Abstract
Members of the sirtuin family including the founding protein Sir2 in Saccharomyces cerevisiae have been linked to lifespan extension in simple organisms. This finding prompted evaluation of the role of Sir2 orthologues in many aging associated conditions including neurodegeneration, type II diabetes and cancer. These studies have demonstrated that genetic and pharmacologic manipulation of sirtuin activity have beneficial effects in a surprisingly broad spectrum of aging-associated conditions suggesting that the Sir2 family of enzymes presents an attractive target for the development of pharmacological agents. While the initial model favored pharmacological activators of sirtuins as calorie restriction mimetics, it now appears that either activation or inhibition of sirtuins may be desirable for ameliorating disease depending on the pathological condition and the target tissue. In this chapter we review the development of pharmacological small molecule activators and inhibitors of the sirtuin family of enzymes.
Extension of human life span through direct intervention has been a goal since the ancient Greeks and probably longer. The explorer Ponce de Leon was just one of many seekers for the magical formula that would extend human life. The latest chapter in this saga is based on the observation that calorie restriction promotes longevity in organisms ranging from yeast to primates. This finding raises the expectation that molecular mechanisms mediating life span extension may also be shared between species. Pharmacological modulation of these mechanisms could potentially yield a fountain of youth. In Saccharomyces cerevisiae, Sir2, the founding member of the sirtuin family of nicotinamide adenine dinucleotide (NAD+)-dependent deacetylases, has been proposed to be a link between metabolism, epigenetic silencing, genome stability and lifespan control [1]. In yeast, the unequal division of “mother” and “daughter” cells makes it amenable to analysis of replicative lifespan (RLS), the number of daughter cells that a mother cell can produce before it dies [2]. One of the events that limits mothers’ lifespan is accumulation of rDNA circles that are excised during recombination at the rDNA locus and are preferentially retained in mother cells during cell division[3]. Although the exact mechanism by which accumulation of rDNA circles decreases lifespan is still an active area of research, the inverse correlation between the amount of rDNA circles and longevity has fueled the search for regulators of rDNA recombination. Increased SIR2 dosage was found to extend the lifespan of mother cells by reducing recombination at the rDNA locus,[3]. This observation led to studies in multicellular eukaryotes such as nematodes [4] and fruit flies [5], which showed that increased gene dosage of the Sir2 orthologues also extends lifespan. Because nematodes and flies do not show accumulation of rDNA circles during aging, and yet Sir2 orthologues promote longevity in these organisms, it has been proposed that the longevity control role of sirtuins has been retained during evolution despite differences in the specific degenerative processes that occur during aging in different species. Since both calorie restriction and increased dosage of Sir2 extend lifespan in the different species, a hypothesis was put forward that the beneficial effects of calorie restriction during aging are mediated through a conserved molecular pathway that involves calorie-restriction-induced increase in sirtuin activity [6]. Despite ongoing controversy whether calorie restriction mediated lifespan extension requires Sir2 [7], this hypothesis generated a high level of enthusiasm for studies from many laboratories that evaluated the role of Sir2 orthologues in various aspects of aging (e.g. neurodegeneration, type II diabetes) in metazoan organisms including mice. Genetic and pharmacologic manipulations of sirtuin activity have shown beneficial effects in a surprisingly broad spectrum of aging associated conditions and diseases suggesting that the Sir2-family of enzymes presents an attractive pharmacological target.
Over the past 10 years, studies in mice have shown that either activation or inhibition of sirtuin activity, depending on the pathological state and the tissue, may be desirable for ameliorating disease state [8]. Accordingly, efforts have been undertaken to identify compounds that can either activate or inhibit specific sirtuins and serve as leads for the development of human therapeutics. Most of these efforts have been focused on developing modulators of major nuclear sirtuin, SIRT1, however as our understanding of the cellular roles of other sirtuins expands, other sirtuins are likely to be targeted as well in the future.
Endogenous modulators of sirtuin activity
Acetylation of lysine residues in proteins by acetyltransferases and deacetylation by deacetylases serves a regulatory function analogous to the way kinases and phosphatases modulate the ionic charge of serine, threonine and tyrosine residues and as a result regulate protein/protein interactions or enzyme function. Acetylation and deacetylation of lysines play similar roles as an ionic switch that regulates protein function. The molecular mechanism of sirtuin activity is well understood. During the sirtuin-mediated deacetylation reaction, cleavage of the glycosidic bond between nicotinamide and ribose of NAD+ is coupled to transfer of an acetyl group from the acetylated lysine residue in the target protein to the ribose moiety of ADP-ribose. The reaction yields deacetylated lysine, O-acetyl-ADP-ribose and nicotinamide as products [9]. Cellular sirtuin activity can be modulated by enzyme abundance, availability of NAD+ as well as the local presence of the nicotinamide, a deacetylation byproduct that has been shown to inhibit sirtuin activity. Furthermore, SIRT1 has been found in different complexes with other proteins which can either directly activate [10] or inhibit [11] SIRT1 activity.
