/− ES cells are CGI promoters, it seemed likely that Cfp1 DNA-binding activity would help target the Set1 complex to these loci. To test this hypothesis, we used an ES cell line expressing Cfp1C169A (C169A rescue), which has a missense mutation within the CxxC motif that abolishes DNA binding (Tate et al. 2009a). This cell line expressed Cfp1C169A at near endogenous levels (Supplemental Fig. S1G), and the mutant protein was shown by immunoprecipitation to interact with components of the Set1 complex as wild-type Cfp1 (Supplemental Fig. S5A). Unexpectedly, H3K4me3 ChIP-seq with this mutant rescue cell line showed restoration of nearly normal H3K4me3 levels at promoters where it was lost in Cfp1−/− ES cells (Fig. 4A; Supplemental Fig. S5B). Both the 20% most affected promoters and promoters of the 10% most active genes showed high levels of H3K4me3 in the C169A rescue cell line, although H3K4me3 levels were consistently slightly lower than in wild-type rescue ES cells (Fig. 4B). We confirmed restoration of H3K4me3 as well as Cfp1 binding at active gene promoters by ChIP-qPCR in the C169A rescue cell line (Fig. 4C,D). Targeting of Cfp1 and H3K4me3 to active promoters by a DNA-binding-deficient form of Cfp1 suggests that transcriptional activity can recruit Cfp1 and Set1. We conclude that transcription itself plays a key role in the targeting of Cfp1 and H3K4me3 to active promoters, as it is observed in the presence of a DNA-binding-deficient Cfp1.
Figure 4.
A Cfp1 DNA-binding mutant can restore H3K4me3 at active promoters. (A) Screenshots representing H3K4me3 read coverage in wild-type (wt), Cfp1−/−, wild-type rescue, and C169A rescue ES cells at selected gene loci (Sox2, Actb, Gapdh, Pou5f1, Nanog, Rpl3, and Ulk1). (B) Composite profile showing H3K4me3 signal for Cfp1−/− (pink), wild-type rescue (orange), and C169A rescue (turquoise) ES cells at all 8527 Ensembl promoters showing decreased H3K4me3 in Cfp1−/− ES cells (left panel), the top 20% decreased H3K4me3 promoters (middle panel), or the top 10% most active genes (right panel). (C,D) Quantification (percent input) of H3K4me3 (C) or Cfp1 (D) using ChIP-qPCR at the promoter and proximal coding region of selected active (Sox2, Actb, Gapdh, Pou5f1, and Nanog) and inactive (Hoxc8, Hoxa10, and Brachyury) gene loci. The negative control region (Control) corresponds to an intergenic region on chromosome 15. Colors are as in B. Error bars show the standard deviation of PCR replicates.
The DNA-binding activity of Cfp1 is required to prevent accumulation of H3K4me3 at regulatory regions outside CGIs
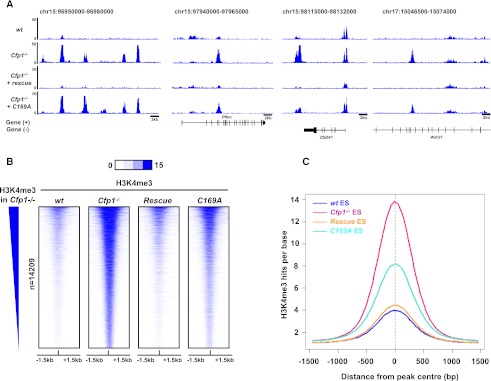
In addition to loss of H3K4me3 at many CGI promoters, Cfp1−/− ES cells also show a second prominent chromatin phenotype; namely, the accumulation of H3K4me3 at discrete regions that are remote from annotated CGIs and TSSs (Fig. 5A). Excluding CGIs and TSSs from the analysis, we detected 14,209 such “ectopic” H3K4me3 sites (Supplemental Table S2). Significantly, ectopic H3K4me3 sites were rescued by wild-type Cfp1 but persisted in cells expressing the C169A DNA-binding mutant (Fig. 5A,B). Ectopic H3K4me3 accumulation did not coincide with CpG-rich regions, although a modestly increased CpG and G+C content was detected (Supplemental Fig. S6A). As a control, significant H3K4me3 enrichment was not detected over 10,000 randomly selected genomic regions (Supplemental Fig. S6B). Aggregation of ChIP DNA sequence reads across all of these sites showed that the DNA-binding mutant of Cfp1 fails to efficiently relieve aberrant H3K4me3 at these regions (Fig. 5C), and this was confirmed by ChIP-qPCR at selected ectopic peaks (Supplemental Fig. S6C). Our results indicate that the DNA-binding activity of Cfp1 prevents aberrant accumulation of H3K4me3 away from CGI promoters, probably due to imprecise targeting of the Set1 complex.
