Abstract
Huntington’s Disease (HD) is a fatally inherited neurodegenerative disorder caused by an expanded glutamine repeat in the N-terminal region of the huntingtin (HTT) protein. The result is a progressively worsening triad of cognitive, emotional, and motor alterations that typically begin in adulthood and end in death 10-20 years later. Autopsy of HD patients indicates massive cell loss in the striatum and its main source of input, the cerebral cortex. Further studies of HD patients and transgenic animal models of HD indicate that corticostriatal neuronal processing is altered long before neuronal death takes place. In fact, altered neuronal function appears to be the primary driver of the HD behavioral phenotype, and dysregulation of glutamate, the excitatory amino acid released by corticostriatal afferents, is believed to play a critical role. Although mutant HTT interferes with the operation of multiple proteins related to glutamate transmission, consistent evidence links the expression of mutant HTT with reduced activity of glutamate transporter 1 (rodent GLT1 or human EAAT2), the astrocytic protein responsible for the bulk of glutamate uptake. Here, we review corticostriatal dysfunction in HD and focus on GLT1 and its expression in astrocytes as a possible therapeutic target.
Keywords: ascorbate, astrocytes, electrophysiology, glutamate transport, huntingtin, oxidative stress, transgenic models
Huntington’s disease (HD) is an autosomal dominant, neurodegenerative disorder that afflicts approximately 1 in 10,000 individuals of European descent but with a slightly lower incidence among other populations [1]. The underlying cause is a mutation in the huntingtin gene (htt, initially termed “IT15”), located in the 4p16.3 region of the short arm of chromosome 4 [2,3]. Although htt contains 67 exons, most research has focused on the first exon because it is the site of the HD mutation [4]. Normally, this exon contains between 3 and 30 repeats of the nucleotide triplet CAG, which is translated into a poly-glutamine peptide segment (poly-Q). The final product is a protein of 3,144 amino acids, with a poly-Q region of variable length at the N-terminal site. In HD, the number of CAG repeats often exceeds 35 with full penetrance occurring above 39 [2]. Generally, as the number of CAG repeats increases, the age of onset decreases. Individuals carrying 40 CAG repeats, for example, are likely to show the first signs of HD at 35-40 years of age, but two or three fewer repeats may delay onset by 20 or more years. Interestingly, a childhood or juvenile variant of HD emerges at more than 50 CAG repeats. In this case, transmission is predominantly paternal with maternal transmission accounting for only ~ 20% of juvenile HD cases [5, 6]. In fact, a progressive increase in CAG repeats across generations, a phenomenon known as anticipation, appears to represent an effect of HD on spermatogenesis [7]. But CAG repeat length is not the sole determinant of age of onset since environmental factors and genetic variations are also involved [8-13].
The physiological role of the huntingtin protein (HTT) is complex owing to its multiple functions, including vesicular transport, modulation of the synthesis and release of brain derived neurotrophic factor (BDNF), as well as transcription factor activity [for review see [14]. HTT is ubiquitously expressed throughout the body, with no difference in expression between peripheral tissues and the central nervous system [15,16]. The most prominent feature of HD, however, is brain degeneration, particularly the caudate nucleus-putamen (striatum) and cerebral cortex (see below).
Juvenile and Adult Huntington’s Disease
Juvenile HD represents <10% of all HD cases and occurs when mutant HTT contains more than 50 glutamine repeats. The first signs are observed before age 20. Carried to an extreme, a very high number of CAG repeats (210-250) has a reported age of onset of 18 months, but such cases are rare [17,18]. Psychiatric disturbances and behavioral problems precede the development of motor alterations, which are mainly manifest as bradykinesia and rigidity, but may also include dysarthria, hyperreflexia and occulomotor disturbances. Juvenile HD patients also show a progressive decline in cognitive abilities and eventually dementia [19,20]. Epileptic seizures are common. Thus, the overall picture of juvenile HD is an accelerated mental and physical deterioration that leads to death between 7-10 years after symptom onset. In contrast, adult HD follows three well-defined phenotypic stages with an approximate duration of 10-20 years, with a typical onset at middle age (~ 40 years old) [20]. In the first stage, HD patients are likely to show depression, mood alterations, behavioral abnormalities, and changes in personality. Mild cognitive and motor abnormalities also begin to emerge and then worsen over time [21]. Cognitive impairments first appear in relation to complex tasks, but eventually include loss of memory, language disturbances, and finally, dementia. During the second or hyperkinetic stage, motor abnormalities, commonly known as “chorea,” become the defining feature of HD and are manifest as exaggerations of gesture and abrupt, spontaneous movements of the trunk, face, and limbs. Choreic movements cannot be voluntarily suppressed and worsen during stress. Skilled movements -- such as gait, speech, and swallowing -- deteriorate; even eating declines, as HD patients show a marked loss of body weight. Approximately 10 years after HD onset, the last or akinetic stage emerges, in which chorea is replaced by progressively worsening bouts of bradykinesia and rigidity. Patients lose the ability to take care of themselves, and their overall health deteriorates [22]. Pneumonia and heart disease become the most common cause of death.
HD brains show a 10-20% reduction in weight compared with age-matched controls. The most striking feature is gross atrophy of the striatum (caudate nucleus and putamen). Neuropathological analysis of HD postmortem brains indicates five grades of damage [23]. In grade 0, no detectable histological neuropathology is observed. In Grade 1, neuropathological changes can be detected microscopically with as much as 50% depletion of striatal neurons. In Grades 2-3, striatal neuronal depletion, marked neuronal atrophy and gliosis are progressively observed. In Grade 4, the last and most severe grade, 90% of striatal neurons are lost. In addition to striatal damage, HD brains show atrophy in cerebral cortex. HD postmortem brains show selective and progressive loss of pyramidal neurons in cortical layers III, V and VI as these layers also show reduced thickness [24]. These alterations are observed broadly across the cerebral cortex, including frontal and parietal areas [24-26].
Neuropathological studies also revealed the presence of intracellular aggregates in HD brains. HTT is cleaved by different proteases, and in HD, cleavage of mutant HTT results in the formation of N-terminal fragments that aggregate and also recruit other proteins, which may disrupt crucial cellular functions. These aggregates are heterogeneously distributed but occur prominently in striatum and cerebral cortex [27].
Huntington’s Disease Transgenic Models
Soon after the HD gene was identified, transgenic models were developed in rodents to identify the mechanisms underlying HD progression. Two broad categories of transgenic models are available: truncated models, created by insertion of the first exon of the human mutant htt gene, and full-length models, in which the entire human mutant htt gene is expressed. In both cases, however, the models also express the endogenous htt alleles. Thus, the ratio of mutant HTT to HTT is not the same as in human HD patients, raising the possibility that some cellular alterations in the models may not be an entirely accurate reflection of human HD. In this regard, the creation of knock-in models in which the CAG repeats were inserted into the first exon of the endogenous Hdh gene (the murine analog of htt) may be more accurate because expression of mutant HTT occurs under the control of endogenous regulatory elements [28].
The first transgenic HD model emerged from the R6 line, which was created by the insertion of the first exon of human mutant htt carrying either 113 or 156 CAG repeats in R6/1 and R6/2 mice, respectively. R6 mice develop an aggressive phenotype, which includes the presence of tremor, reduction in body weight, and susceptibility to epileptic seizures; this phenotype typically survives for only 3-5 (R6/2) or 10-14 months (R6/1) [29]. Because the R6 line expresses a robust HD behavioral phenotype that develops rapidly, R6 mice have been widely used to study the cellular alterations associated with mutant HTT expression. R6/2 brains, for example, show striatal and cortical atrophy along with the presence of intracellular and intranuclear aggregates containing the mutant HTT toxic fragment and several proteins [30]. Other truncated models include the N171-82Q mouse and the transgenic rat model [31,32]. The N171-82Q mouse expresses cDNA encoding 171 amino acids of the first exon of htt with 82 CAG repeats. This mouse develops multiple behavioral abnormalities including loss of coordination, tremor, abnormal gait, and, like the R6 line, premature death. Intracellular and neuritic aggregates are observed in the brain [31]. The transgenic rat model carries the mutant htt fragment with 51 CAG repeats and also shows intracellular aggregates. But unlike the mouse truncated models, the transgenic rat exhibits an adult-onset neurological phenotype that includes slowly progressive motor dysfunction [32].
Two full-length HD transgenic models have been developed: the yeast artificial chromosome (YAC) and the bacterial artificial chromosome (BAC) models. Several varieties of the YAC model have been created based on the number of CAG repeats: YAC46, YAC72 and YAC128. In general, all YAC models develop a slowly progressive behavioral phenotype that varies with the number of CAG repeats, and show progressive changes in motor control and cognitive dysfunction along with striatal atrophy and intracellular and nuclear aggregates [33]. The BAC model, with 97 CAG repeats, exhibits progressive motor deficits, cortical and striatal atrophy and also the presence of mutant HTT aggregation [34,35]. Multiple variants of the knock-in model also have been developed: Hdh/Q72-80 (expressing 72-80 CAG repeats), Hdh Q111 (expressing 111 CAG), and the CAG140 and CAG150 models [for review see [28]. These models develop a slowly progressing phenotype, which includes motor alterations, the presence of intracellular aggregates, and striatal atrophy [28].
Despite the wide range of models and their underlying genetic basis, they all develop phenotypic and neuropathologic alterations similar to those seen in HD patients, including the presence of intracellular and intranuclear mutant HTT aggregates and striatal and cortical atrophy. Likewise, transgenic models also resemble the inverse correlation between the number of CAG repeats and the severity of the disease. In fact, R6/2 mice which carry the mutant htt with the highest number of CAG repeats (150) develop an aggressive HD-like phenotype and premature death, which has been suggested to resemble juvenile HD.
Interestingly, all transgenic models show only a small percentage of neuronal death, which is largely confined to the late stages of HD progression [28-35]. It is likely, therefore, that the HD behavioral phenotype emerges from altered neuronal function, which occurs long before neuronal death (see below).
