ABSTRACT
Though the bacterial opportunist Enterococcus faecalis causes a myriad of hospital-acquired infections (HAIs), including catheter-associated urinary tract infections (CAUTIs), little is known about the virulence mechanisms that it employs. However, the endocarditis- and biofilm-associated pilus (Ebp), a member of the sortase-assembled pilus family, was shown to play a role in a mouse model of E. faecalis ascending UTI. The Ebp pilus comprises the major EbpC shaft subunit and the EbpA and EbpB minor subunits. We investigated the biogenesis and function of Ebp pili in an experimental model of CAUTI using a panel of chromosomal pilin deletion mutants. A nonpiliated pilus knockout mutant (EbpABC− strain) was severely attenuated compared to its isogenic parent OG1RF in experimental CAUTI. In contrast, a nonpiliated ebpC deletion mutant (EbpC− strain) behaved similarly to OG1RF in vivo because it expressed EbpA and EbpB. Deletion of the minor pilin gene ebpA or ebpB perturbed pilus biogenesis and led to defects in experimental CAUTI. We discovered that the function of Ebp pili in vivo depended on a predicted metal ion-dependent adhesion site (MIDAS) motif in EbpA’s von Willebrand factor A domain, a common protein domain among the tip subunits of sortase-assembled pili. Thus, this study identified the Ebp pilus as a virulence factor in E. faecalis CAUTI and also defined the molecular basis of this function, critical knowledge for the rational development of targeted therapeutics.
IMPORTANCE
Catheter-associated urinary tract infections (CAUTIs), one of the most common hospital-acquired infections (HAIs), present considerable treatment challenges for physicians. Inherently resistant to several classes of antibiotics and with a propensity to acquire vancomycin resistance, enterococci are particularly worrisome etiologic agents of CAUTI. A detailed understanding of the molecular basis of Enterococcus faecalis pathogenesis in CAUTI is necessary for the development of preventative and therapeutic strategies. Our results elucidated the importance of the E. faecalis Ebp pilus and its subunits for enterococcal virulence in a mouse model of CAUTI. We further showed that the metal ion-dependent adhesion site (MIDAS) motif in EbpA is necessary for Ebp function in vivo. As this motif occurs in other sortase-assembled pili, our results have implications for the molecular basis of virulence not only in E. faecalis CAUTI but also in additional infections caused by enterococci and other Gram-positive pathogens.
Introduction
In recent decades, Enterococcus faecalis and Enterococcus faecium, commensal gut bacteria, have emerged as human pathogens (1). Enterococci frequently cause hospital-acquired and device-associated infections, including bloodstream infections, infective endocarditis, and catheter-associated urinary tract infections (CAUTIs) (2). As the most common hospital-acquired infection (HAI) (3) and because they are frequently and often unnecessarily treated with antibiotics (4), CAUTIs are a reservoir of nosocomial and antimicrobial-resistant pathogens (5). Due to the tremendous incidence of CAUTI, infrequent but life-threatening sequelae such as bacteremia and urosepsis are significant complications (6). Enterococci are responsible for roughly 15% of CAUTIs (7). The recent surge of enterococcal infections correlates with both their inherent and acquired antimicrobial resistances, including vancomycin resistance (2). Both rising antimicrobial resistance and bacterial biofilm formation on indwelling catheters, abiotic surfaces protected from host immune responses and antibiotics, contribute to the difficulty associated with successful CAUTI treatment. Thus, a better understanding of bacterial pathogenesis in CAUTI is critical for the development of preventative and therapeutic antimicrobial agents.
Among the few described enterococcal virulence factors is the endocarditis- and biofilm-associated pilus (Ebp) (8). The Ebp pilus is an example of the sortase-assembled pilus family, whose members are now described among diverse Gram-positive genera, including Corynebacterium (9), Actinomyces (10), Streptococcus (11–13), Bacillus (14), Enterococcus (8, 15, 16), Lactobacillus (17), and Bifidobacterium (18). Sortase-assembled pili consist of a major pilin subunit and up to two minor subunits, each with a C-terminal cell wall sorting signal (CWSS) that includes an LPXTG-like sortase recognition motif (19). One or more genetically linked membrane-associated transpeptidase enzymes, pilus-associated sortases, catalyze the formation of interpilin isopeptide bonds found in mature pili (20). Repeating, covalently linked major pilin subunits comprise the bulk of the pilus fiber. When present, a second minor subunit is proposed to localize to the fiber tip and a third subunit is proposed to localize to the base (19). Respectively, these ancillary pilins may facilitate interaction with host proteins and the anchoring of pilus fibers to the cell wall via processing by the housekeeping sortase. Though dispensable for major pilin polymerization, minor subunits have been shown to affect aspects of pilus biogenesis, including pilus length, thickness, and subcellular compartmentalization in several sortase-assembled pilus systems (21, 22). Characteristically, the E. faecalis ebp operon encodes three structural proteins, EbpA, EbpB, and EbpC, and the pilus-associated sortase SrtC (or Bps) (8). A housekeeping sortase, SrtA, is encoded elsewhere in the genome (23). Ebp pilus fibers consist largely of the major pilin EbpC, whose polymerization is mediated by SrtC. EbpA and EbpB are covalently incorporated into the cell wall-anchored Ebp pilus fibers as minor components (8), but their roles in pilus biogenesis and pilus function have not been directly investigated previously.
Sortase-assembled pili or their subunits have been reported as virulence factors in vivo in infection models of group B streptococci (GBS; Streptococcus agalactiae) (24–26), group A streptococci (GAS; Streptococcus pyogenes) (27, 28), and the pneumococcus (Streptococcus pneumoniae) (13). A polar, nonpiliated E. faecalis ebp mutant was attenuated in animal models of infective endocarditis (8) and ascending UTI (29), as were E. faecalis mutants lacking SrtC or SrtA (30) and a nonpiliated E. faecium ebp mutant (31). A mutant lacking SrtA was also attenuated in the model of CAUTI used in this study (32), but the role of Ebp pili in CAUTI has not been directly investigated. Though the function of Ebp pili in pathogenesis is unclear, in vitro studies suggested that Ebp pili were important for static biofilm formation (8) and adherence to extracellular matrix (ECM) proteins (33) and human platelets (34). However, the unique contributions of EbpA, EbpB, or EbpC to any described Ebp pilus function have not been explored.
In this report, we hypothesized that the Ebp pilus is a virulence factor in CAUTI. To test this hypothesis, we investigated its importance for E. faecalis pathogenesis in a mouse model that mimics many aspects of human CAUTI (32). We evaluated the role of each Ebp structural subunit in pilus biogenesis and in virulence in CAUTI, revealing that the minor pilins mediated pilus function in vivo. Specifically, the metal ion-dependent adhesion site (MIDAS) motif in EbpA’s predicted von Willebrand factor A (VWA) domain, a common domain among sortase-assembled pili (22), was critical for E. faecalis virulence in CAUTI. Thus, this study elucidated the molecular basis of Ebp pilus function in experimental E. faecalis CAUTI.
RESULTS
Ebp pili are important in a mouse model of CAUTI.
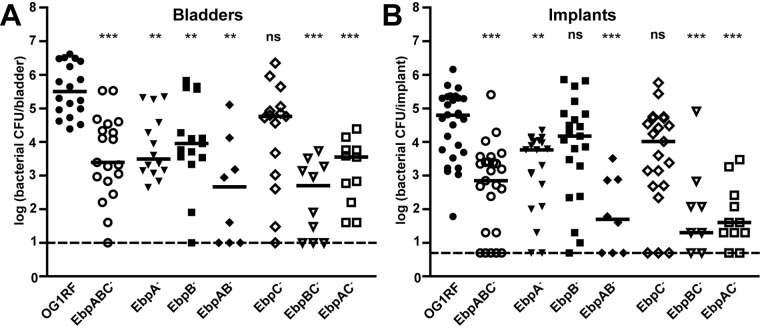
We used a previously described mouse model of E. faecalis CAUTI (32) and a chromosomal deletion mutant lacking all three Ebp structural pilins (EbpABC− strain) to investigate the importance of Ebp pili in pathogenesis. Bladders of female mice were implanted with a short segment of silicone tubing (implant) to mimic urinary catheterization and subsequently infected with the E. faecalis OG1RF or EbpABC− strain. Twenty-four hours postinfection (p.i.), the bacterial burdens in the bladders and those associated with the implants were determined as a measure of virulence. Median CFU recovered from the bladders (Fig. 1A) and implants (Fig. 1B) of mice infected with the EbpABC− strain were significantly lower than those recovered from the bladders and implants of mice infected with OG1RF. These results demonstrated that Ebp pili were important for E. faecalis pathogenesis in experimental CAUTI.
FIG 1 .
Virulence of E. faecalis OG1RF and its isogenic pilin deletion mutants in experimental CAUTI. Twenty-four-hour viable bacterial titers in the bladders (A) and those associated with implants (B) from 2 to 4 independent experiments/strain are shown. Each shape corresponds to one mouse; open shapes represent nonpiliated bacterial strains. Median titers (CFU/bladder, CFU/implant) are shown with a bar: OG1RF (3.20 × 105, 6.25 × 104), EbpABC− strain (2.48 × 103, 7.00 × 102), EbpA− strain (3.10 × 103, 5.82 × 103), EbpB− strain (9.08 × 103, 1.50 × 104), EbpAB− strain (4.60 × 102, 50), EbpC− strain (5.80 × 104, 1.04 × 104), EbpBC− strain (5.00 × 102, 20), and EbpAC− strain (3.56 × 103, 40). Dashed lines are the limits of detection (10 CFU/bladder, 5 CFU/implant). Results of statistical comparisons of each mutant to OG1RF for bladders and implants are shown. There were no significant differences in bladder or implant colonization between any double pilin deletion mutant and the EbpABC− strain (data not shown). P values were adjusted for 10 comparisons (**, P < 0.01; ***, P < 0.001; ns, not significant).
