Abstract
The global transcriptional profile of peripheral blood mononuclear cells (PBMCs) stimulated with HIV candidate vaccine (virus-like particles, VLPs) has been evaluated in HIV-infected patients with low/high viral load compared to healthy volunteers. Baseline activation of chemokine production was observed in PBMC from HIV-infected patients and innate immune stimulation with HIV-VLPs was not blunted. The immune profile among HIV-infected patients was found to be qualitatively similar but quantitatively extremely variable. This diversity was independent of viral load and it might be dependent on individual immunogenetic traits or concurrent immunological status.
This ex vivo screening strategy represents an efficient tool for guiding modifications/optimizations of vaccination strategies and understanding failures in individuals enrolled in clinical trials.
Keywords: Immunogenomics, Vaccine, HIV-1, Peripheral blood mononuclear cell
1. Introduction
HIV-1 Pr55gag virus-like particles (VLPs) [1], [2], [3] and [4] have been extensively characterized by our group for the ability to induce maturation and activation of in vitro cultured monocyte-derived dendritic cells (MDDCs) partly through Toll-like receptor (TLR)-3 and -9 signaling [5]. We observed that the transcriptional profile of VLP-stimulated MDDCs indicates the reduced transcription of genes associated with phagocytosis and pathogen recognition and the activation of genes with function associated with antigen presentation and MDDCs migration [6]. Similar information can be observed also in CD14+ uncultured peripheral blood mononuclear cells (PBMCs) [7].
Recently, we have demonstrated that HIV-1 seropositivity status does not significantly impair the immune activation status and the responsiveness of circulating monocyte CD14+ cell populations to an immunogenic stimulus [8]. Moreover, Th-1 and Th-2 cytokine production was compared among healthy volunteers, HIV patients with low and high viral loads demonstrating higher basal levels of Th2 interleukin (IL)-6 [8].
To better dissect the potential differences in the response of circulating PBMCs from healthy volunteers as well as low and high viremia HIV-positive subjects to VLPs, in the present study we applied transcriptional profiling to three representative samples from each of the three categories and tested baseline transcriptional patterns as well as those after stimulation with VLPs or lipopolysaccharide (LPS) [9].
2. Methods
2.1. Cell culture medium
PBMC culture medium consisted of RPMI 1640 medium (Life Technologies, Carlsbad, CA) supplemented with 2 mM l-glutamine (Sigma), 1% nonessential amino acids (Life Technologies), 1% sodium pyruvate (Life Technologies), 50 µM 2-mercaptoethanol (Sigma), 50 µg of gentamicin (Life Technologies) per ml, and 10% fetal calf serum (Life Technologies).
2.2. Patient and donor cohorts, PBMC preparation and treatment
Human specimens were obtained under informed consent, as approved by the University of Maryland Baltimore Institutional Review Board. Fresh PBMCs were isolated by Ficoll-Hypaque density gradient centrifugation and plated in 6-well plates at a concentration of 1 × 107/well in 3 ml/well volume. PBMCs were stimulated with 6 µg/ml of HIV-VLPs or 1 µg/ml of LPS. Control PBMCs were added with PBS. The residual endotoxin activity possibly left over by the VLP preparation was inhibited by pre-incubation with Polymixin B Sulfate (Sigma) at a concentration of 10 µg/ml. After 4 and 8 h the cells were harvested and processed for total RNA extraction.
2.3. RNA preparation and microarray hybridization
PBMCs were harvested, total RNA and RNA for hybridization were obtained as previously described [10] and [11]. Six micrograms of amplified test samples aRNA were labeled with Cy5 (Amersham) while the same amount of reference sample (pooled normal donor PBMCs) was labeled with Cy3. Test-reference sample pairs were mixed and co-hybridized to a custom-made 37 K oligo-based microarray platform encompassing the whole human genome. Array data consistency was assessed based on the principle of reference concordance as previously described [12].
2.4. Microarrays and statistical analyses
Hybridized arrays were scanned at 10-µm resolution on a GenePix 4000 scanner (Axon Instruments). Resulting jpeg and data files were deposited at microarray database (mAdb) (http://nciarray.nci.nih.gov) and retrieved after median centered, filtering of intensity (>300) and spot elimination (bad and no signal). Data were further analyzed using the NCI/BRB software [13]. Hierarchical cluster analysis was conducted on according to Eisen et al. [14]; differential expressed genes were visualized by Treeview and displayed according to the central method of normalization [15].
Class comparison was performed using paired or unpaired Student’s t test as appropriate. Multivariate analysis was performed on multiple groups based on the F test with a P-value cutoff of 0.001. Assessment of random identification of significance was based on univariate and multivariate permutation test as previously described [16]. Boolean comparisons were based on the BRBArray Software. Fisher’s exact test or χ2-test was used to assess the significance of sample distribution among self-organizing classes of samples as appropriate.