The concentration of cellular NAD+ is maintained by balancing NAD+ biosynthesis with cellular NAD+ utilization. In humans, the dietary sources used for synthesis of NAD+ include tryptophan, nicotinic acid, nicotinamide and a newly discovered precursor, nicotinamide ribose (reviewed in [12]). De novo NAD+ biosynthesis from tryptophan through the kynurenin pathway requires eight enzymatic steps that are highly conserved in evolution. In yeast, expression of the enzymes in the kynurenine pathway is controlled by a sirtuin, Hst1, which functions as a sensor for cellular NAD+ level that represses the expression of de novo biosynthesis enzymes according to the availability of NAD+ [13]. SIRT1 dependent control of the genes in a salvage pathway has been recently shown in mammals [18, 19] (see bellow). Besides de novo biosynthesis, NAD+ can be synthesized from its breakdown product, nicotinamide, through a salvage pathway. The biggest consumers of cellular NAD+ are mono-ADP ribose and poly-ADP ribose transferases. These enzymes cleave the glycosidic bond in NAD+ and transfer or polymerize ADP onto other proteins. DNA double strand breaks activate poly-ADP ribose polymerase (PARP) and may result in catastrophic depletion of cellular NAD+ [14]. In addition to regenerating NAD+, the salvage pathway also reutilizes and thus removes the deacetylase inhibitor, nicotinamide. Both of these outcomes can promote sirtuin activity [15–17]. The key enzyme in the NAD salvage pathway is nicotinamide phosphoribosyl transferase (NAMPT) whose activity has been shown to modulate NAD+ levels and SIRT1 cellular activity [15–17]. The expression of NAMPT was recently shown to be a target of the circadian transcription factors that induce diurnal oscillations in NAD+ levels and thus sirtuin activity [18, 19].
Besides reactions that consume NAD+ such as those involving PARPs and sirtuins, NAD+ and acts a co-factor for hydrogen transfer enzymes and results in interconversion of NAD+ and NADH. The redox state in cells and concomitant alterations of NAD+/NADH ratio have been implicated in the regulation of cellular sirtuin activity and control of several processes including calorie restriction-induced life span extension in yeast [20], muscle differentiation [21] and neurogenesis [22]. The NAD+/NADH ratio has been proposed to influence sirtuin activity in two ways. A decreased NAD+/NADH ratio may result from a decreased amount of NAD+, a sirtuin activator, or conversely from an increase in cellular NADH, a sirtuin inhibitor. Although this model is appealing for a variety of reasons, a detailed characterization of coenzyme specificity of Sir2-proteins failed to generate biochemical support for the role of cellular NAD+/NADH alterations as cellular regulators of sirtuin activity [23]. Two key observations undermine this hypothesis. First, given that only a fraction of total cellular NAD+ and NADH pool is in the reduced state (NADH) (i.e. NAD+/NADH ratio is estimated to be very high) even if the total pool were converted to NAD+, the result would be only a minor increase in the available NAD+. Second, NADH has been shown to be extremely inefficient sirtuin inhibitor (IC50 15 mM) [23]. Because cellular NADH levels are at least two orders of magnitude lower than the measured IC50 of NADH it is unlikely that cellular NADH level can increase sufficiently to have a significant direct influence on sirtuin enzymatic activity.
While the alterations in NAD+/NADH ratio are unlikely to directly influence sirtuin enzymatic activity, there is evidence that cellular redox state regulates SIRT1 protein level at the transcriptional and posttranscriptional level. Cellular NADH levels and thus NAD+/NADH ratio have been proposed to control SIRT1 transcription though a regulatory circuit that involves redox sensor CtBP and HIC1, an inhibitor of SIRT1 transcription [24]. Furthermore, a separate study suggested that glucose deprivation and cellular pyruvate control SIRT1 protein even though the level of SIRT1 mRNA does not change [25]. These studies suggest that overall cellular SIRT1 activity may be influenced by NAD+/NADH ratio through alterations SIRT1 protein level rather than through direct control of SIRT1 enzymatic activity.
Sirtuin Inhibitors
Since the discovery of sirtuin’s enzymatic activity 10 years ago several compound that inhibit this class of enzymes have been described. Both whole cells and biochemical screens have been employed for identification of these inhibitors. More recently, crystal structures of human sirtuins and homology models have allowed for structure-based design of more potent and selective Sirtuin inhibitors. Among the seven human homologs, SIRT1 and SIRT2 have been exploited the most for drug discovery due to recognition of their therapeutic potential in diabetes and in neurodegenerative diseases. The inhibitors can be classified into categories based on their pharmacophore.
β-napthols
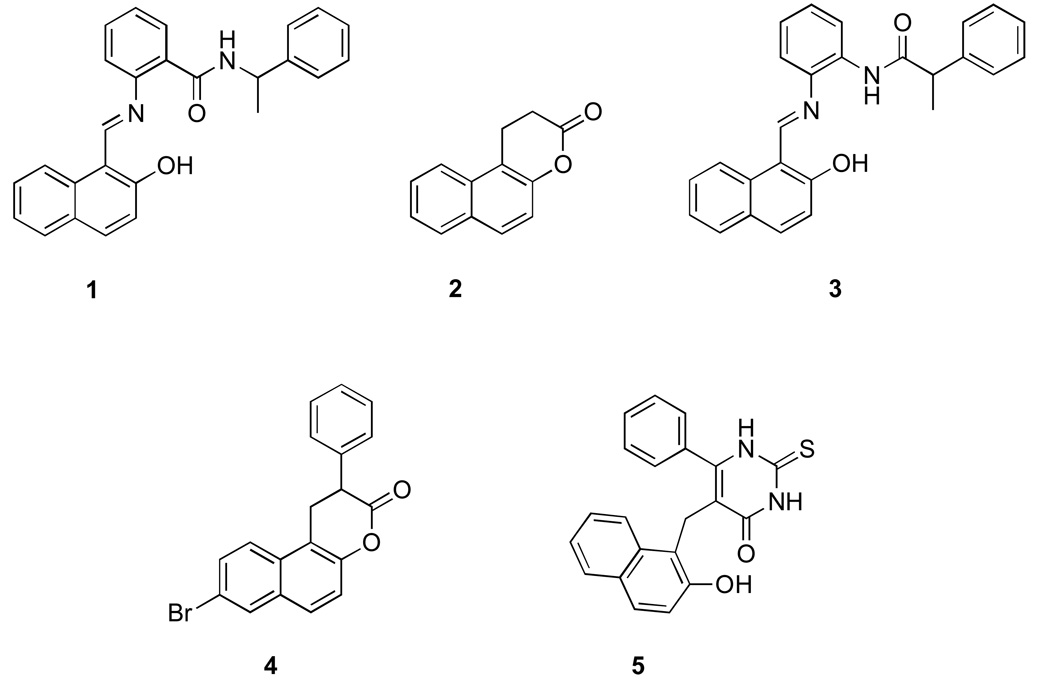
The β-naphtol nucleus is a key group for several sirtuin inhibitors. Both sirtinol (1) and splitomicin (2) [26, 27] (Fig. 1), were identified through cell-based screens in yeast Saccharomyces cerevisiae for compounds that abrogate telomeric silencing (see Figure 1). We observed limited activity of sirtinol in vivo against SIRT1 and SIRT2 enzymes judged by acetylation of the known SIRT1 and SIRT2 cellular targets. Structure activity relationship (SAR) studies on sirtinol resulted in improved analogs such as salermide (3), which has been shown to induce apoptosis in cancer cells [28].