To identify the functional subunit critical for pilus-mediated virulence in CAUTI, we created a panel of unmarked, in-frame single and double pilin deletion mutants. In the initial steps of this analysis, we evaluated (i) EbpC assembly and expression in minor pilin deletion mutants (EbpA−, EbpB−, and EbpAB− strains) and (ii) expression of minor pilin(s) in nonpiliated ebpC deletion mutants (EbpC−, EbpBC−, and EbpAC− strains).
Expression of EbpA and EbpB in the absence of pilus fibers.
EbpC is the major pilus subunit necessary for fiber polymerization (8). Thus, we evaluated minor pilin expression in the absence of EbpC in Western blot analyses using anti-EbpA and anti-EbpB sera after reducing SDS-PAGE. In EbpC− cell lysates, we observed two EbpA doublets migrating at approximately 140 kDa and 100 kDa (Fig. 2A, open arrowhead and asterisk, respectively). We observed only one EbpB species migrating at a size similar to the larger ~140-kDa EbpA species (Fig. 2B, open arrowhead). Because the ~140-kDa species recognized by both anti-EbpA and anti-EbpB sera was larger than the predicted molecular masses for either EbpA or EbpB monomer after cleavage of its signal sequence and CWSS (~120 and ~46 kDa, respectively), we hypothesized that this species likely represents an EbpA-EbpB heterodimer. As formation of autocatalytic intramolecular isopeptide bonds has been shown to change the mobility of other sortase-assembled pilus subunits on SDS-PAGE (35), we predict that the ~100-kDa EbpA species is the fully processed EbpA monomer.
FIG 2 .
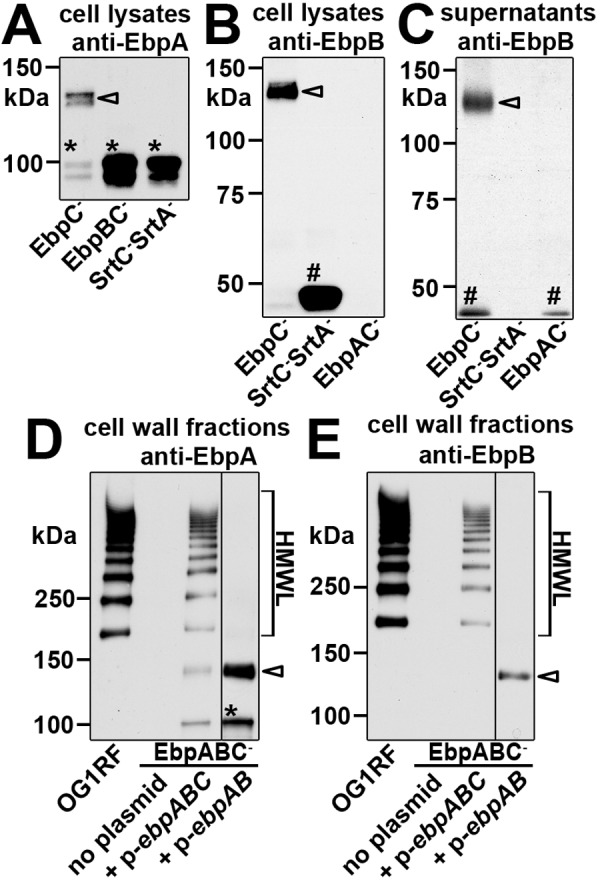
EbpA and EbpB expression in nonpiliated strains. Western blot analyses were performed after SDS-PAGE of the indicated bacterial strains and fractions using anti-EbpA (A and D) or anti-EbpB (B, C, and E) sera. Open arrowheads indicate the ~140-kDa EbpA and EbpB species observed in the EbpC− strain (A to C) and the EbpABC−/p-ebpAB strain (D and E). Asterisks show the ~100-kDa EbpA monomer in EbpC−, EbpBC−, and SrtC− SrtA− strains (A) and in the EbpABC−/p-ebpAB strain (D). Hash marks show the EbpB monomer in EbpC−, EbpAC−, and SrtC− SrtA− strains (B and C) and in the EbpABC−/p-ebpAB strain (E). Brackets indicate pilus HMWLs observed in OG1RF and the EbpABC−/p-ebpABC strain.
To test this hypothesis, we analyzed EbpA and EbpB expression in a mutant lacking all sortase genes (SrtC− SrtA− strain) to prevent formation of interpilin isopeptide bonds. In Western blots of SrtC− SrtA− cell lysates, the ~100-kDa EbpA species predominated (Fig. 2A, asterisk). The same result was obtained with our SrtC− mutant (data not shown), consistent with prior reports for OG1RF (8) and E. faecalis OG1X (36). Furthermore, we observed the ~100-kDa EbpA species in cell lysates of the EbpBC− strain (Fig. 2A, asterisk).
The ~140-kDa species was also absent when anti-EbpB sera were used to probe Western blots of SrtC− SrtA− (Fig. 2B and 2C), SrtC− (data not shown), and EbpAC− (Fig. 2B and 2C) strains. In these strains, EbpB migrated just below the 50-kDa protein marker, consistent with its predicted monomer size (Fig. 2B and 2C, hash marks). Thus, formation of the ~140-kDa EbpA and EbpB species observed in the EbpC− strain depended on EbpB and EbpA, respectively, and SrtC, consistent with the formation of an EbpA-EbpB heterodimer. Interestingly, we found EbpB monomers in the culture supernatants of OG1RF (Fig. 3B) and pilin deletion mutants: EbpA− (Fig. 3B) and EbpC− and EbpAC− (Fig. 2C) strains. However, in the SrtC− SrtA− mutant, EbpB monomers were observed only in cell lysates (Fig. 2B), suggesting sortase-dependent dissociation of EbpB monomers from bacterial cells.
FIG 3 .
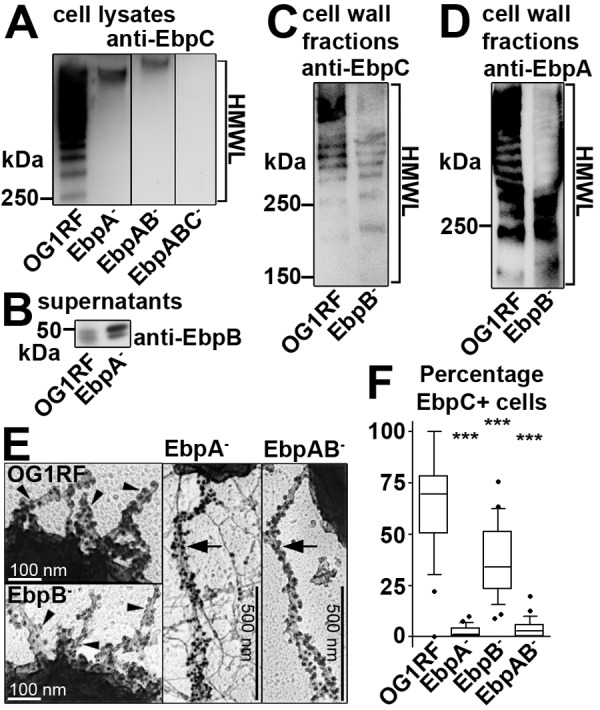
Minor pilin deletions affect pilus biogenesis. Anti-EbpC sera were used to assess pilus assembly by Western blot analysis (A to D), to visualize pilus morphology with deep-etch EM (E), and to evaluate population piliation dynamics by IFM (F). (A) Western blot analysis of EbpA− and EbpAB− cell lysates showed condensed EbpC bands with reduced mobility on SDS-PAGE compared to OG1RF EbpC HMWL. (B) EbpB is expressed in OG1RF and EbpA− culture supernatants. (C and D) EbpC HMWL (C) and EbpA HMWL (D) in EbpB− and OG1RF strains were similar. HMWLs (brackets) indicate pilus polymerization (anti-EbpC) or minor pilin incorporation (anti-EbpA). (E) Pilus morphology was altered in EbpA− and EbpAB− strains. Arrowheads point to gold bead-labeled pilus fibers in OG1RF and EbpB− strains. Large arrows indicate gold bead-labeled long EbpC fibers in EbpA− and EbpAB− strains. (F) The percentage of bacterial cells expressing EbpC (EbpC+ cells) was quantified in 3 independent experiments. The median percentages of EbpC+ cells for each strain were determined: OG1RF (69.6), EbpA− strain (1.1), EbpB− strain (33.9), and EbpAB− strain (2.8). Whiskers show the 10th and 90th percentiles; dots show outliers. Statistically significant differences between OG1RF and each mutant strain are shown; P values were adjusted for 3 comparisons (***, P < 0.001).
Finally, complementation of the EbpABC− strain with ebpA, ebpB, and ebpC provided in trans resulted in incorporation of EbpA and EbpB into high-molecular-weight pilus ladders (HMWLs) in cell wall fractions, similar to those seen in OG1RF (Fig. 2D and 2E, respectively). However, when the EbpABC− strain was provided with only ebpA and ebpB in trans, EbpA and EbpB expression mirrored that seen in the EbpC− strain: ~140-kDa and ~100-kDa EbpA species (Fig. 2D, open arrowhead and asterisk) and ~140-kDa EbpB species (Fig. 2E, open arrowhead). Taken together, these data suggest that an EbpA-EbpB heterodimer is formed by SrtC and anchored to the cell wall in the absence of EbpC.