3. Results
3.1. Clinical parameters of the subjects included in the analysis
Eleven subjects were enrolled in the study. Eight were HIV-1-seropositive, of whom four showed a low HIV-1 viremia (<2.6 log RNA copies/ml) and four a high viremia (>4.69 log RNA copies/ml) (SI Table 1). The first group was under ART therapy, the latter enrolled at their first visit and naïve for ART treatment. Three healthy seronegative volunteers were enrolled as controls [8].
3.2. LPS- and VLP-induced activation of PBMC in healthy volunteers
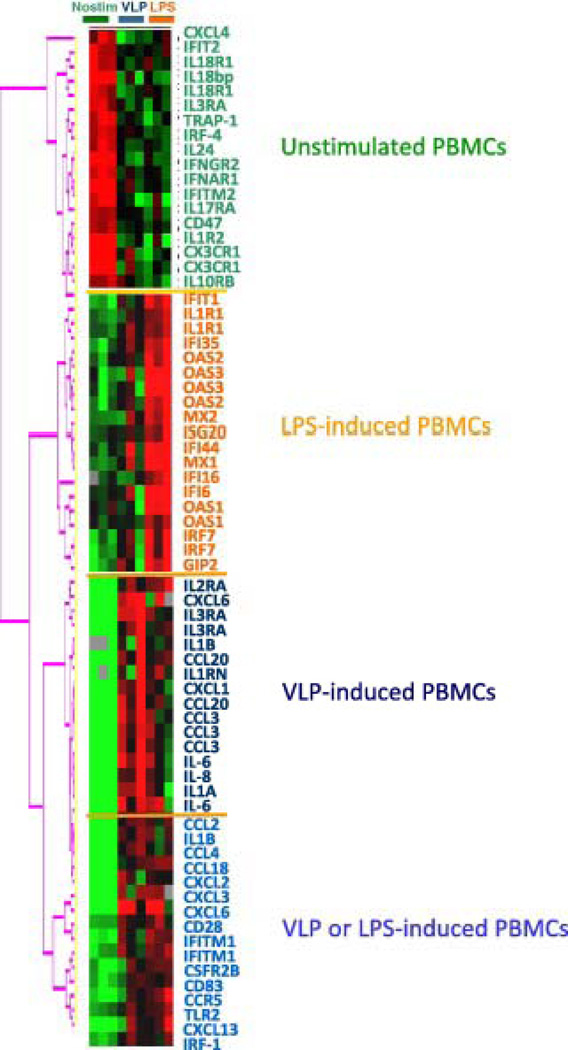
The effect of VLPs and LPS on the activation of PBMCs obtained from healthy volunteers identified 560 genes down-regulated and 596 up-regulated by either treatment (multivariate permutation test < 0.001), with a broad overlap among genes modulated by LPS and VLPs (Fig. 1).
Fig. 1.
Self-organizing heat map based on genes with immune annotation according to Gene Ontology, most differentially expressed in unstimulated PBMCs from normal volunteers compared to PBMCs stimulated with VLPs or LPS in the same cohort.
3.3. Baseline transcriptional patterns of PBMCs
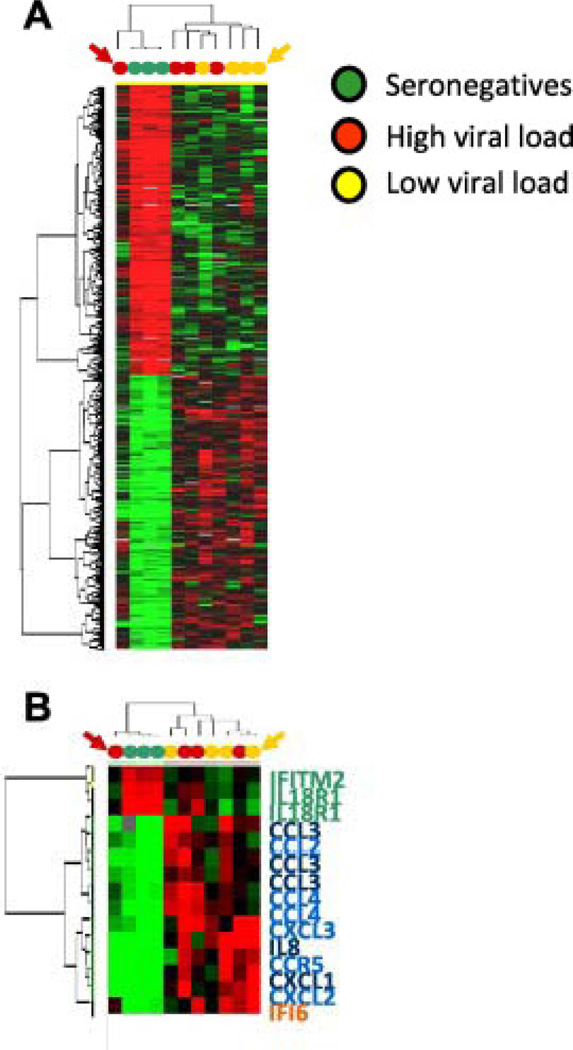
Nine hundred and seventy eight up-regulated genes and 960 down-regulated genes were found differentially expressed in unstimulated PBMCs from HIV-infected individuals compared to healthy volunteers. PBMCs of healthy volunteers demonstrated a highly consistent pattern while PBMCs from HIV-infected individuals displayed a heterogeneous transcriptional patterns and samples from low- and high-viremia group do not neatly segregate into two independent clusters (Fig. 2A). Patients 05 and 12 were excluded from this global analysis, considering that they have been found anergic