Overview of the corticostriatal pathway
The cerebral cortex sends glutamatergic projections to the striatum, the main information processing unit of the basal ganglia. Corticostriatal projections are massive and broad, arising from all cortical regions [36-38]. Pyramidal neurons in layer V provide the bulk of cortical input; their axons terminate primarily on the spines of medium (soma size of 15-20 μm) spiny neurons (MSNs), which account for roughly 95% of the striatal neuronal population; the remainder includes a wide variety of interneurons, which modulate MSN activity [36]. The striatum also receives glutamatergic projections from thalamus and dopaminergic input from substantia nigra pars compacta [36]. MSNs release gamma-aminobutyric acid (GABA) and give rise to the so-called direct and indirect striatal output systems. The direct system projects to the internal segment of globus pallidus (GPi) and the substantia nigra pars reticulata (SNr), both of which comprise the output structures of the basal ganglia and project to select targets in brainstem and thalamus. The indirect system also targets the GPi and SNr but through synaptic connections in the external segment of the globus pallidus (GPe) and the subtalamic nucleus (STN). The direct and indirect systems also can be distinguished on the basis of the neuropeptides and the type of dopamine receptor expressed by MSN populations. MSNs that form the direct pathway contain substance P and dynorphin and preferentially express D1-type dopamine receptors, whereas enkephalin and D2-type dopamine receptors are expressed in MSNs projecting to GPe [37].
The canonical model of basal ganglia function posits that the direct and indirect pathways act in opposite and parallel ways to adjust the magnitude of GPi/SNr output in order to inhibit or release movement [38]. According to this model, activation of the direct pathway leads to disinhibition of the thalamocortical projection and facilitation of motor activity, whereas activation of the indirect pathway results in a net inhibition of thalamocortical projections and attenuates movement [38]. Unbalanced activity in these pathways has been invoked to explain the motor abnormalities associated with basal ganglia damage such as bradykinesia in Parkinson’s disease or choreic movements in HD. In an admittedly oversimplified view, an over-active indirect pathway would lead to bradykinesia, whereas choreic movements would result from an over-active direct pathway [38,39]. Interestingly, although both pathways eventually degenerate in HD, MSNs that contribute to the indirect pathway appear to be affected first (see below) [40].
Four major subtypes of striatal interneurons have been identified: (1) large aspiny neurons (soma size 20-50 m), which use acetylcholine as a neurotransmitter, (2) medium-sized GABA/parvalbumin interneurons (soma size 10-25 m), (3) GABA/calretinin interneurons (soma size 7-20 m), and (4) somatostatin/neuropeptide Y/nitric oxide synthase neurons (soma size 12-35 m), that possibly use GABA as well [for review see, 41]. Although interneurons constitute a small percentage of the striatal neuronal population, they branch extensively within the striatum to exert strong control over the excitability and spike activity of MSNs [for review see, 42]. While GABA-containing interneurons produce a strong inhibition thought to influence the timing of neuronal firing in either individual or ensembles of MSNs [for review see [43,44], cholinergic interneurons exert neuromodulatory effects. For example, it has been shown that nitric oxide activates cholinergic interneurons, which in turn modify MSN activity associated with neuronal plasticity [45,46]. Thus, interneurons can exert powerful control over striatal output, and their dysfunction also may contribute to movement disorders. In fact, selective inhibition of fast-spiking interneurons induces dyskinesia [47].
Glial cells also constitute a major part of the striatal cellular population. A ratio of 3.35 glial cells per neuron has been described for the human striatum [48]. A much higher ratio has been reported for rat striatum, though it is unclear if this difference has functional implications [49]. Initially, astrocytes were thought to equilibrate the extracellular ionic environment after neuronal activity, but recent data indicate that astrocytes participate in many essential mechanisms. During glutamate transmission, astrocytes contribute to the de novo synthesis of glutamine, a glutamate precursor [50]. There also is evidence of energetic coupling between neurons and astrocytes; astrocytes are the only cell type in the brain for storage of glycogen, an energy precursor that can support high levels of neuronal activity [51]. Moreover, astrocytes supply the precursor for glutathione, an antioxidant synthesized by neurons [51]. In fact, growing evidence suggests that glial mechanisms are critically involved in the neuropathology of HD (see below).
Corticostriatal pathway in Huntington’s Disease
Electroencephalogram (EEG) studies of HD patients provided the first evidence that expression of mutant HTT leads to altered brain activity [52]. Specifically, a reduction of alpha (8-12.5 Hz) and increase of theta (4-7.5 Hz) and delta rhythms (1.5-3.5 Hz) have been described [53-55]. It is unclear, however, whether the abnormal EEG activity correlates with the severity of the neurological impartment. Although De Tommaso et al. [53] did not find an association between EEG activity and the severity of the HD phenotype, Bylsma et al. [54] reported a significant correlation. Similarly, Streletz et al. [56] reported a correlation between increased theta and reduced alpha power and signs of dementia. More recently, EEG low-resolution brain electromagnetic (LORETA) tomography shows that decreased power in theta and alpha-2 (10-12 Hz) frequency bands correlates with worsening of motor symptoms. Also, low power in the alpha-1 (8-10 Hz) frequency band correlates with a decline in mental performance. Interestingly, LORETA tomography indicates that altered EEG patterns appear in patients at early stages of HD [57]. In HD transgenic models, abnormal EEG also has been reported in R6/2 mice at 14 weeks of age [58]. However, detailed studies evaluating the relationship between EEG parameters and HD phenotype progression have not been carried out on HD transgenic models.
Intra-cellular aggregates of mutant HTT are found in both the cytoplasm and nucleus [59]. In neurons, mutant HTT aggregates are also located in axon terminals, where the presence of aggregates has been correlated with decreased density of synaptic vesicles and decreased release of glutamate in striatum of HD mice [60-62]. Altered glutamate release at corticostriatal synapses has been observed in slice preparations, which show that paired-pulse facilitation induces a greater response in HD than control animals [63-66]. In vivo experiments also indicate altered corticostriatal function. In a microdialysis study, R6/2 mice showed decreased glutamate release in response to depolarization of striatal axon terminals [67]. Collectively, these results indicate that mutant HTT alters the function of the corticostriatal projection.
Neuronal dysfunction
In vitro electrophysiological recordings of cortical pyramidal neurons in R6/2, YAC128, and knock-in HD transgenic models show altered membrane properties and a shift in the balance of excitatory and inhibitory inputs, leading to a hyperexcitable cortex [68]. Likewise, cortical neurons from BACHD mice show decreased pyramidal inhibition, suggesting increased cortical excitability [35,69]. Besides altering the activity of cortical pyramidal neurons in this model, mutant HTT also alters the activity of cortical parvalbumin-containing interneurons, which may explain the loss of synchronized firing in cortical pyramidal neurons observed in HD (see below) [35]. Cortical cultured neurons expressing the first 171 amino acids of mutant HTT grown on microelectrode arrays show altered network-wide spiking activity, as evidenced by decreases in the number of spikes, population bursts, and interburst intervals [70]. The decline in burst firing is especially interesting because of the importance of spike bursts for information transmission and synaptic plasticity [71,72]. Altered cortical processing in HD also has been described in vivo. Electrophysiological recordings of cortical neurons in freely behaving R6/2 mice show reduced bursting and a decrease in the number of spikes that participate in a burst [73]. These changes in cortical activity were observed early (7-9 weeks) in the disease process. Interestingly, although the 140CAG knock-in model did not show a change in the burst properties of individual neurons, simultaneously recorded neuron pairs showed significantly less correlated firing, including fewer coincident bursts. A decline in correlated firing also has been reported for R6/2 mice [73]. Thus, a loss of correlated or synchronous firing between cortical neurons appears to be a common feature of HD. This is a key point because cooperative interactions among functionally related neurons, which are often manifest as synchronous oscillatory activity, shape behavioral output [74-76]. In fact, dysfunction of prelimbic neurons, which comprise a subregion of prefrontal cortex, is observed in R6/2 mice during extinction of fear conditioning [77].
Altered electrophysiological properties also have been reported for MSNs. Intracellular recordings from R6/2 and knock-in MSNs indicate a depolarized resting membrane potential [78] and enhanced sensitivity of the N-methyl–D-aspartate (NMDA) glutamate receptor relative to wild-type [66,79]. Thus, HD appears to increase MSN excitability [33,64,80]. In accord with this view, MSNs recorded from R6/2s require lower current intensity to reach an action potential than age-matched controls [66]. Recordings from behaving, symptomatic HD models indicate increased MSN activity in both R6 lines, but not in 140CAG knock-ins, which express a relatively mild behavioral phenotype relative to the R6s [81-83]. Burst activity, which corresponds to periods of high-frequency firing, is decreased in all three models [82,83]. Moreover, reduced correlated firing and coincident bursting between pairs of simultaneously recorded MSNs is a consistent finding across mouse models as well the behaving transgenic HD rat [83,84]. Thus, as in cortex, disrupted MSN processing occurs across multiple HD models [81,85]. Furthermore, altered firing properties in cortex and striatum are associated with learning deficits in pre-symptomatic R6/1s during a procedural learning task [86]. Similarly, electrophysiological changes are observed before the appearance of pronounced HD motor signs and during cognitive tasks in the transgenic HD rat [87].
Recently, electrophysiological evaluation of striatal output pathways show an agedependent imbalanced activity in the neurons of the direct and indirect pathway in HD models. In YAC128 mice, at early stages of the disease (1.5 months), D1-positive neurons show increased excitatory and inhibitory neurotransmission, while no changes were observed in D2-positive cells. At 12 months, D1- and D2- positive cells show decrease excitatory neurotransmission, while D2-positive cells show increased inhibitory neurotransmission as compared with wild-type cells. Similar alterations were described in BACHD transgenic models. Together, these data suggest that imbalanced glutamatergic and dopaminergic input may underlie the progression of the HD behavioral phenotype [88-91].