Chromosomal deletion of minor pilins perturbs Ebp biogenesis.
Others have observed that deletion of minor subunits from other sortase-assembled pilus islands can affect aspects of pilus biogenesis, including morphology and population piliation dynamics. Thus, we analyzed pilus biogenesis in the EbpA−, EbpB−, and EbpAB− minor pilin deletion mutants using anti-EbpC sera. Western blots showed that deletion of either ebpA or ebpB did not prevent expression of EbpC (Fig. 3A and 3C, respectively) or expression of the remaining minor subunit (Fig. 3B and 3D, respectively). However, the EbpC species detected in EbpA− and EbpAB− cell lysates was a compressed band that barely migrated into gels (Fig. 3A). This differed from the EbpC pilus HMWL observed in OG1RF. In contrast, EbpC HMWLs from the EbpB− and OG1RF strains were indistinguishable (Fig. 3C). To test whether ebpA provided in trans could restore OG1RF-like EbpC HMWLs in EbpA− and EbpAB− strains, ebpA was placed under the control of a tetracycline-inducible promoter on a plasmid. High- and low-level EbpA expression was observed from this plasmid with induction (100 µg ml−1 anhydrotetracycline) and no induction, respectively, in the EbpABC− strain. However, EbpA expressed from this plasmid with or without induction was not incorporated into and did not affect compressed EbpC bands in the EbpA− and EbpAB− strains (data not shown). Thus, compressed EbpC bands caused by deletion of ebpA indicated altered pilus morphology that could not be complemented in trans.
We next investigated pilus morphology in minor pilin deletion mutants using deep-etch immunogold electron microscopy (EM). In the OG1RF and EbpB− strains, multiple pilus fibers several hundred nanometers long were observed associated with bacterial cells (Fig. 3E, arrowheads). In contrast, in cultures of the EbpA− and EbpAB− strains, we observed fewer but much longer EbpC fibers (greater than 1 µm) associated with some cells (Fig. 3E, large arrows). An analysis of population piliation dynamics using immunofluorescence microscopy (IFM) also revealed defects of the minor pilin deletion strains in pilus biogenesis. A significantly smaller median proportion of EbpA−, EbpB−, and EbpAB− bacterial cells (1.1%, 33.9%, and 2.8%, respectively) expressed EbpC at their surface than did OG1RF (65.8%) (Fig. 3F). Thus, deletion of minor pilin coding sequences in OG1RF affected Ebp pilus biogenesis.
Minor pilin deletion mutants are attenuated in experimental E. faecalis CAUTI.
We then investigated the minor pilin deletion mutants in experimental CAUTI. EbpA−, EbpB−, and EbpAB− strains were all significantly attenuated in bladder colonization compared to OG1RF (Fig. 1A). EbpA− and EbpAB− strains were also attenuated in implant colonization (Fig. 1B). The defect of these strains in vivo may have been due to the lack of EbpA and/or EbpB or to the effects of the minor pilin deletions on pilus biogenesis described in the studies above.
Expression of EbpA and EbpB in the EbpC− strain is sufficient for pilus-mediated virulence in CAUTI.
Interestingly, the nonpiliated EbpC− mutant and OG1RF colonized the bladders and implants of infected mice to similar levels (Fig. 1A and 1B), demonstrating that EbpC and pilus fibers were dispensable for virulence in experimental E. faecalis CAUTI. This result suggested that the EbpA-EbpB ~140-kDa species, the ~100-kDa EbpA monomer, or the ~50-kDa EbpB monomer expressed in the EbpC− strain (Fig. 2A to 2C) was sufficient to mediate E. faecalis bladder and implant colonization. To investigate this further, we tested our nonpiliated mutants that expressed pilin monomer(s) but not the EbpA-EbpB heterodimer. In contrast to the EbpC− strain, median bladder and implant titers from mice infected with EbpBC− and EbpAC− strains were significantly lower than those from mice infected with OG1RF (Fig. 1A and 1B), showing that neither the EbpA nor the EbpB monomer alone was sufficient for pilus-mediated virulence in experimental CAUTI. Furthermore, we observed a similar defect in bladder and implant colonization by the SrtC− strain (Fig. 4A and 4B), which expresses both EbpA and EbpB monomers but lacks the EbpA-EbpB ~140-kDa species, further supporting a role for the EbpA-EbpB complex in pilus function in CAUTI.
FIG 4 .
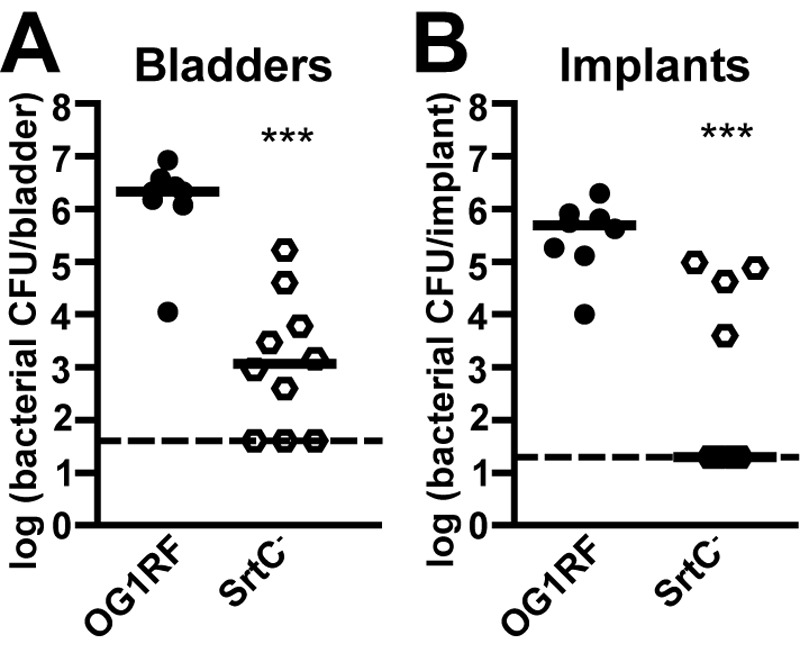
The SrtC− mutant is attenuated in experimental CAUTI. Mice were infected with ~2 × 107 CFU of OG1RF (closed circles) or the nonpiliated SrtC− mutant (open hexagons). Bacterial titers 24 h p.i. in the bladders (A) and implants (B) from 2 independent experiments are shown. Each shape corresponds to one mouse. Median titers (CFU/bladder, CFU/implant) are shown with a bar: OG1RF (2.16 × 106, 4.90 × 105) and SrtC− strain (1.18 × 103, 20). Dashed lines are limits of detection (40 CFU/bladder; 20 CFU/implant). Statistically significant differences between OG1RF and SrtC− titers are shown (***, P < 0.001).
Mutation of EbpA’s MIDAS motif does not affect pilus biogenesis.
Protein domain prediction revealed Cna B domains in the minor pilins and a von Willebrand factor A (VWA) domain with a metal ion-dependent adhesion site (MIDAS) motif (Asp-Xaa-Ser-Xaa-Ser… Thr… Asp) (37) in EbpA. To explore the importance of this motif in Ebp pilus function, we mutated predicted metal ion-coordinating residues of EbpA’s MIDAS motif (Asp315-Trp-Ser317-Gly-Ser319) to Ala (Ala315-Trp-Ala317-Gly-Ala319) and assessed the effect of the mutations on pilus biogenesis and virulence in experimental CAUTI. When a mutant lacking all pilus subunits and srtC (EbpABC− SrtC− strain) was transformed with a plasmid carrying the ebpABCsrtC locus with a mutated MIDAS motif (p-ebpAAWAGABCsrtC), EbpC pilus fibers expressed by the resultant strain were similar to those observed from the EbpABC− SrtC− strain complemented with the unmutated OG1RF locus (p-ebpABCsrtC) as determined by negative-stain immunogold EM (Fig. 5A). We next introduced the mutant MIDAS motif into the chromosomal ebpA locus in OG1RF. Pili produced by this MIDAS mutant (EbpAAWAGA) and by OG1RF were also similar when examined with negative-stain immunogold EM (Fig. 5B). Similarly, no difference between EbpAAWAGA and OG1RF was observed in an IFM analysis of population piliation dynamics (Fig. 5C). Furthermore, HMWLs of EbpAAWAGA and OG1RF were indistinguishable on Western blots probed with any antipilin immune serum (Fig. 5D). Thus, mutating EbpA’s MIDAS motif did not affect pilus biogenesis.
FIG 5 .