Figure 1. Sirtuin inhibitors with β-napthol pharmacophore.
1.Sirtinol 2. Splitomicin 3. Salermide 4. β-phenylsplitomicin 5. Cambinol
The lactone in splitomicin was found to be essential for activity but at the same time conferred instability at physiological pH [29]. Replacement of the lactone with a lactam resulted in analogs with improved pH stability and efficacy. Further studies on splitomicin led to identification of β-phenylsplitomicins (4) with low micromolar inhibition against SIRT1 [30]. Phenyl splitomicins with substitutions on the 8-position of naphthalene ring were found to have increased selectivity for SIRT2 over SIRT1. Point mutations within the small helical domain close to substrate binding site resulted in loss of splitomicin Sir2 inhibitory activity in cell-based assays identifying a potential binding pocket close to the substrate-binding site [26, 31].
Cambinol (5), a β-naphthol derivative with a substituted thiouracil ring represents the most promising sirtuin inhibitor in this class of compounds. It is a non–selective SIRT1 (56 µM) and SIRT2 (59 µM) inhibitor but shows no inhibitory activity against other human sirtuins and HDACs. In contrast to sirtinol and splitomicin, cambinol is stable and highly effective in vivo. It has shown to induce hyperacetylation of SIRT1 and SIRT2 substrates such p53, BCL6, α-tubulin in cells and inhibited growth of lymphoma xenograft in mice [32]. Several analogs of cambinol have been developed with low micromolar activity and improved selectivity for SIRT1 over SIRT2 [33]. Cambinol shows competitive inhibition with the acetyl-peptide, suggesting it binds close to the substrate-binding site similarly to splitomicin. The fact that β-naphthol class of compounds bind to a site other than the NAD+ binding site make them potentially less toxic due to off-target interactions.
Indoles
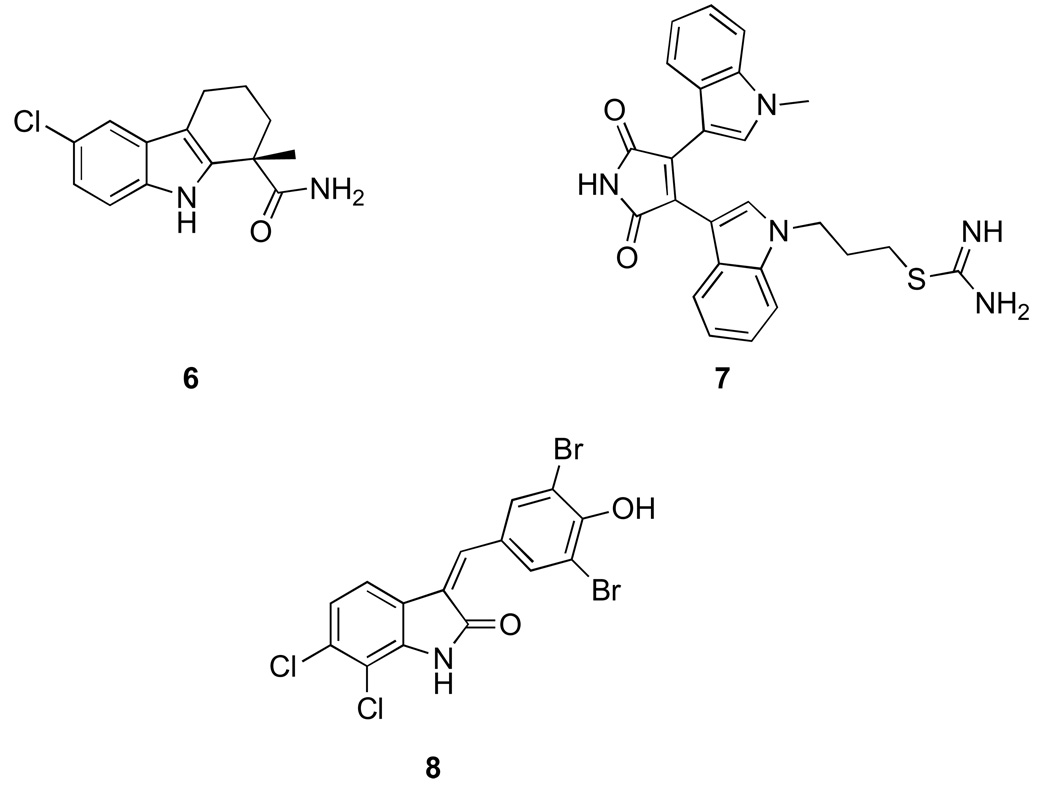
Similar to the β-naphthol scaffold, the indole ring has been exploited in development of potent sirtuin inhibitors. The most notable in this class is EX-527 (6) (Fig. 2), which was identified by high throughput screening [34]. The compound was initially reported to inhibit SIRT1 in the low nanomolar range (60–100 nM), however higher IC50 values have been reported depending on the assay used for analysis. The compound has been proven to be highly useful in understanding the role of SIRT1 in cell survival and its interaction with p53. Docking studies and nicotinamide release-based assays suggest that indoles such as EX-527 bind to NAD+ binding site unlike the β-napthols [30]. A distinct set of inhibitors containing an indole nucleus but potentially binding to adenine part of the ATP binding pocket have been identified. These compounds, referred as bis(indolyl)maleimides (BIMs, 7) resemble kinase inhibitors and are selective SIRT2 inhibitors in the low micromolar range (e.g. Ro31–8220) [35]. Recently, additional compounds that resemble ATP-competitive kinase inhibitors have been identified. An interesting compound belonging to this indole class is the oxyindole (8) which is a hybrid consisting of structural features of a kinase inhibitor and an anti-proliferative natural product bauerine C. The compound has good selectivity for SIRT2 in vitro and was shown to inhibit deacetylation of α-tubulin in MCF-7 cells [36].