Network dysfunction
Neurons tend to operate in a coordinated way within a given network. Motor episodes are driven by oscillations in large populations of striatal neurons recorded as local field potentials (LFPs) [92]. Spectral analysis of LFPs recorded from the globus pallidus of a HD patient revealed an increase in power in the theta/alpha (4-12 Hz) and low gamma (35-45 Hz) bands [93]. Likewise, dysregulated LFP oscillations in theta (7-14 Hz) and gamma (30-40 Hz) activity have been observed in the striatum of R6/2 mice when they engage in the same open-field behaviors (e.g., grooming, locomotion) as wild-type [83]. When tested in a plus maze, R6/2 mice show an inflexible behavior pattern characterized by a decreased likelihood to turn right or left into a perpendicular arm [94]. Although both R6/2 and wild-type mice show similar LFP patterns prior to entering and after leaving the center or choice point of the maze, the R6/2 pattern is significantly more variable at the choice point [95]. Thus, mutant HTT expression causes changes in corticostriatal function that may promote the development of the HD behavioral phenotype. In support of this view, Díaz-Hernández et al. reported that after mutant HTT expression is switched off in a conditional HD model, animals show a full recovery of motor function despite striatal neuronal loss and the presence of mutant HTT aggregates [96]. Because cortical pyramidal neurons send glutamatergic projections to MSNs, altered glutamate transmission may underlie dysregulated corticostriatal function in HD. Among the glutamate changes elicited by mutant HTT, a decline in glutamate uptake caused by decreased GLT1 activity has received considerable attention.
Glutamate transmission and glutamate transporter 1 (GLT1)
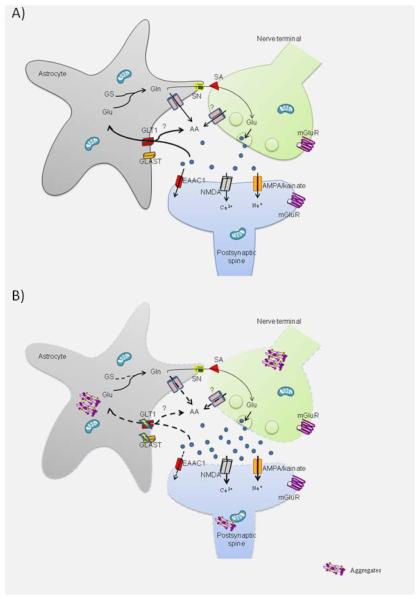
As shown schematically in Figure 1A, effective glutamate transmission requires the coordinated operation of proteins located pre- and post-synaptically and also in surrounding astrocytes [97]. Depolarization of the presynaptic terminal induces calcium influx followed by the mobilization and docking of synaptic vesicles to the plasma membrane to release glutamate into the synaptic space. Glutamate activates -amino-3-hydroxy-5-methyl-4-iso-xazolepropionic acid (AMPA) and kainate receptors, both of which lead to sodium influx and depolarization of the postsynaptic membrane. This effect promotes extrusion of the magnesium ion that normally blocks the NMDA receptor channel, allowing this receptor to be fully activated provided that glycine, an obligatory co-agonist, is also present. Full NMDA activation results in the influx of calcium and sodium. In addition, glutamate can bind to three groups of metabotropic receptors (mGluR), differentiated on the basis of their signal transduction pathways and sequence homologies. Group I includes mGluR1 and mGluR5, which are coupled to protein G q and the production of diacylglycerol and inositol 1,4,5-triphosphate (IP3). Group II includes mGluR2 and mRGlu3, which are coupled to G i/o protein to inhibit adenylate cyclase and decrease the formation of cAMP [98]. Finally, group III are coupled to G i/o protein, and thus their activation also lowers cAMP levels [98]. In addition to their localization on pre- and post-synaptic terminals, glutamate receptors also have been found on astrocytes [99], and like neurons, astrocytes may release vesicular-stored glutamate [100, 101]. Therefore, astrocytes are likely to play a more active role in glutamate transmission than previously believed, but their precise role is far from understood.
Figure 1A.
Schematic illustration of presumed transmission at a glutamatergic synapse in normal conditions (A) and in HD (B). A) Depolarization of the axon terminal allows the fusion of the presynaptic vesicles with the plasma membrane and release of glutamate (blue dots) into the synaptic cleft. On the postsynaptic dendritic spine, glutamate activates ionotropic receptors, AMPA/kainate, which allows the influx of sodium, and NMDA, which when sufficiently depolarized leads to calcium influx, and metabotropic glutamate receptors (mGluR), which occupy pre- and postsynaptic sites. Glutamate is removed from the synaptic space by transporters located in neurons (EAAC1) and astrocytes (GLT1, GLAST). When glutamate is taken up by astrocytes, it is metabolized to glutamine (Gln) by glutamine synthetase (GS). Eventually, glutamine will be transported to the extracellular space by glutamine transporters (SN) and then taken up (SA) by neurons. Glutamine is used as a precursor in the synthesis of glutamate. In parallel with glutamate uptake by GLT1, ascorbate (AA) is released into the extracellular space. Although the precise mechanism by which AA is released is unknown, three possibilities have emerged: release via a specific channel in astrocytes, co-transport by GLT1, or release from glutamate axon terminals. B) Mutant HTT aggregates are located in axon terminals, dendritic spines and in astrocytes. Morphological changes are induced by the expression of mutant HTT; also intracellular aggregates are observed at all components of the glutamatergic synapse. In the axon terminal, expression of mutant HTT alters vesicular glutamate release; in the dendritic spine, mutant HTT alters the function of glutamatergic receptors and decreases function of the neuronal transporter EAAC1. In astrocytes, there is decreased glutamate uptake and reduced expression of GLT1 and GLAST. Finally, along with dysfunctional GLT1 uptake resulting in elevated extracellular glutamate, there is a corresponding decrease in ascorbate release in the HD glutamatergic synapse.
After its release, glutamate is removed from the synaptic space by specific Na+-dependent high affinity transporters located mainly on astrocytes, but neuronal glutamate transporters also have been described [102]. Five glutamate transporters have been reported (known as excitatory amino acid transporters or EAAT in humans): EAAT1/GLAST (Glutamate aspartate transporter); GLT1 (Glutamate transporter 1)/EAAT2; EAAC1 (excitatory amino acid carrier 1)/EAAT3; EAAT4; and EAAT5 [103]. Although GLAST and GLT1 are mainly located on astrocytes, neuronal isoforms of GLT, GLT1a and GLT1b, have been reported in synaptic terminals in hippocampus and somatosensory cortex, respectively [104,105]. EAAC1 is a neuronal glutamate transporter. The distribution of EAAT4 and EAAT5 is restricted to cerebellum and retina, respectively [102]. Studies based on glutamate transporter knockout mice indicate that astrocytes play the most important role in regulating extracellular glutamate concentrations. EAAC1 has been associated with the uptake of precursors for the synthesis of the antioxidant, glutathione [106,107]. Extracellular glutamate removed by astrocytes is metabolized to glutamine, which is released to the extracellular space and taken up by neurons, where it is again metabolized to glutamate to replenish the transmitter pool [50].
Glutamate transporters are transmembrane proteins that, according to crystallographic studies performed on the GLT1 homologue from Pyrococcus horikoshii, operate as a bowl-shaped trimer with a concave aqueous basin facing the extracellular space and a pointed base facing the cytoplasm. The glutamate binding site is located beneath the bottom of the basin [108]. When glutamate and three Na+ ions bind to the extracellular site, the transporter internalizes them while it extrudes one K+ ion [108-110]. Glutamate uptake increases the intracellular concentration of sodium regulated by the activity of Na+/K+ATPase, which generates the Na+ transmembrane gradient, essential for the function of the glutamate transporter. Recent evidence suggests that GLT1 co-localizes with N+/K+ATPase, glycolytic proteins, and even with mitochondria, which collectively constitutes a large protein complex that could support the energy demands of glutamate uptake [111,112].
The coordinated operation of glutamate receptors and transporters is essential for normal excitatory transmission and rapid clearance of glutamate from the synapse. If glutamate is not removed efficiently, the extracellular concentration could approach 1 mM, triggering an excitotoxic cascade. Excitotoxicity is triggered by the over-stimulation of glutamate receptors, leading to calcium-dependent protease activity, oxidative damage, and eventually, death of the neuron. In fact, glutamate uptake failure and excitotoxicity are associated with several neuropathological conditions and with neurodegenerative diseases, including HD [113].
GLT1 and neuronal vulnerability in HD
Consistent evidence shows a link between altered GLT1 function and mutant HTT expression. Evaluation of HD postmortem tissue shows reduced binding of D-[3H] aspartic acid to high-affinity binding sites and diminished mRNA of GLT1 [114,115]. Immunohistochemical detection of GLT1 in the caudate nucleus of HD patients showed a reduction in GLT1 in a grade-dependent manner, which occurs in spite of increased labeling of the glial fibrillary acidic protein (GFAP) specific for astrocytes, suggesting that mechanisms other than loss of astrocytes are associated with the reduction of GLT1 protein [116]. Decreased glutamate uptake also has been described for prefrontal cortex of HD postmortem tissue, and this effect is observed in the early stages of HD [117]. Similar alterations have been described for transgenic models. Reduced GLT1 mRNA and decreased GLT1 protein content have been consistently described in R6/2 and other models expressing mutant HTT [58,116,118-120]. A corresponding reduction in glutamate uptake has been described in the striatum of R6/2 mice in early symptomatic stages [121]. Similarly, decreased glutamate uptake occurs in R6/1 mice at 16 weeks of age when the behavioral phenotype may not yet emerge [122]. Interestingly, the reduction in glutamate uptake in HD models can occur without a reduction in protein content, suggesting that GLT1 function can be affected independently of GLT1 expression (Figure 1B). Although multiple mechanisms regulate GLT1 function, a key mechanism affected by HD is palmitoylation, the post-translational addition of the saturated 16-carbon lipid palmitate to specific cysteine residues on GLT1 [123]. Reduced GLT1 palmitoylation has been reported to occur in YAC128 mice along with decreased GLT1-mediated uptake activity [123].
In fact, GLT1 dysfunction may explain, at least in part, the direct involvement of astrocytes in HD neuropathology. Glutamate-induced neurotoxicity in R6/2 striatum correlates with decreased levels of GLT1 [58]. Moreover, the molecular mechanisms altered by mutant HTT in astrocytes are closely associated with neuronal dysfunction and the development of phenotypic alterations in HD. Consistent with this view, Bradford et al. [124] restricted the expression of mutant HTT to astrocytes and found decreased levels of GLT1, reduced glutamate uptake, and most importantly, emergence of the HD behavioral phenotype.