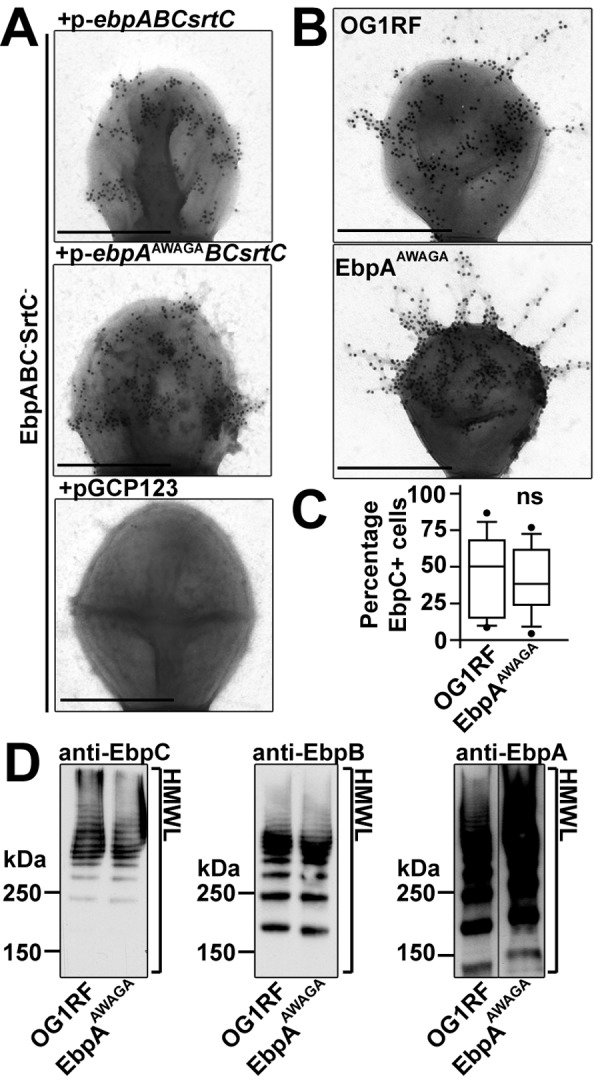
Mutation of EbpA’s MIDAS motif does not affect pilus biogenesis. Mouse anti-EbpC polyclonal sera were used to assess pilus biogenesis by negative-stain immunogold EM (A and B), IFM (C), and Western blot analysis (D) after SDS-PAGE of the indicated cell fractions. (A and B) Piliation of the MIDAS motif mutant strains (EbpABC− SrtC−/p-ebpAAWAGABCsrtC strain and EbpAAWAGA) was similar to that of control strains (EbpABC− SrtC−/p-ebpABCsrtC strain and OG1RF, respectively). Bars, 500 nm. (C) Comparison of the median percentages of EbpC+ bacterial cells in OG1RF (50%) and EbpAAWAGA (38%) from 2 independent experiments revealed no significant differences in population piliation dynamics. Whiskers show the 10th and 90th percentiles; dots show outliers (ns, not significant). (D) Pilus HMWLs on Western blots of OG1RF and EbpAAWAGA probed with anti-EbpC (left), anti-EbpB (middle), and anti-EbpA (right) sera were indistinguishable.
EbpA MIDAS motif mutants are attenuated in experimental CAUTI.
We next examined MIDAS motif mutants in experimental E. faecalis CAUTI. There were no significant differences between the median 24-h bladder or implant bacterial titers of mice infected with strains expressing pili with unmodified MIDAS motifs: OG1RF/pGCP123 (empty vector) and EbpABC− SrtC−/p-ebpABCsrtC strain. Twenty-four-hour bacterial titers from mice infected with the nonpiliated EbpABC− SrtC−/pGCP123 strain and with the EbpABC− SrtC−/p-ebpAAWAGABCsrtC MIDAS motif mutant also did not differ significantly from each other. However, infections with either of these strains resulted in significantly lower bladder and implant bacterial burdens 24 h p.i. than those of OG1RF/pGCP123 or the EbpABC− SrtC−/p-ebpABCsrtC strain (Fig. 6A and 6B). Thus, supplying the OG1RF ebpABCsrtC locus to the EbpABC− SrtC− strain in trans fully complemented its virulence defect in CAUTI, while provision of an ebpAAWAGABCsrtC mutant MIDAS motif locus in trans resulted in attenuation in CAUTI similar to that seen with provision of the empty vector alone.
FIG 6 .
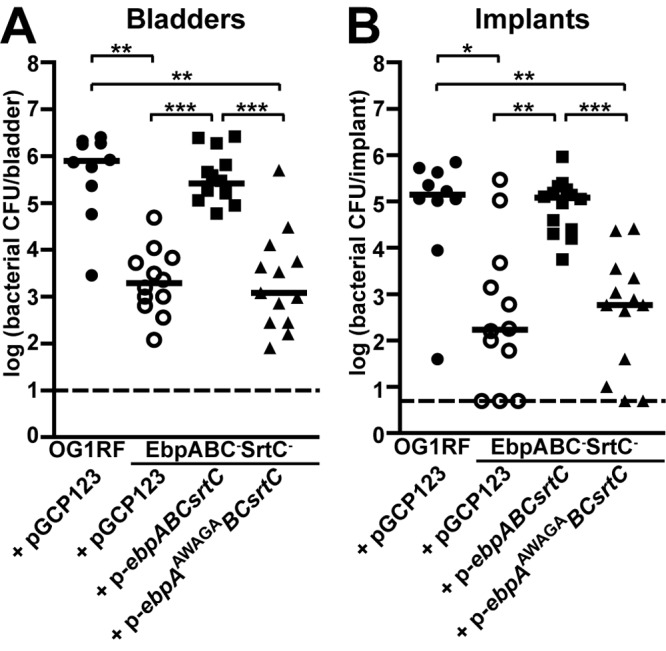
Transformation of EbpABC− SrtC− strain with p-ebpABCsrtC but not p-ebpAAWAGABCsrtC or pGCP123 complemented its virulence defect in experimental CAUTI. Bacterial titers 24 h p.i. of the bladders (A) and implants (B) from 2 independent experiments are shown. Each shape corresponds to one mouse; open shapes represent a nonpiliated bacterial strain. Median titers (CFU/bladder, CFU/implant) are shown with a bar: OG1RF/pGCP123 (5.92 × 105, 1.96 × 105), EbpABC− SrtC−/pGCP123 strain (1.94 × 103, 1.72 × 102), EbpABC− SrtC−/p-ebpABCsrtC strain (2.60 × 105, 1.20 × 105), and EbpABC− SrtC−/p-ebpAAWAGABC strain (1.20 × 103, 5.80 × 102). Dashed lines are the limits of detection (10 CFU/bladder, 5 CFU/implant). All possible strain pairs were compared statistically. P values were adjusted for 6 comparisons. Significant differences are shown (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Similarly, when we implanted and infected mice with the chromosomal MIDAS motif mutant (EbpAAWAGA) and monitored the bacterial burdens both 24 h (Fig. 7A and 7B) and 7 days (Fig. 7C and 7D) p.i., the EbpAAWAGA and EbpABC− strains were similarly attenuated in both bladder (Fig. 7A and 7C) and implant (Fig. 7B and 7D) colonization at each time point tested compared to OG1RF, revealing that EbpA’s MIDAS motif is necessary for Ebp pilus function in experimental E. faecalis CAUTI.
FIG 7 .
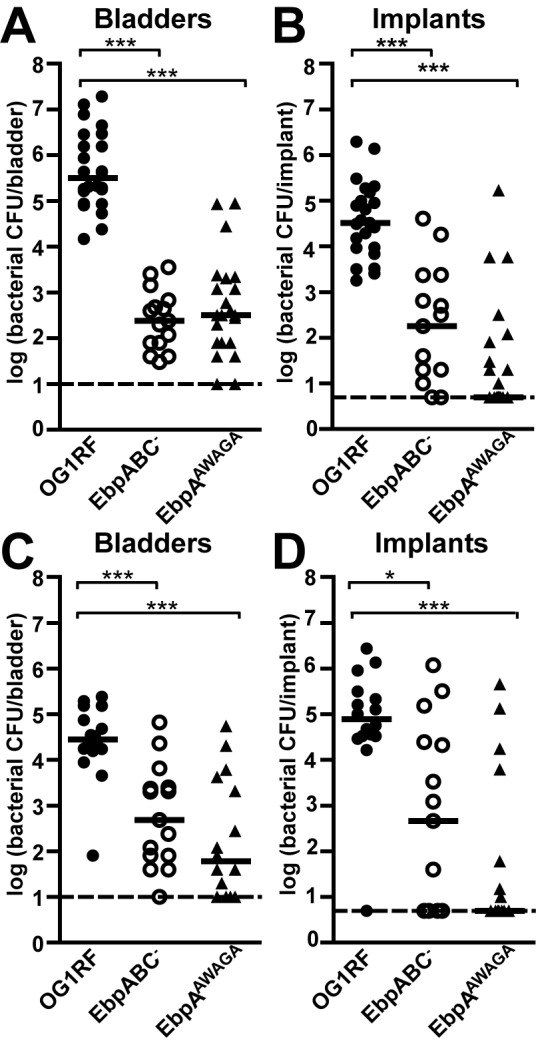
The chromosomal MIDAS mutant (EbpAAWAGA) was as attenuated as the EbpABC− strain in experimental CAUTI. Twenty-four-hour (A and B) or 7-day (C and D) viable bacterial counts from the bladders (A and C) and implants (B and D) of mice infected with E. faecalis were pooled from 2 to 3 independent experiments for each strain. Each shape corresponds to one mouse; open shapes represent a nonpiliated bacterial strain. Median bacterial titers (CFU/bladder, CFU/implant) were determined at 24 h p.i. for OG1RF (3.16 × 105, 3.24 × 104), EbpAAWAGA (2.40 × 102, 1.80 × 102), and EbpABC− (3.20 × 102, 5) strains and at 7 days for OG1RF (2.82 × 104, 7.86 × 104), EbpAAWAGA (4.80 × 102, 4.60 × 102), and EbpABC− (60, 5) strains. Bars are medians; dashed lines are the limits of detection (10 CFU/bladder, 5 CFU/implant). All possible strain combinations were compared statistically; P values were adjusted for 3 comparisons. Significant differences are shown (*, P < 0.05; ***, P < 0.001).