Figure 5.
H3K4me3 accumulates at ectopic sites in the absence of Cfp1 or its DNA-binding activity. (A) Screenshots showing H3K4me3 signal read coverage in wild-type (wt), Cfp1−/−, wild-type rescue, and C169A rescue ES cells at selected genomic regions on chromosomes 15 and 17. (B) Heat map representing ChIP-seq signal for H3K4me3 (blue) in wild-type, Cfp1−/−, wild-type rescue, and C169A rescue ES cells for each of 14,209 ectopic peaks. Peaks are rank-ordered from the highest to the lowest H3K4me3 signal in Cfp1−/− ES cells. Signal is displayed from −1.5 kb to +1.5 kb surrounding the center of each peak. (C) Composite profile showing H3K4me3 signal at ectopic peak sites in wild-type (blue), Cfp1−/− (pink), wild-type rescue (orange), and C169A rescue (turquoise) ES cells.
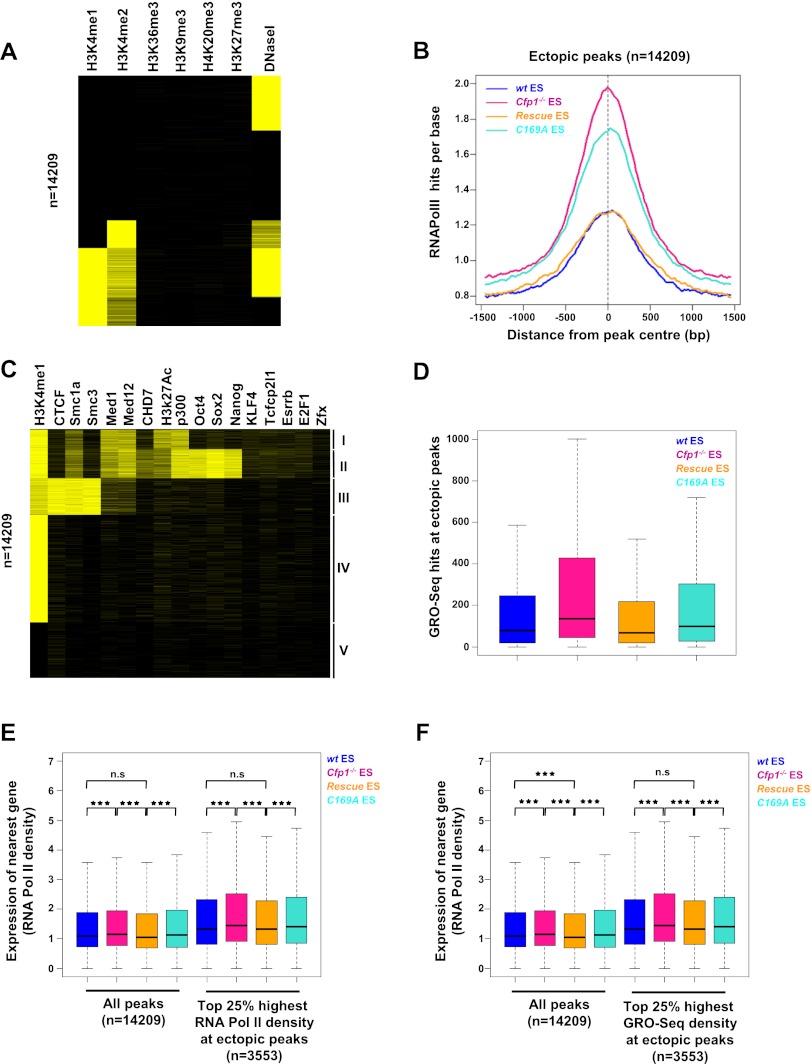
To investigate the possibility that these sites coincide with functional features of the genome, we aligned ectopic peaks with published ES cell data sets for H3K4me1, H3K4me2, H3K27me3, H3K36me3, H3K9me3, and H4K20me3 in ES cells (Mikkelsen et al. 2007; Ku et al. 2008; Meissner et al. 2008), and a collection of DNase I-hypersensitive sites (http://www.ensembl.org/index.html). Cluster analysis revealed that ectopic H3K4me3 sites tend to overlap with H3K4me1, H3K4me2, and DNase I-hypersensitive sites (Fig. 6A). In contrast, ectopic peaks colocalized poorly with peaks of the “heterochromatic” histone modifications H3K9me3 and H4K20me3, the transcriptional elongation mark H3K36me3, or the Polycomb-associated mark H3K27me3. We performed H3K4me1 ChIP-seq at increased sequence depth for each cell line, and this revealed that most ectopic peaks (>70%) are positive for H3K4me1 in wild-type ES cells (Supplemental Fig. S7A,C). We conclude that ectopic H3K4me3 peaks align with chromatin features typically found at transcriptional enhancers (Birney et al. 2007; Heintzman et al. 2007). As recent reports have detected RNA Pol II at enhancers (De Santa et al. 2010; Kim et al. 2010), we tested for bound RNA Pol II at ectopic sites using our ChIP-seq data sets for all four cell lines. RNA Pol II occupancy was seen in wild-type and wild-type rescue ES cells (Fig. 6B; Supplemental Fig. S7B,E), and levels of RNA Pol II were significantly higher in both Cfp1−/− and C169A rescue ES cells. Therefore, elevated H3K4me3 is accompanied by increased recruitment of RNA Pol II. The data argue strongly that many sites of ectopic H3K4me3 accumulation correspond to bona fide gene regulatory regions.