according to functional analyses described in our recent paper [8]. However, patient 12 (a high-viremia sample) segregated in between experimental cohorts (red arrow, Fig. 2A), while patient 05 (a low-viremia sample) segregated with the other samples from low-viral load patients but displayed a relatively reduced induction of about 50% of HIV-specific genes (yellow arrow, Fig. 2A).
Fig. 2.
(A) Self-organizing heat map representing 1097 genes differentially expressed in PBMC. The red and yellow arrows points to subjects #12 and #5, respectively, who were found anergic to VLP-stimulation among the high and low-viremia patients. (B) Immune genes selected, among those identified according to the criteria in panel A, according the list in Fig. 1.
Unstimulated PBMCs from HIV-infected individuals largely reflected in baseline conditions the transcriptional profile of PBMCs from healthy volunteers stimulated ex vivo with VLPs. No transcripts specifically induced by LPS in vitro in healthy volunteers were found to be constitutively expressed in vivo by HIV-infected patients (Fig. 2B).
3.4. Transcription pattern of stimulated PBMCs
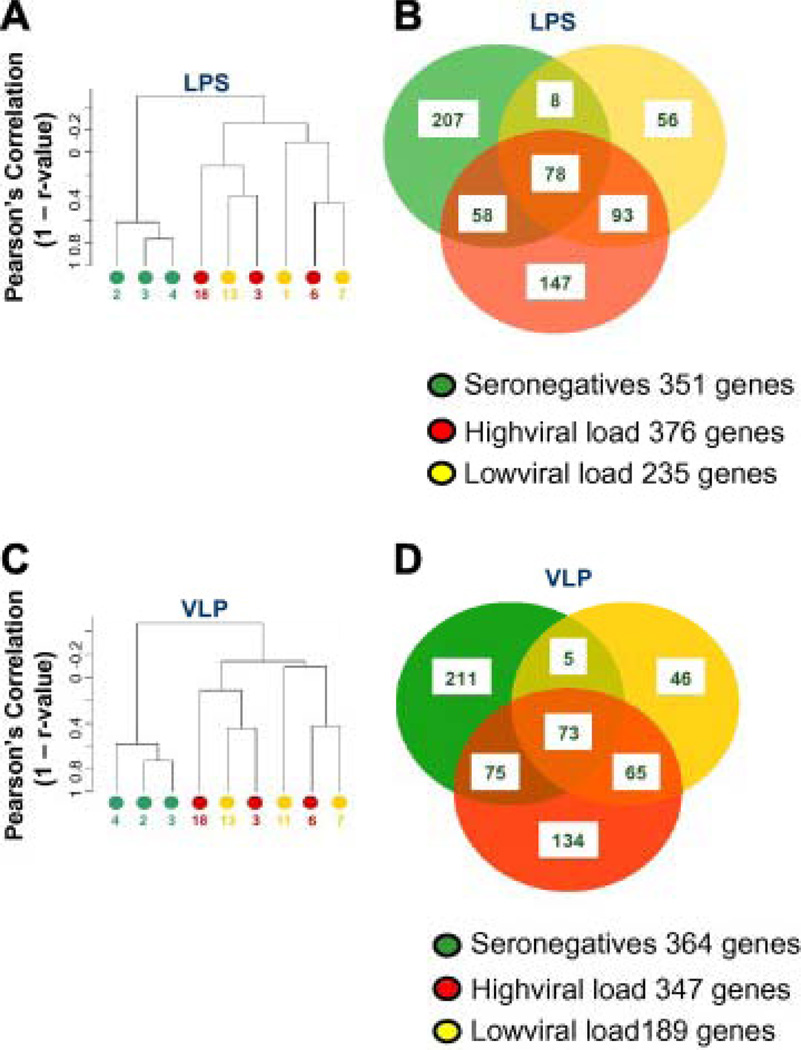
The transcriptional profile of PBMCs stimulated with LPS from healthy volunteers shows significant differences with those from HIV-infected patients, independent of viral load (Fig. 3A). Boolean comparison of genes differentially expressed in each cohort identified predominantly overlapping transcriptional patterns consistent among the three groups with the least overlap between PBMCs from healthy volunteers and those from HIV-infected, low-viral load patients (Fig. 3B) [17]. The ISGs modulated by LPS in the different experimental are shown in SI Table 2.