Ebp pilus importance in UTI is tissue and model specific.
There were no significant differences between the median bacterial kidney titer of mice infected with any pilin mutant and that of mice infected with OG1RF at any time point in experimental CAUTI (see Fig. S1 in the supplemental material). Thus, the importance of Ebp pili in this model was specific to implants and implanted bladders. To further investigate the tissue specificity of Ebp pili, we compared the EbpABC− and EbpC− strains to OG1RF in a previously described mouse model of ascending UTI (38) in which E. faecalis displays kidney tropism (see Text S1 for methods). Since growth in serum was reported to increase expression of Ebp pili (8), we tested two inoculum preparation methods: bacterial subculture in brain heart infusion (BHI) broth alone or that in BHI broth with 40% horse serum. Each mutant fared as well as or better than OG1RF in bladders and kidneys at both 6 h and 48 h p.i. using either inoculum preparation method (see Fig. S2 in the supplemental material). Thus, Ebp pili were dispensable for E. faecalis virulence in our model of ascending UTI using mice of the same age and genetic background as those used in our model of CAUTI (7- to 8-week-old C57BL/6 mice). Consistent with the model specificity of Ebp pili in UTI pathogenesis observed here, Singh et al. previously showed a defect for a polar, nonpiliated ebp mutant in a different model of ascending UTI using younger, outbred mice, different bacterial inoculum preparation methods, and distinct virulence metrics (29).
DISCUSSION
In this study, we demonstrated a specific role for Ebp pili in a newly characterized mouse model of E. faecalis CAUTI. To investigate the molecular basis of pilus function in vivo, we described the effects of an extensive panel of pilus structural subunit deletions on both pilus biogenesis and pilus function in experimental CAUTI. Finally, we showed that EbpA and the MIDAS motif encoded by its predicted VWA domain were critical for pilus-mediated virulence in vivo, thus defining the molecular basis of pilus function in experimental E. faecalis CAUTI.
The EbpABC− mutant lacking all structural subunits was severely attenuated in bladder and implant colonization in experimental CAUTI, showing that the Ebp pilus was an important virulence factor in this model. The residual bladder and implant colonization by the EbpABC− strain suggests that additional bacterial factors may play a role in E. faecalis CAUTI. To investigate the molecular basis of pilus function in vivo, we characterized a panel of pilin deletion mutants. We found that deletion of ebpA altered pilus morphology, leading to extended EbpC fibers. Similarly, deletion of either the pilA or pilC minor pilin gene from the GBS NEM316 pilus island 2A (PI-2A) led to longer pilus fibers (22). In Corynebacterium diphtheriae, mutants lacking the minor anchor pilin (SpaB) produced longer pilus fibers, presumably because cell wall anchoring achieved via SpaB processing by the housekeeping sortase prevents further polymerization by the pilus-associated sortase (21, 39). However, pili of our mutant lacking EbpB, the predicted Ebp base pilin, were not appreciably different from pili of OG1RF in our study. As overexpression of the major pilin has also been shown to increase pilus length (40), it is possible that the ebpA deletion affected relative EbpC levels.
We demonstrated that the deletion of either or both minor pilins reduced the proportion of piliated E. faecalis cells in a population by an unknown mechanism. Many sortase-assembled pilus islands include a divergently transcribed, upstream positive regulator, ebpR in the case of the E. faecalis ebp operon (41). Additionally, deletion of rnjB, a putative RNase J2, reduced pilus expression and levels of ebpABC mRNA transcript (42). Interestingly, it was recently reported that RrgA, the tip pilin of the S. pneumoniae rlrA pilus islet, interacts with RlrA, the upstream positive regulator of pilus expression, to exert a negative effect on population piliation dynamics of S. pneumoniae (43), presenting a mechanism whereby a structural pilin affected population piliation dynamics. Future studies will determine how the ebpA and ebpB deletions affected pilus biogenesis and whether they interacted with ebpR or rnjB to do so. Not surprisingly, the minor pilin deletion mutants (EbpA−, EbpAB−, and EbpB− strains), which all exhibited perturbed pilus biogenesis, were attenuated in experimental CAUTI.
Interestingly, the nonpiliated EbpC− mutant behaved similarly to OG1RF in experimental CAUTI, showing that the major polymerizing EbpC subunit and pilus fibers were dispensable for E. faecalis virulence. Similarly, the major subunits, but not the minor RrgA and Cpa tip pilins, were dispensable for pneumococcal mouse upper airway colonization (44) and GAS skin colonization in a humanized mouse model (27), respectively. Indeed, minor pilins expressed in the absence of pilus fibers have been shown to govern several sortase-assembled pilus functions in vitro, including adherence to cell lines and static biofilm formation (22, 44, 45). It has thus been suggested that pilus fibers serve to extend a minor functional pilin beyond the bacterial capsule where it can interact with host molecules (22). In this case, since OG1RF does not produce the E. faecalis capsular polysaccharide (46), EbpC pilus fibers, but not a functional minor subunit, would be dispensable for pilus function in OG1RF, just as we observed in experimental CAUTI.
Our analysis of minor pilin expression in ebpC and sortase mutants argued that a sortase-assembled EbpA-EbpB heterodimer was expressed in the absence of pilus fibers in the EbpC− strain. Similarly, the C. diphtheriae SpaC and SpaB minor pilins heterodimerized and anchored to the cell wall in the absence of the major pilin in a sortase-dependent fashion (47). The nonpiliated EbpAC−, EbpBC−, and SrtC− mutants that expressed mainly EbpA and/or EbpB monomers, but not the putative EbpA-EbpB heterodimer, were severely attenuated in experimental UTI, suggesting that sortase-assembled EbpA and/or EbpB mediated Ebp pilus function.
To directly test the importance of the minor pilins in CAUTI, we sought to create mutations in functional domains that did not affect pilus biogenesis. EbpA contains a predicted Cna B domain, not investigated here, and a VWA domain. VWA domains, named for their role in platelet adhesion to damaged vascular endothelium by the human plasma protein von Willebrand factor (48), are widely distributed among archaea, bacteria, and eukaryotes. Well-studied examples occur in some integrins, ECM proteins, and magnesium (Mg) chelatases and perform diverse functions, usually protein-protein interaction or cell adhesion (49). Coordination of a divalent cation by a MIDAS motif, present in almost half of all VWA domains, is critical for the function of some VWA domain-containing proteins (49). Most prokaryotic VWA domains have not been investigated in detail. However, Konto-Ghiorghi et al. showed that the GBS PilA tip pilin VWA domain was important for pilus-mediated bacterial adhesion to human alveolar and intestinal epithelial cells in vitro (22). Furthermore, the crystal structure of the pneumococcal RrgA tip pilin modeled an Mg2+ ion coordinated by the MIDAS motif of its VWA domain (50). We therefore hypothesized that EbpA’s MIDAS motif would be important for Ebp pilus function in our model of CAUTI. Indeed, MIDAS motif mutants were as attenuated in vivo as were the relevant nonpiliated control strains, showing that an intact MIDAS motif is necessary for Ebp pilus function in bladder and implant colonization in experimental E. faecalis CAUTI. To our knowledge, this is the first study ascribing a sortase-assembled pilus function in vivo in a disease model to a specific protein domain. The importance of a MIDAS motif for the function of a prokaryotic VWA domain-containing protein has otherwise been shown only for the Rhodobacter capsulatus Mg chelatase BchD subunit (51), a member of an evolutionarily distinct family of VWA domain-containing proteins (49).
The functional role of the Ebp pilus governed by EbpA’s VWA domain and MIDAS motif in bladder and implant colonization in experimental CAUTI remains to be determined. However, this function was tissue and model specific since pilin mutants colonized kidneys similarly to OG1RF in experimental CAUTI and both kidneys and bladders in our mouse model of ascending UTI. MIDAS motifs in the integrin beta and some alpha subunits are involved in integrin binding to ECM proteins (49). The VWA domain- and MIDAS motif-containing tip pilins RrgA and PilA have both been reported to bind ECM proteins such as collagen (26, 52). Furthermore, PilA’s interaction with collagen is a critical component of GBS virulence in a mouse model of hemorrhagic meningitis (26). Crude cell wall extracts of a distinct nonpiliated E. faecalis mutant demonstrated reduced adherence to purified human collagens and fibrinogen compared to those of OG1RF, implicating Ebp pili in adhesion to these ECM molecules (33). Binding of bacteria to ECM proteins exposed by damage to vascular endothelium initiates infective endocarditis, an enterococcal disease in which Ebp pili are also implicated (8). In our CAUTI model, implantation leads to physiological changes in the bladder epithelium and induction of inflammation (32), potentially revealing host binding partners, such as ECM proteins, for recognition by EbpA. Colonization of implants may proceed by the same mechanism, as urinary catheters become coated with host proteins and components (53). Alternatively, EbpA’s VWA domain may perform a distinct behavior that facilitates in vivo biofilm formation on implants. Polar ebp disruption mutants showed reduced static biofilm formation in vitro (8), suggesting that Ebp pili may be involved in adherence to abiotic surfaces or bacterial surface components.