Figure 6.
H3K4me3 ectopic peaks in Cfp1−/− ES cells coincide with regulatory regions and correlate with enhanced expression of neighboring genes. (A) Cluster analysis showing colocalization of ectopic peaks with regions enriched with histone modifications using published data sets. Yellow bars represent overlap of an ectopic peak with a region enriched for that histone modification. (B) Composite profile showing RNA Pol II ChIP-seq signal for wild-type (wt) (blue), Cfp1−/− (pink), wild-type rescue (orange), and C169A rescue (turquoise) ES cells at all ectopic peaks. (C) As A using published binding sites for various factors. The H3K4me1 data used were generated in the present study. (D) Transcription as measured by GRO-seq read density at ectopic peaks (n = 14,209) in wild-type (blue), Cfp1−/− (pink), wild-type rescue (orange), and C169A rescue (turquoise). Box plots show the central 50% of the data (filled box), the median (central bisecting line), and 1.5× the interquartile range (whiskers). Read density was calculated over the central 500 bp of each peak. (E) Expression (measured by RNA Pol II density over the gene body) of the nearest Ensembl gene to each ectopic peak or to the top 25% ectopic peaks with the highest RNA Pol II occupancy. Box plots show the central 50% of the data (filled box), the median (central bisecting line), and 1.5× the interquartile range (whiskers). P-values were calculated using the nonpaired Wilcoxon test; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (F) Expression (measured by RNA Pol II density over the gene body) of the nearest Ensembl gene to each ectopic peak or to the top 25% ectopic peaks with the highest transcriptional activity (by GRO-seq). Box plots show the central 50% of the data (filled box), the median (central bisecting line), and 1.5× the interquartile range (whiskers). P-values were calculated using the nonpaired Wilcoxon test; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
To further annotate ectopic H3K4me3 peaks, we tested for colocalization with a panel of factors known to be associated with enhancers or other regulatory sequences in ES cells (Chen et al. 2008; Creyghton et al. 2010; Kagey et al. 2010; Schnetz et al. 2010; Ong and Corces 2011; Rada-Iglesias et al. 2011). We also compared ectopic peaks with binding sites for cohesin (Smc1a and Smc3), which contribute to promoter/enhancer interactions (Kagey et al. 2010), and CTCF, whose diverse regulatory functions include organization of global chromatin architecture and chromatin loops (Phillips and Corces 2009). Ectopic peaks grouped into classes that reflect known relationships between the tested factors (Fig. 6C). One class is characterized by the enrichment of p300, H3K27ac, Mediator, Smc1a, and H3K4me1, all of which are typical of active enhancers (Creyghton et al. 2010; Kagey et al. 2010; Schnetz et al. 2010). A second group is characterized by enrichment for binding sites for the core ES cell pluripotency factors Oct4, Sox2, and Nanog together with p300, all of which can occupy active and inactive enhancers (Creyghton et al. 2010; Rada-Iglesias et al. 2011). A third separate class is dominated by simultaneous binding of cohesin and CTCF (Kagey et al. 2010). A fourth class is enriched only by H3K4me1 based on our data set. Finally, a fifth class contains regions that do not strongly enrich for any of the tested features that could represent uncharacterized regulatory regions in ES cells or, alternatively, belong to a separate group of currently nonannotated elements. Many ectopic peaks evidently have the characteristics of regulatory regions, including enhancers, transcription factor-binding sites, or potential chromatin loops (CTCF/cohesin-binding sites). More detailed analysis suggested that CTCF/Smc1a sites as a whole are affected by the absence of Cfp1, as H3K4me3 levels at a large set of CTCF/Smc1a promoter distal sites were increased in both Cfp1−/− and C169A rescue cells (Supplemental Fig. S8A,B). A similar increase in H3K4me3 was observed at CTCF and cohesin sites, regardless of their overlap (Supplemental Fig. S8C–F; Kagey et al. 2010), but only a subset of all p300 sites is affected by Cfp1 deficiency (Supplemental Fig. S8G,H). As both CTCF and cohesin are postulated to mediate chromatin looping (Ong and Corces 2011), we speculate that Cfp1 prevents H3K4me3 from invading chromatin loops by a mechanism that requires its ability to bind to nonmethylated CpG.