Fig. 3.
Unsupervised hierarchical clustering of PBMC samples stimulated with LPS (A) or VLPs (B). High stringent filter was applied resulting in the inclusion of 4,529 cDNA clones. Venn Diagram comparing the overlap of genes differentially expressed in PBMCs upon stimulation with LPS (B) or VLPs (D).
Similarly, a spontaneous segregation is observed between the transcriptional profile of PBMCs from healthy volunteers and HIV-infected individuals stimulated with VLPs (Fig. 3C). The transcriptional patterns of individuals with HIV shows a close overlap in correlative distance (Spearman’s correlation; 1 − R2 values) among patients stimulated with LPS (Fig. 3A) or VLPs (Fig. 3C). Boolean comparison of genes differentially expressed in response to VLPs identified overlapping transcriptional patterns among the three cohorts with the least overlap between PBMCs from normal individuals and those from HIV-infected, low-viral load patients (Fig. 3D).
In particular, considering specifically the ISG genes, a multiple dimensional scaling based on IFNa2b-induced 453 ISGs genes identified by our group (Pos et al., manuscript in preparation) clearly shows a distinct baseline pattern in unstimulated PBMCs from healthy seronegative and HIV-seropositive subjects as well as a distinct pattern in VLP-induced PBMCs from the three different groups (SI Fig. 1). The genes, including ISGs, modulated by VLPs in the different experimental are shown in SI Table 3.
3.5. Comparison of genes induced by LPS and VLPs
The expression of distinct ISGs genes was stimulated by LPS and VLPs, while the down regulation of some interferon regulated transcripts occurred in common with that induced by VLPs (see SI Table 4). On the contrary, the expression of several genes with immune function was significantly affected by LPS and VLP stimulation and are listed according to experimental cohorts (see SI Table 5).
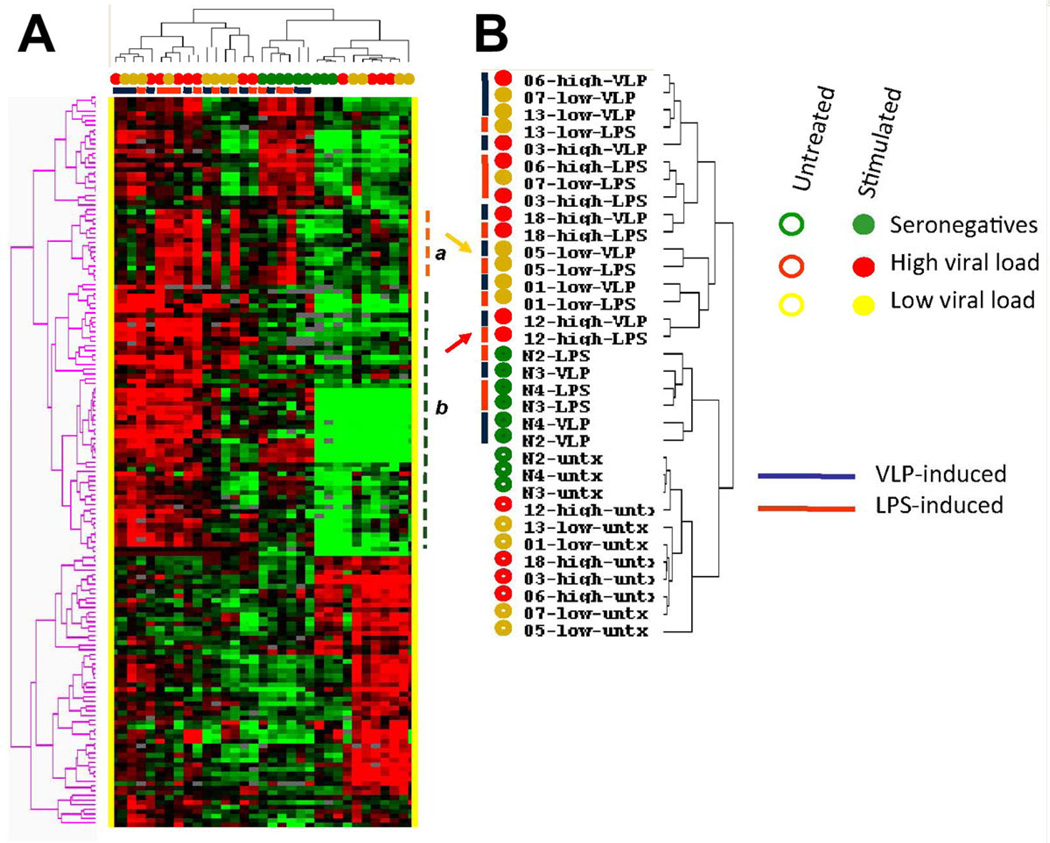
In addition, self-organizing maps based on immune genes differentially expressed upon LPS and VLP stimulation by the three groups identified a cluster of transcripts that included a specific sub-group of ISGs (Fig. 4Aorange dashed line a). Reshuffling of patients’ and donors’ sample based on this specific ISG cluster clearly predicted a class enriched of LPS-stimulated samples (10 of 11 samples) compared with VLP-stimulated samples (3 of 11 samples) or unstimulated samples (2 of 11 samples; χ2P-value < 0.001, SI Fig. 2).