The diversity of bacterial species with sortase-assembled pili is matched only by the variety of potential niches and disease processes in which these pili function. However, a particular pilin or pilin domain is implicated for only a few of these behaviors. By mutating the predicted metal ion-coordinating amino acids of the MIDAS motif in EbpA’s VWA domain, we preserved pilus biogenesis and showed a clear role for this motif in the function of the Ebp pilus in vivo. Future studies will determine whether the VWA domains of EbpA and other sortase-assembled tip pilins function similarly to those of the integrin subunits or other characterized MIDAS motif-containing VWA domains, allowing the development of structure-function correlates for pilus-mediated virulence in a wide variety of diseases.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains are listed in Table S1 in the supplemental material. Unless otherwise noted, we grew Escherichia coli in Difco LB (Luria-Bertani) broth at 37°C with agitation and E. faecalis OG1RF (ATCC 47077) and its derivative strains statically at 37°C in Bacto BHI broth with rifampin (Rif; 25 to 100 µg ml−1). Plasmid-containing strains were grown with appropriate antibiotics (see Table S2 in the supplemental material). For E. coli, erythromycin (Erm) was added at 500 µg ml−1, kanamycin (Kan) at 50 µg ml−1 (25 µg ml−1 for pREP4), and ampicillin (Amp) at 100 µg ml−1. For E. faecalis, Erm was added at 25 µg ml−1 and Kan at 500 µg ml−1. All media were purchased from BD (Becton, Dickinson and Company, Franklin Lakes, NJ). Antibiotics were purchased from Sigma-Aldrich Corporation (St. Louis, MO).
General cloning techniques.
DNA and amino acid sequences were retrieved and analyzed as described in Text S1 in the supplemental material. Bacterial genomic DNA (gDNA) was isolated with the Wizard Genome DNA purification kit (Promega Corp., Madison, WI). Plasmids are listed in Table S2 in the supplemental material. Aside from pABG5, pGCP123, and their derivatives, which were purified with the Hurricane Maxi Prep kit (Gerard Biotech LLC, Oxford, OH), plasmid DNA was isolated with the Wizard Plus SV Minipreps DNA purification system (Promega Corp.). Primers are listed in Table S3 in the supplemental material. PCR was performed with Phusion DNA polymerase from Finnzymes (Thermo Fisher Scientific, Inc., Rockford, IL). Site-directed mutagenesis (SDM) was carried out with QuikChange or QuikChange II kits (Stratagene, La Jolla, CA). Restriction endonucleases and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA). Ligations were transformed into E. coli TOP10, NovaBlue, or XL1-Blue cells. Plasmids derived here were confirmed by sequencing of inserts.
Generation of chromosomal deletion strains.
Deletion of pilin coding sequences from the OG1RF chromosome was accomplished by allelic replacement as described previously using the temperature-sensitive shuttle vector pJRS233 (36). The ΔebpA, ΔebpB, ΔebpC ΔebpAB, ΔebpBC, ΔebpABC, ΔebpABCsrtC, and ΔsrtC deletion alleles comprised upstream (US) and downstream (DS) DNA fragments (~1,000 bp each) of the region to be deleted that were spliced by overlap extension PCR (SOE-PCR) (54). Single gene deletion alleles removed protein-coding open reading frame (ORF) sequences of the deleted gene only, preserving any overlapping ORF or intergenic sequence. Alleles for multiple gene deletions preserved any overlapping and intergenic sequence between remaining ORFs and deleted ORFs but removed any intergenic sequence between consecutively deleted ORFs. Table S4-A in the supplemental material details allele construction. Each allele was cloned into pJRS233 as described for each resultant plasmid listed in Table S2-G. When noted in Table S4-A, alleles were first blunt end ligated into a commercial vector. The EbpA−, EbpB−, EbpC−, EbpAB−, EbpBC−, EbpABC−, and SrtC− strains were created by transformation of OG1RF with pSJH-529, pSJH-530, pSJH-523, pSJH-531, pSJH-532, pSJH-524, and pSJH-189, respectively, by electroporation. The EbpAC−, EbpABC− SrtC−, and SrtC− SrtA− strains were created by transformation of EbpC−, EbpABC−, and SrtA− strains with pSJH-529, pSJH-279, and pSJH-189, respectively. Transformants were selected with Erm at 30°C and then passaged at the nonpermissive temperature (42°C) with Erm to select for chromosomal integration of the plasmid. Subsequent plasmid excision was allowed by passage at 30°C without Erm. Erm-sensitive colonies with double-crossover events that integrated deletion alleles were selected by PCR screening. Deletions were confirmed by PCR with gDNA.
Chromosomal MIDAS motif mutant construction.
Mutation of EbpA’s MIDAS motif on the OG1RF chromosome was accomplished by allelic replacement with the ebpAAWAGA allele (coding for Ala315-Trp-Ala317-Gly-Ala319 in EbpA; see Table S4-B in the supplemental material) using pGCP213 instead of pJRS233. Creation of the pGCP213 temperature-sensitive shuttle vector is described in Text S1 in the supplemental material. The ebpAAWAGA allele was introduced into pGCP213 as described for pSJH-509 in Table S2-H. An extraneous nucleotide added by primers HVN228 and HVN229 was deleted by SDM using primers HVN241 and HVN242, resulting in pSJH-509. OG1RF was transformed with pSJH-509 and passaged as described above. We screened for incorporation of the mutant allele by BspEI digestion of internal ebpA colony PCR products. A BspEI restriction site in the OG1RF allele is absent in the ebpAAWAGA allele. The resultant EbpAAWAGA chromosomal MIDAS motif mutant was confirmed by gDNA sequencing.
Generation of E. faecalis expression strains.
Plasmids were created for expression of ebp genes in trans in E. faecalis using the Gram-positive expression vector pGCP123 (derived here as described in Text S1 in the supplemental material). All pGCP123 derivatives included the region 500 bp upstream of EbpA’s translational start codon as the putative ebpA promoter (ebpAp). See Table S2-I for construction details of the following plasmids: pSJH-491 (p-ebpABC) encodes all structural pilins, pSJH-492 (p-ebpAB) encodes just EbpA and EbpB, pSJH-496 (p-ebpABCsrtC) encodes all structural pilins and SrtC, and pSJH-559 (p-ebpAAWAGABCsrtC) encodes the ebpAAWAGA allele of EbpA and OG1RF alleles of EbpB, EbpC, and SrtC. EbpABC− and EbpABC− SrtC− strains were transformed with empty pGCP123 (see Table S1-C) and the plasmids derived above (see Table S1-D).
Generation of polyclonal antisera.
Expression constructs for each Ebp pilin lacking the signal sequence and CWSS (EbpA-X, EbpB-X, and EbpCA-X) were created as described in Table S4-D in the supplemental material and cloned into the isopropyl-d-1-thiogalactopyranoside (IPTG)-inducible expression vector pQE-30Xa (Qiagen Inc., Valencia, CA), resulting in the addition of an N-terminal RGS-6×His tag. The EbpA-X construct comprised roughly the C-terminal half of EbpA. When noted in Table S4-D, constructs were first blunt end ligated into a commercial vector. The resultant pQE-30Xa-derived plasmids encoding recombinant EbpA, EbpB, and EbpC (pSJH-541, pSJH-547, and p-SJH-550, respectively) were used to transform the E. coli expression strain M15/pREP4 or SG13009/pREP4, resulting in strains SJH1987, SJH1988, and SJH1985. Text S1 describes the purification of recombinant EbpA, EbpB, and EbpC. Polyclonal antisera were generated commercially by immunization of New Zealand White rabbits with purified, recombinant EbpA or EbpB (New England Peptide, Gardner, MA) and by immunization of mice with purified, recombinant EbpC (Agro-Bio, La Ferté Saint-Aubin, France). Specificities of the immune sera were confirmed by a lack of signal on Western blots of cell lysates from the appropriate deletion mutants. No reactivity of preimmune sera to Ebp pili was observed on Western blots of OG1RF (data not shown).
Western blots.
Bacterial cell fractions (prepared as described in Text S1 in the supplemental material) were boiled for at least 10 min in β-mercaptoethanol-containing loading buffer, and SDS-PAGE was performed with NuPAGE Novex 3 to 8% Tris-acetate gels in NuPAGE Tris-acetate SDS running buffer (Life Technologies Corp., Carlsbad, CA). Membranes were probed with antipilin sera as indicated and Pierce stabilized horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse IgG (Thermo Fisher Scientific, Inc.). Blots were developed with SuperSignal West Femto chemiluminescent substrate (Thermo Fisher Scientific, Inc.); film was processed with a Kodak X-Omat processor. Precision Plus Protein Kaleidoscope Standards (Bio-Rad Laboratories, Inc., Hercules, CA) are indicated. Distinct blots or exposures are separated by white space in the figures; lines represent an irrelevant gel lane that was removed using Adobe Photoshop CS2 (Adobe Systems Inc., Mountain View, CA).
IFM.
IFM was performed as described previously (36). Slides were labeled with mouse anti-EbpC sera at a 1:1,000 dilution, Molecular Probes Alexa Fluor 594 anti-mouse IgG (Life Technologies Corp.), and Hoechst stain. Imaging was performed with AxioVision software and a Zeiss Axioskop 2 MOT Plus wide-field fluorescence microscope at the Department of Molecular Microbiology Imaging Facility of Washington University in St. Louis, MO. Quantification of EbpC-expressing cells is described in Text S1 in the supplemental material.
Deep-etch immunogold EM.