A critical question, therefore, is whether an increase in glutamate uptake is a viable therapeutic target for HD. It is interesting in this regard that ceftriaxone, a beta-lactam antibiotic that crosses the blood-brain barrier, increases brain GLT1 expression in mice [125]. To determine if ceftriaxone could have value as an HD therapeutic, Miller et al. treated early symptomatic R6/2 mice and wild-type controls with ceftriaxone for five consecutive days [126]. This treatment not only increased striatal GLT1 expression, but also increased glutamate uptake, indicating that the GLT1 up-regulation was functional. Ceftriaxone, moreover, improved several aspects of the HD behavioral phenotype, including increased turning in the plus maze, increased climbing behavior in the open field, and a decrease in clasping [126]. Thus, despite the down-regulation of GLT1 in HD, it is possible to increase the expression of functional GLT1 and improve the HD phenotype.
Although an improved phenotype follows GLT1 up regulation, the potential use of ceftriaxone in clinical trials requires caution because of some noteworthy side effects. For example, the ceftriaxone-induced up regulation of GLT1 impairs mGluR-dependent long-term depression (LTD) at mossy fibre-CA3 synapses in hippocampus [127], which suggests a decline in synaptic plasticity. Ceftriaxone treatment also leads to an impairment of prepulse inhibition [128], further suggesting a change in cognitive processing. Recently, Bellesi et al. show that ceftriaxone treatment resulted in a delayed reduction in EEG theta (7-9 Hz), which occurred along with increased motor activity [129]. Thus, even though a loss of GLT1 function may underlie the corticostriatal communication problem in HD, and GLT1 up regulation improves the HD phenotype, long-term exposure to ceftriaxone may compromise key neuronal operations. Further study of this and other GLT1 activators is warranted.
Glutamate uptake has long been linked to the release of ascorbate, the deprotonated form of vitamin C found in high concentrations in the mammalian forebrain [130-132, Figure 1A, B]. In fact, extracellular levels of striatal ascorbate are low in R6/2 and other HD mouse models relative to wild-type [133-136], and this effect cannot be explained by an overall loss of ascorbate since cellular levels of ascorbate do not decline in HD [137]. It appears, therefore, that the mechanism responsible for ascorbate release is dysfunctional. To determine if the mechanism involves GLT1, R6/2 mice were treated with ceftriaxone and striatal ascorbate release was assessed in response to cortical activation, which triggers glutamate release from corticostriatal terminals [126, 135]. Ceftriaxone, but not saline vehicle, was found to increase cortically evoked ascorbate release, and this effect was blocked by pharmacological inhibition of GLT1 [126]. These results further confirm that the GLT1 up-regulation induced by ceftriaxone is functional and also suggest that an increase in ascorbate release is part of this process. Although glutamate uptake via GLT1appears necessary for cortically evoked ascorbate release, it is not clear if the source of ascorbate release is the corticostriatal projection itself or striatal astrocytes or both.
Ample evidence links glutamate uptake with ascorbate release. For example, infusion of L-glutamate, the naturally occurring isomer, promotes the rapid release of ascorbate in rat striatal tissue [138] and in synaptosomal fractions [139], whereas D-glutamate, which has less affinity for transport, fails to alter ascorbate release. Similarly, global blockade of glutamate uptake using non-selective uptake inhibitors diminishes extracellular ascorbate in striatum [139, 140]. Thus, the level of striatal ascorbate in extracellular fluid appears to depend on the extent of glutamate uptake, but the precise molecular mechanism by which GLT1 function leads to ascorbate release remains to be determined (Figure 1A).
Nevertheless, because GLT1 is oxidation sensitive [141], it is tempting to speculate that the failure to release striatal ascorbate, which can act as an extracellular antioxidant under most conditions [for review see 142], may contribute to GLT1 dysfunction in HD. Ascorbate and glutamate also are linked in their effects on MSN activity. An increase in extracellular ascorbate, for example, can either enhance or suppress glutamate-evoked activation of striatal neurons depending on how much ascorbate is present [143]. Interestingly, in R6/2 mice, which have low extracellular ascorbate and a high rate striatal neuronal firing, an ascorbate treatment regimen that elevates the level of extracellular ascorbate in striatum and also lowers MSN activity [81]. Conceivably, therefore, the corticostriatal dysfunction underlying HD may involve a dysregulation of both ascorbate and glutamate owing to a down-regulation of GLT1.
Ascorbate release also may be modulated by glutamate receptors. Blockade of NMDA receptors with either the non-selective antagonist amantadine or the non-competitive antagonist MK-801 decreases striatal ascorbate release [144,145]. Release of ascorbate in retina, moreover, is mediated by activation of AMPA receptors [146]. Although activation of postsynaptic glutamate receptors may modulate striatal ascorbate release, it is unlikely that they play a major role since destruction of striatal neurons fails to alter the extracellular level of ascorbate [147].
Although the precise mechanism by which mutant HTT leads to decreased function and expression of GLT1 is unknown, it has been suggested that mutant HTT fragments form insoluble inclusions in the cytoplasm and nucleus that alter GLT1 transcription [148,149]. It also is possible that glutamate transporter function is inhibited by reactive oxygen species [150], which are known to accumulate in HD and trigger oxidative damage [151,152]. Alternatively, because glutamate uptake is an energy-demanding process, deficient ATP production, which is known to occur in HD, may play a role in the loss of glutamate uptake [153]. In fact, inhibition of the glycolitic enzyme, glyceraldehyde-3-phospohate dehydrogenase (GAPDH), decreases glutamate transporter levels and enhances glutamate-induced damage in the striatum of the R6/2 model [154].
Although the contribution of GLAST and EAAC1 in HD has received less attention than GLT1, reduced GLAST protein levels have been observed in late stages of the disease in R6/2 mice [58,155]. Likewise, expression of mutant HTT reduces protein levels of GLAST in astrocytes in vivo [116]. Altered function of EAAC1 has been described for cultured cortical neurons expressing exon 1 of the mutant htt gene, which is associated with deficient glutathione synthesis and increased oxidative damage [156]. More research is required to determine if GLAST and EAAC1 are associated with neuronal dysfunction and neuronal vulnerability in HD.
Conclusion
Despite the increase in research that followed identification of the underlying genetic mutation, an effective treatment for HD remains elusive. As described here, altered corticostriatal neuronal processing is the primary driver of the HD behavioral phenotype, and impaired operation of GLT1 appears to play a critical role. This conclusion is supported by several lines of evidence indicating that deficient GLT1-mediated glutamate uptake occurs early in HD progression, which in turn might lead to altered neuronal function and the development of HD phenotype. Further evidence for GLT1 as a possible therapeutic target comes from experiments showing that increased expression of GLT1 by ceftriaxone, restores glutamate uptake, and improves the HD behavioral phenotype. In addition to its role in regulating extracellular glutamate, GLT1 activity may be associated with neuronal survival, and thus GLT1 down regulation may promote the large-scale neuronal loss identified in HD patients at autopsy. Further understanding of GLT1 involvement in HD will require new information on astrocytes, where most GLT1 is expressed. In fact, selective expression of mutant HTT in astrocytes is sufficient to down regulate GLT1 and to induce the HD behavioral phenotype in mice. Extending this line of research will have important implications for HD treatment development.
Acknowledgments
Research by the authors is supported by USPHS grants NS035663 and AG039818, the Indiana METACyt Initiative of Indiana University funded in part by a major grant from the Lilly Endowment, Inc., and a contract with the Cure Huntington’s Disease Initiative (CHDI). We also gratefully acknowledge Faye Caylor for administrative support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Harper PS. The epidemiology of Huntington’s disease. In: Bates G, Harper PS, Jones L, editors. Huntington’s disease. 3rd ed Oxford University Press; New York: 2001. pp. 159–97. [Google Scholar]
- [2].Gusella JF, Wexler NS, Conneally PM, Naylor SL, Anderson MA, Tanzi RE, et al. A polymorphic DNA marker genetically linked to Huntington’s disease. Nature. 1983;306:234–8. doi: 10.1038/306234a0. [DOI] [PubMed] [Google Scholar]
- [3].The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–83. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- [4].Ambrose CM, Duyao MP, Barnes G, Bates PG, Lin SC, Srinidhi J, et al. Structure and expression of the Huntington’s disease gene: evidence against simple inactivation due to an expanded CAG repeat. Somat Cell Mol Genet. 1994;20:27–38. doi: 10.1007/BF02257483. [DOI] [PubMed] [Google Scholar]