Does aberrant H3K4me3 accumulation at promoter-distal regions have functional consequences? As RNA Pol II occupies many ectopic peaks, we looked for evidence of active transcription by measuring GRO-seq read density over the central 500 base pairs (bp) of each peak. Indeed, ectopic peaks showed increased transcription in both Cfp1−/− and C169A rescue ES cells, where H3K4me3 is the highest, when compared with wild-type and wild-type rescue lines (Fig. 6D). We conclude that increased H3K4me3 is matched by increased transcription at ectopic peaks. As active enhancers generally regulate the nearest flanking genes (Visel et al. 2009; Creyghton et al. 2010; De Santa et al. 2010; Kim et al. 2010; Rada-Iglesias et al. 2011), we asked whether the presence of ectopic H3K4me3 peaks affects nearby genes. A small yet significant increase in expression of the gene proximal to each ectopic peak was detected in both Cfp1−/− and C169A rescue ES cells, as assayed by RNA Pol II occupancy (Fig. 6E) and GRO-seq read density (Supplemental Fig. S9A). These data suggest that aberrant H3K4me3 is associated with increased expression of the neighboring gene. RNA Pol II occupancy and transcriptional activity at enhancers are an indication of their activity (De Santa et al. 2010; Kim et al. 2010; Bonn et al. 2012). We therefore selected a subset of ectopic peaks (25%) with the highest RNA Pol II occupancy based on ChIP read density in Cfp1−/− ES cells and investigated the effect of transcription of the proximal gene. As expected, higher RNA Pol II occupancy was linked with increased expression of the nearest gene in Cfp1−/− and C169A rescue ES cells (Fig. 6E; Supplemental Fig. S9A). A similar effect was observed at 25% of ectopic peaks showing the highest transcriptional activity by GRO-seq (Fig. 6F; Supplemental Fig. S9B). We conclude that aberrant H3K4me3 accumulation at regulatory regions in the absence of Cfp1 or its DNA-binding activity is functionally linked to increased expression of the neighboring gene in a manner dependent on RNA Pol II occupancy and transcriptional activity at those sites. This confirms that the DNA-binding activity of Cfp1 restricts H3K4me3 to its proper genomic compartment in ES cells, thereby preventing misregulation of local gene expression.
Discussion
Cfp1 function linked to transcription
The dinucleotide CpG is highly concentrated in CGIs and can attract CxxC domain-containing chromatin-modifying complexes (Blackledge et al. 2010; Thomson et al. 2010). In a simple model, the CxxC protein Cfp1 could recruit the Set1 histone methyltransferase and establish H3K4me3 at CGI promoters whether or not they are transcriptionally active (Deaton and Bird 2010; Blackledge and Klose 2011). In support of this idea, insertion of promoter-less CpG-rich DNA can mediate de novo accumulation of H3K4me3 in ES cells (Mendenhall et al. 2010; Thomson et al. 2010). This basal level of H3K4me3 at CGIs might then be enhanced by transcription of the associated gene; for example, via recruitment of chromatin-modifying activities by RNA Pol II itself (Smith and Shilatifard 2010). We evaluated this two-component model by studying mutant ES cells that lack Cfp1. Our results show that Cfp1 is not essential for basal H3K4me3 at CGIs in mouse ES cells, but is an important contributor to transcription-coupled deposition of this mark. We found that Cfp1 deficiency preferentially decreased H3K4me3 at active gene promoters, whereas nonproductive (or “poised”) genes, which do not produce detectable mature mRNAs, were not affected. In mouse brains or fibroblasts, most H3K4me3 at CGIs seems to rely on the Cfp1/Set1 pathway (Thomson et al. 2010). Our results suggest that a separate activity can maintain basal H3K4me3 levels at poised promoters in ES cells. Mll1 and Mll2, two CxxC domain H3K4 methyltransferases, are present in ES cells (Glaser et al. 2006; Jiang et al. 2011). Depletion of subunits shared by Set1 and Mll1/2 complexes leads to reduction in H3K4me3 at both active and “poised” genes, unlike deficiency of Cfp1 alone (Ang et al. 2011; Jiang et al. 2011). We propose that in ES cells, establishment of H3K4me3 at poised promoter CGIs (or artificial GC-rich insertions) depends on CxxC domain proteins other than Cfp1, likely Mll1 and/or Mll2.