Fig. 4.
(A) Self-organizing heat map based on 156 genes with immune annotations according to gene ontology modulated at high degree of significance by stimulation with either VLPs or LPS. The orange dashed line emphasizes the ISGs cluster (a); the blue dashed line emphasizes the lymphokine pattern (b). (B) The yellow and red arrows point to patient 05 and 12, respectively.
3.6. Patterns of gene expression
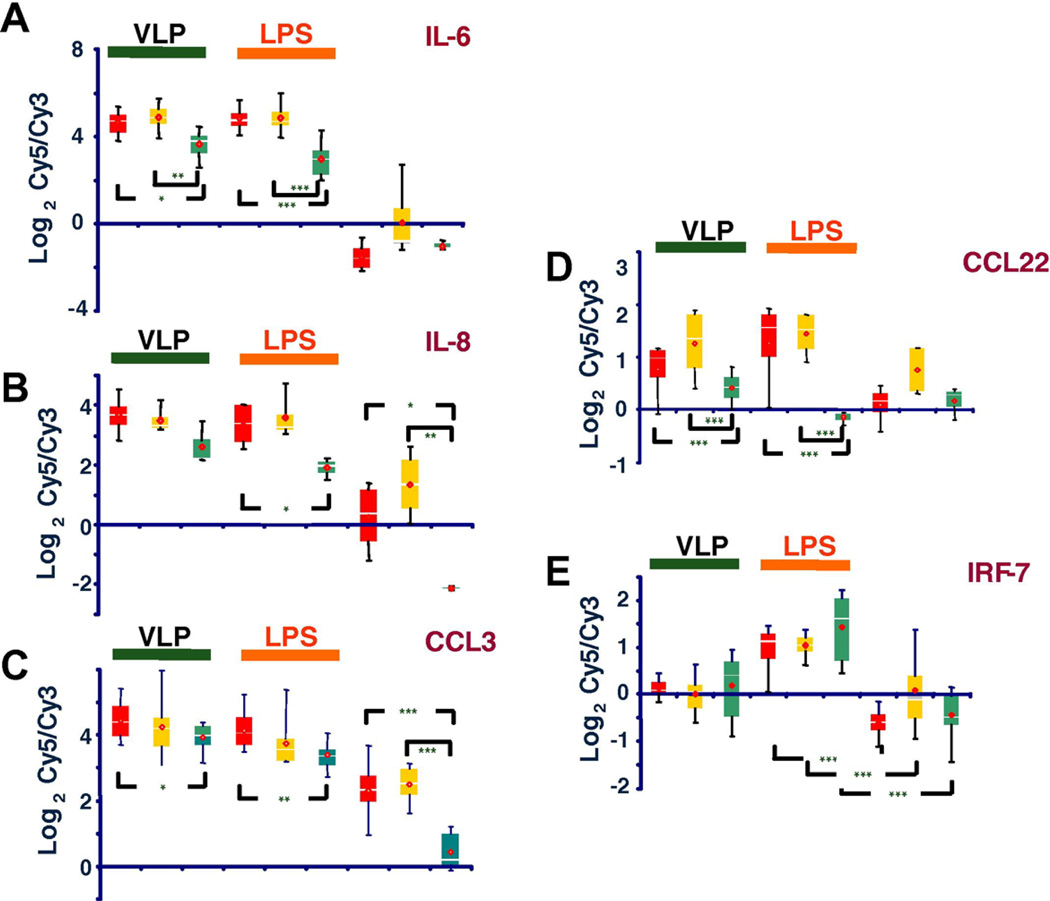
The expression of several genes with immune function was significantly affected by LPS and VLP and five distinct patterns of gene expression were identified. (1) Genes like IL-6 were not differentially expressed in baseline conditions among experimental cohorts, but their response to either VLP or LPS was strongly affected by HIV-status (Fig. 5A). (2) Genes like IL-8 that were very strongly differentially expressed between HIV patients (up-regulated) compared to healthy volunteers in baseline conditions and whose post-stimulation levels were partially affected by the HIV-status (Fig. 5B). (3) Genes like CCL3 whose expression was significantly higher in baseline conditions in HIV patients compared to healthy volunteers and experienced strong up-regulation in response to VLP, more than to LPS, in HIV-infected patients more than in healthy volunteers (Fig. 5C). (4) Genes like CCL22 whose expression was not altered by LPS or VLP stimulation in healthy volunteers but it was strongly induced by LPS and VLPs in HIV-infected patients independent of viral load (Fig. 5D). (5) Genes like IRF-7, whose expression was only induced by LPS and it was comparable among all experimental cohorts independent of HIV infection status (Fig. 5E).