- [5].Telenius H, Kremer HP, Theilmann J, Andrew SE, Almqvist E, Anvret M, et al. Molecular analysis of juvenile Huntington disease: The major influence on (CAG)n repeat length is the sex of the affected parent. Hum Mol Genet. 1993;2:1535–40. doi: 10.1093/hmg/2.10.1535. [DOI] [PubMed] [Google Scholar]
- [6].Nahhas FA, Garbern J, Krajewski KM, Roa BB, Feldman GL. Juvenile onset Huntington disease resulting from a very large maternal expansion. Am J Med Genet A. 2005;137:328–31. doi: 10.1002/ajmg.a.30891. [DOI] [PubMed] [Google Scholar]
- [7].Zühlke Ch, Reiss O, Bockel B, Lange H, Thies U. Mitotic stability and meiotic variability of the (CAG)n repeat in the Huntington disease gene. Hum Mol Genet. 1993;2:2063–7. doi: 10.1093/hmg/2.12.2063. [DOI] [PubMed] [Google Scholar]
- [8].Rubinsztein DC, Leggo J, Chiano M, Dodge A, Norbury G, Rosser E, et al. Genotypes at the GluR6 kainate receptor locus are associated with variation in the age of onset of Huntington disease. Proc Natl Acad Sci USA. 1997;94:3872–6. doi: 10.1073/pnas.94.8.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].MacDonald ME, Vonsattel JP, Shrinidhi J, Couropmitree NN, Cuples LA, Bird E, et al. Evidence for the GluR6 gene associated with younger onset age of Huntington’s disease. Neurology. 1999;53:1330–2. doi: 10.1212/wnl.53.6.1330. [DOI] [PubMed] [Google Scholar]
- [10].The U.S.-Venezuela collaborative research project. Wexler N. Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington’s disease age of onset. Proc Natl Acad Sci USA. 2004;101:3498–03. doi: 10.1073/pnas.0308679101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Arning L, Kraus PH, Valentin S, Saft C, Andrich J, Epplen JT. NR2A and NR2B receptor gene variations modify age at onset in Huntington disease. Neurogenetics. 2005;6:25–28. doi: 10.1007/s10048-004-0198-8. [DOI] [PubMed] [Google Scholar]
- [12].Arning L, Saft C, Wieczorek S, Andrich J, Kraus PH, Epplen JT. NR2A and NR2B receptor gene variations modify age at onset in Huntington disease in a sex-specific manner. Hum Genet. 2007;122:175–182. doi: 10.1007/s00439-007-0393-4. [DOI] [PubMed] [Google Scholar]
- [13].Lee JM, Gillis T, Mysore JS, Ramos EM, Myers RH, Hayden MR. Common SNP-based haplotype analysis of the 4p16.3 Huntington disease gene region. Am J Hum Genet. 2012;90:434–44. doi: 10.1016/j.ajhg.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cattaneo E, Zuccato C, Tartari M. Normal huntingtin function: an alternative approach to Huntington’s disease. Nat Rev Neurosci. 2005;6:919–30. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- [15].Li SH, Schilling G, Young WS, Li XJ, Margolis RL, Stine OC, et al. Huntington’s Disease gene (IT15) is widely expressed in human and rat tissue. Neuron. 1993;11:985–93. doi: 10.1016/0896-6273(93)90127-d. [DOI] [PubMed] [Google Scholar]
- [16].Trottier Y, Devys D, Imbert G, Saudou F, An I, Lutz Y. Huntington’s disease protein and discrimination of the normal and mutated form. Nature Genetics. 1995;10:104–10. doi: 10.1038/ng0595-104. [DOI] [PubMed] [Google Scholar]
- [17].Nicolas G, Devys D, Goldenberg A, Maltete D, Hervé C, Hannequin D. Juvenile Huntington disease in an 18-month-old boy revealed by global developmental delay and reduced cerebellar volume. Am J Med Genet A. 2011;155:815–8. doi: 10.1002/ajmg.a.33911. [DOI] [PubMed] [Google Scholar]
- [18].Rasmussen A, Macias R, Yescas P, Ochoa A, Davila G, Alonso E. Huntington disease in children: Genotype–phenotype correlation. Neuropediatrics. 2000;31:190–4. doi: 10.1055/s-2000-7461. [DOI] [PubMed] [Google Scholar]
- [19].Goebel HH, Heipertz R, Scholz W, Iqbal K, Tellez-Nagel I. Juvenile Huntington chorea: clinical, ultrastructural, and biochemical studies. Neurology. 1978;28:23–31. doi: 10.1212/wnl.28.1.23. [DOI] [PubMed] [Google Scholar]
- [20].Huntington G. On Chorea. The Medical and Surgical Reporter. 1872;26:317–21. [Google Scholar]; J Neuropsychiatry Clin Neurosci. 2003;15:109–12. doi: 10.1176/jnp.15.1.109. Re-Edited: [DOI] [PubMed] [Google Scholar]
- [21].Lemiere J, Decruyenaere M, Evers-Kiebooms G, Vandenbusschet E, Dom R. Cognitive changes in patients with Huntington’s disease (HD) and asymptomatic carriers of the HD mutation: a longitudinal follow-up study. J Neurol. 2004;251:935–42. doi: 10.1007/s00415-004-0461-9. [DOI] [PubMed] [Google Scholar]
- [22].Kremer B. Clinical neurology of Huntington’s disease. In: Bates G, Harper P, Jones L, editors. Huntington’s disease. 3rd ed Oxford University Press; New York: 2001. pp. 28–61. [Google Scholar]
- [23].Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP. Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- [24].Cudkowicz M, Kowall NW. Degeneration of pyramidal projection neurons in Huntington’s disease cortex. Ann Neurol. 1990;27:200–4. doi: 10.1002/ana.410270217. [DOI] [PubMed] [Google Scholar]
- [25].Sotrel A, Paskevich PA, Kiely DK, Bird ED, Williams RS, Myers RH. Morphometric analysis of the prefrontal cortex in Huntington’s disease. Neurology. 1991;41:1117–23. doi: 10.1212/wnl.41.7.1117. [DOI] [PubMed] [Google Scholar]
- [26].Macdonald V, Halliday GM, Trent RJ, McCusker EA. Significant loss of pyramidal neurons in the angular gyrus of patients with Huntington’s disease. Neuropathol Appl Neurobiol. 1997;23:492–5. doi: 10.1111/j.1365-2990.1997.tb01326.x. [DOI] [PubMed] [Google Scholar]
- [27].Gutekunst CA, Li SH, Yi H, Mulroy JS, Kuemmerle S, Jones R, et al. Nuclear and neuropil aggregates in Huntington’s disease: relationship to neuropathology. J Neurosci. 1999;19:2522–34. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Menalled LB. Knock-in mouse models of Huntington’s disease. NeuroRx. 2005;2:465–470. doi: 10.1602/neurorx.2.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–06. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- [30].Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–48. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- [31].Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, Kotzuk JA, et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet. 1999;8:397–07. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- [32].Von Hörsten S, Schmitt I, Nguyen HP, Holzmann C, Schmidt T, Walther T, et al. Transgenic rat model of Huntington’s disease. Hum Mol Genet. 2003;12:617–24. doi: 10.1093/hmg/ddg075. [DOI] [PubMed] [Google Scholar]
- [33].Hodgson JG, Apopyan N, Gutekunts CA, Leavitt BR, LePiane F, Singaraja R. A YAC mouse model for Huntington’s disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron. 1999;23:181–92. doi: 10.1016/s0896-6273(00)80764-3. [DOI] [PubMed] [Google Scholar]
- [34].Gray M, Shirasaki DI, Cepeda C, André VM, Wilburn B, Lu XH, et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 2008;28:6182–95. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Spampanato J, Gu X, Yang XW, Mody I. Progressive synaptic pathology of motor cortical neurons in a BAC transgenic mouse model of Huntington’s disease. Neuroscience. 2008;157:606–20. doi: 10.1016/j.neuroscience.2008.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bolam JP, Hanley JJ, Booth PA, Bevan MD. Synaptic organization of the basal ganglia. J Anat. 2000;196:527–42. doi: 10.1046/j.1469-7580.2000.19640527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Smith Y, Bevan MD, Shink E, Bolam JP. Microcircuitry of the direct and indirecty pathways of the basal ganglia. Neuroscience. 1998;86:353–87. doi: 10.1016/s0306-4522(98)00004-9. [DOI] [PubMed] [Google Scholar]
- [38].Albin RL, Young AB, Penney JB. The functional anatomy of basal ganglia disorders. Trends Neurosci. 1989;12:366–75. doi: 10.1016/0166-2236(89)90074-x. [DOI] [PubMed] [Google Scholar]
- [39].Gerfen CR, Surmeier DJ. Modulation of striatal projection systems by dopamine. Annu Rev Neurosci. 2011;34:441–66. doi: 10.1146/annurev-neuro-061010-113641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Reiner A, Albin RL, Anderson KD, D’Amato CJ, Penney JB, Young AB. Differential loss of striatal projection neurons in Huntington disease. Proc Natl Acad Sci U S A. 1988;85:5733–7. doi: 10.1073/pnas.85.15.5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kawaguchi Y, Wilson CJ, Augood SJ, Emson PC. Striatal interneurons: chemical, physiological and morphological characterization. Trends Neurosci. 1995;18:527–35. doi: 10.1016/0166-2236(95)98374-8. [DOI] [PubMed] [Google Scholar]
- [42].Fino E, Venance L. Spike-timing dependent plasticity in striatal interneurons. Neuropharmacology. 2011;60:780–8. doi: 10.1016/j.neuropharm.2011.01.023. [DOI] [PubMed] [Google Scholar]
- [43].Tepper JM, Koós T, Wilson CJ. GABAergic microcircuits in the neostriatum. Trends Neurosci. 2004:27662–9. doi: 10.1016/j.tins.2004.08.007. [DOI] [PubMed] [Google Scholar]
- [44].Tepper JM, Abercrombie ED, Bolam JP. Basal ganglia macrocircuits. Prog Brain Res. 2007;160:3–7. doi: 10.1016/S0079-6123(06)60001-0. [DOI] [PubMed] [Google Scholar]
- [45].Centonze D, Gubellini P, Pisani A, Bernardi G, Calabresi P. Dopamine, acetylcholine and nitric oxide systems interact to induce corticostriatal synaptic plasticity. Rev Neurosci. 2003;14:207–16. doi: 10.1515/revneuro.2003.14.3.207. [DOI] [PubMed] [Google Scholar]