Bacterial cells grown in TSBG (BBL Trypticase soy broth with 0.25% glucose) were deposited onto glass slides, fixed, and labeled as described previously (55) using mouse anti-EbpC sera and 18-nm gold bead-conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories Inc., West Grove, PA). Samples were freeze-dried and imaged as described elsewhere (55, 56). The Shadows/Highlights function of Adobe Photoshop CS2 (Adobe Systems) was applied to Fig. 3E as a whole.
Negative-stain immunogold EM.
Bacteria grown overnight were diluted 1:1,000 into TSBG and grown for ~14 h. Cells were harvested by centrifugation (5,000 × g, 5 min), washed in phosphate-buffered saline (PBS), and resuspended in PBS-5% calf serum. Cells were adsorbed to grids, labeled with anti-EbpC, negatively stained with uranyl acetate, and imaged as described previously (36) using goat anti-rabbit IgG conjugated to 18-nm colloidal gold particles (Jackson ImmunoResearch Laboratories).
Mouse model of CAUTI.
All mouse CAUTI experiments were carried out in compliance with protocols approved by the Washington University in St. Louis Animal Studies Committee. C57BL/6 female mice purchased from the National Cancer Institute (Frederick, MD) were acclimated in our animal facility for 1 week. Experiments were performed with 7- to 8-week-old mice as described previously (32). Briefly, silicone implants were inserted transurethrally, and mice were infected with ~4 × 107 CFU (unless otherwise noted) of E. faecalis. Twenty-four hours or 7 days p.i., mice were sacrificed; bladders, kidneys, and implants were harvested; and bacterial burdens were determined by viable counting on Rif- and fusidic acid (Fus)-containing media. The CFU values of samples from which no colonies were recovered were set to the limit of detection. Samples from infections with plasmid-containing strains were also plated on Kan-containing media. No significant differences between the titers from Kan-containing and Kan-free media were observed (data not shown). Data from mice that lost their implant before sacrifice were excluded. Three to eight mice were included for each bacterial strain at each time point in each experiment.
Statistical analyses.
Data from multiple experiments were pooled. Two-tailed Mann-Whitney U tests were performed with GraphPad Prism 5 software (GraphPad Software, San Diego, CA) for all comparisons described in CAUTI, ascending UTI, and IFM experiments. When noted in the figure legends, Bonferroni’s adjustment for multiple comparisons was performed manually. An adjusted P value of <0.05 was considered statistically significant.
SUPPLEMENTAL MATERIAL
OG1RF and pilin mutants colonize kidneys to similar levels in CAUTI. Viable bacterial counts from the kidney pairs of mice infected with the indicated E. faecalis strain were determined at 24 h (A and B) or 7 days (C) p.i. in 2 to 4 independent experiments for each strain. Each shape corresponds to one mouse; open shapes represent nonpiliated bacterial strains. Dashed lines are the limit of detection (10 CFU/kidney pair). Median titers (CFU/kidney pair) for each strain in each set of experiments are shown with a bar: OG1RF (40), EbpABC− strain (5.20 × 102), EbpA− strain (10), EbpB− strain (80), EbpAB− strain (20), EbpC− strain (10), EbpBC− strain (10), EbpAC− strain (10) (A); OG1RF (30), EbpABC− strain (2.40 × 102), EbpAAWAGA (2.40 × 102) (B); OG1RF (10), EbpABC− strain (10), and EbpAAWAGA (10) (C). No significant differences were observed when each strain was compared to OG1RF and P values were adjusted for 7 multiple comparisons (A) or when all possible strain combinations were compared and P values were adjusted for 3 multiple comparisons (B and C). Download Figure S1, TIF file, 1.4 MB.
Ebp pilus mutants are not attenuated in a mouse model of ascending UTI. Bacterial titers 6 h or 48 h p.i. from the bladders (A and C) and kidney pairs (B and D) of mice infected with ~1.0 × 107 CFU of E. faecalis subcultured in BHI broth alone (A and B) or BHI broth with 40% serum (C and D). Each shape corresponds to one mouse; open shapes represent nonpiliated bacterial strains. Median titers for the BHI-only subculture experiment were determined 6 h p.i. (CFU/bladder, CFU/kidney pair) for the OG1RF (3.00 × 102, 1.96 × 105), EbpABC− (1.94 × 103, 2.38 × 105), and EbpC− (5.36 × 103, 1.84 × 105) strains and 48 h p.i. for the OG1RF (65, 1.28 × 105), EbpABC− (80, 2.62 × 105), and EbpC− (2.20 × 102, 6.24 × 104) strains. For the BHI/serum subculture experiment, median titers were determined 6 h p.i. for the OG1RF (1.00 × 103, 3.68 × 105), EbpABC− (1.12 × 104, 3.42 × 105), and EbpC− (1.08 × 103, 2.32 × 105) strains and 48 h p.i. for the OG1RF (8.60 × 102, 2.54 × 104), EbpABC− (1.56 × 104, 8.50 × 103), and EbpC− (3.18 × 103, 1.74 × 104) strains. Bars are medians; dashed lines are limits of detection (10 CFU/bladder; 10 CFU/kidney pair). Each mutant was statistically compared to OG1RF in each tissue at each time point, P values were adjusted for 2 comparisons, and significant differences are shown (*, P < 0.05; **, P < 0.01). Download Figure S2, TIF file, 2.6 MB.
Supplemental materials and methods used in this study. Download Text S1, PDF file, 0.3 MB.
Bacterial strains used in this study.
Plasmids used in this study.
Primers used in this study.
Alleles and constructs used in this study.
ACKNOWLEDGMENTS
This work was supported by the NIDDK grant DK51406 and its ARRA supplement DK51406-12S1 awarded to S.J.H., the American Heart Association Midwest Affiliate Predoctoral Fellowship 10PRE2640099 to H.V.N., an ASM Robert D. Watkins Graduate Research Fellowship Award to P.S.G., and a W. M. Keck Postdoctoral Fellowship to G.C.P.
We thank D. K. Robertson, R. Roth, and J. E. Heuser for assistance with EM studies. We are grateful to K. W. Dodson for critical reading of the manuscript and to members of the Hultgren, Caparon, and Normark laboratories for helpful insight and discussion.
Footnotes
Citation Nielsen HV, et al. 2012. The metal ion-dependent adhesion site motif of the Enterococcus faecalis EbpA pilin mediates pilus function in catheter-associated urinary tract infection. mBio 3(4):e00177-12. doi:10.1128/mBio.00177-12.
REFERENCES
- 1. Murray BE. 1990. The life and times of the enterococcus. Clin. Microbiol. Rev. 3:46–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10:266–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Edwards JR, et al. 2009. National Healthcare Safety Network (NHSN) report: data summary for 2006 through 2008, issued December 2009. Am. J. Infect. Control 37:783–805 [DOI] [PubMed] [Google Scholar]
- 4. Cope M, et al. 2009. Inappropriate treatment of catheter-associated asymptomatic bacteriuria in a tertiary care hospital. Clin. Infect. Dis. 48:1182–1188 [DOI] [PubMed] [Google Scholar]
- 5. Wagenlehner FM, et al. 2002. Epidemiological analysis of the spread of pathogens from a urological ward using genotypic, phenotypic and clinical parameters. Int. J. Antimicrob. Agents 19:583–591 [DOI] [PubMed] [Google Scholar]
- 6. Warren JW, et al. 1987. Fever, bacteremia, and death as complications of bacteriuria in women with long-term urethral catheters. J. Infect. Dis. 155:1151–1158 [DOI] [PubMed] [Google Scholar]
- 7. Hidron AI, et al. 2008. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect. Control Hosp. Epidemiol. 29:996–1011 [DOI] [PubMed] [Google Scholar]
- 8. Nallapareddy SR, et al. 2006. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J. Clin. Invest. 116:2799–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ton-That H, Schneewind O. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 50:1429–1438 [DOI] [PubMed] [Google Scholar]
- 10. Mishra A, Das A, Cisar JO, Ton-That H. 2007. Sortase-catalyzed assembly of distinct heteromeric fimbriae in Actinomyces naeslundii. J. Bacteriol. 189:3156–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lauer P, et al. 2005. Genome analysis reveals pili in group B Streptococcus. Science 309:105 [DOI] [PubMed] [Google Scholar]
- 12. Mora M, et al. 2005. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. U. S. A. 102:15641–15646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barocchi MA, et al. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. U. S. A. 103:2857–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Budzik JM, Marraffini LA, Schneewind O. 2007. Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol. Microbiol. 66:495–510 [DOI] [PubMed] [Google Scholar]
- 15. Sillanpää J, et al. 2008. Identification and phenotypic characterization of a second collagen adhesin, Scm, and genome-based identification and analysis of 13 other predicted MSCRAMMs, including four distinct pilus loci, in Enterococcus faecium. Microbiology 154:3199–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hendrickx AP, et al. 2008. Expression of two distinct types of pili by a hospital-acquired Enterococcus faecium isolate. Microbiology 154:3212–3223 [DOI] [PubMed] [Google Scholar]
- 17. Kankainen M, et al. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc. Natl. Acad. Sci. U. S. A. 106:17193–17198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Foroni E, et al. 2011. Genetic analysis and morphological identification of pilus-like structures in members of the genus Bifidobacterium. Microb. Cell Fact. 10(Suppl. 1):S16 http://dx.doi.org/10.1186/1475-2859-10-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kline KA, Dodson KW, Caparon MG, Hultgren SJ. 2010. A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol. 18:224–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ton-That H, Schneewind O. 2004. Assembly of pili in gram-positive bacteria. Trends Microbiol. 12:228–234 [DOI] [PubMed] [Google Scholar]