Fig. 5.
(A–E) Relative Cy5/Cy3 values of IL-6, IL-8, CCL3, CCL22 and IRF-7 in different patient cohorts and type of stimulation. Data represent cumulative Cy5/Cy3 values for each experimental category. Red, yellow and green boxes indicate high–low-viral load and healthy normal controls, respectively. (*, **, *** = P-value <0.05, <01 and <0.001, respectively) [15].
3.7. Analysis of range of gene induction
We finally investigated the dynamic range of gene induction in response to VLP-stimulation independent of HIV infections status. This analysis identified 133 transcripts whose dynamic range in response to VLPs differed significantly between HIV-infected patients and healthy volunteers. A selection of lymphokines among these genes is shown in SI Fig. 3. Although all patients and donors experienced increases in the stimulated values of each cytokine, the dynamic range was quite different and was strongly affected by the HIV status independent of viral load. This was particularly evident for IL-1β and the IL-2R α-chain which were more strongly up-regulated in HIV-infected patients while the dynamic range was reversed for several lymphokines. In spite of this significance difference in Δ-VLP between healthy donors and HIV-infected individuals, the cumulative Cy5/Cy3 values in response to VLP stimulation were consistently higher in HIV-infected patients due to their higher baseline expression.
4. Discussion
The relevance of innate immunity in the natural history of HIV infection is becoming increasingly recognized [18], [19], [20], [21], [22] and [20]. To address the baseline profile of innate immunity and its modulation, we studied the ex vivo responsiveness of PBMCs to the HIV-VLP vaccine molecule and the lipopolysaccharide (LPS), as example of pathogen associated molecular pattern (PAMP), comparing HIV-infected individuals to normal, healthy volunteers.
According to previous observations [6], the transcriptional activation of PBMCs in baseline conditions or in response to stimulation with VLPs or LPS recapitulated that observed in MDDCs; this has important practical implications because ex vivo stimulation provides a simplified tool for the analysis of individual patient’s responsiveness to VLP that does not require extensive in vitro manipulation. This may be of particular importance considering the current lack of clinical immunological/biological correlates of responsiveness of HIV-infected individuals, which may be extremely relevant within the conduction of vaccination protocols. Transcriptional patterns of PBMC from HIV-infected patients clearly differed from those of healthy volunteers and signatures consistent with profound activation of chemokine production were observed. Interestingly, the RNA expression levels did not always correlate with those protein expression, reported in our recent paper [8], which are dependent also on external metabolic pathways affecting either their half life or their compartmentalized production.
Unexpectedly, in spite of extensive heterogeneity among individual patients, several consistent immunological patterns could be identified distinguishing HIV-infected individuals from healthy volunteers that may shed important insights about the immune biology of HIV infection.
An unexpected observation was the finding that the IFN-independent innate immune response of HIV patients to LPS and VLPs is not hampered but rather enhanced. In particular, stimulation of PBMCs from HIV-infected patients with VLPs further enhanced the innate immune response already ongoing in HIV-infected individuals. A distinct pattern of ISGs induced by VLPs and LPS in PBMCs from HIV seronegative as well as seropositive subjects may suggest the need to combine the VLPs with adjuvanting “LPS-like” molecules for an optimal transient induction of the innate immunity and, downstream, of the adaptive anti-HIV-1 responses aiming to an effective viral clearance.