Our findings may explain why ES cells and somatic cells differ significantly in their requirements for specific CxxC proteins. Loss of the Cfp1 gene in mice results in early embryonic lethality (Carlone and Skalnik 2001). Moreover Cfp1 deficiency in somatic cell lines is toxic (Young and Skalnik 2007; Thomson et al. 2010). ES cells, on the other hand, are robustly viable without Cfp1, although their inability to differentiate suggests that Cfp1 is required upon lineage commitment (Carlone et al. 2005). Similarly, gene ablation studies have revealed changing roles for Mll2 during mouse development (Glaser et al. 2006; Andreu-Vieyra et al. 2010) because in oocytes, it is required for global H3K4me3, whereas later in development, its role is much reduced. Others also report that Mll proteins have specific roles during development, with Set1 becoming the dominant H3K4 methyltransferase activity at later developmental stages (Wu et al. 2008; Wang et al. 2009; Ardehali et al. 2011; Mohan et al. 2011). Thus, it appears that the contribution of CxxC proteins in shaping chromatin varies during cellular differentiation.
Targeting Set1 and H3K4me3 at promoters
The dependency of highly transcribed genes on Cfp1 is reminiscent of the role of Set1 in yeast, where RNA polymerase is thought to directly recruit the complex (Krogan et al. 2003; Ng et al. 2003). Accordingly, the Cfp1 ortholog in Drosophila, an organism that lacks CGIs, is necessary to recruit dSet1 to transcription puffs on polytene chromosomes (Ardehali et al. 2011). We found that transcription-dependent enhancement of H3K4me3 by Cfp1 does not require its DNA-binding domain, as it is restored in Cfp1−/− cells by expression of CpG-binding-deficient Cfp1. It follows that Cfp1 functions to link Set1 and the transcriptional apparatus in a manner that primarily depends on gene activity rather than CGI binding. It is unclear which region of Cfp1 mediates this function, although Cfp1 contains redundant functional domains that can rescue several phenotypic features of Cfp1−/− ES cells (Tate et al. 2009a). The ability to bind DNA and interact with Set1 appears to be key in this respect (Tate et al. 2009a). Cfp1 also contains a PHD finger, which in its yeast ortholog can bind H3K4me3 (Shi et al. 2007) and may therefore assist targeting to low-level H3K4me3 found at active promoters in the absence of Cfp1. Other mechanisms in play to ensure that the Set1 complex is recruited to chromatin include monoubiquitylation of histone H2B (JS Lee et al. 2007), perhaps mediated by the Wdr82 protein (Lee and Skalnik 2008; Wu et al. 2008) or the elongation factor Paf1 (Krogan et al. 2003). It is apparent that proper establishment of the H3K4me3 signature at active promoters can be achieved by redundant mechanisms, several of which involve domains of Cfp1.
H3K4me3 is not required for active transcription
Given the ubiquity of H3K4me3 as a mark of transcriptional activity, we were surprised to find that drastic decreases in this chromatin mark did not necessarily lead to reduced gene expression. It was previously reported that depletion of Dpy-30, which leads to global H3K4me3 reduction in ES cells, had minimal effects on expression of most genes (Jiang et al. 2011). We significantly extend this observation by specifically interrogating genes that show decreased H3K4me3 at their promoters using three independent methods (expression arrays, GRO-seq, and RNA Pol II ChIP-seq). Although several studies report that H3K4me3 can attract specific PHD domain proteins to promoters (Wysocka et al. 2006; Vermeulen et al. 2007; Gaspar-Maia et al. 2009), we did not identify a specific requirement for H3K4me3 in regulating transcriptional activity. In yeast, Set1 is the only H3K4 histone methyltransferase, and its deletion abolishes global H3K4me1, H3K4me2, and H3K4me3 (Schneider et al. 2005). Nevertheless, yeast Set1 is not an essential gene, as mutants are viable, albeit slow-growing (Briggs et al. 2001; Miller et al. 2001), and do not display widespread transcriptome alterations (Miller et al. 2001; Guillemette et al. 2011). H3K4me3 function may be gene-specific or may play alternative transcription-related roles. For example, H3K4me3 may modulate the kinetics of RNA Pol II elongation to facilitate transcript processing (Terzi et al. 2011) or affect antisense transcription of regulatory RNAs (van Dijk et al. 2011). Alternatively, as we rarely observe a complete lack of H3K4me3 upon Cfp1 deletion, it remains possible that low levels can be sufficient to mediate the role of H3K4me3 in transcription and splicing.