- [46].Centonze D, Pisani A, Bonsi P, Giacomini P, Bernardi G, Calabresi P. Stimulation of nitric oxide-cGMP pathway excites striatal cholinergic interneurons via protein kinase G activation. J Neurosci. 2001;21:1393–400. doi: 10.1523/JNEUROSCI.21-04-01393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gittis AH, Leventhal DK, Fensterheim BA, Pettibone JR, Berke JD, Kreitzer AC. Selective inhibition of striatal fast-spiking interneurons causes dyskinesias. J Neurosci. 2011;31:15727–31. doi: 10.1523/JNEUROSCI.3875-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Heinsen H, Strik M, Bauer M, Luther K, Ulmar G, Gangnus D, et al. Cortical and striatal neurone number in Huntington’s disease. Acta Neuropathol. 1994;88:320–33. doi: 10.1007/BF00310376. [DOI] [PubMed] [Google Scholar]
- [49].Gomide V, Bibancos T, Chadi G. Dopamine cell morphology and glial cell hypertrophy and process branching in the nigrostriatal system after striatal 6-OHDA analyzed by specific sterological tools. Int J Neurosci. 2005;115:557–82. doi: 10.1080/00207450590521118. [DOI] [PubMed] [Google Scholar]
- [50].Schousboe A, Sickmann HM, Bak LK, Schousboe I, Jajo FS, Faek SA, et al. Neuron-glia interactions in glutamatergic neurotransmission: roles of oxidative and glycolytic adenosine triphosphate as energy source. J Neurosci Res. 2011;89:1926–34. doi: 10.1002/jnr.22746. [DOI] [PubMed] [Google Scholar]
- [51].Fernandez-Fernandez S, Almeida A, Bolaños JP. Antioxidant and bioenergetic coupling between neurons and astrocytes. Biochem J. 2012;443:3–11. doi: 10.1042/BJ20111943. [DOI] [PubMed] [Google Scholar]
- [52].Scott DF, Heathfield KWG, Toone B, Margerison JH. The EEG in Huntington’s chorea: a clinical and neuropathological study. J Neurol Neurosurgery and Psych. 1972;35:97–102. doi: 10.1136/jnnp.35.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].De Tommaso M, De Carlo F, Difruscolo O, Massafra R, Sciruicchio V, Bellotti R. Detection of subclinical brain electrical activity changes in Huntington’s disease using artificial neural networks. Clin Neurophysiol. 2003;114:1237–45. doi: 10.1016/s1388-2457(03)00074-9. [DOI] [PubMed] [Google Scholar]
- [54].Bylsma FW, Peyser CE, Folstein SE, Folstein MF, Ross C, Brandt J. EEG power spectra in Huntington’s disease: clinical and neuropsychological correlates. Neuropsychologia. 1994;32:137–50. doi: 10.1016/0028-3932(94)90001-9. [DOI] [PubMed] [Google Scholar]
- [55].Painold A, Anderer P, Holl AK, Letmaier M, Saletu-Zyhlarz GM, Saletu B, et al. Comparative EEG mapping studies in Huntington’s disease patients and controls. J Neural Transm. 2010;117:1307–18. doi: 10.1007/s00702-010-0491-7. [DOI] [PubMed] [Google Scholar]
- [56].Streletz LJ, Reyes PF, Zalewska M, Katz L, Fariello RG. Computer analysis of EEG activity in dementia of the Alzheimer’s type and Huntington’s disease. Neurobiol Aging. 1990;11:15–20. doi: 10.1016/0197-4580(90)90057-7. [DOI] [PubMed] [Google Scholar]
- [57].Painold A, Anderer P, Holl AK, Letmaier M, Saletu-Zyhlarz GM, Saletu B, et al. EEG low-resolution brain electromagnetic tomography (LORETA) in Huntington’s Disease. J Neurol. 2011;258:840–54. doi: 10.1007/s00415-010-5852-5. [DOI] [PubMed] [Google Scholar]
- [58].Estrada-Sánchez AM, Montiel T, Segovia J, Massieu L. Glutamate toxicity in the striatum of the R6/2 Huntington’s disease transgenic mice is age-dependent and correlates with decreased levels of glutamate transporters. Neurobiol Dis. 2009;34:78–6. doi: 10.1016/j.nbd.2008.12.017. [DOI] [PubMed] [Google Scholar]
- [59].Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–58. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- [60].Li H, Li SH, Cheng AL, Mangiarini L, Bates GP, Li XJ. Ultrastructural localization and progressive formation of neuropil aggregates in Huntington’s disease transgenic mice. Hum Mol Genet. 1999;8:1227–36. doi: 10.1093/hmg/8.7.1227. [DOI] [PubMed] [Google Scholar]
- [61].Li H, Li SH, Johnston H, Shelbourne PF, Li XJ. Amino-terminal fragments of mutant huntingtin show selective accumulation in striatal neurons and synaptic toxicity. Nat Genet. 2000;25:385–9. doi: 10.1038/78054. [DOI] [PubMed] [Google Scholar]
- [62].Li H, Wyman T, Yu ZX, Li SH, Li XJ. Abnormal association of mutant huntingtin with synaptic vesicles inhibits glutamate release. Hum Mol Genet. 2003;12:2021–30. doi: 10.1093/hmg/ddg218. [DOI] [PubMed] [Google Scholar]
- [63].Cepeda C, Hurst RS, Calvert CR, Hernández-Echeagaray E, Nguyen OK, Jocoy E, et al. Transient and progressive electrophysiological alterations in the corticostriatal pathway in a mouse model of Huntington’s disease. J Neurosci. 2003;23:961–9. doi: 10.1523/JNEUROSCI.23-03-00961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Milnerwood AJ, Raymond LA. Corticostriatal synaptic function in mouse models of Huntington’s disease: early effects of huntingtin repeat length and protein load. J Physiol. 2007;585:817–31. doi: 10.1113/jphysiol.2007.142448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Höhn S, Dallérac G, Faure A, Urbach YK, Nguyen HP, Riess O, et al. Behavioral and in vivo electrophysiological evidence for presymptomatic alteration of prefrontostriatal processing in the transgenic rat model for Huntington disease. J Neurosci. 2011;31:8986–97. doi: 10.1523/JNEUROSCI.1238-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Klapstein GJ, Fisher RS, Zanjani H, Cepeda C, Jokel ES, Chesselet MF, et al. Electrophysiological and morphological changes in striatal spiny neurons in R6/2 Huntington’s disease transgenic mice. J Neurophysiol. 2001;86:2667–77. doi: 10.1152/jn.2001.86.6.2667. [DOI] [PubMed] [Google Scholar]
- [67].Traficante A, Riozzi B, Cannella M, Rampello L, Squitieri F, Battaglia G. Reduced activity of cortico-striatal fibers in the R6/2 mouse model of Huntington’s disease. Neuroreport. 2007;18:1997–2000. doi: 10.1097/WNR.0b013e3282f262ca. [DOI] [PubMed] [Google Scholar]
- [68].Cummings DM, André VM, Uzgil BO, Gee SM, Fisher YE, Cepeda C. Alterations in cortical excitation and inhibition in genetic mouse models of Huntington’s disease. J Neurosci. 2009;29:10371–86. doi: 10.1523/JNEUROSCI.1592-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Gu X, Li C, Wei W, Lo V, Gong S, Li SH, et al. Pathological cell-cell interactions elicited by a neuropathogenic form of mutant Huntingtin contribute to cortical pathogenesis in HD mice. Neuron. 2005;5(46):433–44. doi: 10.1016/j.neuron.2005.03.025. [DOI] [PubMed] [Google Scholar]
- [70].Gambazzi L, Gokce O, Seredenina T, Katsyuba E, Runne H, Markram H, et al. Diminished activity-dependent brain-derived neurotrophic factor expression underlies cortical neuron microcircuit hypoconnectivity resulting from exposure to mutant huntingtin fragments. J Pharmacol Exp Ther. 2010;335:13–22. doi: 10.1124/jpet.110.167551. [DOI] [PubMed] [Google Scholar]
- [71].Izhikevich EM, Desai NS, Walcott EC, Hoppensteadt FC. Bursts as a unit of neural information: selective communication via resonance. Trends Neurosci. 2003;26:161–7. doi: 10.1016/S0166-2236(03)00034-1. [DOI] [PubMed] [Google Scholar]
- [72].Lisman JE. Bursts as a unit of neural information: making unreliable synapses reliable. Trends Neurosci. 1997;20:38–43. doi: 10.1016/S0166-2236(96)10070-9. [DOI] [PubMed] [Google Scholar]
- [73].Walker AG, Miller BR, Fritsh JN, Barton SJ, Rebec GV. Altered information processing in the proforntal cortex of Huntington’s Disease mouse models. J Neurosci. 2008;28:8973–82. doi: 10.1523/JNEUROSCI.2804-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Berke JD, Okatan M, Skurski J, Eichenbaum HB. Oscillatory entrainment of striatal neurons in freely moving rats. Neuron. 2004;43:883–96. doi: 10.1016/j.neuron.2004.08.035. [DOI] [PubMed] [Google Scholar]
- [75].Burns SP, Xing D, Shapley RM. Comparisons of the dynamics of local field potential and multiunit activity signals in macaque visual cortex. J Neurosci. 2010;30:13739–49. doi: 10.1523/JNEUROSCI.0743-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Buzsáki G, Chrobak JJ. Synaptic plasticity and self-organization in the hippocampus. Nat Neurosci. 2005;8:1418–20. doi: 10.1038/nn1105-1418. [DOI] [PubMed] [Google Scholar]
- [77].Walker AG, Ummel JR, Rebec GV. Reduced expression of conditioned fear in the R6/2 mouse model of Huntington’s Disease is related to abnormal activity in prelimbic cortex. Neurobiol Dis. 2011;43:379–87. doi: 10.1016/j.nbd.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Levine MS, Klapstein GJ, Koppel A, Gruen E, Cepeda C, Vargas ME, et al. Enhanced sensitivity to N-methyl-D-aspartate receptor activation in transgenic and knockin mouse models of Huntington’s disease. J Neurosci Res. 1999;58:515–32. [PubMed] [Google Scholar]
- [79].Cepeda C, Ariano MA, Calvert CR, Flores-Hernández J, Chandler SH, Leavitt BR, et al. NMDA receptor function in mouse models of Huntington disease. J Neurosci Res. 2001;66:525–39. doi: 10.1002/jnr.1244. [DOI] [PubMed] [Google Scholar]
- [80].Laforet GA, Sapp E, Chase K, McIntyre C, Boyce FM, Campbell M, et al. Changes in cortical and striatal neurons predict behavioral and electrophysiological abnormalities in a transgenic murine model of Huntington’s disease. J Neurosci. 2001;21:9112–23. doi: 10.1523/JNEUROSCI.21-23-09112.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Rebec GV, Conroy SK, Barton SJ. Hyperactive striatal neurons in symptomatic Huntington R6/2 mice: variations with behavioral state and repeated ascorbate treatment. Neuroscience. 2006;137:327–36. doi: 10.1016/j.neuroscience.2005.08.062. [DOI] [PubMed] [Google Scholar]