Furthermore, the immune profile among HIV-infected patients was found to be qualitatively similar but quantitatively extremely variable and this diversity was independent of viral load suggesting that such differences may be more likely related either to immunogenetic traits, as recently discussed by Singh et al. [23], or to concurrent immunological status [24]. This striking variation should be taken into account when interpreting correlative studies related to the natural history of the disease or its responsiveness to therapy.
In summary, this study indicates that this screening approach is able to identify specific genomic signatures induced by two distinct immune activators (HIV-VLPs and LPS) and provides a road map for the study of HIV-infected patients based on HIV-specific signatures, their heterogeneity among patients, their response to exogenous LPS and VLPs.
Supplementary Material
Fig. S1. Multiple dimensional scaling plot representing gene expression profile of 453 ISGs (interferon-stimulated genes) in PBMCs as affected by HIV infection, HIV viral load and VLP treatment.
View thumbnail images
View high quality image (78K)
Fig. S2. (A) Self-organizing heat map based on genes with immune annotations included in cluster a (orange dashed line, ISGs cluster) of SI Fig 1. (B) Self-organizing heat map based on genes with lymphokine annotations included in cluster (b) of SI Fig 1.
Fig. S3. Dynamic range of expression in response to VLPs stimulation (D-VLPs) of distinct lymphokines in healthy donors and HIV-infected individuals.
Acknowledgements
This study was supported by grants from the Ministero Italiano Università e Ricerca (MIUR, 2004), the Ministero Italiano della Sanità (Ricerca Corrente and Progetto Finalizzato AIDS 2006), and the Institute of Human Virology and the NIH (NIH grants to G.K.L.).
References
- 1.Buonaguro L, Buonaguro FM, Tornesello ML, Mantas D, Beth-Giraldo E, Wagner R, Michelsonr S, Prevost M-C, Wolf H, Giraldo G. High efficient production of Pr55gag Virus-like Particles expressing multiple HIV-1 epitopes, including a gp120 protein derived from an Ugandan HIV-1 isolate of subtype A. Antiviral Res. 2001;49:35–47. doi: 10.1016/s0166-3542(00)00136-4. [DOI] [PubMed] [Google Scholar]
- 2.Buonaguro L, Racioppi L, Tornesello ML, Arra C, Visciano ML, Biryahwaho B, Sempala SDK, Giraldo G, Buonaguro FM. Induction of neutralizing antibodies and CTLs in Balb/c mice immunized with Virus-like Particles presenting a gp120 molecule from a HIV-1 isolate of clade A (HIV-VLPAs) Antiviral Res. 2002;54:189–201. doi: 10.1016/s0166-3542(02)00004-9. [DOI] [PubMed] [Google Scholar]
- 3.Buonaguro L, Visciano ML, Tornesello ML, Tagliamonte M, Biryahwaho B, Buonaguro FM. Induction of systemic and mucosal cross-clade neutralizing antibodies in BALB/c mice immunized with human immunodeficiency virus type 1 clade A virus-like particles administered by different routes of inoculation. J. Virol. 2005;79:7059–7067. doi: 10.1128/JVI.79.11.7059-7067.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buonaguro L, Devito C, Tornesello ML, Schroder U, Wahren B, Hinkula J, Buonaguro FM. DNA-VLP prime-boost intra-nasal immunization induces cellular and humoral anti-HIV-1 systemic and mucosal immunity with cross-clade neutralizing activity. Vaccine. 2007;25:5968–5977. doi: 10.1016/j.vaccine.2007.05.052. [DOI] [PubMed] [Google Scholar]
- 5.Buonaguro L, Tornesello ML, Tagliamonte M, Gallo RC, Wang LX, Kamin-Lewis R, Abdelwahab S, Lewis GK, Buonaguro FM. Baculovirus-derivedhumanimmunodeficiencyvirustype1virus-likeparticlesactivatedendriticcellsandinduceexvivoT-cellresponses. JVirol. 2006;80:9134–9143. doi: 10.1128/JVI.00050-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aricò E, Wang E, Tornesello ML, Tagliamonte M, Lewis GK, Marincola FM, Buonaguro FM, Buonaguro L. Immature monocyte derived dendritic cells gene expression profile in response to Virus-Like Particles stimulation. J. Transl. Med. 2005;3:45. doi: 10.1186/1479-5876-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buonaguro L, Monaco A, Arico E, Wang E, Tornesello ML, Lewis GK, Marincola FM, Buonaguro FM. Gene expression profile of peripheral blood mononuclear cells in response to HIV-VLPs stimulation. BMC Bioinformatics. 2008;9(Suppl. 2):S5. doi: 10.1186/1471-2105-9-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buonaguro L, Tornesello ML, Gallo RC, Marincola FM, Lewis GK, Buonaguro FM. Th2 polarization in peripheral blood mononuclear cells from human immunodeficiency virus (HIV) infected subjects, as activated by HIV Virus-Like Particles. J. Virol. 