The CxxC domain of Cfp1 restricts Set1 activity to promoters
In addition to its effects at promoters, Cfp1 deficiency caused increased H3K4me3 at many discrete sites across the genome. Ectopic H3K4me3 peaks were not random, but often coincided with regulatory regions, including active and inactive enhancers and potential regulatory chromatin loops defined by CTCF and cohesin. Unlike H3K4me3 deficiency at promoters, ectopic peaks of H3K4me3 were not abolished by expression of the DNA-binding mutant of Cfp1. Cfp1C169A associated with the Set1 complex and is shown here to restore appropriate levels of H3K4me3 at multiple TSSs. We conclude that Cfp1 DNA-binding activity is essential for appropriate deposition of H3K4me3, as it restricts the action of the Set1 complex to its appropriate genomic compartment (Fig. 7). The Cfp1 DNA-binding domain has been previously implicated by fluorescence microscopy in restricting Set1 to euchromatin (Tate et al. 2009b). Here, we show that Cfp1 is also required for proper Set1 targeting within euchromatin. By binding preferentially to CGIs, Cfp1 biases H3K4 methyltransferase activity toward promoters and impedes unsupervised acquisition of this promoter-like feature in regions of the genome that do not define the 5′ ends of genes. Thus, Cfp1 can integrate both CpG content and gene activity in order to establish proper H3K4me3 throughout the genome.
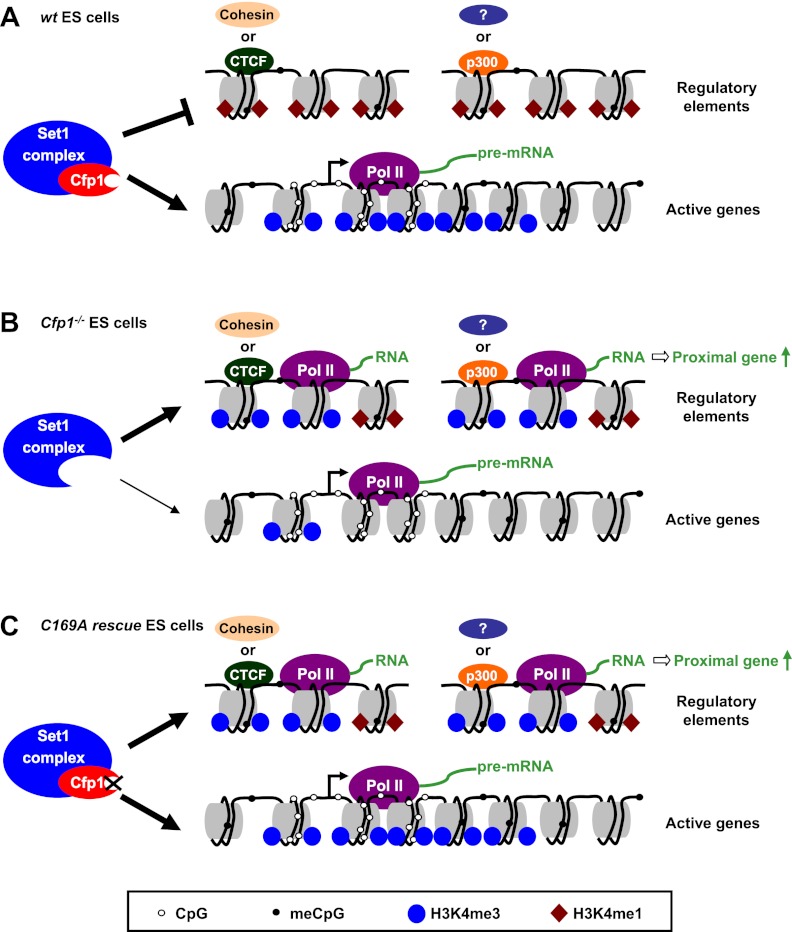
Figure 7.
Model for the role of Cfp1 in regulating genome-wide H3K4me3. (A) Multiple Cfp1-dependent and Cfp1-independent mechanisms contribute to targeting of the Set1 complex to transcriptionally active CGI promoters in wild-type (wt) mouse ES cells. Cfp1 binding to CpG-rich DNA minimizes Set1 conversion of H3K4me1 to H3K4me3 (brown diamonds and blue circles, respectively) in CpG-poor regulatory regions, including binding sites for CTCF, cohesin, or enhancers (marked by p300 binding) and uncharacterized regions bound by unknown factors (marked by a question mark). (B) In the absence of Cfp1, H3K4me3 is reduced at active CGI promoters due to incorrect targeting of the Set1 complex and accumulates inappropriately at regulatory regions. This is accompanied by RNA Pol II binding and transcription at these sites and increased expression of the proximal gene. (C) Cfp1 that is unable to bind CpG cannot prevent aberrant H3K4me3 accumulation at regulatory regions but is able to correctly target the Set1 complex to transcriptionally active promoters.