- [82].Miller BR, Walker AG, Shah AS, Barton SJ, Rebec VG. Dysregulated information processing by medium spiny neurons in striatum of freely behaving mouse models of Huntington’s disease. J Neurophysiol. 2008;100:2205–16. doi: 10.1152/jn.90606.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Miller BR, Walker AG, Barton SJ, Rebec GV. Dysregulated neuronal activity patterns implicate corticostriatal circuit dysfunction in multiple rodent models of Huntington’s Disease. Frontiers in Systems Neuroscience. 2011;5:1–10. doi: 10.3389/fnsys.2011.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Miller BR, Walker AG, Fowler SC, von Hörsten S, Riess O, Johnson MA, et al. Dysregulation of coordinated neuronal firing patterns in striatum of freely behaving transgenic rats that model Huntington’s disease. Neurobiol Dis. 2010;37:106–13. doi: 10.1016/j.nbd.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Miller BR, Bezprozvanny I. Corticostriatal circuit dysfunction in Huntington’s disease: intersection of glutamate, dopamine and calcium. Future Neurol. 2010;5:735–756. doi: 10.2217/fnl.10.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Cayzac S, Delcasso S, Paz V, Jeantet Y, Cho YH. Changes in striatal procedural memory coding correlate with learning deficits in a mouse model of Huntington disease. Proc Natl Acad Sci U S A. 2011;108:9280–5. doi: 10.1073/pnas.1016190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Höhn S, Dallérac G, Faure A, Urbach YK, Nguyen HP, Riess O, et al. Behavioral and in vivo electrophysiological evidence for presymptomatic alteration of prefrontostriatal processing in the transgenic rat model for huntington disease. J Neurosci. 2011;31:8986–97. doi: 10.1523/JNEUROSCI.1238-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Joshi PR, Wu NP, André VM, Cummings DM, Cepeda C, Joyce JA, et al. Age-dependent alterations of corticostriatal activity in the YAC128 mouse model of Huntington disease. J Neurosci. 2009;29:2414–27. doi: 10.1523/JNEUROSCI.5687-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].André VM, Cepeda C, Levine MS. Dopamine and glutamate in Huntington’s disease: A balancing act. CNS Neurosci Ther. 2010;16:163–78. doi: 10.1111/j.1755-5949.2010.00134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].André VM, Cepeda C, Fisher YE, Huynh M, Bardakjian N, Singh S, et al. Differential electrophysiological changes in striatal output neurons in Huntington’s disease. J Neurosci. 2011;31:1170–82. doi: 10.1523/JNEUROSCI.3539-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].André VM, Fisher YE, Levine MS. Altered balance of activity in the striatal direct and indirect pathways in mouse models of Huntington’s Disease. Front Syst Neurosci. 2011;5:46. doi: 10.3389/fnsys.2011.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].DeCoteau WE, Thorn C, Gibson DJ, Courtemanche R, Mitra P, Kubota Y, et al. Oscillations of local field potentials in the rat dorsal striatum during spontaneous and instructed behaviors. J Neurophysiol. 2007;97:3800–5. doi: 10.1152/jn.00108.2007. [DOI] [PubMed] [Google Scholar]
- [93].Groiss SJ, Elben S, Reck C, Voges J, Wojtecki L, Schnitzler A. Local field potential oscillations of the globus pallidus in Huntington’s disease. Mov Disord. 2011;26:2577–8. doi: 10.1002/mds.23914. [DOI] [PubMed] [Google Scholar]
- [94].Rebec GV, Barton SJ, Marseilles AM, Collins K. Ascorbate treatment attenuates the Huntington behavioral phenotype in mice. Neuroreport. 2003;14:1263–5. doi: 10.1097/00001756-200307010-00015. [DOI] [PubMed] [Google Scholar]
- [95].Hong SL, Barton SJ, Rebec GV. Altered Neural and Behavioral Dynamics in Huntington’s Disease: An Entropy Conservation Approach. PLoS One. 2012;7:e30879. doi: 10.1371/journal.pone.0030879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Díaz-Hernández M, Torres-Peraza J, Salvatori-Abarca A, Morán MA, Gómez-Ramos P, Alberch J, et al. Full motor recovery despite striatal neuron loss and formation of irreversible amyloid-like inclusions in a conditional mouse model of Huntington’s disease. J Neurosci. 2005;25:9773–81. doi: 10.1523/JNEUROSCI.3183-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Hassel B, Dingledine R, Siegel GJ, Albers RW, Brady ST, Price DL. Basic neurochemistry: molecular, cellular and medical aspects. 7th ed Elsevier Academic Press; Canada: 2006. Glutamate; pp. 267–290. [Google Scholar]
- [98].Kim CH, Lee J, Lee J-Y, Roche KW. Metabotropic glutamate receptors: phosphorylation and receptor signaling. J Neurosci Res. 2008;86:1–10. doi: 10.1002/jnr.21437. [DOI] [PubMed] [Google Scholar]
- [99].Palygin O, Lalo U, Verkhratsky A, Pankratov Y. Ionotropic NMDA and P2X1/5 receptors mediate synaptically induced Ca2+ signalling in cortical astrocytes. Cell Calcium. 2010;48:225–31. doi: 10.1016/j.ceca.2010.09.004. [DOI] [PubMed] [Google Scholar]
- [100].Bezzi P, Gundersen V, Galbete JL, Seifert G, Steinhäuser C, Pilati E, et al. Astrocytes contain a vesicular compartment that is competent for regulated exocytosis of glutamate. Nature Neurosci. 2004;7:613–20. doi: 10.1038/nn1246. [DOI] [PubMed] [Google Scholar]
- [101].Hamilton NB, Attwell D. Do astrocytes really exocytose neurotransmitters? Nat Rev Neurosci. 2010;11:227–38. doi: 10.1038/nrn2803. [DOI] [PubMed] [Google Scholar]
- [102].Danbolt NC. Glutamate uptake. Prog Neurobiol. 2001;65:1–105. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- [103].Arriza JL, Fairman WA, Wadiche JI, Murdoch GH, Kavanaugh MP, Amara SG. Functional comparisons of three glutamate transporter subtypes cloned from human motor cortex. J Neurosci. 1994;14:5559–69. doi: 10.1523/JNEUROSCI.14-09-05559.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Chen W, Aoki C, Mahadomrongkul V, Gruber CE, Wang GJ, Blitzblau R, et al. Expression of a variant form of the glutamate transporter GLT1 in neuronal cultures and in neurons and astrocytes in the rat brain. J Neurosci. 2002;22:2142–52. doi: 10.1523/JNEUROSCI.22-06-02142.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Chen W, Mahadomrongkul V, Berger UV, Bassan M, DeSilva T, Tanaka K, et al. The glutamate transporter GLT1a is expressed in excitatory axon terminals of mature hippocampal neurons. J Neurosci. 2004;24:1136–48. doi: 10.1523/JNEUROSCI.1586-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Rothstein JD, Dykes-Hober M, Pardo CA, Bristol LA, Jin L, Kuncl RW, et al. Knock out of glutamate transporters reveals a major role for astroglial transport in excitotoxicity and clearance of glutamate. Neuron. 1996;16:675–86. doi: 10.1016/s0896-6273(00)80086-0. [DOI] [PubMed] [Google Scholar]
- [107].Chen Y, Swanson R. The glutamate transporter EAAT2 and EAAT3 mediates cysteine uptake in cortical neurons cultures. J Neurochem. 2003;84:1332–39. doi: 10.1046/j.1471-4159.2003.01630.x. [DOI] [PubMed] [Google Scholar]
- [108].Yernool D, Boudker O, Jin Y, Gouaux E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature. 2004;431:811–8. doi: 10.1038/nature03018. [DOI] [PubMed] [Google Scholar]
- [109].Watzke N, Bamberg E, Grewer C. Early intermediates in the transport cycle of the neuronal excitatory amino acid carrier EAAC1. J Gen Physiol. 2001;117:547–62. doi: 10.1085/jgp.117.6.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Boudker O, Ryan RM, Yernool D, Shimamoto K, Gouaux E. Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature. 2007;445:387–93. doi: 10.1038/nature05455. [DOI] [PubMed] [Google Scholar]
- [111].Rose EM, Koo JC, Antflick JE, Ahmed SM, Angers S, Hampson DR. Glutamate transporter coupling to Na,K-ATPase. J Neurosci. 2009;29:8143–55. doi: 10.1523/JNEUROSCI.1081-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Genda EN, Jackson JG, Sheldon AL, Locke SF, Greco TM, O’Donnell JC, et al. Cocompartmentalization of the astroglial glutamate transporter, GLT-1, with glycolytic enzymes and mitochondria. J Neurosci. 2011;31:18275–88. doi: 10.1523/JNEUROSCI.3305-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Sánchez AM Estrada, Mejía-Toiber J, Massieu L. Excitotoxic neuronal death and the pathogenesis of Huntington’s disease. Arch Med Res. 2008;39:265–76. doi: 10.1016/j.arcmed.2007.11.011. [DOI] [PubMed] [Google Scholar]
- [114].Cross AJ, Slater P, Reynolds GP. Reduced high-affinity glutamate uptake sites in the brains of patients with Huntington’s disease. Neurosci Lett. 1986;67:198–02. doi: 10.1016/0304-3940(86)90397-6. [DOI] [PubMed] [Google Scholar]
- [115].Arzberger T, Krampfl K, Leimgruber S, Weindl A. Changes of NMDA receptor subunit (NR1, NR2B) and glutamate transporter (GLT1) mRNA expression in Huntington’s disease-an in situ hybridization study. J Neuropathol Exp Neurol. 1997;56:440–54. doi: 10.1097/00005072-199704000-00013. [DOI] [PubMed] [Google Scholar]
- [116].Faideau M, Kim J, Cormier K, Gilmore R, Welch M, Auregan G, et al. In vivo expression of polyglutamine-expanded huntingtin by mouse striatal astrocytes impairs glutamate transport: a correlation with Huntington’s disease subjects. Hum Mol Genet. 2010;19:3053–67. doi: 10.1093/hmg/ddq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Hassel B, Tessler S, Faull RL, Emson PC. Glutamate uptake is reduced in prefrontal cortex in Huntington’s disease. Neurochem Res. 2008;33:232–7. doi: 10.1007/s11064-007-9463-1. [DOI] [PubMed] [Google Scholar]
- [118].Liévens JC, Woodman B, Mahal A, Spasic-Boscovic, Samuel D, Kerkerian-Le Goff, et al. Impaired glutamate uptake in the R6 Huntington’s disease transgenic mice. Neurobiol Dis. 2001;8:807–21. doi: 10.1006/nbdi.2001.0430. [DOI] [PubMed] [Google Scholar]
- [119].Behrens PF, Franz P, Woodman B, Lindenberg KS, Landwehrmeyer GB. Impaired glutamate transport and glutamate-glutamine cycling: downstream effects of the Huntington mutation. Brain. 2002;125:1908–22. doi: 10.1093/brain/awf180. [DOI] [PubMed] [Google Scholar]