Figure 2. Sirtuin inhibitors based on indole.
6. EX-527 7. Bis(indolyl)maleimides, Ro31-8220 8. Oxyindole
Ureas and thioureas
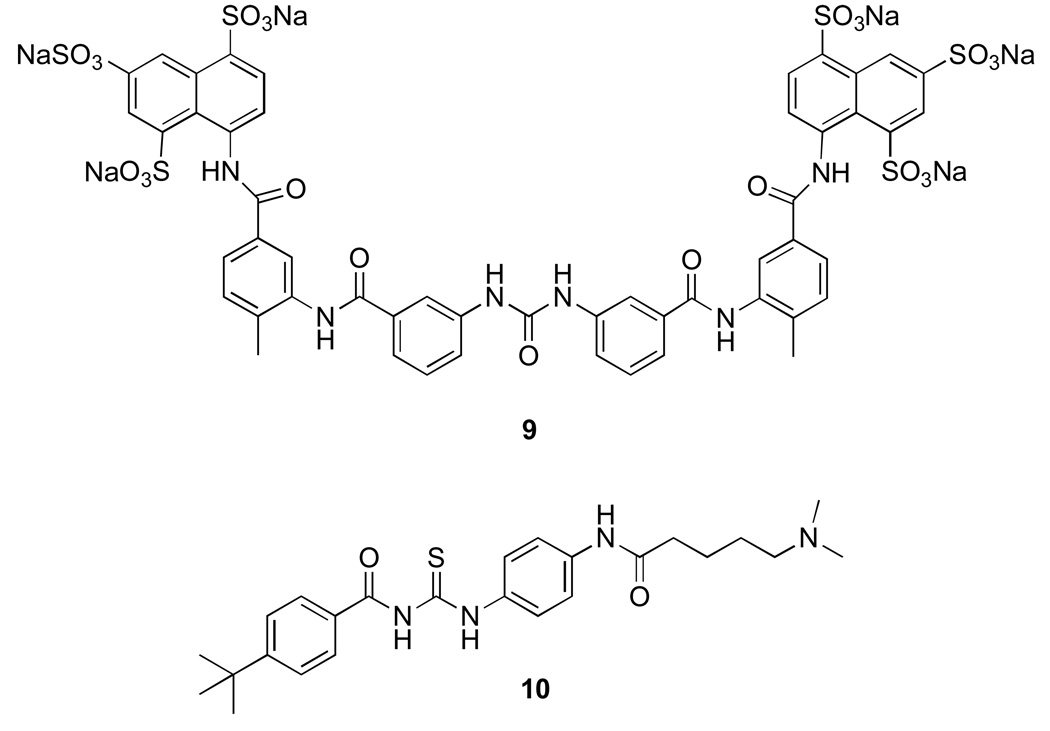
The compounds in this class were primarily identified during screening for different targets and disease states. Subsequent studies revealed that they exhibit sirtuin inhibition. One such compound is suramin (9) (Fig. 3), a polyanionic urea derivative, which is used in treatment of trypanosomiasis and has antiviral and anticancer activity [37]. It was later found to be a potent sirtuin inhibitor with IC50 of 297 nM and 1150 nM for SIRT1 and SIRT2 respectively [38]. Synthesis of suramin analogs has resulted in more selective SIRT1 inhibitors. The compounds show non-competitive inhibition with both NAD+ and acetylated peptide substrate suggesting that their binding site spans these two sites. Binding mode analysis based on suramin co-crystallized with SIRT5 and docking studies using SIRT2 model also suggest that the suramin binding site exist between NAD+ and the peptide substrate [39]. Similar to suramin-related compounds, the thiourea-based tenovins (10) were initially identified in a cell based screen for p53 activators [40]. Further studies revealed that tenovins are low micromolar inhibitors of SIRT1 and SIRT2. Their high hydrophobicity precluded any in vivo use even though they showed decreased tumor growth in all the major tumor cell lines. Tenovin-6 a more water soluble analog has shown to be effective in reducing tumor growth in mouse model of melanoma thereby showing promise for this class of compounds.
Figure 3. Sirtuin inhibitors based on urea and thiourea moiety.