It is of interest that TAF3, a component of the basal transcription factor TFIID, is found at nonpromoter regions occupied by CTCF/cohesin and is implicated in gene regulation (Liu et al. 2011). TAF3 is a PHD finger protein known to bind H3K4me3 (Vermeulen et al. 2007), raising the possibility that H3K4me3 outside promoters is normally functional. Disturbance of nonpromoter H3K4me3 levels in the absence of Cfp1 may therefore be partly responsible for the inability of Cfp1−/− ES cells to differentiate. Overall, our results implicate Cfp1 in the maintenance of histone modification patterns. Lack of Cfp1—or impairment of its DNA-binding activity—undoubtedly affects the subdivision of chromatin into discrete “states” that distinguish, for example, active promoters and regulatory regions (Filion et al. 2010; Kharchenko et al. 2010; Ernst et al. 2011). Cfp1 and, perhaps, other CxxC proteins evidently play a central role in maintaining the balance between the alternative conditions of chromatin.
Materials and methods
Antibodies
Anti-H3K4me3 was obtained from Millipore (07-473). All other antibodies are listed in the Supplemental Material.
ES cell culture
ES cells were grown in gelatinized dishes in Glasgow MEM (Gibco) supplemented with 10% fetal bovine serum (Hyclone), 1× MEM nonessential amino acids, 1 mM sodium pyruvate, 50 μM 2-mercaptoethanol (Gibco), and LIF. The C169A rescue cell line was grown in the presence of 50 μg/mL hygromycin B (Invitrogen). E14TG2a ES cells were used as wild-type ES cells. Other ES cell lines (Cfp1−/−, wild-type rescue, and C169A rescue) have been described previously (Carlone et al. 2005; Tate et al. 2009a).
ChIP and GRO
Detailed protocols can be found in the Supplemental Material.
MAP and CxxC affinity purification (CAP)
Sonicated genomic DNA from ES cells was processed for MAP and CAP as previously described (Illingworth et al. 2010).
High-throughput sequencing
A detailed description of library preparation for high-throughput sequencing as well as data analysis, generation of composite profiles and heat maps, and cluster analysis of ectopic peaks can be found in the Supplemental Material. Data sets are described in Supplemental Table S3.
Illumina BeadChip arrays
Total RNA was labeled using the TotalPrep RNA amplification kit (Ambion) and was hybridized to Illumina MouseWG-6 BeadChip arrays. Three biological replicates were carried out for each cell type. Bead level data were summarized using Illumina BeadStudio, and data were normalized using the cubic spline normalization method in BeadStudio. Subsequent analysis was carried out using GeneSpring GX11 (Agilent Technologies). Data were quantile-normalized between arrays, and the average value for each replicates was given to each probe in each cell line. Fold change and statistical significance were computed for each probe using Genespring.
Accession numbers
Data have been deposited under ArrayExpress accession numbers E-ERAD-79, E-ERAD-80, E-ERAD-53, and E-MTAB-1198.
Acknowledgments
We thank L. Gibson, J. Burton, and their teams, and K. Auger and S. Messager at the Wellcome Trust Sanger Institute for managing and performing the DNA sequencing. Illumina BeadChip experiments were performed at the Wellcome Trust Clinical Research Facility, Edinburgh, by L. Evenden and L. Murphy. We also thank M. Koerner, M. Lyst, and S. Lagger for a critical reading of the manuscript. This work was funded by the Wellcome Trust (091580), The Medical Research Council UK (G0800401), and Cancer Research UK (C1295/A9590).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.194209.112.