- [120].Shin JY, Fang ZH, Yu ZX, Wang CE, Li SH, Li XJ. Expression of mutant huntingtin in glial cells contributes to neuronal excitotoxicity. J Cell Biol. 2005;171:1001–12. doi: 10.1083/jcb.200508072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Miller BR, Dorner JL, Shou M, Sari Y, Barton SJ, Sengelaub DR, et al. Up-regulation of GLT-1 expression increases glutamate uptake and attenuates the Huntington’s disease phenotype in the R6/2 mouse. Neuroscience. 2008;153:329–37. doi: 10.1016/j.neuroscience.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Nicniocaill B, Haraldsson B, Hansson O, O’Connor WT, Brundin P. Altered striatal amino acid neurotransmitter release monitored using microdialysis in R6/1 Huntington transgenic mice. Eur J Neurosci. 2001;13:206–10. doi: 10.1046/j.0953-816x.2000.01379.x. [DOI] [PubMed] [Google Scholar]
- [123].Huang K, Kang MH, Askew C, Kang R, Sanders SS, Wan J, et al. Palmitoylation and function of glial glutamate transporter-1 is reduced in the YAC128 mouse model of Huntington disease. Neurobiol Dis. 2010;40:207–15. doi: 10.1016/j.nbd.2010.05.027. [DOI] [PubMed] [Google Scholar]
- [124].Bradford J, Shin JY, Roberts M, Wang CE, Li XJ, Li S. Expression of mutant huntingtin in mouse brain astrocytes causes age-dependent neurological symptoms. Proc Natl Acad Sci USA. 2009;106:22480–5. doi: 10.1073/pnas.0911503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Rothstein JD, Patel S, Regan MR, Haenggeli C, Huang YH, Bergles DE, et al. Beta-lactam antibiotics offer neuroprotection by increasing glutamate transporter expression. Nature. 2005;433:73–7. doi: 10.1038/nature03180. [DOI] [PubMed] [Google Scholar]
- [126].Miller BR, Dorner JL, Bunner KD, Gaither TW, Klein EL, Barton SJ, et al. Up-regulation of GLT1 reverses the deficit in cortically evoked striatal ascorbate efflux in the R6/2 mouse model of Huntington’s disease. J Neurochem. 2012;121:629–38. doi: 10.1111/j.1471-4159.2012.07691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Omrani A, Melone M, Bellesi M, Safiulina V, Aida T, Tanaka K, et al. Up-regulation of GLT-1 severely impairs LTD at mossy fibre--CA3 synapses. J Physiol. 2009;587:4575–88. doi: 10.1113/jphysiol.2009.177881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Bellesi M, Melone M, Gubbini A, Battistacci S, Conti F. GLT-1 upregulation impairs prepulse inhibition of the startle reflex in adult rats. Glia. 2009;57:703–13. doi: 10.1002/glia.20798. [DOI] [PubMed] [Google Scholar]
- [129].Bellesi M, Vyazovskiy VV, Tononi G, Cirelli C, Conti F. Reduction of EEG theta power and changes in motor activity in rats treated with ceftriaxone. PLoS One. 2012;7:e34139. doi: 10.1371/journal.pone.0034139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Chinoy NJ. Ascorbic acid levels in mammalian tissues and its metabolic significance. Comp Biochem Physiol A Comp Physiol. 1972;42:945–52. doi: 10.1016/0300-9629(72)90400-8. [DOI] [PubMed] [Google Scholar]
- [131].Schreiber M, Trojan S. Ascorbic acid in the brain. Physiolog Res. 1991;40:413–8. [PubMed] [Google Scholar]
- [132].Rebec GV, Pierce RC. A vitamin as neuromodulator: ascorbate release into the extracellular fluid of the brain regulates dopaminergic and glutamatergic transmission. Prog Neurobiol. 1994;43:537–65. doi: 10.1016/0301-0082(94)90052-3. [DOI] [PubMed] [Google Scholar]
- [133].Rebec GV, Barton SJ, Ennis MD. Dysregulation of ascorbate release in the striatum of behaving mice expressing the Huntington’s disease gene. J Neurosci. 2002;22:RC202. doi: 10.1523/JNEUROSCI.22-02-j0006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Dorner JL, Miller BR, Barton SJ, Brock TJ, Rebec GV. Sex differences in behavior and striatal ascorbate release in the 140 CAG knock-in mouse model of Huntington’s disease. Behav Brain Res. 2007;178:90–7. doi: 10.1016/j.bbr.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Dorner JL, Miller BR, Klein EL, Murphy-Nakhnikian A, Andrews RL, Barton SJ, et al. Corticostriatal dysfunction underlies diminished striatal ascorbate release in the R6/2 mouse model of Huntington’s disease. Brain Res. 2009;1290:111–20. doi: 10.1016/j.brainres.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Rebec GV. From interferant anion to neuromodulator: ascorbate oxidizes its way to respectability. In: Michael AC, Borland LM, editors. Electrochemical Methods for Neuroscience. CRC Press; Boca Raton, FL: 2009. pp. 149–165. [PubMed] [Google Scholar]
- [137].Tkac I, Dubinsky JM, Keene CD, Gruetter R, Low WC. Neurochemical changes in Huntington R6/2 mouse striatum detected by in vivo 1H NMR spectroscopy. J Neurochem. 2007;100:1397–406. doi: 10.1111/j.1471-4159.2006.04323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Ghasemzadeh B, Cammack J, Adams RN. Dynamic changes in extracellular fluid ascorbic acid monitored by in vivo electrochemistry. Brain Res. 1991;547:162–166. [PubMed] [Google Scholar]
- [139].Grunewald RA, Fillenz M. Release of asco rbate from synaptosomal fraction of rat brain. Neurochem Int. 1984;6:491–500. doi: 10.1016/0197-0186(84)90120-7. [DOI] [PubMed] [Google Scholar]
- [140].Miele M, Boutelle MG, Fillenz M. The physiologically induced release of ascorbate in rat brain is dependent on impulse traffic, calcium influx and glutamate uptake. Neuroscience. 1994;62:87–91. doi: 10.1016/0306-4522(94)90316-6. [DOI] [PubMed] [Google Scholar]
- [141].Trotti D, Rizzini BL, Rossi D, Haugeto O, Racagni G, Danbolt NC, et al. Neuronal and glial glutamate transporters possess an SH-based redox regulatory mechanism. Eur J Neurosci. 1997;9:1236–43. doi: 10.1111/j.1460-9568.1997.tb01478.x. [DOI] [PubMed] [Google Scholar]
- [142].Harrison FE, May JM. Vitamin C function in the brain: vital role of the ascorbate transporter SVCT2. Free Radic Biol and Med. 2009;46:719–30. doi: 10.1016/j.freeradbiomed.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Kiyatkin EA, Rebec GV. Ascorbate modulates glutamate-induced excitations of striatal neurons. Brain Res. 1998;812:14–22. doi: 10.1016/s0006-8993(98)00814-2. [DOI] [PubMed] [Google Scholar]
- [144].Liu J, Wu C, Liu W, Zhang H, Li CL. Involvement of the corticostriatal glutamatergic pathway in ethanol-induced ascorbic acid release in rat striatum. Addict Biol. 1999;4:273–81. doi: 10.1080/13556219971489. [DOI] [PubMed] [Google Scholar]
- [145].Pierce RC, Rebec GV. Dopamine-, NMDA- and sigma-receptor antagonists exert differential effects on basal and amphetamine-induced changes in neostriatal ascorbate and DOPAC in awake, behaving rats. Brain Res. 1992;579:59–66. doi: 10.1016/0006-8993(92)90741-q. [DOI] [PubMed] [Google Scholar]
- [146].Portugal CC, Miya VS, Calaza Kda C, Santos RA, Paes-de-Carvalho R. Glutamate receptors modulate sodium-dependent and calcium-independent vitamin C bidirectional transport in cultured avian retinal cells. J Neurochem. 2009;108:507–520. doi: 10.1111/j.1471-4159.2008.05786.x. [DOI] [PubMed] [Google Scholar]
- [147].Pierce RC, Miller DW, Reising DB, Rebec GV. Unilateral neostriatal kainate, but not 6-OHDA, lesions block dopamine agonist-induced ascorbate release in the neostriatum of freely moving rats. Brain Res. 1992;597:138–43. doi: 10.1016/0006-8993(92)91515-g. [DOI] [PubMed] [Google Scholar]
- [148].DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–3. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- [149].Liévens JC, Rival T, Iché M, Chneiweiss H, Birman S. Expanded polyglutamine peptides disrupt EGF receptor signaling and glutamate transporter expression in Drosophila. Hum Mol Genet. 2005;14:713–24. doi: 10.1093/hmg/ddi067. [DOI] [PubMed] [Google Scholar]
- [150].Trotti D, Rossi D, Gjesdal O, Levy LM, Racagni G, Danbolt NC, et al. Peroxynitrite inhibits glutamate transporter subtypes. J Biol Chem. 1996;271:5976–79. doi: 10.1074/jbc.271.11.5976. [DOI] [PubMed] [Google Scholar]
- [151].Sanchez AM Estrada, Massieu L. Role of oxidative stress in the pathogenesis of Huntington’s disease. In: Santamaría A, Jiménez-Capdeville ME, editors. New perspective on brain cell damage, neurodegenertation and neuroprotective strategies. Research signpost; Kerala, India: 2007. pp. 21–36. [Google Scholar]
- [152].Chen CM. Mitochondrial dysfunction, metabolic deficits, and increased oxidative stress in Huntington’s disease. Chang Gung Med J. 2011;34:135–52. [PubMed] [Google Scholar]
- [153].Browne SE, Beal MF. The energetics of Huntington’s disease. Neurochem Res. 2004;29:531–46. doi: 10.1023/b:nere.0000014824.04728.dd. [DOI] [PubMed] [Google Scholar]
- [154].Estrada-Sánchez AM, Montiel T, Massieu L. Glycolysis inhibition decreases the levels of glutamate transporters and enhances glutamate neurotoxicity in the R6/2 Huntington’s disease mice. Neurochem Res. 2010;35:1156–63. doi: 10.1007/s11064-010-0168-5. [DOI] [PubMed] [Google Scholar]
- [155].Liu X, Miller BR, Rebec GV, Clemmer DE. Protein expression in the striatum and cortex regions of the brain for a mouse model of Huntington’s disease. J Proteome Res. 2007;6:3134–42. doi: 10.1021/pr070092s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Li X, Valencia A, Sapp E, Masso N, Alexander J, Reeves P, et al. Aberrant rab11-dependent trafficking of the neuronal glutamate transporter EAAC1 causes oxidative stress and cell death in Huntington’s disease. J Neurosci. 2010;30:4552–61. doi: 10.1523/JNEUROSCI.5865-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]