9. Suramin 10. Tenovin
Miscellaneous
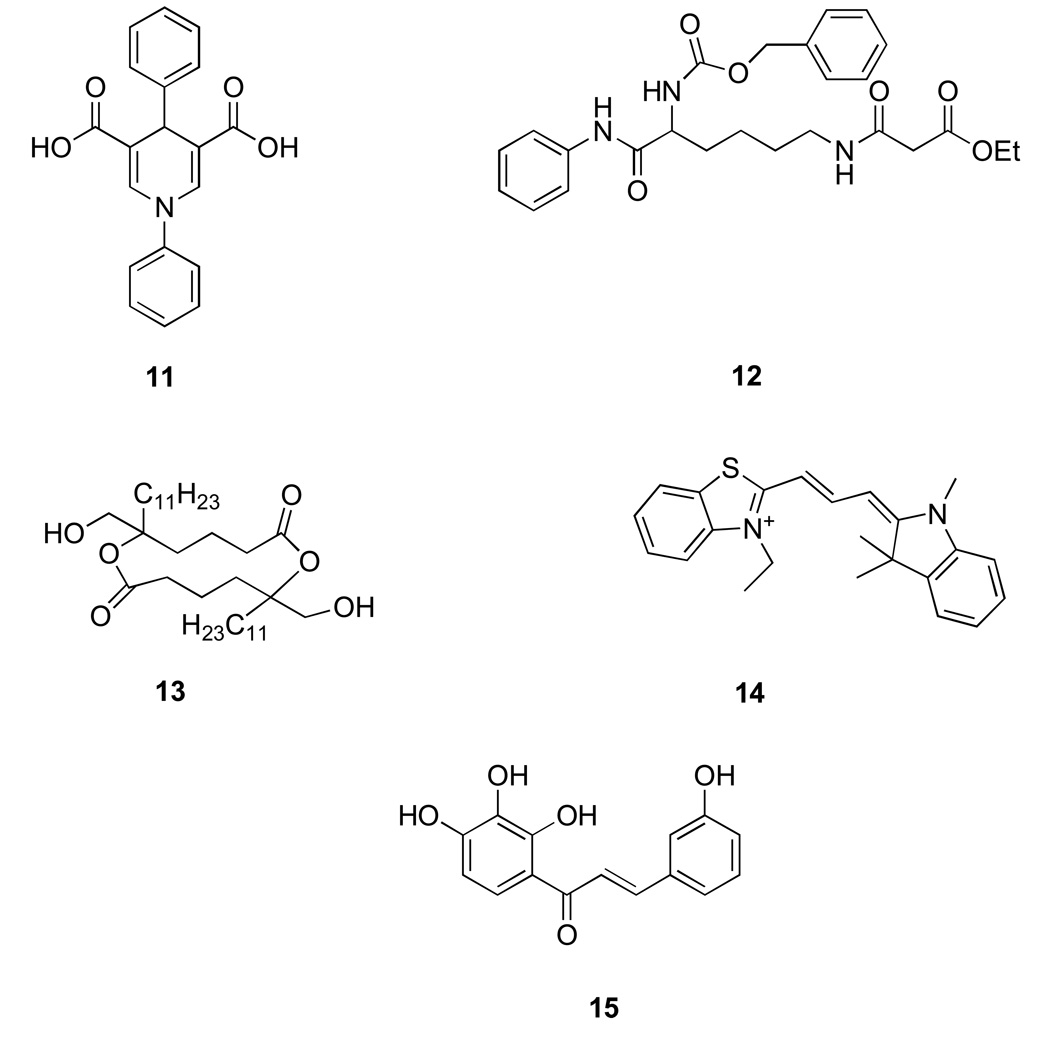
The inhibitors discussed here don’t fall into a particular structural class but hold promise as additional, potent sirtuin inhibitors. The 1,4 dihydropyrdines (11) (Fig. 4) are interesting as a slight variation in substituent pattern results in sirtuin activation rather than inhibition [41]. Compound (12) is novel as it is a mechanism-based inhibitor of sirtuin. The enolate at the lysine mimetic group attacks NAD+ at the active site of sirtuins forming a stable inhibitor-ADP-ribose conjugate that inhibits the enzyme in low micromolar range [42]. Tanikolide dimers (13) and AC-93253 (14) are selective and potent SIRT2 inhibitors (3–6 µM) [43, 44]. Polyphenols such as the chalcone (15) have also shown to inhibit sirtuins both in vitro and in vivo [45].
Figure 3. Miscellaneous Sirtuin inhibitors.
11. Dihydropyridine 12. Lysine-mimetic mechanism based inhibitor 13. Tankolide dimer 14. AC-93253 15. Chalcone
It should be noted that the in vitro IC50 reported for some of the inhibitors discussed above was measured solely using assays that employ fluorophor-containing substrates that do not always faithfully reproduce deacetylation of the native substrates [46, 47]. While the discrepancy between deacetylation of native and flurophore-containing substrates has been particularly evident for putative sirtuin activators (see below), it is not clear yet if this is problem for sirtuin inhibitors. It has therefore become imperative to confirm the IC50 using multiple methods such as the radio-labeled, HPLC or mass spectrometry assays and followed by validation in cell-based assays using acetyl p53 (for SIRT1) or tubulin acetylation (for SIRT2) as a readout. A major drawback is that these alternative methods have limited high throughput capabilities and has therefore thwarted their implementation routinely.