References
- Andreu-Vieyra CV, Chen R, Agno JE, Glaser S, Anastassiadis K, Stewart AF, Matzuk MM 2010. MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS Biol 8: e1000453 doi: 10.1371/journal.pbio.1000453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. 2011. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145: 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T 2011. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J 30: 2817–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. 2006. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538 [DOI] [PubMed] [Google Scholar]
- Bach C, Mueller D, Buhl S, Garcia-Cuellar MP, Slany RK 2009. Alterations of the CxxC domain preclude oncogenic activation of mixed-lineage leukemia 2. Oncogene 28: 815–823 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Birke M, Schreiner S, Garcia-Cuellar MP, Mahr K, Titgemeyer F, Slany RK 2002. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res 30: 958–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Klose R 2011. CpG island chromatin: A platform for gene regulation. Epigenetics 6: 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ 2010. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell 38: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EE 2012. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet 44: 148–156 [DOI] [PubMed] [Google Scholar]
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev 15: 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlone DL, Skalnik DG 2001. CpG binding protein is crucial for early embryonic development. Mol Cell Biol 21: 7601–7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlone DL, Lee JH, Young SR, Dobrota E, Butler JS, Ruiz J, Skalnik DG 2005. Reduced genomic cytosine methylation and defective cellular differentiation in embryonic stem cells lacking CpG binding protein. Mol Cell Biol 25: 4881–4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A 2010. CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G 2010. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 8: e1000384 doi: 10.1371/journal.pbio.1000384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Shilatifard A 2010. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol 339: 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, et al. 2009. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460: 863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser S, Schaft J, Lubitz S, Vintersten K, van der Hoeven F, Tufteland KR, Aasland R, Anastassiadis K, Ang SL, Stewart AF 2006. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development 133: 1423–1432 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette B, Drogaris P, Lin HH, Armstrong H, Hiragami-Hamada K, Imhof A, Bonneil E, Thibault P, Verreault A, Festenstein RJ 2011. H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet 7: e1001354 doi: 10.1371/journal.pgen.1001354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP 2009. CpG islands–‘a rough guide.’ FEBS Lett 583: 1713–1720 [DOI] [PubMed] [Google Scholar]
- Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr AR, James KD, Turner DJ, Smith C, Harrison DJ, Andrews R, Bird AP 2010. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet 6: e1001134 doi: 10.1371/journal.pgen.1001134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG 2011. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell 144: 513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. 2010. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A 2002. COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem 277: 10753–10755 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol Cell 11: 721–729 [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242 doi: 10.1371/journal.pgen.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG 2005. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem 280: 41725–41731 [DOI] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG 2008. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol 28: 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Voo KS, Skalnik DG 2001. Identification and characterization of the DNA binding domain of CpG-binding protein. J Biol Chem 276: 44669–44676 [DOI] [PubMed] [Google Scholar]
- Lee JH, Tate CM, You JS, Skalnik DG 2007. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem 282: 13419–13428 [DOI] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A 2007. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131: 1084–1096 [DOI] [PubMed] [Google Scholar]
- Liu Z, Scannell DR, Eisen MB, Tjian R 2011. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell 146: 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE 2010. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet 6: e1001244 doi: 10.1371/journal.pgen.1001244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A 2001. COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci 98: 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, Florens L, Eissenberg JC, Shilatifard A 2011. The COMPASS family of H3K4 methylases in Drosophila. Mol Cell Biol 31: 4310–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML 2002. A Trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci 99: 90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Ong CT, Corces VG 2011. Enhancer function: New insights into the regulation of tissue-specific gene expression. Nat Rev Genet 12: 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG 2009. CTCF: Master weaver of the genome. Cell 137: 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J 20: 7137–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411 [DOI] [PubMed] [Google Scholar]
- Schneider J, Wood A, Lee JS, Schuster R, Dueker J, Maguire C, Swanson SK, Florens L, Washburn MP, Shilatifard A 2005. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell 19: 849–856 [DOI] [PubMed] [Google Scholar]
- Schnetz MP, Handoko L, Akhtar-Zaidi B, Bartels CF, Pereira CF, Fisher AG, Adams DJ, Flicek P, Crawford GE, Laframboise T, et al. 2010. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet 6: e1001023 doi: 10.1371/journal.pgen.1001023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, Chan SM, Martin DG, Fingerman IM, Briggs SD, et al. 2007. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem 282: 2450–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ III, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D 2007. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 28: 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A 2010. The chromatin signaling pathway: Diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell 40: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate CM, Lee JH, Skalnik DG 2009a. CXXC finger protein 1 contains redundant functional domains that support embryonic stem cell cytosine methylation, histone methylation, and differentiation. Mol Cell Biol 29: 3817–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate CM, Lee JH, Skalnik DG 2009b. CXXC finger protein 1 restricts the Setd1A histone H3K4 methyltransferase complex to euchromatin. FEBS J 277: 210–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzi N, Churchman LS, Vasiljeva L, Weissman J, Buratowski S 2011. H3K4 trimethylation by Set1 promotes efficient termination by the Nrd1–Nab3–Sen1 pathway. Mol Cell Biol 31: 3569–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, et al. 2010. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464: 1082–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk EL, Chen CL, d'Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Ne P, Loeillet S, et al. 2011. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 475: 114–117 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT 2007. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131: 58–69 [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, et al. 2009. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol 29: 6074–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A 2008. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol 28: 7337–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442: 86–90 [DOI] [PubMed] [Google Scholar]
- Young SR, Skalnik DG 2007. CXXC finger protein 1 is required for normal proliferation and differentiation of the PLB-985 myeloid cell line. DNA Cell Biol 26: 80–90 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W, et al. 2010. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res 38: 4246–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]