2009;83:304–313. doi: 10.1128/JVI.01606-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagorsen D, Deola S, Smith K, Wang E, Monsurro V, Zanovello P, Marincola FM, Panelli MC. Polarized monocyte response to cytokine stimulation. Genome Biol. 2005;6:R15. doi: 10.1186/gb-2005-6-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang E, Miller LD, Ohnmacht GA, Liu ET, Marincola FM. High-fidelitymRNAamplificationforgeneprofiling. Nat. Biotechnol. 2000;18:457–459. doi: 10.1038/74546. [DOI] [PubMed] [Google Scholar]
- 11.Wang E. RNA amplification for successful gene profiling analysis. J. Transl. Med. 2005;3:28. doi: 10.1186/1479-5876-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin P, Zhao Y, Ngalame Y, Panelli MC, Nagorsen D, Monsurro V, Smith K, Hu N, Su H, Taylor PR, Marincola FM, Wang E. Selection and validation of endogenous reference genes using a high throughput approach. BMC Genomics. 2004;5:55. doi: 10.1186/1471-2164-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu X, Zhao Y, Simon R. Gene set expression comparison kit for BRB-ArrayTools. Bioinformatics. 2008;24:137–139. doi: 10.1093/bioinformatics/btm541. [DOI] [PubMed] [Google Scholar]
- 14.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman PT, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO. Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genetics. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 16.Wang E, Miller LD, Ohnmacht GA, Mocellin S, Perez-Diez A, Petersen D, Zhao Y, Simon R, Powell JI, Alexander H, Duray PH, Herlyn M, Restifo NP, Liu ET, Rosenberg SA, Marincola FM. Prospective moleculare profiling of melanoma metastases suggests classifiers of immune responsiveness. Cancer Res. 2005;62:3581–3586. [PMC free article] [PubMed] [Google Scholar]
- 17.Jin P, Wang E, Provenzano M, Deola S, Selleri S, Ren J, Voiculescu S, Stroncek D, Panelli MC, Marincola FM. Molecular signatures induced by interleukin-2 on peripheral blood mononuclear cells and T cell subsets. J. Transl. Med. 2006;4:26. doi: 10.1186/1479-5876-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doherty PC, Turner SJ. The challenge of viral immunity. Immunity. 2007;27:363–365. doi: 10.1016/j.immuni.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iqbal SM, Kaul R. Mucosal innate immunity as a determinant of HIV susceptibility. Am. J. Reprod. Immunol. 2008;59:44–54. doi: 10.1111/j.1600-0897.2007.00563.x. [DOI] [PubMed] [Google Scholar]
- 20.Lehner T, Wang Y, Pido-Lopez J, Whittall T, Bergmeier LA, Babaahmady K. The emerging role of innate immunity in protection against HIV-1 infection. Vaccine. 2008;26:2997–3001. doi: 10.1016/j.vaccine.2007.11.060. [DOI] [PubMed] [Google Scholar]
- 21.Lobo PI, Schlegel KH, Yuan W, Townsend GC, White JA. Inhibition of HIV-1 infectivity through an innate mechanism involving naturally occurring IgM anti-leukocyte autoantibodies. J. Immunol. 2008;180:1769–1779. doi: 10.4049/jimmunol.180.3.1769. [DOI] [PubMed] [Google Scholar]
- 22.Shattock RJ, Haynes BF, Pulendran B, Flores J, Esparza J. Improving defences at the portal of HIV entry: mucosal and innate immunity. PLoS Med. 2008;5:e81. doi: 10.1371/journal.pmed.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh P, Kaur G, Sharma G, Mehra NK. Immunogenetic basis of HIV-1 infection, transmission and disease progression. Vaccine. 2008;26:2966–2980. doi: 10.1016/j.vaccine.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 24.Borkow G, Bentwich Z. HIV and helminth co-infection: is deworming necessary? Parasite Immunol. 2006;28:605–612. doi: 10.1111/j.1365-3024.2006.00918.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Multiple dimensional scaling plot representing gene expression profile of 453 ISGs (interferon-stimulated genes) in PBMCs as affected by HIV infection, HIV viral load and VLP treatment.
View thumbnail images
View high quality image (78K)
Fig. S2. (A) Self-organizing heat map based on genes with immune annotations included in cluster a (orange dashed line, ISGs cluster) of SI Fig 1. (B) Self-organizing heat map based on genes with lymphokine annotations included in cluster (b) of SI Fig 1.
Fig. S3. Dynamic range of expression in response to VLPs stimulation (D-VLPs) of distinct lymphokines in healthy donors and HIV-infected individuals.