Despite the progress there is still a great need to develop both more potent non-selective as well a selective sirtuin inhibitors. While different sirtuins and the members of class I and II deacetylases (HDACs) have distinct substrates that define their cellular roles, the division of labor among these enzymes is not absolute. Several acetylated proteins have been found to be deacetylated by more than one member within a family or by the enzymes belonging to different families. For example, lysine 16 of the histone H4 has been shown to be deacetylated both with SIRT1 and SIRT2 [48, 49], whereas tubulin is deacetylated by both SIRT2 and HDAC6 [50, 51]. The functional and substrate overlap among different deacetylases has important implications in designing and using small molecule inhibitors of these enzymes for therapeutic purposes and for dissecting biology as the inhibitors that are highly specific may not fully ablate specific cellular roles of a given deacetylase. Therefore, less selective inhibitors may be advantageous for altering cellular function and bringing therapeutic benefit. By the same token, cancer therapies may involve combination of inhibitors of different deacetylase classes (e.g. sirtuin and HDACs). In contrast, highly selective inhibitors may be advantageous over nonselective ones as their side effects are expected to be lessened by the relatively unperturbed function of the backup systems. Therefore, it will be important to develop both highly selective as well broadly active inhibitors for evaluating sirtuins as therapeutic targets.
Sirtuin Activators
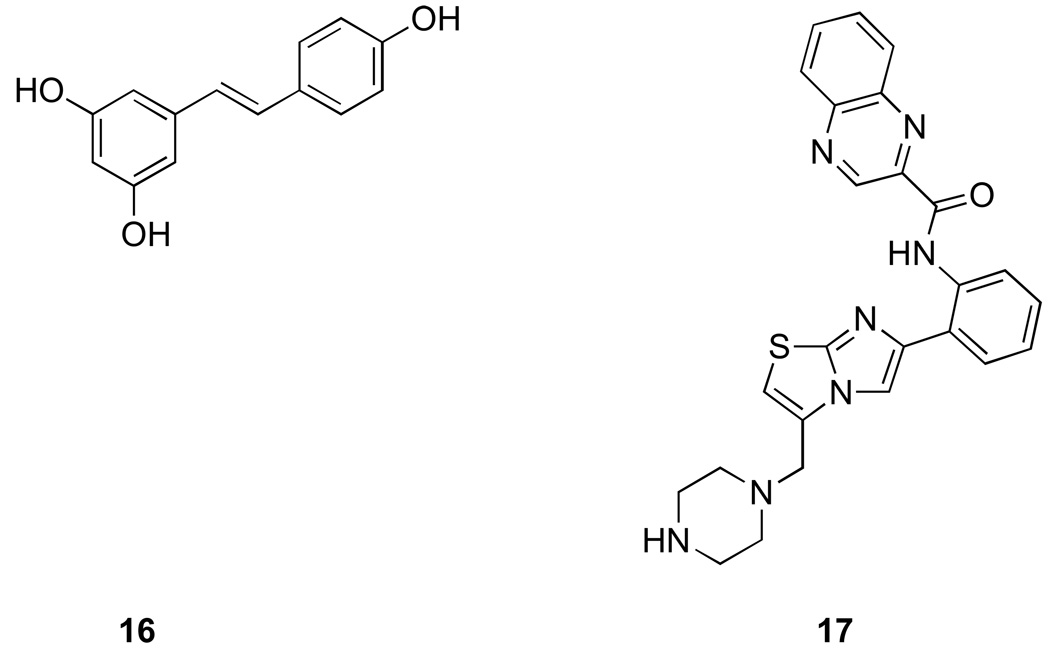
First activators of SIRT1 were identified by high throughput biochemical screening using a commercially available fluorescent deacetylation assay by Howitz et al. in 2003 [52]. The most potent activator identified from this screen was resveratrol (16) (Fig. 5), a polyphenol commonly found in red wine and associated with beneficial effects of wine in general. Resveratrol was shown to decrease the Km of the peptide substrate and also NAD+ without any significant effect on Vmax suggesting that it is an allosteric effector of SIRT1. In the initial report resveratrol was also shown to increase yeast replicative life span in a Sir2 dependent fashion. This work resulted in tremendous excitement and led to many studies that evaluated a resveratrol as an anti-aging drug and calorie restriction mimetic [53–55].
Figure 4. Sirtuin activators.
16. Resveratrol 17. SRT1720
Two independent reports in 2005 raised the possibility that the in vitro effects of resveratrol on SIRT1-catalyzed deacetylation may be related to the substrate that was used in the initial screen fluorophore [46, 47]. The screen that led to identification of activators employed as a substrate peptide containing acetyl-lysine and a coumarin fluorophore [52]. SIRT1 deacetylation of the peptide enabled trypsin to cleave the peptide releasing the fluorophore, which was then capable of light emission. The two reports demonstrated that this substrate peptide has significantly lower affinity for SIRT1 compared to the same peptide without a fluorophore [46, 47]. Resveratrol specifically increases the affinity and deacetylation of the fluorophore containing peptide while having no influence on the same peptide without the fluorophore. Furthermore, increased deacetylation of the fluorophore-containing substrate appeared to be specific to SIRT1 as neither human SIRT2 nor yeast Sir2 were activated by resveratrol even when the fluorophore-containing peptides was used. Consistent with the lack of effect on yeast Sir2 (regardless of the presence or absence of fluorophore) and in contrast to the initial report [52], in a study by Kaberlein et al [46], resveratrol did not extend yeast replicative lifespan. These findings raised the possibility that the activation of SIRT1 by resveratrol may be an artifact of the biochemical screen used for their identification.
In 2007, synthetic compounds structurally unrelated to resveratrol were also reported to activate SIRT1 (e.g. SRT1720, SRT2183) [56]. These compounds were collectively named STACs for SIRT1 activating compounds. STACs were shown activate SIRT1 at submicromolar concentrations and increase SIRT1 catalytic activity several hundred fold [56]. Furthermore, these compounds were orally bioavailable and were reported to lead to improvement of several metabolic parameters in mice fed a high fat diet and other diabetes model systems. Based on these encouraging results, human clinical trials with several synthetic activators as well as with reseveratrol had been initiated. Similarly to resveratrol, these compounds were proposed to act as allosteric enzyme activators by increasing binding of the peptide substrates and mapping studies using truncated forms of enzyme revealed that small helical domain of SIRT1 was required for their activity.