- 21. Mandlik A, Das A, Ton-That H. 2008. The molecular switch that activates the cell wall anchoring step of pilus assembly in gram-positive bacteria. Proc. Natl. Acad. Sci. U. S. A. 105:14147–14152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Konto-Ghiorghi Y, et al. 2009. Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 5(5):e1000422 http://dx.doi.org/10.1371/journal.ppat.1000422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paulsen IT, et al. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074 [DOI] [PubMed] [Google Scholar]
- 24. Maisey HC, et al. 2008. A group B streptococcal pilus protein promotes phagocyte resistance and systemic virulence. FASEB J. 22:1715–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Papasergi S, et al. 2011. The GBS PI-2a pilus is required for virulence in mice neonates. PLoS One 6:e18747 http://dx.doi.org/10.1371/journal.pone.0018747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Banerjee A, et al. 2011. Bacterial pili exploit integrin machinery to promote immune activation and efficient blood-brain barrier penetration. Nat. Commun. 2:462 http://dx.doi.org/10.1038/ncomms1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lizano S, Luo F, Bessen DE. 2007. Role of streptococcal T antigens in superficial skin infection. J. Bacteriol. 189:1426–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Becherelli M, et al. 2012. The ancillary protein 1 of Streptococcus pyogenes FCT-1 pili mediates cell adhesion and biofilm formation through heterophilic as well as homophilic interactions. Mol. Microbiol. 83:1035–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Singh KV, Nallapareddy SR, Murray BE. 2007. Importance of the ebp (endocarditis- and biofilm-associated pilus) locus in the pathogenesis of Enterococcus faecalis ascending urinary tract infection. J. Infect. Dis. 195:1671–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kemp KD, Singh KV, Nallapareddy SR, Murray BE. 2007. Relative contributions of Enterococcus faecalis OG1RF sortase-encoding genes, srtA and bps (srtC), to biofilm formation and a murine model of urinary tract infection. Infect. Immun. 75:5399–5404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sillanpää J, et al. 2010. Characterization of the ebpfm pilus-encoding operon of Enterococcus faecium and its role in biofilm formation and virulence in a murine model of urinary tract infection. Virulence 1:236–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guiton PS, Hung CS, Hancock LE, Caparon MG, Hultgren SJ. 2010. Enterococcal biofilm formation and virulence in an optimized murine model of foreign body-associated urinary tract infections. Infect. Immun. 78:4166–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nallapareddy SR, Singh KV, Sillanpää J, Zhao M, Murray BE. 2011. Relative contributions of Ebp pili and the collagen adhesin Ace to host extracellular matrix protein adherence and experimental urinary tract infection by Enterococcus faecalis OG1RF. Infect. Immun. 79:2901–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nallapareddy SR, et al. 2011. Conservation of Ebp-type pilus genes among enterococci and demonstration of their role in adherence of Enterococcus faecalis to human platelets. Infect. Immun. 79:2911–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. El Mortaji L, Terrasse R, Dessen A, Vernet T, Di Guilmi AM. 2010. Stability and assembly of pilus subunits of Streptococcus pneumoniae. J. Biol. Chem. 285:12405–12415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kline KA, et al. 2009. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J. Bacteriol. 191:3237–3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee JO, Rieu P, Arnaout MA, Liddington R. 1995. Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18). Cell 80:631–638 [DOI] [PubMed] [Google Scholar]
- 38. Kau AL, et al. 2005. Enterococcus faecalis tropism for the kidneys in the urinary tract of C57BL/6J mice. Infect. Immun. 73:2461–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Swaminathan A, et al. 2007. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol. Microbiol. 66:961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Swierczynski A, Ton-That H. 2006. Type III pilus of corynebacteria: pilus length is determined by the level of its major pilin subunit. J. Bacteriol. 188:6318–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bourgogne A, et al. 2007. EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis OG1RF. J. Bacteriol. 189:6490–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gao P, et al. 2010. Enterococcus faecalis rnjB is required for pilin gene expression and biofilm formation. J. Bacteriol. 192:5489–5498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Basset A, et al. 2011. Expression of the type 1 pneumococcal pilus is bistable and negatively regulated by the structural component RrgA. Infect. Immun. 79:2974–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nelson AL, et al. 2007. RrgA is a pilus-associated adhesin in Streptococcus pneumoniae. Mol. Microbiol. 66:329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mandlik A, Swierczynski A, Das A, Ton-That H. 2007. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol. Microbiol. 64:111–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thurlow LR, Thomas VC, Hancock LE. 2009. Capsular polysaccharide production in Enterococcus faecalis and contribution of CpsF to capsule serospecificity. J. Bacteriol. 191:6203–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chang C, Mandlik A, Das A, Ton-That H. 2011. Cell surface display of minor pilin adhesins in the form of a simple heterodimeric assembly in Corynebacterium diphtheriae. Mol. Microbiol. 79:1236–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sadler JE. 1998. Biochemistry and genetics of von Willebrand factor. Annu. Rev. Biochem. 67:395–424 [DOI] [PubMed] [Google Scholar]
- 49. Whittaker CA, Hynes RO. 2002. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 13:3369–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Izoré T, et al. 2010. Structural basis of host cell recognition by the pilus adhesin from Streptococcus pneumoniae. Structure 18:106–115 [DOI] [PubMed] [Google Scholar]
- 51. Lundqvist J, et al. 2010. ATP-induced conformational dynamics in the AAA+ motor unit of magnesium chelatase. Structure 18:354–365 [DOI] [PubMed] [Google Scholar]
- 52. Hilleringmann M, et al. 2008. Pneumococcal pili are composed of protofilaments exposing adhesive clusters of RrgA. PLoS Pathog. 4(3):e1000026 http://dx.doi.org/10.1371/journal.ppat.1000026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Habash M, Reid G. 1999. Microbial biofilms: their development and significance for medical device-related infections. J. Clin. Pharmacol. 39:887–898 [DOI] [PubMed] [Google Scholar]
- 54. Horton RM, Cai ZL, Ho SN, Pease LR. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528–535 [PubMed] [Google Scholar]
- 55. Hanson PI, Roth R, Lin Y, Heuser JE. 2008. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 180:389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Heuser JE, et al. 1979. Synaptic vesicle exocytosis captured by quick freezing and correlated with quantal transmitter release. J. Cell Biol. 81:275–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
OG1RF and pilin mutants colonize kidneys to similar levels in CAUTI. Viable bacterial counts from the kidney pairs of mice infected with the indicated E. faecalis strain were determined at 24 h (A and B) or 7 days (C) p.i. in 2 to 4 independent experiments for each strain. Each shape corresponds to one mouse; open shapes represent nonpiliated bacterial strains. Dashed lines are the limit of detection (10 CFU/kidney pair). Median titers (CFU/kidney pair) for each strain in each set of experiments are shown with a bar: OG1RF (40), EbpABC− strain (5.20 × 102), EbpA− strain (10), EbpB− strain (80), EbpAB− strain (20), EbpC− strain (10), EbpBC− strain (10), EbpAC− strain (10) (A); OG1RF (30), EbpABC− strain (2.40 × 102), EbpAAWAGA (2.40 × 102) (B); OG1RF (10), EbpABC− strain (10), and EbpAAWAGA (10) (C). No significant differences were observed when each strain was compared to OG1RF and P values were adjusted for 7 multiple comparisons (A) or when all possible strain combinations were compared and P values were adjusted for 3 multiple comparisons (B and C). Download Figure S1, TIF file, 1.4 MB.
Ebp pilus mutants are not attenuated in a mouse model of ascending UTI. Bacterial titers 6 h or 48 h p.i. from the bladders (A and C) and kidney pairs (B and D) of mice infected with ~1.0 × 107 CFU of E. faecalis subcultured in BHI broth alone (A and B) or BHI broth with 40% serum (C and D). Each shape corresponds to one mouse; open shapes represent nonpiliated bacterial strains. Median titers for the BHI-only subculture experiment were determined 6 h p.i. (CFU/bladder, CFU/kidney pair) for the OG1RF (3.00 × 102, 1.96 × 105), EbpABC− (1.94 × 103, 2.38 × 105), and EbpC− (5.36 × 103, 1.84 × 105) strains and 48 h p.i. for the OG1RF (65, 1.28 × 105), EbpABC− (80, 2.62 × 105), and EbpC− (2.20 × 102, 6.24 × 104) strains. For the BHI/serum subculture experiment, median titers were determined 6 h p.i. for the OG1RF (1.00 × 103, 3.68 × 105), EbpABC− (1.12 × 104, 3.42 × 105), and EbpC− (1.08 × 103, 2.32 × 105) strains and 48 h p.i. for the OG1RF (8.60 × 102, 2.54 × 104), EbpABC− (1.56 × 104, 8.50 × 103), and EbpC− (3.18 × 103, 1.74 × 104) strains. Bars are medians; dashed lines are limits of detection (10 CFU/bladder; 10 CFU/kidney pair). Each mutant was statistically compared to OG1RF in each tissue at each time point, P values were adjusted for 2 comparisons, and significant differences are shown (*, P < 0.05; **, P < 0.01). Download Figure S2, TIF file, 2.6 MB.
Supplemental materials and methods used in this study. Download Text S1, PDF file, 0.3 MB.
Bacterial strains used in this study.
Plasmids used in this study.
Primers used in this study.
Alleles and constructs used in this study.