However, because STACs were also identified using a fluorophore-containing substrate via fluorescence polarization assay and characterized by mass spectrometry with the same substrate, there was a possibility that these compounds may suffer from the same shortcomings as resveratrol. Indeed, in 2010 a detailed characterization of these compounds using biochemical assays was published by Pachelec et al [57], and revealed that apparent activation of SIRT1 enzymatic activity was completely dependent on the presence of the fluorophore in the substrate. While extremely effective in deacetylating acetyl-TAMRA-p53 peptide, SRT1720 (17) and related compounds failed to activate deacetylation of any substrates that did not contain a fluorophore including native acetylated p53 peptide, full length acetylated p53 protein or acetyl-CoA synthetase 1. In the same report, the authors went on to demonstrate, using NMR and Surface Plasmon Resonance, that SRT1720 binds directly to TAMRA peptide (but not to native peptides) and that binding occurs in the absence of SIRT1. These findings argue against a model that putative SIRT1 activators bind directly to SIRT1 and act as allosteric activators that are specific for deacetylation of fluorophore containing substrates (an increase in deacetylation of native substrates has never been demonstrated). Rather, the most plausible model, for which the study by Pacholec et al provides solid experimental support, is that STATs bind to the substrate in a fluorophore dependent fashion, which promotes substrate deacetylation. The fluorophore-dependent binding of the STATs to the substrate provides a more straightforward explanation for the substrate selectivity than the allostery model in which binding of the putative activators would specifically promote deacetylation of the fluorophore but not the native substrates. Regardless of the model, it appears that apparent SIRT1 activation by small molecule ligands including resveratrol is a screening artifact.
If resveratrol and STATs do not affect sirtuin activity, what accounts for the reported in vivo effects of these agents and resveratrol? First, as discussed above, the initial in vivo result that resveratrol extends yeast lifespan has been called into question [46]. Likewise, the reexamination of STATs activity in mouse diabetes models by Pacholec et al. did not reproduce glucose-lowering activity or improvements in mitochondrial capacity [57] that were shown in the initial report [56]. At the present time, the reasons for the discrepancy for the in vivo results are not clear. However, it is an important issue that will need to be resolved, particularly in light of clinical trials that have been initiated with this class of compounds. Second, resveratrol has many other proposed cellular targets that could account for the various metabolic benefits observed with this compound [58]. While it has been proposed that some of these effects (e.g. stimulation of AMP-activated protein kinase) could indirectly promote sirtuin activity [59], the multitude of cellular targets makes it difficult to determine which of these is mediating the desired biological effects.
Beside resveratrol and other compounds that were thought to activate SIRT1 by promoting peptide binding, a different mechanism has been described for isonicotinamide (11), which was shown to activate yeast Sir2 through relief of nicotinamide inhibition [60]. As discussed previously nicotinamide is an endogenous sirtuin inhibitor that promotes the chemical reversal of the covalent reaction intermediates and generation of NAD+ and acetyl lysine resulting in nicotinamide exchange. Isonicotinmide is competitive with nicotinamide in the exchange reaction thus promoting deacetylation both in vitro and in vivo.
The multitude of the protein targets and physiological roles that SIRT1 has in different tissues as well as complex regulation of SIRT1 protein level by nutritional cues makes it difficult to predict the net benefit of promoting or inhibiting SIRT1 activity on metabolism in diabetes. It should be noted however that, while extremely valuable, rodent models have only limited value in prediction the utility of drug targets in metabolic diseases in humans. The role of SIRT1 inhibitors and activators in the treatment of diabetes and obesity associated metabolic syndrome will ultimately be decided though clinical trials.
Ackowledgments
This work was supported by the National Institute of Health grant CA129132 to AB.
References
- 1.Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14(9):1021–1026. [PubMed] [Google Scholar]
- 2.Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183(4677):1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- 3.Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91(7):1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 4.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410(6825):227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 5.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101(45):15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005;6(4):298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 7.Kaeberlein M, et al. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2(9):E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci. 2010;31(5):212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanner KG, et al. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product-1-O-acetyl-ADP-ribose. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(26):14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim EJ, et al. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;28(2):277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 11.Zhao W, et al. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451(7178):587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogan KL, Brenner C. Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu Rev Nutr. 2008;28:115–130. doi: 10.1146/annurev.nutr.28.061807.155443. [DOI] [PubMed] [Google Scholar]
- 13.Bedalov A, et al. NAD-dependent deacetylase Hst1p controls biosynthesis and cellular NAD levels in Saccharomyces cerevisiae. Mol Cell Biol. 2003;23(19) doi: 10.1128/MCB.23.19.7044-7054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ha HC, Snyder SH. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(24):13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson RM, et al. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. Journal of Biological Chemistry. 2002;277(21):18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 16.Anderson RM, et al. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423(6936):181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279(49):50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 18.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324(5927):651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakahata Y, et al. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324(5927):654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin SJ, et al. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18(1):12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fulco M, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003 doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 22.Prozorovski T, et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat Cell Biol. 2008;10(4):385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt MT, et al. Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J Biol Chem. 2004;279(38):40122–40129. doi: 10.1074/jbc.M407484200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q, et al. Metabolic regulation of SIRT1 transcription via a HIC1:CtBP corepressor complex. Proc Natl Acad Sci U S A. 2007;104(3):829–833. doi: 10.1073/pnas.0610590104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434(7029):113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 26.Bedalov A, et al. Identification of a small molecule inhibitor of Sir2p. Proc Natl Acad Sci U S A. 2001;98:15113–15118. doi: 10.1073/pnas.261574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grozinger CM, et al. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J Biol Chem. 2001;276(42):38837–38843. doi: 10.1074/jbc.M106779200. [DOI] [PubMed] [Google Scholar]
- 28.Lara E, et al. Salermide, a Sirtuin inhibitor with a strong cancer-specific proapoptotic effect. Oncogene. 2009;28(6):781–791. doi: 10.1038/onc.2008.436. [DOI] [PubMed] [Google Scholar]
- 29.Posakony J, et al. Inhibitors of Sir2: evaluation of splitomicin analogues. J Med Chem. 2004;47(10):2635–2644. doi: 10.1021/jm030473r. [DOI] [PubMed] [Google Scholar]
- 30.Neugebauer RC, et al. Structure-activity studies on splitomicin derivatives as sirtuin inhibitors and computational prediction of binding mode. J Med Chem. 2008;51(5):1203–1213. doi: 10.1021/jm700972e. [DOI] [PubMed] [Google Scholar]
- 31.Hirao M, et al. Identification of selective inhibitors of NAD+-dependent deacetylases using phenotypic screens in yeast. J Biol Chem. 2003;278(52):52773–52782. doi: 10.1074/jbc.M308966200. [DOI] [PubMed] [Google Scholar]
- 32.Heltweg B, et al. Antitumor activity of a small molecule inhibitor of human Sir2 enzymes. Cancer Res. 2006;66(8):4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 33.Medda F, et al. Novel cambinol analogs as sirtuin inhibitors: synthesis, biological evaluation, and rationalization of activity. J Med Chem. 2009;52(9):2673–2682. doi: 10.1021/jm8014298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Napper AD, et al. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J Med Chem. 2005;48(25):8045–8054. doi: 10.1021/jm050522v. [DOI] [PubMed] [Google Scholar]
- 35.Trapp J, et al. Adenosine mimetics as inhibitors of NAD+-dependent histone deacetylases, from kinase to sirtuin inhibition. J Med Chem. 2006;49(25):7307–7316. doi: 10.1021/jm060118b. [DOI] [PubMed] [Google Scholar]
- 36.Huber K, et al. Novel 3-arylideneindolin-2-ones as inhibitors of NAD+ -dependent histone deacetylases (sirtuins) J Med Chem. 2010;53(3):1383–1386. doi: 10.1021/jm901055u. [DOI] [PubMed] [Google Scholar]
- 37.Voogd TE, et al. Recent research on the biological activity of suramin. Pharmacol Rev. 1993;45(2):177–203. [PubMed] [Google Scholar]
- 38.Trapp J, et al. Structure-activity studies on suramin analogues as inhibitors of NAD+-dependent histone deacetylases (sirtuins) ChemMedChem. 2007;2(10):1419–1431. doi: 10.1002/cmdc.200700003. [DOI] [PubMed] [Google Scholar]
- 39.Schuetz A, et al. Structural basis of inhibition of the human NAD+-dependent deacetylase SIRT5 by suramin. Structure. 2007;15(3):377–389. doi: 10.1016/j.str.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Lain S, et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell. 2008;13(5):454–463. doi: 10.1016/j.ccr.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mai A, et al. Study of 1,4-dihydropyridine structural scaffold: discovery of novel sirtuin activators and inhibitors. J Med Chem. 2009;52(17):5496–5504. doi: 10.1021/jm9008289. [DOI] [PubMed] [Google Scholar]
- 42.Asaba T, et al. Inhibition of human sirtuins by in situ generation of an acetylated lysine-ADP-ribose conjugate. J Am Chem Soc. 2009;131(20):6989–6996. doi: 10.1021/ja807083y. [DOI] [PubMed] [Google Scholar]
- 43.Gutierrez M, et al. Structural and synthetic investigations of tanikolide dimer, a SIRT2 selective inhibitor, and tanikolide seco-acid from the Madagascar marine cyanobacterium Lyngbya majuscula. J Org Chem. 2009;74(15):5267–5275. doi: 10.1021/jo900578j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, et al. Identification of a small molecule SIRT2 inhibitor with selective tumor cytotoxicity. Biochem Biophys Res Commun. 2009;386(4):729–733. doi: 10.1016/j.bbrc.2009.06.113. [DOI] [PubMed] [Google Scholar]
- 45.Kahyo T, et al. A novel chalcone polyphenol inhibits the deacetylase activity of SIRT1 and cell growth in HEK293T cells. J Pharmacol Sci. 2008;108(3):364–371. doi: 10.1254/jphs.08203fp. [DOI] [PubMed] [Google Scholar]
- 46.Kaeberlein M, et al. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280(17):17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 47.Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005;280(17):17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 48.Vaquero A, et al. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16(1):93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 49.Vaquero A, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20(10):1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hubbert C, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417(6887):455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 51.North BJ, et al. The human Sir2 ortholog, SIRT2, is an NAD dependnet tubulin deacetylase. Mol Cell. 2003;11(2):437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 52.Howitz KT, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425(6954):191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 53.Wood JG, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430(7000):686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 54.Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127(6):1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 55.Baur JA, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444(7117):337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Milne JC, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450(7170):712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pacholec M, et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2010;285(11):8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harikumar KB, Aggarwal BB. Resveratrol: a multitargeted agent for age-associated chronic diseases. Cell Cycle. 2008;7(8):1020–1035. doi: 10.4161/cc.7.8.5740. [DOI] [PubMed] [Google Scholar]
- 59.Feige JN, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8(5):347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 60.Sauve AA, et al. Chemical activation of Sir2-dependent silencing by relief of nicotinamide inhibition. Mol Cell. 2005;17(4):595–601. doi: 10.1016/j.molcel.2004.12.032. [DOI] [PubMed] [Google Scholar]